Abstract
The search for prognostic markers in breast cancer has bumped into a typical feature of these tumors, intra and intertumoral heterogeneity. Changes in the expression profile, localization of these proteins or shedding to the surrounding stroma can be useful in the search for new markers. In this context, classification by molecular subtypes can bring perspectives for both diagnosis and screening for appropriate treatments. However, the Triple Negative (TN) subtype, which is already the one with the worst prognosis, lacks appropriate and consistent molecular markers. In this work, we analyzed 346 human breast cancer samples in tissue microarrays (TMA) from cases diagnosed with invasive breast carcinoma to assess the expression and localization pattern of Maspin and their correlation with clinical parameters. To complement our findings, we also used TCGA data to analyze the mRNA levels of these respective genes. Our data suggests that the TN subtype demonstrates a higher level of cytoplasmic Maspin compared to the other subtypes. Maspin transcript levels follow the same trend. However, TN patients with lower Maspin expression tend to have worse overall survival and free-survival metastasis rates. Finally, we used Maspin expression data to verify possible relationships with the clinicopathological information of our cohort. Our univariate analyses indicate that Maspin is related to the expression of estrogen receptor (ER) and progesterone receptor (PR). Furthermore, Maspin expression levels also showed correlation with Scarff-Bloom-Richardson (SBR) parameter, and stromal Maspin showed a relationship with lymph node involvement. Our data is not consistently robust enough to categorize Maspin as a prognostic marker. However, it does indicate a change in the expression profile within the TN subtype.
Keywords: Biomarkers, Triple negative, Breast cancer, Tissue microarray, Maspin
Subject terms: Biochemistry, Biomarkers, Molecular medicine
Introduction
Breast cancer is the most diagnosed malignancy and the one with the highest mortality rates in women worldwide1,2. In Brazil, not unlike global data, it is the most incident tumor with an estimate of approximately 66,280 new cases in 20223,4. Breast cancer is a complex, heterogeneous and multifactorial disease, and can be classified by the profile of gene expression or immunophenotyping in different intrinsic subtypes: Luminal A (ER+ PR+ HER2− Ki-67 < 14%), Luminal B (ER+ PR+/− HER2 ± Ki-67 ≥ 14%), HER2+ (ER− PR− HER2+ Ki-67 > 20%) and Triple Negative (TN) (ER− PR− HER2− Ki-67 > 30%)5,6.
Despite great advances in the molecular classification of breast cancer, a major challenge in clinical oncology has been the complete understanding of the mechanisms of intertumoral and mainly intratumoral heterogeneity7,8. In addition, multiple drug resistance (MDR) has been considered the biggest obstacle in the systemic treatment of breast cancer, making the disease often uncontrollable and leading to high mortality rates9. Consequently, hundreds of other candidates for biomarkers have been investigated for potential implications for diagnosis, prognosis, and prediction of therapeutic response10.
The SERPIN superfamily comprises serine protease inhibitors and is divided into 16 different classes11. The gene SERPINB5 encodes the protein Maspin, Mammary serine protease inhibitor, that can act in different cellular processes, such cell adhesion, migration, epithelial mesenchymal transition (EMT) and modulation of gene transcription, depending on different ligands and subcellular locations12–14. Described as a tumor suppressor due to experimental evidence of a relationship between its expression and inhibition of tumor growth, invasion, metastasis, and a better prognosis in different cancer types15–18. On one hand, in the cell membrane it can inhibit invasion and motility of tumor cells19,20, yet translocation to nucleus can also regulate gene transcription and favor tumor suppression21–24.
In the present study, we evaluated Maspin expression immunohistochemically, associated its expression levels with clinicopathological features and prognosis of resected breast cancer patients and performed in silico analyses using breast cancer patient’s public data.
Material and methods
Patients and tumor specimens
This study was based on a cohort of 346 breast tumor samples, fixed in formalin and embedded in paraffin, obtained from the archive of the Department of Pathological Anatomy of the A.C. Camargo Cancer Center, São Paulo, Brazil, between 1980 and 2005. Inclusion criteria were as follows: Confirmatory histological diagnosis of invasive ductal or lobular breast carcinoma, female gender, availability of demographic data, clinical and pathological information, details of treatment and clinical follow-up. Supplementary Table 1 shows the main clinical characteristics of the studied cohort. Cases that exhibited only lesions in situ, tissue nuclei with < 10% of representative tumor cells, worn or nonexistent spots, patients who received neoadjuvant chemotherapy combined or not with radiotherapy, in addition to samples from male patients were excluded. This study was evaluated and approved by the Research Ethics Committee (CIPE—Centro Internacional de Pesquisa e Ensino—AC Camargo Cancer Center) with protocol number 1822/13. All patients agreed with an informed consent document.
Construction of tissue microarrays (TMAs)
Briefly, the specimens of breast tumor tissues embedded in paraffin were recovered, sectioned, and stained with H&E (hematoxylin–eosin) to select viable and morphologically representative areas, as described previously25. 1 mm cylindrical samples were extracted from the donor blocks using a manual tissue matrix (Tissue microarrayer—Beecher Instruments, Silver Spring, MD, USA) and inserted in order in the recipient blocks. Subsequently, 4 μm sections were cut with a microtome from each TMA block (Tissue Microarray) on adhesion slides for subsequent IHC staining.
Immunohistochemistry (IHC) staining
Immunohistochemistry (IHC) for detection of candidate proteins for biomarkers in breast cancer was performed as previously described25–28. Maspin IHC staining were performed after confection of TMAs. Briefly, the slides were deparaffinized and rehydrated with a decreasing concentration of ethanol, and antigenic recovery was performed using sodium citrate buffer pH 6.0 in 95 °C steam, as previously described by Norton29. Then, the slides were treated with proteinase K and blocked with 3% hydrogen peroxide in phosphate buffered saline (PBS) at pH 7.4 for 5 min and washed 3 times with PBS, before adding the primary antibodies of interest. To standardize IHC staining, the dilution of antibodies and the visualization system were optimized in tissue sections archives. TMA’s slides were stained with the following primary antibody anti-Maspin, (BD, #554,292, 1:250 dilution). After, mouse secondary antibody HRP (Horseradish peroxidase) coupled dextran polymer detection system (Advance TM HRP link—Dako) was incubated for 30 min at room temperature after standard washes accordingly to manufacturer recommendations25. The presence of Maspin was finally detected by addition of 3,3-diaminobenzine tetrachloride (FLEX DAB + Substrate Chromogen System, Dako) to the samples, as previously described26–29. All slides were contrasted with Harris Hematoxylin. All immunostained slides were digitized on the Aperio ScanScope CS platform (Aperio Technologies, Inc., Vista, CA, USA) with 20 × magnification.
