ABSTRACT
The Solar Lake in Taba, Egypt, encompasses one of the few modern-day microbial mats’ systems metabolically analogous to Precambrian stromatolites. Solar Lake benthic communities and their adaptation to the Lake’s unique limnological cycle have not been described for over two decades. In this study, we revisit the flat mat and describe the summer’s shallow water versus exposed microbial community; the latter occurs in response to the seasonal partial receding of water. We employed metagenomic NovaSeq-6000 shotgun sequencing and 16S rRNA, mcrA, and dsrB quantitative PCR. A total of 292 medium-to-high-quality metagenome-assembled genomes (MAGs) were reconstructed. At the structural level, Candidatus Aenigmatarchaeota, Micrarchaeota, and Omnitrophota MAGs were exclusively detected in the shallow-water mats, whereas Halobacteria and Myxococcota MAGs were specific to the exposed microbial mat. Functionally, genes involved in reactive oxygen species (ROS) detoxification and osmotic pressure were more abundant in the exposed than in the shallow-water microbial mats, whereas genes involved in sulfate reduction/oxidation and nitrogen fixation were ubiquitously detected. Genes involved in the utilization of methylated amines for methane production were predominant when compared with genes associated with alternative methanogenesis pathways. Solar Lake methanogen MAGs belonged to Methanosarcinia, Bathyarchaeia, Candidatus Methanofastidiosales, and Archaeoglobales. The latter had the genetic capacity for anaerobic methane oxidation. Moreover, Coleofasciculus chthonoplastes, previously reported to dominate the winter shallow-water flat mat, had a substantial presence in the summer. These findings reveal the taxonomic and biochemical microbial zonation of the exposed and shallow-water Solar Lake flat mat benthic community and their capacity to ecologically adapt to the summer water recession.
IMPORTANCE
Fifty-five years ago, the extremophilic “Solar Lake” was discovered on the Red Sea shores, garnering microbiologists’ interest worldwide from the 1970s to 1990s. Nevertheless, research on the lake paused at the turn of the millennium. In our study, we revisited the Solar Lake benthic community using a genome-centric approach and described the distinct microbial communities in the exposed versus shallow-water mat unveiling microbial zonation in the benthic communities surrounding the Solar Lake. Our findings highlighted the unique structural and functional adaptations employed by these microbial mat communities. Moreover, we report new methanogens and phototrophs, including an intriguing methanogen from the Archaeoglobales family. We describe how the Solar Lake’s flat mat microbial community adapts to stressors like oxygen intrusion and drought due to summer water level changes, which provides insights into the genomic strategies of microbial communities to cope with altered and extreme environmental conditions.
KEYWORDS: Solar Lake, hypersaline, community genomics, methanogenesis, candidate phyla, flat microbial mat
INTRODUCTION
Hypersaline lakes are water bodies of significant ecological importance due to their classification as “poly-extreme ecosystems.” These lakes inhabit harsh environmental conditions, including high salinity levels above 35 g/L, exposure to UV radiation, and low dissolved oxygen (1). Hypersaline lakes harbor distinctive microbial mat ecosystems characterized by high microbial diversity, environmental resilience, and adaptability. These microbial mats have been previously studied in Shark Bay, Australia, Lake Fryxell, the Arctic, Guerrero Negro, Mexico, Kiritimati Atoll, Pacific Ocean (2–6), and the Solar Lake, Egypt (7–11).
The hypersaline Solar Lake in Taba, Egypt, is a heliothermal lake (12). The lake is 140 m × 70 m and is 4–6 m deep (11). The Solar Lake undergoes seasonal salinity stratification; in summer, holomixis occurs due to evaporation, the lake water becomes completely oxic, and the salinity can reach 18% (11). In fall, the existing highly saline water and the introduction of fresh seawater from the Red Sea create a salinity gradient and water stratification (10, 11). The effect of the limnological cycle on the Solar Lake microbial communities remains largely unexplored. The lake is known to harbor a 1-mm thick lithifying cyanobacterial mat, and it is considered among the few mat ecosystems that are analogous to Precambrian stromatolites (7, 8). Studies on the Solar Lake benthic microbial communities ceased in the late 1990s, leaving a significant knowledge gap (7, 8, 13–16). Earlier studies on the Solar Lake benthic microbial communities indicated that there are four types of mats, including the shallow-water flat mat surrounding the lake (8, 17). Between the late 1970 and 1990s, studies on shallow-water flat microbial mat showed the prevalence of Cyanobacteria belonging to Coleofasciculus chthonoplastes (previous name: Microcoleus chthonoplastes), Oscillatoria salina, Oscillatoria limnetica, Spirulina labyrinthiformis, Spirulina sp., and Aphanothece halophytica. Sulfate reducers belonging to Desulfovibrio and Desulfonema along with methanogens thrived in the shallow-water flat mat (13, 16). The Solar Lake benthic methanogens were enriched in the presence of methylated amines and H2/CO2 rather than acetate (13).
Recent studies using bacterial 16S rRNA gene sequencing of the Solar Lake sediment showed that 72% of the sediments’ bacterial community were unclassified at the genus level (18, 19). As expected, sulfur oxidizing and sulfate-reducing communities were abundant in the lake sediments (18, 19). The lake limnological cycle has been shown to affect the shallow-water microbial mat (8). During the summer season and due to the high evaporation rate, the water recedes partially from the flat shallow-water mat, which becomes exposed (8, 20). The exposed mat is likely to undergo heightened salinity and increased oxygen infiltration due to drought, as observed in similar ecosystems (21, 22). Studying the microbial communities in the Solar Lake shallow-water versus exposed mats can provide valuable insights into the mechanisms of adaptation employed by microbial communities to cope with environmental stressors caused by the reduction in summer water level.
In this study, we revisited the Solar Lake flat microbial mats during the summer of 2021 to reveal the identity and functional potential of the benthic archaeal and bacterial communities with a predominant interest in understanding the genetic adaptation of the exposed versus shallow-water community to summer partial water recession, specifically phototrophic and methanogenic communities. We anticipated that the microbial community within the flat mats would experience a change in the genetic makeup causing taxonomic and biochemical zonation in the exposed versus shallow water in response to summer exposure. Hence, we sampled from both shallow water and exposed mat sites and applied in-depth sequencing using Illumina Novaseq followed by extensive analyses of raw reads, metagenome assembly, and metagenome-assembled genomes (MAGs) binning. Additionally, the absolute abundance of bacterial, archaeal, methanogenic, and sulfate-reducing communities was investigated.
MATERIALS AND METHODS
Sample collection and measuring mats physicochemical parameters
Triplicate sediment samples were collected from two shallow water and two exposed flat microbial mats in July 2021 (Fig. S1). The N-SOshallow-water and N-SOexposed samples were collected from sediments in the northern mountainside region covered by a small mountain shade during field sampling (12:00–13:00), and the E-SOshallow-water and E-SOexposed samples were collected from sediments in the eastern seaside part of the Lake, exposed to sunlight and closer to the Red Sea (Fig. S1). The shallow-water mat was covered by ~10 cm of lake water.
Samples were collected using handmade cores (5 cm diameter and 10 cm length) and were kept at 4°C until they reached the American University in Cairo (AUC) laboratories. The cores were made of polypropylene 50 mL syringe, cut after the tip by the university workshop, and autoclaved before use. The sediment cores were homogenized and then separated into 1.5 mL screw-capped tubes, under sterile conditions at AUC, then shock-frozen for later processing. The homogenized samples are referred to as “mats” throughout the paper.
The Hanna Combo pH/EC/TDS/Temp tester (Model HI98129) (Hanna instrument, Woonsocket, RI, USA) was used to measure pH and temperature in the field.
A subset of the homogenized sediments was dried at 60°C for 72 h (according to MEDAC LTD instructions). Each dried replicated sediment sample was equally mixed into one composite sediment and sent to MEDAC LTD (Chobham, UK) for total carbon, hydrogen, nitrogen, sulfur, and anions (sulfate, nitrate, and nitrite) quantification. The sediments’ gravimetric water contents were measured by drying ~1 g at 60°C for 72 h (23).
DNA extraction and NGS sequencing
Frozen sediments (0.2–0.4 g) were washed three times with phosphate buffer saline (PBS) buffer (ratio 1:5) and centrifuged at 3,000 RPM for 3 min. Washed sediment samples were then treated with lysozyme (100 mg/mL) for 1 h at 37°C, followed by proteinase K (20 mg/mL) for 30 min at 56°C. Treated samples were then processed with the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany). The DNeasy PowerSoil Kit was used per the manufacturer’s protocol, except for an hour bead beating step using a vortex adapter.
Solar Lake shallow-water and exposed microbial mats were sequenced in triplicate at Novogene (Cambridge, UK) using the Illumina NovaSeq 6000 (PE150). Each replicate had a sequencing depth of 20 M reads (Table S1).
Absolute abundance of bacterial, archaeal, methanogenic, and sulfate-reducing communities
Gene copies (qPCR) of bacterial and archaeal (16S) rRNA, dsrB, and mcrA genes were quantified using the Sybr Green-based assays as previously described (24, 25). The calibration curve for 16S rRNA and mcrA genes was created as previously described (24), whereas the dsrB gene calibration curve was made using a PCR amplicon of dsrB from Desulfovibrio vulgaris (10–105 dsrB copies). Microbial absolute abundance quantification was carried out using CFX Connect Real-Time PCR detection system (Bio-Rad, CA, USA). qPCR reaction efficiency was at least 80% (R2 > 0.98). Melting curve analysis was used to assess the presence of unspecific products.
