ABSTRACT
Rivers are critical ecosystems that impact global biogeochemical cycles. Nonetheless, a mechanistic understanding of river microbial metabolisms and their influences on geochemistry is lacking. Here, we announce metaproteomes of river sediments that are paired with metagenomes and metabolites, enabling an understanding of the microbial underpinnings of river respiration.
KEYWORDS: microbial ecology, river microbiology, metagenomics, metaproteomics, metabolomics, genomes, hyporheic zone
ANNOUNCEMENT
Rivers are vital ecosystems with global-scale consequences. While they have economic value for practices like agriculture, they are also a link between terrestrial and aquatic ecosystems. In fact, rivers transport, mineralize, and bury ~2.7 Pg of carbon per year (1). The hyporheic zone (HZ) is a saturated transitional space between river surface water and underlying sediments, which greatly contributes to river respiration (2, 3). Within HZs, microbial metabolisms significantly contribute to the biogeochemical transformations that ultimately modulate river respiration (4). Currently, a lack of genome-resolved, multi-omic data sets makes the linkages between microbial metabolism and river biogeochemistry opaque, leading to uncertainties in ecological model constraints. Here, we present a metaproteomic data set from Columbia River sediments with paired metagenomes and metabolites and Hyporheic Uncultured Microbial and Viral (HUM-V), a genome-resolved database of river microorganisms (5), providing a multi-omics infrastructure to decode microbial contributions to river carbon and nutrient processing.
We collected six HZ sediment cores of the Columbia River (46°22′15.80″N, 119°16′31.52″W) and subsampled each core into six 10 cm depth sections (0–60 cm) (5, 6). Liquid nitrogen frozen sediment profiles were collected across two transects (170 m apart), yielding 33 samples that were processed for metaproteomics, metagenomics, Fourier-transform ion cyclotron resonance mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Sediment was prepared for metaproteomic analysis as described previously in detail (6) with a modified application of the MPLEx protocol (7).
Briefly (6), extractions were analyzed on a Q-Exactive Plus Orbitrap mass analyzer (Thermo Electron, Waltham, MA, USA) coupled to a Waters NanoAcquity high-performance liquid chromatography system (Waters Corporation, Milford, MA, USA) through 75 µm × 70 cm columns packed with Phenomenex Jupiter C18 3 µm beads (Phenomenex, Torrance, CA, USA). Samples were loaded onto columns with 0.05% formic acid in water and eluted with 0.05% formic acid in acetonitrile over 100 min. Ten data-dependent MS/MS scans (17.5 K resolution, centroided) were recorded for each survey MS scan (35K resolution) using a normalized collision energy of 30, isolation width of 2.0 m/z, and rolling exclusion window of ±1 m/z lasting 30 s before previously fragmented MS1 signals were eligible for re-analysis. All liquid chromatography-tandem mass spectrometry data sets were converted to ASCII text using MSConvert (8), and the files were interrogated with a target-decoy approach to reach a ~1% false detection rate with formula [100 × 191 decoy peptide-spectrum matches (PSMs)/19,084 total filter passing PSMs] (9) and using MSGF+ (10). Our metaproteome contained 10,063,272 protein entries from 1,299,102,456 amino acids (6).
Spectra were searched against files that included (i) 55 dereplicated bacterial and archaeal metagenome-assembled genome (MAG) and (ii) 111 clustered viral MAG (vMAG) amino acid sequences. Due to functional conservation across closely related MAG strains, peptide recruitments were divided into three categories: (i) unique: peptides with hits to a single protein, (ii) non-unique specialized: peptides with hits to multiple amino acid sequences that shared functional annotation using multiple annotation databases by DRAM (5, 11) and MAG taxonomy with the genome-taxonomy database toolkit (12), and (iii) non-unique: peptides with hits to multiple amino acid sequences with different annotation or taxonomy. Microbial metaproteomes were converted to spectral abundance factors by calculating the length of each protein and normalizing by total counts per peptide mapped to each protein per sample.
Our genome-resolved strategies assigned gene expression to all 55 dereplicated, quality-verified MAGs in HUM-V. These MAGs recruited 13,102 total peptides to 1,313 proteins. The “unique” peptides represented 67% of genes expressed in our proteome. Interestingly, non-unique peptides with identical genus assignment and protein annotation accounted for 14% of the total genes expressed. The addition of this specialized non-unique category prevented the exclusion of ecologically relevant data due to strain overlap. In addition to our microbial results, 66% of our identified vMAGs uniquely recruited peptides (5).