Immunomarking evaluation
All image analyses were performed using the digital pathology software QuPath (Quantitative Pathology & Bioimage Analysis, v0.2.0-m1, University of Edinburgh, United Kingdom)30. The quantitative analysis of nuclear and cytoplasmic expression for Maspin were evaluated and scored on tumor cells, in addition, thinking that this protein can be secreted into the extracellular environment, we carry out additional analyses evaluating the tumor microenvironment, in this way, we evaluate and score this marker in the surrounding stroma31. All analyses of the immunohistochemical expression of Maspin were performed without prior knowledge of the clinical details of our cohort and the clinical outcome of patients, according to REMARK guidelines32. In summary, the flow of our work comprised the estimation of scores for each marker, TMA design, cell segmentation, computation of characteristics and identification of cell type with intensity classification, and all stages were assisted by an experienced pathologist. All TMAs were identified using the “dearrayer” tool with the attribution of the grids and the map with the ID of each imported case. All tissue nuclei were detected using the “simple tissue detection” tool so that all tissues considered insufficient (< 10% of representative tumor cells in the core), with dominant artifacts or consisting only of ductal carcinoma in situ, were removed from our analysis25. In addition, for cases that were considered evaluable, but which presented confusing objects and folds of tissue, they were removed manually at this stage. The cells were identified using the custom “cell detection” algorithm31,33. A cell classifier was trained separately for each marker evaluated in the study, defining the tumor cell class, stroma, necrosis, and inflammatory infiltrate31. The expression rates of each marker were calculated as the percentage of the number of positive cells in relation to the total number of tumor cells31,34. The detection classifier for our marker was applied to all datasets. At the end, the samples were divided into two groups (low and high) based on the median values obtained from the H-score for cytoplasmic and nuclear labeling in tumor cells and stromal labeling of Maspin IHC. The following H-score medians were obtained for the Maspin: Cytoplasm 71.07, Nucleus 145.84 and Stroma 19.32, and subsequently, Kaplan Meyer (KM) curves were elaborated according to follow up patients’ data.
In silico analyses
For compare expression of SERPINB5 gene, that express Maspin protein, in cancer and normal samples we use the online tool UALCAN (http://ualcan.path.uab.edu)35. The Firehose Legacy TCGA public data was accessed through cBioPortal platform (https://www.cbioportal.org/)36,37 and mRNA Maspin levels of 1108 patients were downloaded. Excluded from the study were cases of male patients (16 cases) and cases that did not have expression information (4 cases), resulting in a total of 1088 evaluable patients. After performing the exclusion criteria, the expression data were cross-checked with the clinical-pathological data accessed through the UCSC Xena platform (http://xena.ucsc.edu/)38 to analyze the association of SERPINB5 gene expression with different clinical and pathological parameters. The integration of data from both databases was achieved by leveraging the patients' ID/barcode, a shared identifier across the spreadsheets. Median gene expressions were computed, serving as the basis to classify groups as Low (≤ median) or High (> median). Subsequently, the contingency table was formulated, employing the chi-squared test or Fisher's exact test as appropriate. The survival curve adopted the same "high" and "low" stratification, utilizing the Kaplan–Meier (KM) method and log-rank test. For a comprehensive quantitative analysis of SERPINB5 expression profiles, an initial assessment of Gaussian distribution was conducted. Following this, the variables underwent either the t-test or Mann–Whitney test for paired variables, or ANOVA or Kruskal–Wallis for three or more groups. The beta value (HM450) refers to the DNA methylation level of individual CpG sites in comparison to the corresponding mRNA expression. The presentation of Maspin levels involved log-transformed mRNA expression z-scores, compared to the expression distribution of all samples (RNA Seq V2 RSEM) as previously described36,37.
Survival curves were also constructed using downloaded TCGA data. The median expression of the studied genes was used as a cut-off for classification into low or high expression.
Cell cultures and western blot
The human breast cancer cell lines MCF-7, (luminal) and MDA-MB-231 (Triple Negative) and HS578T were grown in Dulbecco’s Modified Eagle Medium, (DMEM Invitrogen) with 10% fetal bovine serum (Invitrogen) and the non-malignant MCF-10A cells were grown in DMEM/F-12 (Invitrogen) with 10 µg/mL insulin, 100 ng/mL cholera toxin, 500 ng/mL hydrocortisone, 20 ng/mL Epidermal growth factor (Sigma), and 5% horse serum (Invitrogen). All cells were kept in a humidified 5% CO2 incubator at 37 °C and cultured until 70% confluence before subculture or protein extraction. Protein extraction, SDS-PAGE and Western blot were previously described25.
Statistical analysis
Categorical data were assessed using Pearson's chi-square test or Fisher's exact test. Fisher was applied when 25% or more of the expected value was less than 5. Statistical analyzes of the box plots were performed using the t-student or Mann–Whitney, depending on whether the data had a normal distribution or not, respectively. Cumulative survival probabilities were estimated using the Kaplan–Meier method and survival curves were compared using the log-rank test. The survival times for overall survival (OS) were calculated from the date of surgery until the moment of death or last follow-up and for disease-free survival (DFS), the outcome was at the date of cancer recurrence after surgery or end of the follow-up. A statistically significant difference was predetermined for values of p < 0.05. IBM SPSS Statistics (version 23.0; SPSS Inc., Chicago, IL, USA) was used for our analyses.
Ethics approval and consent
This study was evaluated and approved by the Research Ethics Committee (CIPE—Centro Internacional de Pesquisa e Ensino—AC Camargo Cancer Center) with protocol number 1822/13 and registered at the research regulatory platform of the Brazilian Ministry of Health (CAAE: 20373713.8.0000.5432). Clinical samples used in this study were retrieved from the A.C. Camargo Cancer Center biorepository and all participants signed an approved informed consent form.
Good practices statement
We attest that all methods performed were performed in accordance with institutional guidelines and regulations, as well as good laboratory practices.
Results
Expression profile of maspin in human breast tumors
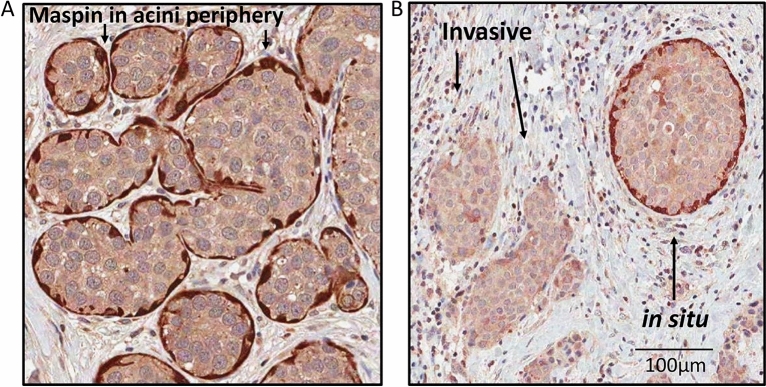
We evaluated the expression levels and subcellular location of the Maspin protein in our panel of primary human breast cancer tumors. The immunoreactivity of Maspin was identified predominantly in the cytoplasm and nucleus of breast cancer cells (Fig. 1). In addition, these target proteins were not restricted to tumor cells only since they were also observed in surrounding stroma and in the nuclei of inflammatory stromal cells (Fig. 1). In our visual inspection, we also noticed high Maspin expression in normal acini and hyperplasic with a preserved myoepithelial cell layer in the contour of the acini (Fig. 2A and Supp. Figure 1). In some cases, we observed Ductal Carcinoma in Situ (DCIS) components present in lesions diagnosed as Invasive Ductal Carcinoma (IDC). DCIS components were positively stained for Maspin at the periphery of the tumor cell clusters, which is consistent with the fact that the myoepithelial cells layer is preserved in these structures (Fig. 2B). In contrast, this staining pattern is lost in invasive components (Fig. 2B).
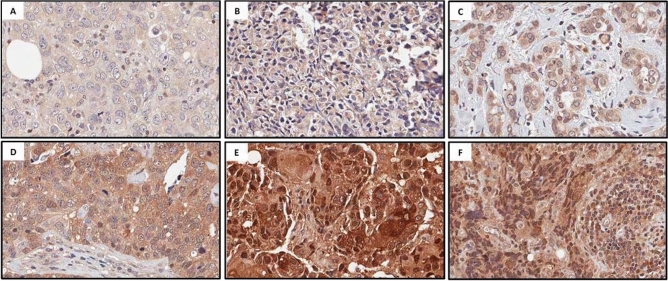
Figure 1.
