Abstract
Background
Monitoring genotypes of HIV infections in blood donors may provide insights into infection trends in the general population.
Methods
HIV RNA was extracted from plasma samples of blood donors confirmed as HIV positive by blood screening nucleic acid and antibody tests. HIV genome target regions were amplified using nested real time-polymerase chain reaction followed by next-generation sequencing. Sequences were compared to those in the Los Alamos National Laboratory (LANL) database. Sequences were also assessed for drug resistance mutations (DRM) using the Stanford HIV DRM Database.
Results
From available HIV-positive donations collected between 1 September 2015 and 31 December 2020, 563 of 743 (75.8%) were successfully sequenced; 4 were subtype A, 543 subtype B, 5 subtype C, 1 subtype G, 5 circulating recombinant forms (CRF), and 2 were subtype B and D recombinants. Overall, no significant differences between blood donor and available LANL genotypes were found, and the genotypes of newly acquired versus prevalent HIV infections in donors were similar. The proportion of non-B subtypes and CRF remained a small fraction, with no other subtype or CRF representing more than 1% of the total. DRM were identified in 122 (21.6%) samples with protease inhibitor, nucleoside reverse transcriptase inhibitor and non-nucleoside reverse transcriptase inhibitor DRMs identified in 4.9%, 4.6% and 14.0% of samples, respectively.
Conclusions
HIV genetic diversity and DRM in blood donors appear representative of circulating HIV infections in the US general population and may provide more information on infection diversity than sequences reported to LANL, particularly for recently transmitted infections.
Keywords: blood donors, blood screening, genetic sequence analysis, HIV, phylogenetics
The Transfusion Transmissible Infections Monitoring System (TTIMS) was established by the Food and Drug Administration (FDA), National Heart Lung and Blood Institute of the National Institutes of Health, and the Office of the Assistant Secretary for Health of the US Department of Health and Human Services to evaluate bloodborne infections in blood donors and changes in risk of transfusion transmission of those infections to blood recipients over time [1]. Reported rates of infections in donors from the TTIMS program show an overall HIV prevalence of 2.6 per 100 000 donations in 2015–2019, with nucleic acid test (NAT)-only RNA-reactive donations, representing very recently acquired infections, of 0.05 per 100 000 donations or approximately 2% [2]. The TTIMS program also estimates HIV incidence based on new infections in repeat donors and the ratio of NAT-only reactive donations in repeat to first-time donors to estimate overall HIV incidence as well as using other incidence estimation methods [3]. For the period 2015–2019, overall incidence declined from 4.09 per 100 000 person-years to 2.96 per 100,000 person-years following the adoption of the 12-month deferral for men who have sex with men [4]. Although prevalence and incidence rates of HIV infections in blood donors are low compared to the general population, molecular characterization and surveillance of infections in blood donors following routine donation testing is valuable to monitor for genetic changes in HIV [5]. Transmitted or acquired mutations and recombinant viruses could impact detection by blood donor molecular and serological screening assays, with false-negative results leading to increased risk of transmission to recipients [6]. In addition, molecular surveillance of infections in blood donors in the TTIMS program may provide public health insights into trends in the genetic evolution of these infections in the broader US population. Studies of the phylogenetic similarities, including drug resistance mutation (DRM) patterns, among HIV in donors compared to those reported for the general population are rare; such studies can highlight changes in the epidemic, particularly as treatment and prevention practices and blood donor eligibility policies evolve to be more inclusive of all members of the population [7, 8]. Here, we evaluate HIV phylogenetic relationships and DRM prevalence in HIV-positive donations collected by 4 large US blood collection organizations over a 5-year period.
METHODS
The TTIMS program is a collaborative multicenter research group comprised of 4 large US blood centers (American Red Cross, New York Blood Center, OneBlood, and Vitalant) and government agencies. The program established procedures for the collection and storage of donation samples that have tested confirmed positive for known transfusion-transmissible infections [9]. The TTIMS risk factor coordinating center includes a laboratory and biorepository component that allows for investigation of donor and infection characteristics in addition to molecular surveillance of HIV infections.
Recently acquired infections, at the time of the blood donation, were defined as either confirmed NAT-reactive, serology negative, or confirmed NAT-reactive and serology reactive with recent antibody seroconversion. We used the LAg avidity assay to identify recently acquired infections by measuring antibody avidity (which matures during the early months of HIV infection) based on a limiting concentration of recombinant HIV-1 antigen [3]. LAg avidity normalized optical density results <2.0 triggered triplicate retesting, and the median value of triplicate repeat tests constituted the final result. For this analysis, normalized optical density values ≤1.5 were considered recently acquired HIV infections and are estimated to have a mean duration of recent infection of approximately 130 days for subtype B infections [10].
