Abstract
Clostridioides difficile causes a wide range of intestinal diseases through the action of two main cytotoxins, TcdA and TcdB. Ingested spores germinate in the intestine establishing a population of cells that produce toxins and spores. The pathogenicity locus, PaLoc, comprises several genes, including those coding for TcdA/B, for the holin-like TcdE protein, and for TcdR, an auto-regulatory RNA polymerase sigma factor essential for tcdA/B and tcdE expression. Here we show that tcdR, tcdA, tcdB and tcdE are expressed in a fraction of the sporulating cells, in either the whole sporangium or in the forespore. The whole sporangium pattern is due to protracted expression initiated in vegetative cells by σD, which primes the TcdR auto-regulatory loop. In contrast, the forespore-specific regulatory proteins σG and SpoVT control TcdR production and tcdA/tcdB and tcdE expression in this cell. We detected TcdA at the spore surface, and we show that wild type and ΔtcdA or ΔtcdB spores but not ΔtcdR or ΔtcdA/ΔtcdB spores are cytopathic against HT29 and Vero cells, indicating that spores may serve as toxin-delivery vehicles. Since the addition of TcdA and TcdB enhance binding of spores to epithelial cells, this effect may occur independently of toxin production by vegetative cells.
Subject terms: Pathogens, Microbiology
This study shows that the genes coding for the two main toxins produced by Clostridioides difficile are expressed during sporulation and that the toxins associate with the mature spores, which have a cytopathic effect on human intestinal cell lines.
Introduction
Clostridioides difficile is a major nosocomial pathogen and the leading cause of intestinal diseases that range from mild diarrhea to life-threatening illnesses, linked to the use of antibiotics1,2. In the last two decades, the emergence and spreading of epidemic clones of ribotype 027 was responsible for worldwide outbreaks associated with more severe disease symptoms, recurrence rates, morbidity and mortality1,2. The epidemiology of C. difficile is however changing, with new ribotypes disseminating both in healthcare units and at the community level, and with increased incidence among groups not previously considered at risk3. Moreover, the prevalence of some ribotypes in animals used for human consumption raises serious concerns of widespread dissemination through the food chain4,5.
Infection is initiated by the ingestion of spores when gut dysbiosis, most frequently due to continued antibiotic treatment, allows spores to germinate in the intestine6,7. For many strains, the resulting cells produce two cytotoxins, TcdA and TcdB, which are the main factors responsible for the disease symptoms, and spores1,2,8. Spores are highly resistant dormant cells, hard to eradicate, that allows dissemination of this strict anaerobic pathogen6,7,9,10. Spores also allow C. difficile to persist in the environment and in the host and are linked to disease recurrence6,7,11,12. A recent study shows that spores enter intestinal epithelial cells and may persist in this intracellular niche contributing to disease recurrence13.
Spores are formed during the stationary phase of growth. Initially, an asymmetric division partitions the rod-shaped cell into a larger mother cell and a smaller forespore, the future spore (Fig. 1a)6,7. Soon after division, the mother cell begins to engulf the forespore, which eventually becomes isolated from the external medium. Several protective layers are then deposited around the forespore, including a peptidoglycan layer known as the cortex essential for heat resistance, and two proteinaceous layers, the coat and a more external exosporium. These two structures contribute to spore resistance against noxious chemicals and peptidoglycan-breaking enzymes, are required for proper germination, binding to host cells and dramatically influence colonization and disease-causing ability13–19. Finally, lysis of the mother cell releases the spore into the environment6,7 (Fig. 1a). Although there are differences in the morphology of different spore layers among species, the main morphological stages of sporulation are conserved among spore-formers6,7.
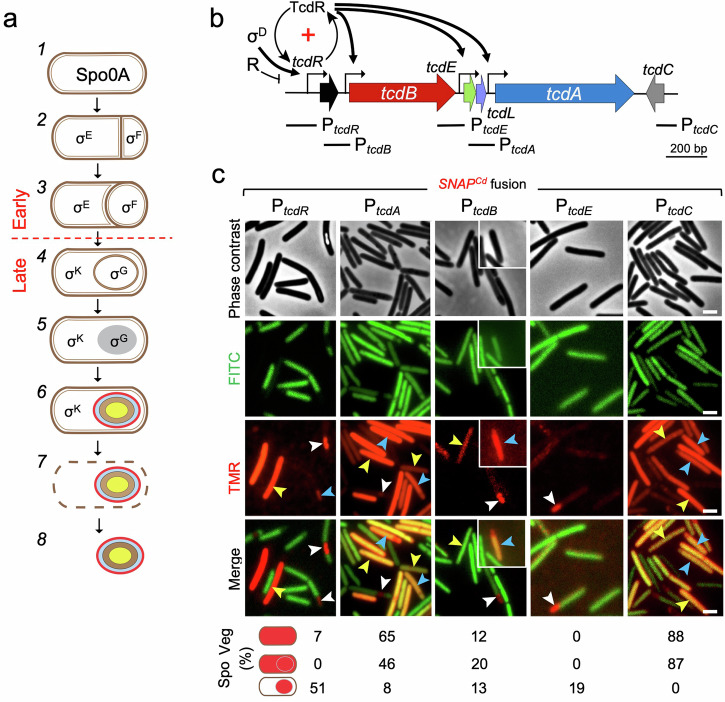
Fig. 1. Expression of the PaLoc genes during C. difficile growth and sporulation.
a Shown is the pathway of spore differentiation starting with vegetative (pre-divisional cells, (1), asymmetric division (2), a stage in engulfment (3), engulfment completion (4), synthesis of the spore protective layers (5 and 6) and free mature spores (8), resulting from mother cell lysis (7). Spo0A controls several stationary phase processes and is also essential for entry into sporulation. Cell type-specific gene expression results from the action of the indicated σ factors. Early and late stages in development are defined as those occurring prior to or following engulfment completion, as indicated. b Genetic organization of the PaLoc. Broken arrows represent promoters in the region; the TcdR positive auto-regulatory loop (“+” sign) and the role of σD in its priming are indicated. The black arrows represent direct regulation of the indicated promoters by TcdR. Other direct or indirect regulatory factors that impinge onto the expression of the PaLoc-encoded genes are collectively represented by “R”. The lines below the genetic map show the position and extent of the DNA fragments used to create the indicated transcriptional SNAPCd fusions. Note that the PtcdR fragment includes two tcdR-dependent promoters (P1 and P2), the σD-dependent promoter and a σA-type promoter (see also Fig. 3). c Cell type-specific expression patterns of PtcdR-, PtcdA-, PtcdB-, PtcdE- and PtcdC-SNAPCd transcriptional fusions in strain 630Δerm. The cells were collected 24 h after inoculation in TY liquid medium, labeled with TMR-Star and examined by phase contrast and fluorescence microscopy to monitor SNAPCd production. The merged images show the overlap between the TMR-Star (red) and the auto-fluorescence (green) channels. The images are representative of the expression patterns observed for the different fusions in three independent experiments. Yellow arrowheads point to vegetative cells with expression, white arrowheads point to sporulating cells with forespore-specific expression and blue arrowheads point to sporulating cells with a whole sporangium expression pattern. The various cellular patterns of SNAPCd production were scored and their percentage relative to the total number of vegetative (Veg) or sporulating cells (Spo) is shown. The images are representative of the expression patterns observed for the different fusions in three independent experiments (see also Fig. S1 and the Methods section). For sporulating cells the scoring includes a whole sporangium expression pattern and a forespore-specific pattern. The number of cells analyzed for each fusion, n, is as follows: PtcdA-SNAPCd, n = 245; PtcdB-SNAPCd, n = 410; PtcdC-SNAPCd, n = 400; PtcdE-SNAPCd, n = 579; PtcdR-SNAPCd, n = 353. Scale bar, 1 μm.
Spo0A is a response regulator and the master regulatory protein governing entry into sporulation20–22. Spo0A is essential for the asymmetric division of the rod-shaped cell into a smaller forespore and a larger mother cell23,24 (reviewed by refs. 6,7,9; Fig. 1a). Once the forespore and the mother cell are formed, σF is activated in the forespore and σE is soon after activated in the mother cell. At later stages in development, σK is the main factor driving developmental regulated gene expression in the mother cell, whereas σG replaces σF in the forespore23–25 (Fig. 1a). While gene expression during sporulation is mainly governed by the cell type-specific sigma factors, ancillary transcription factors sub-divide the sigma regulons into several temporal and epistatic classes20–22,26. For example, SpoVT is produced in the forespore under the control of both σF and σG, and positively regulates the expression of a subset of σG-controlled genes while repressing genes under the control of σF9,25. Importantly, the mother cell-specific σE and σK are the key players in the morphogenesis of the spore surface layers23–25,27.
The genes coding for TcdA and TcdB are located in a pathogenicity locus, or PaLoc1,2,8 (Fig. 1b). TcdA and TcdB are glucosyltransferases that belong to the family of Large Clostridial Toxins (LCTs) (reviewed in8,28). The toxins enter host cells via receptor-mediated endocytosis and several TcdA receptors have been proposed8,28,29. The PaLoc carries four other genes, tcdR, tcdE, tcdL, and tcdC (Fig. 1b). tcdR codes for an RNA polymerase sigma factor, TcdR, required for the transcription of tcdA, tcdB and tcdE30,31 (Fig. 1b). tcdC codes for a small acidic transmembrane protein thought to be a negative regulator of toxin production32, although its role is still unclear33–38. The tcdE gene codes for a holin-like protein; expression of tcdE in Escherichia coli complements a lambda S mutant, and under certain conditions causes cell lysis39. Although neither TcdA nor TcdB have recognizable secretion signals, at least in some strains TcdE appears to be required for TcdA and TcdB release39–42 in a process proposed to be partially redundant with stationary phase autolysis mediated by a lytic transglycosylase43. Finally, tcdL codes for a 43 amino acid-long polypeptide with structural similarity to a non-catalytic fragment of a peptidoglycan endolysin and it binds to TcdB suggesting a role in toxin transport44,45.
Expression of tcdA and tcdB is strongly induced early in stationary phase and persists during this phase46,47. Toxin production is subject to the action of several regulatory proteins and signals that converge to limit expression of the toxin-encoding genes during growth (Fig. 1b, collectively denoted as R; reviewed in ref. 48). The sigma factor σD, in turn, a central regulator of flagellar assembly49,50 is a positive regulator of toxin production; it uses a promoter upstream of tcdR to prime a positive auto-regulatory loop involving two TcdR-dependent tcdR promoters51,52 (Fig. 1b). The TcdR auto-regulatory loop establishes a bistable switch that results in the expression of tcdA in only a fraction of the population52,53.
Some of the regulators and signals that repress toxin production also repress sporulation initiation (reviewed in ref. 48). For instance, the regulatory protein RstA, promotes sporulation while simultaneously acting as a repressor of toxin production, by binding to a site upstream of and overlapping the σD-dependent promoter of the tcdR gene54–56. The regulatory systems that influence both sporogenesis and toxinogenesis may act mainly to bias the early stationary phase cell population towards toxin production or sporulation, in response to nutritional and other signals (52–54,56 reviewed in ref. 48). The two processes are not antithetical, however, but partially overlap as illustrated by the observation that a fraction of sporulating cells express tcdA52,53. Expression of tcdA was detected in the mother cell52,53, but it was unclear whether expression also occurred in the forespore, whether other PaLoc genes were expressed in sporulating cells, their regulation during sporulation and whether the toxins associated with spores. These are important questions, however, since binding of spores to intestinal epithelial cells, their internalization and contribution to disease recurrence is promoted by TcdA and TcdB13,57.
Using single-cell analysis, we show here that in a fraction of the sporulating cells, tcdR, tcdA, tcdB and tcdE exhibit forespore-specific expression, that tcdA and tcdB additionally show a whole-sporangium pattern, and that tcdC only shows this latter pattern. We found that expression of the PaLoc genes in the mother cell is σD-dependent and most likely results from protracted σD- and TcdR-dependent expression by vegetative cells. In contrast, we found that tcdR expression is specifically induced in the forespore under the joint control of σG and the ancillary transcription factor SpoVT. Together with SpoVT, σG utilizes a promoter in the tcdR regulatory region that partially overlaps the σD promoter, leading to the expression of tcdA, tcdB and tcdE in the forespore at a late stage in development. We demonstrate that TcdA associates with the spore surface layers and we show that wild-type spores, including those of an epidemic strain of ribotype 027, have a cytopathic effect on monolayers of HT29 and Caco-2 intestinal cells. Thus, at least TcdA is associated with mature spores in an active form and the infectious spores of C. difficile are toxin delivery vehicles.
Results
Expression of the PaLoc genes in vegetative cells
Previous work has shown that in strain 630Δerm a substantial fraction of the sporangia of phase bright spores, i.e., at a late stage in development, expressed a PtcdA-rfp fusion52. In a more recent study, and using a dual reporter system in which tcdA expression was monitored using a transcriptional fusion to mNeon Green (mNG) and sporulating cells were identified using a transcriptional fusion of the σE-controlled sipL promoter to mScarlet (mSc), the simultaneous expression of both fusions was detected in 11% of the sporulating cells53. While the two studies showed that sporulation and tcdA expression overlap to some extent, the regulation of tcdA expression in sporulating cells was not directly addressed, and the expression of the other PaLoc genes during sporulation was not reported52,53. Moreover, in the study of Ransom and co-authors, some free spores exhibited red fluorescence but whether tcdA was also expressed in the forespore or whether the reporter, produced in the mother cell, associated with the developing spore was unclear52. Both studies found that the auto-fluorescent proteins used were not sensitive forespore-specific reporters52,53.
