Abstract
The African swine fever virus protein A238L inhibits activation of NFAT transcription factor by binding calcineurin and inhibiting its phosphatase activity. NFAT controls the expression of many immunomodulatory proteins. Here we describe a 14-amino-acid region of A238L that is needed and sufficient for binding to calcineurin. By introducing mutations within this region, we have identified a motif (PxIxITxC/S) required for A238L binding to calcineurin; a similar motif is found in NFAT proteins. Peptides corresponding to this domain of A238L bind calcineurin but do not inhibit its phosphatase activity. Binding of A238L to calcineurin stabilizes the A238L protein in cells. Although A238L-mediated suppression of NF-κB-dependent gene expression occurs by a different mechanism, the A238L-calcineurin interaction may be required to stabilize A238L.
Many large DNA viruses encode proteins that help the virus to evade the host immune response (1, 9, 24). African swine fever virus (ASFV), the prototypic member of the African swine fever-like virus genus (10), encodes a potent immunosuppressive protein, A238L. A238L inhibits activation of the NF-κB transcription factor (25, 28, 32) and the activity of the calcium/calmodulin-regulated phosphatase calcineurin (CaN) (23). The virus, therefore, has the potential to inhibit transcriptional activation of immunomodulatory genes dependent on these pathways in infected macrophages.
NF-κB is retained in the cytoplasm in a complex with the inhibitor protein, IκB, in resting cells. Following activation, IκB is phosphorylated at two key Ser residues (Ser32 and Ser36 in IκB-α) and is then ubiquitinated and targeted for degradation by the 26S proteasome (for a review, see reference 16). This process exposes nuclear localization signals within nuclear factor κB (NF-κB) which subsequently translocates to the nucleus and binds to promoters with the appropriate DNA recognition sequence. A238L coprecipitates with the p65 subunit of NF-κB (32) and therefore might function as an IκB mimic which does not respond to signal-induced degradation.
The second function of A238L, inhibition of CaN phosphatase activity, is mediated by direct binding to the catalytic subunit of CaN [CaN(A)] (23). CaN is activated following calcium release and binding of calmodulin, which results in displacement of the autoinhibitory (AI) domain from the enzyme active site. CaN regulates a number of different pathways, including activation of the NFAT family of transcription factors, which are present in a phosphorylated form in the cytoplasm in resting cells. Activation of CaN results in dephosphorylation and nuclear translocation of NFAT factors which, in cooperation with other transcription factors, play an essential role in transcriptional activation of cytokine and other immunomodulatory genes (8, 29, 30). The immunosuppressive drugs cyclosporin A (CsA) and FK506 bind to CaN in a complex with the immunophilin proteins cyclophilin A (CypA) and FKBP12, respectively (34); this drug-immunophilin complex inhibits CaN phosphatase activity (30). The effective immunosuppression induced by these drugs demonstrates the critical role played by CaN and NFAT in regulating the immune response. The crystal structure of the FK506-FKBP12 complex bound to CaN has been solved, indicating that the complex blocks access to the CaN active site (15, 18, 35). Through inhibition of CaN phosphatase activity, A238L enables the virus to downregulate NFAT-dependent expression of immunomodulatory genes (23).
Recent studies have identified a number of cellular proteins that bind to CaN and inhibit its activity, presumably providing a level of regulatory control on CaN-mediated pathways. Cain and Cabin1 are 88% identical over an overlap of 2,220 amino acid residues; the CaN binding domain was shown to map within a 38-amino-acid domain near the COOH terminus (19, 36). AKAP79 (A kinase anchoring protein), a protein involved in anchoring protein kinase A (PKA), PKC, and CaN in specific subcellular locations, also binds to and inhibits CaN, via a 180-amino-acid central region of the protein (6, 12, 17). A fourth protein that binds and inhibits CaN phosphatase is CHP, a ubiquitous protein that shares significant homology with CaN(B) and calmodulin and presumably binds to CaN(A) by a mechanism similar to that used by these proteins (20, 21). The most recently identified cellular calcineurin binding/inhibiting protein is myocyte-enriched calcineurin-interacting protein (MCIP), which binds to calcineurin via sequence motifs similar to those in NFAT (33). The roles these proteins play in regulating CaN-mediated pathways are not well understood, although Cabin1 regulates activity of the CaN-activated MEF2 family of transcription factors (4, 5, 40).
The ability of the A238L protein to inhibit both activation of NF-κB and CaN phosphatase activity, two key signaling pathways involved in activating expression of immunomodulatory genes, has not been described for any other single protein. In this work we have investigated the mechanism by which A238L inhibits CaN and have defined a motif in A238L that is required for binding to CaN. Interestingly, this domain is similar to the CaN docking motif of NFAT proteins, suggesting that the two proteins bind to CaN at the same site. We show that although there is a correlation between the ability of A238L proteins to bind CaN and their ability to inhibit CaN phosphatase activity, neither a 14-amino-acid A238L CaN-binding peptide nor a similar NFAT peptide can inhibit CaN phosphatase activity. We predict that this sequence acts as a docking domain which binds CaN with high affinity; a second A238L domain then contacts CaN and inhibits its activity. The identification of a conserved CaN binding motif in two otherwise unrelated proteins suggests that this motif may be conserved in other CaN binding proteins, including both substrates of the enzyme and inhibiting proteins.
MATERIALS AND METHODS
A238L deletion analysis.
Segments of the A238L open reading frame (ORF) were amplified by PCR, incorporating an EcoRI site at the 5′ end and a PstI site at the 3′ end of each of the gene fragments, and fused in frame with the Gal4 DNA binding domain in pGBT9 (Clontech). Clones were transformed into yeast strain Y190 together with the porcine CaN(A) cloned in pACT2 (23) and tested for activation of the lacZ (β-galactosidase) reporter gene.
CaN deletion analysis.
Segments of the CaN(A) ORF were amplified by PCR, incorporating a BamHI site at the 5′ end and a XhoI site at the 3′ end of each gene fragment, and fused in frame with the Gal4 activation domain in pACT2 (11). Clones were transformed into yeast strain Y190 together with A238L cloned in pGBT9 and then tested for activation of the reporter gene as before.
Random mutagenesis of A238L.
The NH2 terminus (residues 1 to 196) of the A238L ORF was amplified; PCR was used to incorporate an AatII site (boldface in the sequences below) in amino acid residues Asp195 and Val196. Primers were 5′-CGG GAA TTC ATG GAA CAC ATG TTT CCA GAA AG and 5′-AAA GAC GTC CAG CTT GTA AAG AGG GAA ATG C. The COOH terminus (residues 195 to 238) was amplified, also incorporating an AatII site in Asp195 and Val196 and degeneracy spanning residues 200 to 213. The primers were 5′-AAA GAC GTC TTC CAC CGG (GT)GG (GT)TT A(AC)G A(AC)A A(AC)G (CT)CC A(AC)A A(CT)T A(CT)T A(CT)T (AG)CT G(GC)C T(GC)T A(AC)A AAT AAT G (parentheses denote positions of degeneracy) and 5′-AAA CTG CAG AGA TTA CTT TCC ATA CTT GTT CAG. The NH2 and COOH termini of A238L were fused at the AatII site and ligated into pGBT9 (Clontech) in frame with the Gal4 DNA binding domain. The pool of mutants was tested for CaN binding activity by assaying for β-galactosidase. The nucleotide sequence of A238L genes was determined for a selection of CaN-interacting and non-CaN-interacting clones.
