Abstract
Adeno-associated virus (AAV) type 2 Rep78 is a multifunctional protein required for AAV DNA replication, integration, and gene regulation. The biochemical activities of Rep78 have been described, but the effects of Rep proteins on the cell have not been characterized. We have analyzed Rep-mediated cytotoxicity. We demonstrated that Rep78 expression is sufficient to induce cell death and disruption of the cell cycle. Cell death was found to be mediated by apoptosis. Rep78 expression resulted in the activation of caspase-3, a terminal caspase directly involved in the execution of cell death. A peptidic inhibitor of caspase-3, Z-Asp-Glu-Val-Asp-fluoromethylketone (Z-DEVD-FMK), abrogated Rep78-induced apoptosis, indicating that Rep78-mediated apoptosis is caspase-3 dependent. Rep78 induced apoptosis in wild-type p53-containing human embryonal carcinoma NT-2 cells and in p53-null promyelocytic human HL-60 cells, indicating that at least one pathway of Rep78-induced apoptosis is p53 independent. Apoptosis was shown to occur during the G1 and early S phases of the cell cycle. By analyzing the effects of Rep78 mutations on cell viability, the cause of cell death was attributed in part to two biochemical activities of Rep78, DNA binding and ATPase/helicase activity. The endonuclease activity of Rep78 did not contribute to apoptosis induction.
Adeno-associated virus type 2 (AAV-2) is classified as a member of the family Parvoviridae and has been assigned to the genus Dependovirus because efficient replication typically requires a coinfecting helper virus (5, 54). Under nonpermissive conditions, AAV-2 establishes a latent infection by locus-specific integration into the q arm of chromosome 19 of the infected cell (32, 33, 51). AAV-2 is a small, nonenveloped, icosahedral virus with a linear, single-stranded DNA genome of 4,680 nucleotides (nt). The coding region of AAV-2 is flanked by inverted terminal repeat (ITR) sequences of 145 nt each. The ITRs contain all of the cis-acting sequences required for DNA replication, packaging, and integration. The AAV genome harbors two open reading frames (ORFs) termed rep and cap that encode the nonstructural and structural proteins, respectively. The cap ORF encodes the viral capsid proteins (VP-1, VP-2, and VP-3). The rep ORF is expressed from promoters at map positions 5 (p5) and 19 (p19) and is translated into four overlapping nonstructural proteins by alternative splicing (35, 49). The p5 proteins, Rep78 and Rep68, have been shown to be required for AAV DNA replication, integration, and gene regulation. The p5 Rep proteins possess site- and strand-specific endonuclease, DNA ligase, ATPase, and helicase activities, as well as the ability to specifically bind the viral ITR in either a duplex or single-stranded conformation (26, 27, 55, 68). The p19 proteins, Rep52 and Rep40, lack the 224 N-terminal residues of Rep78 and Rep68, retain ATP-dependent helicase activity (56), and appear to be involved in the encapsidation of virus genomes (11, 18). While the activities of Rep proteins that are required for viral replication are well described, still relatively little is known regarding the mechanisms by which Rep affects the cell. Although interaction of several cellular proteins—such as the transcription factor Sp1 (24), the transcription cofactor PC4 (61), high-mobility group nonhistone protein 1 (16), the oncosuppressor p53 (4), and cyclic AMP-dependent protein kinases PrKX and PKA (12, 17)—with Rep proteins has been described, the consequences of these interactions for the cell remain obscure. The effects of Rep expression on the phenotype of the cell have been described anecdotally and include both retardation of cell growth and cell death. Rep expression has been shown to inhibit transformation by cellular and viral oncogenes and to repress cellular and viral DNA replication through unknown processes (23, 29, 64). Studies with a cell line that inducibly expresses Rep have demonstrated inhibition of cellular DNA synthesis and distortion of the cell cycle (64, 65). The combination of Rep expression with UV irradiation or incubation with cadmium induced cell death that exhibited some of the characteristics of apoptosis (66, 67).
Apoptosis, or programmed cell death, is a highly regulated cellular suicide process. It is involved in normal tissue development as well as in the response of the cell to stress, growth factor deprivation, and DNA damage. Morphological changes typical for apoptotic cells include nuclear and cytoplasmatic condensation, which is followed by the fragmentation of the cell into apoptotic bodies. In vivo, these cell fragments are consumed by macrophages without the elicitation of an inflammatory response (57, 60). Biochemical characteristics include fragmentation of DNA, partial loss of plasma membrane asymmetry, and reduction of the mitochondrial transmembrane potential. Apoptosis can be triggered by external or cellular signals. Stimuli from both pathways are integrated and amplified by a family of cysteine proteases with aspartate specificity, referred to as caspases. Caspases are expressed as zymogens, inactive proenzymes which are proteolytically activated during the transduction of death signals. Caspases by proteolytically cleaving vital cellular proteins, are also involved in the execution of cell death. Among the caspase targets are actin, lamins, and poly(ADP-ribose) polymerase (8, 19).
In this study, we showed that AAV-2 Rep78 expression is sufficient to induce apoptosis as well as an accumulation of cells in the G1 phase of the cell cycle. Apoptosis was mediated by caspase-3 and was shown to be independent of p53. By analyzing the effects of Rep mutations on cell viability, it was determined that cell death was attributable in part to the DNA binding and ATPase-helicase activities of Rep78. Surprisingly, the endonuclease activity of Rep78 did not contribute to apoptosis induction.
MATERIALS AND METHODS
Plasmids.
A plasmid expressing a nucleus-localized green fluorescent protein (GFP), pNLS-GFP, was constructed by N-terminal fusion of a sequence coding for the simian virus 40 large T antigen nuclear localization signal PKKKRKV (28) to the GFP ORF of pEGFP-N1 (Clontech, Palo Alto, Calif.) by PCR cloning. The GFP gene was amplified using pEGFP-N1 as template and 5′-GAG AAG ATC TCA CCA TGG GTC CTA AGA AGA AGC GTA AGG TGA GCA AGG GCG AGG AGC TGT-3′ and 5′-CCT CTA CAA ATG TGG TAT GGC T-3′ as primers. The PCR product was digested with BglII and NotI and inserted into BglII- and NotI-cut pEGFP-N1. The plasmid pRep78-GFP, expressing a Rep78-GFP fusion protein, was constructed by amplifying the Rep78 ORF from pAV2, a plasmid containing the AAV-2 genome (34), using primers 5′-GAG GCT AGC CAC CAT GGC CCC GGG GTT TTA CGA GAT T-3′ and 5′-ACT GCT CGA GTT GTT CAA AGA TGC AGT CAT C-3′. The PCR product was digested with NheI and XhoI and inserted into NheI- and XhoI-cut pEGFP-N1. pRep52-GFP was constructed accordingly, using primers 5′-GAG GCT AGC CAC CAT GGA GCT GGT CGG GTG GCT C-3′ and 5′-ACT GCT CGA GTT GTT CAA AGA TGC AGT CAT C-3′ to amplify the Rep52 ORF from the pAV2 template. pRep78-GFP(K340H) and pRep78-GFP(Y156F) were constructed by replacing the 1.2-kbp NruI-KpnI fragment of pRep78-GFP with the corresponding fragment of pMBP-Rep78 (NTP) (56), bearing a K340H mutation, and pMBP-Rep78(Y156F) (55), respectively.