Here, we have constructed derivatives of strain 630Δerm bearing transcriptional fusions of the tcdA, tcdB, tcdC, tcdE and tcdR promoter regions to the SNAPCd reporter58,59 and used phase contrast and fluorescence microscopy to monitor the expression of the PaLoc genes across the cell population during sporulation. Note that no promoter has been identified between the tcdE and tcdL genes44,45. Also of note, the tcdA promoter fragment used for the construction of the SNAPCd transcriptional fusion is similar to that used in the two studies mentioned above52,53 (Fig. 1b; see also the Supplementary Information). The PtcdR-SNAPCd transcriptional fusion is termed full-length to distinguish it from a shorter version, described in a section below.
Samples were collected from cultures of strains bearing the various fusions 24 h after inoculation in TY, a medium that supports both sporulation and toxin production46,52,53. The cells were labeled with the red-fluorescent SNAP substrate TMR-Star and processed for phase contrast and fluorescence microscopy. Sporulation was evaluated by both the accumulation of partially or fully phase bright spores and by the pattern of green auto-fluorescence characteristic of C. difficile60; auto-fluorescence allows division septa and the forespore, which shows reduced auto-fluorescence relative to the mother cell, to be identified (27,60; Supplementary Fig. S1a). For reference, we also scored expression of the PaLoc genes in vegetative cells52,53. Expression of PtcdA-SNAPCd was detected in 65% of the vegetative cells (TcdA-ON cells; Fig. 1c, yellow arrowheads) while the remaining cells showed no signal. This bimodal pattern of PtcdA-SNAPCd expression is consistent with earlier results: PtcdA-rfp expression was detected in 85% of the vegetative cells in the study of Ransom and co-authors52, and in 61% or 37% of the cells as assessed with the mNG or mSc reporters, respectively, by Donnelly and co-authors53. Transcription of tcdR or tcdB was not detected using the RFP reporter in 630Δerm; in a congenic ΔcodY mutant, however, tcdB expression was detected and was bimodal52. In the present study, expression of PtcdB-SNAPCd was detected in 12% of the vegetative cells of the 630Δerm and was thus bimodal (Fig. 1c, yellow arrowheads). Previous work has shown that tcdB has around 10-100-fold lower expression levels when compared with tcdA33,61,62. It thus seems possible that the lower fraction of TcdB-ON cells, relative to the fraction of TcdA-ON cells, is because the signal from PtcdB-SNAPCd is, in a fraction of cells, too close to the background to be detected. Expression of PtcdR-SNAPCd itself, was only detected in a sub-population of about 7% of the vegetative cells (Fig. 1c, yellow arrowheads). The lower fraction of cells may again reflect our detection limit and/or the rate of transcription initiation from the tcdR promoter, which is lower than from the tcdA or tcdB promoters46.
The bimodal pattern of tcdA expression has its origin in the TcdR auto-regulatory loop and one prediction was that the expression of tcdB and of tcdR itself could also be bimodal52. Here, we confirm this expectation, in that as for tcdA, only a fraction of the cells are in a TcdB-ON or TcdR-ON state. In contrast, we did not detect expression of tcdE in vegetative cells (Fig. 1c). Finally, tcdC expression was detected in 88% of the vegetative cells; this gene, however, is not known to be under TcdR control (Fig. 1c, yellow arrowheads). In all cases, complete labeling of the SNAPCd reporter was achieved (Supplementary Figs. S2 and S9). Although there was some variation in the percentage of ON/OFF cells between experiments, the general pattern of bimodal transcription, verified for the PtcdR and PtcdA promoters is maintained (7 ± 3% for TcdR-ON cells; 60 ± 12% for TcdA-ON cells, as assessed in 8 independent experiments; Supplementary Fig. S1b).
Expression of the PaLoc genes in sporulating cells
We then examined the expression of the PaLoc genes during sporulation. Control experiments showed very similar sporulation kinetics and efficiencies (all below 10%) and kinetics for the strains bearing the various transcriptional fusions as measured 12, 24, and 48 h after inoculation (Supplementary Fig. S1c). The exception was the PtcdR-SNAPCd-bearing strains, which showed an efficiency of sporulation below 10% at hour 12 but greater than 50% at hours 24 and 48 (Supplementary Fig. S1c). The reason for this behavior is not presently understood but one possibility is that the DNA fragment fused to the SNAPCd reporter contains a site that titrates out a negative regulator of sporulation. In any event, it seems unlikely that the increased sporulation efficiency of the PtcdR-SNAPCd-bearing strain introduces a bias in our scoring of the fraction of tcdR-ON cells.
During sporulation, expression of tcdR was detected in 51% of the sporulating cells, specifically in the forespore (Fig. 1c, white arrowheads). The forespore-specific expression of tcdR was mainly seen in sporangia that had completed the engulfment process and thus, at a late stage in development (Fig. 1c). Expression of tcdA was also detected in sporulating cells, but with a more complex pattern than that found for tcdR; while expression was confined to the forespore in 8% of the sporangia scored (Fig. 1c, white arrowheads), 46% showed a whole sporangia pattern, i.e., expression in both the forespore and the mother cell (Fig. 1c, blue arrowheads). Importantly, we did not detect expression of tcdA only in the mother cell; in the study of Ransom and co-authors, what seems to be mother cell-specific tcdA expression may result from the low sensitivity of the RFP reporter in the forespore (see also above52;). SNAPCd, however, is an efficient reporter for gene expression in the forespore24,25,27,58 .
As for tcdA, expression of tcdB was also detected in sporulating cells; of these, 13% showed expression only in the forespore (Fig. 1c, white arrowheads), whereas 20% showed a whole sporangium pattern (blue arrowhead). Contrasting with the vegetative cells, expression of tcdE was only detected, albeit weakly, in sporulating cells and only in the forespore (19% of the sporangia scored; Fig. 1c, white arrowheads). Finally, 87% of the sporulating cells showed a whole sporangia pattern of PtcdC-SNAPCd expression (Fig. 1c, blue arrowheads), But unlike the other PaLoc genes, expression of tcdC was not detected in the forespore only. As also shown for the expression in vegetative cells (above), there was some variation between experiments in the percentage of cells showing PtcdR- and PtcdA-SNAPCd expression in whole-sporangia (52 ± 10% for tcdA, less that 3% for tcdR) or the forespore (50 ± 10% for tcdR; 5 ± 3% for tcdA) (Supplementary Fig. S1b).
In summary, in line with earlier work52, we detected expression of tcdA in sporulating cells. Moreover, we found that tcdR, tcdA, tcdB and tcdE exhibit forespore-specific expression, while tcdA and tcdB additionally show a whole-sporangium pattern, and tcdC only shows this latter pattern. Regardless of the pattern, forespore or whole-sporangium, the bimodality in expression of the PaLoc genes seen in vegetative cells is also observed in sporulating cells.
Expression of the PaLoc genes during sporulation is TcdR-dependent
TcdR is required for expression of the PaLoc genes tcdA, tcdB, and tcdE30,31,63. To determine whether TcdR was required for the expression of the PaLoc genes during sporulation, we first constructed a tcdR in-frame deletion mutant using Allelic Coupled Exchange64. The insertional inactivation of tcdR in strain 630Δerm was reported to cause a small, two-fold increase in sporulation65, a result that we also obtained (Supplementary Table 1).
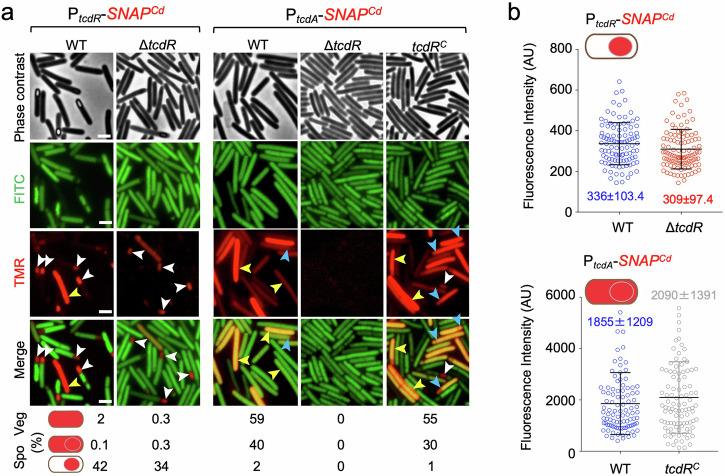
We then examined expression of PtcdR- and PtcdA-SNAPCd during sporulation in a ΔtcdR mutant, the latter as a proxy for the expression of the toxin-encoding genes. Surprisingly, expression of PtcdR-SNAPCd during sporulation, was still detected in the ΔtcdR mutant, both in whole sporangia (Fig. 2a, 0.3%) and mostly in the forespore (white arrowheads; 34% of the sporangia scored). Moreover, the average intensity of the fluorescence signal in the forespore (Fig. 2b; 309 ± 97.4 AU) did not differ significantly from the WT (Fig. 2b; 336 ± 103.4 AU). Thus, TcdR is not required for the forespore-specific expression of tcdR. In sharp contrast, expression of PtcdA-SNAPCd was not detected in sporulating cells of the ΔtcdR mutant but was restored when a tcdR was inserted at the pyrE locus in single copy (Fig. 2, tcdRC strain). Complementation could be observed for the whole sporangia pattern, both in the percentage of cells (Fig. 2a; 40% in the WT, 30% in the tcdRC strain), as well as in the average intensity of the fluorescence signal from PtcdA-SNAPCd (Fig. 2b; 1855 ± 1209 AU in the WT, 2090 ± 1391 in the tcdRC strain).
Fig. 2. The role of TcdR in regulating toxin production.
a Microscopy analysis of C. difficile cells carrying transcriptional fusions of the tcdR and tcdA promoters to SNAPCd in strain 630Δerm (WT), in the ΔtcdR mutant and in the ΔtcdR mutant complemented with the wild-type allele at the pyrE locus (tcdRC). The cells were collected after 24 h of growth in TY liquid medium, labeled with TMR-Star and examined by phase contrast and fluorescence microscopy to monitor SNAP production. The merged images show the overlap between the TMR-Star (red) and the auto-fluorescence (green) channels. The images are representative of the expression patterns observed for the different fusions in three independent experiments (see also Fig. S1b and the Methods section). Yellow arrowheads point to vegetative cells with expression, white arrowheads point to sporulating cells with forespore-specific expression and blue arrowheads point to sporulating cells with whole sporangium SNAPCd production. The numbers below the panels show the percentage of vegetative (Veg) cells and sporulating cells (Spo) with the represented patterns. The number of cells analyzed for each strain, n, is as follows: WT with PtcdA-SNAPCd, n = 918; tcdRC with PtcdA-SNAPCd, n = 820; WT with PtcdR-SNAPCd, n = 1456; ΔtcdR with PtcdR-SNAPCd, n = 2246; tcdRC with PtcdR-SNAPCd, n = 2688. Scale bar, 1 μm. b Quantitative analysis of the fluorescence intensity (in Arbitrary Units, AU) of the SNAPCd signal per forespore for the tcdR fusion and in whole sporangia for the tcdA fusion, in the WT or in the ΔtcdR and tcdRC strains; the data refers to the experiments described in (a). The numbers inside the graphs represent the mean value ± the standard deviation. All pairwise comparisons were non-significant (see Methods).
Since tcdR expression in sporulating cells does not require TcdR whereas tcdA expression is tcdR-dependent, we infer that a factor other than TcdR most likely drives tcdR expression in sporulating cells.
Expression of the PaLoc genes in whole sporangia, but not in the forespore, is σD-dependent
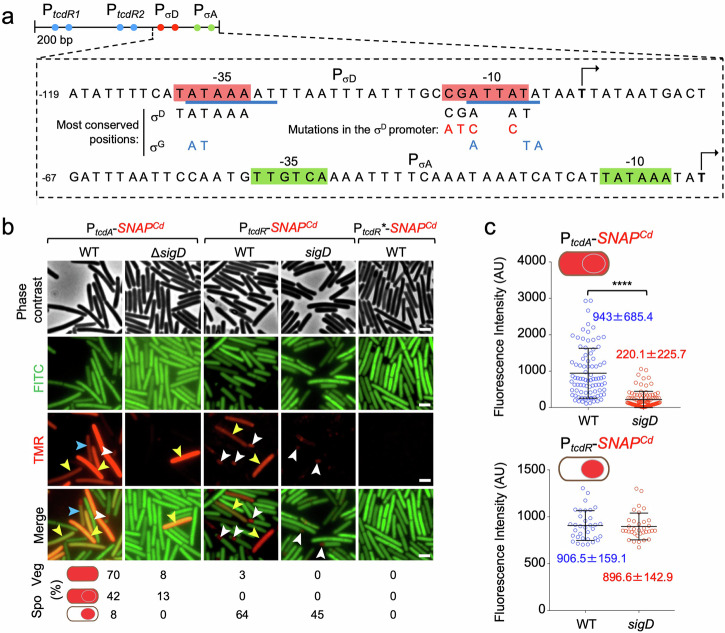
A σD-dependent promoter was previously mapped upstream of tcdR49,51,66 (Fig. 3a). To explore the relevance of this promoter for tcdR expression in the forespore we fused a fragment from the tcdR regulatory region containing the σD promoter to the SNAPCd reporter; this transcriptional fusion is called PtcdR-D-SNAPCd (Supplementary Fig. S3a). In sporulating cells, no whole-sporangium pattern was detected using PtcdR-D (Supplementary Fig. S3b). The percentage of forespores showing PtcdR-D expression (44%), however, did not differ significantly from that obtained with the full-length fusion (52%; Supplementary Fig. S3b). Moreover, the intensity of the fluorescence signal in the forespore also did not differ significantly between the two fusions (Supplementary Fig. S3c). Together, these results indicate that the region containing the σD promoter is sufficient for expression of tcdR in the forespore. σD, however, is not known to be directly involved in sporulation48, leading us to hypothesize that other, sporulation-specific factor could recognize sequences within the PtcdR-D fragment.