Representative images of Maspin IHC staining in human breast cancer tissue samples. The image panel shows different subcellular locations of Maspin as well as different immunostaining intensities. (A) and (D) cytoplasmic immunostaining, (B) and (E) nuclear immunostaining, C and F stromal immunostaining. (A), (B) and (C) low immunoreactivity and (D), (E) and (F) high immunoreactivity.
Figure 2.
Maspin in myoepithelial cells layer. (A) Photomicrographs showing hyperplasic acini in breast tissue surrounded by myoepithelial cells, which express more Maspin than luminal cells. (B) Typical in situ component (indicated by an arrow) outlined by Maspin αV staining, whereas invasive component shows almost complete absence of peripheral Maspin. Scale bar = 100 µm. See supplementary Fig. 1 for an additional.
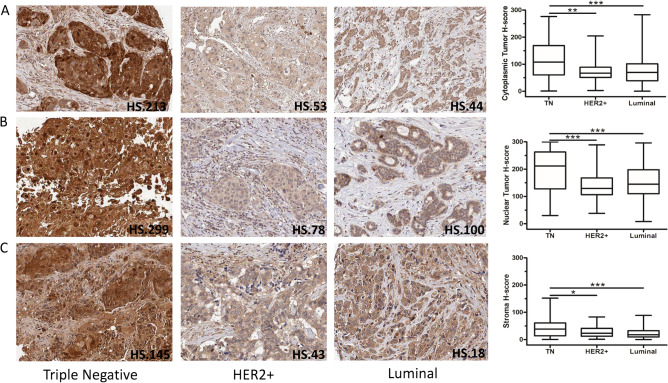
Next, we investigated the Maspin expression levels according to the molecular subtype. In the studied cohort, Luminal represents 67.9% of cases (n = 235), HER2+, 8.7% (n = 30), and Triple Negative (Basal Like), 17.1% (n = 59). Non characterized cases are 6.4% (n = 22) as described in Supplementary Table 1. Expression data were obtained using the QuPath program and we discriminated expression according to the cytoplasmic, nuclear compartment or considering the adjacent stroma. According to the molecular subtype, Maspin protein levels were higher in both the nucleus and cytoplasm of breast cancer cells belonging to the TN tumor group compared to the other subgroups. (Cytoplasmic marking: TN vs HER2+ p = 0.0012; TN vs Luminal p < 0.0001/Nuclear Marking: TN vs HER2+ p = 0.0006; TN vs Luminal p < 0.0001) (Fig. 3). Additionally, we also detected higher protein levels of Maspin in surrounded stroma in TN tumors, in relation to the other molecular subtypes (TN vs HER2+ p = 0.0344; TN vs Luminal p < 0.0001). (TN vs HER2+ p = 0.0344; TN vs Luminal p < 0.0001) (Fig. 3). Despite comparing distinct molecular approaches.
Figure 3.
IHC staining of Maspin in samples of primary breast cancer tumors and its expression profile in different molecular subgroups. Image above representative photomicrographs of tumor tissues classified as high and low for Maspin in the TN (A and D, respectively), HER2+ (B and E, respectively) and Luminal (C and F, respectively) subtypes. Image below IHC expression levels of Maspin between the different molecular groups and according to the cytoplasmic and nuclear location in breast cancer cells (A and B, respectively). IHC expression levels of Maspin between the different molecular groups and according to the protein present in the tumor microenvironment (C). Statistical analyzes of the box plots were performed using the t-student or Mann–Whitney test as appropriate, with data expressed as mean ± s.e.m. (standard error). The following H-score medians were obtained for the Maspin: Cytoplasm 71.07, Nucleus 145.84, and Stroma 19.32, and subsequently, Kaplan Meyer (KM) curves were elaborated according to follow up patients’ data. *p < 0.05; **p < 0.01; ***p < 0.001.
Association of maspin protein levels with clinical and pathological characteristics
To investigate Maspin expression in tumor cancer progression, we assessed the correlation between clinical and anatomopathological characteristics of cohort. To access Maspin expression levels in human breast cancer, we performed IHC using a tissue microarray (TMA). Expression data obtained using the QuPath program, was categorized according to subcellular location: cytoplasmic, nuclear, or secreted into the surrounding stroma. We determined a cutoff using the mean of H-score values to group samples in low (H-score under the mean) or high (H-score above mean) Maspin levels, and as described in Material and Methods. The median age of patients at the time of diagnosis was 55 years (ranging from 26 to 96 years). The tumor size in most patients was a maximum of 5 cm (72%), and the histological classification according to the Scarff-Bloom-Richardson (SBR) grading system was 13.8% grade 1, 56.9% grade 2, and 26% grade 3. In the studied cohort, 33.2% showed metastasis and the frequency of cases by molecular subtype of our cohort is comparable to a large study of distribution by molecular entities of the Brazilian female population with breast cancer (Supp. Table 1)39.
High Maspin protein levels were linked to increased severity in the SBR classification, specifically showing significance in Cytoplasmic marking (p = 0.047) and Stromal marking (p = 0.008). Additionally, greater lymph node involvement was associated with Stromal marking (p = 0.05) (Table 1). In addition, there were statistically significant associations between the differential expression of Maspin with the status of ER (Cytoplasmic marking: p = 0.028; Stromal marking: p < 0.0001) and PR (Nuclear marking: p = 0.025; Stromal marking: p = 0.001) and regarding the molecular subtype (Cytoplasmic marking: p = 0.037; Nuclear marking: p = 0.017; Stromal marking: p = 0.008) (Table 1).
Table 1.
Association of the immunohistochemical expression of Maspin according to clinical-pathological data.
