Abstract
Normal herpesvirus assembly and egress depend on the correct intracellular localization of viral glycoproteins. While several post-Golgi transport motifs have been characterized within the cytoplasmic domains of various viral glycoproteins, few specific endoplasmic reticulum (ER)-to-Golgi transport signals have been described. We report the identification of two regions within the 125-amino-acid cytoplasmic domain of Varicella-Zoster virus gB that are required for its ER-to-Golgi transport. Native gB or gB containing deletions and specific point mutations in its cytoplasmic domain was expressed in mammalian cells. ER-to-Golgi transport of gB was assessed by indirect immunofluorescence and by the acquisition of Golgi-dependent posttranslational modifications. These studies revealed that the ER-to-Golgi transport of gB requires a nine-amino-acid region (YMTLVSAAE) within its cytoplasmic domain. Mutations of individual amino acids within this region markedly impaired the transport of gB from the ER to the Golgi, indicating that this domain functions by a sequence-dependent mechanism. Deletion of the C-terminal 17 amino acids of the gB cytoplasmic domain was also shown to impair the transport of gB from the ER to the Golgi. However, internal mutations within this region did not disrupt the transport of gB, indicating that its function during gB transport is not sequence dependent. Native gB is also transported to the nuclear membrane of transfected cells. gB lacking as many as 67 amino acids from the C terminus of its cytoplasmic domain continued to be transported to the nuclear membrane at apparently normal levels, indicating that the cytoplasmic domain of gB is not required for nuclear membrane localization.
Varicella-zoster virus (VZV), one of eight herpesviruses known to infect humans, is classified along with herpes simplex virus types 1 and 2 (HSV-1 and 2) as an alphaherpesvirus based on its growth characteristics and ability to become latent in the nervous system of the host (28). Unlike the other alphaherpesviruses, however, VZV induces syncytia and is highly cell associated when grown in cultured cells, indicating that its cellular egress pathway differs from that of the other alphaherpesviruses.
Herpesvirus virions acquire their initial envelope upon budding through the inner nuclear membrane into the perinuclear space (8, 11, 22, 30). Following envelopment at the inner nuclear membrane, the route taken by virions as they transit through and ultimately exit the cell is less certain and may vary between different herpesviruses (5, 20, 34). Regardless of the site at which the final envelope is acquired, however, the importance of proper glycoprotein transport and processing during egress has been demonstrated experimentally. When cells defective for the intracellular transport or glycosylation of viral glycoproteins are infected with HSV-1, human cytomegalovirus (HCMV), or pseudorabies virus, the resultant virions fail to egress normally and instead accumulate in cytoplasmic vacuoles (2, 12, 35).
All known herpesviruses encode a glycoprotein B (gB) homolog. VZV gB contains 868 amino acids (aa) and is a type I membrane protein consisting of a large ectodomain, a hydrophobic transmembrane region, and a cytoplasmic domain. The cytoplasmic domain of VZV gB is predicted to contain 125 aa, making it by far the longest cytoplasmic domain of any VZV membrane proteins (6). In the alphaherpesviruses, gB is present in the virion envelope, where it is thought to play a role in fusion of the virion and plasma membranes following viral attachment (3, 26, 31). gB also plays an important role in viral egress, as evidenced by the existence of numerous HSV-1 gB mutants that promote syncytium formation in cultured cells (1, 26). Many of the syncytium-generating mutations are in the cytoplasmic domain of HSV-1 gB, suggesting that this domain may be particularly important for viral egress. In addition, Epstein-Barr virus (EBV) gB, while unlikely to be essential for viral entry because little, if any, is present in the virion envelope (10), is nonetheless required for the production of infectious virions, suggesting a function primarily during viral egress (16, 17).
The cytoplasmic domains of many membrane proteins, including VZV gE and gI, contain specific signal sequences that mediate their post-Golgi intracellular transport (14, 16, 24, 25, 32, 37). In addition, the efficient endoplasmic reticulum (ER)-to-Golgi transport of HSV-1 gB requires an intact cytoplasmic domain. HSV-1 containing large C-terminal deletions in its cytoplasmic domain (66 to 109 aa) was transported to the Golgi more slowly than native gB (27), and linker insertion mutations at various places in the cytoplasmic domain of HSV-1 gB also slowed ER-to-Golgi transport (3). While several specific sequence motifs that mediate the post-Golgi transport of membrane proteins have been identified (14, 32), ER-to-Golgi transport signal sequences are less well characterized. Recently, a signal sequence that mediates the transport of vesicular stomatitis virus G protein (VSV-G) from the ER to the Golgi was identified within the cytoplasmic domain of this type I transmembrane protein (23). This sequence, YTDIE, contains a YXXφ motif and two acidic amino acids (in the pattern DXE), which are essential for its function. Several other transmembrane proteins have been identified that contain YXXφ motifs followed at variable distances (2 to 6 aa) by acidic amino acids in the pattern (D/E)X(D/E). However, it is not known whether these motifs function during protein transport.
The cytoplasmic domain of VZV gB contains two YXXφ motifs, one (underlined) followed by a single acidic residue (YSRVSRE, aa 857 to 863) and one (underlined) followed by two acidic residues in the pattern EXXE (YMTLVSAAERQE, aa 818 to 829). To determine whether these or other regions of the VZV gB cytoplasmic domain mediate the ER-to-Golgi transport of gB, we introduced deletions and specific point mutations into the cytoplasmic domain of gB, expressed the mutated proteins in mammalian cells, and assessed their transport to the Golgi. The C-terminal 17 aa of VZV gB (residues 852 to 868) and the 9-aa region between residues 818 and 826 were found to be critical for the transport of VZV gB from the ER to the Golgi.
MATERIALS AND METHODS
Cell culture and virus propagation.
MeWo, an immortalized human melanoma cell line, and HEp-2, a human epithelial cell line, were grown in Eagle's minimum essential medium (EMEM) containing 10% fetal bovine serum (FBS; Bio-Whittaker) and 2 mM l-glutamine (Quality Biological). Recombinant vaccinia virus vTF7-3 (21) was obtained from the American Type Culture Collection, and viral stocks were prepared and titered in BSC-40 African green monkey kidney cells that were also grown in EMEM containing 10% FBS and l-glutamine.
Immune reagents and intracellular markers.