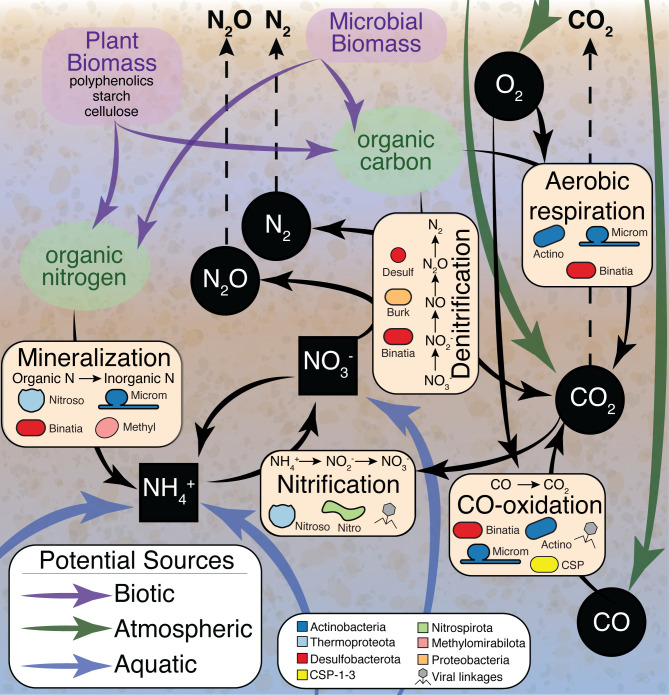
Leveraging HUM-V, we unveiled a myriad of microbial and viral metabolisms contributing to carbon and nitrogen cycling in river sediments (Fig. 1) (5). Our co-expression analyses revealed heterotrophic microbial members (e.g., Actinobacteriota and Nitrososphaeraceae) that expressed enzymes for the degradation of complex carbon polymers like starch and cellulose to generate monomers that could be utilized by saccharolytic microorganisms (e.g., Thermoplasmatota and Proteobacteria) (5). We also identified organisms (e.g., Binatia and Nitrospiraceae) that expressed nitrogen mineralization genes, supplying a source of ammonium that could sustain coupled nitrification and denitrification (5). Furthermore, viral metaproteomics demonstrated that viruses likely influence the biogeochemical cycling of river carbon and nitrogen both by active predation of microbial members and through the expression of auxiliary metabolic genes like glycoside hydrolases (5). Ultimately, our paired multi-omic data set enabled the description of a mechanistic, conceptual model of Columbia River microbial biogeochemical cycling.
Fig 1.
Conceptual model summarizing the microbial and viral contributions to HZ carbon and nitrogen cycling identified in this study. This figure is taken from the original publication of this data set in reference (5) and is included here to demonstrate the value of our metaproteomics approach. Black arrows signify microbial transformations uncovered in our MAG-resolved metaproteomic data. Specific processes (e.g., mineralization, nitrification, CO oxidation, denitrification, and aerobic respiration) are highlighted in beige boxes, with microorganisms inferred to carry out these processes denoted by overlaid cell shapes colored by phylum. Possible biotic, atmospheric, and aquatic carbon and nitrogen sources are indicated by purple, green, and blue arrows, respectively. Inorganic carbon and nitrogen sources are shown by black squares (aqueous) and black circles (gaseous), with white text and dashed arrows indicating possible gases that could be released to the atmosphere. Processes that could be impacted by viruses are marked with gray virus symbols.
ACKNOWLEDGMENTS
This work was supported by the Subsurface Biogeochemical Research (SBR) program (DE-SC0018170); the National Sciences Foundation Division of Biological Infrastructure (#1759874); and the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Environmental System Science (ESS) program through a subcontract from the River Corridor Scientific Focus Area project at Pacific Northwest National Laboratory. The NMR data, FTICR-MS data, and MS-proteomics data in this work were collected using instrumentation in the Environmental Molecular Science Laboratory (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. Pacific Northwest National Lab is operated by Battelle for the DOE under Contract DE-AC05-76RL01830. Metagenomic sequencing for this research was performed by the Joint Genome Institute via a large-scale sequencing award (Award 1781) and at the Genomics Shared Resource Core at The Ohio State University Comprehensive Cancer Center supported by P30 CA016058.