Directed mutagenesis of A238L.
PCR was used to create point mutations in the A238L ORF by amplifying the COOH terminus (residues 195 to 238), incorporating mutations where required. An AatII site was incorporated in residues Asp195 and Val196, and single changes were incorporated in residues 205 (Pro to Ser), 207 (Ile to Thr), 209 (Ile to Thr), 210 (Thr to Ala), 211 (Gly to Ala), and 212 (Cys to Ser or Gly). The PCR primers used were 5′-AAA GAC GTC TTC CAC CGG TGG TTT AAG AAA AAG C(TSer205)CC AAA AT(CThr207)T ATT AT(CThr209)T G(AAla210)CT GG(CAla211)C TG(CSer212 or GGly212)T AAA AAT AAT G. The altered nucleotide is in parentheses after the wild-type residue. These mutated COOH termini were each fused to the wild-type NH2 terminus in pGBT9 and tested for CaN binding as before.
Cloning and expression of the porcine macrophage NFAT gene.
A gene fragment from porcine macrophage NFAT corresponding to amino acids 437 to 556 of human NFAT2 was labeled with [32P]dATP and used as a probe to screen 2 × 105 plaques from our porcine macrophage cDNA library (23). We detected one plaque that hybridized with the NFAT probe; plasmid (pACT2-NFAT) was excised using Escherichia coli strain BNN132 (11), and the nucleotide sequence of the corresponding insert was determined. Porcine macrophage NFAT was cloned in frame with the c-Myc epitope tag in pCDNA3 (Invitrogen) and transfected, either alone or together with constitutively active CaN, into 106 Vero cells using Lipofectin (Life Technologies). Cell extracts were analyzed by Western blotting with anti-c-Myc monoclonal antibody (Santa Cruz).
Surface plasmon resonance.
Surface plasmon resonance was carried out using a Biacore X machine. Biotinylated peptides (A238Lwt14 [biotin-AAAWFKKKPKIIITGCK], A238Lscram14 [biotin-AAAKCIKGIKPTKFIWK], A238LThr20714 [biotin-AAAWFKKKPKTIITGCK], and NFATwt14 [biotin-AAAPALESPRIEITSYL]) were bound to the surface of a streptavidin-coated chip (sensor chip SA; Biacore). Purified bovine CaN (20 to 400 nM; Sigma), in buffer containing 20 mM TES [N-tris (hydroxymethyl)methyl-2-aminoethanesulfonic acid; pH 7.5], 120 mM NaCl, 5 mM MgCl2, 300 μM CaCl2, 100 μM EGTA, and 0.005% Biacore surfactant, was passed over both flow cells in the absence or presence of competitor peptides. Competitor peptide (A238Lwt14 [AAAWFKKKPKIIITGCK], A238Lwt10 [KPKIIITGCK], or NFATwt14 [AAAPALESPRIEITSYL]) was mixed with CaN prior to passing over both flow cells. The amount of CaN bound to each flow cell and the difference between flow cells 1 and 2 (FC1 and FC2) under different conditions was measured in response units.
In vitro CaN phosphatase assays.
Purified CaN (100 nM; Sigma) and calmodulin (600 nM; Sigma) were incubated with 32P-labeled RII phosphopeptide labeled with PKA catalytic subunit (Sigma) for 30 min at 30°C in 60 μl of 12 mM Tris-Cl (pH 7.5)–3.5 mM MgCl2–8 mM 2-mercaptoethanol–58 μg of bovine serum albumin ml−1–17 mM CaCl2 (2). Where indicated, CaN/calmodulin was incubated with 200 μM peptide (A238Lwt14, A238Lwt10, A238Lscram14, A238Lscram10 [TICKKIPKIG], NFATwt14, or CaN AI [Sigma]) or complexes of CsA (15 μM) and CypA (5 μM). CaN phosphatase activity was assayed (13).
Transient expression of epitope-tagged A238L.
Vero cells (106) were infected with modified vaccinia virus Ankara expressing T7 RNA polymerase (MVA-T7) (37) and transfected with pT7-SV5-A238L (23), pT7-SV5-IκB (a gift from Ron Hay), pT7-SV5-A238L(W13), pT7-SV5-A238L(W20), pT7-SV5-A238L(B19), or pT7-SV5-A238L(Thr207). Cells were radiolabeled with [35S]cysteine/methionine, washed with phosphate-buffered saline, harvested in ice-cold buffer (500 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.05% Nonidet P-40, 0.05% deoxycholate, aprotinin [2 μg ml−1], leupeptin [1 μg ml−1], pepstatin A [1 μg ml−1], soybean trypsin inhibitor [4 μg ml−1]), and lysed by multiple passage through a 26-gauge needle. Lysates were clarified by centrifugation, preabsorbed with protein A-Sepharose (Sigma), and immunoprecipitated with 2 μg of monoclonal anti-simian virus 5 (SV5) PK tag (Serotec). Immune complexes were analyzed by autoradiography or Western blotting with monoclonal anti-CaN(B) (Sigma). Purified CaN was run in parallel as a control.
Construction of recombinant viruses.
SV5-tagged A238L(W13), SV5-tagged A238L(W20), SV5-tagged A238L(B19), and SV5-tagged A238L(Thr207) were cloned downstream from the A238L promoter, and ASFV recombinants expressing SV5 PK epitope-tagged mutant A238L were isolated by purification of recombinant virus from plaques expressing the β-galactosidase gene product as previously reported (23).
Expression of A238L from ASFV.
Vero cells (106) (10 were infected with ASFV (Ba71V, ΔA238L [23], SV5-A238L [23], SV5-W13, SV5-W20, SV5-B19, or SV5-Thr207) at a multiplicity of infection of 2. After 3.5 h the cells were pulse-labeled with [35S]Met/Cys for 1 h. Cells were washed, harvested, and lysed (as described above for transient expression) either immediately or after a 1-h chase period in nonradioactive medium. Samples were taken prior to immunoprecipitation and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with monoclonal anti-SV5 PK tag (Serotec) or monoclonal anti-ASFV P30. Proteins were immunoprecipitated with 2 μg of monoclonal anti-SV5 PK tag (Serotec) and analyzed by autoradiography or Western blotting with monoclonal anti-CaN(B) (Sigma).
CaN phosphatase assays of ASFV-infected cells.