Cell lines.
HL60 human promyelocytic leukemia cells were grown in suspension at densities between 2 × 105 and 1 × 106 cells/ml. The cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. NTERA-2 (NT2) cells, a pluripotent human embryonal carcinoma cell line, were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 1 mM pyruvate, 2 mM l-glutamine, 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. Cells were maintained at 37°C in a 5% CO2 humidified atmosphere. Both cell lines were obtained from the American Type Culture Collection (Manassas, Va.). Tissue culture media and supplements were purchased from Life Technology (Gaithersburg, Md.) and HyClone (Logan, Utah).
DNA transfection.
DNA transfection was performed by electroporation with a 600R Electro Cell Manipulator (BTX, San Diego, Calif.). Cells (2 × 106) were resuspended in 400 μl of RPMI 1640 medium supplemented with 20% FCS and 40 μg of plasmid DNA. Cells were pulsed in 0.4-cm-gap electrode cuvettes (Bio-Rad, Hercules, Calif.) at 300 V and 1,050 μF (HL-60) or at 200 V and 1,050 μF (NT2).
Flow cytometric analysis of viability.
Cells were washed with phosphate-buffered saline (PBS) and resuspended in 200 to 500 μl of PBS containing 50 μg of propidium iodide (PI)/ml for staining of dead cells. Cells were incubated for at least 10 min at room temperature and analyzed using an Epics XL flow cytometer (Beckman-Coulter, Fullerton, Calif.).
Flow cytometric analysis of apoptosis.
The quantitative determination of cells undergoing apoptosis was done by an Annexin V staining assay (BD PharMingen, San Diego, Calif.) according to the manufacturer's instructions. Briefly, 5 × 105 cells were washed with PBS and resuspended in 400 μl of Annexin V binding buffer (10 mM HEPES-NaOH, pH 7.4; 140 mM NaCl; 2.5 mM CaCl2), and 15 μl of Annexin V-phycoerythrin was added. The vital dye ViaProbe (BD PharMingen; 15 μl/400 μl) was used to discriminate apoptotic cells (which are Annexin V positive and ViaProbe negative) from necrotic and dead cells (which are Annexin V positive and ViaProbe positive). Stained cells were analyzed by flow cytometry after a 15-min incubation at room temperature. During data collection, cells were gated by forward- and side-scatter analysis to exclude cell debris. A minimum of 10,000 events were collected for each sample.
Flow cytometric analysis of DNA content.
DNA content of live cells was determined by staining with the DNA dye Hoechst 33342 (Molecular Probes, Eugene, Oreg.) at a concentration of 10 μg/ml in cell culture medium for 30 min at 37°C. After the incubation, cells were washed in PBS and resuspended in 400 μl of Annexin V binding buffer. A 15-μl aliquot of ViaProbe was added to discriminate living from dead cells. DNA content analysis was done by flow cytometry (Coulter EPICS Elite ESP). Cell debris and dead cells were excluded from the analysis by forward- and side-scatter analysis and exclusion of ViaProbe-positive (dead) cells. Data were analyzed with the ModFit software (Verity Software House, Topsham, Maine).
Caspase-3 assay.
Cells transfected with pNLS-GFP or pRep78-GFP were stained with 50-μg/ml PI and sorted by fluorescence-activated cell sorting, using an EPICS Elite ESP flow cytometer. Cells which were GFP positive (i.e., NLS-GFP- or Rep78-GFP-expressing cells) and PI negative (living cells) were collected. Sorted cells were washed in PBS and analyzed for caspase-3 activity by using a CPP32/caspase-3 colorimetric protease assay kit (Chemicon, Temecula, Calif.) according to the instructions of the manufacturer. Briefly, cells were lysed in 150 μl of cell lysis buffer provided in the kit. Protein concentrations of the lysates were determined by using the bicinchoninic acid assay reagent (Pierce, Rockford, Ill.). Equal amounts of lysates were incubated with the caspase-3 substrate, 200 μM DEVD-pNA, at 37°C for 3 h. Absorbances of samples were read every 60 min in a Spectramax 250 (Molecular Dynamics) microplate reader at 405 nm. Caspase-3 activity was proportional to the optical density at 405 nm.
Inhibition of caspase-3.
The caspase-3 inhibitor Z-Asp-Glu-Val-Asp-fluoromethylketone (Z-DEVD-FMK) was obtained from Calbiochem (San Diego, Calif.) and used at a final concentration of 100 μM. The inhibitor was added to the medium 1 h after transfection. Half of the medium was exchanged every 24 h with fresh medium containing 100 μM Z-DEVD-FMK.
RESULTS
AAV-2 Rep78 is cytotoxic.
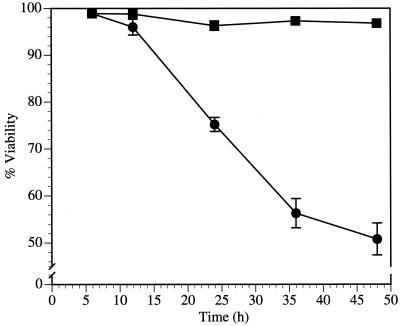
AAV-2 Rep78 is a multifunctional protein involved in AAV DNA replication, integration, and gene regulation. Recent studies using an inducible Rep-expressing cell line demonstrated that UV irradiation, incubation, with a heavy metal, and Rep78 expression in various combinations resulted in cell death with some characteristics of apoptosis and perturbation of the cell cycle (65–67). The goal of the study reported here was to analyze the effect of Rep78 on the cell in the absence of toxic chemicals and other stressful condition. Rep78 was expressed transiently as an N-terminal fusion protein with the GFP encoded by the plasmid pRep78-GFP. The GFP tag allowed tracking of Rep78-expressing cells at the single-cell level. Rep78-GFP retained the biochemical activities necessary for AAV replication. We were able to complement a Rep78 frame shift mutant with Rep78-GFP for the production of recombinant AAV (data not shown). To determine if Rep78 expression was sufficient to induce cytotoxicity, pRep78-GFP and the control plasmid pNLS-GFP, encoding a nucleus-localized GFP, were transfected separately into HL-60 cells, a human promyelocytic leukemia cell line. Expression of Rep78-GFP and NLS-GFP was detectable by fluorescence microscopy as early as 3 h posttransfection; the transfection efficiency was typically found to be in the range of 20 to 60% (data not shown). At various time points, cells were analyzed for viability by PI staining and flow cytometry (Fig. 1). The staining of cells with the vital dye PI is commonly used to quantify cell death. This method is based on the ability of live cells with intact cytoplasmic membranes to exclude PI while dead cells, with compromised cytoplasmic membranes, are unable to exclude this dye and are stained by the formation of a fluorescent PI-DNA complex (36). Figure 1 shows that about 25% of Rep78-GFP-expressing cells were dead at 24 h and that about 50% were dead at 48 h posttransfection. The observed cytotoxicity was not due to the GFP tag of Rep78-GFP, since only a minor reduction in cell viability (4% at 48 h posttransfection) was detected in cells expressing NLS-GFP.