| Variables | Category | Maspin cytoplasmic expression | p value | Maspin nuclear expression | p value | Maspin stromal expression | p value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | |||||
| Age, n (%) | < 40 | 15 (8.7) | 20 (11.6) | 0.34 | 13 (7.5) | 22 (12.7) | 0.218 | 16 (9.2) | 19 (11) | 0.864 |
| 41–59 | 92 (53.2) | 79 (45.7) | 91 (52.6) | 80 (46.2) | 86 (49.7) | 85 (49.1) | ||||
| ≥ 60 | 66 (38.2) | 74 (42.7) | 69 (39.9) | 71 (40.1) | 71 (41.1) | 69 (39.9) | ||||
| Hormonal Status, n (%) | Premenopause | 111 (64.2) | 107 (61.8) | 0.656 | 109 (63) | 109 (63) | 0.098 | 109 (63) | 109 (63) | 0.0001 |
| Postmenopause | 62 (35.8) | 66 (38.2) | 64 (37) | 64 (37) | 64 (37) | 64 (37) | ||||
| Tumor Size, n (%) | ≤ 2 cm | 31 (18.1) | 30 (17.5) | 0.883 | 34 (20) | 27 (15.7) | 0.375 | 36 (20.9) | 25 (14.7) | 0.256 |
| 2.1 cm–5 cm | 99 (57.9) | 96 (56.1) | 98 (57.6) | 97 (56.4) | 97 (56.4) | 98 (57.7) | ||||
| > 5 cm | 41 (24) | 45 (26.4) | 38 (22.4) | 48 (27.9) | 39 (22.7) | 47 (27.6) | ||||
| Lymph node Involvement, n (%) | pN0 | 61 (35.3) | 60 (35.1) | 0.717 | 57 (32.9) | 64 (37.4) | 0.112 | 68 (39.3) | 53 (31) | 0.05* |
| pN1 | 56 (32.4) | 57 (33.3) | 64 (37) | 49 (28.7) | 60 (34.7) | 53 (31) | ||||
| pN2 | 40 (23.1) | 33 (19.3) | 39 (22.6) | 34 (19.9) | 33 (19.1) | 40 (23.4) | ||||
| pN3 | 16 (9.2) | 21 (12.3) | 13 (7.5) | 24 (14) | 12 (6.9) | 25 (14.6) | ||||
| Presence of Metastasis, n (%) | pM0 | 165 (95.4) | 163 (94.8) | 0.794 | 164 (95.3) | 164 (94.8) | 0.813 | 166 (96) | 162 (94.2) | 0.448 |
| pM+ | 8 (4.6) | 9 (5.2) | 8 (4.7) | 9 (5.2) | 7 (4) | 10 (5.8) | ||||
| Scarff-Bloom-Richardson, n (%) | Grade 1 | 29 (17.4) | 17 (10.3) | 0.047* | 30 (17.9) | 16 (9.7) | 0.097 | 27 (16.1) | 19 (11.5) | 0.008** |
| Grade 2 | 101 (60.4) | 96 (57.8) | 95 (56.5) | 102 (61.8) | 108 (64.3) | 89 (53.9) | ||||
| Grade 3 | 37 (22.2) | 53 (31.9) | 43 (25.6) | 47 (28.5) | 33 (19.6) | 57 (34.5) | ||||
| Nuclear Grade, n (%) | Grade 1 | 2 (1.2) | 1 (0.6) | 0.412 | 2 (1.2) | 1 (0.6) | 0.195 | 2 (1.2) | 1 (0.6) | 0.319 |
| Grade 2 | 60 (36.6) | 49 (30.1) | 62 (37.6) | 47 (29) | 62 (37.1) | 47 (29.4) | ||||
| Grade 3 | 102 (62.2) | 113 (69.3) | 101 (61.2) | 114 (70.4) | 103 (61.7) | 112 (70) | ||||
| Clinical Internship, n (%) | Stage I | 9 (5.20 | 10 (5.8) | 0.983 | 7 (4) | 12 (6.9) | 0.356 | 9 (5.2) | 10 (5.8) | 0.09 |
| Stage II | 93 (53.8) | 90 (52) | 99 (57.2) | 84 (48.6) | 103 (59.5) | 80 (46.2) | ||||
| Stage III | 63 (36.4) | 64 (37) | 59 (34.1) | 68 (39.3) | 54 (31.3) | 73 (42.2) | ||||
| Stage IV | 8 (4.6) | 9 (5.2) | 8 (4.7) | 9 (5.2) | 7 (4) | 10 (5.8) | ||||
| Estrogen Receptor, n (%) | Negative | 43 (25.7) | 61 (37) | 0.028* | 45 (27.1) | 59 (35.5) | 0.098 | 37 (22.3) | 67 (40.4) | < 0.0001**** |
| Positive | 124 (74.3) | 104 (63) | 121 (72.9) | 107 (64.5) | 129 (77.7) | 99 (59.6) | ||||
| Progesterone Receptor, n (%) | Negative | 76 (45.2) | 91 (54.5) | 0.09 | 73 (43.7) | 94 (56) | 0.025* | 68 (40.7) | 99 (58.9) | < 0.001*** |
| Positive | 92 (54.8) | 76 (45.5) | 94 (56.3) | 74 (44) | 99 (59.3) | 69 (41.1) | ||||
| HER2, n (%) | Negative | 121 (85.2) | 126 (84.6) | 0.878 | 114 (82) | 133 (87.5) | 0.192 | 125 (88.7) | 122 (81.3) | 0.082 |
| Positive | 21 (14.8) | 23 (15.4) | 25 (18) | 19 (12.5) | 16 (11.3) | 28 (18.7) | ||||
| Molecular Subtypes | Triple Negative | 20 (14.3) | 39 (26.5) | 19 (13.9) | 40 (26.7) | 0.017* | 20 (14.4) | 39 (26.4) | ||
| HER2+ | 16 (11.4) | 14 (9.5) | 0.037* | 18 (13.1) | 12 (8) | 11 (7.9) | 19 (12.8) | 0.008** | ||
| Luminal | 104 (74.3) | 94 (64) | 100 (73) | 98 (65.3) | 108 (77.7) | 90 (60.8) | ||||
HER2, human epidermal growth factor receptor 2.
p-values obtained by Pearson’s chi-square test or Fisher's exact test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Fisher was applied when 25% or more of the expected value was less than 5.
Significant values are in bold.
Prognostic value of maspin protein level on breast cancer patients' survival
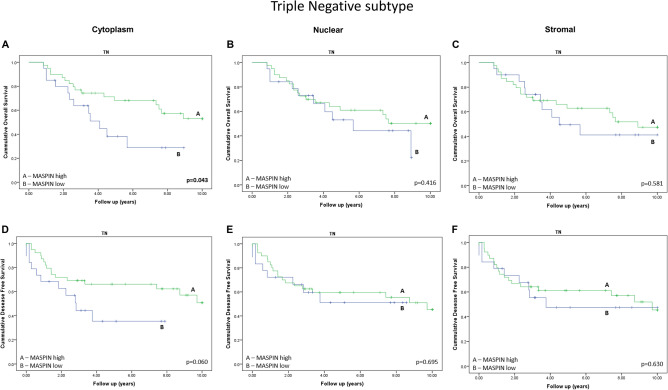
Next, we assessed the prognostic value of Maspin protein level in relation of breast cancer patient’s outcome data in Kaplan Meyer curves analysis. Initially, we did not detect any significant differences in the protein levels of Maspin affecting the survival of breast cancer patients when analyzing all molecular subtypes simultaneously (Supp. Figure 2). Likewise, we also did not find significant differences in Maspin expression in the Luminal and HER2 + subtypes. (Supp. Figure 3 and Supp. Figure 4). However, in TN subtype tumors, we observed that patients with lower Maspin expression had a worse prognosis for cancer specific survival (Cytoplasmic marking: OS p = 0.043; DFS p = 0.06) (Fig. 4). We found no significant correlation with the expression of nuclear Maspin or surrounding stroma (Fig. 4).
Figure 4.
Kaplan–Meier curves for overall survival (OS) and disease-free survival (DFS) of patients with breast cancer of the TN subtype stratified according to the Maspin expression. The OS curves (A, B and C) and DFS curves (D, E and F) were calculated for each group of patients. All cases were classified as high or low expression for Maspin. (A) and (D) Evaluation of cytoplasmic immunostaining. (B) and (E) Evaluation of nuclear immunostaining. (C) and (F) Stromal immunostaining assessment.