Metagenomes, assembly, binning, and MAGs refinement
Quality control of the raw reads was performed using fastp (version 0.23.2) default parameters (26). High-quality paired-end reads were taxonomically classified using Kaiju (version 1.9.2) and the nr database (27).
High-quality reads from the triplicate sequencing files were co-assembled using Megahit (version 1.2.9) with minimum kmer 27, maximum kmer 127, and 10 kmer step (28). Individual replicate files from each sample were mapped to the sample’s assembled contigs using BBMap version 38.94 (options minid = 0.9, covstats, and scafstats were used) (29). MAGs were binned using MetaBAT2 (Version 2.12.1) (30). CheckM2 (version 1.0.0) was used to examine MAGs’ quality (completeness and contamination levels) (31). Any MAG failing the SAG/MAG current community standards (32), with >10% contamination, was refined by mdmcleaner (version 0.8.3) (33) and then subjected to another round of quality checks using CheckM2 (31). All high-quality (>70% completeness & <5% contamination) and medium-quality (>50% completeness and < 10% contamination) bins were further analyzed.
MAGs taxonomic classification, phylogeny, relative abundance, beta-diversity, and PTR calculation
The genome taxonomy database (GTDB) was used to taxonomically classify MAGs generated from each metagenome as a part of the mdmcleaner pipeline (33, 34). The CheckM utility command “profile” was used to assess the MAGs’ relative abundance within each metagenome; the data presented are based on the percent community estimation (35).
The draft MAGs maximum likelihood phylogenomic tree was generated using the GTOTree workflow (version 1.7.05) based on the alignment of 25 archaeal and bacterial single-copy housekeeping genes (SCGs) (36). Prodigal (version 2.6.3) was used to predict the open reading frames (ORFs), and then, target SCGs were identified with HMMER3 (version 3.3.2) (37, 38). SCGs were then individually aligned with muscle (version 5.1), trimmed with trimal (version 1.4.1), and FastTree2 (version 2.1.11) was used to concatenate SCGs and perform the phylogenetic tree (39–41). Only 227 out of 292 MAGs are displayed on the tree, since 69 MAGs contained few SCGs or had >10% redundancy with one or more represented MAG/s.
A Solar Lake archaea-specific tree was created to understand the relation of our archaeal MAGs with previously described taxa, based on 16S rRNA gene fragments from water samples from the Solar Lake (42). The CheckM utility command “ssu finder” was deployed on all archaeal MAGs to retrieve the 16S rRNA gene, when applicable (35). Water-column archaeal 16S rRNA partial genes were retrieved from the GeneBank database (43). Cytryn et al. (42) was the only study that analyzed the Solar Lake archaeal 16S rRNA, to date (42) . Water-column and MAGs 16S rRNA genes were aligned with muscle and manually trimmed (39, 41). The Maximum likelihood tree was generated using PhyML using 1,000 bootstraps (44). Trees were visualized using the Interactive Tree Of Life (iTOL) server (45). Beta-diversity analysis of shallow-water and exposed prokaryotic community and Compute Peak-to-Trough ratio (PTR) of MAGs calculation methods are presented in the supplementary file. Additionally, codes and scripts that were used to generate the metagenomic assemblies and MAG annotation are included in the supplementary file.
Metagenomes assemblies and MAGs annotations
Metabolic reconstruction of metagenome contigs and MAGs was carried out using the Distilled and Refined Annotation of Metabolism (DRAM, version 1.4.3) tool (46). MAGs’ ORFs were called using Prodigal (version 2.6.3) and were annotated using KOfam, Uniref90, Pfam, and dbCAN databases (access date: 4 January 2023) (47–50). The metagenome contigs were annotated using the KOfam, pfam, and dbCAN databases (48–50). All annotations presented in this study were retrieved from either distillate or product files generated by DRAM, except the environmental adaptation gene annotation which were retrieved from the DRAM raw annotation file. Genes would be considered correctly annotated if detected by at least two out of four databases.
For each sample, contigs coverage was extracted from the covstats files generated by BBMap. Contig coverage was used as an indication of gene copy numbers. The count per million (CPM) of functional genes, pathways, and Kyoto Encyclopedia of Genes and Genomes (KEGG) modules presented in the study was calculated according to the following formula for each replica:
Where contig coveragex is the contig coverage of all contigs containing the functional gene in the sample.
Statistical analysis
The Kruskal–Wallis test was applied to all qPCR data (51). Followed by an ad-hoc multiple comparison adjustment/correction test (Dunn test) (52). Deseq2 differential abundance pipeline was applied on N-SOshallow-water versus N-SOexposed and E-SOshallow-water versus E-SOexposed pairs to infer the significant difference between taxonomic assignments of exposed versus shallow-water microbial mats (53). The significant difference in the functional pathways, modules, and genes CPM between the exposed and shallow-water mats was tested using an unpaired-t-test followed by a two-stage step-up P-value false discovery rate (FDR) correction using the Benjamini, Krieger, and Yekutieli methods (54).
RESULTS
Physicochemical characteristics of Solar Lake microbial flat mats
The exposed and shallow-water microbial mats had neutral pH (6.78–7.63), except for N-SOexposed, which was mildly alkaline (pH 8.00). The temperature ranged between 35.26°C and 38.00°C in the shallow-water mats and between 39.73°C and 39.86°C in the exposed mats (Table 1). As expected, the water content in the exposed (31%–28%) was lower than the shallow-water mats (54%–55%). Nitrogen content ranged between 0.29% and 0.31%. Nitrate concentration was the lowest in the N-SOexposed mat (100 ppm) (Table 1).
TABLE 1.
Physicochemical characteristics of Solar Lake sediments
| Temperature (C°) | Water content (%) | pH | H % a | C %a | N %a | S %a | Nitrite (ppm)a | Nitrate (ppm)a | Sulfate (ppm)a | |
|---|---|---|---|---|---|---|---|---|---|---|
| N-SOshallow-water | 35.26 (±0.25) | 55 (±10) | 7.61 (±0.28) | 4.52 | 1.09 | 0.31 | 1.05 | 900 | 3,400 | 7,500 |
| N-SOexposed | 39.86 (±1.36) | 31 (±2) | 8.00 (±0.00) | 2.80 | 1.13 | 0.28 | 0.77 | 500 | 100 | 9,400 |
| E-SOshallow-water | 38.00 (±0.82) | 54 (±11) | 6.78 (±0.27) | 5.15 | 1.09 | 0.29 | 0.79 | 100 | 2,300 | 30.800 |
| E-SOexposed | 39.73 (±0.87) | 28 (±3) | 7.63 (± 0.26) | 3.21 | 1.00 | 0.29 | 1.14 | 100 | 1,700 | 11,900 |
per gram dry weight.
Metagenome-driven taxonomic and functional annotation of Solar Lake shallow-water and exposed flat mat microbiome
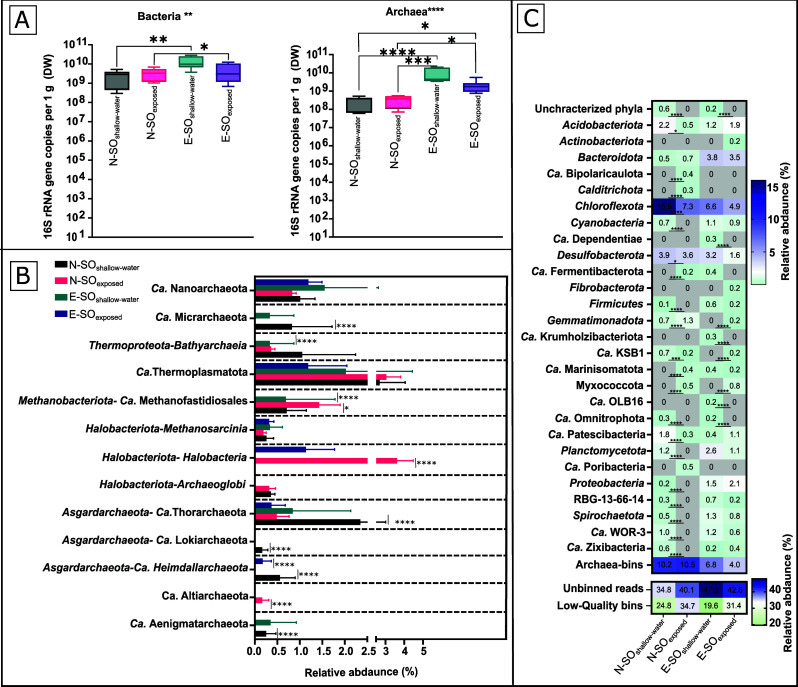
The raw reads and the assembled metagenomes were annotated to deduce the taxonomic distribution and identify specific potential functional characteristics, respectively. The bacterial community in the Solar Lake mats had an absolute abundance ranging between 2.34E+10 (N-SOshallow-water) and 1.32E+11 (E-SOshallow-water), whereas archaeal absolute abundance ranged between 2.72E+09 (N-SOshallow-water) and 8.38E+10 (E-SOshallow-water) (Fig. 1). Taxonomic classification of the raw reads showed that the archaeal microbial community was mainly composed of Euryarchaeota, Asgardarchaeota, Candidatus Thermoplasmatota, Bathyarchaeota, and members of the DPANN (acronym of Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanohaloarchaeota, and Nanoarchaeota). Meanwhile, the bacterial community was mainly composed of Pseudomonadota (Proteobacteria), Chloroflexota, Bacteroidota, and Cyanobacteria (Fig. S2). Raw reads taxonomic classification agrees with the Solar Lake MAGs taxonomic assignments (Fig. 1B and C).