FIG. 1.
Rep78 expression is cytotoxic. The viability of pNLS-GFP- or pRep78-GFP-transfected HL-60 cells was determined by PI staining (50 μg/ml) and flow cytometry. Percent viability represents the fraction of cells that excluded PI times 100. The abscissa is the time after electroporation. ■, pNLS-GFP; ●, pRep78-GFP.
Rep78-mediated cytotoxicity results from apoptosis.
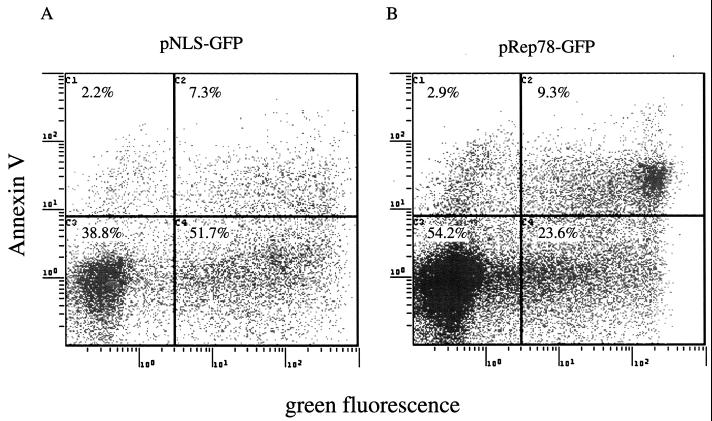
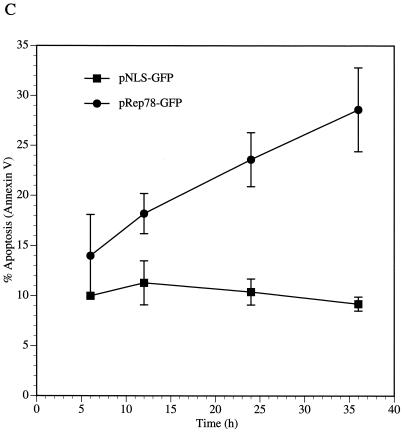
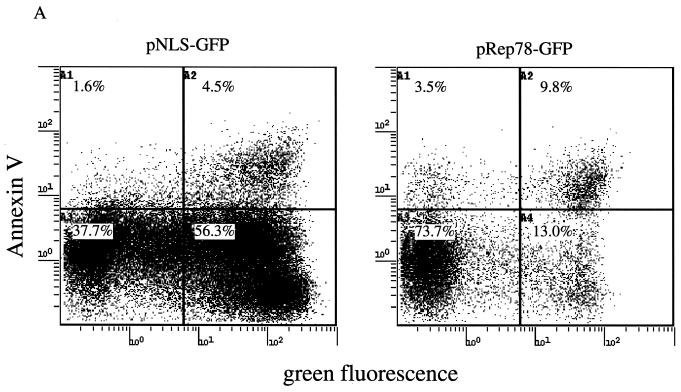
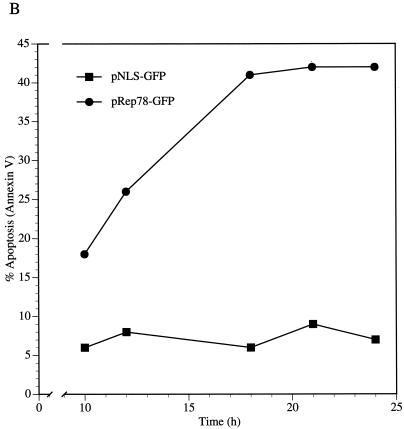
Our results indicate that Rep78 expression is sufficient to cause cytotoxicity. Cell death occurs by either necrotic or apoptotic pathways (38). Necrosis is a passive process usually observed after chemical or mechanical injury of the cell. It is characterized by cell swelling and early membrane rupture. Apoptosis is an active process that is highly regulated and is characterized by cellular shrinkage and nuclear condensation. An early event in cells undergoing apoptosis is the partial loss of phospholipid asymmetry, leading to the translocation of phosphatidylserine (PS) from the inner to the outer leaflet of the plasma membrane. Since the membrane integrity is not affected by this rearrangement, apoptotic cells exclude vital dyes. The exposure of PS can be detected with fluorescently labeled Annexin V, a phospholipid binding protein that has a high affinity for PS (31). To discriminate between apoptotic and necrotic cells, ViaProbe was used as a costain. ViaProbe is a cell membrane-impermeable nucleic acid dye which stains only dead and necrotic cells with compromised membrane integrity (52). Annexin V analysis by flow cytometry of pRep78-GFP- or pNLS-GFP-transfected HL-60 cells showed a correlation between Annexin V-positive cells and Rep78-expressing cells, indicating that Rep78 induced cell death by apoptosis (Fig. 2A and B). In a time course experiment, we observed an increase of apoptotic Rep78-expressing cells from 14% at 6 h posttransfection to 29% at 36 h (Fig. 2C). During this time period, we noticed an increase of nonviable Rep78-GFP-expressing cells of from 1 to 48% (Fig. 1). The percentage of Annexin V-positive, NLS-GFP-expressing cells remained relatively constant over time at about 10% in the pNLS-GFP-transfected cultures (Fig. 2C), while we noticed a slight (1 to 3.7%) increase in dead cells (Fig. 1).
FIG. 2.
Rep78 induces apoptosis. Shown are representative diagrams of Annexin V staining of pNLS-GFP-transfected (A) or pRep78-GFP-transfected (B) HL-60 cells at 24 h posttransfection. The x axis represent the green fluorescence intensity (NLS-GFP expression in panel A and Rep78-GFP expression in panel B). The fluorescence intensity of Annexin V-PE is represented on the y axis. The populations in the upper quadrants are apoptotic cells, while cells in the lower quadrants are nonapoptotic. Left quadrants are GFP-negative cells, while cells in the right quadrants are those expressing NLS-GFP (A) or Rep78-GFP (B). Cells displayed were gated for the exclusion of ViaProbe (i.e., dead cells). The percentage of Rep78-mediated apoptosis was calculated by division of the percentage of Annexin V-positive, Rep78-GFP-expressing cells by the percentage of the total Rep78-GFP subpopulation [9.3%/(9.3% + 23.6%) = 28.2%]. (C) Time course of induction of apoptosis by Rep78. Data are means ± standard errors of values from three experiments. Percent apoptosis was determined as in described for panel B.
Rep78 expression activates caspase-3.