Fig. 3. σD regulates toxin production in whole sporangia but not in the forespore.
a Shows the regulatory region of the tcdR gene, with the −35 and −10 elements of the tandem tcdR-dependent promoters (PtcdR1 and PtcdR2, blue dots) and the σD- and σA-dependent promoters (red and green dots). The bases underlined in blue indicate a putative σG-dependent promoter. The most conserved positions for σD and σG-dependent promoters are shown as well as the point mutations introduced in the −10 region of the σD promoter (letters in red). Transcriptional start sites are indicated by broken arrows. Numbering is relative to the start site downstream of the σA-type promoter. b Microscopy analysis of C. difficile cells carrying fusions of the tcdA, tcdR and tcdR* (with point mutations in the σD-dependent promoter) promoters to SNAPCd in strain 630Δerm (WT) and in the ΔsigD mutant. The cells were collected after 24 h of growth in TY liquid medium, labeled with TMR-Star and examined by phase contrast and fluorescence microscopy to monitor SNAPCd production. The merged images show the overlap between the TMR-Star (red) and the auto-fluorescence (green) channels. The images are representative of the expression patterns observed for the different fusions in three independent experiments (see Methods). Yellow arrowheads point to vegetative cells with SNAPCd expression, white arrowheads point to sporulating cells with forespore-specific expression and blue arrowheads point to sporulating cells with whole sporangium expression. To score the indicated patterns in vegetative (Veg) or sporulating cells (Spo) the number of cells analyzed for each strain, n, was as follows: WT with PtcdA-SNAPCd, n = 856; sigD with PtcdA-SNAPCd, n = 1550; WT with PtcdR-SNAPCd, n = 670; sigD with PtcdR-SNAPCd, n = 3902; WT with PtcdR*-SNAPCd, n = 268. c Fluorescence intensity (in Arbitrary Units, AU) of the SNAPCd signal per sporangia for the tcdA fusion and in the forespore for the tcdR fusion, in the WT or in the ΔsigD mutant. The numbers in the panels represent the mean value ± the standard deviation. ****, p < 0.0001; no stars, non-significant differences (see Methods). Scale bar, 1 μm.
Although our PtcdR-D fusion includes the σA-type promoter located just downstream of the σD promoter (Supplementary Fig. S3a), the former shows weak activity (56; see also below). To test whether the σD promoter was involved in the forespore-specific expression of tcdR, we introduced point mutations in the -10 region of the promoter in the context of the full length of PtcdR-SNAPCd fusion. The most conserved positions in the σD promoter51,67 are shown in Fig. 3a, together with the substitutions introduced in the -10 region of the promoter. The new fusion, carrying the mutations in the -10 region of the σD promoter was designated PtcdR*-SNAPCd (Fig. 3a). Additionally, we introduced the PtcdR- and PtcdA-SNAPCd fusions into a sigD mutant51.
Single-cell analysis showed that in a sigD mutant, the fraction of vegetative cells expressing tcdA was reduced from 70% to 8% (Fig. 3b, yellow arrowheads). Moreover, TcdR-ON cells were not detected. This is consistent with the role of σD in activating the TcdR auto-regulatory loop52.
In sporulating cells, the whole-sporangia pattern of PtcdA-SNAPCd was reduced from 42% to 13% (Fig. 3b, blue arrowheads) and the average intensity of the signal measured in the mother cell decreased significantly relative to the WT (Fig. 3c; from 943 ± 685.4 AU to 220.1 ± 225.7 AU in the sigD mutant). Importantly, the forespore-specific expression of tcdR was still detected in 45% of the forespores scored, as compared to 65% for the WT (Fig. 3b, white arrowheads), and the average intensity of the fluorescence signal per forespore did not differ significantly from the WT (Fig. 3c; 906.5 ± 159.1 AU for the WT and 896.6 ± 142.9 AU for the sigD mutant).
The results suggest that the whole sporangia pattern of expression results mostly from either persistent activity of σD in the mother cell and the forespore following asymmetric division, or that TcdR, produced under σD control in pre-divisional cells partitions between the two cells following asymmetric division.
The point mutations introduced in the −10 region of the σD promoter, abolished expression of PtcdR*-SNAPCd in vegetative cells, again consistent with the role of σD in driving production of TcdR and arguing in favor of a minor role for the σA-type promoter in tcdR expression (Fig. 3b). Strikingly, however, the forespore-specific expression of PtcdR*-SNAPCd was also abolished (Fig. 3b). Since expression of PtcdR-SNAPCd in the forespore is not affected by deletion of sigD (above), we infer that the point mutations affect sequences that are also recognized by a forespore-specific regulatory factor other than σD. The target for this factor must at least partially overlap the σD-dependent promoter.
Expression of the PaLoc genes in the forespore is under σG control
Having established that the region required for forespore-specific expression of tcdR overlaps the σD-recognized promoter, we next aimed at identifying the forespore-specific factors involved. We started by evaluating tcdR expression in sigF, sigE, sigG and sigK mutants, all of which are unable to sporulate, and in spo0A cells, for reference. In the spo0A mutant, vegetative expression of PtcdR-SNAPCd is maintained, although the number of cells expressing tcdR was reduced to 0.6% as compared to 7% for the WT (Supplementary Fig. S4). Thus, under our conditions, Spo0A seems to positively control toxin production, at least in a fraction of vegetative cells, consistent with previous reports53,68–72. Unlike the pleiotropic spo0A mutation, deletion of sigF only affects sporulation. In the sigF mutant, which is arrested in development just after asymmetric division23,24, PtcdR-SNAPCd expression was not detected in either the mother cell and/or the forespore (Supplementary Fig. S4). The absence of vegetative cells expressing tcdR in the sigF mutant (7% in the WT; Supplementary Fig. S4) seems to suggest that a contribution from cells outgrowing from germinated spores is lacking (see also below). Importantly, no forespore-specific expression of PtcdR-SNAPCd was observed in a sigF mutant, indicating the involvement of σF or a σF-dependent factor (Supplementary Fig. S4). Since tcdR expression is mostly detected following engulfment completion (above), we anticipated that the late σG factor could be required for the forespore-specific expression of tcdR. Consistent with this inference, in a sigG mutant, which completes the engulfment process but does not proceed further into development24, expression of tcdR was only detected in 28% of the forespores, as compared with 49% for the WT (Supplementary Fig. S4). We presume that expression of tcdR is reduced but not abolished in the sigG mutant because under certain conditions σF can utilize promoters that are normally recognized by σG, the two sigma factors having very similar recognition sequences23,24,67. In cells unable to produce the early mother cell regulator σE, tcdR expression was reduced to 9% of the sporangia (Supplementary Fig. S4) consistent with a requirement for σE for the full activity of σG24,25,27,73. In cells unable to produce the late, mother cell-specific regulatory protein σK, the forespore-specific expression of tcdR was reduced (to 29% of the sporangia, similar to the sigG mutant; Supplementary Fig. S4). This suggests that σK influences late gene expression in the forespore (see also the Discussion).
The sequences for promoter recognition by σG are very similar in B. subtilis and C. difficile, and include the −35 and −10 elements, GAATAAAAT and ATAATA, with a spacing of 15 bp25. While sequences that approach the consensus for σG-recognized -10 element overlap the -10 region of the σD promoter upstream of tcdR, the −35 element is less conserved (Fig. 3a, nucleotides underlined in blue; the most conserved bases are also indicated). Therefore, σG may drive the post-engulfment expression of tcdR in the forespore by recognizing sequences that partially overlap the σD promoter.
σG and SpoVT control expression of the PaLoc genes in the forespore
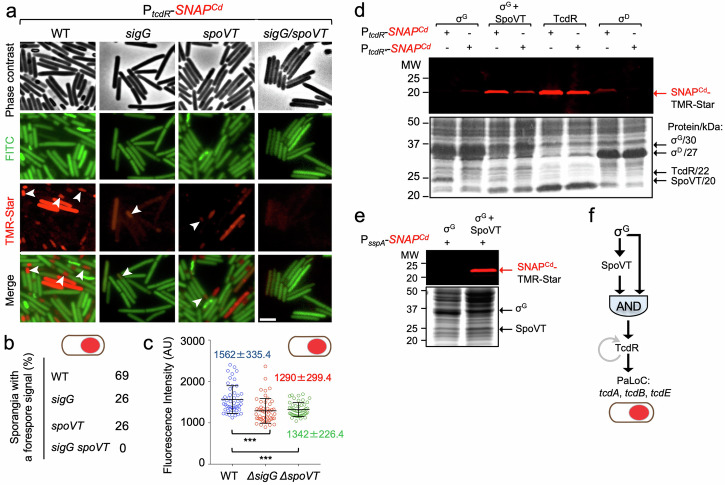
The results suggest the involvement of σG in the forespore-specific expression of tcdR, and a possible σG-dependent promoter overlaps the σD -recognized promoter (Fig. 3a and Supplementary Fig. S3a). Since the expression of a subset of σG-dependent genes additionally requires SpoVT9,25 we wanted to monitor expression of tcdR in a spoVT mutant, and in a sigG/spoVT double mutant. Single-cell analysis revealed forespore-specific expression of tcdR in 26% of the spoVT sporangia scored (Fig. 4a, white arrowheads), similar to the fraction seen for the sigG mutant but lower than for the WT (69%; Fig. 4b).
Fig. 4. Toxin production in the forespore is dependent on σG and SpoVT.
a Strains of C. difficile carrying a fusion of the tcdR regulatory region to SNAPCd in strain 630Δerm (WT) and in the sigG, spoVT and sigG/spoVT were grown in TY medium and cells collected after 24 h of growth. The cells were labeled with TMR-Star and examined by phase contrast and fluorescence microscopy. The merged images show the overlap between the TMR-Star (red) and the auto-fluorescence (green) channels. The images are representative of the PtcdR-SNAPCd expression patterns seen in three independent experiments, with white arrowheads identifying forespore-specific expression. Scale bar, 1 μm. b Shows the percentage of sporangia with a forespore-specific signal for the indicated strains. The number of cells analyzed for each strain, n, was: WT, n = 450; sigG, n = 878; spoVT, n = 540; sigG/spoVT, n = 500. c Fluorescence intensity (in Arbitrary Units, AU) of the SNAPCd signal in the forespore for the PtcdR-SNAPCd fusion in the WT or congenic sigG and spoVT mutants. Note that no signal was detected for the sigG/spoVT double mutant. The numbers in the panels represent the mean value ± the standard deviation. ***, p < 0.001 (see Methods); d E. coli BL21(DE3) derivatives with plasmids carrying PtcdR- or PtcdR*-SNAPCd fusions, as indicated, were transformed with compatible plasmids for the induction of σG, σG, and SpoVT, TcdR or σD. e As in (d), except that E. coli BL21(DE3) containing a plasmid with a PsspA-SNAPCd fusion was transformed with plasmids for the induction of σG alone or together with SpoVT, as indicated. In (d) and (e), the various proteins were produced through auto-induction, whole cell extracts prepared, labeled with TMR-Star, proteins resolved by SDS-PAGE, and the gels scanned on a fluoroimager (top) before Coomassie staining (bottom). The position of the various regulatory proteins is indicated by black arrows and their molecular weights are given on the right side of (d). The position of the SNAPCd-TMR-Star complex is indicated by the red arrows. The regions shown in panels d and e were cropped from original gels shown in Supplementary Fig. S10. f Representation of the coherent feed-forward loop formed by σG and SpoVT, with an AND gate logic, though to lead to delayed production of TcdR in the forespore, and to the expression of the PaLoc genes tcdA, tcdB, tcdR, and tcdE in this cell.
Importantly, in the sigG/spoVT double mutant no signal for tcdR expression could be detected (Fig. 4b). Moreover, the average intensity of the fluorescence signal per forespore in the spoVT mutant (1342 ± 226.4 AU) was lower than in the WT (1562 ± 335.4 AU) and similar to the sigG mutant (1290 ± 299.4 AU) (Fig. 4c). These results support the idea that SpoVT, together with σG, is involved in regulating the expression of tcdR in the forespore.