Fig 1.
Relative and absolute abundance of the Solar Lake microbial shallow-water and exposed benthic communities. (A) Archaea and bacteria absolute abundance based on 16S rRNA gene copy numbers per gram dry weight. (B) Archaeal MAGs relative abundance. (C) Bacterial MAGs relative abundance. The relative abundance of MAGs reported is based on the proportion of a bin relative to the number of reads mapped to assembled contigs and adjusted for the size of the bin (CheckM “% community”). P-values indicate the statistical difference between samples (P-value ≤ 0.05 = *, P-value ≤ 0.01 = **, P-value ≤ 0.001 = ***, P-value ≤ 0.0001 = ****). In panels B and C, DESeq2 was used to infer the significantly different abundant phyla between each exposed and shallow-water sample pair (N-SOshallow-water versus N-SOexposed and E-SOshallow-water versus E-SOexposed). For qPCR data, Kruskal–Wallis test was used for the analysis of variance between samples (depicted above the qPCR graph), and the Dunn test was used for multiple comparison correction (P-values are depicted above the data points).
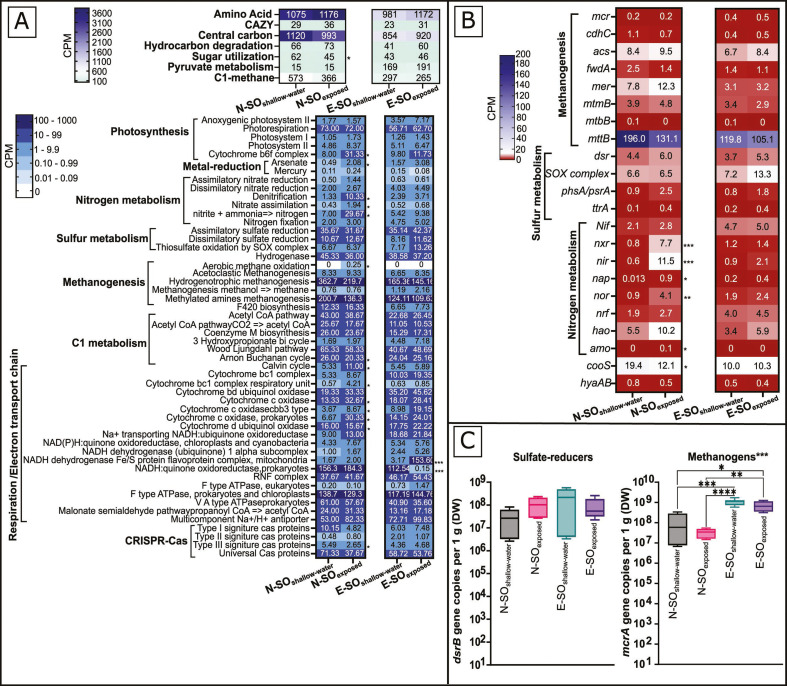
Annotations of the Solar Lake assemblies were performed to determine the genetic capacity of the microbial communities in the exposed and shallow-water mats (Fig. 2A and B; Table S2).
Fig 2.
Distribution of functional pathways, KEGG modules, and key functional genes across the shallow-water and exposed Solar Lake microbial mats metagenome assemblies. (A) Heat map showing the CPM per functional pathway or KEGG module. (B) Heat map showing CPM of the key functional genes involved in methanogenesis, sulfur, and nitrogen metabolism, in the assemblies. Numbers shown in each cell represent CPMs for each pathway, module, or functional gene. (C) Box plots showing the number of gene copies per gram dry weight (DW) of mcrA and dsrB, respectively. P-values (FDR corrected) indicate statistical differences between exposed and shallow water communities (P-value ≤ 0.05 = *, P-value ≤ 0.01 = **, P-value ≤ 0.001 = ***, P-value ≤ 0.0001 = ****). In panel C, the Kruskal–Wallis P-values are depicted above the graphs, whereas Dunn test P-values are depicted between each significantly different pair of data.
Genes responsible for photosystem I (psaA) and II (psbA and psbD) and anoxygenic photosystem II (pufL and pufM) were detected in all samples. The gene content of the Solar Lake mats is suggestive of autotrophic communities that depend on the Wood-Ljungdahl cycle (40–65 CPM), Arnon-Buchanan cycle (20–26 CPM), and Calvin cycle (5–11 CPM) (Fig. 2A).
The relative abundance of genes classified by the CAZy database responsible for the degradation of complex carbohydrates ranged between 29 and 36 CPM (Fig. 2A). The AA1 (laccases/ferroxidases) carbohydrate-active enzyme (Cazyme) family was significantly enriched (10 CPM, P-value < 0.05) in the N-SOexposed mat (Fig. S3).
The genes encoding for the methanogenesis key enzyme, methyl coenzyme M reductase (mcrAB), were detected in all samples. The mcrAB had comparable abundance (CPM) (Fig. 2B) with the mcrA absolute abundance (qPCR) (Fig. 2C), with N-SOshallow-water and N-SOexposed pair having lower CPM compared with E-SOshallow-water and E-SOexposed (Fig. 2B and C). The four Solar Lake metagenomes had the key genes involved in the production of methane from acetate (cdhc and acs), monomethylamine (mtmB), trimethylamine (mttB), and H2/CO2 (mer and fwdA) (Fig. 2B). The genetic potential for aerobic methane oxidation was detected in the N-SOexposed sample only (Fig. 2A).
The solar lake shallow-water and exposed microbial mats have the genetic potential to reduce sulfate as well as tetrathionate and oxidizing/reducing thiosulfate (Fig. 2A). The dsrB gene, encoding sulfate reduction, had a similar relative (CPM) and absolute (qPCR) abundance (Fig. 2B and C). All Solar Lake mat samples had a similar nitrogen fixation key gene abundance (nif). In contrast, genes involved in nitrification (amo and hao) and denitrification (nir) were enriched (P ≤ 0.05 and 0.001, respectively) in N-SOexposed (Fig. 2A and B).
An overview of bacterial and archaeal MAGs
A total of ~240 M high-quality raw reads (~60 M for each metagenome) were assembled and binned to generate a total of 108 archaeal and 601 bacterial MAGs (Table S1). This study investigated predominantly medium-to-high-quality MAGs, which included 72 archaeal and 220 bacterial MAGs (Table S3), with the exception of eight archaeal MAGs with ≥50% completeness and ≤10% contamination criteria according to mdmcleaner, and not CheckM2, and the only Candidatus Altiarchaeota MAG (S-SO2-bin.64) with 49% completeness (Table S3)
MAGs summary, including but not limited to MAGs completeness, contamination, GTDB taxonomic assignment, and the number of predicted genes, is presented (Table S3).
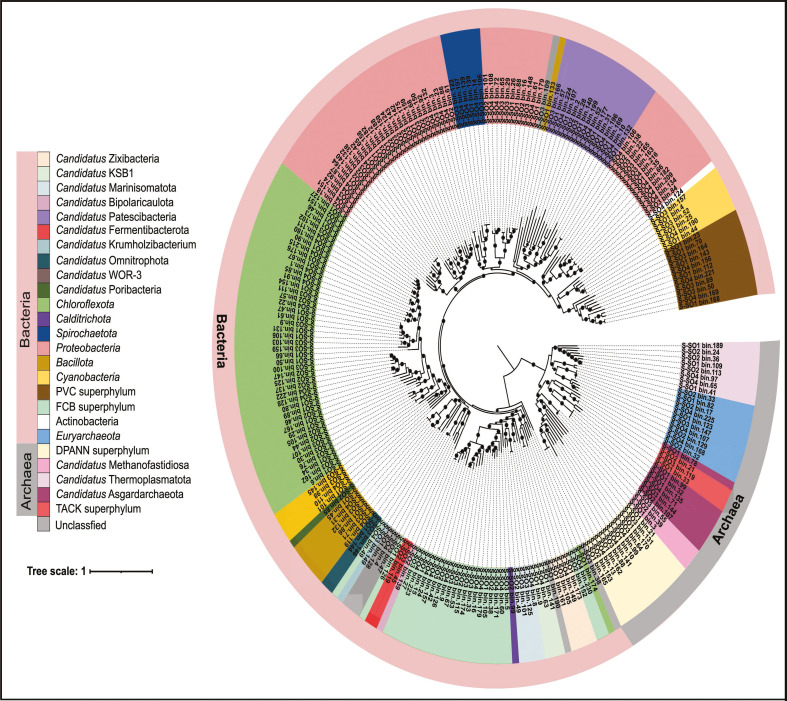
The Solar Lake MAGs spanned nine archaeal and 28 bacterial phyla based on GTDB taxonomy (Fig. 3; Table S3). The archaeal MAGs assigned to Candidatus Thermoplasmatota had the highest relative abundance in all the samples (Fig. 1B). We also detected MAGs belonging to the deeply branching Asgardarchaeota superphylum in all mats.
Fig 3.