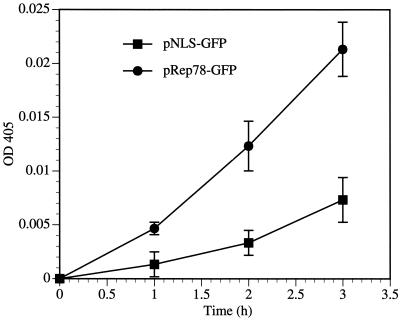
Apoptotic signals from membrane-associated receptors and mitochondrial sensors converge on a common pathway, the caspases (8). Caspases are a family of cysteine proteases which integrate and amplify death signals and also mediate cell death. Caspase-3 is a terminal caspase involved in the execution of cell death by proteolytic cleavage of essential cellular proteins. Among the targets of caspase-3 are poly(ADP-ribose) polymerase, lamins, and gesolin (19). Caspase-3 activation during apoptosis induced by infection with human immunodeficiency virus type 1 (HIV-1) (2), Sendai virus (6), parvovirus B19 (42), and adenovirus (13) has been described. We investigated the involvement of caspase-3 in Rep78-induced apoptosis. Caspase-3 activity in the lysates of pNLS-GFP- or pRep78-GFP-transfected cells was measured. At 12 h posttransfection, a substantial increase of caspase-3 activity in Rep78-expressing cells relative to NLS-GFP-expressing cells was observed (Fig. 3). These results indicate that Rep78 expression activates caspase-3 and that Rep78-induced apoptosis is mediated at least in part by caspase-3.
FIG. 3.
Rep78 expression results in activation of caspase-3. pNLS-GFP- or pRep78-GFP transfected HL-60 cells were stained with PI and sorted by flow cytometry for the expression of GFP or Rep78-GFP and exclusion of PI (i.e., living cells) at 12 h posttransfection. Extracts of sorted cells were assayed for caspase-3 activity toward the substrate DEVD-pNA. Caspase-3 activity is proportional to the optical density (OD) at 405 nm. Data are means ± standard errors of values from three experiments.
Inhibition of Rep78-induced apoptosis by inhibition of caspase-3.
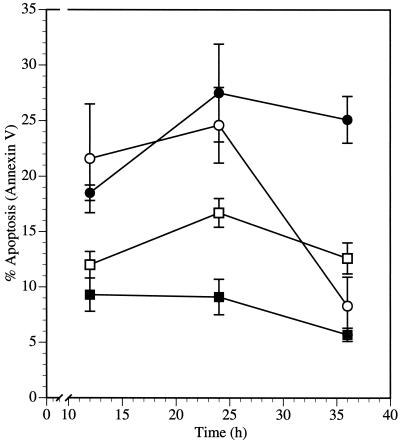
Since Rep78 expression resulted in activation of caspase-3, we explore whether caspase-3 activation was essential for Rep78-induced apoptosis by studying the effects of a caspase-3 inhibitor. HL-60 cells transfected with pRep78-GFP or pNLS-GFP were treated with the peptidic inhibitor of caspase-3, Z-DEVD-FMK (45). Cells were assayed for apoptosis by costaining with Annexin V-PE and ViaProbe prior to flow cytometry. While incubation with Z-DEVD-FMK resulted in a slight increase in the number of apoptotic cells in pNLS-GFP-transfected cells (Fig. 4), we noted a decrease of Annexin V-positive, Rep78-GFP-expressing cells after treatment with Z-DEVD-FMK, to the level of NLS-GFP-expressing cells at 36 h posttransfection. Thus, incubation with Z-DEVD-FMK efficiently inhibited Rep78-mediated apoptosis. From these results we conclude that Rep78-induced apoptosis in HL-60 cells is dependent on caspase-3 activity. Incubation with Z-DEVD-FMK also led to an increase in expression levels in pNLS-GFP- and in pRep78-GFP-transfected cells (data not shown), which might have led to a slightly increased toxicity compared to that of non-Z-DEVD-FMK-treated cells observed at early time points.
FIG. 4.
The synthetic peptide inhibitor Z-DEVD-FMK reduced levels of Rep78-induced apoptosis. HL-60 cells were transfected with pNLS-GFP or pRep78-GFP. The caspase-3 inhibitor Z-DEVD-FMK (100 μM) was added 1 h after transfection and was present throughout the experiment. Cells were analyzed for apoptosis by Annexin V staining and flow cytometry. Data are means ± standard errors of values from three experiments. ■, pNLS-GFP; □, pNLS-GFP plus Z-DEVD-FMK; ●, pRep78-GFP; ○, pRep78-GFP plus Z-DEVD-FMK.
Rep78 expression results in an accumulation of cells in G1.
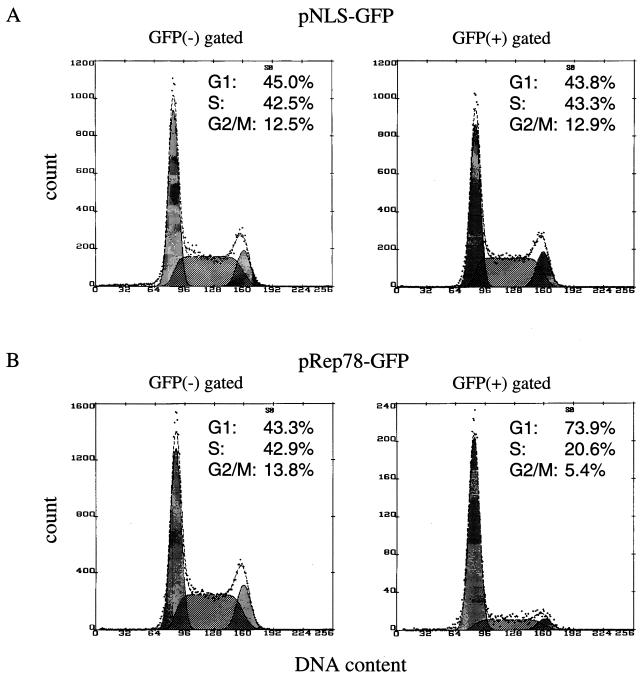
Since deregulation of the cell cycle may be involved in the induction of apoptosis (30), we investigated the effects of Rep78 expression on cell cycle progression in HL-60 cells. The DNA content of cells transfected with pNLS-GFP or pRep78-GFP was determined 12 h posttransfection by staining with Hoechst 33342 and performing flow cytometry (Fig. 5). Hoechst 33342 is a cytoplasmic membrane-permeative DNA dye which can be used for DNA staining without fixation of the cells (53). Analysis of the DNA content of pNLS-GFP-transfected cells showed that the patterns of cell cycle distribution were similar in NLS-GFP-expressing cells (Fig. 5A, right panel) and in non-NLS-GFP-expressing cells (Fig. 5A, left panel), indicating that NLS-GFP expression had no effect on cell cycle progression. In contrast, Rep78 expression resulted in a dramatic alteration of the cell cycle. We observed an accumulation of cells in G1 (Fig. 5B, right panel) in the Rep78-GFP-expressing subpopulation of pRep78-GFP-transfected cells (73.9% of cells in G1, versus 43.3% in pRep78-GFP-transfected, non-Rep78-GFP-expressing cells) that was accompanied by a reduction of cell numbers in the S and G2/M phases.
FIG. 5.
