ABSTRACT
Transmissible gastroenteritis virus (TGEV)-induced enteritis is characterized by watery diarrhea, vomiting, and dehydration, and has high mortality in newborn piglets, resulting in significant economic losses in the pig industry worldwide. Conventional cell lines have been used for many years to investigate inflammation induced by TGEV, but these cell lines may not mimic the actual intestinal environment, making it difficult to obtain accurate results. In this study, apical-out porcine intestinal organoids were employed to study TEGV-induced inflammation. We found that apical-out organoids were susceptible to TGEV infection, and the expression of representative inflammatory cytokines was significantly upregulated upon TGEV infection. In addition, retinoic acid-inducible gene I (RIG-I) and the nuclear factor-kappa B (NF-κB) pathway were responsible for the expression of inflammatory cytokines induced by TGEV infection. We also discovered that the transcription factor hypoxia-inducible factor-1α (HIF-1α) positively regulated TGEV-induced inflammation by activating glycolysis in apical-out organoids, and pig experiments identified the same molecular mechanism as the ex vivo results. Collectively, we unveiled that the inflammatory responses induced by TGEV were modulated via the RIG-I/NF-κB/HIF-1α/glycolysis axis ex vivo and in vivo. This study provides novel insights into TGEV-induced enteritis and verifies intestinal organoids as a reliable model for investigating virus-induced inflammation.
IMPORTANCE
Intestinal organoids are a newly developed culture system for investigating immune responses to virus infection. This culture model better represents the physiological environment compared with well-established cell lines. In this study, we discovered that inflammatory responses induced by TGEV infection were regulated by the RIG-I/NF-κB/HIF-1α/glycolysis axis in apical-out porcine organoids and in pigs. Our findings contribute to understanding the mechanism of intestinal inflammation upon viral infection and highlight apical-out organoids as a physiological model to mimic virus-induced inflammation.
KEYWORDS: TGEV, apical-out intestinal organoids, inflammation, NF-κB, HIF-1α
INTRODUCTION
Transmissible gastroenteritis virus (TGEV), a member of the genus Alphacoronavirus (family Coronaviridae, order Nidovirales), is a single-stranded, positive-sense RNA virus (1). TGEV primarily attacks small intestinal epithelial cells and causes acute watery diarrhea, vomiting, dehydration, and anorexia, with high morbidity and mortality, particularly in nursing piglets (2). Enteritis caused by TGEV results in high mortality in piglets less than 2 weeks old (3), suggesting that the inflammation of the small intestine might be instrumental in the pathogenesis of TGEV infection. Wang et al. reported that TGEV nonstructural protein 2 (Nsp2) contributes to inflammation via NF-κB activation in ST cells and IPEC-J2 cells (4). However, ST cells and IPEC-J2 cells are immortalized single-cell lines, which may not reveal the actual inflammatory responses occurring in vivo. Therefore, a more physiological culture system is urgently needed for investigating TGEV-induced inflammation.
Intestinal organoids were differentiated from Lgr5+ stem cells and first reported from mice in 2009 (5). Intestinal organoids include many intestinal cell types, such as stem cells, goblet cells, Paneth cells, enterocytes, and so on, and can better mimic the real gut environment (5). Recently, our laboratory developed an apical-out porcine intestinal organoid culture system and intestinal organoid monolayer to explore virus–host interactions and found that TGEV can infect organoid models and induce immune responses effectively (6, 7). Apical-out organoids, which are a three-dimensional (3D) culture model, are reported to be more likely to benefit viral infection and the differentiation of intestinal cell types, suggesting that they are a more physiological model for exploring inflammatory responses to TGEV infection (6).
Inflammatory responses are critical effectors of host responses against pathogen invasion, but excessive inflammatory responses can be harmful (8). Coronavirus triggers inflammatory responses through a complex signal cascade (9). In detail, pattern recognition receptors (PRRs) are appointed to sense pathogen-associated molecular patterns (PAMPs) (10). PRRs related to coronavirus mainly include Toll-like receptor 3 (TLR3), Toll-like receptor 7 (TLR7), Toll-like receptor 8 (TLR8), retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated gene 5 (MDA5), which all recruit adaptor molecules to mediate the signaling cascade (11). Specifically, the Toll-like receptors recruit TIR-domain-containing adaptor-inducing IFN-β (TRIF) and myeloid differentiation primary response gene (MyD88) to activate downstream pathways. Meanwhile, RIG-I and MDA5 belong to the RIG-like receptors (RLRs), which interact with mitochondrial antiviral signaling protein (MAVS) to deliver the signal (12, 13). The production of inflammatory cytokines can be induced through the activation of various pathways such as NF-κB and mitogen-activated protein kinases (MAPK)-activating protein 1 (AP-1) pathways (14). The classical NF-κB activation cascade is initiated by stimulus-induced ubiquitinated degradation of IκBα, releasing NF-κB dimers and promoting their nuclear translocation (15). The AP-1 pathway is composed of JUN, FOS, or ATF (activating transcription factor) subunits, and activation of the pathway is characterized by the phosphorylation of JUN (16).
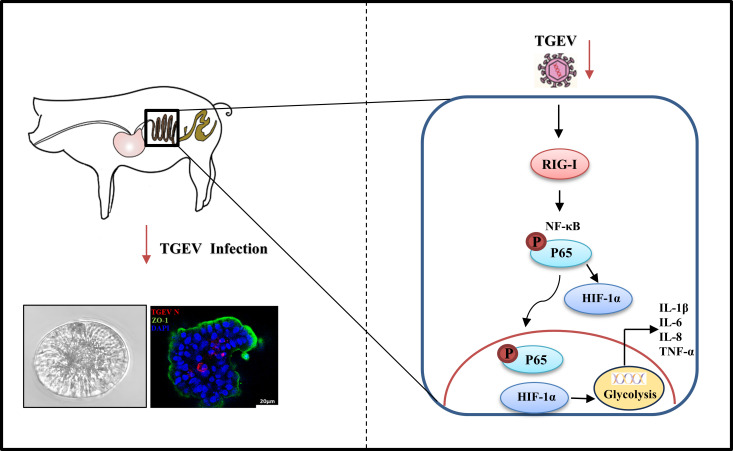
In this study, apical-out intestinal organoids were established to explore TGEV-induced inflammation. Using this model, we found that, TGEV can effectively infect apical-out organoids and induce the production of inflammatory cytokines, such as tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-1β (IL-1β), and interleukin-18 (IL-18). In addition, we found that RIG-I, but not MDA5, can positively activate the NF-κB pathway to regulate TGEV-induced inflammation. Furthermore, hypoxia-inducible factor-1α (HIF-1α) was demonstrated to regulate TGEV-induced inflammation by activating glycolysis downstream of the RIG-I–NF-κB pathway. Finally, animal experiments showed the same molecular mechanism for TGEV-induced inflammation. Collectively, TGEV induces inflammatory responses via the RIG-I/NF-κB/HIF-1α/glycolysis axis in apical-out intestinal organoids and pigs.
RESULTS
Apical-out porcine intestinal organoids are susceptible to TGEV
Porcine intestinal crypts were isolated from the intestinal follicle-associated epithelium of the ileum according to a previous protocol (6) and were cultured in Matrigel supplemented with organoid growth medium (OGM) (Fig. S1A). The formation of crypt-villus structures was observed from 1 to 5 days (Fig. S1B). To better mimic the physiological environment, apical-out intestinal organoids were established with zonula occludens-1 (ZO-1) in the outer membrane of the organoids, which means that apical-out organoids were successfully generated (Fig. S1C and D). In addition, different intestinal epithelial cell subsets, including absorptive enterocytes (Villin-positive), enteroendocrine cells (CGA-positive), stem cells (SOX9-positive), goblet cells (MUC2-positive), and Paneth cells (LYZ-positive), were successfully detected in the apical-out organoids (Fig. S1E). This finding illustrated that the apical-out organoids possessed complex multicellularity and thus were a more physiologically relevant research model.