Maspin mRNA levels and breast cancer patients' survival
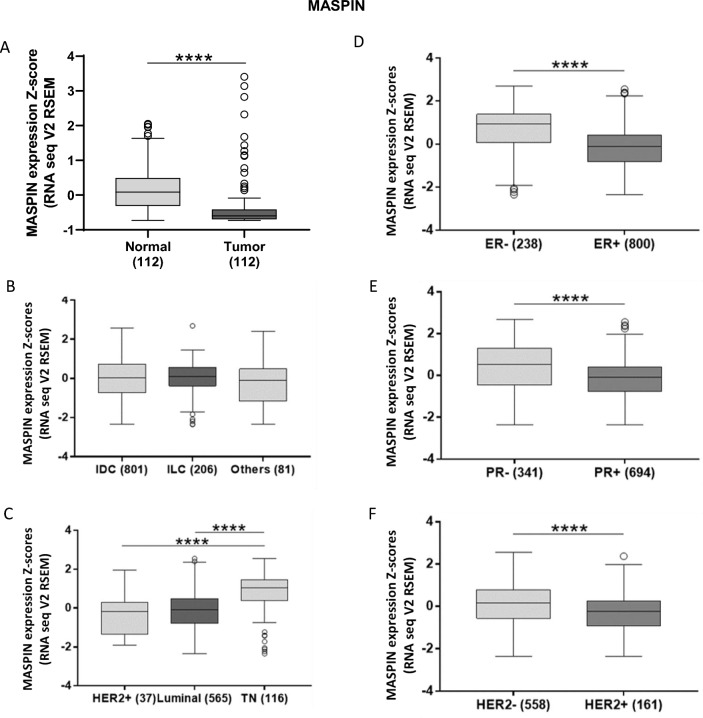
In silico analyses were conducted using TCGA RNA-seq data to evaluate the expression of SERPINB5 gene, official name of the gene that express Maspin protein, in tumor samples and normal tissue. It was observed that this gene is differentially expressed in tumor tissue compared to normal breast samples, while Maspin mRNA levels were significantly reduced in tumors (p < 0.0001) (Fig. 5A). Corroborating the in silico data, our Western Blot evaluation revealed that Maspin mRNA levels was significantly lower in breast tumor cell lines when compared to normal cell lines (Supp. Figure 4). No significant differences were observed between the different histological subtypes and the mRNA levels of Maspin (Fig. 5B). The stratification of tumors in the different subtypes demonstrated that Maspin mRNA levels has a significant association with breast cancer subtypes and that TN tumors is the group with the highest expression levels (p < 0.0001) (Fig. 5C).
Figure 5.
Expression profile of Maspin in different tissue and molecular contexts. (A) Differential expression of Maspin in normal and tumoral breast tissue, (B) in different histopathological subtypes and (C) in different molecular subtypes of breast cancer. On the right, expression profile of Maspin in breast cancer according to classical breast cancer biomarkers. (D) Expression of Maspin and the estrogen receptor. (E) Maspin and the progesterone receptor, and (F) expression of Maspin and HER2, respectively. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Results based on TCGA data.
Then, we also classified the patients according to different clinicopathological parameters and based on estrogen, progesterone and HER2 receptors status. Maspin mRNA levels were significantly reduced in patients with ER (p < 0.0001), PR (p < 0.0001) and HER2 (p < 0.0001) positive tumors (Fig. 5D, E and F) and there was a significant association between Maspin mRNA levels and age (p < 0.0001), hormonal status (p < 0.0001), TP53 status (p = 0.002) and TN status (p < 0.0001) (Supplementary Table 2).
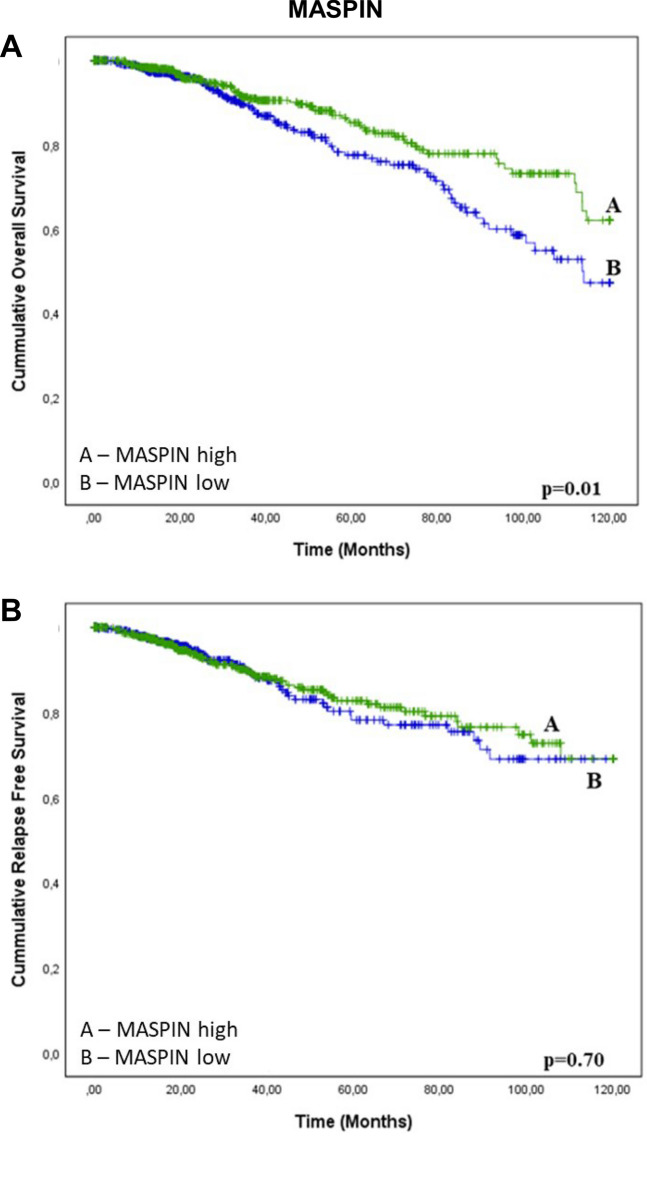
We assessed the prognostic significance of Maspin mRNA level in 1088 patients from TCGA database. The results show that low mRNA levels of Maspin are associated to a reduced overall survival conferring a worse prognosis (Fig. 6A). There was no association between Maspin mRNA levels and relapse free survival (Fig. 6B).
Figure 6.
Kaplan–Meier curves for breast cancer patient survival as a function of Maspin mRNA expression. Kaplan–Meier curves for overall survival (OS) and relapse-free survival (RFS) of breast cancer patients stratified according to mRNA of Maspin. All patients are included in this analysis. The analyses are conducted using the on TCGA Firehose Legacy breast cancer patients. ****p < 0.0001. Results based on TCGA data.
Discussion
Cancer biomarkers are valuable clinical tools that can be used for diagnostic and prognostic purposes, as well as predictive therapeutic approaches. Despite advances in cancer biology research, coupled with the use of "omic" technologies, only a few candidates have been successfully adopted in the routine clinic40. For breast cancer, this problem is even greater due to the surprising inter- and intratumoral genetic heterogeneity41,42.
Maspin is a member of the Serpin family, which has pleiotropic action and a great diversity of molecular targets43,44. Despite initially being found in breast tissue45, hence its name Mammary serine protease inhibitor, it is widely expressed in different types of homeostatic epithelial cells44,46. Among the various cellular processes influenced by Maspin, adhesion, migration, invasion, proliferation, cell death and oxidative stress deserve special attention, because are intricate to neoplastic progression43,44,47. In breast tissues, Maspin is highly expressed in normal epithelial cells, especially in myoepithelial cells47–49. However, Maspin expression levels in neoplasms show contradictory findings. If on the one hand, there is data in the literature demonstrating that Maspin expression is down-regulated in breast45, prostate50, gastric15 and melanoma51 cancers46–49, other evidence demonstrated over-expressed in pancreatic52, gallbladder, colorectal, and thyroid cancers50–53. The controversy may even be greater considering that some studies show that Maspin expression may be biphasic, being silenced at early stages of tumorigenesis and re-expressed in the metastatic phase47, suggests that Maspin has versatile biological functions under different pathophysiological context, which does not make its suggestion as a biomarker pragmatic. In this work, our findings showed no correlation between Maspin protein level and survival of breast cancer patients in the studied cohort, when analyzed collectively. What had also been reported in the literature53. However, our TCGA analyses demonstrated that Maspin mRNA levels in neoplastic breast tissues are lower when compared to their non-tumor counterparts. We could not perform a similar analysis at the protein level using our TMA cohort because the tissue deposited in the blocks did not comprise samples of normal breast tissue from the respective patients. Despite comparing distinct molecular approaches—using mRNA or protein data for patient stratification in survival curve analysis from two different populations—, the observed discrepancy between protein and mRNA expression levels can potentially be attributed to variations in the dynamics of RNA expression and protein translation, degradation, as well as different post-transcriptional regulatory mechanisms. Should this pattern persist within the same population when comparing mRNA/protein parameters, it highlights the independence of these processes and their susceptibility to distinct regulatory influences54,55. Furthermore, it must be considered that these analyses were carried out on different cohorts, which may reflect heterogeneous genetic backgrounds.