Phylogenomic tree of the Solar Lake shallow-water and exposed mat MAGs. The maximum likelihood phylogenomic tree of up to 25 archaeal and bacterial single-copy housekeeping genes (SCGs) was generated using the GoToTree workflow. Nodes with bootstrap higher than 50% are represented by a closed circle. MAGs assigned to different superphyla or phyla when no superphylum is available are highlighted by different colors. MAGs assignment to a superphylum or phylum was based on GTDB taxonomic assignment.
MAGs assigned to Candidatus Aenigmatarchaeota, and Candidatus Micrarchaeota were shallow-water microbial mat-specific MAGs. On the contrary, MAGs assigned to Halobacteriota (class: Halobacteria) were only detected in the exposed microbial mat (Fig. 1B; Table S3).
Shallow-water flat mat-specific bacterial MAGs were assigned to uncharacterized phyla (2 MAGs) and Candidatus Omnitrophota (2 MAGs), and MAGs assigned to Myxococcota were detected only in the exposed mats (Fig. 1C).
MAGs and reads assigned to the Chloroflexota phylum had the highest abundance across all samples (Fig. 1C; Fig. S2). Sulfate-reducing bacteria assigned to the Desulfobacterota phylum represented 1.6-3.9% of the total Solar Lake benthic community (Fig. 1C). Cyanobacterial MAGs had a relative abundance of 0.7%–1.1%.
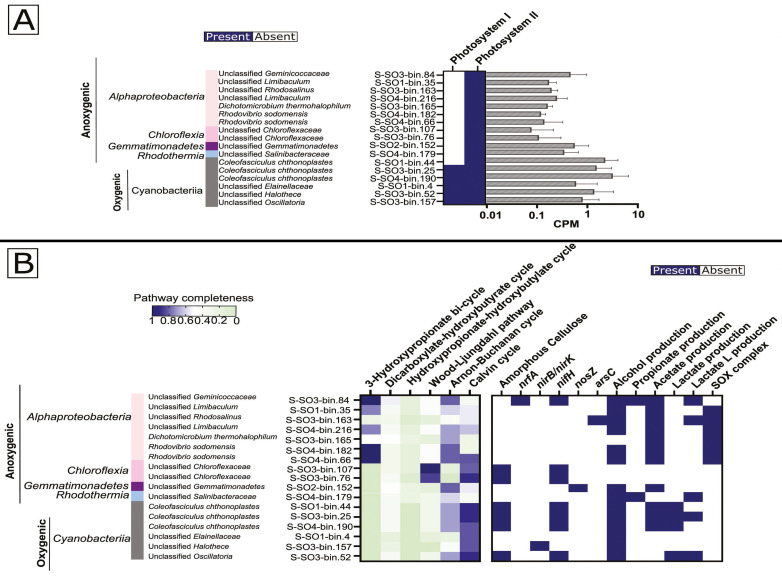
Phototrophic potential of Solar Lake MAGs
We detected genes encoding for phototrophy in a total of 17 Solar Lake microbial mat MAGs, with six cyanobacterial and 11 anoxygenic phototrophic MAGs. Three of the six cyanobacterial MAGs belong to C. chthonoplastes (Fig. 4A and B; Table S3). The highest relative abundance for C. chthonoplastes was detected in E-SOexposed (~0.87%), followed by N-SOshallow-water (~0.56%) and E-SOshallow-water (0.39%) (Table S4). The genes encoding photosystem I and II in the C. chthonoplastes MAGs showed higher CPMs by trend than other photosynthetic Cyanobacteria (Fig. 4A). C. chthonoplastes MAGs possessed a nearly complete set of genes for the Calvin cycle and had the genetic potential to degrade amorphous cellulose, fix nitrogen (nifH), and produce lactate, acetate, and alcohol (Fig. 4B).
Fig 4.
The functional genetic potential of the recovered phototroph MAGs in the Solar Lake mats. (A) Photosystem I and II in phototrophic bacterial MAGs, (B) Carbon fixation and other metabolic pathways in phototrophic MAGs. The genetic potential for phototrophy in MAGs is indicated by the presence or absence and the CPM of photosystem I and II. Carbon fixation pathways are indicated by percent completeness in the Solar Lake MAGs. One indicates the detection of a complete pathway.
The other cyanobacterial MAGs were only detected in shallow-water mats (Fig. 4A and B; Table S4) and included an Elainellaceae MAG (National Center for Biotechnology Information [NCBI] taxonomy: unclassified Leptolyngbya) assembled from the N-SOshallow-water metagenome and Oscillatoria and Halothece unclassified species from the E-SOshallow-water (Fig. 4A and B; Table S4). Similar to C. chthonoplastes, shallow-water mats cyanobacterial MAGs encoded a nearly complete set of genes for the Calvin cycle (Fig. 4B). The Oscillatoria MAG additionally showed the genetic potential to fix nitrogen and degrade amorphous cellulose (Fig. 4B).
Of the 11 anoxygenic phototrophic MAGs, only one MAG (S-SO2-bin.152) showed the genetic potential for anoxygenic phototrophy in the N-SOexposed sample and was taxonomically assigned to an unclassified class within Gemmatimonadetes. The former had genes involved in the Arnon-Buchanan cycle for carbon fixation and alcohol and acetate production (Fig. 4B). In E-SOshallow-water, two MAGs assigned to an unknown genus within Chloroflexaceae had the genetic potential for phototrophy, carbon fixation via the Wood-Ljungdahl pathway, and the ability to fix nitrogen (Fig. 4B).
Seven of the 11 MAGs belonged to the Alphaproteobacteria. Five of these MAGs contained genes for the 3-hydroxypropionate bi-cycle for carbon fixation (Fig. 4B), whereas six MAGs had the genetic capacity for thiosulfate oxidation. One MAG assigned to an unknown genus within the Geminicoccaceae family had the genetic potential to reduce nitrite to ammonia (nrfA) and fix nitrogen (nifH).
Methanogenesis potential of Solar Lake MAGs
Genes encoding the methanogenesis pathway were detected in different archaeal MAGs. A complete methanogenesis pathway was detected in four nearly complete MAGs across the four Solar Lake mat samples (Fig. 5A). These MAGs were taxonomically classified within the Methanosarcinia class (species: Methanohalobium evestigatum). All M. evestigatum MAGs had the key genes responsible for acetolactic (acs), methyleamine (mtmB), and methanogenesis (Fig. 5A).
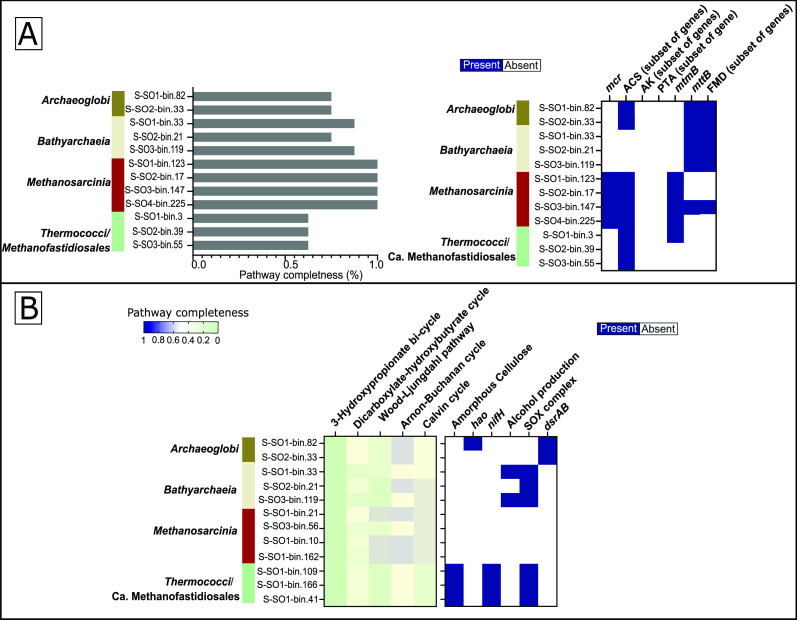
Fig 5.
The functional genetic potential of the recovered methanogen MAGs in the Solar Lake mats. (A) Methanogenesis pathway completeness and key functional genes in the Solar Lake MAGs. (B) Carbon fixation and selected metabolic pathway key genes in the putative methanogen MAGs. The genetic potential of each MAG to produce methane is indicated by methanogenesis pathway completeness and the presence of key methanogenesis. Carbon fixation and methanogenesis pathways are indicated by percent completeness. One indicates the detection of a complete pathway.
Additionally, more than half of the genes involved in acetolactic methanogenesis were detected in three MAGs that belonged to the Candidatus Methanofastidiosales (Fig. 5A). These three MAGs showed the genetic potential to degrade amorphous cellulose, fix nitrogen, and oxidize thiosulfate (Fig. 5B).
Moreover, three MAGs affiliated with the Bathyarchaeia class encoded a nearly complete set of methanogenesis genes (Fig. 5A). The functional annotation analysis suggests that the microorganisms represented by these MAGs are capable of producing methane from trimethylamine (mttB) and CO2 (FMD) (Fig. 5A). Besides methanogenesis, the Bathyarchaeia MAGs had the genetic potential to oxidize thiosulfate and produce alcohol.
In addition, two Archaeoglobaceae MAGs had most of the genes involved in acetoclastic and trimethylamine-driven methanogenesis. These two Archaeoglobaceae MAGs also showed the genetic potential to perform dissimilatory sulfate reduction (Fig. 5A and B).