To test whether σG could, together with SpoVT, directly utilize a promoter in the tcdR regulatory region, σG was overproduced alone or in combination with SpoVT, from the IPTG-inducible T7lac promoter, in E. coli cells bearing a compatible plasmid carrying either the wild-type PtcdR-SNAPCd fusion or the version with the point mutations in the −10 region of the σG promoter, PtcdR*-SNAPCd. Additionally, expression of the fusions was also monitored in E. coli cells overproducing either TcdR or σD (Fig. 4d; see also Supplementary Fig. S10a). Accumulation of SNAPCd was used as a proxy for the utilization of the WT or mutant forms of the tcdR promoter by the various regulatory proteins. As expected, induction of tcdR resulted in high-level production of SNAPCd (Fig. 4d). Also as expected, induction of sigD expression led to SNAPCd production from the WT but not from the PtcdR* mutant promoter (Fig. 4d). When sigG only was induced, SNAPCd was not detected from neither the WT nor the mutant promoter. Strikingly, however, when sigG and spoVT were co-expressed, SNAPCd production was detected from the WT PtcdR promoter, but at much lower levels from the mutant promoter, PtcdR* (Fig. 4d). In a control for the E. coli induction assay, we used a PsspA-SNAPCd fusion, as expression of sspA is known to be under the joint control of σG and SpoVT23,25. The PsspA-driven production of SNAPCd required the induction of both σG and SpoVT (Fig. 4e and Supplementary Fig. S10b). Thus, our in vitro assay suggests that together with SpoVT, σG is able to induce PtcdR-SNAPCd expression by recognizing a promoter that overlaps the one utilized by σD in vegetative cells. σG and SpoVT thus establish a coherent feed forward loop that controls tcdR expression in the forespore (Fig. 4f).
TcdA associates with the spore surface
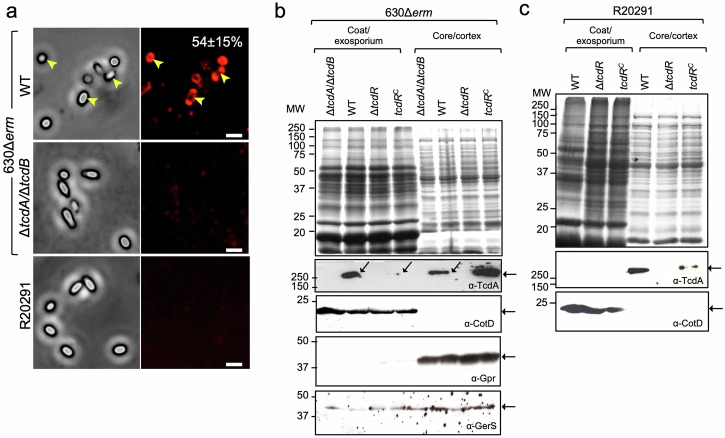
Since, with the exception of tcdC, expression of the PaLoc genes was detected in sporulating cells, we wanted to test whether the toxins associated with mature spores. C. difficile spores were purified and utilized in immunofluorescence assays using an anti-TcdA monoclonal antibody; the presence of TcdB in spores was not assessed as we do not have an antibody with sufficient specificity and/or sensitivity. We found that an anti-TcdA antibody strongly decorated a fraction of the purified WT spores (Fig. 5a; 54 ± 15%) but not those of a ΔtcdA/ΔtcdB double mutant. The accessibility of TcdA to the anti-TcdA antibody in spores of strain 630Δerm suggests that the toxin is associated with the coat and/or exosporium, the outermost layers of spores; protein components of these structures have been detected by immunofluorescence18,19,74. In the epidemic ribotype 027, strain R20291, however, spore-associated TcdA was not detected by immunofluorescence, suggesting that the toxin either does not associate with the spore or is not accessible to the antibody (Fig. 5a).
Fig. 5. TcdA localization in mature spores.
a The accessibility of TcdA to an anti-TcdA antibody was examined in spores of strains 630Δerm, ΔtcdA/ΔtcdB and R20291 by immunofluorescence. The numbers in the top right image represent the percentage of 630Δerm spores with a fluorescence signal, indicated by yellow arrowheads (average of three independent experiments ± standard deviation). Fractionation of 630Δerm (b) or R20191 (c) spores, their congenic ΔtcdAΔtcdB and ΔtcdR derivatives and a ΔtcdR mutant complemented with the wild-type allele at the pyrE locus, tcdRC. Intact mature spores were fractionated into a coat/exosporium and a core/cortex fraction. The proteins in the various fractions were resolved by SDS-PAGE (top) and subject to immunoblotting (bottom) with anti-TcdA, and anti-CotD antibodies (CotD is a bona fide coat protein). For 630Δerm spores, the extracts were also probed with anti-GPR (GPR is a germination protease localized in the spore core) and anti-GerS antibodies (GerS is a cortex-modifying protein involved in spore germination). The regions shown in panels b and c were cropped from the original immunoblots shown in Supplementary Figs. S11 and S12.
TcdA associates with different spore layers
To independently test the association of TcdA with spores and its localization within spores, we used biochemical fractionation. Spores formed by strains 630Δerm and R20291 and spores formed by their ΔtcdR derivatives were boiled in the presence of SDS and DTT to extract proteins present in the coat and exosporium layers18,24,73–77. These proteins define a coat/exosporium fraction (Fig. 5b, c; see also Supplementary Figs. S11, S12).
An earlier study reported that the insertional inactivation of tcdR in strain R20291 resulted in a three-fold reduction in spore heat resistance65. Under our culturing conditions, however, the in-frame deletion of tcdR in R20291 or 630Δerm, caused a small, three-fold increase in spore heat resistance (Table S1). Spores of all strains remained phase bright after extraction, indicating that the cortex and core did not suffer major structural alterations24.
Following extraction of the coat and exosporium, the spores were treated with lysozyme, leading to at least the partial release of proteins that are in proximity to or within the cortex peptidoglycan78, and most likely also of spore core proteins. The lysozyme digestion defined a core/cortex fraction (Fig. 5b, c). Proteins in the two fractions were then resolved by SDS-PAGE and the gels stained with Coomassie (Fig. 5b, c, top panels; loading control) and subject to immunoblotting analysis with the anti-TcdA antibody. TcdA was detected in both the coat/ exosporium and core/cortex fractions of 630Δerm spores, although to a higher level in the coat/ exosporium (Fig. 5b, middle panel). We then extended this analysis to the fractions obtained from R20291 spores. We detected TcdA in the core/cortex fraction but not in the coat/ exosporium fraction (Fig. 5c; middle panel; see also Supplementary Fig. S12). Importantly, for both 630Δerm and R20291 spores, TcdA was not detected in the fractions prepared from ΔtcdR spores, but complementation with the wild-type allele at the pyrE locus partially restored its association with the coat/exosporium and core/cortex for 630Δerm (Fig. 5b) and with the core/cortex fraction for R20291 spores (Fig. 5c).
We used several antibodies to control for the fraccionation procedure using 630Δerm spores. CotD was only detected in the coat/ exosporium fraction, consistent with its reported association with both the coat and exosporium structures74,77. On the other hand, GerS, required for cortex modification during spore germination79, could be detected in both fractions (Fig. 5b, bottom panel). It seems possible that GerS associates with both the coat/exosporium and the cortex, but that during the initial steps of germination it re-localizes exclusively to the cortex79. Finally, GPR, a known core protein23–25,80, could only be detected in the lysozyme-treated fraction, suggesting that this fraction indeed includes core-associated proteins (Fig. 5b, bottom panel; see also Supplementary Fig. S11). Overall, this analysis suggests that our fractionation procedure enriched for coat/exosporium proteins such as CotD, which are not detected in the core/cortex fraction (Fig. 5b, c).
Together, these results do not exclude the presence of TcdA in the spore core, but they indicate that TcdA associates with the coat/exosporium layers and is accessible to antibodies in 630Δerm spores, whereas in spores of the epidemic strain R20291, TcdA has a more internal localization, within the core and/or cortex, or the epitope recognized by the anti-TcdA antibody is not exposed in the coat/exosporium.
To estimate the total amount of toxin extractable from spores, spores formed by strains 630Δerm and R20291 were biochemically fractionated and TcdA was immunodetected as described above. The signal was compared to a standard curve obtained using a range of purified TcdA, from 0 to 0.5 μg (Supplementary Figs. S5a, S13) and normalized to the number of viable spores used. Taking into consideration that TcdA was only detected in about 54% of the 630Δerm spores the amount of extractable, full-length TcdA per spore, on average, was estimated at a minimum of 9.9 × 102 molecules (Supplementary Figs. S5 and S13). A minimum of about 7.3 × 102 full-length TcdA molecules were extractable from the same number of R20291 spores (Supplementary Figs. S5, S13) but since TcdA was not detected by immunofluorescence in R20291 spores, the average number of molecules per spore could not be estimated.
We also extended our analysis to a group of characterized clinical strains of ribotypes 126 (strain E1), 053 (strain E7), 106 (strain E12), 014 (E14), 001/072 (strain E23), and 005 (strain E25)81. Spores produced by these strains were purified, fraccionated as above and the core/cortex and cortex/coat/exosporium proteins extracted, resolved by SDS-PAGE and the gels were subject to immunoblotting with the anti-TcdA antibodies. Spores of the non-toxinogenic E13 strain (RT017)81 were included as a specificity control for the antibody; as expected TcdA was mainly detected in the cortex/coat/exosporium fraction in spores of strain 630Δerm whereas the toxin was not detected in any of the fraction prepared from E13 spores (Supplementary Figs. S6, S14), as expected from its genome sequence81. In contrast, TcdA was detected mainly in the core/cortex fraction of spores purified from strains E12, E14, E23, and E25 (Supplementary Figs. S6,S14). This analysis shows that the association of at least TcdA with spores is not specific to strains 630Δerm and R20291, but also occurs in clinical isolates of several other ribotypes.
The forespore or mother cell-specific expression of tcdR results in the association of TcdA with spores
That tcdA expression was detected both in the forespore and in the whole sporangium, suggested that expression during sporulation was required for the association of the toxin with spores. Because of protracted expression of tcdA in the mother cell and whole sporangia, however, to eliminate expression of the gene specifically in sporulating cells did not seem feasible. Therefore, we tested whether expression of tcdA in the forespore or the mother cell would be sufficient for the association of TcdA with spores, the tcdR deletion mutant was complemented in trans at the pyrE locus using a tcdR allele expressed from the forespore specific sspA promoter, or from the mother cell-specific spoIIIAA promoter23–25. We first examined expression of the PtcdA-SNAPCd fusion in the PsspA-tcdR (tcdRFS for simplicity in Supplementary Figs. S7 and S15) and PspoIIIAA-tcdR (tcdRMC) complementation strains by fluorescence microscopy. In the tcdRFS strain, expression of the fusion was detected in the forespore (Supplementary Fig. S7a, white arrowheads) whereas for the tcdRMC strain a mother cell-only pattern of PtcdA-SNAPCd expression was detected (Supplementary Fig. S7a, blue arrowheads). Note that in the WT, a mother cell-only pattern of PtcdA-SNAPCd expression was not detected (see also above).
Biochemical fractionation of mature spores produced by the tcdRMC strain shows that TcdA is present in the coat/exosporium fraction, at a level slightly higher than for the WT (Supplementary Figs. S7b and S15). TcdA is also present in the core/cortex fraction of tcdRMC spores, but at levels lower than for the WT (Supplementary Figs. S7b and S15). In spores of the tcdRFS strain, however, TcdA is present in the core/cortex fraction, but less extractable (or less abundant) than for WT or tcdRMC spores, and absent or undetected in the coat/exosporium fraction (Supplementary Figs. S7b and S15). This suggests that forespore-produced TcdA is unable to reach the coat/exosporium (or it does so but below our detection level). It also suggests that most of the toxin detected in spores by immunofluorescence (Fig. 5a; see also above), results from the fraction of sporulating cells in which the σD-dependent expression of tcdA persists in the mother cell. We further infer that the production of TcdA in the mother cell is sufficient for the association of the toxin not only with the coat/exosporium but also with the more internal core and/or cortex layers of spores.
The spore-associated toxins remain functional
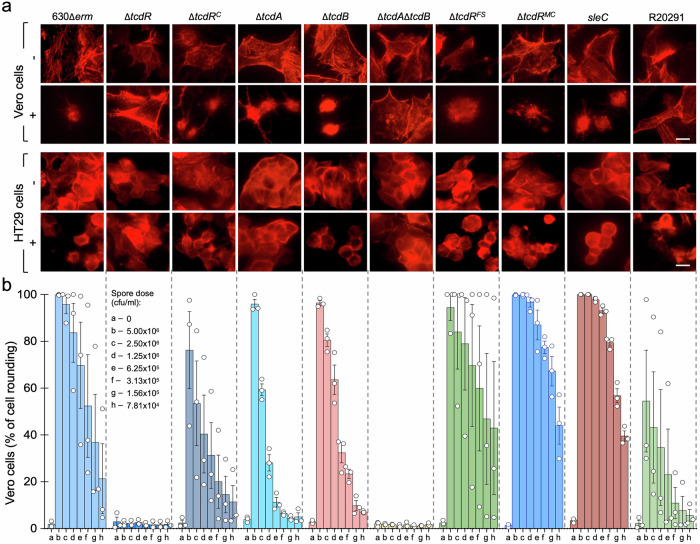
The finding that at least TcdA associates with mature spores begged the question of its functionality. To test whether TcdA and TcdB, the latter of which we were not able to detect in spores by immunoblotting (above), remained functional, we conducted cytopathic assays using HT29 and Vero cell lines82,83. The toxins induce cell rounding, which can be monitored by microscopy82,83. HT29 and Vero cells, grown as monolayers, were exposed to different amounts of WT or ΔtcdR spores and cell rounding was monitored as a function of the spore concentration. We found that WT spores, but not those of the ΔtcdR mutant, caused cell rounding of both HT29 and Vero cells (Fig. 6a and Supplementary Fig. S8). This effect was dose-dependent for both cell lines (Supplementary Fig. S8) and shown in graphical form for Vero cells (Fig. 6b). Complementation with the wild-type tcdR allele at pyrE locus partially restored the cell-rounding ability to ΔtcdR spores (Fig. 6 and Supplementary Fig. S8). Thus, TcdA associates with spores in an active form. R20291 spores have a weaker cytopathic effect than 630Δerm spores (Fig. 6 and Supplementary Fig. S8), possibly because of the more internal localization or lower abundance of TcdA, found mainly in the core/cortex fraction (Fig. 5c).