Rep78 expression results in an accumulation of cells in the G1 phase of the cell cycle. Twelve hours after transfection of HL-60 cells with pNLS-GFP (A) or pRep78-GFP (B), the cell cycle distribution was analyzed by staining for DNA content with the dye Hoechst 33342. Diagrams on the right show cells [GFP(+)] which have been gated for GFP expression (i.e., NLS-GFP in panel A and Rep78-GFP in panel B, whereas those on the left [GFP(−)] show data for GFP-negative cells. Dead cells detected by ViaProbe staining were excluded from this analysis.
Rep78-induced apoptosis is cell cycle specific.
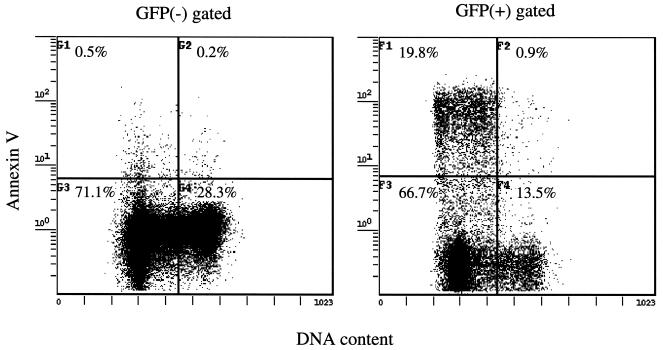
To determine whether the observed Rep78-induced accumulation of cells in G1 was due to cell cycle arrest at the G1/S checkpoint or to a selective killing of cells in the S or G2/M phase of the cell cycle, we analyzed whether apoptosis upon Rep78 expression was cell cycle specific. pRep78-GFP-transfected HL-60 cells were analyzed 12 h posttransfection for apoptosis by Annexin V staining and, simultaneously, for DNA content by incubation with the dye Hoechst 33342 (44). Figure 6 shows that apoptotic cells accumulated in the G1 and early S phases of Rep78-GFP-expressing cells. The observed accumulation of cells in G1 after Rep78 expression cannot be explained by a selective killing of cells in G2; a Rep78-induced cell cycle block at the G1/S checkpoint therefore appears more likely to be the cause of accumulation of cells in G1.
FIG. 6.
Rep78-induced apoptosis is G1/early-S-phase specific. HL-60 cells were transfected with pRep78-GFP. Twelve hours after transfection, cells were analyzed for DNA content by staining with the dye Hoechst 33342 (x axis) and for apoptosis by Annexin V staining (y axis). Cells analyzed were gated for expression of Rep78-GFP [GFP(+); right panel) or nonexpression [GFP(−); left panel). Cells in the left quadrants of the panels are in G1 or early S phase, and cells in the right quadrants are in late S or G2/M phase of the cell cycle. The upper quadrants of each panel represent apoptotic cells, while the lower quadrants show nonapoptotic cells. Dead cells were excluded from this analysis by costaining with ViaProbe. Data shown are for cells which had been gated for size, granularity, and exclusion of ViaProbe.
The ATPase activity of Rep78 is critical for the cytotoxic effect.
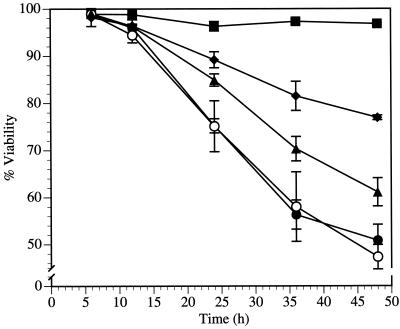
The biochemical properties of Rep78 include DNA binding, endonuclease, ATPase, and helicase activities. To investigate the contributions of these activities to cytotoxicity, we analyzed the abilities of a series of Rep mutants to induce cell death (Fig. 7). The tyrosine residue at position 156 of Rep78 is necessary for transesterification, and a phenylalanine substitution at this position causes a deficiency in DNA strand cleavage (55). Transfection with pRep78(Y156F)-GFP resulted in no change in cytotoxicity compared to wild-type Rep78-GFP, indicating that DNA cleavage does not contribute significantly to Rep78-induced cytotoxicity. Rep52-GFP, lacking the 224 N-terminal residues of Rep78 including the DNA binding domain, showed an intermediate level of cytotoxicity, while a disruption of the nucleoside triphosphate binding domain of Rep78 in pRep78(K340H)-GFP which resulted in the loss of ATPase and helicase activities (56, 62) was most efficient at suppressing the cytotoxicity of Rep78, although complete abrogation of cell death was not observed. These results are summarized in Table 1. Rep-GFP expression was analyzed by Western blot analysis and by monitoring the GFP fluorescence intensity. Rep78-GFP, Rep78(K340H)-GFP, and Rep78(Y156F)-GFP were expressed at similar levels, while the level of expression of Rep52-GFP was found to be significantly higher (about twofold) than that of Rep78-GFP (data not shown).
FIG. 7.
Mutation of the ATP binding site of Rep78 reduces cytotoxicity. The viability of pNLS-GFP-, pRep78-GFP-, pRep78(Y156F)-GFP-, or pRep78(K340H)-GFP-transfected HL-60 cells was assayed by PI staining (50 μg/ml) and flow cytometry. Data are means ± standard errors of values from three experiments. ■, pNLS-GFP; ●, pRep78-GFP; ○, pRep78(Y156F)-GFP; ⧫, pRep78(K340H)-GFP; ▴, pRep52-GFP.
TABLE 1.
Activities and cytotoxicity of Rep78 and of Rep78 mutants
| Rep protein | Activity expresseda
|
Cytotoxicityb | |||
|---|---|---|---|---|---|
| DNA binding | Endonuclease | ATPase | Helicase | ||
| Rep78 | Y | Y | Y | Y | +++ |
| Rep78(Y156F) | Y | N | Y | Y | +++ |
| Rep78(K340H) | Y | Y | N | N | + |
| Rep52 | N | N | Y | Y | ++ |
Y, activity present; N, no activity.
+, moderate cytotoxicity; ++, intermediate cytotoxicity; +++, high-level cytotoxicity.
Induction of apoptosis by Rep78 is not confined to HL-60 cells and is independent of p53.
The data presented so far demonstrate that Rep78 induces apoptosis in HL-60 cells, a p53-null promyeloid cell line (15). To determine whether this ability is confined to this cell type, we analyzed the effect of Rep78 expression on NT2 cells, a wild-type p53-containing embryonal carcinoma cell line (9). Annexin V staining of pRep78-GFP-transfected cells revealed that 43% of Rep78-GFP-expressing NT2 cells underwent apoptosis by 24 h posttransfection (Fig. 8A). A time course experiment showed that the percentage of Rep78-expressing cells undergoing apoptosis between 10 to 24 h posttransfection increased from 17% to about 40%, while cells transfected with pNLS-GFP showed only background levels of Annexin V-positive cells (Fig. 8B). Apoptosis and viability correlated inversely. Apoptosis in Rep78-GFP-expressing NT2 cells resulted in cell death. These results demonstrate that Rep78 induces apoptosis in cells of different lineage, independently of p53.
FIG. 8.