Differential ROS detoxification, UV radiation resistance, and salt-tolerance genes in the Solar Lake microbial shallow-water versus exposed mats
Both the Solar Lake shallow-water and exposed microbial mats’ metagenomes and MAGs encoded salt tolerance genes such as the osmoprotectant transport systems (opuABCD), the glycine betaine/proline transporter system (proV), and the betaine-aldehyde dehydrogenase (betB). Additionally, the relative abundance (CPM) of the genes encoding salt tolerance was significantly greater (P ≤ 0.05) in the N-SOexposed than in the N-SOshallow-water metagenome (Fig. 6A). All 17 MAGs defined by their genetic potential for phototrophy had at least one copy of these osmoprotectant genes; except S-SO1-bin.4 and S-SO4-bin.179. On the contrary, within the 12 methanogenic archaeal communities, osmoprotectant genes were detected only in four M. evestigatum MAGs and one Archaeoglobaceae MAG (Fig. 6B).
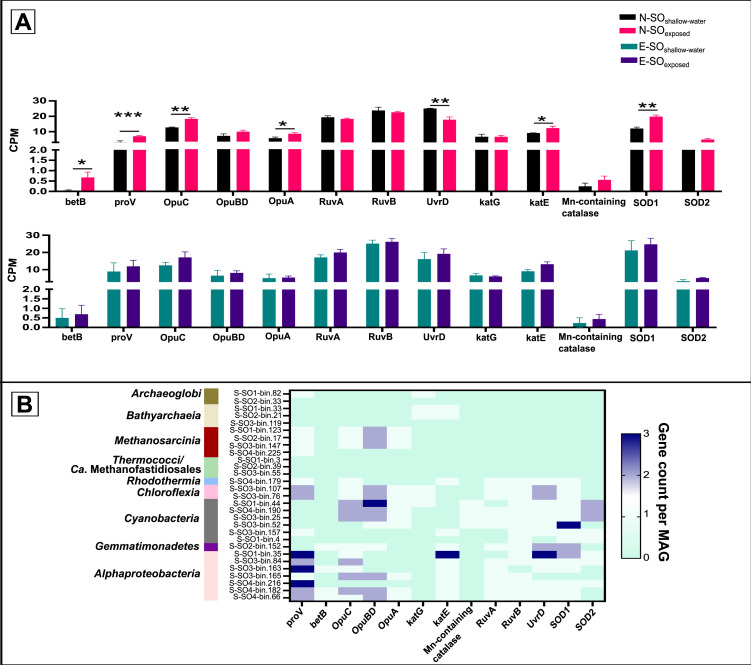
Fig 6.
Environmental adaptation genes in the Solar Lake shallow-water and exposed microbial mats. (A) Relative metagenomic abundance (CPM) of genes encoding stress tolerance. (B) Distribution pattern (CPM) of stress-tolerance genes among MAGs representing phototrophic or methanogenic populations. P-values (FDR corrected) are shown at the top of each comparison. P-values indicate the statistical difference (P-value ≤ 0.05 = *, P-value ≤ 0.01 = **, P-value ≤ 0.001 = ***, P-value ≤ 0.0001 = ****).
Compared with N-SOshallow-water, the relative abundance (CPM) of ROS detoxifying genes, such as catalase-peroxidase (katE) and superoxide dismutase genes (SOD1) in the metagenome, was significantly greater (P ≤ 0.05 and 0.01) in N-SOexposed (Fig. 6A) compared with N-SOshallow-water. Additionally, the E-SOexposed had higher CPM of ROS detoxifying genes compared with E-SOshallow-water, but only by trend (Fig. 6A). Note that all the MAGs representing phototrophic populations had at least one or more genes involved in ROS response (Fig. 6B).
The urvD and the RuvAB genes encoding UV radiation resistance genes were detected in all 17 phototrophic MAGs defined by their genetic potential for phototrophy, except S-SO1-bin.4 and S-S03-bin.165 (Fig. 6B). RuvAB and urvD were absent in all MAGs affiliated with methanogens.
DISCUSSION
The heliothermal Solar Lake in Taba, Egypt, is expected to host distinctive microbial communities able to adapt to the lake’s limnological cycle (11). Our present study is the first to revisit the Solar Lake benthic microbial communities since 1998 (16). We used a genome-centric approach to highlight the structural and functional adaptations occurring in exposed versus shallow-water mats in response to the summer’s partial water receding. We focused on the cyanobacterial and methanogenic communities. This study is a logical continuation of previous reports from 1970s and 1980s, which relied on conventional microbiology techniques (8, 13).
Bacterial and archaeal absolute abundance was comparable in the exposed (N-SOexposed and E-SOexposed) versus shallow water (N-SOshallow-water and E-SOshallow-water) (Fig. 1). Unlike the absolute abundance, archaeal and bacterial phyla-relative abundance and beta-diversity significantly shifted between the exposed and shallow-water flat microbial mats (Fig. 1; Fig. S4).
Archaeal benthic community in the shallow versus exposed Solar Lake microbiome
MAGs assigned to the symbiotic Candidatus Micrarchaeota and Candidatus Aenigmarchaeota were uniquely detected in the shallow-water microbial mats. It is unclear why Ca. Aenigmarchaeota and Bathyarchaeia (order B26-1) MAGs were specifically detected in the shallow-water microbial mat (S-SO1_bin.160 and S-SO3_bin.10). However, horizontal gene transfer events between the two phyla were previously reported, hence emphasizing the symbiotic relationship between Bathyarchaeia and Ca. Aenigmarchaeota members (55, 56). Both Ca. Aenigmarchaeota and Bathyarchaeota were detected in other microbial mats (57), deep-sea (56) freshwater sediments (58), coastal sediments (59), and groundwater sediments (60).
The aerobic heterotrophic Halobacteria class (1), previously reported to be the most abundant archaeal member in the Solar Lake water during summer holomixis (42), was not detected in the shallow-water microbial mats community (Fig. 1B). This could be driven by the high salinity and density of the water creating a sub-oxic/anoxic environment disfavoring Halobacteria growth (11, 18). On the contrary, Halobacteria MAGs were enriched in the Solar Lake exposed samples (N-SOexposed and E-SOexposed) (Fig. 1B) and denitrifying, possessing nir gene, Halobacteria (2.5%) and Anaerolineae (3.3%) MAGs were only enriched in N-SOexposed.(Table S5). The latter bacterial genus is included because it has been previously shown to be involved in denitrification in hypersaline ecosystems (61–64). Nevertheless, Anaerolineae has not been associated with nitrite reduction and nitric oxide generation (nir genes) (64). The relative depletion of both nitrate and nitrite in N-SOexposed (Table 1) supports the presence of active denitrification.
Similar to hypersaline marsh in Tristomo bay and hypersaline microbial mats in Guerrero Negro (65, 66), Candidatus Thermoplasmatota (class E2) MAGs were detected in the Solar Lake mats. This order was identified in Guaymas Basin hydrothermal vent (58). Asgardarchaeota superphylum MAGs were detected in all Solar Lake microbial mats and were previously reported as major players in the Shark Bay, Australia microbial mats (5, 67, 68). The detection of Asgardarchaeota MAGs carrying complete carbon fixation pathways and several glycoside hydrolases in several microbial mat systems (5, 67, 68) support the hypothesis that they play an important role in carbon cycling in anoxic mat systems (67, 69). Functional prediction of the Solar Lake Asgard MAGs suggests that they play a similar role (Table S6).
Bacterial benthic community in the shallow versus exposed Solar Lake microbiome
Candidatus Omnitrophota MAGs were detected only in the shallow-water flat mats, whereas Myxococcota MAGs were specific to the exposed mats.
Members of Candidatus Omnitrophota are metabolically versatile and detected in diverse environments (70, 71). The majority of the Candidatus Omnitrophota members lack ROS response genes including the two MAGs assembled in our study (71). On the other hand, all Myxococcota MAGs had ROS response genes, explaining their presence in the exposed microbial community (Table S7). Unlike the sulfated exopolysaccharide (EPS) recycling Myxococcota members detected in the cyanobacterial mat ecosystems in Shark Bay (Australia) (72), the Solar Lake Myxococcota members possessed genes responsible for the degradation of cellulose, arabinose, and xylose (Table S7).
It is worth noting that the bacterial phylum Chloroflexota dominated the Solar Lake shallow-water and exposed mats similarly to the Shark Bay, Australian smooth mats (Fig. 1C; Fig. S2) (67). Phototrophic Chloroflexota MAGs (family: Chloroflexaceae) genetic makeup suggests that they play a role in anoxygenic phototrophy and carbon fixation using the Wood-Ljungdahl pathway. Since previous studies have shown that Anaerolineae are chemoorganotrophs and play a role in carbon degradation and mat stability (73, 74), Anaerolineae MAGs within Chloroflexota are likely to play a similar role in the Solar Lake.