Intriguingly, evidence in the literature indicating that, more than the absolute expression level of Maspin, its subcellular location correlates better with tumor suppression and prognosis22,56,57. Maspin protein has already been found in the cytoplasmic and nuclear compartment23,58, as well as a cell surface-associated protein20. Furthermore, it has also been documented to be secreted extracellularly46,59. Surprisingly, subcellular location seems to have different prognostic values, that in the nucleus it may be associated with good prognosis60–62, whereas cytoplasmic localization correlates with poor prognosis63–65. This discrepancy can be explained by Maspin’s interaction with different molecular targets, acting either as a suppressor or promoter of the neoplastic process depending on the cellular circumstances43,66,67. In our work, by visually inspecting the images relating to the cases in our TMA cohort, Maspin was detected by IHC in the nucleus, cytoplasm, as well as secreted in the tumor microenvironment of these tissues, corroborating with previous published data23,46,59. Interestingly, in cases that presented mammary gland components (or DCIS component associated) with preserved epithelial architecture, Maspin protein level appears to be juxtaposed with the myoepithelial cell layer, corroborating with previous publications49–51. Despite this, our results did not demonstrate statistically significant correlations between nuclear, cytoplasmatic or stromal Maspin protein level and the survival rates or incidence of metastases in the cohort patients analyzed collectively. So, we proceeded to analyze according to the molecular subtype.
Classification of breast carcinomas into subtypes according to the expression of molecular markers, mainly ER, PR and HER-2, brought highly significant correlations with clinical outcomes, including overall survival and recurrence free survival68,69. For decades HER-2 has been used as a breast cancer biomarker associated with alteration in gene expression, and by extension, predictor of Trastuzumab efficacy70. However, only a limited proportion of breast cancer patients are HER-2+, and even in these, there is a chance that the expression will lose consistency and they will become refractory to treatment71,72. Likewise, hormone therapy is conditioned on the detection of ER and PR receptors in luminal subtypes73. For the triple-negative breast cancer subtype, which is known to have worse outcome compared to other breast cancer subtypes74, remains without specific treatment guidelines75 and identifying markers as well69. Our clinicopathological evaluation demonstrated that positive tumor expression of both ER and PR was significantly associated with lower Maspin expression, and it is precisely these markers that define the molecular subtypes. This corroborates previous data in the literature, since ER has been described to suppress Maspin expression64,65. Furthermore, our results shows that Maspin is more expressed in the TN subtype when compared to Luminal and HER2+ subtypes. Corroborating this finding, our silico analyses using TCGA RNA-seq data show higher levels of Maspin mRNA in TN subtypes when compared to other subtypes. If Maspin protein and transcript levels are reduced in the TN subtype, we next asked whether it could have any prognostic value in this subtype. Our Kaplan–Meier curves indicate that TN patients with lower cytoplasmatic Maspin had a worse prognosis for Overall survival. Even so, we recognize that the value of this observation for the prognosis is limited.
There has been debate about the involvement of the host stroma and a diversity of non-tumor cell populations in tumor progression76. In this context, several mechanisms can contribute synergistically, such as molecules that change the pattern of gene expression in the tumor77 or being improperly secreted78,79, as well as heterotypic cell interactions80 mediated by mediators such as growth factors81, cytokines82, exosomes83 and others84, that contributes for maintenance of immunosuppressive microenvironment, pro-tumor, with high potential to be refractory to conventional therapies85. When analyzing the images of the cases in our cohort, we noticed the immunostaining of both Maspin in regions corresponding to the stroma. When analyzing h-score of Maspin staining in the stroma, our results indicated that is greater in the TN, when compared to the Luminal and HER2 subtypes. Evidence in the literature indicates that Maspin is predominantly cytoplasmic86, with some membrane or nucleus association87, but which may also be partially secreted46,70. Nevertheless, Maspin at the epithelium-stroma interface, which most likely corresponds to myoepithelial cells, is an indicator of a better clinical outcome88. Several evidence in the literature have shown that the subcellular location of Maspin may have greater prognostic value in some types of cancer than its absolute expression measure21,56,67,89,90. On the other hand, it is yet unclear the consequences of the presence of Maspin in the extracellular matrix adjacent to tumors. Although our data do not explain the discrepancy between the highest Maspin expression in TN patients versus the worst prognosis of those with low expression in this subtype, changes in subcellular location or even its secretion could be speculated. Even so, our data suggested that stromal Maspin was related to greater lymph node involvement. Further investigations will be necessary to better elucidate its involvement in tumor progression, as well as its possible use as a prognostic indicator.
Closing remarks
Our TMA expression data did not detect statistically significant relationships between Maspin expression levels and indices of overall survival when to the entire cohort analyzed collectively. However, when divided by molecular subtype, Triple negative (TN) presents alterations in the expression levels of Maspin, in both of its mRNAs, and at the protein level. Although TN breast cancer has Maspin levels increased compared to other Luminal and HER2 subtypes, our results shows that downregulation in TN had a poor prognosis. Collectively, the Maspin expression data are not consistent for indicating a prognostic marker in breast cancer, but they demonstrate that there is a change in the expression profile in the TN subtype.
Supplementary Information
Acknowledgements
The authors thank to Bryan E. Strauss for manuscript review and Suely Nonogaki for assistance and technical support.
Author contributions
R.G.N.: contributed with patient data harvest, conventional microscopy and digital pathology, bioinformatics and statistical analysis, manuscript writing and reviewing; M.P.F.C.: contributed with data analysis and statistics, manuscript writing; D.R.B.: contributed with data analysis and statistics, manuscript writing; C.A.B.T.O.: contributed with supervision in pathology, TMA data generation and analyses; R.V.M.L.: contributed with data analysis and statistics; E.M.R.: contributed with the experimental design and supporting grants; O.L.D.C.: coordinated TMA assays, harvest and analyzed patient data, conventional microscopy and digital pathology, data analysis, the manuscript writing and reviewing.
Funding
OLDC was a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) PhD scholarship recipient during 2009 – 2014 (CAPES, Finance Code 001). However, the work currently has no supporting grants.