Vero cells (106) were not infected or infected for 11 h with wild-type or recombinant ASFV. Cells were treated (where indicated) with CsA for 2 h immediately prior to harvesting. Cells were harvested, and cell extracts were assayed for CaN phosphatase activity as described previously (13).
Nucleotide sequence accession number.
The sequence data shown in Fig. 2A have been submitted to the GenBank database under accession number AF069996.
FIG. 2.
(A) Predicted amino acid sequence of porcine NFAT2 compared with human NFAT2 (NFATc.β). Amino acid residues in human NFAT2 that differ from the porcine sequence are shown. Three SP repeat motifs are underlined. The SPRIEIT CaN docking sequence is double underlined. An asterisk denotes a stop codon. The region flanked by arrows corresponds to the Rel similarity domain. (B) c-Myc epitope-tagged porcine NFAT2 was transiently transfected into Vero cells, either alone (lane 1) or together with constitutively active CaN (lane 2). Proteins were analyzed by SDS-PAGE and Western immunoblotting with anti-c-Myc antibody.
RESULTS
Definition of a 14-amino-acid domain (residues 200 to 213) within the A238L protein that is necessary for binding to CaN(A).
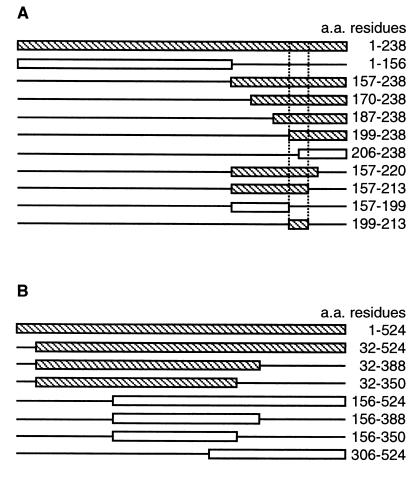
To define the region within A238L that interacts with CaN(A), we constructed a series of 10 NH2- and COOH-terminal deletions in the A238L gene and tested for interaction of the expressed proteins with CaN(A), using the yeast two-hybrid system. Binding of A238L deletion mutants to CaN(A) was indicated by expression of the Gal4-dependent lacZ reporter gene (Fig. 1A). Initial analysis showed that the NH2-terminal 156 residues of A238L did not bind to CaN(A), whereas the COOH-terminal 82 residues (residues 157 to 238) retained the ability to bind CaN(A). Further deletions within this COOH-terminal domain (residues 157 to 238) showed that removal of NH2-terminal residues up to position 199 did not prevent binding of A238L to CaN(A), whereas deletion to residue 206 prevented binding to CaN(A). Deletions from the COOH terminus of the fragment spanning residues 157 to 238 showed that residues 213 to 238 could be deleted without affecting A238L binding to CaN(A), but deletion of residues 199 to 238 abolished binding. This analysis localized the CaN(A) binding domain of A238L to a 14-amino-acid region between residues 200 and 213. The region encoding this fragment (residues 200 to 213) was cloned downstream of the Gal4 DNA binding domain and retained the ability to bind to CaN(A) in the two-hybrid assay (Fig. 1A). The level of expression of the lacZ reporter gene was indistinguishable from that observed with the full-length, wild-type A238L protein, suggesting that the domain between 200 and 213 residues is the only or the major CaN binding sequence in the protein.
FIG. 1.
Deletion analysis and mapping of the A238L and CaN binding domains. (A) NH2- and COOH-terminal A238L deletion mutants were constructed in a Gal4 DNA binding domain vector. Deletions were transformed into yeast strain Y190 together with CaN(A) cloned in a Gal4 activation domain vector. Deletions were analyzed for activation of the Gal4-dependent reporter gene product, β-galactosidase. CaN(A)-binding (shaded) and non-CaN(A)-binding (open) deletion mutants were used to locate the CaN binding domain within A238L. (B) NH2- and COOH-terminal CaN(A) deletion mutants were constructed in a Gal4 activation domain vector. Deletions were transformed into yeast strain Y190 together with A238L cloned in a Gal4 DNA binding domain vector. Deletions were analyzed as before; A238L-binding (shaded) and non-A238L-binding (open) deletion mutants are shown. a.a., amino acid.
A238L binds CaN(A) within its NH2-terminal catalytic domain.
We identified the region within CaN(A) required for interaction with A238L. Deletions constructed in the CaN(A) gene and fused to the activation domain of Gal4 were tested for interaction with full-length A238L fused to the Gal4 DNA binding domain as before. We had previously demonstrated that the 32 NH2-terminal amino acids of CaN(A), containing the poly(Pro) domain, were not required (23). Deletion of the 174 COOH-terminal amino acid residues (corresponding to 350 to 524 of human CaN) had no effect on A238L binding to CaN (Fig. 1B). However, deletion of the 156 NH2-terminal amino acid residues prevented the interaction of CaN with A238L (Fig. 1B). The minimal region tested which bound to A238L corresponded to residues 32 to 350; this region contains the majority of the CaN catalytic domain.
A PxIxITxC/S motif between amino acid residues 200 to 213 is needed for binding of A238L to CaN.
To identify critical residues within the 14-amino-acid CaN binding region of A238L (residues 200 to 213), mutations were constructed in the full-length A238L gene and fused to the Gal4 DNA binding domain. The library of mutants constructed could have either the wild-type or a mutated residue at each of the 14 positions (Table 1). Mutant A238L proteins expressed by plasmids in this library were assayed for binding to CaN(A), using the yeast two-hybrid system. A selection of 19 CaN-interacting and 17 non-CaN-interacting (Table 1) clones were isolated, and the nucleotide and amino acid sequences in the CaN binding region of the encoded A238L genes were determined. In the clones encoding CaN-binding A238L proteins, five amino acid residues did not vary from the wild-type sequence (WFKKKPKIIITGCK; invariant residues in boldface). One CaN-interacting clone had a Cys-Ser substitution at position 212. Since Ser can sometimes substitute for Cys, it remained possible that this residue was also critical for CaN binding. Directed substitutions introduced at this residue showed that Ser could be substituted for Cys without interfering with binding but that substitution of Cys for Gly abolished binding (Table 1). All the non-CaN binding A238L mutants that did not bind CaN were determined; all had mutations at one or more of the key residues required for binding (Table 1).
TABLE 1.