For sequence analysis, HIV RNA was extracted from plasma samples of blood donors confirmed as HIV positive by blood screening NAT and antibody tests, and the target region was amplified using nested real time-polymerase chain reaction (PCR). The primers used in PCR were chosen to amplify the 1712–2986 nt range of the reference genome (NC_001802), which includes the protease and part of the reverse transcriptase genes of HIV [5]. The samples run in next-generation sequencing libraries were selected based on the presence of a positive band of the expected size on gel electrophoresis. The Illumina libraries were produced using the transposon based Nextera XT Sample Preparation Kit with 15 PCR cycles (Illumina, San Diego, CA, USA). The DNA library concentrations were assessed using the Quant-iT DNA HS Assay Kit. Subsequently, libraries consisting of multiple samples were combined at equal concentrations and size-selected within the range of 300–1000 bp using the Pippin Prep system (Sage Science, Beverly, MA, USA). After pooling, quantification of the library was performed using the KAPA library quantification kit for the Illumina platform (Kapa Biosystems, Wilmington, MA, USA). The library, with a concentration of 10 picomolar, was loaded onto the MiSeq sequencing platform for 2 × 250 cycles pair-end sequencing, incorporating dual barcoding. For the genome assembly, raw data obtained from next-generation sequencing were mapped using the HIV reference genome using the Geneious R10 program. First, low sensitivity assembly was used and after the contigs were obtained the raw data were reassembled using the high sensitivity setting.
Multiple sequence alignment was performed using the MUSCLE algorithm (Molecular Evolutionary Genetics Analysis software v11) [11] to identify evolutionary relationships and common patterns between the HIV sequences from donors sequences and among persons with HIV in the broader US population. Sequences for the broader US population were derived from the Los Alamos Laboratory National (LANL) HIV sequence library [12, 13] using the following criteria: from the period 2015–2020, from the United States, subtype B, and including polymerase (pol) coding region sequence data. Recombinants were excluded and only 1 sequence per individual in LANL was used. LANL results were sorted alpha-numerically by accession, so the selected sequence was the first one accessioned in alpha-numeric order. The sequence distance matrix, obtained from pairwise sequence comparison, was computed using the maximum composite likelihood method, assuming an equality of substitution pattern among lineages [14]. The phylogeny, which illustrates the lines of evolutionary descent of different strains from a common ancestor, was reconstructed based on the multiple sequence alignment using the maximum likelihood method in the Tamura-Nei model using Molecular Evolutionary Genetics Analysis [15].
Phylogenetic trees were constructed to depict evolutionary relationships among the donor and LANL sequences [16]. Sequences relative to the broader population of persons with HIV, as well as clustering by various donor characteristics captured by TTIMS, including if the infection was recently acquired, sex, race, age group, US Census Bureau regions (Northeast, Midwest, South, and West), and year of donation, were evaluated to assess evolutionary relatedness. Some groups were combined where small numbers limited ability to conduct meaningful comparisons. We collapsed race/ethnicity groups into White, Black, and other including Hispanic, and collapsed the census regions into South or non-South.
Two statistical tests were employed for testing population substructures of HIV infection. Kruskal-Wallis tests were conducted for population diversity derived from the genetic distance matrix. The genetic diversity for a sample is defined as the mean genetic distance within each subgroup (eg, male or female, race) and visualized as violin plots. The genetic distances were compared across different groups using the Kruskal-Wallis 1-way analysis of variance test for population diversity. Slatkin-Maddison tests were performed for population segregation within the phylogenetic tree by permutation tests using HYPHY 2.5.8 [17]. The Slatkin-Maddison test employs simulation to estimate the minimum number of migration events using maximum parsimony and then evaluates whether this number is lower than would be expected to be found randomly in an unstructured population. The input for this test is a phylogenetic tree with leaf names partitioned into sets of group characteristics (eg, male or female, race).
HIV DRMs in all sequenced samples (B and non-B subtypes) were identified using the Stanford HIV Drug Resistance Database [18]. Phylogenetics, DRM prevalence, specific mutations, and drug sensitivities were evaluated. DRM types were compared between all infections over time and between prevalent and recently acquired HIV infections. Statistical comparisons between demographic and other group characteristic were conducted using standard parametric tests with P values adjusted to account for multiple comparisons.
Patient Research Consent
Blood donors are not patients. Individual donor consent was not required because each donor provides consent for use of donation samples and donor information in research related to blood supply safety as part of consenting to donate blood. This study was approved by Ethics Committees, known as institutional review boards (IRB) in the US, of the participating blood centers. Vitalant and OneBlood obtained approval from the University of California San Francisco IRB, the IRB of record for Vitalant Research Institute. American Red Cross and New York Blood Center Enterprises IRBs also approved this study.
RESULTS
Of 905 confirmed HIV-positive blood donations from 1 September 2015 through 31 December 2020 donated at the participating blood centers, 743 samples were available for analysis and 563 (75.8%) were successfully real-time PCR amplified and sequenced (available in a Supplemenatry Date file). Raw data mapping to reference genomes showed 100% coverage to the targeted genomic regions. By donation screening test classification, 10 of 16 (62.5%) RNA+/Ab− (NAT-only donations) and 553 of 691 (80.0%) RNA+/Ab+ (concordant positive) samples were successfully sequenced (Table 1). Of the concordant RNA+/Ab+ samples, 143 were classified as recently acquired infections using LAg avidity testing. Thus, 153 of 563 (27.2%) sequenced samples represent newly acquired HIV at the time of donation. Among all successfully genotyped samples, 4 were HIV subtype A, 543 subtype B, 5 subtype C, 1 subtype G, 5 circulating recombinant forms (CRFs), and 2 were subtype B and D recombinants (Table 2). The distribution of subtypes of newly acquired and long-standing HIV infections were not different, with >96% subtype B infections in both groups. The proportions of successfully sequenced samples among donors by demographic and donation characteristics were similar, although donors between the ages of 16 and 24 had a higher proportion of successfully sequenced donations (Table 3).
Table 1.