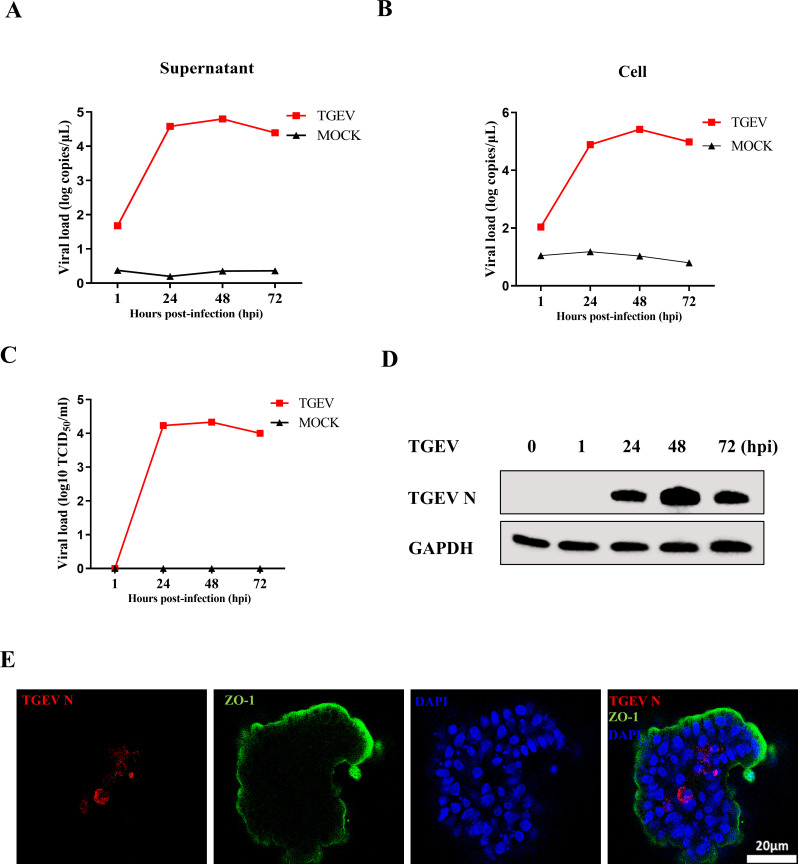
To investigate whether the apical-out intestinal organoids were valid for exploring immune responses to viral infection, TGEV was employed to infect the organoids. Reverse transcription-quantitative PCR (RT-qPCR) and a 50% tissue culture infective dose (TCID50) assay demonstrated that the apical-out organoids were susceptible to TGEV. The viral load in the supernatant and cells peaked at 48 hours post-infection (hpi) and subsequently decreased by 72 hpi (Fig. 1A and B). The viral titer was also peaked at 48 hpi (Fig. 1C). In addition, Western blotting detected the presence of TGEV N protein (Fig. 1D). Consistent with this finding, immunofluorescence assay (IFA) results also showed that the expression of TGEV N in apical-out intestinal organoids detected at 48 hpi (Fig. 1E). Collectively, these data illustrated that the apical-out porcine intestinal organoids were susceptible to TGEV.
Fig 1.
Apical-out porcine intestinal organoids are susceptible to TGEV. (A and B) Apical-out organoids were inoculated with TGEV, and organoids and supernatant were collected at the indicated times for viral load detection by RT-qPCR. (C and D) TGEV titers and N protein at the indicated times were detected by TCID50 and Western blotting, respectively. (E) Apical-out organoids infected with TGEV for 48 h were stained with TGEV N monoclonal antibody; scale bar: 20 µm. All experiments were performed in triplicate.
TGEV infection induces inflammatory responses in apical-out porcine intestinal organoids
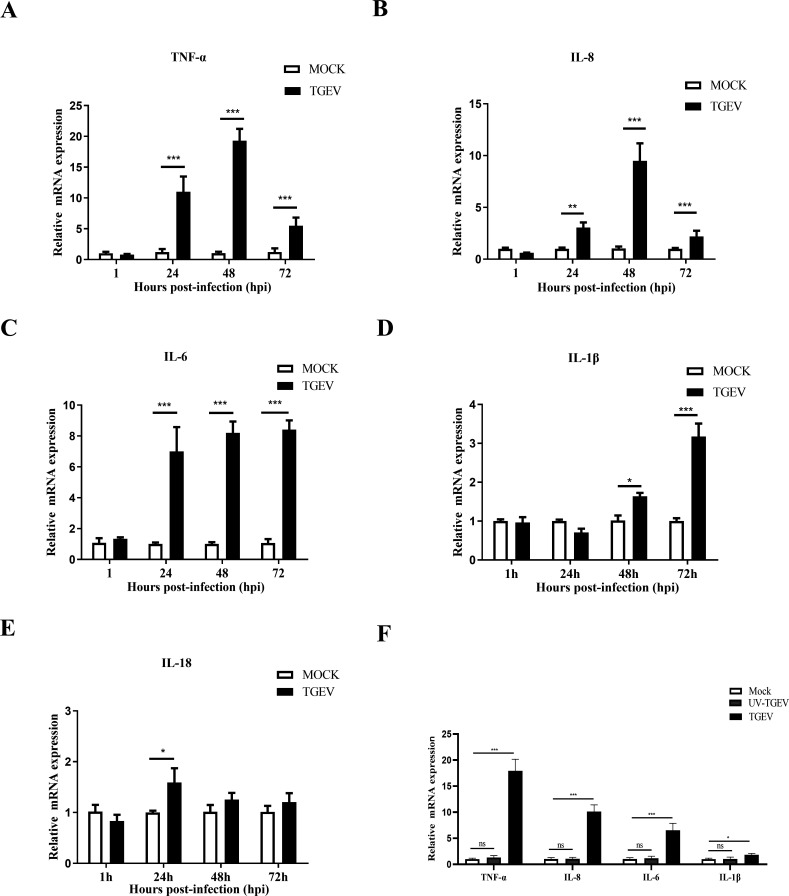
To further investigate the inflammatory responses of apical-out organoids upon TGEV infection, the transcription levels of TNF-α, IL-8, IL-6, IL-1β, and IL-18 were evaluated by RT-qPCR. TNF-α and IL-8 mRNA levels peaked at 48 hpi (Fig. 2A and B). Moreover, the TGEV infection significantly activated the transcription of IL-6 and IL-1β at 72 hpi (Fig. 2C and D). Meanwhile, the level of IL-18 mRNA was also upregulated at 24 hpi (Fig. 2E). These data demonstrated that inflammatory responses can be induced by TGEV infection in apical-out organoids.
Fig 2.
TGEV infection induces inflammatory responses in apical-out porcine intestinal organoids. (A–E) Transcription levels of TNF-α, IL-8, IL-6, IL-1β, and IL-18 at the indicated times post-TGEV infection were evaluated by RT-qPCR. (F) Transcription levels of TNF-α, IL-8, IL-6, and IL-1β at 48 h after TGEV or UV-TGEV infection were measured by RT-qPCR. Results are presented as mean ± SD of data from three independent experiments *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, determined by two-tailed Student’s t test.
Next, to determine whether these TGEV-induced inflammatory responses were caused by TGEV protein or by TGEV nucleic acid, the apical-out organoids were infected with UV-TGEV and TGEV, respectively. The results demonstrated that TGEV, but not UV-TEGV, could upregulate TNF-α, IL-8, IL-6, and IL-1β mRNA levels (Fig. 2F), indicating that TGEV protein, but not nucleic acid, induced inflammation.