Data availability
Please contact corresponding author (Otto Cerqueira: ottoluix@alumni.usp.br) for data requests.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-53870-y.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Orlandini LF, et al. Epidemiological analyses reveal a high incidence of breast cancer in young women in Brazil. JCO Glob. Oncol. 2021;7:81–88. doi: 10.1200/GO.20.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cecilio AP, et al. Breast cancer in Brazil: Epidemiology and treatment challenges. Breast Cancer. 2015;7:43–49. doi: 10.2147/BCTT.S50361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russnes HG, et al. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am. J. Pathol. 2017;187(10):2152–2162. doi: 10.1016/j.ajpath.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Tsang JYS, Tse GM. Molecular classification of breast cancer. Adv. Anat. Pathol. 2020;27(1):27–35. doi: 10.1097/PAP.0000000000000232. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5(2):77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbeck N, et al. Breast cancer. Nat. Rev. Dis. Primers. 2019;5(1):66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 9.Calaf GM, et al. Molecular aspects of breast cancer resistance to drugs (Review) Int. J. Oncol. 2015;47(2):437–445. doi: 10.3892/ijo.2015.3055. [DOI] [PubMed] [Google Scholar]
- 10.Canevari RA, et al. Identification of novel biomarkers associated with poor patient outcomes in invasive breast carcinoma. Tumour Biol. 2016;37(10):13855–13870. doi: 10.1007/s13277-016-5133-8. [DOI] [PubMed] [Google Scholar]
- 11.Law RH, et al. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endsley MP, et al. Maspin, the molecular bridge between the plasminogen activator system and beta1 integrin that facilitates cell adhesion. J. Biol. Chem. 2011;286(28):24599–24607. doi: 10.1074/jbc.M111.235788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenstine TM, et al. Maspin: Molecular mechanisms and therapeutic implications. Cancer Metastasis Rev. 2012;31(3–4):529–551. doi: 10.1007/s10555-012-9361-0. [DOI] [PubMed] [Google Scholar]
- 14.Maass N, et al. Maspin—A novel protease inhibitor with tumor-suppressing activity in breast cancer. Acta Oncol. 2000;39(8):931–934. doi: 10.1080/02841860050215909. [DOI] [PubMed] [Google Scholar]
- 15.Wang MC, et al. Maspin expression and its clinicopathological significance in tumorigenesis and progression of gastric cancer. World J. Gastroenterol. 2004;10(5):634–637. doi: 10.3748/wjg.v10.i5.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maass N, et al. Down regulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin. Biochem. 2001;34(4):303–307. doi: 10.1016/S0009-9120(01)00220-X. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa M, et al. Maspin expression and its clinical significance in non-small cell lung cancer. Ann. Surg. Oncol. 2006;13(11):1517–1523. doi: 10.1245/s10434-006-9030-z. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Expression and localization of maspin in cervical cancer and its role in tumor progression and lymphangiogenesis. Arch. Gynecol. Obstet. 2014;289(2):373–382. doi: 10.1007/s00404-013-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng S, et al. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc. Natl. Acad. Sci. USA. 1996;93(21):11669–11674. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cella N, et al. Maspin is physically associated with [beta]1 integrin regulating cell adhesion in mammary epithelial cells. FASEB J. 2006;20(9):1510–1512. doi: 10.1096/fj.05-5500fje. [DOI] [PubMed] [Google Scholar]
- 21.Goulet B, et al. Nuclear localization of maspin is essential for its inhibition of tumor growth and metastasis. Lab. Investig. 2011;91(8):1181–1187. doi: 10.1038/labinvest.2011.66. [DOI] [PubMed] [Google Scholar]
- 22.Goulet B, et al. An emerging role for the nuclear localization of maspin in the suppression of tumor progression and metastasis. Biochem. Cell Biol. 2012;90(1):22–38. doi: 10.1139/o11-053. [DOI] [PubMed] [Google Scholar]
- 23.Reina J, et al. Identification of a putative nuclear localization signal in the tumor suppressor maspin sheds light on its nuclear import regulation. FEBS Open Bio. 2019;9(7):1174–1183. doi: 10.1002/2211-5463.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vecchi M, et al. Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene. 2008;27(15):2148–2158. doi: 10.1038/sj.onc.1210858. [DOI] [PubMed] [Google Scholar]
- 25.Cerqueira OLD, et al. Prognostic value of integrin αV expression and localization pattern in invasive breast carcinomas. Neoplasia. 2022;30:100803. doi: 10.1016/j.neo.2022.100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes LR, et al. RECK is not an independent prognostic marker for breast cancer. BMC Cancer. 2015;15:660. doi: 10.1186/s12885-015-1666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerqueira OL, et al. CIP4 promotes metastasis in triple-negative breast cancer and is associated with poor patient prognosis. Oncotarget. 2015;6(11):9397–9408. doi: 10.18632/oncotarget.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobba ARM, et al. High CD90 (THY-1) expression positively correlates with cell transformation and worse prognosis in basal-like breast cancer tumors. PLoS ONE. 2018;13(6):e0199254. doi: 10.1371/journal.pone.0199254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norton AJ, Jordan S, Yeomans P. Brief, high-temperature heat denaturation (pressure cooking): A simple and effective method of antigen retrieval for routinely processed tissues. J. Pathol. 1994;173(4):371–379. doi: 10.1002/path.1711730413. [DOI] [PubMed] [Google Scholar]
- 30.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hein AL, et al. QuPath digital immunohistochemical analysis of placental tissue. J. Pathol. Inform. 2021;12:40. doi: 10.4103/jpi.jpi_11_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): An abridged explanation and elaboration. J. Natl. Cancer Inst. 2018;110(8):803–811. doi: 10.1093/jnci/djy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker GM, et al. Immunohistochemistry scoring of breast tumor tissue microarrays: A comparison study across three software applications. J. Pathol. Inform. 2022;13:100118. doi: 10.1016/j.jpi.2022.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts MR, et al. Reliability of a computational platform as a surrogate for manually interpreted immunohistochemical markers in breast tumor tissue microarrays. Cancer Epidemiol. 2021;74:101999. doi: 10.1016/j.canep.2021.101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandrashekar DS, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman MJ, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho FM, et al. Geographic differences in the distribution of molecular subtypes of breast cancer in Brazil. BMC Womens Health. 2014;14:102. doi: 10.1186/1472-6874-14-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mordente A, et al. Cancer biomarkers discovery and validation: State of the art, problems and future perspectives. Adv. Exp. Med. Biol. 2015;867:9–26. doi: 10.1007/978-94-017-7215-0_2. [DOI] [PubMed] [Google Scholar]
- 41.Desmedt C, et al. Uncovering the genomic heterogeneity of multifocal breast cancer. J. Pathol. 2015;236(4):457–466. doi: 10.1002/path.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natrajan RC. Breast cancer heterogeneity: parallel evolution or conscious uncoupling? J. Pathol. 2015;237(1):1–3. doi: 10.1002/path.4557. [DOI] [PubMed] [Google Scholar]
- 43.Bailey CM, et al. Biological functions of maspin. J. Cell Physiol. 2006;209(3):617–624. doi: 10.1002/jcp.20782. [DOI] [PubMed] [Google Scholar]
- 44.Khalkhali-Ellis Z. Maspin: the new frontier. Clin. Cancer Res. 2006;12(24):7279–7283. doi: 10.1158/1078-0432.CCR-06-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou Z, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263(5146):526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 46.Pemberton PA, et al. Maspin is an intracellular serpin that partitions into secretory vesicles and is present at the cell surface. J. Histochem. Cytochem. 1997;45(12):1697–1706. doi: 10.1177/002215549704501213. [DOI] [PubMed] [Google Scholar]
- 47.Lockett J, et al. Tumor suppressive maspin and epithelial homeostasis. J. Cell Biochem. 2006;97(4):651–660. doi: 10.1002/jcb.20721. [DOI] [PubMed] [Google Scholar]