Anti-VZV gB monoclonal antibodies (MAbs) were purchased from Biodesign International (catalog no. C05102M). Tetramethylrhodamine isothiocyanate (TRITC)-conjugated wheat germ agglutinin (WGA) was purchased from EY Laboratories. Rabbit anticalnexin polyclonal antibodies were purchased from StressGen. Goat anti-mouse immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (FITC) and TRITC-conjugated goat anti-rabbit IgG were purchased from Sigma.
Plasmid construction and site-directed mutagenesis of VZV gB.
An 8,131-bp PmeI/SpeI fragment of VZV strain Oka (consensus nucleotides 53875 to 62007 [12]) containing the VZV gB coding sequence (nucleotides 57008 to 59611) was cloned into pNEB193 (New England Biolabs) in which the SmaI site had been replaced with a SpeI linker. The 8,148-bp fragment resulting from HindIII/SpeI digestion of this plasmid (8,131-bp VZV DNA fragment and 17 bp from the pNEB polylinker) was cloned into pBluescript SK+ (Stratagene) at the corresponding restriction sites such that the gB coding sequence was downstream of the T7 promoter to yield pBS-8131. A 2,329-bp DNA fragment between the T7 promoter and the gB start codon was eliminated by digesting pBS-8131 with StuI and HindIII followed by recircularization to yield pBS-5802.
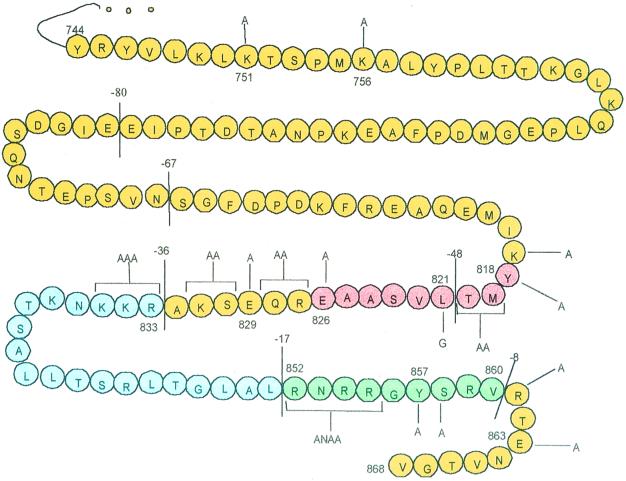
All VZV gB mutations used in this study were derived from pBS-5802 by site-directed mutagenesis using the Bio-Rad Muta-Gene phagemid in vitro mutagenesis kit, which employs the Kunkel method (15). The uracil-containing single-stranded DNA was isolated from Escherichia coli CJ236 after superinfection of M13KO7 helper phage (Promega). To generate truncated forms of VZV gB, oligonucleotides were designed to substitute a stop codon for a native codon at the desired site. Similarly, substitution mutations were introduced by substituting alanine or, in one case, glycine codons for specific native codons. To generate gB containing internal deletion mutations, the oligonucleotides were designed to eliminate the unwanted codons while maintaining the remainder of the coding sequence in frame. The mutagenic oligonucleotides used in this study were purchased from Genosys and are listed in Table 1. Figure 1 graphically depicts the entire cytoplasmic domain of VZV gB, showing the location of each mutation generated for this study. Each mutation was confirmed by sequencing the relevant region of the VZV gB gene by the dideoxy-chain termination method (29).
TABLE 1.
Oligonucleotides used to generate VZV gB mutations
| Mutationa | Comment | Oligonucleotidesb |
|---|---|---|
| Truncations | ||
| R861 to stop | 8-aa truncation | TCC CGT GTT tga ACC GAG AAT |
| R817 to stop | 17-aa truncation | CTT GCT TTA tga AAT CGC CGA |
| R833 to stop | 36-aa truncation | TCT AAA GCC tga AAA AAA AAT |
| L821 to stop | 48-aa truncation | TAT ATG ACG taa GTA TCT GCG |
| S802 to stop | 67-aa truncation | TCG GTA AAT tga GGG TTT GAT |
| Y745 to stop | 125-aa truncation | TTT TTT GCG tag CGG TAC GTG |
| Deletions | ||
| Δ818 to 826 | 9-aa deletion | G GAA ATG ATT AAA / CGC CAA GAA TCT |
| Δ833 to 851 | 17-aa deletion | CAA GAA TCT AAA GCC / CGA AAT CGC CGA GGA |
| Δ852 to 860 | 9-aa deletion | ACC GGC CTT GCT TTA / CGC ACC GAG AAT GTA |
| Point mutations | ||
| K817 to A | aa 817 | GAA ATG ATT gca TAT ATG AGG |
| Y818 to A | aa 818 | ATG ATT AAA gct ATG ACG TTA |
| MT to AA | aa 819–820 | ATT AAA TAT gcg gcg TTA GTA T |
| L821 to G | aa 821 | TAT ATG ACG gga GTA TCT GC |
| E826 to A | aa 826 | GCG GCT gcg CGC CAA |
| RQ to AA | aa 827–828 | GCG GCT GAG gcc gca GAA TCT AAA |
| E829 to A | aa 829 | GAG CGC CAA gca TCT AAA GCC |
| SK to AA | aa 830–831 | CGC CAA GAA gct gca GCC CGC AAA |
| RKK to AAA | aa 833–835 | GAA TCT AAA GCC gcc gca gca AAT AAG ACT AGC |
| RNRR to ANAA | aa 852–854 | GCC CTT GCT TTA gca ATT gcc gca GGA TAC TCC CGT |
| Y857 to A | aa 857 | C CGA GGA gcc TCC CGT G |
| S858 to A | aa 858 | CGA GGA TAC gcc CGT GTT CGC |
| R861 to A | aa 861 | TCC CGT GTT gcc ACC GAG AAT |
| E863 to A | aa 863 | GCG ACC gcg AAT GTA |
Single-letter amino acid abbreviations are used. Numbers represent amino acid positions within VZV gB (868 aa in total).
Lowercase letters represent altered codons, and slash marks represent sites at which native codons have been omitted.
FIG. 1.
VZV gB cytoplasmic domain mutations. The 125-aa residues (from aa 744 to 868) that make up the cytoplasmic domain of VZV gB are shown. The lines bisecting the cytoplasmic domain are located immediately after the terminal amino acids of the truncation mutants used in this study. These mutants are named according to the number of deleted C-terminal amino acids. Substitution mutations are indicated by lines and brackets, and the amino acids replacing the native residue(s) are noted. The regions in red (aa 818 to 826), blue (aa 833 to 851), and green (aa 852 to 860) denote separate internal deletion mutations.