TGEV infection induces inflammatory responses by RIG-I in apical-out porcine intestinal organoids
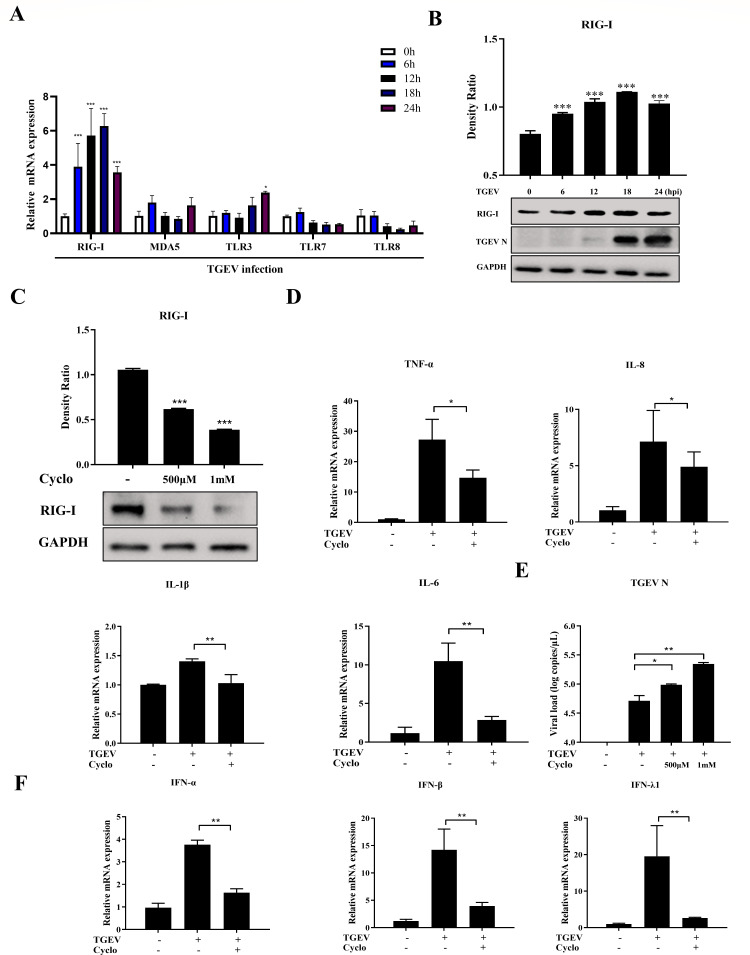
Activation of various PRRs represents the prime step of the inflammatory response to induce cytokine production. RIG-I, MDA5, TLR3, TLR7, and TLR8 have been reported to be involved in the immune response of coronavirus (11). To explore which PRRs have critical roles in recognizing viral components and inducing inflammatory response during TGEV infection in apical-out organoids, the expression of PRRs, namely RIG-I, MDA5, TLR3, TLR7, and TLR8, was analyzed using RT-qPCR. The mRNA level of RIG-I was significantly upregulated, whereas no obvious changes in the expression of other PRRs were observed (Fig. 3A; Fig. S2A). This trend was confirmed by Western blot analysis (Fig. 3B). To elucidate the function of RIG-I in TGEV-induced inflammation, Cyclo (Phe-Pro), a specific inhibitor of RIG-I activation, was added to mock- or TGEV-infected organoids (17). As expected, RIG-I activation was suppressed by Cyclo treatment (Fig. 3C), and levels of inflammatory cytokines were also reduced in Cyclo-treated organoids during TGEV infection (Fig. 3D). Furthermore, TGEV infection was enhanced by Cyclo treatment (Fig. 3E) and IFN responses were significantly decreased (Fig. 3F). Cyclo exhibited no cytotoxicity at the concentrations used in this study (Fig. S3A).
Fig 3.
TGEV infection induces inflammatory responses by RIG-I in apical-out porcine intestinal organoids. (A) Transcription levels of RIG-I, MDA5, TLR3, TLR7, and TLR8 in apical-out organoids at the indicated times post-TGEV infection were evaluated by RT-qPCR. (B) RIG-I expression in apical-out organoids at the indicated times post-TGEV infection was detected by Western blotting and calculated with ImageJ. (C) Apical-out organoids were treated with 500 µM or 1 mM Cyclo for 48 h, then RIG-I expression was measured by Western blotting and calculated with ImageJ. (D–F) Apical-out organoids were infected with TGEV followed by Cyclo treatment (1 mM) for 48 h. Subsequently, mRNA levels of TNF-α, IL-8, IL-6, and IL-1β (D), TGEV viral load (E), and mRNA levels of IFN-α, IFN-β, and IFN-λ1 (F) were determined by RT-qPCR. Results are presented as mean ± SD of data from three independent experiments *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, determined by two-tailed Student’s t test.
TGEV infection triggers inflammatory responses via the NF-κB pathway in apical-out porcine intestinal organoids
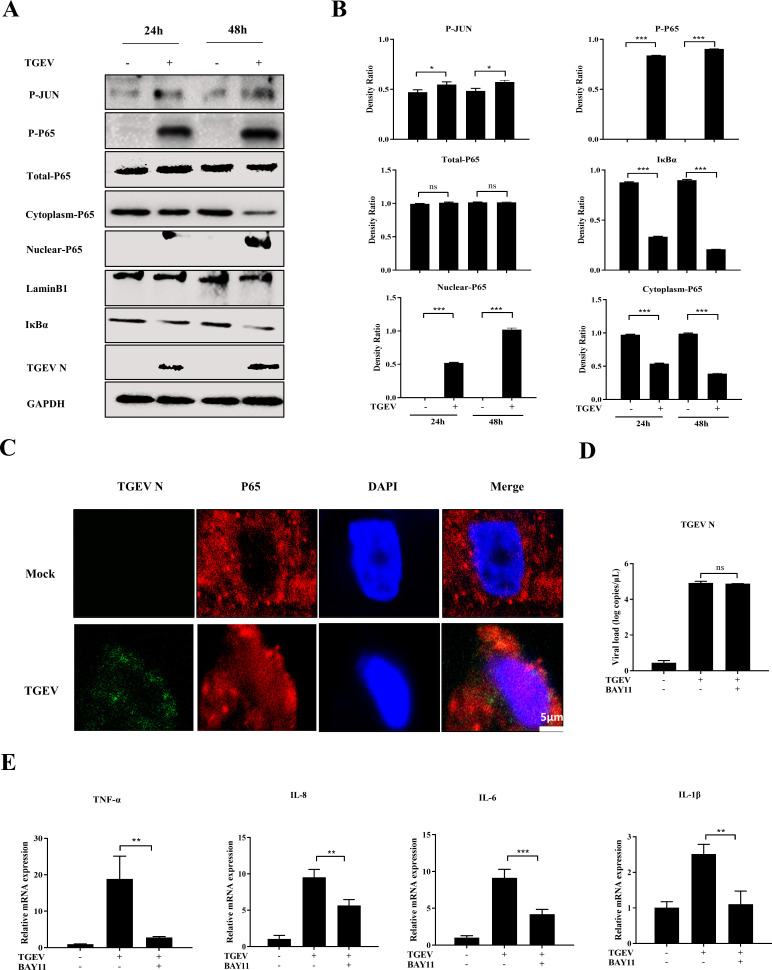
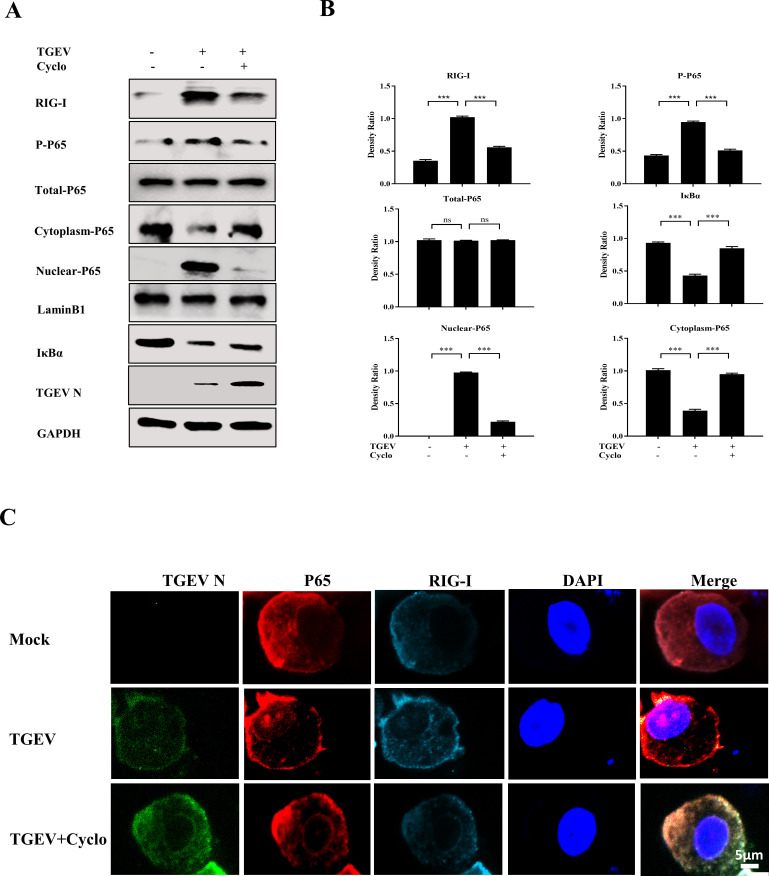
To further explore the role of key pathways of inflammatory cytokines during TGEV infection in apical-out intestinal organoids, the activation of NF-ĸB and AP-1 was determined by Western blotting to detect phosphorylation of P65 and JUN after TGEV infection. The level of phosphorylated P65 was significantly upregulated, whereas the phosphorylation of JUN was slightly increased after TGEV infection, suggesting that TGEV infection can markedly activate the NF-ĸB pathway (Fig. 4A and B). This result was further confirmed by Western blotting, which showed that nuclear P65 expression and IκBα degradation were promoted by TGEV infection (Fig. 4A and B). In addition, IFA results demonstrated that TGEV induced nuclear translocation of P65 in apical-out organoids at 48 hpi (Fig. 4C).
Fig 4.