- 48.Lele SM, Graves K, Gatalica Z. Immunohistochemical detection of maspin is a useful adjunct in distinguishing radial sclerosing lesion from tubular carcinoma of the breast. Appl. Immunohistochem. Mol. Morphol. 2000;8(1):32–36. doi: 10.1097/00129039-200003000-00005. [DOI] [PubMed] [Google Scholar]
- 49.Shao ZM, et al. The human myoepithelial cell exerts antiproliferative effects on breast carcinoma cells characterized by p21WAF1/CIP1 induction, G2/M arrest, and apoptosis. Exp. Cell Res. 1998;241(2):394–403. doi: 10.1006/excr.1998.4066. [DOI] [PubMed] [Google Scholar]
- 50.Pierson CR, et al. Maspin is up-regulated in premalignant prostate epithelia. Prostate. 2002;53(4):255–262. doi: 10.1002/pros.10107. [DOI] [PubMed] [Google Scholar]
- 51.Chua R, et al. Maspin expression, angiogenesis, prognostic parameters, and outcome in malignant melanoma. J. Am. Acad. Dermatol. 2009;60(5):758–766. doi: 10.1016/j.jaad.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 52.Maass N, et al. Expression of the tumor suppressor gene Maspin in human pancreatic cancers. Clin. Cancer Res. 2001;7(4):812–817. [PubMed] [Google Scholar]
- 53.Kim DH, et al. Association of maspin expression with the high histological grade and lymphocyte-rich stroma in early-stage breast cancer. Histopathology. 2003;42(1):37–42. doi: 10.1046/j.1365-2559.2003.01567.x. [DOI] [PubMed] [Google Scholar]
- 54.Brockmann R, et al. Posttranscriptional expression regulation: what determines translation rates? PLoS Comput. Biol. 2007;3(3):e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mata J, Marguerat S, Bähler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 2005;30(9):506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 56.Sood AK, et al. The paradoxical expression of maspin in ovarian carcinoma. Clin. Cancer Res. 2002;8(9):2924–2932. [PubMed] [Google Scholar]
- 57.Sopel M, Surowiak P, Berdowska I. Nuclear maspin expression as a good prognostic factor in human epithelial ovarian carcinoma. Folia Morphol. 2010;69(4):204–212. [PubMed] [Google Scholar]
- 58.Woenckhaus M, et al. Nuclear and cytoplasmic Maspin expression in primary non-small cell lung cancer. J. Clin. Pathol. 2007;60(5):483–486. doi: 10.1136/jcp.2005.033407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katz AB, Taichman LB. A partial catalog of proteins secreted by epidermal keratinocytes in culture. J. Investig Dermatol. 1999;112(5):818–821. doi: 10.1046/j.1523-1747.1999.00572.x. [DOI] [PubMed] [Google Scholar]
- 60.Frey A, et al. Nuclear, compared with combined nuclear and cytoplasmic expression of maspin, is linked in lung adenocarcinoma to reduced VEGF-A levels and in Stage I, improved survival. Histopathology. 2009;54(5):590–597. doi: 10.1111/j.1365-2559.2009.03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lonardo F, et al. Maspin nuclear localization is linked to favorable morphological features in pulmonary adenocarcinoma. Lung Cancer. 2006;51(1):31–39. doi: 10.1016/j.lungcan.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 62.Banias L, et al. Nuclear maspin expression: A biomarker for budding assessment in colorectal cancer specimens. Pathol. Res. Pract. 2017;213(9):1227–1230. doi: 10.1016/j.prp.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 63.Ohno T, et al. Clinical significance of subcellular localization of Maspin in patients with pathological stage IA lung adenocarcinoma. Anticancer Res. 2018;38(5):3001–3007. doi: 10.21873/anticanres.12553. [DOI] [PubMed] [Google Scholar]
- 64.Takagi Y, et al. Cytoplasmic maspin expression is a predictor of poor prognosis in patients with lung adenocarcinoma measuring <3 cm. Histopathology. 2015;66(5):732–739. doi: 10.1111/his.12586. [DOI] [PubMed] [Google Scholar]
- 65.Märkl B, et al. Shift from cytoplasmic to nuclear maspin expression correlates with shorter overall survival in node-negative colorectal cancer. Hum. Pathol. 2010;41(7):1024–1033. doi: 10.1016/j.humpath.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 66.Berardi R, et al. Role of maspin in cancer. Clin. Transl. Med. 2013;2(1):8. doi: 10.1186/2001-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurzu S, Jung I. Subcellular expression of Maspin in colorectal cancer: Friend or foe. Cancers. 2021;13(3):336. doi: 10.3390/cancers13030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alluri P, Newman LA. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014;23(3):567–577. doi: 10.1016/j.soc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheng S. The promise and challenge toward the clinical application of maspin in cancer. Front. Biosci. 2004;9:2733–2745. doi: 10.2741/1432. [DOI] [PubMed] [Google Scholar]
- 71.Wong H, et al. Integrating molecular mechanisms and clinical evidence in the management of trastuzumab resistant or refractory HER-2+ metastatic breast cancer. Oncologist. 2011;16(11):1535–1546. doi: 10.1634/theoncologist.2011-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev. Anticancer Ther. 2011;11(2):263–275. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barchiesi G, et al. Neoadjuvant endocrine therapy in breast cancer: Current knowledge and future perspectives. Int. J. Mol. Sci. 2020;21(10):3528. doi: 10.3390/ijms21103528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bauer KR, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 75.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 76.Mao Y, et al. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32(1–2):303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 78.Skaik Y, et al. Secreted β3-integrin enhances natural killer cell activity against acute myeloid leukemia cells. PLoS ONE. 2014;9(2):e98936. doi: 10.1371/journal.pone.0098936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen D, Gyllensten U. MICA polymorphism: biology and importance in cancer. Carcinogenesis. 2014;35(12):2633–2642. doi: 10.1093/carcin/bgu215. [DOI] [PubMed] [Google Scholar]
- 80.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019;18(2):99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 81.Ros XR, Vermeulen L. Turning cold tumors hot by blocking TGF-β. Trends Cancer. 2018;4(5):335–337. doi: 10.1016/j.trecan.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 82.Seo N, et al. Interleukin-10 expressed at early tumour sites induces subsequent generation of CD4(+) T-regulatory cells and systemic collapse of antitumour immunity. Immunology. 2001;103(4):449–457. doi: 10.1046/j.1365-2567.2001.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sazeides C, Le A. Metabolic relationship between cancer-associated fibroblasts and cancer cells. Adv. Exp. Med. Biol. 2018;1063:149–165. doi: 10.1007/978-3-319-77736-8_11. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, et al. The influence of microenvironment on tumor immunotherapy. FEBS J. 2019;286(21):4160–4175. doi: 10.1111/febs.15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zitvogel L, et al. Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity. 2013;39(1):74–88. doi: 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 86.Sakabe T, et al. Role of cytoplasmic localization of maspin in promoting cell invasion in breast cancer with aggressive phenotype. Sci. Rep. 2021;11(1):11321. doi: 10.1038/s41598-021-90887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Longhi MT, et al. PI3K-AKT, JAK2-STAT3 pathways and cell-cell contact regulate maspin subcellular localization. Cell Commun. Signal. 2021;19(1):86. doi: 10.1186/s12964-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reis-Filho JS, et al. Maspin expression in myoepithelial tumors of the breast. Pathol. Res. Pract. 2001;197(12):817–821. doi: 10.1078/0344-0338-00165. [DOI] [PubMed] [Google Scholar]
- 89.Matsushige T, Sakabe T, Umekita Y. Investigation of the subcellular localization-dependent anti- or pro-tumor functions of Maspin in human lung adenocarcinoma cell line. Yonago Acta Med. 2022;65(1):44–52. doi: 10.33160/yam.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uchinaka EI, et al. Cytoplasmic-only expression of Maspin predicts unfavorable prognosis in patients with pancreatic ductal adenocarcinoma. Anticancer Res. 2021;41(5):2543–2552. doi: 10.21873/anticanres.15032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Please contact corresponding author (Otto Cerqueira: ottoluix@alumni.usp.br) for data requests.