Defining the A238L CaN binding motif by mutation analysisa
| Sequence | Amino acid residue
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 | 201 | 202 | 203 | 204 | 205 | 206 | 207 | 208 | 209 | 210 | 211 | 212 | 213 | |
| Wild type | W | F | K | K | K | P | K | I | I | I | T | G | C | K |
| Mutant | G | V | T | T | T | S | T | T | T | T | A | A | S | T |
| Non-CaN binding | 8 | 6 | 11 | 11 | 9 | 12 | 7 | 8 | 8 | 6 | 10 | 3 | 7 | 5 |
| CaN binding | 14 | 15 | 9 | 3 | 13 | 0 | 5 | 0 | 5 | 0 | 0 | 0 | 1 | 14 |
| Point −ve | S | T | T | A | G | |||||||||
| Point +ve | A | S | ||||||||||||
| Motifb | P | I | I | T | C/S | |||||||||
The number of times each residue was mutated during the random mutagenesis experiments in non-CaN-binding (n = 17) or CaN-binding (n = 19) mutants (residues not mutated in positive interactors are in boldface) and individual point mutations which prevented CaN binding (Point −ve) or bound CaN (Point +ve) are shown.
The deduced minimal CaN binding motif from A238L.
To confirm the results from random mutagenesis, we constructed a series of individual point mutations in each of the residues predicted to be essential for CaN binding: Pro205 (mutated to Ser), Ile207 (mutated to Thr), Ile209 (mutated to Thr), Thr210 (mutated to Ala), and Gly211 (mutated to Ala). Clones encoding the full-length A238L gene with these defined point mutations were transformed into yeast and tested for the ability to bind to CaN(A) by assaying expression of the Gal4-dependent lacZ reporter gene. This showed that mutation of Pro205, Ile207, Ile209, and Thr210 abolished binding of A238L to CaN; replacement of the Gly at position 211 with Ala had no effect on binding of A238L to CaN, showing that this residue was not essential for binding (Table 1). The combined data from the random and directed mutagenesis defined the sequence PxIxITxC/S as critical for A238L binding to CaN (Table 1). This PxIxITxC motif is conserved in A238L genes from different ASFV isolates; the Cys212 residue is not substituted for Ser in any of the isolates sequenced (25).
The porcine macrophage NFAT gene contains a CaN binding motif similar to that in A238L.
The domain of NFAT proteins that is responsible for docking of NFAT to CaN also contains an essential PxIxIT motif (2, 3, 14). A PxIxITxC/S motif is present in murine and human NFAT1 and human NFAT2, human NFAT3/4 has an Ile residue in the position of the motif's Cys/Ser residue (Table 2). These species of NFAT contain a Ser residue in the position immediately after the relative position of Cys212 of A238L. The similarity of the NFAT motif suggests that the A238L protein may bind to CaN at the same site as NFAT proteins and hence inhibit their interaction with CaN.
TABLE 2.
Amino acid sequences of known CaN binding domains
| Identity | Accession no. | Amino acid sequence
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| Porcine NFAT2 | AF069996 | P | A | L | E | S | P | R | I | E | I | T | S | Y | L | G | L |
| A238L | Q65142 | W | F | K | K | K | P | K | I | I | I | T | G | C | K | N | N |
| VIVIT peptide | M | A | G | P | H | P | V | I | V | I | T | G | P | H | E | E | |
| Human NFAT1 | Q60591 | A | S | G | P | S | P | R | I | E | I | T | P | S | H | E | L |
| Human NFAT2 | U08015 | P | A | L | E | S | P | R | I | E | I | T | S | C | L | G | L |
| Human NFAT3 | Q14934 | R | V | L | E | C | P | S | I | R | I | T | S | I | S | P | T |
| Human NFAT4 | Q60896 | K | P | F | E | C | P | S | I | Q | I | T | S | I | S | P | N |
ASFV infects predominantly macrophages in vivo; to confirm that NFAT is expressed in porcine macrophages and identify the predicted CaN docking sequence, we isolated a clone encoding a full-length NFAT gene from a porcine macrophage cDNA library. This clone contained an insert of 2.8 kbp and encoded a protein of 822 amino acid residues that was most similar to human NFAT2 (NFATc.β isoform [U59736] [26]), with 89% amino acid similarity and 82% identity (Fig. 2A). We refer to this porcine macrophage NFAT gene as porcine NFAT2. Figure 2A shows the predicted amino acid sequence of the porcine NFAT2 ORF. The first ATG codon was located approximately 100 bp from the 5′ end of the clone, and the NH2-terminal predicted amino acid sequence was highly conserved compared to human NFAT2. Porcine NFAT2 contained a stop codon within 11 amino acid residues of the human NFAT2 stop codon, indicating that it was likely to be a full-length cDNA. Porcine NFAT2 contained the amino acid sequence SPRIEITSY in its putative CaN docking region. We cloned the 14-amino-acid region corresponding to residues 105 to 118 of the porcine polypeptide (sequence PALESPRIEITSYL) and residues 100 to 113 of NFAT2 (sequence PALESPRIEITSCL) into a Gal4 DNA binding domain vector; the fusions were analyzed for binding to CaN(A) in the yeast two-hybrid system. Both of these NFAT SPRIEIT fusions bound to CaN; this suggests that porcine NFAT2 protein binds to and is regulated by CaN, as occurs for NFAT1 to -4 (30). Transfected Myc-tagged porcine NFAT2 was detected by Western blotting and was partially dephosphorylated when cotransfected with constitutively active CaN (Fig. 2B). In contrast NFAT5, which is located in the nucleus in resting cells and is not regulated by CaN, does not have the CaN docking sequence (22).
A 14-amino acid-peptide encoding the CaN binding domain from A238L binds to CaN with high affinity.
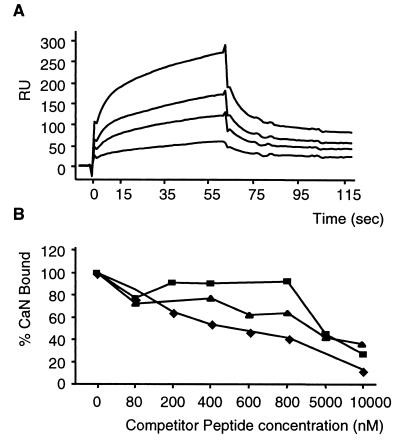
To confirm that the 14-amino-acid CaN binding motif within A238L was sufficient for binding to CaN and that other proteins were not required, we examined the binding of CaN to immobilized synthetic peptides by surface plasmon resonance. Binding of CaN to the A238Lwt14-biotin peptide was observed in a dose-dependent manner at CaN concentrations ranging from 40 to 240 nM (Fig. 3A). No significant binding to the control A238Lscram14-biotin peptide was observed even at high concentrations of ligand peptide and CaN. This showed that the 14-amino-acid domain from A238L was sufficient for binding of A238L to CaN, that other proteins were not required, and that the binding was high affinity (in the nanomolar range). A second control peptide (A238LThr20714 biotin) also showed no detectable CaN binding, confirming that mutating Ile to Thr at this position was sufficient to prevent the A238L-CaN interaction.
FIG. 3.