Fig. 6. Spores produced by the 630Δerm and R20291 strains have a cytopathic effect on epithelial cell lines.
a Immunostaining of monolayers of Vero and HT29 cells. The cells were exposed to 5 × 106 spores purified from sporulating cultures of the indicated strains. Following incubation for 24 h with spores, the cells were immunostained with Texas Red-Phalloidin to detect cell rounding. Scale bar, 10 μm. b Spores cause a dose-dependent cytopathic effect. Monolayers of Vero cells were exposed to the indicated number of spores (in CFU/ml) purified from each of the strains specified at the bottom. Following incubation, the cells were examined by phase contrast microscopy and the percentage of cell rounding scored for 220 cells (n). The experiment was performed in triplicate and error bars indicate the standard error.
Both cell lines were exposed to spores produced by ΔtcdA and ΔtcdB single mutants (Fig. 6 and Supplementary Fig. S8). The ΔtcdA spores have a cytopathic effect in Vero cells, although lower than WT spores, since TcdB is present and the latter is known to cause cell rounding in this cell line84. However, these spores do not have a cytopathic effect in HT29 cells since this cell line is mainly susceptible to TcdA (Fig. 6)84. ΔtcdB spores have a cytopathic effect, again lower than WT spores, in both Vero and HT29 cells, consistent with the presence of TcdA in these spores and its reported cytopathic effect in both cell lines84.
Importantly, cell culture medium and cell culture supernatant did not induce spore germination, indicating that the toxin-induced cell rounding is caused by the spores. In line with this inference, sleC mutant spores were equally toxic against both HT29 and Vero cells when compared with WT 630Δerm spores. Although they initiate germination, spores of a sleC mutant are not able to degrade the spore cortex and to complete the process85. Thus, germination does not need to be completed for the spore-associated cytopathic activity.
Spores produced by the tcdRFS and tcdRMC strains had a similar cytopathic effect against Vero cells and HT29 cells as those of the WT strain (Fig. 6). This suggests that when produced either in the forespore or the mother cell the toxins remain active when spore associated. That the cytopathic effect observed for tcdRFS spores was the same as for tcdRMC or WT spores is intriguing since in this strain TcdA shows a more internal, core and/or cortex localization. It seems likely, however, that differences in the structure of the spore surface layers between 630Δerm and R20291 spores influence the exposure and activity of the spore-associated toxins.
Discussion
The tcdA gene was found before to be expressed in the mother cell in a fraction of the sporulating cells52,53. We now show that other PaLoc genes are also expressed in sporulating cells according to different spatial patterns and regulatory schemes. We show that in a fraction of the sporulating cells, tcdR, tcdA, tcdB and tcdE are expressed only in the forespore, that tcdA and tcdB additionally show a whole-sporangium pattern of expression, i.e., both in the mother cell and in the forespore, and that the expression of tcdC only exhibits this latter pattern. We did not observe a mother cell-specific pattern of expression for any of the genes examined. In previous work, tcdA expression seemingly exhibited a mother cell-specific pattern but this could correspond to the whole sporangium pattern herein reported, because the fluorescence reporters used before, RFP, mSc and mNG, generally perform less well than the SNAP reporter in vegetative cells59 and possibly also in the forespore52,53. For this reason, it was also unclear from those studies whether forespore-specific expression of the PaLoc genes did occur.
We show that the whole sporangium pattern is both σD- and TcdR-dependent, because the first triggers the auto-regulatory production of the second52,53. Expression of tcdR, and also of tcdA, which initially takes place in pre-divisional cells, persists in whole sporangia following asymmetric division, either because σD remains active and/or because sufficient TcdR is partitioned into the forespore and the mother cell. Heterogeneity in the population of sporulating cells with respect to whole sporangium tcdA expression most likely results from the TcdR auto-regulatory loop that operates in pre-divisonal cells52 (Fig. 2). In other words, the fraction of sporulating cells expressing the PaLoc genes reflects, at least in part, the fraction of ON vegetative cells and the bistable switch that controls their expression is propagated to whole sporangia. Thus, expression of the toxin-encoding genes in whole sporangia relies on the memory of a previous state, the vegetative state, referred to as hysteresis86.
We have shown that expression of tcdR is specifically induced in the forespore under the control of both σG and SpoVT and that tcdA, and presumably tcdB and tcdE, also shows TcdR-dependent, forespore-specific, expression. SpoVT enables σG to utilize a promoter that partially overlaps the σD-dependent promoter. Expression of tcdA, tcdB and tcdE was only detected in a fraction of the forespores and TcdA was only detected in a fraction of spores. Possibly, the bistable switch controlling the expression of the PaLoc genes in vegetative cells is reproduced in the forespore with σG replacing σD in priming the auto-regulatory expression of tcdR and the expression of the PaLoc genes in a fraction of the forespores (Fig. 4f).
SpoVT is conserved among spore formers20–22 and positively regulates a subset of σG-dependent genes in both B. subtilis, in B. cereus in C. difficile25,87,88. In B. subtilis, σG and SpoVT define coherent and incoherent feed-forward loops with AND gate logic that result in a pulse of expression of early σG-dependent genes, and delayed expression of late genes, respectively89. σG and SpoVT may function in a similar way in C. difficile forming a coherent feed-forward loop with AND gate logic (Fig. 4f), that delays toxin production in the forespore until a late stage in development. SpoVT is a dimer of dimers, with each monomer formed by an N-terminal DNA binding domain and a C-terminal GAF (cGMP-specific and cGMP-stimulated phosphodiesterases, Anabaena adenylate cyclases, and Escherichia coli FhlA) domain. GAF domains act as sensory modules that can bind linear and cyclic nucleotides, porphyrin rings, as well as small signaling molecules such as homoserine lactones90. Whether SpoVT binds a specific ligand is unclear, as some GAF domains may function exclusively to mediate dimerization90. As yet unknown signals may control the activity of SpoVT and thus the level of toxin production in the forespore. Expression of tcdR and tcdA in the forespore additionally appears to be controlled through the activity of the late mother cell-specific regulator σK. Yet, no cell-cell signaling pathway is known that links the activity of σK to that of σG in the forespore. One possibility is that the activity of σK is necessary to convey to the forespore signals that influence the activity of SpoVT.
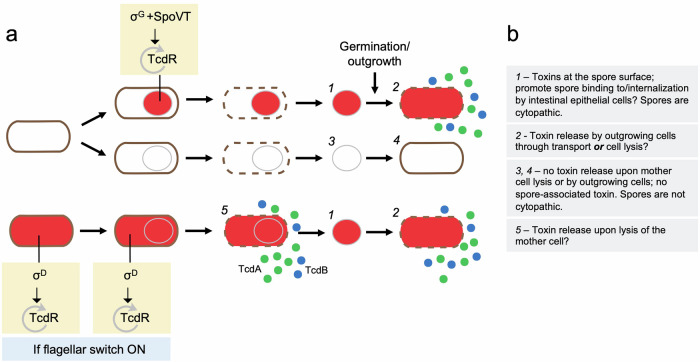
We show that TcdA associates with spores and that expression of tcdA in sporulating cells, from either a forespore or a mother cell-specific promoter, is sufficient for this association (Supplementary Fig. S7). A study by Hong and co-authors showed that antibodies raised against a fragment of TcdA (residues 26-39 within the CROPs region) recognized species of about 100, 60, 50, and 20 kDa at the surface of C. difficile spores, and two of these species were identified as the aldehyde-alcohol dehydrogenase AdhE1 and the exosporium protein CdeC91. This study raised the possibility that coat/exosporium proteins cross-reacted with anti-TcdA antibodies. In our study, however, in which we used a monoclonal antibody that recognizes the TcdA CROPS region92, we detected a species of about 250 kDa, close to the expected size for TcdA, in WT but not in ΔtcdA/ΔtcdB mutant spores. Moreover, spores of both 630Δerm and R20291 have a cytopathic effect when assayed against Vero or HT29 cells. Thus, TcdA associates in an active form with mature spores. In particular, fractionation and immunofluorescence studies showed the association of TcdA with the coat/exosporium and core/cortex of 630Δerm spores, whereas in spores of the epidemic R20291 strain, TcdA is mainly associated with the spore core/cortex fraction. Importantly, TcdA was also found mainly in the core/cortex fraction of spore produced by clinical isolates of ribotypes 106, 014, 001/072, and 005 (Supplementary Fig. S6). This indicates that the association of the toxins with spores may be a general phenomenon. TcdA production in the mother cell results from protracted expression of σD, which primes the TcdR auto-regulatory loop; σD production, in turn, is subject to a phase variation mechanism that controls the production of flagella and toxins93. The flagella switch involves inverted repeats that are conserved in all C. difficile strains that have been sequenced and carry the flagellar genes, but maybe locked in the ON state in strain 630Δerm, in which the inverted repeats are shorter93. Since assembly of both the cortex and coat layers is mainly a function of the mother cell it is conceivable that some TcdA can associate with these layers during their formation but only in strains, such as 630Δerm in which production of σD and TcdR is maintained in the mother cell. In the forespore, however, the TcdR auto-regulatory loops are primed by σG and SpoVT (Fig. 7). This may explain why in R20291 and the additional epidemic strains analyzed herein, TcdA is not detected in the cortex/coat/exosporium fraction while detected in the core/cortex fraction of all strains (Fig. 7).
Fig. 7. Functionally differentiated cell and spore populations.
a Schematic representation of vegetative cells expressing the toxin-encoding genes (red filling) or not and their patterns of expression during sporulation (whole sporangium or forespore-specific). Five different possible functional classes (1 to 5) are highlighted with respect to the expression pattern of the toxin-encoding genes, the association of the toxins with spores and their release through lysis of the mother or following spore germination and outgrowth. The main regulatory proteins involved in the control of TcdR production are shown. If the flagellar switch is in the ON state93 σD primes the TcdR auto-regulatory loop in vegetative cells and in the mother cell during sporulation. In the forespore, however, the TcdR auto-regulatory loop is primed by σG together with SpoVT. b The expected properties of classes 1 to 5 are summarized in the panel.
Since that maintains expression of in the mother cell. Perhaps suggestively, TcdA has an affinity for glycan motifs with the core structure Gal β1-4 GlcNAc8,28,94 and it seems possible that TcdA associates with the cortex peptidoglycan via its CROPs region. Although we were unable to directly detect TcdB at the spore surface, the cytopathic effect of ΔtcdA spores on Vero cells, together with the lack of effect of ΔtcdA/ΔtcdB and ΔtcdR spores, suggests that TcdB also associates in active form with spores. Our finding that spores have a cytopathic effect on both Vero and HT29 cells is in line with the observation that spores induce the production of inflammatory cytokines and cause a cytotoxic effect on macrophages95.
The different patterns of tcdR and tcdA expression in sporulating cells indicate the existence of functionally different populations of cells and spores (Fig. 7). For spores that are formed in the host, it seems likely that mother cell expression provides an important route for toxin release upon lysis of this cell, coupling toxin release to the completion of spore development. The forespore-specific expression, of tcdR and tcdA, in turn, will likely result in the accumulation of the toxins in the forespore cytoplasm and presumably also in the cytoplasm of the outgrowing cell. Thus, for the fraction of forespores that produce them, the toxins could be quickly released upon spore outgrowth and since, as we show, tcdE is also expressed in the forespore, we speculate that tcdE-mediated release is involved (Fig. 7). It has been proposed that low levels of toxin cause inflammation, which in turn promotes colonization by vegetative cells during infection96. Thus, toxin release from mature spores or during spore germination and outgrowth might play a role in colonization. Cytopathic assays with a sleC null mutant suggest that germination does not need to be completed for spores to have a cytopathic effect (Fig. 6 and Supplementary Fig. S8). Still, we cannot rule out the possibility that the initial stages of germination85, with the consequent modifications of the spore surface, lead to some toxin release. In mice, the TcdA lethal dose is in the order of 50 ng and in challenge experiments using animal model around 100 spores are used97–99. It is difficult to correlate the amount of spore-associated toxin with the TcdA lethal dose and the amount of spore-delivered toxin will depend on the number of spores ingested. Low quantities of toxin, however, may still affect host cells; even sub-lethal concentrations of TcdA were shown to affect cell polarity, which in turn appears to facilitate access of the toxin to the host cell receptors, and therefore, colonization (96, reviewed in refs. 8,28).