Rep78 induces apoptosis in NT2 cells. NT2 cells were transfected with pNLS-GFP or pRep78-GFP and analyzed for apoptosis by Annexin V staining. (A) Representative data for cells at 24 h posttransfection. The x axis represents the green fluorescence intensity (i.e., NLS-GFP or Rep78-GFP). The fluorescence intensity of Annexin V-PE is represented on the y axis. The populations in the upper quadrants show apoptotic cells, while cells in the lower quadrants are nonapoptotic. Cells were gated for the exclusion of ViaProbe (i.e., dead cells). (B) Time course of the experiment shown in panel A.
DISCUSSION
In this study, we demonstrated that AAV Rep78 is cytotoxic and cytostatic. Cell death is induced by apoptosis, independently of p53 and mediated by caspase-3. Rep78 expression was also accompanied by an accumulation of cells in the G1 phase of the cell cycle.
The majority of experiments analyzing the cytotoxicity of Rep78 have been done with inducible stable cell lines which either require a toxic heavy metal for induction of expression (65–67) or have shown low-level expression (25). We established a new system to study the effects of Rep78 on the cell by fusing the ORF of the autofluorescent peptide GFP to Rep78. This allowed tracking of Rep78 expression on a single-cell level. Using this system, we were able to demonstrate that Rep78 is highly cytotoxic: 50% of Rep78-expressing cells died within 48 h posttransfection. Cell death can be caused by two different pathways, apoptosis and necrosis (38). Apoptosis, or programmed cell death, is an active, highly regulated process characterized by morphological and biochemical changes. In an early event, an outer-membrane leaflet inversion leads to exposure of PS. Nuclear and cytoplasmic condensation is followed by fragmentation of the cell and breakdown of the nuclear membrane (57, 60). Necrosis is characterized by a rapid loss of membrane integrity and cell swelling followed by a collapse of the cell. In vivo, apoptotic cell death is reported to be noninflammatory while necrotic death is typically accompanied by inflammation (41). In HL-60 cells, a p53-null promonocyte cell line, we observed a correlation between Rep78 expression and detection of the apoptosis-specific loss of membrane asymmetry. Apoptosis in Rep78-expressing cells was detectable very early after transfection, and its level increased over time. During the same time period we noticed an increase of nonviable Rep78-expressing cells. Induction of apoptosis by Rep78 was not restricted to the p53-negative cell line HL-60 but was also observed in the wild-type p53-containing embryonal carcinoma cell line NT2. These results demonstrate that Rep78 is cytotoxic, inducing apoptosis independently of p53.
In this study we addressed the characteristics and molecular mechanism of Rep-induced cell death. Caspases are involved in the transduction and amplification of apoptotic signals from different pathways as well as in the execution of cell death. We analyzed the function of caspase-3 in Rep-induced apoptosis. Caspase-3 is a terminal caspase which is activated by a variety of different stimuli and is involved in the execution of cell death via cleavage of critical cellular proteins such as poly(ADP-ribose) polymerase, lamins, and gelsolin (19). Expression of Rep78 in HL-60 cells resulted in the activation of caspase-3. Furthermore, incubation of Rep78-expressing cells with the peptidic caspase-3 inhibitor Z-DEVD-FMK efficiently inhibited apoptosis. These findings demonstrate that Rep78 induces apoptosis by a caspase-3-dependent pathway. The involvement of caspase-3 activation in the apoptosis induced by infection with HIV (2), adenovirus (13) and Sendai virus (6) has also been shown.
Recent studies suggest that apoptosis and the cell cycle are connected (7, 22). Apoptotic stimuli affect both cell proliferation and death (30). We therefore analyzed the effect of Rep78 on cell cycle progression. We observed an accumulation of cells in the G1 phase upon Rep78 expression compared to nontransfected or control cells and a decrease of cells in S and G2/M. Rep78-induced apoptosis was shown to be specific for cells in the G1 and early S phases of the cell cycle by staining of Rep-expressing cells simultaneously with Hoechst 33342 for DNA content and with Annexin V for detection of apoptosis. This result indicates that Rep-induced apoptosis may be dependent on factors specific for these cell cycle phases. It also implies that the accumulation of Rep78-expressing cells in G1 is due to a cell cycle block at the G1/S checkpoint rather than to a selective killing of cells in S and G2. The observed effects of Rep78 on cell survival and cell cycle progression in this transient system differ from the data obtained with stable Rep-inducible cell lines. Holscher et al. (25) described a stable HeLa cell line expressing Rep under the control of the glucocorticoid-responsive mouse mammary tumor virus promoter. Upon induction, no cytotoxic or antiproliferative effect of Rep was detected. Yang et al. (65) described 293-based cell lines that express the AAV Rep proteins under the control of an inducible mouse metallothionein transcription promoter. Upon induction with heavy metals, an accumulation of Rep-expressing cells in S phase was observed, but no toxicity was reported. However, Rep enhanced the toxicity of UV irradiation and incubation with cadmium (66, 67). These differences in the effect of Rep on the cell may be due to the cell types used for the studies, expression efficiencies of the rep gene, and adaptation of the cell lines to background levels of Rep expression or alterations in the genomic rep gene. 293 cells contain a colinear segment from human adenovirus type 5, from nt 1 to 4344, which is integrated into chromosome 19 (19q13.2) (37). This segment contains the E1A-E1B region of the adenovirus genome. The E1A gene products bind to key elements such as members of the pRB family, inducing unscheduled cell cycle progression. The E1B-55K protein was shown to bind and inhibit p53, while E1B-19K acts as a Bcl-2 homolog in inhibiting apoptosis (3). HeLa cells are a cervical-carcinoma-derived cell line containing multiple copies of integrated human papillomavirus (HPV) type 18 DNA (40). HPV type 18 encodes regulators of cell cycle progression and apoptosis which act similarly to the adenovirus-encoded E1 proteins (3). The viral gene products of both cell lines are likely to modulate the effect of Rep on the cell cycle and apoptosis and may explain the differences observed in the different systems. In addition, an effect of the inducers of the stable Rep-expressing cell lines on cell growth and death is possible. Yang et al. used zinc and cadmium to induce Rep expression. Cadmium is a highly toxic heavy metal that has been shown to induce distortion of the cell cycle (48) and apoptosis by causing oxidative stress and DNA damage (1); zinc, in contrast, is efficient at inhibiting apoptosis (39, 58). The transient Rep expression system described in this study may be advantageous for profiling the effects of Rep on the cell compared to the stable Rep-expressing cell line, since no chemical inducers are required and alterations of cell physiology due to background levels of Rep expression and clonal selection can be excluded.
Our findings show an interesting analogy between the effects of AAV-2 Rep78 and the nonstructural protein (NS-1) of the autonomous parvoviruses on the cell. Both proteins possess cytostatic and cytotoxic potential. Cell death caused by NS-1 expression was induced by apoptosis (10) which was mediated by and dependent on caspase-3 (42). The cytostatic potential of NS-1 correlated with an accumulation of cells in G2 (46) and a block in cellular DNA replication which was proposed to be a consequence of NS-1-induced DNA damage (47).