Biacore analysis of the interaction between CaN and A238L or NFAT peptides. (A) CaN binding to biotinylated wild-type A238L peptide (FC1) over a range of CaN concentrations (40, 80, 120, 240, and nM). Biotinylated scrambled peptide was bound to FC2. Kinetics of the interaction were calculated using the data from FC1 minus FC2 and the Biaevaluation program. RU, response units. (B) Effects of various concentrations of competitor peptides on the binding of CaN (80 nM) to biotinylated A238Lwt14 or the control peptide biotinylated A238Lscram14. The amount of CaN remaining bound to FC1 minus FC2 was calculated at 30 s postinjection and plotted as percentage of CaN bound in the absence of competitor peptide. Competitor peptides were A238Lwt14 (⧫; AAAWFKKKPKIIITGCK), NFATwt14 (▴; AAAPALESPRIEITSYL), and A238Lwt10 (■; KPKIIITGCK).
Although the CaN binding motifs from A238L and NFAT proteins contain the same critical residues, other residues in the domain differ considerably. The domain from A238L contains several basic and hydrophobic residues, whereas the NFAT sequences contain several acidic residues (Table 2). Since A238L acts as an inhibitor of CaN, it may bind with higher affinity to CaN than NFAT proteins, which are substrates for the enzyme and may form less tightly associated complexes. We wanted to determine if this was true for peptides containing the CaN binding domain. We compared the kinetics of CaN binding to A238Lwt14-biotin and NFATwt14-biotin peptides, using limiting amounts of peptide to minimize mass transport effects; A238Lscram14-biotin peptide was used as a control. Comparison of the kinetics of CaN binding to peptides over a range of flow rates (5 to 60 μl min−1) confirmed that mass transport effects were minimal. Values for the on and off rates and affinity constant were calculated over a range of CaN concentrations. The affinity constant of the interaction of CaN with the A238L peptide (1.22 to 3.11 e−8) was similar to that with the NFAT peptide (1.94 to 3.03 e−8), but the A238L peptide had a faster on rate (1.2 to 2.8 e6) than the NFAT peptide (3.7 to 7.0 e5). The off rate of the A238L peptide was also faster (1.6 to 3.3 e−2) than that of the NFAT peptide (8.9 e−3 to 1.1 e−2 [data not shown]). If the characteristics of the peptides reflect those of the full-length proteins, this would support our hypothesis that A238L binds CaN at a faster rate than NFAT proteins.
Different concentrations of nonbiotinylated competitor peptides were tested for the ability to inhibit the interaction between CaN and A238Lwt14-biotin (Fig. 3B). The A238Lwt14 peptide was the most effective inhibitor; a concentration of 600 nM was sufficient to reduce binding of CaN to 50% of that in the absence of competitor peptide. In comparison, 5 μM NFATwt14 peptide was needed for similar inhibition. A238Lwt10 peptide was less effective at inhibiting the interaction, possibly because the peptide was too short to mimic the three-dimensional structure of the CaN binding motif of A238L. These results show that a peptide containing the CaN binding motif from NFAT can compete with the A238L peptide for binding to CaN. This suggests that the two peptides bind at the same or overlapping sites on CaN and that the A238L peptide binds more effectively than the NFAT peptide to CaN. The interaction between A238L peptide and CaN was not inhibited by CsA-CypA complexes, indicating that these interact with CaN at a different site (data not shown).
A peptide containing the CaN binding domain from the A238L protein does not inhibit CaN phosphatase activity.
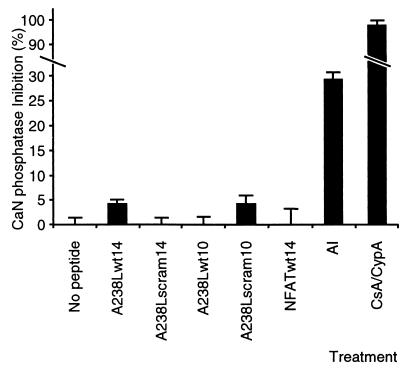
Our previous results show that expression of A238L protein inhibits CaN phosphatase activity in virus-infected cells (23). Our data suggest that A238L initially binds to CaN at the same site as NFAT proteins. Docking of NFAT with CaN via its binding motif brings a second site in the NFAT protein in proximity with the CaN active site and results in dephosphorylation of residues in this hyperphosphorylated domain by CaN (14). Peptides containing the NFAT CaN docking sequence do not inhibit CaN phosphatase activity, although they can block interaction of NFAT protein with CaN (2). We predicted that the A238L CaN binding sequence would also function to dock A238L on CaN but not inhibit its phosphatase activity. Docking might then allow another domain of the A238L protein to block the active site. We used an in vitro CaN phosphatase assay to determine if peptides containing the A238L CaN binding sequence inhibited CaN phosphatase activity. The A238L (A238Lwt14, A238Lwt10, A238Lscram14, and A238Lscram10) and NFAT (NFATwt14) peptides tested showed no significant inhibition of CaN phosphatase activity in this assay (Fig. 4). As expected, the control CaN AI peptide and CsA-CypA complexes inhibited CaN phosphatase activity by 29 and 98%, respectively.
FIG. 4.
Effects of peptides on CaN phosphatase activity in vitro. In vitro CaN phosphatase assays were carried out by incubating purified CaN and calmodulin with 32P-labeled RII phosphopeptide. Where indicated, peptides (A238Lwt14, A238Lwt10, A238Lscram14, A238Lscram10, NFATwt14, or CaN AI or complexes of CsA (15 μM) and CypA (5 μM) were added. Results shown mean percentage (± standard error of the mean) inhibition of CaN phosphatase activity in the presence of the indicated peptide or immunosuppressant complexes relative to untreated CaN and calmodulin. CaN phosphatase activity was assayed by measuring 32P released from the peptide substrate.
The PxIxITxC/S CaN binding motif from A238L is needed for interaction with CaN in mammalian cells.
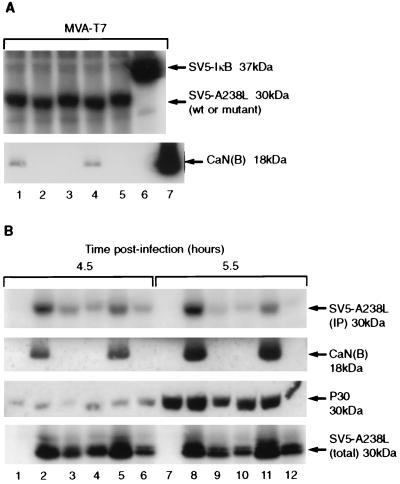
To confirm that our results using yeast to assay for CaN binding were valid for interactions of the two proteins in mammalian cells, A238L mutant proteins were expressed in mammalian cells and assayed for CaN binding. Three mutant non-CaN-binding A238L mutants (W13, W20, and Thr207) and one CaN-binding mutant (B19) were chosen for study (Table 3). The mutants were tagged at the NH2 terminus with the SV5 PK epitope tag and cloned downstream of a T7 promoter. These clones were transfected into Vero cells infected with MVA-T7. Expressed proteins were radiolabeled, immunoprecipitated with anti-PK tag antibody, blotted onto membranes, and reacted with anti-CaN(B) antibody to test for CaN coprecipitation with the A238L proteins. Approximately equal amounts of the A238L proteins and the control protein (SV5 PK epitope-tagged IκB) were immunoprecipitated from transfected cell extracts (Fig. 5A). CaN coprecipitated with the wild-type SV5-A238L and SV5-B19 mutant proteins but not with the SV5-W13, SV5-W20, SV5-Thr207 A238L proteins or with the SV5-IκB protein (Fig. 5A). These results demonstrated that the CaN binding motif is required for interaction of A238L with CaN in mammalian cells and that other ASFV-encoded proteins were not required for the interaction to occur.