TGEV infection triggers inflammatory responses via the NF-κB pathway in apical-out intestinal organoids. (A) Apical-out organoids were infected with TGEV for 24 and 48 h, then P-JUN, P-P65, total P65, cytoplasmic P65, nuclear P65, IκBα, and TGEV N were detected by Western blotting. (B) The density ratios of P-JUN, P-P65, total P65, cytoplasmic P65, nuclear P65, and IκBα were calculated with ImageJ. (C) TGEV-infected or mock-infected apical-out organoids at 48 h were stained with TGEV N and P65 and analyzed by confocal microscopy; scale bar: 5 µm. (D and E) Apical-out organoids were infected with TGEV followed by BAY11 (2 µM) treatment for 48 h. Subsequently, TGEV viral load (D) and mRNA levels of TNF-α, IL-8, IL-6, and IL-1β (E) were detected by RT-qPCR. Results are presented as mean ± SD of data from three independent experiments *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, determined by two-tailed Student’s t test.
To further investigate the effect of NF-ĸB activation on TGEV-induced inflammation, BAY11-7082 (BAY11), which is a specific inhibitor of NF-ĸB, was added into mock- or TGEV-infected organoids. This inhibitor did not have any effect on viral load and cytotoxicity in organoids (Fig. 4D; Fig. S3B). As expected, mRNA levels of inflammatory cytokines, including TNF-α, IL-8, IL-6, and IL-1β, were reduced in BAY11-treated organoids during TGEV infection (Fig. 4E). This finding indicated that NF-ĸB signaling plays a critical role in the TGEV-induced inflammatory response in apical-out intestinal organoids.
RIG-I controls NF-κB pathway activation upon TGEV infection in apical-out porcine intestinal organoids
PRRs have been reported to regulate the NF-κB pathway to induce inflammation (18). Therefore, we hypothesized that RIG-I controls NF-κB pathway activation after TGEV infection. To verify this hypothesis, RIG-I, the phosphorylation of P65, nuclear P65, cytoplasmic P65, and IκBα expression were measured after Cyclo treatment. Western blotting showed that when RIG-I activation was repressed by Cyclo, the phosphorylation of P65 and nuclear P65 were reduced and cytoplasmic P65 expression and IκBα degradation were restored compared to the TGEV-infected group (Fig. 5A and B). Furthermore, TGEV-induced nuclear translocation of P65 was abolished by RIG-I inhibition (Fig. 5C). These results illustrated that NF-κB pathway activation can be regulated by RIG-I upon TGEV infection.
Fig 5.
RIG-I controls NF-κB pathway activation upon TGEV infection in apical-out porcine intestinal organoids. (A) Apical-out organoids were infected with TGEV followed by Cyclo (1 mM) treatment for 48 h, then RIG-I, P-P65, total P65, cytoplasmic P65, nuclear P65, IκBα, and TGEV N were detected by Western blotting. (B) The density ratios of RIG-I, P-P65, total P65, cytoplasmic P65, nuclear P65, and IκBα were calculated using ImageJ. (C) Apical-out organoids were infected with TGEV followed by Cyclo (1 mM) treatment for 48 h, and then the organoids were stained with TGEV N, P65, and RIG-I and analyzed by confocal microscopy; scale bar: 5 µm. Results are presented as mean ± SD of data from three independent experiments ***, P ≤ 0.001; ns, not significant, determined by two-tailed Student’s t test.
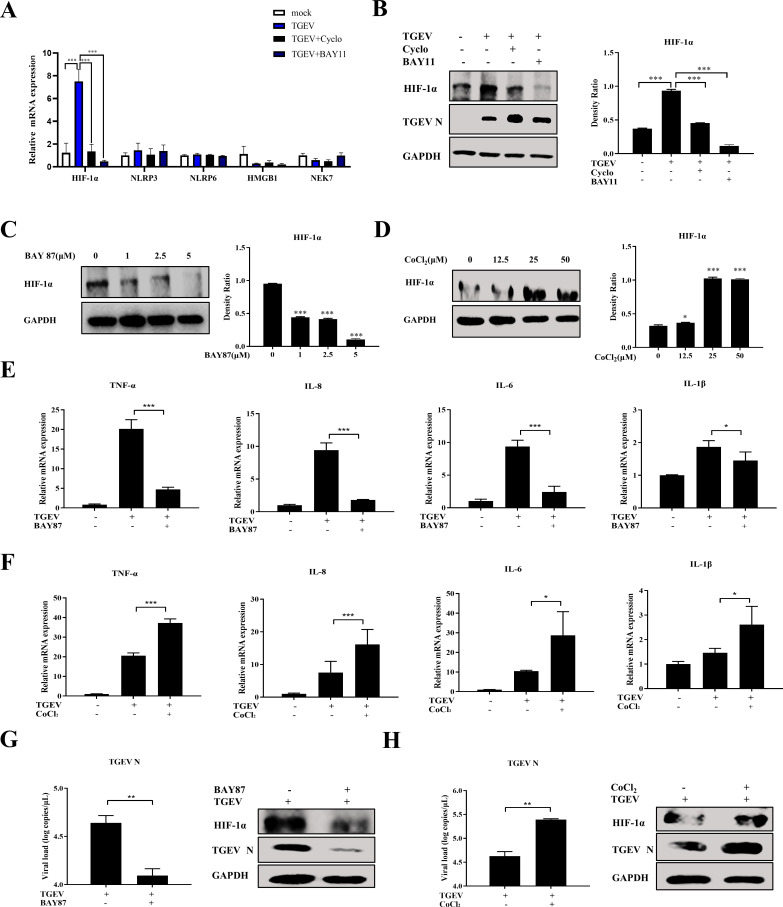
HIF-1α positively regulates TGEV-induced inflammation downstream of the RIG-I–NF-κB pathway
The above investigations demonstrated that TGEV induced inflammatory responses via the RIG-I–NF-κB pathway. We hypothesized that key factors participate in TGEV-induced inflammation downstream of the RIG-I–NF-κB pathway. This hypothesis was explored by screening some key proinflammatory proteins related to coronavirus, namely HIF-1α, NOD-like receptor family pyrin domain-containing 3 (NLRP3), NOD-like receptor family pyrin domain-containing 6 (NLRP6), high mobility group box-1 (HMGB1), and NIMA-related kinase 7 (NEK7). HIF-1α was significantly induced by TGEV infection, but inhibited after downregulation of RIG-I expression and NF-κB pathway by specific inhibitors, respectively, suggesting that the RIG-I–NF-κB pathway positively regulates HIF-1α expression upon TGEV infection (Fig. 6A and B; Fig. S4A). Those inhibitors showed no cytotoxicity at the concentrations used in this study (Fig. S3 and S4B). To further validate the role of HIF-1α on TGEV-induced inflammation, BAY 87-2243 (BAY87, a HIF-1α inhibitor) and CoCl2 (a HIF-1α agonist) were employed. HIF-1α expression was inhibited by BAY87 treatment (Fig. 6C), whereas CoCl2 treatment significantly induced HIF-1α expression (Fig. 6D) in apical-out organoids. BAY87 and CoCl2 exhibited no cytotoxicity at the concentrations used in this study (unpublished data). As expected, TGEV-induced inflammatory cytokines were reduced after BAY87 treatment (Fig. 6E), whereas the same cytokines were promoted by CoCl2 treatment (Fig. 6F), suggesting that HIF-1α positively controls TGEV-induced inflammation. In addition, pharmacological inhibition of HIF-1α could restrict TGEV infection (Fig. 6G), whereas pharmacological upregulation of HIF-1α could promote TGEV infection (Fig. 6H). Taken together, the proposed molecular mechanism is that TGEV induced inflammation via the RIG-I/NF-κB/HIF-1α axis in apical-out intestinal organoids.
Fig 6.
HIF-1α positively regulates TGEV-induced inflammation downstream of the RIG-I–NF-κB pathway. (A and B) Apical-out organoids were infected with TGEV followed by Cyclo (1 mM) or BAY11 (2 µM) treatment for 48 h. Subsequently, transcription levels of HIF-1α, NLRP3, NLRP6, HMGB1, and NEK7 in the apical-out organoids post TGEV infection were measured by RT-qPCR (A) and HIF-1α protein expression was determined by Western blotting and analyzed using ImageJ (B). (C and D) Apical-out organoids were treated with the indicated concentrations of BAY87 or CoCl2 for 48 h, then HIF-1α protein expression was detected by Western blotting and analyzed by ImageJ. (E and F) Apical-out organoids were infected with TGEV followed by BAY87 (5 µM) or CoCl2 (25 µM) treatment for 48 h, then transcription levels of TNF-α, IL-8, IL-6, and IL-1β were determined by RT-qPCR. (G and H) Apical-out organoids were infected with TGEV followed by BAY87 (5 µM) or CoCl2 (25 µM) treatment for 48 h, then TGEV viral load was detected by RT-qPCR, and TGEV N and HIF-1α protein expressions were measured by Western blotting. Results are presented as mean ± SD of data from three independent experiments *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, determined by two-tailed Student’s t test.