Apoptosis can be induced by a variety of stimuli, including DNA damage, nutrient deprivation, hypoxia, and cytokines of the tumor necrosis factor family (8, 63). In addition, virus infection may trigger apoptosis of the infected cell (50). Several viral gene products are associated with apoptosis induction; adenovirus E1A, simian virus 40 large T antigen, and HPV type 18 E7 protein mediate apoptosis by inducing unscheduled DNA synthesis (59). HIV-1 gp120 and Sindbis virus E2 cause programmed cell death by receptor signaling, and human T-cell leukemia virus type 1 Tax causes apoptosis via transcriptional dysregulation of the cell (20). In addition, oxidative stress and overload of the endoplasmic reticulum as a consequence of viral replication have been shown to be inducers of cell death (20, 50). Apoptosis induced by viruses can be seen as a mechanism for viral release while limiting the inflammatory and immune responses (59) or as an innate immune response of the cell to the viral infection (14). We started to determine the mechanism of Rep-induced apoptosis. The biochemical activities of Rep are characterized as ATPase, helicase, endonuclease, ligase, and DNA binding protein. These functions are required for viral DNA replication, integration, and gene regulation. In addition, Rep has been shown to interact with the cellular proteins SP-1, PC4, p53, and high-mobility group nonhistone protein 1 (4, 16, 24, 61). Rep78 and Rep52 bind and inhibit protein kinases PKA and PrKX (12, 17). The mechanism of Rep-mediated cell death may involve any one or a combination of Rep's biochemical activities and interactions with cell proteins. In this study, by analyzing the cytotoxicity of Rep mutants, we examined the biochemical activities of Rep that contribute to cell death. The mutation of the endonuclease activity in pRep78(Y156F)-GFP resulted in no change in toxicity compared to that of wild-type Rep78. This indicates that Rep78 is not inducing apoptosis directly by causing DNA damage. Rep52, which lacks the 224 N-terminal residues of Rep78, is an ATP-dependent helicase but lacks specific DNA binding activity; its toxicity was 23% lower than that of p78, while the mutation of the nucleotide binding site in pRep78(K340H)-GFP resulted in a 53% lower level of induction of cell death. The mutation in Rep78(K340H) resulted in a loss of ATPase and helicase activities, while the specific DNA binding activity and the endonuclease activity were not affected. Taken together, these results suggest that Rep78 induces apoptosis by multiple mechanisms. The major determinant of toxicity appears to be linked to the ability of Rep to hydrolyze ATP. The ATPase activity of Rep has been shown to be constitutively active and independent of a DNA substrate (68). Overexpression of Rep may therefore lead to a partial depletion of the cellular ATP pool, an occurrence that is known to trigger apoptosis (21). The helicase activity may adversely affect DNA synthesis or transcription. Interestingly, the toxicity of the related NS-1 protein of human parvovirus B19 has been demonstrated to be dependent on ATPase activity as well (43). The remaining toxicity of Rep78(K340H) suggests the existence of an additional ATPase- and helicase-independent mechanism of induction of cell death which appears to be coupled to the specific DNA binding and gene regulation domain of Rep. Further studies will analyze the mechanism and pathway of Rep-induced apoptosis in more detail.
ACKNOWLEDGMENTS
We thank Martha Kirby for expert assistance with flow cytometry and helpful discussions and Richard Smith for a critical review of the manuscript.
REFERENCES
- 1.Bagchi D, Joshi S S, Bagchi M, Balmoori J, Benner E J, Kuszynski C A, Stohs S J. Cadmium- and chromium-induced oxidative stress, DNA damage, and apoptotic cell death in cultured human chronic myelogenous leukemic K562 cells, promyelocytic leukemic HL-60 cells, and normal human peripheral blood mononuclear cells. J Biochem Mol Toxicol. 2000;14:33–41. doi: 10.1002/(sici)1099-0461(2000)14:1<33::aid-jbt5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Banki K, Hutter E, Gonchoroff N J, Perl A. Molecular ordering in HIV-induced apoptosis. Oxidative stress, activation of caspases, and cell survival are regulated by transaldolase. J Biol Chem. 1998;273:11944–11953. doi: 10.1074/jbc.273.19.11944. [DOI] [PubMed] [Google Scholar]
- 3.Barry M, McFadden G. Apoptosis regulators from DNA viruses. Curr Opin Immunol. 1998;10:422–430. doi: 10.1016/s0952-7915(98)80116-7. [DOI] [PubMed] [Google Scholar]
- 4.Batchu R B, Shammas M A, Wang J Y, Munshi N C. Interaction of adeno-associated virus Rep78 with p53: implications in growth inhibition. Cancer Res. 1999;59:3592–3595. [PubMed] [Google Scholar]
- 5.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitzer M, Prinz F, Bauer M, Spiegel M, Neubert W J, Gregor M, Schulze-Osthoff K, Lauer U. Sendai virus infection induces apoptosis through activation of caspase-8 (FLICE) and caspase-3 (CPP32) J Virol. 1999;73:702–708. doi: 10.1128/jvi.73.1.702-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady H J, Gil-Gomez G. The cell cycle and apoptosis. Results Probl Cell Differ. 1999;23:127–144. doi: 10.1007/978-3-540-69184-6_7. [DOI] [PubMed] [Google Scholar]
- 8.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 9.Burger H, Nooter K, Boersma A W, Kortland C J, Stoter G. Expression of p53, Bcl-2 and Bax in cisplatin-induced apoptosis in testicular germ cell tumour cell lines. Br J Cancer. 1998;77:1562–1567. doi: 10.1038/bjc.1998.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chejanovsky N, Carter B J. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology. 1989;171:239–247. doi: 10.1016/0042-6822(89)90531-x. [DOI] [PubMed] [Google Scholar]
- 12.Chiorini J A, Zimmermann B, Yang L, Smith R H, Ahearn A, Herberg F, Kotin R M. Inhibition of PrKX, a novel protein kinase, and the cyclic AMP-dependent protein kinase PKA by the regulatory proteins of adeno-associated virus type 2. Mol Cell Biol. 1998;18:5921–5929. doi: 10.1128/mcb.18.10.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiou S K, White E. Inhibition of ICE-like proteases inhibits apoptosis and increases virus production during adenovirus infection. Virology. 1998;244:108–118. doi: 10.1006/viro.1998.9077. [DOI] [PubMed] [Google Scholar]
- 14.Clouston W M, Kerr J F. Apoptosis, lymphocytotoxicity and the containment of viral infections. Med Hypotheses. 1985;18:399–404. doi: 10.1016/0306-9877(85)90107-0. [DOI] [PubMed] [Google Scholar]
- 15.Collins S J. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood. 1987;70:1233–1244. [PubMed] [Google Scholar]
- 16.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Pasquale G, Stacey S N. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J Virol. 1998;72:7916–7925. doi: 10.1128/jvi.72.10.7916-7925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubielzig R, King J A, Weger S, Kern A, Kleinschmidt J A. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J Virol. 1999;73:8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earnshaw W C, Luis L M, Martins M, Kaufmann S H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 20.Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7:160–165. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 21.Feldenberg L R, Thevananther S, del Rio M, de Leon M, Devarajan P. Partial ATP depletion induces Fas- and caspase-mediated apoptosis in MDCK cells. Am J Physiol. 1999;276:F837–F846. doi: 10.1152/ajprenal.1999.276.6.F837. [DOI] [PubMed] [Google Scholar]