Blood Screening Results and Samples Available for Testing and Successfully Sequenced for the Period 1 September 2015 to 31 December 2020
| HIV Donation Screening Classification | Donations Submitted for Molecular Surveillance Testing | Donations Successfully Sequenced N (%) | |
|---|---|---|---|
| NAT Reactive/serology Negative | 18 | 16 | 10 (62.5) |
| Concordant NAT and serology reactive | 841 | 691 | 553 (80.0) |
| LAg recent | 191 | 179 | 143 (79.9) |
| viral load negative or low-level NAT/serology positive | 46 | 36 | 0 |
| Total | 905 | 743 | 563 (75.8) |
Abbreviation: NAT, nucleic acid test.
Table 2.
Genetic Subtype of Sequenced HIV Infections From Blood Donors by Duration of Infections at Time of Donation, 2015–2020
| Subtype | Overall Frequency N = 563 n (%) |
Recently Acquired/Incident N = 153 n (%) |
Longstanding/Prevalent N = 410 n (%) |
|---|---|---|---|
| A | 3 (0.5) | 1 (0.7) | 2 (0.5) |
| A1 | 1 (0.2) | - | 1 (0.2) |
| B | 543 (96.5) | 149 (97.4) | 394 (96.1) |
| B-like | 2 (0.4) | 1 (0.7) | 1 (0.2) |
| BD_recombinant | 2 (0.4) | - | 2 (0.5) |
| C | 5 (0.9) | 2 (1.3) | 3 (0.7) |
| CRF01_AE | 3 (0.5) | - | 3 (0.7) |
| CRF18_cpx | 1 (0.2) | - | 1 (0.2) |
| CRF19_cpx | 1 (0.2) | - | 1 (0.2) |
| CRF20_BG | 1 (0.2) | - | 1 (0.2) |
| G | 1 (0.2) | - | 1 (0.2) |
| Total | 563 (100) | 153 (27.2) | 410 (72.8) |
Table 3.
Characteristics of Blood Donors With HIV With Sequenced and not Sequenced Samples
| Characteristic | Group | Sequenced N = 563 n (%) |
Not Sequenced N = 342 n (%) |
Total N = 905 n (%) |
|---|---|---|---|---|
| Sex | Male | 456 (81.0) | 284 (83.0) | 740 (81.8) |
| Female | 107 (19.0) | 58 (17.0) | 165 (18.2) | |
| Age | 16–24 | 211 (37.5) | 98 (28.7) | 309 (34.1) |
| 25–34 | 150 (26.6) | 86 (25.1) | 236 (26.1) | |
| 35–44 | 91 (16.2) | 72 (21.1) | 163 (18.0) | |
| 45–54 | 70 (12.4) | 50 (14.6) | 120 (13.3) | |
| 55–64 | 34 (6.0) | 32 (9.4) | 66 (7.3) | |
| 65+ | 7 (1.2) | 4 (1.2) | 11 (1.2) | |
| Race/ethnicity | American Indian | 2 (0.4) | 6 (1.8) | 8 (0.9) |
| Asian | 13 (2.3) | 7 (2.0) | 20 (2.2) | |
| Black | 217 (38.5) | 138 (40.4) | 355 (39.2) | |
| Hispanic/Latino | 106 (18.8) | 57 (16.7) | 163 (18.0) | |
| More than 1 race | 4 (0.7) | 9 (2.6) | 13 (1.4) | |
| White | 203 (36.1) | 116 (33.9) | 319 (35.2) | |
| Other | 4 (0.7) | 4 (1.2) | 8 (0.9) | |
| Not available | 14 (2.5) | 5 (1.5) | 19 (2.1) | |
| HHS census regiona | Midwest | 55 (9.8) | 45 (13.2) | 100 (11.0) |
| Northeast | 44 (7.8) | 35 (10.2) | 79 (8.7) | |
| South | 400 (71.1) | 205 (59.9) | 605 (66.9) | |
| West | 63 (11.2) | 56 (16.4) | 119 (13.1) | |
| Not available | 1 (0.2) | 1 (0.3) | 2 (0.2) | |
| Year | 2015b | 32 (5.7) | 25 (7.3) | 57 (6.3) |
| 2016 | 114 (20.2) | 63 (18.4) | 177 (19.6) | |
| 2017 | 134 (23.8) | 58 (17.0) | 192 (21.2) | |
| 2018 | 124 (22.0) | 51 (14.9) | 175 (19.3) | |
| 2019 | 108 (19.2) | 73 (21.3) | 181 (20.0) | |
| 2020 | 51 (9.1) | 72 (21.5) | 123 (13.5) | |
| Donation history | First-time | 394 (70.0) | 228 (66.7) | 612 (67.6) |
| Repeat | 169 (30.0) | 114 (33.3) | 293 (32.4) |
Not sequenced includes those samples in which amplification was unsuccessful and infections where plasma was not available for sequencing.
aCensus region.
b1 September 2015–31 December 2015.