Phototrophy and carbon fixation in the Solar Lake exposed versus shallow-water microbiome
The taxonomic composition of Cyanobacteria within the shallow-water and exposed microbial mats exhibited similarities to the 1977 study by Krumbein and colleagues, where C. chthonoplastes and Aphanothece halophytica (current name: Halothece sp. PCC 7418) were detected in both studies (8). Nevertheless, Krumbein and colleagues showed that the summer shallow-water mats were predominantly occupied by coccoid Cyanobacteria, specifically A. halophytica (Halothece sp. PCC 7418) (8). However, our analysis detected only one MAG affiliated with Halothece (with a relative abundance of 0.46%) in the E-SOshallow-water sample (Table S4). Surprisingly, the filamentous C. chthonoplastes that were previously detected mainly in winter shallow-water mats (8) were detected in our summer shallow-water (N-SO and E-SO) and exposed (E-SO) mats with relative abundance ranging from 0.87% to 0.39%. Additionally, MAGs assigned to C. chthonoplastes had one of the highest PTR (peak-to-trough ratio) values in the studied Solar Lake benthic community, suggesting active replication in comparison to other community members (Fig. S5). Previous reports documented that C. chthonoplaste was able to resist 700 µ E m−2 S−1 and produce preoxidase and superoxide dismutase (75), and several other genes were reported to be upregulated (76) to alleviate photooxidative stress. Such genes, glutathione peroxidase, carotenoid (crtI and crtB), and σ-factor/σE were detected in Solar Lake C. chthonoplastes MAGs (Fig. 6; Table S8). Our analysis challenges the previous notion, primarily based on visualization and cyanobacterial cell morphology, that filamentous Cyanobacteria are exclusively abundant in the Solar Lake winter microbial mats because of their sensitivity to photooxidative stress (8).
As expected, cyanobacterial MAGs possess genes for the Calvin cycle. Nonetheless, two Chloroflexota MAGs had the genetic potential to fix carbon using the Wood-Ljungdahl pathway. This is in contrast to most Chloroflexota that are known to use HP3 bi-cycle; the pathway by which Chloroflexota members usually fix carbon (77). The Solar Lake E-SOexposed aerobic anoxygenic photrophic MAG (AAP) R. sodomensis MAGs had a nearly complete 3-Hydroxypropionate bi-cycle gene set and the SOX complex operon, suggesting its autotrophic potential (78). Previous studies reported that R. sodomensis cannot grow autotrophically (79).
Methanogenesis in the Solar Lake exposed and shallow-water flat mats communities
Methanogens and methanogenesis-related genes were detected in the four microbial mats samples. The absolute and relative abundance of mcrA followed similar trends. The number of mcrA copies was 2–3 times higher than methanogenic cell counts previously reported in the Solar Lake sediments (13). Most probably this is due to the presence of more than one mcrA copy in the genomes of the Solar Lake methanogens, and a limitation in the detection protocol of the previous study (13).
The identity of the Solar Lake benthic methanogens has not been previously explored (13). In this study, we defined methanogen MAGs as those MAGs that harbored ≥60% of the genes involved in methanogenesis based on DRAM annotation. We were unable to define methanogens based on the presence or absence of mcrA because this gene was poorly covered in our metagenomes and the metagenomes of similar environments (5). Putative methanogen MAGs were assigned to M. evestigatum, Archaeoglobales, Candidatus Methanofastidiosales orders, and an unclassified class within the phylum Bathyarchaeia (80–85). Similarly, the methanogenic community members Methanosarcinales and Candidatus Methanofastidiosales have been identified in the hypersaline microbial mats of Guerrero Negro desert region (66). Intriguingly, our study revealed that the previously reported Solar Lake 16S rRNA gene of Archaeal Cluster II (42) formed a cluster (bootstrap: 87%) with the 16S rRNA genes of Archaeoglobales MAGs in this study. This finding provides the first evidence that Archaeoglobales is one of the methanogenic lineages present in both the planktonic and shallow-water mat benthic communities of the Solar Lake (Fig. S6).
Giani and colleagues have shown that the major substrate used for methanogenesis by the Solar Lake benthic community is methylated amines, explaining their coexistence with sulfate reducers (13). Our data support this hypothesis based on (i) high relative abundance (CPM) of genes encoding methylotrophic methanogenesis via the utilization of methylated amines, with mttb as the specific biomarker, and (ii) all Solar Lake methanogen MAGs, except the MAG affiliated to Candidatus Methanofastidiosales, encoded the genes highly indicative of monomethylamine- or trimethylamine-based methanogenesis. In addition to methylotrophic methanogenesis, methanogens in the exposed and shallow-water flat mats have the genetic potential to perform either acetoclastic (M. evestigatum and Candidatus Methanofastidiosales) or hydrogenotrophic (Archaeoglobales and Bathyarchaea) methanogenesis. This is in agreement with the previous hypothesis that methylotrophic methanogenesis is the predominant methanogenic pathway but not the sole one, in hypersaline microbial mats (66, 86). Interestingly, besides methanogenesis, the methanogenic community in the Solar Lake had the genetic potential to reduce sulfate, oxidize thiosulfate, and fix nitrogen. The presence of dsrA/B genes in the Solar Lake Archaeoglobales MAGs might suggest single-cell sulfate-dependent anaerobic methane oxidation (AMO) as previously described with Candidatus Methanomixophus dualitatem (82–85, 87).
The Solar Lake exposed samples (N-SOexposed and E-SOexposed) had lower moisture content (Table 1), and it is anticipated that more oxygen would penetrate the exposed sediments compared with the suboxic shallow-water sediments (18). Hence, it is likely that aerobic methane oxidation would be present in both exposed samples; however, we only detected it in N-SOexposed. The inability to detect aerobic methanotroph (pmoA) in E-SOexposed is likely attributed to their low abundance, which often goes unnoticed in shotgun sequencing (5).
Solar Lake-exposed and shallow-water mat communities’ adaptation to summer exposure
The Solar Lake is characterized by a water salinity reaching 18% during the summer season (11). Hence, the Solar Lake benthic microbial communities are anticipated to possess salinity stress genes analogous to similar ecosystems (5, 88). Environmental changes such as water subsiding from microbial mats can profoundly affect microbial community functional capacity due to oxygen intrusion, higher salinity, and drought. Few studies looked at the effect of tidal-driven desiccation or salinity gradients on the structure of similar cyanobacterial mats, but not on their function (21, 22).
Here, we showed that exposed flat microbial mats specifically N-SOexposed were enriched in dihydrogeodin oxidases/laccases (AA1), a Cazyme gene involved in the oxidation of recalcitrant organic matter such as polyphenolic. This is likely due to drought and oxygen intrusion, into this sample, as previously shown in other environments (24, 89). Moreover, N-SOexposed and E-SOexposed assemblies contained a higher number of ROS response genes. The low number of ROS detoxification genes in the flat shallow-water mats may be supported by the shallow-water suboxic conditions (DO = 2.7 mg/l) previously reported in the lake sediments (18). Moreover, the enrichment of genes involved in ROS response and the genetic potential for osmoprotectants production in the exposed mat N-SO could indicate that this mat is going through oxygen and drought stress (Fig. 6). To our knowledge, the effect of drought was not previously studied on hypersaline stromatolite-like systems. Nevertheless, the reported stress response employed by the Solar Lake microbial mat community, evident in the ROS and salinity response gene enrichment, is similar to other environments such as vegetated mudflat ecosystems (90), paddy soil (24), grassland rhizosphere (91), and alkaline wetland (92).
Our data show that there is an altered genetic makeup of the Solar Lake benthic microbial mat communities in the exposed versus shallow-water zones during the summer. Nevertheless, more sampling sites throughout the lake of exposed and shallow water are needed to ensure the robustness of our observations.
Conclusion
In summary, our study sheds light on the benthic microbial communities thriving in the Solar Lake, Taba, Egypt. The microbial mats in this unique ecosystem exhibit remarkable structural and functional complexity. We stressed on the significant contribution of phototrophy and methanogenesis. Here, we unveiled key findings that collectively contribute to a deeper understanding of the Solar Lake microbial flat mat benthic community. These findings include an increase in ROS response genes and the unique identification of Halobacteriota, Candidatus Altiarchaeota, and Myxococcota in the exposed mats. This iterates the functional and structural adaptation to external environmental stressors such as oxygen intrusion and drought caused by the partial receding of water.
Moreover, we showed that the abundance and computed active replication of C. chthonoplaste indicate that this Cyanobacteria is not only a major player in the winter, as previously reported (8), but in the summer community as well. Additionally, methylotrophic methanogenesis is the most abundant methanogenic pathway in the Solar Lake mats, and the identified Archaeoglobales genetic makeup can contribute to AMO. Further studies are needed to understand the seasonal variation and microbial stratification in this unique Solar Lake ecosystem.
Given the current climate crisis, and the anticipated prolonged drought events in Sinai, Egypt, and the Middle East (93, 94), it is crucial to understand how partially exposed microbial mats respond to drought and oxygen intrusion. There is limited research on the effects of climate change on microbial mats ecosystems (95–97), and none has specifically addressed drought effects. Decoding the hypersaline microbial mats’ genetic adaptation, particularly the exposed mats that are subjected to partial drought, might aid in developing adaptive management plans for maintaining or restoring such ecosystems.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the British Ecological Society “Ecologist in Africa” grant (Project number: EA19/1215).
Mr. Ayman Mohamed is acknowledged for his help during sample collection.
R.Z.A. and R.S. designed the study. R.Z.A., A.A., and M.M. collected the samples. R.Z.A. and S.F.A. carried out laboratory experiments. R.Z.A. and A.H.A.E. analyzed and interpreted the data. R.Z.A. performed statistical analyses and generated figures. R.Z.A. and R.S. wrote the manuscript. R.S. funded the project, except for the qPCR experiments that were undertaken in the W.L. laboratory. All authors discussed the results and commented on the manuscript at all stages.