In vitro expression of VZV gB.
VZV gB was expressed in vitro by the method of Fuerst et al. (7), modified as follows. DNA for transfection was column purified (Qiagen) and transfected using Lipofectin (Gibco-BRL). Cells at 75 to 90% confluence were infected with recombinant vaccinia virus vTF7-3 at a multiplicity of infection of 10 and then transfected with 7 μg of DNA and 12 μl of Lipofectin in 10-cm2 wells or 3.5 μl of DNA and 6 μl of Lipofectin in 1.2-cm2 wells. Cells were incubated at 37°C and 5% CO2 for 16 h prior to metabolic labeling or fluorescent staining.
Radiolabeling and immunoprecipitation of proteins.
All metabolic labeling was performed at 37°C in 5% CO2. For steady-state labeling of in vitro-expressed gB, transfected cells were incubated for 1 h in EMEM lacking cysteine and methionine (Cys−, Met− EMEM) (starvation period) and then incubated in Cys−, Met− EMEM containing 125 μCi of Tran35S-label (ICN) per ml for 4 h. To pulse-label in vitro-expressed gB, transfected cells were incubated for 1 h in Cys−, Met− EMEM containing 125 μCi of Tran35S-label without prior starvation. Following the 1-h labeling period, cells were washed and incubated for 4 h in chase medium (EMEM containing 10% FBS, 24 μg of cysteine per ml, and 15 μg of methionine per ml). Labeled cells were washed extensively in phosphate-buffered saline (PBS) at 4°C and lysed in PBS containing 1% Triton X-100, 0.5% deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). VZV gB was immunoprecipitated by incubating the cell lysates with anti-VZV gB MAbs overnight at 4°C followed by incubation with Staphylococcus protein G (Pharmacia Biotech) for 1 h at 4°C. After washing, precipitated proteins were eluted in sample buffer containing 2% SDS (Bio-Rad) and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on 8% gels. For reducing gels, 40 mM dithiothreitol was added to the sample buffer prior to elution. The gels were dried, and the labeled immunoprecipitated proteins were visualized by autoradiography.
Carbohydrate analysis.
Radiolabeled proteins were immunoprecipitated as described above and then heated at 98°C for 3 min in 10 μl of 0.2% SDS–50 mM Tris-HCl (pH 6.8). After cooling to room temperature, 10 μl of 0.15 M sodium citrate (pH 5.3) and 1 μl (0.005 U) of endoglycosidase H (endo H; Boehringer Mannheim) were added. The reaction mixtures were incubated overnight at 37°C. Sample buffer was added to the endo H-treated proteins, and they were resolved by SDS-PAGE on 8% gels.
Quantitation of immunoprecipitated proteins.
Following autoradiography of immunoprecipitated gB, the proportion of endo H-resistant gB (high-molecular-weight [high-MW] form) relative to endo H-sensitive gB (low-MW form) was quantitated for each sample by measuring the intensities of the corresponding bands using a Molecular Dynamics densitometer. All reported values represent the averages of at least four independent experiments.
Immunofluorescence and confocal microscopy.
VZV gB expressed in transfected cells was detected by indirect immunofluorescence assays using anti-VZV gB MAbs. At 16 h after transfection and infection with vTF7-3, cells on glass coverslips were fixed for 1 h at 4°C in 2% paraformaldehyde. To permeabilize cellular membranes in experiments designed to visualize intracellular structures, 0.1% Triton X-100 was added to the fixative. Fixed cells were blocked by incubation with PBS containing 1% goat serum (Sigma) for 1 h at room temperature, then incubated overnight at 4°C with the primary antibodies (mouse anti-VZV gB MAbs diluted 1:500 in PBS and/or rabbit anticalnexin polyclonal antibody diluted 1:200 in PBS), washed in PBS, and incubated for 1 h at room temperature with the appropriate secondary antibodies (goat anti-mouse IgG FITC diluted 1:1,000 in PBS and/or goat anti-rabbit IgG-TRITC diluted 1:100 in PBS). In colocalization experiments requiring Golgi visualization, WGA-TRITC diluted to 5 μg/ml in PBS was incubated with the fixed cells for 20 min concurrent with the secondary antibody incubation. After washing in PBS, stained cells were examined for immunofluorescence with a Bio-Rad MRC1024 scanning confocal microscope.
RESULTS
VZV gB at steady state concentrates in the Golgi, the nuclear membrane, and the plasma membrane of both infected and transfected cells.
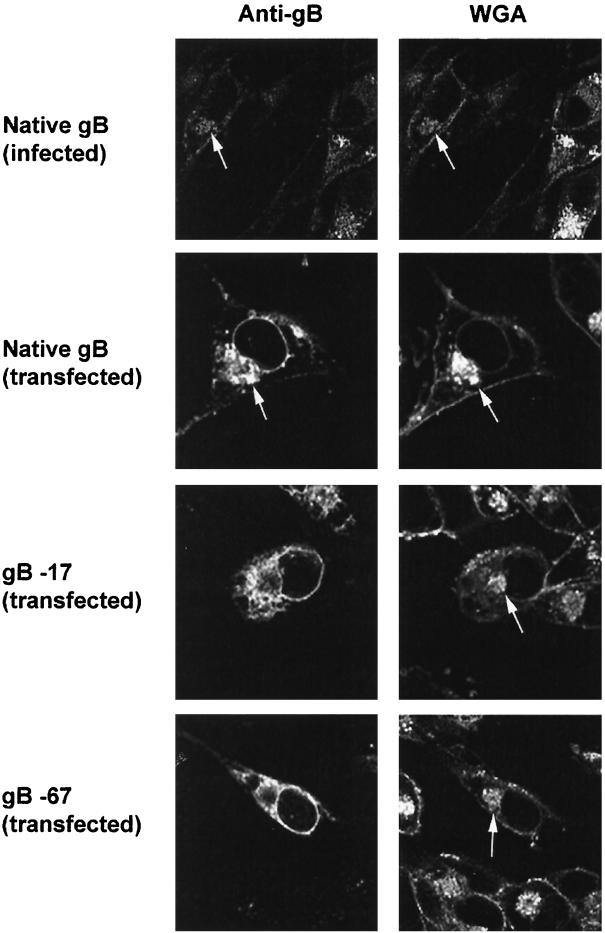
We studied the intracellular localization of VZV gB in mammalian cells by indirect immunofluorescence. VZV gB expressed following either infection or transfection of MeWo and HEp-2 cells was detected using anti-VZV gB MAbs and then visualized with FITC-conjugated secondary antibodies. The Golgi apparatus was identified in the same cells by costaining with TRITC-conjugated WGA, a lectin that binds to complex oligosaccharides that are present in high abundance in the Golgi (4, 8). The nuclear and plasma membranes were also stained by TRITC-WGA. The cells were examined by laser scanning confocal microscopy. Figure 2 (top row) shows that native gB expressed early following VZV infection (at 24 h after inoculation) concentrated in the Golgi based on its colocalization with WGA. It also appeared in the nuclear membrane although some of this staining may represent gB in the ER, as these structures are contiguous. We could not determine from these studies whether gB was present in only the inner or outer nuclear membrane or in both. Later in VZV infection, the internal structures of infected cells became disrupted, making the identification of specific organelles difficult (data not shown). Native gB transiently expressed in cultured cells following transfection localized to the Golgi, the nuclear membrane, and the plasma membrane similarly to gB expressed during infection with VZV (Fig. 2, second row). This demonstrates that VZV gB localizes to the Golgi as well as to the nuclear and plasma membranes independently of interactions with other viral proteins.