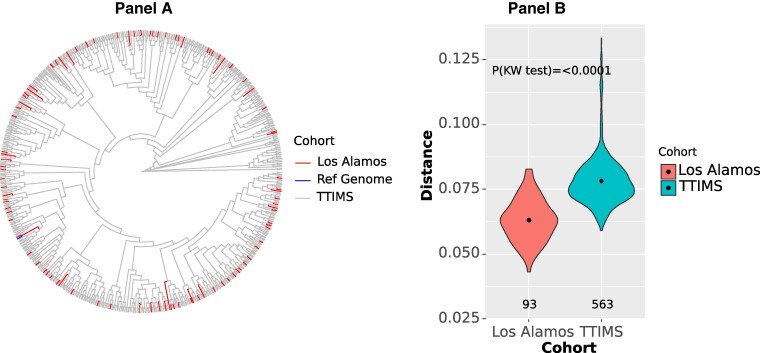
Restricted to subtype B infections, comparing the phylogenetic relationships of donor infections and the reference infections obtained from LANL (Figure 1), LANL and TTIMS populations did not show significant segregation, meaning both sets of sequences come from the same underlying HIV-infected population. However, the 93 available LANL reference sequences covering the same portion of the HIV genome and period were significantly less genetically diverse than the TTIMS sequences (Kruskal-Wallis (KW) P < .001), (Supplementary Table 1). This indicates that more diverse sequences, likely because of the total number samples, but not genotypes are evident in the blood donor samples compared to those we identified for the reference sequence population in LANL.
Figure 1.
Only subtype B infections are included. (A) Phylogenetic tree comparison of HIV of protease and reverse transcriptase gene sequences from the TTIMS programs and selected LANL reference sequences from 2015–2020. (B) Mean genetic distances comparing TTIMS sequences and LANL references sequences. The violin plot and results of the Kruskal-Wallis (KW) 1-way analysis of variance test show the diversity of the TTIMS HIV sequences are significantly greater than the diversity of available reference sequences from LANL. Reference Genome: GenBank ID K03455.1 Human immunodeficiency virus type 1 (HXB2) complete genome HIV1/HTLV-III/LAV reference genome.
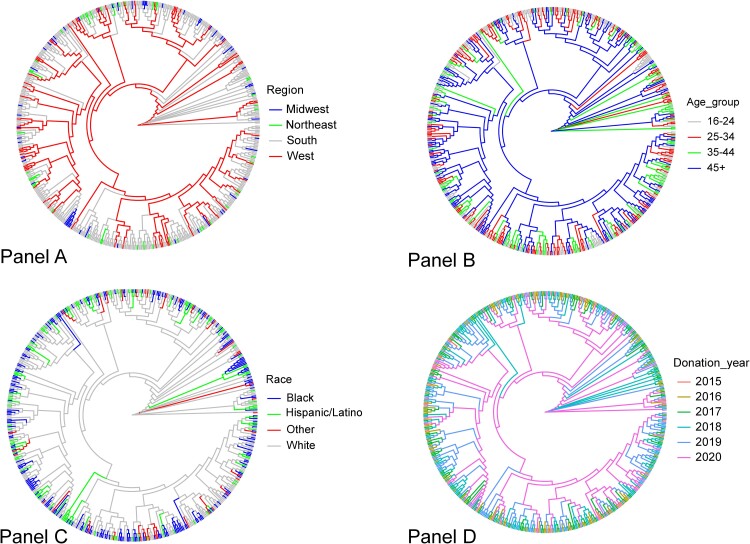
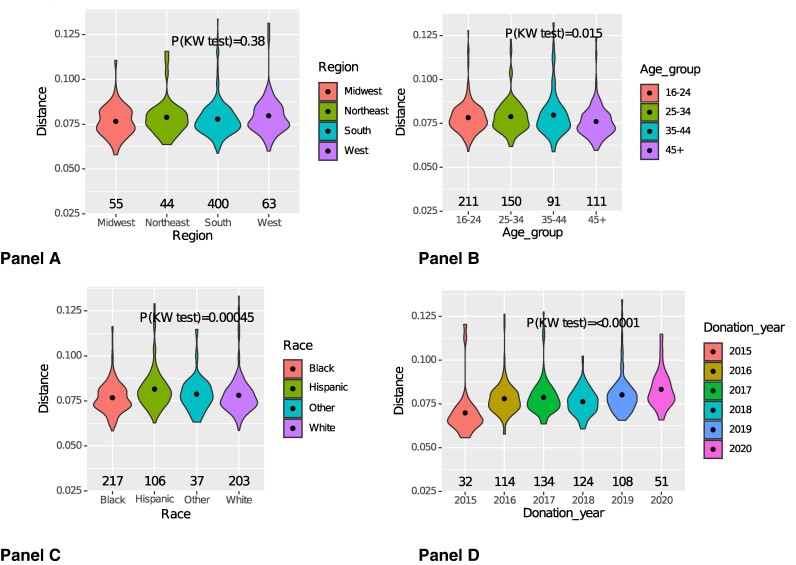
The tree-based Slatkin-Maddison analyses showed no significant segregation of donor sequences compared to LANL sequences from the same period, nor demographic grouping (age group, sex, race/ethnicity, region of the country, donation year, or recently acquired infection status). Some regional clustering of HIV infections was observed, including strong suggestion of local clustering from the Midwest region (Figure 2). Restricted to the blood donor sequences, genetic distance-based tests showed that age, race/ethnicity, and donation year were associated with sequence diversity, but region of the country, donor sex, and recently acquired infection were not (Figure 3, Supplementary Table 1, and Supplementary Figure 1). Older donors (>45 years) had lower sequence diversity than younger donors, and donations in 2020 had higher sequence diversity than those from earlier donations years.
Figure 2.
Only subtype B infections are included. Phylogenetic tree comparison of HIV protease and reverse transcriptase gene sequences from the TTIMS program according to donor demographic and donation characteristics from 2015–2020. (A) Census region, (B) age group, (C) race/ethnicity, and (D) donation year.