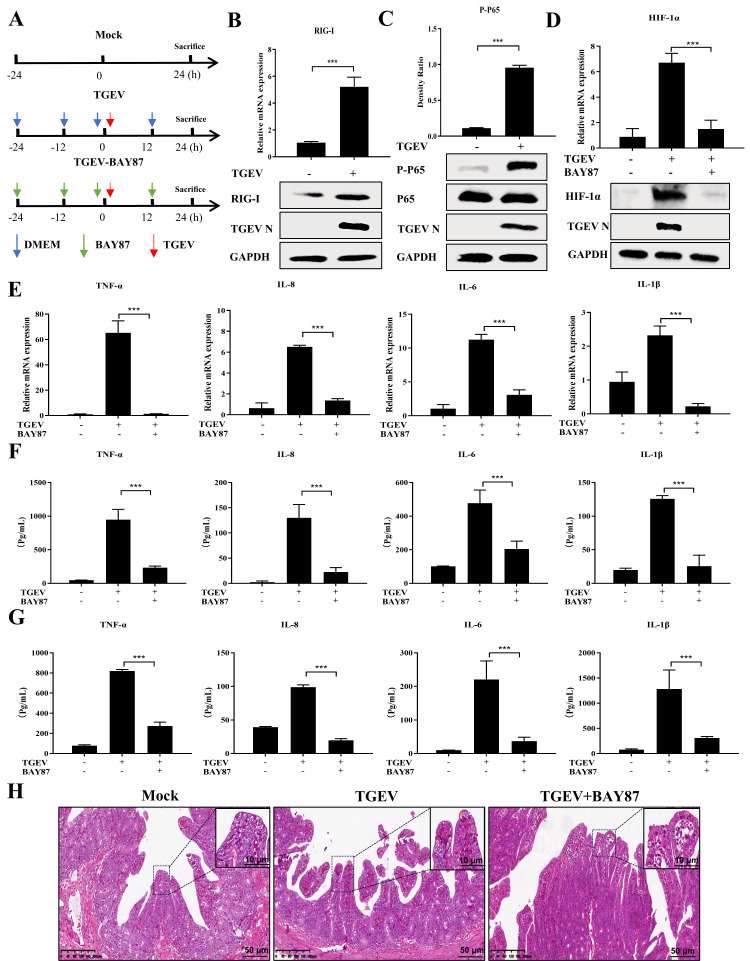
TGEV infection triggers inflammatory responses by the RIG-I/NF-κB/HIF-1α pathway in pigs
Given the above results ex vivo, we hypothesized that TGEV induces inflammation via the RIG-I/NF-κB/HIF-1α axis in vivo. To verify this hypothesis, experiments were performed to evaluate how TGEV induces inflammatory responses in pigs. Groups of pigs were treated with Dulbecco’s modified Eagle’s medium (DMEM) or BAY87 and then inoculated with TGEV individually by oral administration (Fig. 7A). First, RIG-I and MDA5 expressions in the ileum of the mock and TGEV groups were measured. TGEV infection significantly upregulated RIG-I expression but not MDA5 expression (Fig. 7B; Fig. S2B), which was consistent with ex vivo results. In addition, P65 was markedly phosphorylated after TGEV infection, suggesting that the NF-κB pathway is instrumental in TGEV-induced inflammation in pigs (Fig. 7C). Furthermore, HIF-1α expression in the ileum was markedly induced by TGEV infection, but decreased after BAY87 treatment, and this treatment also inhibited TGEV infection in vivo (Fig. 7D; Fig. S5A and S5B).
Fig 7.
TGEV infection triggers inflammatory responses via the RIG-I/NF-κB/HIF-1α pathway in vivo. (A) Experimental schemes in three groups of piglets. (B) The transcription level of RIG-I in the ileum from piglets sacrificed at 24 hpi was detected by RT-qPCR, and RIG-I and TGEV N protein expressions in the ileum were determined by Western blotting. (C) Phosphorylated P65, P65, and TGEV N in the ileum from piglets sacrificed at 24 hpi were detected by Western blotting and analyzed by ImageJ. (D) The transcription level of HIF-1α in the ileum was detected by RT-qPCR, and HIF-1α and TGEV N in the ileum were measured by Western blotting. (E) The mRNA levels of TNF-α, IL-8, IL-6, and IL-1β in the ileum were determined by RT-qPCR. (F and G) A protein microarray was performed to detect TNF-α, IL-8, IL-6, and IL-1β in intestinal digesta (F) and serum (G). (H) Hematoxylin and eosin staining of ilea from piglets sacrificed at 24 hpi; scale bar: 10 or 50 µm. Results are presented as mean ± SD of data from three independent experiments ***, P ≤ 0.001, determined by two-tailed Student’s t test.
To further confirm the relationship between HIF-1α and TGEV-induced inflammation in vivo, the expression of TNF-α, IL-8, IL-6, and IL-1β was determined by RT-qPCR and protein microarray. We found that TGEV-induced cytokines in the ileum, intestinal digesta, and serum were downregulated after oral administration of BAY87 in pigs (Fig. 7E through G). In addition, alanine aminotransferase (ALT) and aminotransferase (AST) were not changed by BAY87 treatment, meaning that oral administration of BAY87 was not cytotoxic in our animal model (Fig. S6A). To further characterize the effect of HIF-1α on inflammatory infiltration caused by TGEV, ileum samples were paraffin-embedded, followed by slicing and staining with hematoxylin and eosin (H&E). Pharmaceutical inhibition of HIF-1α almost reversed the inflammatory infiltration of the ileum (Fig. 7H). These results again suggested that TGEV triggers inflammatory responses predominantly via the RIG-I/NF-κB/HIF-1α axis in vivo.
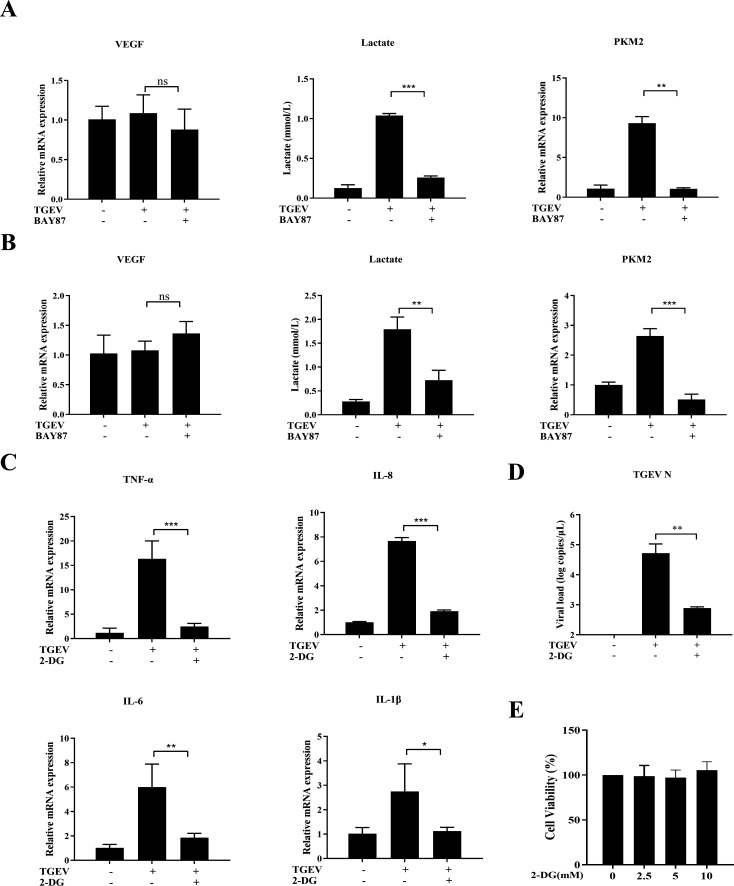
HIF-1α promotes TGEV-induced inflammatory responses by activating glycolysis
Although HIF-1α was proved to be an indispensable factor in TGEV-induced inflammation, the mechanism of HIF-1α-mediated inflammation upon TGEV infection was unclear and warranted further exploration. HIF-1α is known to positively regulate inflammatory responses mainly by upregulating vascular endothelial growth factor (VEGF) or activating glycolysis (19, 20). Therefore, in this study, VEGF and glycolysis were screened after TGEV infection. The results demonstrated that TGEV infection could not regulate VEGF expression, but could significantly upregulate lactate production and PKM2 expression, which are key markers of glycolysis activation. Moreover, the TGEV-induced lactate production and PKM2 expression were markedly decreased by BAY87 treatment, indicating that HIF-1α can positively regulate TGEV-induced glycolysis ex vivo (Fig. 8A) and in vivo (Fig. 8B). To further verify the effect of HIF-1α-mediated glycolysis on TGEV-induced inflammation, 2-DG (a glycolysis inhibitor) was employed to treat TGEV-infected organoids. As expected, TGEV-induced inflammation was obviously decreased and TGEV infection was also inhibited by pharmaceutical inhibition of glycolysis (Fig. 8C and D). In addition, 2-DG presented no cytotoxicity at the concentrations used in this experiment (Fig. 8E). Collectively, the proposed molecular mechanism is that TGEV induces inflammation via the RIG-I/ NF-κB/HIF-1α/glycolysis axis (Fig. 9).