FIG. 2.
Microscopic localization of VZV gB. Native gB or gB containing cytoplasmic domain C-terminal truncations was expressed in mammalian cells by transfection. Cells were also infected with native VZV as positive control. MeWo cells are shown; similar results were obtained with HEp-2 cells. Transfected or infected cells were incubated with both anti-VZV gB MAbs and WGA (a lectin that concentrates in the Golgi). The intracellular localization of gB and WGA was analyzed by laser-scanning confocal microscopy. Arrows mark the location of the Golgi in selected cells.
The cytoplasmic tail of VZV gB is required for Golgi localization.
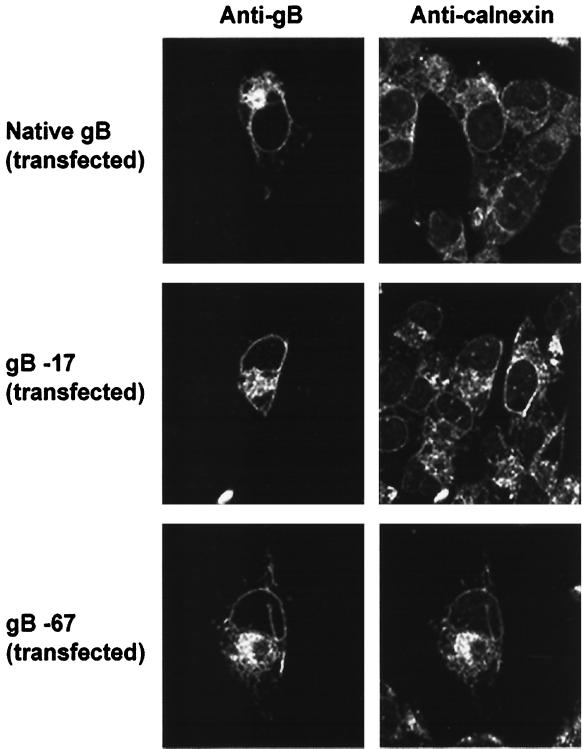
To determine whether the cytoplasmic domain of gB is required for its Golgi localization, we expressed gB containing mutations in its cytoplasmic domain in both MeWo and HEp-2 cells as described above. VZV gB was detected in these cells using anti-VZV gB MAbs and visualized by confocal microscopy. TRITC-conjugated WGA was again used as a colocalization marker for the Golgi. VZV gB lacking 17 or 67 aa from its C terminus (Fig. 1) failed to accumulate within the Golgi (Fig. 2). However, expression of gB in the nuclear envelope was seemingly unaffected by these C-terminal truncations (Fig. 2), and even removal of the entire 125-aa cytoplasmic domain of gB had no apparent effect on its nuclear membrane expression (data not shown). Several other gB cytoplasmic domain amino acid substitutions eliminating single or multiple residues did not alter the Golgi or nuclear envelope localization of gB (data not shown). These included mutations R751A, R756A, RKK-AAA (aa 833 to 835), and RNRR-ANAA (aa 851 to 855) (Fig. 1). To determine the intracellular location of the truncated forms of gB that failed to accumulate in the Golgi, cultured cells expressing gB lacking its C-terminal 17 or 67 aa were costained with anticalnexin antibodies as a marker for the ER. VZV gB containing both the 17- and 67-aa C-terminal truncations colocalized with calnexin, indicating that the mutated forms of gB are concentrated in the ER (Fig. 3). It should be noted that while the C-terminal truncations shown in Fig. 2 greatly diminish the transport of gB to the Golgi, the blockage is not absolute; therefore, some gB lacking the C-terminal 17 or 67 aa may be present in post-Golgi compartments as well. Together, these data indicate that the cytoplasmic domain of gB is required for the efficient transport of gB from the ER to the Golgi.
FIG. 3.
Microscopic localization of VZV gB. Native gB or gB containing cytoplasmic domain C-terminal truncations was expressed in mammalian cells by transfection. Transfected cells were coincubated with anti-VZV gB MAbs and anticalnexin antibodies (a marker of the ER). The intracellular localization of gB and calnexin was analyzed by laser-scanning confocal microscopy.
The C-terminal 17 aa of the VZV gB cytoplasmic domain are required for transport of gB from the ER to the Golgi.
To define specific domains within the cytoplasmic domain of gB that mediate its ER-to-Golgi transport, we expressed gB or gB containing mutations in its cytoplasmic domain in cultured cells and evaluated their transport to the Golgi by assaying for the acquisition of two distinct Golgi-dependent posttranslational modifications: (i) proteolytic cleavage and (ii) conversion of N-linked oligosaccharides from high-mannose to complex forms.