The interaction of spores with E-cadherin promotes attachment to the colonic mucosa, and spore internalization, but whether additional spore receptors exist is unknown13. In contrast, several TcdA and TcdB receptors have been identified (29, reviewed by ref. 8). We speculate that spore-associated toxins may interact with their cognate receptors, thus contributing to spore binding to intestinal epithelial cells. TcdA and TcdB cause the redistribution of E-cadherin and increase spore binding to adherent junctions57. Thus, spore-associated TcdA and TcdB may promote spore binding to intestinal epithelial cells, in which case spore binding would be, to some extent, auto-regulatory. Presumably spore binding would also be independent of toxin production by a population of vegetative cells resulting from spore germination, which could be important for the initial interaction of infectious spores with the intestinal epithelial cells.
Importantly, in spores of the epidemic strain R20291, TcdA may have a more internal localization as compared to 630Δerm spores (Fig. 5) perhaps explaining their lower cytopathic effect (Fig. 6 and Supplementary Fig. S8).
Toxins are known to be produced by sporulating cells in other pathogenic spore-formers: production of enterotoxin by C. perfringens, for example, is under the control of σE and σK, which leads to accumulation of the toxin in the mother cell and its release into the intestinal lumen when the sporulating cells lyse100–103; also, one study reports on the association of a heat-resistant form of the botulin toxin with spores of C. botulinum104 and in several spore-forming entomopathogenic species such as B. thuringiensis, toxins are produced during sporulation and the insecticidal proteins associate with the exosporium as parasporal crystals (105 and references therein); in addition, and even though the toxin production factor AtxA negatively controls sporulation106, the protective antigen component of the anthrax toxin also associates with the exosporium and coat layers of B. anthracis spores107. These observations suggest that spore-forming pathogens, the expression of toxin genes by sporulating cells and the association of toxins with spores may represent a widespread strategy for both toxin release through lysis of the mother cell and for early delivery of the toxin.
Methods
Growth conditions and general methods
The Escherichia coli strain DH5α (Bethesda Research Laboratories) was used for molecular cloning, while HB101 (RP4) was used as the donor strain in C. difficile conjugation experiments108. Luria-Bertani medium was routinely used for growth and maintenance of E. coli. When appropriate, ampicillin (100 µg/mL) or chloramphenicol (15 µg/mL) was added to the culture medium. The C. difficile strains used in this study are congenic derivatives of the wild-type strain 630Δerm108 or R20291 (Anaerobe Reference Laboratory, Cardiff, Wales, United Kingdom) and were routinely maintained anaerobically (5% H2, 15% CO2, 80% N2) at 37 °C in brain heart infusion (BHI) medium (Difco)109. For toxin assays and for sporulation assays, tryptone yeast extract (30 g/L tryptone; 20 g/L yeast extract) and sporulation medium (90 g/L tryptone; 5 g/L peptone; 1 g/L (NH4)2SO4; 1.5 g/L Tris base, pH 7.4) were used supplemented with cefoxitin (25 µg/mL) or thiamphenicol (15 µg/mL) when necessary. A defined minimal media (CDMM)110 with 1% agar was used as uracil-free medium when performing genetic selections. The minimal medium was supplemented with 5-Fluoroorotic acid (2 mg/mL) and uracil (5 µg/mL) when appropriate.
SNAPCd transcriptional fusions and strain construction
The construction of the plasmids required for producing transcriptional SNAPCd fusions to the PaLoc promoters, deletion and mutational analysis of the tcdR promoter, tcdR, spoVT, tcdA, tcdB single and a tcdA/tcdB double mutant, to place tcdR under the control of the sspA or spoIIIAA promoters and for the overproduction of TcdR, σD, σG and SpoVT in E. coli is described in the Supplementary Methods. Primers used for cloning or genome analysis are listed in Table 1. All plasmids are listed in Supplementary Table S2; all strains and their relevant properties are listed in Table S3.
Table 1.
Oligonucleotides used in this work
| Primer | Sequence (5′ to 3′)a |
|---|---|
| PtcdR-SNAP-EcoRI-Fw | CCCTTAAGCGAATTCTAAAAGTAAACG |
| PtcdR-SNAP-XhoI-Rev | CATAAAATCCTCGAGTCTTATATTTATAATG |
| PtcdA-SNAP-EcoRI-Fw | CACAAAGATGAATTCTGGTCAGTTGGT |
| PtcdA-SNAP-XhoI-Rev | GTATTATTACTCGAGATAATAAATCCAC |
| PtcdB-SNAP-EcoRI-Fw | TATCAAAGTGAATTCGTTTTTGAGGAAG |
| PtcdB-SNAP-XhoI-Rev | CTATAATACTCGAGCATCTAAATGCTAAAAC |
| PtcdC-SNAP-EcoRI-Fw | ACTTCACCTGAATTCTGGTATATTC |
| PtcdC-SNAP-XhoI-Rev | CATAATACAATCCTCGAGTTATTAGAT |
| PtcdE-SNAP-EcoRI-Fw | CAATTGGAATTCGATGGATATATGATATG |
| PtcdE-SNAP-XhoI-Rev | CATTCATCATAGCTCGAGTTTTTATTG |
| PtcdRA-SNAP-EcoRI-Fw | CCCCCCGAATTCTACTTTATTTATTAGAAAAA |
| PsigDmut-Fw | GCATATTTTCATATAAAATTTAATTTATTTGCATCTTCTATAATTATAATG |
| PsigDmut-Rev | CATTATAATTATAGAAGATGCAAATAAATTAAATTTTATATGAAAATATGC |
| TcdR-RBSopt-Fw | GCGCGCCTCGAGGGAGGAACTACTATGCAAAAGTCTTTTTATG |
|
TcdR-comp-HindIII-Rev PsspA-BamHI-Fw |
CCCAAGCTTATTAATTTGCTCTTC AGATGAGGAGGATCCGGATAAAAGAGTTC |
| PsspA-SNAP-XhoI-Rev | CTTCCTTCTCTCGAGTTTATTTTGTGTTTGC |
| PspoIIIAA-BamHI-Fw | TAGATGGTGGGATCCCTAGGGCTTACCAAAAAAC |
| PspoIIIAA-SNAP-XhoI-Rev | GTTTATTCATCTCTTGCTCGAGTCCTTG |
| TcdR-comp-XhoI-Rev | CGCGCTCGAGATTAATTTGCTCTTC |
| TcdR-NcoI-Fw | CCCCCATGGAAAAGTCTTTTTATG |
| TcdR-SalI-Rev | CCCGTCGACCAAGTTAAAATAATTTTC |
| CDSigGpET28a-Fw | TGCCTCGAGTACATATTTTCTCATATTTTTTAAAGC |
| CDSigGpET28a-Rev | AGGGGGTGACCCCATGGCAGCTCTTAAATC |
| SigD-NcoI-Fw | CCCCCATGGATAGAGAAGAATTAATAAAAG |
| SigD-XhoI-Rev | CCCCTCGAGTATAGAATATTTAAGTTC |
| SpoVT-NdeI-Fw | CCCCATATGAAAGCAACAGGTATAGTTAG |
| SpoVT-XhoI-Rev | CCCCTCGAGTTATTGAACTTGTTTTCC |
| tcdR-AscI-Fw | CCCCGGCGCGCCATTATCTTAAGAGAGGAG |
| tcdR-SOE-Rev | CATAAATAAAATTTCTTGCAAATCATC |
| tcdR-SOE-Fw | TTGCAAGAAATTTTATTTATGGAAAATTATTTTAACTTG |
| tcdR-SbfI-Rev | CCCCCCTGCAGGTATCTATATAAATATCTG |
| tcdR-vef-Fw | GTATCATTTCACGAAGAGG |
| tcdR-vef-Rev | GGGTCATTTAAGTTTTCTC |
| tcdR-comp-BamHI-Fw | CCCGGATCCTAAAAATATTTTGATATG |
| tcdR-comp-HindIII-Rev | CCCAAGCTTATTAATTTGCTCTTC |
| tcdA-AscI-Fw | CCCCGGCGCGCCGGTAGTATATCAAACATTGG |
| tcdA-SOE-Rev | CTCATTTTCTCTTGGTCTAATGCTATATGCGAG |
| tcdA-SOE-Fw | CCAAGAGAAAATGAGCCTGGGATATATGGC |
| tcdA-SbfI-Rev | CCCCCCTGCAGGGATAAGGTTGTACTATGTAG |
| tcdB-AscI-Fw | CCCCGGCGCGCCCAAAGTAAGTCTGTTTTTGAGG |
| tcdB-SOE-Rev | CAATATTGCAACATATTCATCTTCTTG |
| tcdB-SOE-Fw | GAATATGTTGCAATATTGCAATTAGTG |
| tcdB-SbfI-Rev | CCCCCCTGCAGGGTCTTAAAAAATTGATAC |
| tcdA-vef-Fw | GATGGTGCATGGTCAGTTGG |
| tcdA-vef-Rev | GAAGATGGTGATGAGGTGC |
| tcdB-vef-Fw | GACAAGCTGTTAATAAGGC |
| tcdB-vef-Rev | CTGGTAATCCACATAAGCAC |
| YN3-vef-Fw | CATCAAGAAGAGCGACTTCG |
| YN3-vef-Rev | TTCTTTCTATTCAGCACTGTTATGC |
| pyrE-vef-Fw | CAATAATTTTATAACATTAACATGG |
| pyrE-vef-Rev | GTGTTACTTAAAAAATGTAAAT |
| YN4-vef-Fw | CAAGAAGAGCGACTTCGCGGAGCTGG |
| YN4-vef-Rev | CCATTACAGACTTATCCAGGG |
| SpoVT_sgRNA_Fw | TTTTCGTCGACAAGAATAGATGATCTTGGAAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGC |
| sgRNA_Rev | GCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACGTCGACGAAAA |
| spoVT-AscI-Fw | CCCCGGCGCGCCGAGAAAGATTTAGCAATG |
| spoVT-SOE-Rev | CTTTGAAACAACTACAATTGAACCCTCTTTTGGGATAACTACCCTTCC |
| spoVT-SOE-Fw | GGAAGGGTAGTTATCCCAAAAGAGGGTTCAATTGTAGTTGTTTCAAAG |
| spoVT-AsiSI-Rev | GATGGCGATCGCCTGCAACTTGAGACACAG |
| spoVT-vef-Fw | CGCGGATCCGATGAGTTTTTAAGAGAC |
| spoVT-vef-Rev | CGCGCTCGAGCAAAAGTCTGACCTAGAC |
| PsspA-EcoRI-FW | AGATGAGGAGAATTCGGATAAAAGAGTTCA |
| PsspA-SNAP-SOE-Rev | CATTTCACAATCTTTATCCATGTTGATTACCTTCCTTC |
| SNAP-SOE-Fw | ATGGATAAAGATTGTGAAATGAAGAGAACC |
| SNAPC-XhoI-Rev | CCCCTCGAGTTACCCAAGTCCTGGTTTCCCCAAACG |
aEngineered restriction sites are underlined.
SNAP induction assays in E. coli
In order to test for the ability of TcdR, σD, σG, or SigG together with SpoVT to induce transcription from the tcdR promoter, E. coli BL21 (DE3) strains bearing the plasmids for the overexpression of each of the proteins were co-transformed with either pMS464 or pCC27 (see Tables S2 and S3 and the Supplementary Information). The following strains were obtained that carry: pCC17 and pMS464 (AHED317) or pCC27 (AHEC319); pCC30 and pMS464 (AHEC320) or pCC27 (AHEC322); pFT36 and pMS464 (AHEC290), pCC27 (AHEC291) or pCC32 (AHEC853); pCC29 and pMS464 (AHEC314), pCC27 (AHEC316) or pCC32 (AHEC859). All strains were subjected to an auto-induction regime for the over-production of each protein or protein combinations111. The cultures were collected by centrifugation (4000 × g, for 10 min at 4 °C) and the cell sediment was suspended in PBS with 1 mM DTT. The cells were lysed using a French pressure cell (18,000 lb/in.2) and the extracts were incubated with 250 nM TMR‐Star substrate (New England Biolabs), for 30 min, in the dark. Proteins in the extracts were resolved by SDS-PAGE and labeled proteins were detected by fluoroimaging.
SNAP labeling in C. difficile extracts
Samples (10 mL) were withdrawn from C. difficile TY cultures and the cells labeled with the TMR‐Star substrate (New England Biolabs), at a final concentration of 250 nM, for 30 min, in the dark. Following labeling, the cells were collected by centrifugation (4000 × g, for 5 min at 4 °C), the cell sediment was washed with phosphate-buffered saline (PBS) and suspended in 1 mL French press buffer (10 mM Tris pH 8.0, 10 mM MgCl2, 0.5 mM EDTA, 0.2 mM NaCl, 10% Glycerol, 1 mM PMSF). The cells were lysed using a French pressure cell (at 18,000 lb/in.2). Proteins in the extracts were resolved by SDS-PAGE (15% gels). The gels were first scanned in a TLA-510 fluorimager (Fuji), and then subject to immunoblotting with an anti-SNAP antibody (New England Biolabs) at a 1:1000 dilution; a rabbit secondary antibody conjugated to horseradish peroxidase (Sigma) was used at a dilution 1:10,000. The immunoblots were developed with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Fluorescence microscopy and image analysis
For SNAPCd labeling, the TMR-Star substrate was added to cells in culture samples to a final concentration of 250 nM (New England Biolabs) and the mixture incubated for 30 min in the dark. Following labeling, the cells were collected by centrifugation (4000 × g for 5 min), washed four times with 1 mL of PBS, and finally suspended in 0.5 mL of PBS. For phase contrast and fluorescence microscopy, cells were mounted on 1.7% agarose-coated glass slides and observed on a Leica DM6000B microscope equipped with a phase contrast Uplan F1 100× objective and captured with a CCD Andor Ixon camera (Andor Technologies). Images were acquired and analyzed using the Metamorph software suite (version 5.8; Universal Imaging), cropped and adjusted using Adobe Photoshop.