TABLE 3.
Amino acid sequences of mutant A238L constructs W13, W20, B19, and Thr207 in the 14-amino-acid CaN binding regiona
| Mutant | Amino acid residue
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 | 201 | 202 | 203 | 204 | 205 | 206 | 207 | 208 | 209 | 210 | 211 | 212 | 213 | |
| W13 | G | F | T | T | T | S | K | I | I | I | T | G | S | K |
| W20 | W | V | K | K | K | S | K | I | T | I | A | G | S | K |
| B19 | G | V | T | T | T | P | K | I | I | I | T | G | C | T |
| Thr207 | W | F | K | K | K | P | K | T | I | I | T | G | C | K |
Mutations are in boldface.
FIG. 5.
Analysis of A238L mutants in cells. (A) Transient transfection analysis of A238L wild-type and mutant proteins to detect CaN coprecipitation. Vero cells were infected with MVA-T7 and transfected with pT7-SV5-A238L (lane 1), pT7-SV5-W13 (lane 2), pT7-SV5-W20 (lane 3), pT7-SV5-B19 (lane 4), pT7-SV5-Thr207 (lane 5), or pT7-SV5-IκB (lane 6). Bovine CaN was run as a control (lane 7). Radiolabeled cell extracts were immunoprecipitated with monoclonal anti-SV5 PK tag, and immune complexes were separated by SDS-PAGE. Immunoprecipitated SV5 PK-tagged proteins were detected by autoradiography. Coprecipitation of CaN was detected by protein immunoblot analysis of immune complexes with monoclonal anti-CaN(B). wt, wild type. (B) Vero cells were infected with wild-type ASFV (Ba71V; lanes 1 and 7), recombinant ASFV expressing SV5-tagged wild-type A238L (lanes 2 and 8), or SV5-tagged mutant W13 (lanes 3 and 9), W20 (lanes 4 and 10), B19 (lanes 5 and 11), or Thr207 (lanes 6 and 12). After 3.5 h, cells were pulse-labeled with [35S]Cys/Met for 1 h and harvested either immediately (lanes 1 to 6) or after a 1-h chase period with nonradioactive medium (lanes 7 to 12). Radiolabeled cell extracts were immunoprecipitated (IP) with monoclonal anti-SV5 PK tag antibody, and immune complexes were separated by SDS-PAGE. Radiolabeled immunoprecipitated SV5 PK-tagged A238L proteins were detected by autoradiography (top). Coprecipitation of CaN was detected by protein immunoblot analysis of immune complexes with monoclonal anti-CaN(B). Samples of cell extracts prior to immunoprecipitation were analyzed by SDS-PAGE and Western blot analysis with monoclonal anti-ASFV P30 antibody or monoclonal anti-SV5 PK tag antibody to detect total cellular A238L protein (bottom).
Non-CaN-binding mutants of A238L protein are turned over more rapidly than CaN-binding A238L proteins when expressed by recombinant ASFV.
To analyze expression of the mutant A238L proteins in ASFV-infected cells, we constructed ASFV recombinants in which the wild-type ASFV gene was replaced with mutated genes expressing PK epitope-tagged mutant forms of the A238L protein. The recombinant viruses expressed A238L mutants W13, W20, B19, and Thr207. Vero cells were infected with these viruses and pulse-labeled for 1 h at 3.5 h postinfection. PK-tagged A238L proteins were immunoprecipitated from cell extracts with anti-PK antibody, blotted, and probed with anti-CaN(B) antiserum. CaN coprecipitated with A238L from cells infected with ASFV expressing wild-type A238L or the B19 mutant but not with the other three mutant A238L proteins (W13, W20, and Thr207) (Fig. 5B).
Unexpectedly, in cells infected with ASFV recombinants, the [35S]Met/Cys labeling of CaN-binding A238L proteins (wild type and B19 mutant) during 1 h was considerably greater than that of the three non-CaN-binding mutants of A238L (Fig. 5B, top). The recombinant viruses expressed similar levels of another virus-encoded early protein, P30 (Fig. 5B), and showed similar kinetics of virus production (data not shown), suggesting that the A238L proteins had varied stability. The relative stabilities of wild-type and mutant A238L proteins were compared. Cells infected with mutant ASFV were radiolabeled for 1 h at 3.5 h postinfection and either harvested immediately or chased with nonradioactive medium for 1 h. PK-tagged A238L proteins were immunoprecipitated from cell extracts and analyzed by SDS-PAGE and fluorography. The CaN-binding species of A238L (wild type and B19) were labeled at comparable levels when immunoprecipitated immediately or after a 1-h chase period. The three non-CaN-binding A238L proteins (W13, W20, and Thr207) were less efficiently labeled when harvested immediately after labeling (Fig. 5B, top, compare lanes 3, 4, and 6 with lanes 2 and 5). After a 1-h chase period in nonradioactive medium, the amount of radiolabeled non-CaN-binding A238L protein was less than in the samples collected immediately after labeling (Fig. 5B, top, compare lanes 9, 10, and 12 with lanes 3, 4, and 6). The total amounts of wild-type and mutant A238L proteins in these cell extracts were assessed by SDS-PAGE followed by blotting and reaction with anti-PK antibody. Densitometric analysis showed there was approximately 58% as much non-CaN-binding as CaN-binding A238L (Fig. 5B, bottom). The results suggest that in ASFV-infected cells, A238L is stabilized because of its interaction with CaN. The changes in stability are not likely to result from changes in protein structure, as Thr207 differed from wild-type A238L in a single amino acid residue. In the transient expression experiments similar levels of all mutant proteins were detected, possibly because the A238L expression level is higher in individual transfected cells than in ASFV-infected cells (data not shown). Thus, the amount of A238L protein may be in excess of the amount of enzymes mediating its degradation.
Non-CaN-binding A238L mutants do not inhibit CaN phosphatase activity.