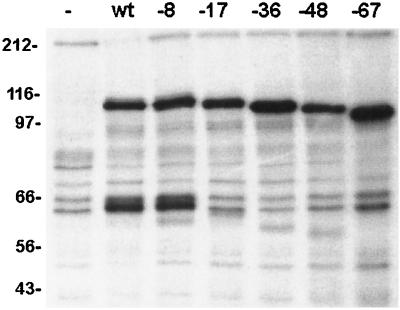
Mature VZV gB is largely, but not completely, posttranslationally cleaved into two disulfide-linked subunits of approximately equal sizes (13, 19); thus, under reducing conditions, gB migrates as an approximately 110-kDa protein representing the uncleaved form of gB and as a doublet of 60 to 68 kDa representing the two proteolytic fragments. As proteolytic cleavage of gB occurs in the Golgi, failure of gB to transit from the ER to the Golgi results in the absence of the cleaved forms of gB. Native gB or gB containing C-terminal truncations within its cytoplasmic domain (Fig. 1) was expressed in MeWo cells and, after metabolic labeling, immunoprecipitated with anti-VZV gB MAbs. As a negative control, mock-transfected MeWo cells were also labeled and incubated with anti-VZV gB MAbs. The immunoprecipitated forms of gB were resolved by SDS-PAGE under reducing conditions (Fig. 4). As expected, native gB migrated as a 110-kDa species and as a doublet of about 60 kDa, while the mock-transfected cells showed only nonspecific background. VZV gB lacking the C-terminal 8 aa from its cytoplasmic domain was cleaved similarly to native gB. However, gB containing larger (17-, 36-, 48-, and 67-aa) C-terminal truncations of its cytoplasmic domain exhibited greatly reduced levels of the cleaved forms of gB relative to the uncleaved species. This indicates that deletion of 17 or more aa from its C terminus impairs gB transport to the Golgi.
FIG. 4.
Proteolytic cleavage of VZV gB cytoplasmic domain truncation mutants. Native gB (wt [wild type]) or gB containing cytoplasmic domain C-terminal truncation mutations was expressed in mammalian cells, immunoprecipitated, and resolved by SDS-PAGE under reducing conditions. Truncation mutants are designated by the number of amino acids removed from the C terminus. Mock-transfected cells (−) were included as a negative control. Native gB migrates as a 110-kDa species and as a doublet of about 60 kDa (resulting from its proteolytic cleavage in the Golgi). Molecular masses are given in kilodaltons.
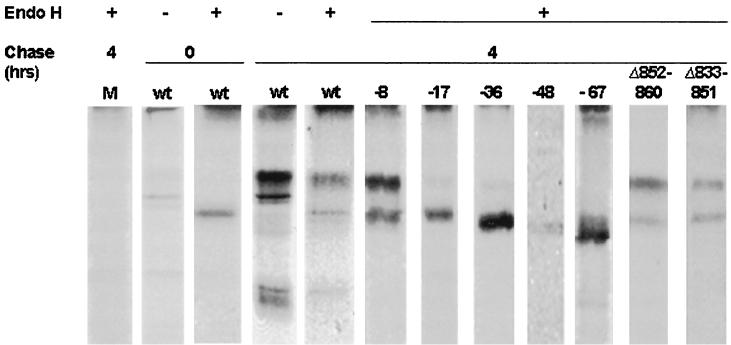
VZV gB has several N-linked oligosaccharides in its ectodomain that are processed in the Golgi from their high-mannose (endo H-sensitive) to complex (endo H-resistant) forms (19). Thus, acquisition of this posttranslational modification serves as a marker for the transport of gB from the ER to the Golgi. Figure 5 compares the endo H sensitivity of native gB to that of gB containing various cytoplasmic domain truncations and internal deletions. After a 1-h labeling period with no chase, native gB existed almost exclusively as a single endo H-sensitive form, indicating that it had not yet been processed in the Golgi (Fig. 5, second and third panels). Consistent with previous studies (19), most native gB transited to the Golgi during a 4-h chase interval, where its oligosaccharides were processed to their complex forms as evidenced by the appearance of a slower-migrating, endo H-resistant species (Fig. 5, fourth and fifth panels). A small proportion of native gB, however, remained in its endo H-sensitive form. Most gB truncated 8 aa from its C terminus acquired endo H resistance during the 4-h chase interval, indicating that gB containing this mutation is efficiently transported to the Golgi. In contrast, all of the gB cytoplasmic domain mutations resulting in truncations of 17 or more aa exhibited greatly reduced levels of the endo H-resistance at 4 h, indicating that these mutations impair gB transport to the Golgi. Note also that the larger truncations (of 48 and 67 aa) imparted a higher level of resistance to endo H than did the smaller truncations (of 17 and 36 aa). C-terminal truncations of 80 and 125 aa also severely impaired ER-to-Golgi transport of gB (data not shown). Mock-transfected MeWo cells (Fig. 5, lane M) were processed identically to a negative control.
FIG. 5.
ER-to-Golgi transport of VZV gB containing deletions in its cytoplasmic domain. Native gB (wt [wild type]) or gB containing cytoplasmic domain deletion mutations was expressed in mammalian cells, pulse-labeled, and then immunoprecipitated immediately (0-h chase) or after a 4-h chase. Treatment of immunoprecipitated gB with endo H is denoted by +. Mock-transfected cells (M) were processed identically as a negative control. The immunoprecipitated proteins were resolved by nonreducing SDS-PAGE on 8% gels.
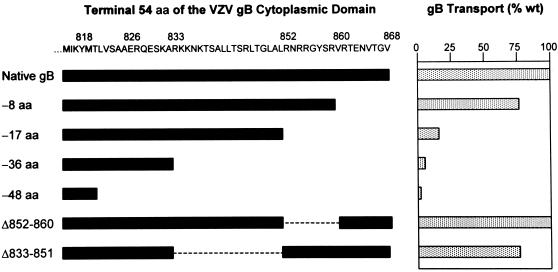
The degree to which specific cytoplasmic domain mutations impaired the transport of VZV gB to the Golgi was quantitated by dividing the amount of gB present as the higher-MW (endo H-resistant) species by the total amount of gB (the sum of the higher- and lower-MW species) as determined by scanning densitometry (Fig. 6). In every case, the quantitations represent the means of at least four independent experiments, and the efficiency of ER-to-Golgi transport is expressed as a percentage of native gB transport. VZV gB lacking the C-terminal 8 aa of its cytoplasmic domain was transported to the Golgi at 77% of the level of native gB, whereas truncation of the C-terminal 17 aa decreased its transport to 15% of that of native gB. VZV gB containing larger C-terminal truncations was transported to the Golgi at less than 5% of the normal level (data for the truncated forms of gB lacking 67, 80, and 100 aa are not shown). These data, in agreement with the proteolytic cleavage data (Fig. 4), indicate that the C-terminal 17 aa of the gB cytoplasmic domain are essential for the efficient transport of gB from the ER to the Golgi.