Figure 3.
Only subtype B infections are included. Mean genetic distances of HIV protease and reverse transcriptase gene sequences from the TTIMS program according to donor demographic and donation characteristics from 2015–2020. Violin plots and results of the Kruskal-Wallis (KW) 1-way analysis of variance test show significant differences in genetic diversity by race/ethnicity, age, and donation year. (A) Census region, (B) age group, (C) race/ethnicity, (D) donation year.
DRMs were identified in 121 (21.5%) of all sequenced samples (Table 4 and Supplementary Tables 2–5) with PI, nucleoside reverse transcriptase inhibitor (NRTI), and non-NRTI (NNRTI) DRMs identified in 5.0%, 4.6%, and 13.9% of sequenced samples, respectively; 9 samples (1.6%) and 1 sample (0.2%) had DRMs to 2 and 3 drug classes, respectively. The most prevalent DRM was K103N (8.3%), followed by V106I (1.6%), M41L (1.6%), and E138A (1.2%); 2 samples (0.4%) had M184V, including 1 that also had K65R. Among the 153 recently acquired infections, DRMs were less frequent overall (17.5%) and for individual classes (3.9%, 1.9%, and 12.3% for PI, NRTI, and NNRTI DRMs, respectively) for recently acquired/incident infections although these differences were not statistically significant. The most notable finding was evidence of reduced prevalence of DRMs for the NNRTI drugs efavirenz and nevirapine in recently acquired compared to long-standing/prevalent HIV infections. No other DRMs were significantly different between prevalent and recently acquired HIV.
Table 4.
HIV Drug Resistance Mutations (DRM) in Blood Donors by Year (N = 563 HIV Sequences)
| DRM | 2015 N = 32 (%) |
2016 N = 114 (%) |
2017 N = 134 (%) |
2018 N = 124 (%) |
2019 N = 108 (%) |
2020 N = 51 (%) |
Overall N = 563 (%) |
Recently Acquired/Incident N = 153 (%) |
Prevalent N = 410 (%) |
|---|---|---|---|---|---|---|---|---|---|
| Any drug class | 8 (25.0) | 20 (17.5) | 22 (16.4) | 27 (21.8) | 31 (28.7) | 13 (25.5) | 121 (21.5) | 27 (17.6) | 94 (22.9) |
| 1 drug class | 7 | 18 | 19 | 26 | 30 | 11 | 111 (19.7) | 26 (17.0) | 85 (20.7) |
| 2 drug classes | 1 | 2 | 2 | 1 | 1 | 2 | 9 (1.6) | 1 (0.7) | 8 (2.0) |
| 3 drug classes | 0 | 0 | 1 | 0 | 0 | 0 | 1 (0.2) | 0 | 1 (0.2) |
| PI | 2 | 4 | 7 | 8 | 5 | 2 | 28 (5.0) | 6 (3.9) | 22 (5.4) |
| NRTI | 1 | 4 | 7 | 7 | 4 | 3 | 26 (4.6) | 3 (2.0) | 23 (5.6) |
| NNRTI | 6 | 14 | 12 | 13 | 23 | 10 | 78 (13.9) | 19 (12.4) | 59 (14.4) |
Abbreviations: NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor .
DISCUSSION
In this study, we sequenced pol regions from >550 HIV-infected donation-derived plasma samples collected from US blood donors between 2015 and 2020. The distribution of HIV subtypes, with >96% subtype B infections and few CRFs identified, were consistent with previously reported US blood donor sequence analyses from earlier times [5, 6, 19, 20]. The proportion of non-B subtypes and CRFs among the donor samples remained a small fraction, with no other subtype or CRFs representing more than 1% of the total sample.
Our results among blood donors are consistent with the known epidemiology and genetic diversity of HIV infections reported in specific areas of the country. For example, our findings parallel those recently reported for San Francisco, California, where non-B and CRF variants remain a small proportion (3%) of all HIV [21]. In addition, among HIV subtype B, we found evidence of strong HIV genetic similarity within small clusters of specific demographic groups. These results match recent analyses of sequencing data generated as part of routine clinical care in the mid-Atlantic region, where males, younger individuals, and those with recently acquired HIV, were more likely to represent clusters of infection [22]. All these findings are consistent with studies reporting that HIV diversity is associated with assortativity (selective mixing) within race/ethnicity groups and geography in the larger US population [23–25]. Note, in our study, as a group, “Other including Hispanic” donors had higher sequence diversity than Black and White donors, but this diversity is attributable to our post hoc grouping of Hispanic and non-White, non-Black groups together because of their small numbers.
Unlike stable subtype diversity, the overall DRM prevalence of >20% in HIV infections was higher during 2015–2020 compared to early periods (9.1% during 1999–2007; 12% during 2006–2009). The prevalence of DRMs overall and for individual drug classes are comparable to those from public health surveillance data from similar periods. Among 28 states during 2014–2018, of 50 747 persons with HIV, 9616 (18.9%) had ≥1 transmitted drug resistance-associated mutation, but although patterns of specific DRMs may vary by year, overall the rate of DRM did not change [26], consistent with our findings. Over longer periods, DRM may be increasing. Continued monitoring of HIV DRMs in blood donors could complement genotyping efforts for people with HIV at entry to care in informing appropriate population-level treatment and prevention strategies.