Contributor Information
Rania Siam, Email: rsiam@aucegypt.edu.
Ryan J. Newton, University of Wisconsin-Milwaukee, Milwaukee, Wisconsin, USA
Ana Camila Gonzalez-Nayeck, Harvard University, Cambridge, Massachusetts, USA.
DATA AVAILABILITY
The metagenome shotgun sequencing raw data were deposited in the NCBI BioProject database under the accession numbers PRJNA1004913. Metagenome assemblies and metagenome assembled genomes (MAGs) used in this study were deposited in Zenodo repository at https://doi.org/10.5281/zenodo.10521873 (98). The data files names corresponding to each sample are indicated in Table S1.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00095-24.
Supplemental materials and methods and Figures S1 to S6.
Solar Lake shallow-water and exposed mat metagenome statistics.
Solar Lake shallow-water and exposed mat metagenome DRAM annotations.
Solar Lake shallow-water and exposed mat MAG summary.
The cyanobacterial MAG relative abundance in the Solar Lake shallow-water and exposed mats.
Nitrogen cycling genes distribution across the Solar Lake shallow-water and exposed archaeal and bacterial MAGs and the relative abundance of nir gene-containing MAGs.
Carbon fixation pathways, Cazyme, and short-chain fatty acids and alcohol conversion genes in Asgardarchaota.
Solar Lake flat mat Myxococcota MAGs annotation using DRAM.
Photooxidative stress gene in Coleofasciculus chthonoplastes MAGs.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Saccò M, White NE, Harrod C, Salazar G, Aguilar P, Cubillos CF, Meredith K, Baxter BK, Oren A, Anufriieva E, Shadrin N, Marambio-Alfaro Y, Bravo-Naranjo V, Allentoft ME. 2021. Salt to conserve: a review on the ecology and preservation of hypersaline ecosystems. Biol Rev Camb Philos Soc 96:2828–2850. doi: 10.1111/brv.12780 [DOI] [PubMed] [Google Scholar]
- 2. Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, von Mering C, Bebout BM, Pace NR, Bork P, Hugenholtz P. 2008. Millimeter‐scale genetic gradients and community‐level molecular convergence in a hypersaline microbial mat. Mol Syst Biol 4:198. doi: 10.1038/msb.2008.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE, Hugenholtz P, Goodrich J, McDonald D, Knights D, Marshall P, Tufo H, Knight R, Pace NR. 2013. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J 7:50–60. doi: 10.1038/ismej.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dillon ML, Hawes I, Jungblut AD, Mackey TJ, Eisen JA, Doran PT, Sumner DY. 2020. Energetic and environmental constraints on the community structure of benthic microbial mats in Lake Fryxell, Antarctica. FEMS Microbiol Ecol 96:fiz207. doi: 10.1093/femsec/fiz207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong HL, White RA, Visscher PT, Charlesworth JC, Vázquez-Campos X, Burns BP. 2018. Disentangling the drivers of functional complexity at the metagenomic level in Shark Bay microbial mat microbiomes. ISME J 12:2619–2639. doi: 10.1038/s41396-018-0208-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schneider D, Arp G, Reimer A, Reitner J, Daniel R. 2013. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, central Pacific. PLoS One 8:e66662. doi: 10.1371/journal.pone.0066662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jørgensen BB, Cohen Y. 1977. Solar Lake (Sinai). 5. the sulfur cycle of the bcnthic cyanobacterial mats. Limnol Oceanogr 22:657–666. doi: 10.4319/lo.1977.22.4.0657 [DOI] [Google Scholar]
- 8. Krumbein WE, Cohen Y, Shilo M. 1977. Solar Lake (Sinai). 4. stromatolitic cyanobacterial mats. Limnol Oceanogr 22:635–656. doi: 10.4319/lo.1977.22.4.0635 [DOI] [Google Scholar]
- 9. Cohen Y, Krumbein WE, Shilo M. 1977. Solar Lake (Sinai). 3. bacterial distribution and production. Limnol Oceanogr 22:621–634. doi: 10.4319/lo.1977.22.4.0621 [DOI] [Google Scholar]
- 10. Cohen Y, Krumbein WE, Shilo M. 1977. Solar Lake (Sinai). 2. distribution of photosynthetic microorganisms and primary production. Limnol Oceanogr 22:609–620. doi: 10.4319/lo.1977.22.4.0609 [DOI] [Google Scholar]
- 11. Cohen Y, Krumbein WE, Goldberg M, Shilo M. 1977. Solar Lake (Sinai). 1. physical and chemical limnology. Limnol Oceanogr 22:597–608. doi: 10.4319/lo.1977.22.4.0597 [DOI] [Google Scholar]
- 12. Por FD. 1968. Solar Lake on the shores of the Red sea. Nature 218:860–861. doi: 10.1038/218860a0 [DOI] [Google Scholar]
- 13. Giani D, Giani L, Cohen Y, Krumbein WE. 1984. Methanogenesis in the hypersaline Solar Lake (Sinai). FEMS Microbiol Lett 25:219–224. doi: 10.1111/j.1574-6968.1984.tb01460.x [DOI] [Google Scholar]
- 14. Conrad R, Frenzel P, Cohen Y. 1995. Methane emission from hypersaline microbial mats: lack of aerobic methane oxidation activity. FEMS Microbiol Ecol 16:297–306. doi: 10.1111/j.1574-6941.1995.tb00294.x [DOI] [Google Scholar]
- 15. Jorgensen BB, Revsbech NP, Cohen Y. 1983. Photosynthesis and structure of benthic microbial mats: microelectrode and SEM studies of four cyanobacterial communities. Limnol Oceanogr 28:1075–1093. doi: 10.4319/lo.1983.28.6.1075 [DOI] [Google Scholar]
- 16. Teske A, Ramsing NB, Habicht K, Fukui M, Küver J, Jørgensen BB, Cohen Y. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl Environ Microbiol 64:2943–2951. doi: 10.1128/AEM.64.8.2943-2951.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohen Y, Aizenshtat Z, Stoler A, Jørgensen BB. 1980. The microbial geochemistry of Solar Lake, Sinai, p 167–172. In Trudinger PA, Walter MR, Ralph BJ (ed), Biogeochemistry of ancient and modern environments, 1st ed. Springer, Berlin, Heidelberg. [Google Scholar]
- 18. Mustafa GA, Abd-Elgawad A, Abdel-Haleem AM, Siam R. 2014. Egypt’s Red sea coast: phylogenetic analysis of cultured microbial consortia in industrialized sites. Front Microbiol 5:363. doi: 10.3389/fmicb.2014.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mustafa GA, Abd-Elgawad A, Ouf A, Siam R. 2016. The Egyptian Red sea coastal microbiome: a study revealing differential microbial responses to diverse anthropogenic pollutants. Environ Pollut 214:892–902. doi: 10.1016/j.envpol.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 20. Friedman GM, Amiel AJ, Braun M, Miller DS. 1973. Generation of carbonate particles and laminites in algal mats—example from sea-marginal hypersaline pool, Gulf of Aqaba, Red sea 57:541–557. doi: 10.1306/819A4302-16C5-11D7-8645000102C1865D [DOI] [Google Scholar]
- 21. Rothrock Jr MJ, Garcia-Pichel F. 2005. Microbial diversity of benthic mats along a tidal desiccation gradient. Environ Microbiol 7:593–601. doi: 10.1111/j.1462-2920.2005.00728.x [DOI] [PubMed] [Google Scholar]
- 22. Abed RMM, Kohls K, de Beer D. 2007. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ Microbiol 9:1384–1392. doi: 10.1111/j.1462-2920.2007.01254.x [DOI] [PubMed] [Google Scholar]
- 23. Sponseller RA. 2007. Precipitation pulses and soil CO2 flux in a Sonoran desert ecosystem. Glob Change Biol 13:426–436. doi: 10.1111/j.1365-2486.2006.01307.x [DOI] [Google Scholar]
- 24. Abdallah RZ, Wegner C-E, Liesack W. 2019. Community transcriptomics reveals drainage effects on paddy soil microbiome across all three domains of life. Soil Biol Biochem 132:131–142. doi: 10.1016/j.soilbio.2019.01.023 [DOI] [Google Scholar]
- 25. Foti M, Sorokin DY, Lomans B, Mussman M, Zacharova EE, Pimenov NV, Kuenen JG, Muyzer G. 2007. Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline Soda lakes. Appl Environ Microbiol 73:2093–2100. doi: 10.1128/AEM.02622-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. 1. Nat Commun 7:11257. doi: 10.1038/ncomms11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 29. Bushnell B. 2014. BBmap. Sourceforge. Available from: https://sourceforge.net/projects/bbmap
- 30. Kang DD, Froula J, Egan R, Wang Z. 2015. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. 2023. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. 8. Nat Methods 20:1203–1212. doi: 10.1038/s41592-023-01940-w [DOI] [PubMed] [Google Scholar]
- 32. Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vollmers J, Wiegand S, Lenk F, Kaster A-K. 2022. How clear is our current view on microbial dark matter? (Re-)assessing public MAG & SAG datasets with MDMcleaner. Nucleic Acids Res 50:e76. doi: 10.1093/nar/gkac294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee MD. 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35:4162–4164. doi: 10.1093/bioinformatics/btz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. doi: 10.1093/bioinformatics/bts429 [DOI] [PubMed] [Google Scholar]