FIG. 6.
Quantitation of ER-to-Golgi transport of VZV gB cytoplasmic domain deletion mutants. The C-terminal 54 aa of the VZV gB cytoplasmic domain (aa 815 to 868) are listed. Native (wild-type [wt]) gB contains this entire region, whereas the segments of this region retained by the various deletion mutants are represented by the black bars. The proportion of each mutant transported to the Golgi (as a percentage of native gB transport) was quantitated and is depicted by the stippled bars.
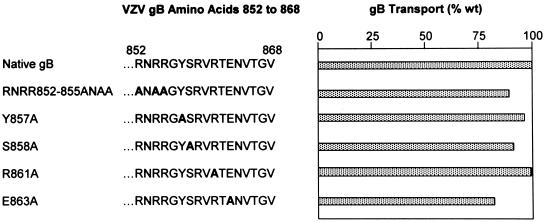
The C-terminal 17 aa of VZV gB includes two particularly notable features. First, it is highly basic, containing six arginine residues and a single acidic residue; second, it contains a consensus YXXφ motif (underlined) followed by an acidic residue (boldface) (YSRVRTE). To test whether these sequences contribute to the ER-to-Golgi transport of gB, we introduced several mutations into gB in which alanine was substituted for one or more of the acidic residues or for key amino acids in the YSRVRTE motif (Fig. 1). In all cases, these mutated forms of gB were transported to the Golgi similarly to native gB (Fig. 7). Therefore, neither the basic residues nor the YSRVRTE sequence contained within the C-terminal 17 aa of gB is specifically required for the transport of gB to the Golgi. To further define the role specific sequences within the C-terminal 17 aa of gB may play in its transport, we introduced an internal deletion into gB that eliminated the proximal 9 aa within this region (Δ852–860 [Fig. 1]). VZV gB lacking aa 852 to 860 was transported to the Golgi as efficiently as native gB (Fig. 5 and 6). Taken together, these data indicate that no specific sequences within the C-terminal 17 aa of the gB cytoplasmic domain are required for its ER-to-Golgi transport.
FIG. 7.
Quantitation of ER-to-Golgi transport of VZV gB mutants containing substitutions in the C-terminal 17 aa of its cytoplasmic domain. The C-terminal 17 aa of the VZV gB cytoplasmic domain (aa 852 to 868) are listed. Boldface letters represent substitution mutations within this region. The proportion of each mutant transported to the Golgi (as a percentage wild-type [wt] gB transport) was quantitated and is shown by the stippled bars.
Our inability to identify specific transport signal sequences in the C-terminal 17 aa of gB raised the possibility that any major perturbation of the gB cytoplasmic domain, as may have resulted from the 17-aa truncation, will impair gB transport to the Golgi through nonspecific conformational changes. To address this question, we introduced mutation Δ833–851 (Fig. 1). gB containing this mutation retains its C-terminal 17 aa but lacks the 19 aa immediately proximal to this region. If nonspecific conformational changes in its cytoplasmic domain were responsible for the impaired transport of gB lacking the C-terminal 17 aa, then deletion of the adjacent upstream 19 aa would seem likely to have a similar detrimental effect on gB transport. However, Δ833–851 was transported to the Golgi at levels similar to that of native gB (Fig. 5 and 6). This observation is most consistent with the conclusion that the C-terminal 17 aa gB performs some specific function during ER-to-Golgi transport rather than serving merely to retain the global conformation of the gB cytoplasmic domain.
The YXXφ-containing region from aa 818 to 826 within the cytoplasmic domain of gB is required for the transport of gB from the ER to the Golgi.
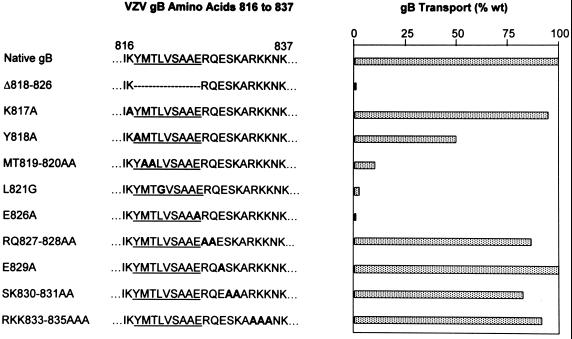
The second YXXφ motif (underlined) in the cytoplasmic domain of VZV gB is followed by two acidic residues (boldface) in the pattern EXXE (YMTLVSAAERQE; aa 818 to 829) and thus closely resembles the predicted ER-to-Golgi transport signal sequences identified in other glycoproteins (23). We introduced mutations within and adjacent to this region (Fig. 1) to determine if it participates in the ER-to-Golgi transport of VZV gB. The efficiency of gB transport to the Golgi was again determined by quantitating the proportion gB acquiring complex oligosaccharides during a 4-h chase period following a 1-h labeling period. Several alanine substitution mutations within this region dramatically inhibited the transport of gB to the Golgi (Fig. 8; note that L821 was changed to glycine to avoid replacing one hydrophobic amino acid with another). The mutation Y818A resulted in only 50% native transport of gB to the Golgi, and mutations MT819–820AA, L821G, and E826A all resulted in less than 10% of the native level of gB transport from the ER to the Golgi (Fig. 8). By contrast, gB containing mutations proximal to this region (K817A) or carboxyl to aa 826 (RQ827–828AA, E829A, SK830–831AA, and RKK833–835AAA) was transported to the Golgi at levels similar to that of native VZV gB (Fig. 8). These data indicate that the region of the gB cytoplasmic domain between aa 818 and 826 is required for the efficient transport of gB from the ER to the Golgi, whereas the sequences immediately adjacent to this region do not contribute significantly to this process. To confirm this result, we deleted the entire region from aa 818 to 826 (YMTLVSAAE [Fig. 1]) from the gB cytoplasmic domain. This mutated form of gB, Δ818–826, was transported to the Golgi at less than 5% of the native level (Fig. 8). This is in sharp contrast to the nearby 19-aa deletion Δ833–851, which, as discussed previously, had little impact on the ER-to-Golgi transport of gB. These data indicate that the 9-aa region of the gB cytoplasmic domain between aa 818 and 826 is required for the efficient transport of gB from the ER to the Golgi. Moreover, several specific amino acids within this region are required for the ER-to-Golgi transport of gB, including those within the YMTL motif and the first of two acidic residues carboxyl to this motif, E826. Therefore, the region between aa 818 and 826, unlike the C-terminal 17-aa domain, participates in the ER-to-Golgi transport of gB through a sequence-dependent mechanism.