- 22.Gil-Gomez G, Berns A, Brady H J. A link between cell cycle and cell death: Bax and Bcl-2 modulate Cdk2 activation during thymocyte apoptosis. EMBO J. 1998;17:7209–7218. doi: 10.1093/emboj/17.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermonat P L. The adeno-associated virus Rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 24.Hermonat P L, Santin A D, Batchu R B. The adeno-associated virus Rep78 major regulatory/transformation suppressor protein binds cellular Sp1 in vitro and evidence of a biological effect. Cancer Res. 1996;56:5299–5304. [PubMed] [Google Scholar]
- 25.Hölscher C, Hörer M, Kleinschmidt J A, Zentgraf H, Bürkle A, Heilbronn R. Cell lines inducibly expressing the adeno-associated virus (AAV) rep gene: requirements for productive replication of rep-negative AAV mutants. J Virol. 1994;68:7169–7177. doi: 10.1128/jvi.68.11.7169-7177.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 27.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 29.Khleif S N, Myers T, Carter B J, Trempe J P. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology. 1991;181:738–741. doi: 10.1016/0042-6822(91)90909-u. [DOI] [PubMed] [Google Scholar]
- 30.King K L, Cidlowski J A. Cell cycle regulation and apoptosis. Annu Rev Physiol. 1998;60:601–617. doi: 10.1146/annurev.physiol.60.1.601. [DOI] [PubMed] [Google Scholar]
- 31.Koopman G, Reutelingsperger C P, Kuijten G A, Keehnen R M, Pals S T, van Oers M H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- 32.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 33.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laughlin C A, Tratschin J D, Coon H, Carter B J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 35.Laughlin C A, Westphal H, Carter B J. Spliced adenovirus-associated virus RNA. Proc Natl Acad Sci USA. 1979;76:5567–5571. doi: 10.1073/pnas.76.11.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loo D T, Rillema J R. Measurement of cell death. Methods Cell Biol. 1998;57:251–264. doi: 10.1016/s0091-679x(08)61583-6. [DOI] [PubMed] [Google Scholar]
- 37.Louis N, Evelegh C, Graham F L. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology. 1997;233:423–429. doi: 10.1006/viro.1997.8597. [DOI] [PubMed] [Google Scholar]
- 38.McConkey D J. Biochemical determinants of apoptosis and necrosis. Toxicol Lett. 1998;99:157–168. doi: 10.1016/s0378-4274(98)00155-6. [DOI] [PubMed] [Google Scholar]
- 39.Meerarani P, Ramadass P, Toborek M, Bauer H C, Bauer H, Hennig B. Zinc protects against apoptosis of endothelial cells induced by linoleic acid and tumor necrosis factor alpha. Am J Clin Nutr. 2000;71:81–87. doi: 10.1093/ajcn/71.1.81. [DOI] [PubMed] [Google Scholar]
- 40.Meissner J D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 41.Melcher A, Gough M, Todryk S, Vile R. Apoptosis or necrosis for tumor immunotherapy: what's in a name? J Mol Med. 1999;77:824–833. doi: 10.1007/s001099900066. [DOI] [PubMed] [Google Scholar]
- 42.Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol. 1998;72:3018–3028. doi: 10.1128/jvi.72.4.3018-3028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momoeda M, Wong S, Kawase M, Young N S, Kajigaya S. A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity. J Virol. 1994;68:8443–8446. doi: 10.1128/jvi.68.12.8443-8446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore A, Donahue C J, Bauer K D, Mather J P. Simultaneous measurement of cell cycle and apoptotic cell death. Methods Cell Biol. 1998;57:265–278. doi: 10.1016/s0091-679x(08)61584-8. [DOI] [PubMed] [Google Scholar]
- 45.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 46.Op De Beeck A, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995;6:781–787. [PubMed] [Google Scholar]
- 47.Op De Beeck A, Caillet-Fauquet P. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J Virol. 1997;71:5323–5329. doi: 10.1128/jvi.71.7.5323-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Payette Y, Lachapelle M, Daniel C, Bernier J, Fournier M, Krzystyniak K. Decreased interleukin-2 receptor and cell cycle changes in murine lymphocytes exposed in vitro to low doses of cadmium chloride. Int J Immunopharmacol. 1995;17:235–246. doi: 10.1016/0192-0561(95)00001-i. [DOI] [PubMed] [Google Scholar]
- 49.Redemann B E, Mendelson E, Carter B J. Adeno-associated virus Rep protein synthesis during productive infection. J Virol. 1989;63:873–882. doi: 10.1128/jvi.63.2.873-882.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roulston A, Marcellus R C, Branton P E. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 51.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. . (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid I, Krall W J, Uittenbogaart C H, Braun J, Giorgi J V. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 53.Shapiro H M. Flow cytometry of DNA content and other indicators of proliferative activity. Arch Pathol Lab Med. 1989;113:591–597. [PubMed] [Google Scholar]
- 54.Siegl G, Bates R C, Berns K I, Carter B J, Kelly D C, Kurstak E, Tattersall P. Characteristics and taxonomy of Parvoviridae. Intervirology. 1985;23:61–73. doi: 10.1159/000149587. [DOI] [PubMed] [Google Scholar]
- 55.Smith R H, Kotin R M. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J Virol. 2000;74:3122–3129. doi: 10.1128/jvi.74.7.3122-3129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 58.Sunderman F W., Jr The influence of zinc on apoptosis. Ann Clin Lab Sci. 1995;25:134–142. [PubMed] [Google Scholar]
- 59.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaux D L, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weger S, Wendland M, Kleinschmidt J A, Heilbronn R. The adeno-associated virus type 2 regulatory proteins Rep78 and Rep68 interact with the transcriptional coactivator PC4. J Virol. 1999;73:260–269. doi: 10.1128/jvi.73.1.260-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyllie A H, Kerr J F, Currie A R. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 64.Yang Q, Chen F, Ross J, Trempe J P. Inhibition of cellular and SV40 DNA replication by the adeno-associated virus Rep proteins. Virology. 1995;207:246–250. doi: 10.1006/viro.1995.1072. [DOI] [PubMed] [Google Scholar]
- 65.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou C, Trempe J P. Induction of apoptosis by cadmium and the adeno-associated virus Rep proteins. Virology. 1999;261:280–287. doi: 10.1006/viro.1999.9882. [DOI] [PubMed] [Google Scholar]
- 67.Zhou C, Yang Q, Trempe J P. Enhancement of UV-induced cytotoxicity by the adeno-associated virus replication proteins. Biochim Biophys Acta. 1999;1444:371–383. doi: 10.1016/s0167-4781(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 68.Zhou X, Zolotukhin I, Im D-S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]