With a few caveats, the DRM patterns among blood donors in this study are less concerning than reported for specific populations. From 4 sites during 2016–2017 of 142 individuals in a cohort including 92 treatment-experienced individuals who were not virally suppressed, 44 (30.9%) had high-level resistance to at least 1 antiretroviral (ARV) drug [27]. Similarly, for Washington, DC, and Maryland from 1987 to 2015, of 1995 individuals (including 1005 on antiretroviral therapy), 37.9% had DRMs [28, 29]. Blood donor data may provide a somewhat more balanced assessment of the trends in DRM in persons with HIV in the United States because most blood donors are not aware of their infection status and hence likely not on ARVs with the DRM mostly representing transmitted (rather than selected) resistance. However, when HIV-positive donor samples are tested for ARV in their plasma, approximately 15% have been identified as taking ARVs at the time of donation [30]. Donors who are taking ARVs may represent cases of either acquired or selected resistance. The samples successfully sequenced had sufficient RNA levels for successful amplification and so would not include those individuals who had achieved control of viremia using antiretroviral therapy.
The findings from this study suggest that HIV infections from blood donors are broadly representative of circulating HIV infections in the United States and may even provide a better snapshot of the overall diversity of infections and DRMs in the US population than the sequences thus far reported to LANL. Interestingly, we had hypothesized that recently acquired HIV among donors might differ genetically than from prevalent infections, but the results do not support this hypothesis. The phylogenetic diversity of subtype B infections were not different for these 2 groups. We believe this finding indicates the HIV strains that are circulating within the US population did not substantially change over the relatively short period of this study (2015–2020). We do not know the times of acquisition for donors with prevalent infections. We speculate that these infections were mostly acquired in the 10-year period before 2015. Thus, we interpret the lack of diversity between recently acquired and prevalent HIV infections to mean during the 15-year period when most infections included in this study would have been acquired, there were not substantial changes in the phylogenetics of HIV in the US general population.
The study has limitations. First, a highly mutated HIV infection may not be identified with the NAT and serological assays used for blood donation screening. Such hypothetical infections would have been missed by donation testing and would not be included as HIV infections in the TTIMS biorepository and thus not sequenced. If a highly modified recombinant HIV infection was in a donated unit, its potentially higher level of genetic diversity would not be evident or accounted for in our study. However, documented cases of transfusion-transmitted HIV are rare, with the last known case in the United States reported in 2007, and these historical transfusion-transmitted infections were from seronegative donors with very low HIV RNA levels in the early infection window period and not attributed to viral variants [31]. Second, we did not sequence the int nor env region of the HIV genome and so do not know if any of the sequenced samples included DRMs for drugs that target the integrase or envelope glycoprotein. Third, a subset of HIV-positive samples could not be PCR amplified. This was most likely because of very low viral loads, but some samples with measurable viral loads were still not amplified. We do not know the reasons for unsuccessful amplification nor if these infections have similar sequences to those samples that were successfully sequenced. Because 75% of samples were successfully sequenced, we assume the sequenced samples are representative of the HIV infections in US blood donors during the study period. Fourth, although 48 states are represented (no samples from Hawaii or Alaska were available in the TTIMS repository), the available samples do not include HIV from all geographic locations or blood donors in the United States during the study period.
The sequence analyses reported here further demonstrate that, even though donors are selected for donation based on health status questions, HIV in blood donors can provide public health insights on the broader HIV epidemic including subtype distributions and DRM patterns. Subtype B infections remain the predominant types of infection found in the United States. Our results are consistent with other publications reporting sequence data from similar years showing that non-B variants are not widely established in the United States. When non-B variants are identified, they are likely acquired outside the United States, and these variants have not yet been established in US HIV transmission networks. The findings also indicate the US blood donor likely share the same transmission networks as the larger population of persons with HIV, and thus provide generally representative molecular surveillance information on the HIV epidemic in the United States.
Supplementary Material
Contributor Information
Brian Custer, Vitalant Research Institute, San Francisco, California, USA; Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, USA.
Eda Altan, Vitalant Research Institute, San Francisco, California, USA.
Leilani Montalvo, Vitalant Research Institute, San Francisco, California, USA.
Alison Coyne, Vitalant Research Institute, San Francisco, California, USA.
Eduard Grebe, Vitalant Research Institute, San Francisco, California, USA.
Xutao Deng, Vitalant Research Institute, San Francisco, California, USA; Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, USA.
Mars Stone, Vitalant Research Institute, San Francisco, California, USA; Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, USA.
Eric Delwart, Vitalant Research Institute, San Francisco, California, USA; Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, USA.
Sonia Bakkour, Global Medical Affairs Donor Screening, Grifols Diagnostic Solutions, Emeryville, California, USA.
Benyam Hailu, Division of Blood Diseases and Resources, National Heart, Lung, and Blood Institute, National Institutes of Health, Rockville, Maryland, USA.
Rita Reik, Scientific, Medical, Technical and Research, OneBlood, St. Petersburg, Florida, USA.
Debra Kessler, Medical Programs and Services, New York Blood Center, New York, New York, USA.
Susan L Stramer, Infectious Disease Consultant, North Potomac, Maryland, USA.