Fig 8.
HIF-1α promotes TGEV-induced inflammatory responses by activating glycolysis. (A) Intestinal organoids were infected with TGEV followed by BAY87 (5 µM) for 48 h, then VEGF and PKM2 mRNA levels, and lactate production were determined by RT-qPCR. (B) Transcription levels of VEGF and PKM2 and lactate production in the ileum were detected by RT-qPCR. (C and D) Intestinal organoids were infected with TGEV followed by 2-DG (5 mM) for 48 h, then transcription levels of TNF-α, IL-8, IL-6, and IL-1β (C) and TGEV viral load (D) were determined by RT-qPCR. (E) Intestinal organoids were incubated with different concentrations of 2-DG for 48 h and cell viability was assessed using a Cell Counting Kit-8. Results are presented as mean ± SD of data from three independent experiments *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, determined by two-tailed Student’s t test.
Fig 9.
Proposed model of TGEV-induced inflammation via the RIG-I/NF-κB/HIF-1α/glycolysis pathway in intestinal organoids and in vivo.
DISCUSSION
Sensitive cell lines have long been used for viral infections, even though cell lines do not mimic the physiological states in the body. Organoids are more acceptable for the establishment and application of models exploring viral infection and immune responses because the origin and composition of organoids are closer to that of organ systems. Intestinal organoids derived from Lgr5+ stem cells were first proposed in 2009 (5). Organoids have proved useful in investigations of the production, signaling, and function of innate immunity in response to human enteric viruses, such as human norovirus, rotavirus, and reovirus (21). Previously, our laboratory developed an apical-out porcine intestinal organoid culture system for swine enteric virus infection and immune response investigations. Apical-out organoids facilitate most enteric virus infections and the differentiation of intestinal cell types, and thus are physiological research models for virus–epithelial interaction investigations (6). In this study, our results demonstrated that TGEV was able to infect the apical-out intestinal organoids and triggers TGEV-induced inflammatory factors, especially TNF-α, IL-8, and IL-6, which were markedly upregulated. We also found that TGEV protein, but not nucleic acid, induced inflammation in the apical-out organoids. We infer that TGEV nonstructural protein participates in TGEV-induced inflammation, but further studies are needed to understand the details.
Activation of PRRs is an important strategy utilized by coronaviruses for manipulating inflammatory responses (10). TGEV infection is reported to trigger inflammation by RIG-I- and MDA5-mediated signaling in ST cells (22). Here, we found that TGEV induces inflammation by upregulating RIG-I expression in apical-out organoids and pigs, rather than activating MDA5. A potential reason for these discrepancies may be that apical-out organoids are more likely to mimic in vivo environments compared to conventional cell lines. Moreover, TGEV infection can be enhanced by Cyclo (a RIG-I inhibitor) treatment, suggesting that RIG-I not only regulates TGEV-induced inflammation but also may affect interferon production which has an antiviral function (23). Some studies have reported that TGEV induces inflammatory responses by the NF-κB pathway in conventional cell lines (4, 22). Our results provide a similar trend in apical-out organoids and in pigs, indicating that the NF-κB pathway could be a reliable target to control TGEV-induced enteritis. In addition, our results suggested that NF-κB signaling pathway activation had no remarkable effects on TGEV replication, which was consistent with the results of previous studies (4, 22). We infer that NF-κB may play a critical role in many cellular processes to regulate viral replication, including cell proliferation, survival, and differentiation except to inflammation. Therefore, NF-κB not only regulates the expression of HIF-1α but also participates in the regulation of other genes related to viral infection. We infer that there are more host factors involved in the relationship between NF-κB and TGEV infection. Further studies are required to understand this in more detail.
There are many proinflammatory factors involved in coronavirus-induced inflammation. It is reported that HIF-1α promotes SARS-CoV-2 infection and aggravates inflammatory responses in COVID-19 (24); and HMGB1 enhances porcine epidemic diarrhea virus-induced inflammation by binding to TLR4-induced activation of p38 MAPK (25). The activation of NLRP3 and NLRP6 inflammasomes also triggers coronavirus-induced inflammatory responses by releasing IL-1β and IL-18 (26, 27). In addition, NLRP3 activation can be mediated by NEK7 downstream of potassium efflux (28). In this study, we found that HIF-1α expression can be regulated by the RIG-I–NF-κB pathway upon TGEV infection. Moreover, HIF-1α had higher expression in intestinal organoids and tissues compared to conventional cell lines, like ST cells (data not shown). We inferred that HIF-1α may be involved in TGEV-induced inflammation. Our subsequent results demonstrated that HIF-1α was actually a key mediator of TGEV-induced inflammatory responses in intestinal organoids and the porcine ileum, further clarifying that intestinal organoids are more likely to mimic in vivo environments than conventional cell lines. However, the detailed molecular mechanism of regulation of NF-κB and HIF-1α remains unclear. We infer that p50 may stabilize HIF-1α protein upon TGEV infection (29), but this theory requires further study.
HIF-1α is reported to positively regulate inflammation by different mechanisms. Codo et al. reported that SARS-CoV-2 stabilized HIF-1α expression, which induced monocyte-derived cytokines by activating glycolysis (20). HIF-1α also promotes the H1N1-induced host inflammatory response by regulating glycolysis (30). In contrast, HIF-1α promotes experimental acute ocular inflammation by stimulating VEGF signaling (19). In our study, VEGF and glycolysis were screened after TGEV infection or inhibition of HIF-1α to further explore the potential mechanism of HIF-1α promoting TGEV-induced inflammation. We found that HIF-1α facilitated TGEV-induced inflammation by regulating glycolysis. It can actually present novel mechanistic insight for this trend. In addition, similar to HIF-1α, activation of glycolysis can also upregulate TGEV infection. It is reported that H1N1-induced HIF-1α can promote glycolysis to produce lactate which reduces accumulation of the RIG-I–MAVS complex and IFN responses to promote viral infection (30). We hypothesize that HIF-1α promotion of TGEV infection may be associated with glycolysis and IFN production, and this theory needs to be explored in detail in future studies.
In conclusion, our research is the first to employ apical-out porcine intestinal organoids and pigs for exploring TGEV-induced inflammation and revealed that TGEV induces inflammatory responses via the RIG-I/NF-κB/HIF-1α/glycolysis axis. Our study provides a novel insight into potential therapeutic targets for TGEV-caused swine enteritis and verifies apical-out organoids as a more physiological model for mimicking enteric virus-induced inflammation.
MATERIALS AND METHODS
Cell culture, virus, and animals
ST cells for TCID50 detection were maintained in DMEM (Sigma-Aldrich, USA, D6429) with 10% fetal bovine serum (Invigentech, Brazil, A6901). The cells were incubated at 37°C in a humidified incubator with 5% CO2. The TGEV Miller strain was maintained in our laboratory and the titer was 107.25 TCID50/mL. Porcine intestines for crypt isolation were obtained from Luoniushan Co., Ltd (Hainan Province, China).
Porcine intestinal 3D organoids culture
Porcine ileum crypts were isolated from pigs and cultured in Matrigel (Corning, USA, 356231) and OGM (Stem Cell, Canada, 06010) containing 10 µM ATP-competitive inhibitor of Rho-associated kinases (Y-27632; CST, USA, 72302) according to a published protocol (6).
Establishment of apical-out porcine intestinal organoids
Porcine 3D ileum organoids cultured with Matrigel for 5 days were dissociated by incubation with 5 mM cold EDTA buffer on a rotating platform for 1 h at 4°C. The organoids were then harvested by centrifugation at 250 × g for 5 min, washed with ice-cold DMEM/F12 (Sigma-Aldrich, USA, D0697), and cultured in ultralow-attachment 24-well tissue culture plates (Corning, USA, 3473) in OGM supplemented with 10 µM Y-27632 at 37°C and 5% CO2. According to the previously published protocol, the apical-out organoids were generated after 3 days (6).