FIG. 8.
Quantitation of ER-to-Golgi transport of VZV gB mutants containing deletions or substitutions in the region between aa 816 and 837 of the cytoplasmic domain. Amino acid residues 816 to 837 of the VZV gB cytoplasmic domain are listed (Native gB). Amino acids 818 to 826 are underlined. Boldface letters represent substitution mutations within this region. The proportion of each mutant transported to the Golgi (as a percentage wild-type [wt] gB transport) was quantitated and is shown by the stippled bars.
DISCUSSION
We have shown that the cytoplasmic domain of VZV gB is critical for gB transport from the ER to the Golgi, and we have identified two specific regions within the gB cytoplasmic domain that are required for this process, the C-terminal 17 aa and an internal sequence consisting of aa 818 to 826. In addition, we have shown that VZV gB localization to the nuclear membrane cells occurs independently of its cytoplasmic domain.
Deletion of either the C-terminal 17 aa (aa 852 to 868) or aa 818 to 826 of VZV gB greatly diminishes the transport of gB from the ER to the Golgi. However, these regions apparently mediate the ER-to-Golgi transport of gB by different mechanisms. The C-terminal 17-aa domain functions independently of its amino acid sequence. Complete deletion of this region results in an almost complete loss of ER-to-Golgi transport; however, neither multiple substitution mutations nor internal deletions within this region had an impact on the ER-to-Golgi transport of gB. One possible explanation is that deletion of the C-terminal 17 aa of gB alters the conformation of its ER luminal domain, resulting in the retention of gB in the ER through its interaction with ER-resident chaperone proteins. It has, for example, been reported that mutations within the transmembrane domain of HCMV gB results in its ER retention by allowing the binding of specific chaperone proteins within the ER (36). This possibility, however, is rendered less likely for VZV gB by the observation that deletion of the adjacent 19 aa has little impact on the ER-to-Golgi transport of gB despite the expectation that this larger deletion would similarly alter the conformation of gB. Another possibility is that the C-terminal 17-aa domain functions through electrostatic rather than sequence-specific interactions with cellular factors. This model is attractive since the C-terminal 17-aa domain of gB contains five basic amino acids and a single acidic residue. Again, our experimental data fail to support this hypothesis, since gB in which three of these five basic residues are replaced by a neutral amino acid is transported to the Golgi as efficiently as native gB. We therefore consider it most likely that deletion of the C-terminal 17 aa affects the accessibility of an ER-to-Golgi translocation signal located elsewhere within the cytoplasmic domain itself such as the aa 818–826 domain.
In contrast to the C-terminal 17-aa domain, the VZV gB aa 818–826 domain functions in a sequence-dependent manner. This domain contains several individual amino acids that are required for the efficient ER-to-Golgi transport of gB, while numerous point mutations elsewhere in the cytoplasmic domain of gB, including those in which charged residues are eliminated, have no impact on transport. These data strongly suggest that the aa 818–826 domain functions as an independent translocation signal sequence that acts by a sequence-specific mechanism, perhaps through specific interactions with one or more cellular factors, rather than by nonspecific mechanisms based on protein conformation or electrostatic interactions.
Perhaps the most interesting sequence feature of the VZV gB aa 818–826 domain is its similarity to a known ER-to-Golgi transport signal, the YTDIE sequence found in the cytoplasmic domain of the VSV-G protein. Both the VZV gB aa 818–826 domain (YMTLVSAAE) and the VZV-G signal sequence contain YXXφ motifs and at least one acidic amino acid residue. Mutation of the acidic residue in the VZV gB aa 818–826 domain markedly impairs the transport of gB from the ER to the Golgi; however, mutation of a second downstream acidic residue, E829, has no impact on the transport of VZV gB. In contrast, mutation of either of the acidic residues in the VSV-G signal sequence greatly reduces its transport to the Golgi. Also, mutations within the YXXφ sequence itself in VZV gB aa 818 to 826 disrupt the ER-to-Golgi transport of gB, while only mutations in the acidic residues of the VSV-G signal sequence have a significant impact on the transport of VSV-G.
As expected, VZV gB that failed to be transported to the Golgi due to mutations in its cytoplasmic domain largely accumulated instead in the ER. However, gB containing large C-terminal truncations was transported to the nuclear membrane at apparently normal levels. These data indicate that the cytoplasmic domain of gB does not contain specific nuclear membrane transport signals. This is consistent with findings for HSV-1 gB, which requires only a portion of its transmembrane domain and none of its cytoplasmic domain for localization to the nuclear membrane (9). While specific nuclear membrane transport signals may be located in the luminal or transmembrane domains of VZV gB, it seems equally plausible that the transport of gB from the ER to the nuclear membrane occurs through passive diffusion of gB between these two contiguous structures.
Both the microscopic colocalization and posttranslational modification data presented here provide clear evidence that VZV gB is transported to the Golgi in the absence of other VZV-encoded proteins. It has, however, previously been reported that VZV gB cannot be detected in the trans-Golgi network of transfected cells (33). This seeming discrepancy may be due to the different cell types used in these studies. The studies which failed to show gB in the trans-Golgi network following transfection were done in Cos cells, whereas we used MeWo and HEp-2 cells. Cos cells, unlike MeWo and HEp-2 cells, do not support the growth of VZV, leading to the intriguing possibility that this deficiency may be due to the inability of Cos cells to correctly mediate the intracellular transport of VZV gB.
It is not known whether the cytoplasmic domain of other herpesvirus gBs contain specific ER-to-Golgi signal sequences analogous to those identified in VZV gB. A region closely related to the VZV gB aa 818–826 transport signal sequence exists in the cytoplasmic domain of HSV-1 gB. This 9-aa sequence, YMALVSAME, contains 7 aa that are identical to the VZV gB aa 818–826 domain and includes a YXXφ motif followed by an acidic residue. However, the VZV gB C-terminal 17-aa domain shown to be necessary for the transport of gB from the ER to the Golgi has little similarity to the C terminus of HSV-1 gB. Notably, HSV-1 gB contains five acidic residues among its C-terminal 7 aa, whereas VZV gB contains a single acidic residue among its C-terminal 7 aa.