Immunofluorescence microscopy
For immunofluorescence analysis, the spores were fixed with 3% paraformaldehyde (pH 7.4) for 20 min in poly-L-lysine-coated glass cover slides. The fixed spores were rinsed three times with PBS and blocked with 1% bovine serum albumin (BSA) for 30 min. The slides were incubated overnight at 4 oC with a monoclonal anti-TcdA primary antibody (Santa Cruz Biotechnology) (at a dilution of 1:5000). The slides were then incubated for 2 h at room temperature with Alexa Fluor 594 goat anti‐mouse IgG secondary antibody (Life Technologies) (1:500) in PBS–1% BSA, washed three times with PBS and once with distilled water (adapted from ref. 75).
Spore production, purification, and fractionation
For spore production 10 mL of BHI media was inoculated with an isolated colony of C. difficile and cultured overnight at 37 oC under anaerobic conditions. The next day, 150 mL of fresh BHI media was inoculated with 1.5 mL of the overnight culture and the new culture was incubated for 7 days at 37 oC under anaerobic conditions. Cells were collected by centrifugation at 4800 × g, suspended in cold water and stored for 24 to 48 h at 4 oC. The sediment was suspended in PBS with 0.1% Tween-20, and the spores were purified with a 42% Renografin (Bayer) step gradient1. The sediment, containing the spores, was suspended in PBS with 0.1% Tween-20, washed twice with the same buffer and twice with cold water. The final spore suspension was stored at −20 oC until use. For spore fractionation, the spore coat was removed by suspending an amount of purified spores corresponding to an OD580 nm of 2.0 (about 108 spores), in 50 μL of decoating buffer (10% glycerol, 4% SDS, 10% β-mercaptoethanol, 1 mM DTT, 250 mM Tris pH 6,8). The spores were then boiled for 5 min and collected by centrifugation. The supernatant, corresponds to the coat/exosporium fraction. The spore sediment was washed twice with PBS with 0.1% Tween-20, and incubated with 50 mM Tris-HCl pH 8,0 with 2 mg/mL lysozyme for 2 h at 37 oC to digest the spore cortex peptidoglycan and to release proteins associated with the spore core and cortex (core/cortex fraction). Proteins in the coat/exosporium and core/cortex fractions were resolved by SDS-PAGE (15% gels) and subject to immunoblotting with anti-CotD5, anti-TcdA (Santa Cruz Biotechnology), anti-GPR6, and anti-GerS7 antibodies.
Sporulation assays
Overnight cultures grown at 37oC in BHI were used to inoculate BHI (at a dilution of 1:200). Once the OD600 reached 0.4, 100 μL the cultures were plated in TY plates and incubated for 24 h. After this incubation, the cells were scarped from the plates and suspended in 1 mL of TY. The suspension was serially diluted in TY and plated before and after heat treatment (30 min at 70 oC), to determine the total and heat resistant colony forming units. The samples were plated as 20 μL spots in triplicate onto TY plates supplemented with 0.1% taurocholate (Carl Roth), to promote efficient spore germination.
Toxin quantification by immunoblotting
For the quantification of the amount of TcdA associated with 630Δerm and R20291 spores, samples of spore suspensions, corresponding to 108 spores were fractionated into a coat/exosporium and core/cortex fractions as described in the preceding section. Samples were boiled for 5 min in 50 μL of decoating buffer (10% glycerol, 4% SDS, 10% β-mercaptoethanol, 1 mM DTT, 250 mM Tris pH 6,8) and loaded in the same gel together with purified TcdA (0.5 μg; 0.25 μg; 0.1 μg and 005 μg). TcdA was purified by Ni2+-affinity chromatography from cultures of B. megaterium carrying pHis-TcdA (8; the B. megaterium strain was a gift from Lacy Borden). TcdA was kept in 1X Phosphate buffer, pH 7.4. The gels were analyzed by immunobloting with an anti-TcdA antibody (Santa Cruz Biotechnology, as above). Pixels were quantified using Fiji-Image J and a linear regression was performed to calculate the amount of toxin in both the coat/exosporium and the core/cortex fractions. The experiment was performed in triplicate.
Cell culture and cytopathic assays
Vero and HT29 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) with GlutaMAXTM supplemented with, 4.5 g/L Glucose, 5 or 10% (v/v) of bovine fetal serum for Vero or HT29 cells, respectively, and 1% (v/v) Pen-Strep (Sigma), at 37 oC in a 5% CO2 atmosphere. Cells were grown until confluence in 96-wells plates and incubated with twofold serially diluted spores in Dulbecco’s modified Eagle’s medium, starting by 5×106 purified spores24. After 24 h of incubation at 37 oC in a 5% CO2 atmosphere, the cytopathic effect was monitored by phase contrast microscopy using a Nikon HCS microscope equipped with a 10× objective and an AndorZyla 4.2 sCMOS 4.2Mpx camera. The percentage of cell rounding was quantified on the phase contrast images.
Actin immunostaining
For actin staining, Vero and HT29 cells with or without added spores were fixed in PBS containing 4% (w/v) paraformaldehyde for 15 min. Texas Red-conjugated Phalloidin (Thermo Fisher Scientific) was then added to a dilution of 1:100 in PBS-0.1% saponin containing 10% (v/v) horse serum. After staining, the cells were consecutively washed with PBS-0.1% saponin, PBS and ddH2O. The coverslips were assembled using Aqua-poly/Mount (Polysciences) on microscopy glass slides and the cells were examined by fluorescence microscopy on a Leica DM6000B microscope equipped with a phase contrast Uplan F1 63× objective and captured with a CCD Andor Ixon camera (Andor Technologies).
Statistical analysis
Distributions obtained from quantifications of the SNAPCd labeling were statistically analyzed using the non-parametric Kolmogorov-Smirnov test. For the quantification of the fluorescence signal in different types of cells, three independent experiments were conducted. Only the results statistically significant (***, p > 0.001; ****, p < 0.0001) in all three experiments were considered statistically relevant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
cassona et al supplemental information revised
Description of additional supplementary files
Acknowledgements
We thank Simon Cutting for the gift of the anti-CotD antibody and Aimee Shen for the gift of the anti-GerS and anti-GPR antibodies. We thank Georges Haustant for help with the spore cytopathic assays. This work was supported by grant PEst-OE/EQB/LA0004/2011 from the “Fundação para a Ciência e a Tecnologia” (FCT) to A.O.H. This work was also financially supported by Project LISBOA-01-0145-FEDER-007660 (“Microbiologia Molecular, Estrutural e Celular”) funded by FEDER funds through COMPETE2020 - “Programa Operacional Competitividade e Internacionalização” (POCI), by national funds through the FCT (“Fundação para a Ciência e a Tecnologia”), and by program IF of the FCT (IF/00268/2013/CP1173/CT0006) to M.S. C.C. (PD/BD/52212/2013), S.R. (SFRH/BD/45459/08) and K.A. (PD/BD/148398/2019) were the recipients of doctoral fellowships from the FCT.
Author contributions
A.O.H., M.S., C.P.C., J.M., C.J.-J., and B.D. conceived the project; C.P.C., together with S.R., K.A., T.C., I.K., C.D.-L., carried out the experiments; C.P.C., M.S. and A.O.H., carried out the data analysis; C.P.C., M.S., B.D., J.M. and A.O.H. co-wrote and revised the manuscript.
Peer review
Peer review information
Communications Biology thanks I-Hsiu Huang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris.
Data availability
Uncropped and unedited blot/gels are shown as follows: for Fig. 4d, e, see Supplementary Fig. S10; for Fig. 5b, c, see Supplementary Figs. S11 and S12; for Supplementary Fig. S2, see Supplementary Fig. S10; for Supplementary Fig. S5a, b, see Supplementary Fig. S13; for Supplementary Fig. S6a, b, see Supplementary Fig. S14; for Supplementary Fig. S7, see Supplementary Fig. S15. The Excel file provided (Supplementary data) contains the data used to make the plots shown in Figs. 2b, 3c, 4c and 6b.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06521-x.
References
- 1.Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 2016;14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat. Rev. Dis. Prim. 2016;2:16020–16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Luo Y, Grinspan AM. Epidemiology of community-acquired and recurrent Clostridioides difficile infection. Ther. Adv. Gastroenterol. 2021;14:175628482110162. doi: 10.1177/17562848211016248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knetsch CW, et al. Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans. J. Clin. Microbiol. 2018;56:e01384-17. doi: 10.1128/JCM.01384-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight DR, Riley TV. Genomic delineation of zoonotic origins of Clostridium difficile. Front. Public Health. 2019;7:164. doi: 10.3389/fpubh.2019.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen, A. Expanding the Clostridioides difficile genetics toolbox. J. Bacteriol.201, 10.1128/jb.00089-19 (2019). [DOI] [PMC free article] [PubMed]
- 7.Shen A. Clostridioides difficile spore formation and germination: new insights and opportunities for intervention. Annu. Rev. Microbiol. 2020;74:545–566. doi: 10.1146/annurev-micro-011320-011321. [DOI] [PubMed] [Google Scholar]
- 8.Kordus SL, Thomas AK, Lacy DB. Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat. Rev. Microbiol. 2022;20:285–298. doi: 10.1038/s41579-021-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saujet L, Pereira FC, Henriques AO, Martin-Verstraete I. The regulatory network controlling spore formation in Clostridium difficile. FEMS Microbiol. Lett. 2014;358:1–10. doi: 10.1111/1574-6968.12540. [DOI] [PubMed] [Google Scholar]
- 10.Swick, M. C., Koehler, T. M. & Driks, A. Surviving between hosts: sporulation and transmission. Virulence Mechanisms of Bacterial Pathogens, Fifth Edition 529–566 10.1128/microbiolspec.VMBF-0029-2015 (Wiley, 216).