Michael P Busch, Vitalant Research Institute, San Francisco, California, USA; Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, USA.
for the Transfusion Transmissible Infections Monitoring System (TTIMS) Program:
E Notari, S Stramer, R Dodd, G Conti, R Fayed, D Nelson, R Townsend, G Foster, J Haynes, E Crawford, E Huseynova, D Krysztof, D Burke, M Lanteri, V Green, S Cyrus, P Williamson, D Kessler, J Gorlin, L Milan-Benson, C DelValle, P Chien, T Brown, R Reik, C Shea, M Lopez, K Richards, T Foster, J Brodsky, M Barr, T Rains, B Custer, R Bruhn, E Grebe, M Busch, M Stone, C Di Germanio, D Hindes, Z Kaidarova, K Zurita, A Tadena, L Montalvo, A Dayana, S Hughes, M Townsend, M Bravo, J Vannoy, S Fallon, S Anderson, B Whitaker, H Yang, A Belov, A Eder, B Hailu, S Zou, J Berger, Brian Custer, Susan Stramer, Debra Kessler, Rita Reik, Phillip Williamson, Steven A Anderson, and Benyam Hailu
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful for the funding support for the Transfusion Transmissible Infections Monitoring System (TTIMS) from the Food and Drug Administration (FDA), the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health (NIH), and the Office of the Assistant Secretary of Health, Health and Human Services (OASH-HHS) via Contract HHSF223201610043I. We are also grateful for the study guidance provided by these agencies.
We acknowledge those individuals who contributed to this study from the TTIMS-Donation and Donor Coordinating Center (DDCC), the TTIMS-Laboratory and Risk Factor Coordinating Center (LRCC), Creative Testing Solutions, and our federal partners without whom this study and program would not be successful.
Author contributions. B.C. designed the study, obtained funding, participated in the interpretation of results, and drafted and finalized the manuscript. E.A. conducted laboratory sequence analyses and results interpretation and provided intellectual content to the draft manuscript. L.M. conducted laboratory sequence and viral load analyses. A.C., E.G., and X.D. conducted statistical, phylogenetic, and drug resistance analyses and provided intellectual content to the manuscript. M.S., E.D., and S.B. provided oversight of laboratory work, results interpretation, and intellectual content to the manuscript. B.H. provided study oversight and intellectual content to the manuscript. R.R., D.K., and S.L.S. provided oversight of study locations and intellectual content to the manuscript. M.P.B. contributed to the study design and results interpretation and provided intellectual content to the manuscript.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the views of the US Food and Drug Administration, National Institutes of Health (NHLBI/NIH), or the United States government, Office of the Assistant Secretary for Health.
Blood center and testing organizations. The American Red Cross: E. Notari, S. Stramer, R. Dodd, G. Conti, R. Fayed, D. Nelson, R. Townsend, G. Foster, J. Haynes, E. Crawford, E. Huseynova, D. Krysztof, D. Burke, Rockville, MD. Creative Testing Solutions: M. Lanteri, V. Green, S. Cyrus, P. Williamson, Tempe, AZ. The New York Blood Center Enterprises: D. Kessler, J. Gorlin, L. Milan-Benson, C. DelValle, P. Chien and T. Brown, New York, NY. OneBlood: R. Reik, C. Shea, M. Lopez, K. Richards, T. Foster, St. Petersburg, FL. Quality Analytics, Inc.: J. Brodsky, M. Barr, T. Rains, Riverwoods, IL. Vitalant Research Institute: B. Custer, R. Bruhn, E. Grebe, M. Busch, M. Stone, C. Di Germanio, D. Hindes, Z. Kaidarova, K. Zurita, A. Tadena, L. Montalvo, A. Dayana, S. Hughes, San Francisco, CA. Vitalant: M. Townsend, M. Bravo, J. Vannoy, S. Fallon, and our other dedicated donor counselors, Scottsdale, AZ.
Federal partners. FDA: S. Anderson, B. Whitaker, H. Yang, A. Belov, A. Eder, Silver Spring, MD. NHLBI NIH: B. Hailu and S. Zou, Bethesda, MD. OASH-HHS: J. Berger, Washington, DC. At the time that the study was conducted, the Transfusion Transmissible Infections Monitoring System 2 (TTIMS2) was the responsibility of the following individuals: Brian Custer, PhD, MPH: Vitalant Research Institute, San Francisco, CA. Susan Stramer, PhD, MS: American Red Cross, Rockville, MD. Debra Kessler, RN, MS: New York Blood Center Enterprises, New York, NY. Rita Reik, MD and Tisha Foster, MD: OneBlood, St. Petersburg, FL. Phillip Williamson, PhD: Creative Testing Solutions, Tempe, AZ. Steven A. Anderson, PhD, MPP: US Food and Drug Administration, Silver Spring MD. Benyam Hailu, MD, MPH: National Institutes of Health, Bethesda, MD.
Financial support. This work was conducted under contract HHSF223201610043I from the US FDA Center for Biologics Evaluation and Research, the National Institutes of Health/National Heart, Lung and Blood Institute (funding provided to the US FDA by interagency agreement Y01 AHL15003001-1-0-1), and the Health and Human Services (HHS) Office of the Assistant Secretary for Health.