Contributor Information
Josué Rodríguez-Ramos, Email: josue.rodriguez@pnnl.gov.
Jennifer Geddes-McAlister, University of Guelph, Guelph, Ontario, Canada.
DATA AVAILABILITY
The data sets supporting the conclusions of this article are publicly available. Sequencing data are available in NCBI under BioProject PRJNA576070, with 16S rRNA amplicon sequences under accession numbers SRX9312157-SRX9312180. Metaproteomics data are deposited in the MassIVE database under accession MSV000087330. Metabolomics data are publicly available and deposited in Zenodo https://doi.org/10.5281/zenodo.5076253.
REFERENCES
- 1. Battin TJ, Luyssaert S, Kaplan LA, Aufdenkampe AK, Richter A, Tranvik LJ. 2009. The boundless carbon cycle. Nature Geosci 2:598–600. doi: 10.1038/ngeo618 [DOI] [Google Scholar]
- 2. Boulton AJ, Findlay S, Marmonier P, Stanley EH, Valett HM. 1998. The functional significance of the Hyporheic zone in streams and rivers. Annu. Rev. Ecol. Syst 29:59–81. doi: 10.1146/annurev.ecolsys.29.1.59 [DOI] [Google Scholar]
- 3. Naegeli MW, Uehlinger U. 1997. Contribution of the Hyporheic zone to ecosystem metabolism in a Prealpine gravel-bed-river. Journal of the North American Benthological Society 16:794–804. doi: 10.2307/1468172 [DOI] [Google Scholar]
- 4. Lewandowski J, Arnon S, Banks E, Batelaan O, Betterle A, Broecker T, Coll C, Drummond JD, Gaona Garcia J, Galloway J, et al. 2019. Is the Hyporheic zone relevant beyond the scientific community? Water 11:2230. doi: 10.3390/w11112230 [DOI] [Google Scholar]
- 5. Rodríguez-Ramos JA, Borton MA, McGivern BB, Smith GJ, Solden LM, Shaffer M, Daly RA, Purvine SO, Nicora CD, Eder EK, Lipton M, Hoyt DW, Stegen JC, Wrighton KC. 2022. Genome-resolved metaproteomics decodes the microbial and viral contributions to coupled carbon and nitrogen Cycling in river sediments. mSystems 7:e0051622. doi: 10.1128/msystems.00516-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham EB, Crump AR, Kennedy DW, Arntzen E, Fansler S, Purvine SO, Nicora CD, Nelson W, Tfaily MM, Stegen JC. 2018. Multi’Omics comparison reveals metabolome biochemistry, not microbiome composition or gene expression, corresponds to elevated biogeochemical function in the Hyporheic zone. Sci Total Environ 642:742–753. doi: 10.1016/j.scitotenv.2018.05.256 [DOI] [PubMed] [Google Scholar]
- 7. Nicora CD, Burnum-Johnson KE, Nakayasu ES, Casey CP, White III RA, Roy Chowdhury T, Kyle JE, Kim Y-M, Smith RD, Metz TO, Jansson JK, Baker ES. 2018. The Mplex protocol for multi-Omic analyses of soil samples. JoVE Journal of visualized experiments:JoVE. doi: 10.3791/57343-v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pwiz: the Proteowizard library is a set of software libraries and tools for rapid development of mass spectrometry and proteomic data analysis software. 2023. Github. Available from: https://github.com/ProteoWizard/pwiz
- 9. Elias JE, Gygi SP. 2010. Target-decoy search strategy for mass spectrometry-based proteomics, p 55–71. In Proteome Bioinformatics. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S, Pevzner PA. 2014. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5:5277. doi: 10.1038/ncomms6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, Liu P, Narrowe AB, Rodríguez-Ramos J, Bolduc B, Gazitúa MC, Daly RA, Smith GJ, Vik DR, Pope PB, Sullivan MB, Roux S, Wrighton KC. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. doi: 10.1093/nar/gkaa621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2020. GTDB-TK: a Toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the conclusions of this article are publicly available. Sequencing data are available in NCBI under BioProject PRJNA576070, with 16S rRNA amplicon sequences under accession numbers SRX9312157-SRX9312180. Metaproteomics data are deposited in the MassIVE database under accession MSV000087330. Metabolomics data are publicly available and deposited in Zenodo https://doi.org/10.5281/zenodo.5076253.