Virus infection on apical-out organoids
Apical-out organoids were harvested from culture suspension by centrifugation at 250 × g for 5 min and inoculated with the TGEV Miller strain (multiplicity of infection = 10) for 1 h at 37°C. The virus residue was removed and washed three times with DMEM/F12 for centrifugation. The organoids were incubated in OGM in a 37°C incubator supplied with 5% CO2. Cells and supernatant were collected at indicated times for determination of viral load and inflammatory responses.
Histopathological and immunofluorescence analysis
Porcine ileum samples were collected, fixed for 24 h in 10% formalin, dehydrated according to the standard protocol, embedded in paraffin, and subjected to H&E staining by standard procedures. For IFA, apical-out porcine intestinal organoids were fixed with 4% paraformaldehyde for 20 min and then were blocked and permeabilized with 10 mM phosphate buffer [with 3% bovine serum albumin (Biofroxx, Germany, 4240GR100) and 1% Triton X-100 (Beyotime, China, ST797)] for 12 h at 4°C. The apical-out organoids were labeled with primary antibodies for 24 h at 4°C. After rinsing, secondary antibodies were incubated for 24 h at 4°C. Next, 4′, 6-diamidino-2-phenylindole (DAPI; Beyotime, China, C1006) was used to stain with the nucleus. After washing, the apical-out organoids were visualized using confocal laser-scanning microscopy (Zeiss LSM 900, Germany).
Nuclear and cytoplasmic extraction, Western blotting, and lactate measurement
Apical-out organoids were collected by NP40 containing PMSF per 20 µL of cell pellet and lysed to nuclear extract and cytoplasmic extract according to the manufacturer’s recommendations (Nuclear and Cytoplasmic Protein Extraction Kit, Beyotime, China, P0027). For the Western blot, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane (GE, USA, 10600023). The membranes were blocked in 5% nonfat milk at room temperature for 2 h and incubated with specific primary antibodies overnight (see Table 1 for antibodies). Subsequently, the secondary antibody was incubated with the membrane for 1 h at room temperature. Finally, the proteins on the membranes were visualized with WesternBright ECL (Advansta, USA, K-12045-D50) (31, 32). The lactate level in supernatant or serum was determined by using a lactate assay kit (Dojindo, China, L256) according to the manufacturer’s instructions.
TABLE 1.
List of antibodies used in this study
| Antibody | Type | Supplier | Product number |
|---|---|---|---|
| ZO-1 | Rabbit | Invitrogen | PA5-85256 |
| Sox9 | Rabbit | CST | 82630 |
| Villin | Mouse | Santa Cruz | SC-58897 |
| CGA | Mouse | Santa Cruz | sc-393941 |
| Muc2 | Rabbit | Abcam | ab134119 |
| LYZ | Rabbit | Invitrogen | PA5-16668 |
| P65 | Rabbit | Proteintech | 10745-1-AP |
| P-JUN | Rabbit | Proteintech | 28891-1-AP |
| P-P65 | Rabbit | CST | 3033 |
| LaminB1 | Rabbit | Proteintech | 12987-1-AP |
| IκBα | Rabbit | Proteintech | 10268-1-AP |
| RIG-I | Rabbit | Proteintech | 20566-1-AP |
| HIF-1α | Rabbit | Proteintech | 20960-1-AP |
| GAPDH | Rabbit | Proteintech | 10494-1-AP |
RNA extraction, real-time quantitative PCR, and protein microarray
Total RNA was extracted using RNAiso reagent (TaKaRa, Japan, 9109) and reverse transcribed into cDNA using HiScript Q RT SuperMix for qPCR (Vazyme, China, R223-01), both following the manufacturer’s recommendations. The TGEV virus copy number was detected by the TaqMan probe-based RT-qPCR developed previously in our laboratory (33). The relative qPCR was performed using the ChamQ SYBR qPCR master mix (Vazyme, China, Q311-02) and calculated with the 2−ΔΔCT method. The primers and probes used in this study are listed in Table 2. For protein microarray, samples of intestinal contents and serum were detected by Quantibody Porcine Cytokine Array 1 (Raybiotech, USA, QAP-CYT-1).
TABLE 2.
Primers for real-time qPCR
| Names | Primer or probe | Sequence (5′−3′) |
|---|---|---|
| TGEV N | Forward | TGCCATGAACAAACCAAC |
| Reverse | GGCACTTTACCATCGAAT | |
| Probe | HEX-TAGCACCACGACTACCAAGC-BHQ1a | |
| TNF-α | Forward | GTCTCAAACCTCAGATAAG |
| Reverse | GTTGTCTTTCAGCTTCAC | |
| IL-8 | Forward | TCCTGCTTTCTGCAGCTCTC |
| Reverse | GGGTGGAAAGGTGTGGAATG | |
| IL-6 | Forward | AATGCTCTTCACCTCTCC |
| Reverse | TCACACTTCTCATACTTCTCA | |
| IL-1β | Forward | AGAGGGACATGGAGAAGCGA |
| Reverse | GCCCTCTGGGTATGGCTTT | |
| IL-18 | Forward | CGATGAAGACCTGGAATCGG |
| Reverse | CATCATGTCCAGGAACACTTCTCTG | |
| RIG-I | Forward | AGAGCAGCGGCGGAATC |
| Reverse | GGCCATGTAGCTCAGGATGAA | |
| IFN-α | Forward | CTGCTGCCTGGAATGAGAGCC |
| Reverse | TGACACAGGCTTCCAGGTCCC | |
| IFN-β | Forward | CCACCACAGCTCTTTCCATGA |
| Reverse | TGAGGAGTCCCAGGCAACT | |
| IFN-λ1 | Forward | CCACGTCGAACTTCAGGCTT |
| Reverse | ATGTGCAAGTCTCCACTGGT | |
| MDA5 | Forward | TCCGGGAAACAGGCAACTC |
| Reverse | CAAAGGATGGAGAGGGCAAGT | |
| TLR3 | Forward | GAGCAGGAGTTTGCCTTGTC |
| Reverse | GGAGGTCATCGGGTATTTGA | |
| TLR7 | Forward | TCTGCCCTGTGATGTCAGTC |
| Reverse | GCTGGTTTCCATCCAGGTAA | |
| TLR8 | Forward | CTGGGATGCTTGGTTCATCT |
| Reverse | CATGAGGTTGTCGATGATGG | |
| HIF-1α | Forward | GGCGCGAACGACAAGAAAAA |
| Reverse | GTGGCAACTGATGAGCAAGC | |
| NLRP3 | Forward | GAGCCAGAATGGGACAATGCAAAT |
| Reverse | CTTTCTTTTTCTTACAAATAGAG | |
| NLRP6 | Forward | CGGGACAATCCCCTAGGACT |
| Reverse | CCTCCCTCCTCGTTCCAAGT | |
| HMGB1 | Forward | ACATCCTGGCCTGTCCATTG |
| Reverse | TCGTATTTTTCCTTCAGCTTCGC | |
| NEK7 | Forward | GGCTGATTCCTGAGAGAACTGT |
| Reverse | AGCCGGCTTTATATCTCGGTG | |
| VEGF | Forward | CAAGATCCGCAGACGTGTAA |
| Reverse | CAACGCGAGTCTGTGTTTCT | |
| PKM2 | Forward | TTCGCATCTTTCATCCGTAA |
| Reverse | CGCCCAATCATCATCTTCT |
Pig experiments
Neonatal pigs spontaneously delivered from sows were confirmed negative for TGEV by RT-qPCR and enzyme-linked immunosorbent assay. The piglets were randomly separated into three groups: mock (three pigs), TGEV (3), and TGEV-BAY87 (3). For the TGEV group, piglets were orally infected 1.245 × 108 PFU TGEV for 24 h. For the TGEV-BAY87 group, piglets were orally administered BAY87 (10 mg/kg) every 12 h from 0 to 48 h and orally infected with 1.245 × 108 PFU TGEV for 24 h. At 24 hpi, all pigs were euthanized, and intestinal tissues were collected for RT-qPCR, Western blot, and pathological examination. ALT and AST in serum were detected by biochemical parameters (HITACHI, Japan, 3110).
Statistical analysis
All data were analyzed using GraphPad Prism 8.0 software (GraphPad, La Jolla, CA, USA) by one- or two-way analysis of variance. Differences between two groups are indicated as *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Every experiment was performed with three biological replicates, and the results were recorded as the mean ± standard deviation (SD).