- 11.Deakin LJ, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu D, Sorg JA, Sun X. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front. Cell. Infect. Microbiol. 2018;8:1–10. doi: 10.3389/fcimb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro-Córdova P, et al. Entry of spores into intestinal epithelial cells contributes to recurrence of Clostridioides difficile infection. Nat. Commun. 2021;12:1140. doi: 10.1038/s41467-021-21355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calderón-Romero P, et al. Clostridium difficile exosporium cysteine-rich proteins are essential for the morphogenesis of the exosporium layer, spore resistance, and affect C. difficile pathogenesis. PLOS Pathog. 2018;14:e1007199–e1007199. doi: 10.1371/journal.ppat.1007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-González F, et al. Protein composition of the outermost exosporium-like layer of Clostridium difficile 630 spores. J. Proteom. 2015;123:1–13. doi: 10.1016/j.jprot.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janoir C, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect. Immun. 2013;81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mora-Uribe, P. et al. Characterization of the adherence of Clostridium difficile spores: the integrity of the outermost layer affects adherence properties of spores of the epidemic strain R20291 to components of the intestinal mucosa. Front. Cell. Infect. Microbiol.6, 99 (2016). [DOI] [PMC free article] [PubMed]
- 18.Phetcharaburanin J, et al. The spore-associated protein BclA1 affects the susceptibility of animals to colonization and infection by Clostridium difficile. Mol. Microbiol. 2014;92:1025–1038. doi: 10.1111/mmi.12611. [DOI] [PubMed] [Google Scholar]
- 19.Pizarro-Guajardo M, et al. Characterization of the collagen-like exosporium protein, BclA1, of Clostridium difficile spores. Anaerobe. 2014;25:18–30. doi: 10.1016/j.anaerobe.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Abecasis AB, et al. A genomic signature and the identification of new sporulation genes. J. Bacteriol. 2013;195:2101–2115. doi: 10.1128/JB.02110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin MY, et al. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 2012;14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin MY, Yutin N, Wolf YI, Vera Alvarez R, Koonin EV. Conservation and evolution of the sporulation gene set in diverse members of the Firmicutes. J. Bacteriol. 2022;204:e00079–22. doi: 10.1128/jb.00079-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fimlaid KA, et al. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 2013;9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira FC, et al. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saujet L, et al. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 2013;9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Hoon MJL, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 2010;20:R735–R745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serrano M, et al. The SpoIIQ-SpoIIIAH complex of C lostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis: The SpoIIQ-SpoIIIAH complex of C. Mol. Microbiol. 2016;100:204–228. doi: 10.1111/mmi.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aktories K, Schwan C, Jank T. Clostridium difficile toxin biology. Annu. Rev. Microbiol. 2017;71:281–307. doi: 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 29.Luo J, et al. TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile. Cell. 2022;185:980–994.e15. doi: 10.1016/j.cell.2022.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl Acad. Sci. USA. 2001;98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moncrief JS, Barroso LA, Wilkins TD. Positive regulation of Clostridium difficile toxins. Infect. Immun. 1997;65:1105–1108. doi: 10.1128/iai.65.3.1105-1108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 2007;64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 33.Bakker D, Smits WK, Kuijper EJ, Corver J. TcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm. PLoS One. 2012;7:e43247. doi: 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter GP, et al. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLOS Pathog. 2011;7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcDc genotype and toxin production. Appl. Environ. Microbiol. 2012;78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curry SR, et al. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 2007;45:215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ. Truncation in the tcdC region of the Clostridium difficilePathLoc of clinical isolates does not predict increased biological activity of Toxin B or Toxin A. BMC Infect. Dis. 2009;9:103. doi: 10.1186/1471-2334-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira Paiva, A. M., de Jong, L., Friggen, A. H., Smits, W. K. & Corver, J. The C-terminal domain of Clostridioides difficile TcdC is exposed on the bacterial cell surface. J. Bacteriol.202, e00771-19 (2020). [DOI] [PMC free article] [PubMed]
- 39.Tan KS, WEE BY, SONG KP. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J. Med. Microbiol. 2001;50:613–619. doi: 10.1099/0022-1317-50-7-613. [DOI] [PubMed] [Google Scholar]
- 40.Govind R, Dupuy B. Secretion of Clostridium difficile Toxins A and B requires the Holin-like protein TcdE. PLOS Pathog. 2012;8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govind R, Fitzwater L, Nichols R. Observations on the Role of TcdE Isoforms in Clostridium difficile Toxin Secretion. J. Bacteriol. 2015;197:2600–2609. doi: 10.1128/JB.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olling A, et al. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb. Pathog. 2012;52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Wydau-Dematteis S, et al. Cwp19 is a novel lytic transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. mBio. 2018;9:e00648-18. doi: 10.1128/mBio.00648-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehner-Breitfeld D, et al. Evidence for an adaptation of a phage-derived holin/endolysin system to toxin transport in Clostridioides difficile. Front. Microbiol. 2018;9:2446. doi: 10.3389/fmicb.2018.02446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monot M, et al. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci. Rep. 2015;5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 47.Hundsberger T, et al. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 1997;244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 48.Martin-Verstraete I, Peltier J, Dupuy B. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins. 2016;8:153. doi: 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger Cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J. Bacteriol. 2013;195:5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 2012;194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Meouche, I. et al. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One8, e83748 (2013). [DOI] [PMC free article] [PubMed]
- 52.Ransom EM, Kaus GM, Tran PM, Ellermeier CD, Weiss DS. Multiple factors contribute to bimodal toxin gene expression in Clostridioides (Clostridium) difficile. Mol. Microbiol. 2018;110:533–549. doi: 10.1111/mmi.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donnelly ML, et al. Development of a dual-fluorescent-reporter system in Clostridioides difficile reveals a division of labor between virulence and transmission gene expression. mSphere. 2022;7:e0013222. doi: 10.1128/msphere.00132-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards, A. N., Anjuwon-Foster, B. R. & McBride, S. M. RstA is a major regulator of Clostridioides difficile toxin production and motility. mBio10, e01991-18 (2019). [DOI] [PMC free article] [PubMed]
- 55.Edwards, A. N. et al. Chemical and stress resistances of Clostridium difficile spores and vegetative cells. Front. Microbiol. 7, 1698 (2016). [DOI] [PMC free article] [PubMed]
- 56.Edwards, A. N., Krall, E. G. & McBride, S. M. Strain-dependent RstA regulation of Clostridioides difficile Toxin Production And Sporulation. J. Bacteriol.10.1128/JB.00586-19 (2019). [DOI] [PMC free article] [PubMed]
- 57.Castro-Córdova P, et al. Redistribution of the novel Clostridioides difficile spore adherence receptor E-cadherin by TcdA and TcdB increases spore binding to adherens junctions. Infect. Immun. 2023;91:e0047622. doi: 10.1128/iai.00476-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cassona, C. P., Pereira, F., Serrano, M. & Henriques, A. O. A Fluorescent reporter for single cell analysis of gene expression in Clostridium difficile. in Clostridium difficile: Methods and Protocols (eds. Roberts, A. P. & Mullany, P.) 69–90 (Springer New York, 2016). [DOI] [PubMed]
- 59.Oliveira Paiva AM, Friggen AH, Douwes R, Wittekoek B, Smits WK. Practical observations on the use of fluorescent reporter systems in Clostridioides difficile. Antonie Leeuwenhoek. 2022;115:297–323. doi: 10.1007/s10482-021-01691-8. [DOI] [PubMed] [Google Scholar]
- 60.Ransom, E. M., Weiss, D. S. & Ellermeier, C. D. Use of mCherryOpt fluorescent protein in Clostridium difficile. in Clostridium difficile (eds. Roberts, A. P. & Mullany, P.) vol. 1476, 53–67 (Springer New York, 2016). [DOI] [PubMed]
- 61.Merrigan M, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vohra P, Poxton IR. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology. 2011;157:1343–1353. doi: 10.1099/mic.0.046243-0. [DOI] [PubMed] [Google Scholar]
- 63.Dupuy B, et al. Regulation of toxin and bacteriocin gene expression in Clostridium by interchangeable RNA polymerase sigma factors. Mol. Microbiol. 2006;60:1044–1057. doi: 10.1111/j.1365-2958.2006.05159.x. [DOI] [PubMed] [Google Scholar]
- 64.Ng YK, et al. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PloS ONE. 2013;8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Girinathan BP, et al. Effect of tcdR mutation on sporulation in the epidemic Clostridium difficile strain R20291. mSphere. 2017;2:e00383-16. doi: 10.1128/mSphere.00383-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aubry A, et al. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 2012;80:3521–3532. doi: 10.1128/IAI.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soutourina O, et al. Genome-wide transcription start site mapping and promoter assignments to a sigma factor in the human enteropathogen Clostridioides difficile. Front Microbiol. 2020;11:1939. doi: 10.3389/fmicb.2020.01939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLOS ONE. 2013;8:e79666. doi: 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettit LJ, et al. Functional genomics reveals that Clostridium difficileSpo0A coordinates sporulation, virulence and metabolism. BMC Genom. 2014;15:160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS ONE. 2012;7:e48608. doi: 10.1371/journal.pone.0048608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, sigh, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 2011;193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Underwood S, et al. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 2009;191:7296–7305. doi: 10.1128/JB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS, Shen A. Regulation of Clostridium difficile spore formation by the SpoIIQ and SpoIIIA proteins. PLoS Genet. 2015;11:e1005562. doi: 10.1371/journal.pgen.1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Permpoonpattana P, et al. Functional characterization of Clostridium difficile spore coat proteins. J. Bacteriol. 2013;195:1492–1503. doi: 10.1128/JB.02104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barra-Carrasco J, et al. The Clostridium difficile exosporium cysteine (CdeC)-rich protein is required for exosporium morphogenesis and coat assembly. J. Bacteriol. 2013;195:3863–3875. doi: 10.1128/JB.00369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawley TD, et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 2009;191:5377–5386. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Permpoonpattana P, et al. Surface layers of Clostridium difficile endospores. J. Bacteriol. 2011;193:6461–6470. doi: 10.1128/JB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandes, C. G., Moran, C. P. Jr & Henriques, A. O. Auto-regulation of SafA assembly through recruitment of a protein cross-linking enzyme. J. Bacteriol.200, e00066-18 (2018). [DOI] [PMC free article] [PubMed]
- 79.Diaz OR, Sayer CV, Popham DL, Shen A. Clostridium difficile lipoprotein GerS is required for cortex modification and thus spore germination. mSphere. 2018;3:e00205–e00218. doi: 10.1128/mSphere.00205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sussman MD, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis GPR gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J. Bacteriol. 1991;173:291–300. doi: 10.1128/jb.173.1.291-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurka H, et al. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLOS ONE. 2014;9:e86535. doi: 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyras D, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Torres J, Camorlinga-Ponce M, Muñoz O. Sensitivity in culture of epithelial cells from rhesus monkey kidney and human colon carcinoma to toxins A and B from Clostridium difficile. Toxicon. 1992;30:419–426. doi: 10.1016/0041-0101(92)90538-G. [DOI] [PubMed] [Google Scholar]
- 84.Huang J-H, et al. Biochemical and immunological characterization of truncated fragments of the receptor-binding domains of C. difficile toxin A. PLoS ONE. 2015;10:e0135045. doi: 10.1371/journal.pone.0135045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burns DA, Heap JT, Minton NP. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J. Bacteriol. 2010;192:657–664. doi: 10.1128/JB.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veening J, et al. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol. Syst. Biol. 2008;4:184. doi: 10.1038/msb.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eijlander, R. T. et al. SpoVT: From fine-tuning regulator in bacillus subtilis to essential sporulation protein in Bacillus cereus. Front. Microbiol. 7, 1607 (2016). [DOI] [PMC free article] [PubMed]
- 89.Wang ST, et al. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 90.Asen I, Djuranovic S, Lupas AN, Zeth K. Crystal structure of SpoVT, the final modulator of gene expression during spore development in Bacillus subtilis. J. Mol. Biol. 2009;386:962–975. doi: 10.1016/j.jmb.2008.10.061. [DOI] [PubMed] [Google Scholar]
- 91.Hong HA, et al. Mucosal antibodies to the C terminus of toxin A prevent colonization of Clostridium difficile. Infect. Immun. 2017;85:e01060–16. doi: 10.1128/IAI.01060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frey SM, Wilkins TD. Localization of two epitopes recognized by monoclonal antibody PCG-4 on Clostridium difficile toxin A. Infect. Immun. 1992;60:2488–2492. doi: 10.1128/iai.60.6.2488-2492.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anjuwon-Foster BR, Tamayo R. A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet. 2017;13:e1006701. doi: 10.1371/journal.pgen.1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gerhard, R. Receptors and binding structures for Clostridium difficile toxins A and B. in Uptake and Trafficking of Protein Toxins (ed. Barth, H.) vol. 406, 79–96 (Springer International Publishing, 2016). [DOI] [PubMed]
- 95.Chiu P-J, et al. Clostridioides difficile spores stimulate inflammatory cytokine responses and induce cytotoxicity in macrophages. Anaerobe. 2021;70:102381. doi: 10.1016/j.anaerobe.2021.102381. [DOI] [PubMed] [Google Scholar]
- 96.Kasendra M, Barrile R, Leuzzi R, Soriani M. Clostridium difficile toxins facilitate bacterial colonization by modulating the fence and gate function of colonic epithelium. J. Infect. Dis. 2014;209:1095–1104. doi: 10.1093/infdis/jit617. [DOI] [PubMed] [Google Scholar]
- 97.Branka JE, et al. Early functional effects of Clostridium difficile toxin A on human colonocytes. Gastroenterology. 1997;112:1887–1894. doi: 10.1053/gast.1997.v112.pm9178681. [DOI] [PubMed] [Google Scholar]
- 98.Lawley TD, et al. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ. Microbiol. 2010;76:6895–6900. doi: 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seregin SS, Aldhamen YA, Rastall DPW, Godbehere S, Amalfitano A. Adenovirus-based vaccination against Clostridium difficile toxin A allows for rapid humoral immunity and complete protection from toxin A lethal challenge in mice. Vaccine. 2012;30:1492–1501. doi: 10.1016/j.vaccine.2011.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 2009;191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li J, McClane BA. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect. Immun. 2010;78:4286–4293. doi: 10.1128/IAI.00528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li, J., Paredes-Sabja, D., Sarker, M. R. & McClane, B. A. Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol. Spectr.4, 10.1128/microbiolspec.TBS-0022-2015 (2016). [DOI] [PMC free article] [PubMed]
- 103.Zhao Y, Melville SB. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 1998;180:136–142. doi: 10.1128/JB.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grecz N, Lin CA, Tang T, So WL, Sehgal LR. The nature of heat resistant toxin in spores of Clostridium botulinum. Jpn. J. Microbiol. 1967;11:384–394. doi: 10.1111/j.1348-0421.1967.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 105.Ammons DR, et al. A novel Bacillus thuringiensis cry-like protein from a rare filamentous strain promotes crystal localization within the exosporium. Appl Environ. Microbiol. 2013;79:5774–5776. doi: 10.1128/AEM.01206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dale JL, Raynor MJ, Ty MC, Hadjifrangiskou M, Koehler TM. A dual role for the Bacillus anthracis master virulence regulator AtxA: control of sporulation and anthrax toxin production. Front. Microbiol. 2018;9:482. doi: 10.3389/fmicb.2018.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cote C, Welkos S. Anthrax toxins in context of Bacillus anthracis spores and spore germination. Toxins. 2015;7:3167–3178. doi: 10.3390/toxins7083167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J. Med. Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 109.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined growth medium for Clostridium difficile. Microbiology. 1995;141:371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- 111.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
cassona et al supplemental information revised
Description of additional supplementary files
Data Availability Statement
Uncropped and unedited blot/gels are shown as follows: for Fig. 4d, e, see Supplementary Fig. S10; for Fig. 5b, c, see Supplementary Figs. S11 and S12; for Supplementary Fig. S2, see Supplementary Fig. S10; for Supplementary Fig. S5a, b, see Supplementary Fig. S13; for Supplementary Fig. S6a, b, see Supplementary Fig. S14; for Supplementary Fig. S7, see Supplementary Fig. S15. The Excel file provided (Supplementary data) contains the data used to make the plots shown in Figs. 2b, 3c, 4c and 6b.