References
- 1. Custer B, Stramer SL, Glynn S, Williams AE, Anderson SA. Transfusion-transmissible infection monitoring system: a tool to monitor changes in blood safety. Transfusion 2016; 56:1499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steele WR, Dodd RY, Notari EP, et al. Prevalence of human immunodeficiency virus, hepatitis B virus, and hepatitis C virus in United States blood donations, 2015 to 2019: the transfusion-transmissible infections monitoring system (TTIMS). Transfusion 2020; 60:2327–39. [DOI] [PubMed] [Google Scholar]
- 3. Grebe E, Busch MP, Notari EP, et al. HIV incidence in US first-time blood donors and transfusion risk with a 12-month deferral for men who have sex with men. Blood 2020; 136:1359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steele WR, Dodd RY, Notari EP, et al. HIV, HCV, and HBV incidence and residual risk in US blood donors before and after implementation of the 12-month deferral policy for men who have sex with men. Transfusion 2021; 61:839–50. [DOI] [PubMed] [Google Scholar]
- 5. Delwart E, Slikas E, Stramer SL, et al. Genetic diversity of recently acquired and prevalent HIV, hepatitis B virus, and hepatitis C virus infections in US blood donors. J Infect Dis 2012; 205:875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Delwart E, Kuhns MC, Busch MP. Surveillance of the genetic variation in incident HIV, HCV, and HBV infections in blood and plasma donors: implications for blood safety, diagnostics, treatment, and molecular epidemiology. J Med Virol 2006; 78(Suppl 1):S30–5. [DOI] [PubMed] [Google Scholar]
- 7. U.S. Department of Health and Human Services . Food and Drug Administration, Center for Biologics Evaluation and Research. Revised recommendations for reducing the risk of human immunodeficiency virus transmission by blood and blood products—guidance for industry. Available at: https://www.federalregister.gov/d/2015-32250. Accessed 16 February 2023.
- 8. U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research . Recommendations for evaluating donor eligibility using individual risk-based questions to reduce the risk of human immunodeficiency virus transmission by blood and blood products. Available at: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/recommendations-evaluating-donor-eligibility-using-individual-risk-based-questions-reduce-risk-human. Accessed 6 August 2023.
- 9. Quiner C, Bruhn R, Grebe E, et al. Recently acquired infection among HIV-seropositive donors in the US from 2010-2018. Transfusion 2020; 60:2340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duong YT, Kassanjee R, Welte A, et al. Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 2015; 10:e0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021; 38:3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alignment of HIV-1/SIVcpz Complete Genomes, HIV Sequence Compendium. Edited by: Thomas Leitner TFB, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, 2015–2021. p. 1–145. https://hfv.lanl.gov/content/sequence/HIV/COMPENDIUM/2021compendium.html. Accessed 28 June 2023.
- 14. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 2004; 101:11030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10:512–26. [DOI] [PubMed] [Google Scholar]
- 16. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kosakovsky Pond SL, Poon AFY, Velazquez R, et al. Hyphy 2.5-A customizable platform for evolutionary hypothesis testing using phylogenies. Mol Biol Evol 2020; 37:295–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. HIV Drug Resistance Database. Stanford, California, USA: Stanford University, 2022. [Google Scholar]
- 19. Delwart EL, Busch MP, Kalish ML, Mosley JW, Mullins JI. Rapid molecular epidemiology of human immunodeficiency virus transmission. AIDS Res Hum Retroviruses 1995; 11:1081–93. [DOI] [PubMed] [Google Scholar]
- 20. Delwart EL, Orton S, Parekh B, Dobbs T, Clark K, Busch MP. Two percent of HIV-positive U.S. blood donors are infected with non-subtype B strains. AIDS Res Hum Retroviruses 2003; 19:1065–70. [DOI] [PubMed] [Google Scholar]
- 21. O'Keefe KJ, Pipkin S, Fatch R, et al. Non-B variants of HIV-1 in San Francisco, California. Infect Genet Evol 2021; 90:104677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kassaye SG, Grossman Z, Vengurlekar P, et al. Insights into HIV-1 transmission dynamics using routinely collected data in the Mid-Atlantic United States. Viruses 2022; 15:68. 10.3390/v15010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oster AM, Pieniazek D, Zhang X, et al. Demographic but not geographic insularity in HIV transmission among young black MSM. Aids 2011; 25:2157–65. [DOI] [PubMed] [Google Scholar]
- 24. Dennis AM, Hué S, Hurt CB, et al. Phylogenetic insights into regional HIV transmission. Aids 2012; 26:1813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ragonnet-Cronin M, Benbow N, Hayford C, et al. Sorting by race/ethnicity across HIV genetic transmission networks in three major metropolitan areas in the United States. AIDS Res Hum Retroviruses 2021; 37:784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McClung RP, Oster AM, Ocfemia MCB, et al. Transmitted drug resistance among human immunodeficiency virus (HIV)-1 diagnoses in the United States, 2014-2018. Clin Infect Dis 2022; 74:1055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fogel JM, Sivay MV, Cummings V, et al. HIV drug resistance in a cohort of HIV-infected MSM in the United States. Aids 2020; 34:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibson KM, Steiner MC, Kassaye S, et al. A 28-year history of HIV-1 drug resistance and transmission in Washington, DC. Front Microbiol 2019; 10:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibson KM, Steiner MC, Kassaye S, et al. Corrigendum: a 28-year history of HIV-1 drug resistance and transmission in Washington, DC. Front Microbiol 2019; 10:2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Custer B, Quiner C, Haaland R, et al. HIV antiretroviral therapy and prevention use in US blood donors: a new blood safety concern. Blood 2020; 136:1351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dwyre DM, Fernando LP, Holland PV. Hepatitis B, hepatitis C and HIV transfusion-transmitted infections in the 21st century. Vox Sang 2011; 100:92–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.