ACKNOWLEDGMENTS
The authors thank Prof. Li Feng from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, for providing a TGEV monoclonal antibody. The authors acknowledge all group members of the Animal Immunology Group of Lanzhou Veterinary Research Institute for helpful discussions and animal care and breeding.
This work was supported by the Natural Science Foundation of Technology Department, Gansu Province, China (20JR10RA020), the National Natural Science Foundation of China (31972689, U22A20522), the CAAS International Exchange Scholarship for graduate students, the Science and Technology Major Project of Gansu Province (22ZD6NA001), and the ULg-CAAS Joint PhD Program and CSC-WBI Joint PhD Program.
G.L. and Y.F. conceived the project. Y.Z., N.Y., Y.L., C.T., Y.C., X.R., and Y.L. performed the experiments. Y.Z., G.L., and Y.F. analyzed the data. Y.Z. drafted the manuscript. G.L. and Y.F. edited the manuscript.
Contributor Information
Yuguang Fu, Email: fuyuguang@caas.cn.
Guangliang Liu, Email: LiuGuangliang01@caas.cn.
Tom Gallagher, Loyola University Chicago - Health Sciences Campus, Maywood, Illinois, USA.
ETHICS APPROVAL
All animals were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. The study was approved by the Animal Administration and Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Permit No. LVRIAEC-2020-030).
DATA AVAILABILITY
The raw data used to generate the figures present within this article are available from the corresponding author upon request.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00461-24.
Figures S1 to S6.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Liu Q, Wang HY. 2021. Porcine enteric coronaviruses: an updated overview of the pathogenesis, prevalence, and diagnosis. Vet Res Commun 45:75–86. doi: 10.1007/s11259-021-09808-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xia L, Yang Y, Wang J, Jing Y, Yang Q. 2018. Impact of TGEV infection on the pig small intestine. Virol J 15:102. doi: 10.1186/s12985-018-1012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang L, Qiao X, Zhang S, Qin Y, Guo T, Hao Z, Sun L, Wang X, Wang Y, Jiang Y, Tang L, Xu Y, Li Y. 2018. Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-κB activation. Virulence 9:1685–1698. doi: 10.1080/21505594.2018.1536632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 6. Li Y, Yang N, Chen J, Huang X, Zhang N, Yang S, Liu G, Liu G. 2020. Next-generation porcine intestinal organoids: an apical-out organoid model for swine enteric virus infection and immune response investigations. J Virol 94:e01006-20. doi: 10.1128/JVI.01006-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang N, Zhang Y, Fu Y, Li Y, Yang S, Chen J, Liu G. 2022. Transmissible gastroenteritis virus infection promotes the self-renewal of porcine intestinal stem cells via WNT/β-catenin pathway. J Virol 96:e0096222. doi: 10.1128/jvi.00962-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salminen A. 2021. Increased immunosuppression impairs tissue homeostasis with aging and age-related diseases. J Mol Med (Berl) 99:1–20. doi: 10.1007/s00109-020-01988-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. 2020. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 20:363–374. doi: 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poeck H, Ruland J. 2012. From virus to inflammation: mechanisms of RIG-I-induced IL-1β production. Eur J Cell Biol 91:59–64. doi: 10.1016/j.ejcb.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 11. Park A, Iwasaki A. 2020. Type I and type III Interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27:870–878. doi: 10.1016/j.chom.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukata M, Arditi M. 2013. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol 6:451–463. doi: 10.1038/mi.2013.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Latz E, Xiao TS, Stutz A. 2013. Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411. doi: 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, Zhang Q, Wu J. 2020. Coronavirus infections and immune responses. J Med Virol 92:424–432. doi: 10.1002/jmv.25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawrence T. 2009. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651. doi: 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karin M, Liu Z g, Zandi E. 1997. AP-1 function and regulation. Curr Opin Cell Biol 9:240–246. doi: 10.1016/s0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- 17. Lee W, Lee SH, Kim M, Moon JS, Kim GW, Jung HG, Kim IH, Oh JE, Jung HE, Lee HK, Ku KB, Ahn DG, Kim SJ, Kim KS, Oh JW. 2018. Vibrio vulnificus quorum-sensing molecule cyclo(Phe-Pro) inhibits RIG-I-mediated antiviral innate immunity. Nat Commun 9:1606. doi: 10.1038/s41467-018-04075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cherry AD, Piantadosi CA. 2015. Regulation of mitochondrial biogenesis and its intersection with inflammatory responses. Antioxid Redox Signal 22:965–976. doi: 10.1089/ars.2014.6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Usta Sofu G, Erzurumlu Y, Karaca U, Candan IA, Savran M, Asci H, Hasseyid N. 2022. Melatonin receptor agonist ramelteon alleviates experimental acute ocular inflammation via HIF-1α/VEGF/E-NOS signaling. Eur J Ophthalmol:11206721221123878. doi: 10.1177/11206721221123878 [DOI] [PubMed] [Google Scholar]
- 20. Codo AC, Davanzo GG, Monteiro L de B, de Souza GF, Muraro SP, Virgilio-da-Silva JV, Prodonoff JS, Carregari VC, de Biagi Junior CAO, Crunfli F, et al. 2020. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab 32:437–446. doi: 10.1016/j.cmet.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nolan LS, Baldridge MT. 2022. Advances in understanding interferon-mediated immune responses to enteric viruses in intestinal organoids. Front Immunol 13:943334. doi: 10.3389/fimmu.2022.943334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding Z, An K, Xie L, Wu W, Zhang R, Wang D, Fang Y, Chen H, Xiao S, Fang L. 2017. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology 507:170–178. doi: 10.1016/j.virol.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue M, Wang W, He H, Li L, Zhang X, Shi H, Liu P, Feng L. 2022. Different mechanisms are utilized by coronavirus transmissible gastroenteritis virus to regulate interferon lambda 1 and interferon lambda 3 production. J Virol 96:e0138822. doi: 10.1128/jvi.01388-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian M, Liu W, Li X, Zhao P, Shereen MA, Zhu C, Huang S, Liu S, Yu X, Yue M, Pan P, Wang W, Li Y, Chen X, Wu K, Luo Z, Zhang Q, Wu J. 2021. HIF-1α promotes SARS-CoV-2 infection and aggravates inflammatory responses to COVID-19. Signal Transduct Target Ther 6:308. doi: 10.1038/s41392-021-00726-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao R, Zhang Y, Kang Y, Xu W, Jiang L, Guo T, Huan C. 2020. Glycyrrhizin inhibits PEDV infection and proinflammatory cytokine secretion via the HMGB1/TLR4-MAPK p38 pathway. Int J Mol Sci 21:2961. doi: 10.3390/ijms21082961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li R, Zhu S. 2020. NLRP6 inflammasome. Mol Aspects Med 76:100859. doi: 10.1016/j.mam.2020.100859 [DOI] [PubMed] [Google Scholar]
- 27. Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, Hauenstein AV, Wu Z, Núñez G, Mao Y, Wu H. 2019. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570:338–343. doi: 10.1038/s41586-019-1295-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y, Zeng MY, Yang D, Motro B, Núñez G. 2016. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530:354–357. doi: 10.1038/nature16959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu J, Huang S, Li Y, Xu J, Chen R, Guo M, Qian X, Li T, Tian Z, Jin H, Huang C. 2022. NF-κB1 p50 stabilizes HIF-1α protein through suppression of ATG7-dependent autophagy. Cell Death Dis 13:1076. doi: 10.1038/s41419-022-05521-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng X, Zhu Y, Yang W, Zhang J, Jin W, Tian R, Yang Z, Wang R. 2024. HIF-1α promotes virus replication and cytokine storm in H1N1 virus-induced severe pneumonia through cellular metabolic reprogramming. Virol Sin 39:81–96. doi: 10.1016/j.virs.2023.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang N, Shi H, Yan M, Liu G. 2021. IFIT5 negatively regulates the type I IFN pathway by disrupting TBK1-IKKε-IRF3 signalosome and degrading IRF3 and IKKε. J Immunol 206:2184–2197. doi: 10.4049/jimmunol.2001033 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Song Z, Wang M, Lan M, Zhang K, Jiang P, Li Y, Bai J, Wang X. 2019. Cholesterol 25-hydroxylase negatively regulates porcine intestinal coronavirus replication by the production of 25-hydroxycholesterol. Vet Microbiol 231:129–138. doi: 10.1016/j.vetmic.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang X, Chen J, Yao G, Guo Q, Wang J, Liu G. 2019. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl Microbiol Biotechnol 103:4943–4952. doi: 10.1007/s00253-019-09835-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S6.
Data Availability Statement
The raw data used to generate the figures present within this article are available from the corresponding author upon request.