The cytoplasmic domains of the beta- and gammaherpesviruses exhibit considerably less sequence homology to the VZV gB cytoplasmic domain. The cytoplasmic domain of HCMV gB contains the sequence YQMLLALAR (aa 845 to 853) at a site corresponding to the location the VZV gB aa 818–826 transport signal sequence. While this sequence contains a YXXφ motif, it has limited (3 of 9 aa) overall homology to the VZV gB aa 818–826 sequence and contains no acidic residues. The cytoplasmic domain of EBV gB contains no region with sequence homology to the VZV gB aa 818–826 domain. Moreover, the function of the EBV gB cytoplasmic domain contrasts sharply with that of the VZV gB cytoplasmic domain. While the VZV gB cytoplasmic domain facilitates the transport of gB from the ER to the Golgi, the EBV gB cytoplasmic domain inhibits the ER-to-Golgi transport of gB by means of an ER retention signal (16). Therefore, the cytoplasmic domains of gB homologs from members of different herpesvirus subfamilies may have in common their function as mediators of gB transport; however, their actual role in the intracellular localization of gB may vary dramatically between viruses from different subfamilies. The degree to which differences in gB localization influence the growth properties of the various herpesviruses remains to be defined.
REFERENCES
- 1.Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas K G. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J Virol. 1993;67:2396–2401. doi: 10.1128/jvi.67.4.2396-2401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banfield B W, Tufaro F. Herpes simplex virus particles are unable to traverse the secretory pathway in the mouse L-cell mutant gro29. J Virol. 1990;64:5716–5729. doi: 10.1128/jvi.64.12.5716-5729.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campadelli G, Brandimarti R, Di Lazzaro C, Ward P L, Roizman B. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 1993;90:2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darlington R W, Moss L H. Herpesvirus envelopment. J Virol. 1968;2:48–55. doi: 10.1128/jvi.2.1.48-55.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison A J, Scott J. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert R, Ghosh K, Rasile L, Ghosh H P. Membrane anchoring domain of herpes simplex virus glycoprotein gB is sufficient for nuclear envelope localization. J Virol. 1994;68:2272–2285. doi: 10.1128/jvi.68.4.2272-2285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong M, Kieff E. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, 350/220 and gp110. J Virol. 1990;64:1507–1516. doi: 10.1128/jvi.64.4.1507-1516.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harson R, Grose C. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J Virol. 1995;69:4994–5010. doi: 10.1128/jvi.69.8.4994-5010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kari B, Radeke R, Gehrz R. Processing of human cytomegalovirus envelope glycoproteins in and egress of cytomegalovirus from human astrocytoma cells. J Gen Virol. 1992;73:253–260. doi: 10.1099/0022-1317-73-2-253. [DOI] [PubMed] [Google Scholar]
- 13.Keller P M, Davison A J, Lowe R S, Bennett C D, Ellis R W. Identification and structure of the gene encoding gpII, a major glycoprotein of varicella-zoster virus. Virology. 1986;152:181–191. doi: 10.1016/0042-6822(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 14.Kirchhausen T, Bonifacino J S, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S K, Compton T, Longnecker R. Failure to complement infectivity of EBV and HSV-1 glycoprotein B (gB) deletion mutants with gBs from different human herpesvirus subfamilies. Virology. 1997;237:170–181. doi: 10.1006/viro.1997.8765. [DOI] [PubMed] [Google Scholar]
- 17.Lee S K, Longnecker R. The Epstein-Barr virus glycoprotein 110 carboxyl-terminal tail domain is essential for lytic virus replication. J Virol. 1998;71:4092–4097. doi: 10.1128/jvi.71.5.4092-4097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little S P, Jofre J T, Courtney R J, Schaffer P A. A virion-associated glycoprotein essential for infectivity of herpes simplex virus type 1. Virology. 1981;115:149–160. doi: 10.1016/0042-6822(81)90097-0. [DOI] [PubMed] [Google Scholar]
- 19.Montalvo E A, Grose C. Assembly and processing of the disulfide-linked varicella-zoster virus glycoprotein gpII(140) J Virol. 1987;61:2877–2884. doi: 10.1128/jvi.61.9.2877-2884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan C, Rose C H, Holden M, Jones E P. Electron microscopic observations on the development of herpes simplex virus. Exp Med. 1959;110:643–656. doi: 10.1084/jem.110.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 22.Nii S, Morgan C, Rose H M. Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol. 1968;2:517–536. doi: 10.1128/jvi.2.5.517-536.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura N, Balch W E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 24.Olson J K, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson J K, Grose C. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J Virol. 1998;72:1542–1551. doi: 10.1128/jvi.72.2.1542-1551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 27.Raviprakash K, Rasile L, Ghosh K, Ghosh H P. Shortened cytoplasmic domain affects intracellular transport but not nuclear localization of a viral glycoprotein. J Biol Chem. 1990;265,No.3:1777–1782. [PubMed] [Google Scholar]
- 28.Roizman B. Herpesviridae: a brief introduction. In: Fields B N, et al., editors. Fields virology. 2nd ed. Philadelphia, Pa: Raven Press; 1990. pp. 1787–1793. [Google Scholar]
- 29.Sanger R, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz J, Roizman B. Concerning the egress of herpes simplex virus from infected cells: electron and light microscope observations. Virology. 1969;38:42–49. doi: 10.1016/0042-6822(69)90126-3. [DOI] [PubMed] [Google Scholar]
- 31.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 32.Stanley K K. Regulation of targeting signals in membrane proteins. Mol Membr Biol. 1996;13:19–27. doi: 10.3109/09687689609160570. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Gershon M D, Lungu O, Panagiotidis C A, Zhu Z, Hao Y, Gershon A A. Intracellular transport of varicella-zoster glycoproteins. J Infect Dis. 1998;178(Suppl. 1):S7–S12. doi: 10.1086/514268. [DOI] [PubMed] [Google Scholar]
- 34.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whealy M E, Robbins A K, Tufaro F, Enquist L W. A cellular function is required for pseudorabies virus envelope glycoprotein processing and virus egress. J Virol. 1992;66:3803–3810. doi: 10.1128/jvi.66.6.3803-3810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, Maidji E, Tugizov S, Periera L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Hao Y, Gershon M D, Ambron R T, Gershon A A. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J Virol. 1996;70:6563–6575. doi: 10.1128/jvi.70.10.6563-6575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]