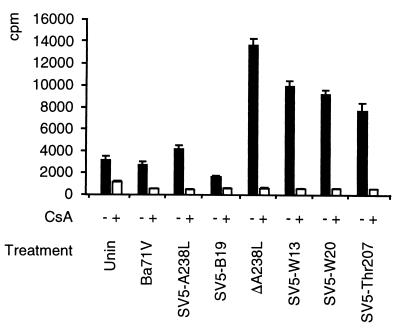
We predict that the CaN binding motif in A238L is needed for the protein to dock with CaN(A). Abolishing A238L binding to CaN should also prevent its inhibition of CaN phosphatase activity. Extracts were prepared from uninfected Vero cells, or cells infected for 11 h with wild-type (Ba71V) or recombinant (ΔA238L [23], SV5-A238L [23], SV5-W13, SV5-W20, SV5-B19, or SV5-Thr207) ASFV, and CaN phosphatase activity was measured (Fig. 6). CaN activity was lower in extracts from cells infected with ASFV expressing wild-type A238L (Ba71V or SV5-A238L) or a mutant of A238L which binds CaN (SV5-B19) than in extracts from cells infected with A238L deletion mutant ΔA238L or ASFV expressing a non-CaN-binding A238L mutant (SV5-W13, SV5-W20, or SV5-Thr207). These assays were specific for CaN activity; CsA reduced the phosphatase activity to background levels. As discussed above, non-CaN-binding mutant A238L proteins are more rapidly turned over than CaN-binding forms of the protein in ASFV-infected cells. Nevertheless, there is 58% as much non-CaN-binding as CaN-binding A238L proteins present in these cells, sufficient to inhibit more than half of the CaN activity if the A238L protein retained this activity. Our results allow us to conclude that there is a direct correlation between the ability of A238L proteins to bind to CaN and their ability to inhibit CaN phosphatase activity.
FIG. 6.
CaN phosphatase assays of extracts from uninfected cells (Unin) or cells infected with wild-type (Ba71V) or recombinant (ΔA238L, SV5-A238L, SV5-W13, SV5-W20, SV5-B19, or SV5-Thr207) ASFV. Cells were untreated (−) or treated with CsA (+). Results are shown as mean (± standard error of the mean) of 32P released from 32P-labeled RII phosphopeptide substrate.
DISCUSSION
We have previously shown that the ASFV A238L protein binds to CaN and inhibits its phosphatase activity (23). In this study we have mapped a 14-amino-acid sequence needed for binding of A238L to CaN and identified a critical motif (PxIxITxC/S) within this sequence. A238L proteins with single amino acid substitutions within these critical residues do not detectably bind to CaN. A similar motif (PxIxIT) required for binding of NFAT proteins to CaN has been identified (2, 3). This suggests that A238L and NFAT proteins bind to CaN at the same site and raises the possibility that other CaN-interacting proteins, including substrates and regulatory proteins, may bind at similar sites and contain similar binding motifs. The same region of CaN(A) is required for binding both A238L and NFAT; CaN(A) residues 32 to 350 bind A238L, and residues 1 to 347 bind NFAT (14). The site(s) on CaN(A) to which A238L and NFAT proteins bind has not been defined, but CsA-CypA complexes do not compete for initial CaN binding with A238L or NFAT protein (2, 23) or with A238L peptide. These results indicate that A238L initially binds to CaN at a site distinct from that bound by the immunosuppressive drug complexes, as was found with NFAT (2, 14). NFAT interacts with CaN via a second site following cell activation (14). It is likely that A238L also has a second site of interaction with CaN, presumably near the CaN active site, and it is through binding at this site that inhibition of CaN phosphatase activity by A238L occurs. We showed that macrophages, the main cells infected by ASFV in vivo, express CaN-regulated NFAT2. We have also demonstrated that A238L inhibits activation of an NFAT-dependent reporter gene in transfected cells (23). Thus, one of the downstream effects of inhibition of CaN by A238L may be to inhibit NFAT-dependent gene transcription in macrophages.
A238L inhibits CaN phosphatase activity, whereas NFAT proteins are substrates for the enzyme. NFAT has presumably evolved a mechanism to bind to CaN with a low to moderate affinity, thus preventing inappropriate NFAT activation at subthreshold levels of stimulus (3). Inhibition of CaN phosphatase activity by A238L may require a higher affinity of binding to CaN than NFAT, enabling A238L to displace the normal cellular substrate(s) of the enzyme. An artificial NFAT-derived peptide sequence (designated VIVIT) was 25 times more effective at inhibiting the NFAT interaction with activated CaN than the wild-type NFAT (SPRIEIT) peptide (3). ASFV has evolved a sequence that resembles the artificial VIVIT more closely than the natural SPRIEIT sequence. Table 2 shows an alignment of the amino acid residues within known CaN binding domains. NFAT1, NFAT2, and porcine macrophage NFAT have a large basic Arg residue at position 7 of the docking site. In this position, A238L has a small, less basic Lys residue, partway to the small uncharged Val residue in the VIVIT peptide. Likewise, the acidic Glu residue at position 9 in the NFAT docking sequences is a hydrophobic residue in A238L (Ile) and the artificial VIVIT sequence (Val). Indeed, A238L peptide binds CaN with faster on rates compared to NFAT peptide, and is more effective at competing for binding sites on CaN than NFAT peptide. Evolution of sequences within CaN docking motifs may determine whether the interactions are weak and transitory (e.g., enzyme-substrate interaction) or strong (e.g., inhibitors or anchoring proteins).
The PxIxIT(x)C/S CaN binding motif may be found in other CaN substrates and regulatory proteins; the FindPatterns algorithm (Genetics Computer Group) was used to search the sequence databases to identify other proteins containing this motif. Examples were gamma isoforms of the mitogen-activated kinase kinase 7 (38) and NPAT (41). It remains to be determined if these are functional CaN binding motifs; the indications are that this may be a useful method to identify novel CaN-binding proteins. Interestingly, two of the characterized calcineurin-binding cellular proteins contain similar motifs; Cain (or Cabin1) contains a PEITVT motif within its 38-amino-acid CaN binding domain (19, 36), and MCIP1 contains a PKIIQT motif which contributes to the interaction with calcineurin (33).
Our observation that A238L proteins are destabilized in ASFV-infected cells when the CaN binding motif is mutated suggests that the A238L protein is normally present in a complex with CaN. It is possible that a number of other proteins, such as NF-κB, are present in these complexes; the P65 subunit of NF-κB binds A238L (32). The importance of multiprotein complexes in the control of signaling pathways is well known; many tyrosine kinases and tyrosine phosphatases are coupled to downstream cytoplasmic enzymes through adapter proteins containing SH2 and SH3 domains (27). It is through these interactions that signaling complexes are formed (7, 12). IκB and the IκB serine kinases are also found in large (500- to 900-kDa) multiprotein complexes in normal cells (16, 31, 39). The A238L interaction with CaN may be important to stabilize the protein; further work is required to determine whether interaction with CaN is required for A238L to inhibit NF-κB activation.
The unique ability of A238L to inhibit both CaN phosphatase activity and NF-κB activation provides ASFV with a powerful and versatile mechanism to evade the host immune responses by inhibiting expression of the many immunomodulatory genes whose transcription is activated by these pathways. Understanding the mechanisms by which A238L carries out these dual functions will enable us to gain new insights into how these pathways are controlled and may provide information relevant to the design of novel immunomodulatory drugs.
ACKNOWLEDGMENTS
We thank Steven Archibald for assistance in preparation of the figures. We also thank Ronald Hay, University of St. Andrews, for his kind gift of pT7-SV5-IκB and E. McKenzie, Yamanouchi Research Institute, for providing a constitutively active CaN clone.
This work was funded by the Biological and Biotechnology Science Research Council and the Ministry of Agriculture Fisheries and Food.
REFERENCES
- 1.Alcami A, Smith G L. Cytokine receptors encoded by poxviruses: a lesson in cytokine biology. Immunol Today. 1995;16:474–478. doi: 10.1016/0167-5699(95)80030-1. [DOI] [PubMed] [Google Scholar]
- 2.Aramburu J, García-Cózar F, Raghavan A, Okamura H, Rao A, Hogan P G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 3.Aramburu J, Yaffe M B, López-Rodriguez C, Cantley L C, Hogan P G, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 4.Blaeser F, Ho N, Prywes R, Chatila T A. Ca2+-dependent gene expression mediated by MEF2 transcription factors. J Biol Chem. 2000;275:197–209. doi: 10.1074/jbc.275.1.197. [DOI] [PubMed] [Google Scholar]
- 5.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coghlan V M, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W M, Scott J D. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 7.Colledge M, Scott J D. AKAPs: from structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree G R. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 9.Davis-Poynter N J, Farrell H E. Masters of deception: a review of herpesvirus immune evasion strategies. Immunol Cell Biol. 1996;74:513–522. doi: 10.1038/icb.1996.84. [DOI] [PubMed] [Google Scholar]
- 10.Dixon L K, Rock D L, Viñuela E. In: African swine fever-like viruses. Murphy F A, et al., editors. 1995. pp. 92–94. (et al.),Virus taxonomy: classification and nomenclature of viruses. Sixth Report of the International Committee on Taxonomy of Viruses.Springer-Verlag, Vienna, Austria. [Google Scholar]
- 11.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 12.Faux M C, Scott J D. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 13.Fruman D A, Pai S-Y, Klee C B, Burakoff S J, Bierer B E. Measurement of calcineurin phosphatase activity in cell extracts. Methods Companion Methods Enzymol. 1996;9:146–154. doi: 10.1006/meth.1996.0020. [DOI] [PubMed] [Google Scholar]
- 14.García-Cózar F J, Okamura H, Aramburu J F, Shaw K T Y, Pelletier L, Showalter R, Villafranca E, Rao A. Two-site interaction of nuclear factor of activated T cells with activated calcineurin. J Biol Chem. 1998;273:23877–23882. doi: 10.1074/jbc.273.37.23877. [DOI] [PubMed] [Google Scholar]
- 15.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzbiggon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 16.Karin M. The beginning of the end: IκB kinase (IKK) and NF-κB activation. J Biol Chem. 1999;274:27339–27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 17.Kashishian A, Howard M, Loh C, Gallatin W M, Hoekstra M F, Lai Y. AKAP79 inhibits calcineurin through a site distinct from the immunophilin-binding region. J Biol Chem. 1998;273:27412–27419. doi: 10.1074/jbc.273.42.27412. [DOI] [PubMed] [Google Scholar]
- 18.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, Gastinel L N, Habuka N, Chen X, Maldonado F, Barker J E, Bacquet R, Villafranca J E. Crystal structures of human calcineruin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 19.Lai M M, Burnett P E, Wolosker H, Blackshaw S, Snyder S H. Cain, a novel physiologic protein inhibitor of calcineurin. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 20.Lin X, Barber D L. A calcineurin homologous protein inhibits GTPase-stimulated Na-H exchange. Proc Natl Acad Sci USA. 1996;93:12631–12636. doi: 10.1073/pnas.93.22.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin X, Sikkink R A, Rusnak F, Barber D L. Inhibition of calcineurin phosphatase activity by a calcineurin B homologous protein. J Biol Chem. 1999;274:36125–36131. doi: 10.1074/jbc.274.51.36125. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Rodriguez C, Aramburu J, Rakeman A S, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miskin J E, Abrams C C, Goatley L C, Dixon L K. A viral mechanism for inhibition of the cellular phosphatase calcineurin. Science. 1998;281:562–565. doi: 10.1126/science.281.5376.562. [DOI] [PubMed] [Google Scholar]
- 24.Mosser D M, Karp C L. Receptor mediated subversion of macrophage cytokine production by intracellular pathogens. Curr Opin Immunol. 1999;11:406–411. doi: 10.1016/s0952-7915(99)80068-5. [DOI] [PubMed] [Google Scholar]
- 25.Neilan J G, Lu Z, Kutish G F, Zsak L, Lewis T L, Rock D L. A conserved African swine fever virus IκB homolog, 5EL, is nonessential for growth in vitro and virulence in domestic swine. Virology. 1997;235:377–385. doi: 10.1006/viro.1997.8693. [DOI] [PubMed] [Google Scholar]
- 26.Park J, Takeuchi A, Sharma S. Characterization of a new isoform of the NFAT (nuclear factor of activated T cells) gene family member NFATc. J Biol Chem. 1996;271:20914–20921. doi: 10.1074/jbc.271.34.20914. , 33705. [DOI] [PubMed] [Google Scholar]
- 27.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 28.Powell P P, Dixon L K, Parkhouse R M E. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J Virol. 1996;70:8527–8533. doi: 10.1128/jvi.70.12.8527-8533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao A. NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol Today. 1994;15:274–281. doi: 10.1016/0167-5699(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 30.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 31.Régnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 32.Revilla Y, Callejo M, Rodriguez J M, Culebras E, Nogal M L, Salas M L, Vinuela E, Fresno M. Inhibition of nuclear factor κB activation by a virus-encoded IκB-like protein. J Biol Chem. 1998;273:5405–5411. doi: 10.1074/jbc.273.9.5405. [DOI] [PubMed] [Google Scholar]
- 33.Rothermel B, Vega R B, Yang J, Wu H, Bassel-Duby R, Williams R S. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J Biol Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber S L, Crabtree G R. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 35.Stoddard B L, Flick K E. Calcineurin-immunosuppressor complexes. Curr Opin Struct Biol. 1996;6:770–775. doi: 10.1016/s0959-440x(96)80006-6. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Youn H-D, Loh C, Stolow M, He W, Liu J O. Cabin1, a negative regulator for calcineurin signaling in T lymphocytes. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- 37.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 38.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol Cell Biol. 1999;19:1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β. NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 40.Youn H-D, Sun L, Prywes R, Liu J O. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–783. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]