- 38. Eddy SR. 2011. Accelerated profile HMM searches. PLOS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar RC. 2021. MUSCLE V5 enables improved estimates of phylogenetic tree confidence by ensemble bootstrapping. bioRxiv. doi: 10.1101/2021.06.20.449169 [DOI] [Google Scholar]
- 40. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cytryn E, Minz D, Oremland RS, Cohen Y. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl Environ Microbiol 66:3269–3276. doi: 10.1128/AEM.66.8.3269-3276.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. 2020. Genbank. Nucleic Acids Res 48:D84–D86. doi: 10.1093/nar/gkz956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:W557–9. doi: 10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Letunic I, Bork P. 2016. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–5. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, Liu P, Narrowe AB, Rodríguez-Ramos J, Bolduc B, Gazitúa MC, Daly RA, Smith GJ, Vik DR, Pope PB, Sullivan MB, Roux S, Wrighton KC. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. doi: 10.1093/nar/gkaa621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium . 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. doi: 10.1093/bioinformatics/btz859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621. doi: 10.1080/01621459.1952.10483441 [DOI] [Google Scholar]
- 52. Dunn OJ. 1964. Multiple comparisons using rank sums. Technometrics 6:241–252. doi: 10.1080/00401706.1964.10490181 [DOI] [Google Scholar]
- 53. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benjamini Y, Krieger AM, Yekutieli D. 2006. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93:491–507. doi: 10.1093/biomet/93.3.491 [DOI] [Google Scholar]
- 55. Li Y-X, Rao Y-Z, Qi Y-L, Qu Y-N, Chen Y-T, Jiao J-Y, Shu W-S, Jiang H, Hedlund BP, Hua Z-S, Li W-J. 2021. Deciphering symbiotic interactions of “Candidatus aenigmarchaeota” with inferred horizontal gene transfers and co-occurrence networks. mSystems 6. doi: 10.1128/mSystems.00606-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y, Zhu X, Zhang W, Zhu D, Zhou X, Zhang L. 2019. Archaeal communities in the deep-sea sediments of the South China sea revealed by Illumina high-throughput sequencing. Ann Microbiol 69:839–848. doi: 10.1007/s13213-019-01477-4 [DOI] [Google Scholar]
- 57. Rodríguez Cruz UEE, Castelán-Sánchez HG, Madrigal-Trejo D, Eguiarte LE, Souza V. n.d. Unveiling local radiation events through metagenome assembled genomes of Archaea and bacteria in Hypersaline microbial mats from the Archean domes site, Cuatro Cienegas, Coahuila, Mexico. Microbiology. doi: 10.1101/2023.08.19.553462 [DOI]
- 58. Liu S, Lin Y, Liu T, Xu X, Wang J, Chen Q, Sun W, Dang C, Ni J. 2023. Planktonic/benthic Bathyarchaeota as a “gatekeeper” enhance archaeal nonrandom co-existence and deterministic assembling in the Yangtze river. Water Res 247:120829. doi: 10.1016/j.watres.2023.120829 [DOI] [PubMed] [Google Scholar]
- 59. Romano RG, Bendia AG, Moreira JCF, Franco DC, Signori CN, Yu T, Wang F, Jovane L, Pellizari VH. 2021. Bathyarchaeia occurrence in rich methane sediments from a Brazilian Ría. Estuar Coast Shelf Sci 263:107631. doi: 10.1016/j.ecss.2021.107631 [DOI] [Google Scholar]
- 60. Ma Z, Gao L, Sun M, Liao Y, Bai S, Wu Z, Li J. 2022. Microbial diversity in groundwater and its response to seawater intrusion in Beihai City, Southern China. Front Microbiol 13:876665. doi: 10.3389/fmicb.2022.876665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miralles-Robledillo JM, Bernabeu E, Giani M, Martínez-Serna E, Martínez-Espinosa RM, Pire C. 2021. Distribution of denitrification among haloarchaea: a comprehensive study. Microorganisms 9:1669. doi: 10.3390/microorganisms9081669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Williams TJ, Allen MA, DeMaere MZ, Kyrpides NC, Tringe SG, Woyke T, Cavicchioli R. 2014. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant haloarchaea. ISME J 8:1645–1658. doi: 10.1038/ismej.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Torregrosa-Crespo J, Miralles-Robledillo JM, Bernabeu E, Pire C, Martínez-Espinosa RM. 2023. Denitrification in hypersaline and coastal environments. FEMS Microbiol Lett 370:fnad066. doi: 10.1093/femsle/fnad066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Feng L, Zhang Z, Yang G, Wu G, Yang Q, Chen Q. 2023. Microbial communities and sediment nitrogen cycle in a coastal eutrophic lake with salinity and nutrients shifted by seawater intrusion. Environ Res 225:115590. doi: 10.1016/j.envres.2023.115590 [DOI] [PubMed] [Google Scholar]
- 65. Pavloudi C, Zafeiropoulos H. 2022. Deciphering the community structure and the functional potential of a hypersaline marsh microbial mat community. FEMS Microbiol Ecol 98:fiac141. doi: 10.1093/femsec/fiac141 [DOI] [PubMed] [Google Scholar]
- 66. García-Maldonado JQ, Latisnere-Barragán H, Escobar-Zepeda A, Cadena S, Ramírez-Arenas PJ, Vázquez-Juárez R, Rojas-Contreras M, López-Cortés A. 2023. Revisiting microbial diversity in hypersaline microbial mats from Guerrero Negro for a better understanding of methanogenic archaeal communities. Microorganisms 11:812. doi: 10.3390/microorganisms11030812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wong HL, MacLeod FI, White RA, Visscher PT, Burns BP. 2020. Microbial dark matter filling the niche in hypersaline microbial mats. Microbiome 8:135. doi: 10.1186/s40168-020-00910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kindler GS, Wong HL, Larkum AWD, Johnson M, MacLeod FI, Burns BP. 2022. Genome-resolved metagenomics provides insights into the functional complexity of microbial mats in Blue Holes, Shark Bay. FEMS Microbiol Ecol 98:fiab158. doi: 10.1093/femsec/fiab158 [DOI] [PubMed] [Google Scholar]
- 69. Banciu HL, Gridan IM, Zety AV, Baricz A. 2022. 123AD. Asgard archaea in saline environments. Extremophiles 26:21. doi: 10.1007/s00792-022-01266-z [DOI] [PubMed] [Google Scholar]
- 70. Williams TJ, Allen MA, Berengut JF, Cavicchioli R. 2021. Shedding light on microbial “dark matter”: insights into novel Cloacimonadota and Omnitrophota from an Antarctic Lake. Front Microbiol 12:741077. doi: 10.3389/fmicb.2021.741077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Perez-Molphe-Montoya E, Küsel K, Overholt WA. 2022. Redefining the phylogenetic and metabolic diversity of phylum Omnitrophota. Environ Microbiol 24:5437–5449. doi: 10.1111/1462-2920.16170 [DOI] [PubMed] [Google Scholar]
- 72. Skoog EJ, Moore KR, Gong J, Ciccarese D, Momper L, Cutts EM, Bosak T. 2022. Metagenomic, (bio)chemical, and microscopic analyses reveal the potential for the cycling of sulfated EPS in Shark Bay pustular mats. ISME Commun 2:43. doi: 10.1038/s43705-022-00128-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Okumura T, Takashima C, Yanagawa K, Harijoko A, Kano A. 2022. Stromatolite formation by Anaerolineae-dominated microbial communities in hot spring travertine in North Sumatra, Indonesia. Sediment Geol 440:106263. doi: 10.1016/j.sedgeo.2022.106263 [DOI] [Google Scholar]
- 74. Yamada T, Sekiguchi Y, Hanada S, Imachi H, Ohashi A, Harada H, Kamagata Y. 2006. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol 56:1331–1340. doi: 10.1099/ijs.0.64169-0 [DOI] [PubMed] [Google Scholar]
- 75. Tovar R, González A, López-Cortés A, Ascencio V, Ochoa J. 1999. Effect of salinity and light intensity on superoxide dismutase and ascorbate peroxidase activity from Microcoleus chthonoplastes strain SC7B9002-1. Pascal and Francis Bibliographic Databases. Monaco: Institut océanographique, Monaco / Musée océanographique, Monaco [Google Scholar]
- 76. Ziegelhoffer EC, Donohue TJ. 2009. Bacterial responses to photo-oxidative stress. Nat Rev Microbiol 7:856–863. doi: 10.1038/nrmicro2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shih PM, Ward LM, Fischer WW. 2017. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. Proc Natl Acad Sci USA 114:10749–10754. doi: 10.1073/pnas.1710798114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Baldani JI, Videira SS, Santos Teixeira KR, Reis VM, Oliveira ALM, Schwab S, Souza EM, Pedraza RO, Baldani VLD, Hartmann A. 2014. The family Rhodospirillaceae, p 533–618. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: alphaproteobacteria and betaproteobacteria. Springer, Berlin, Heidelberg. [Google Scholar]
- 79. Mack EE, Mandelco L, Woese CR, Madigan MT. 1993. Rhodospirillum sodomense, sp. nov., a Dead sea Rhodospirillum species. Arch Microbiol 160:363–371. doi: 10.1007/BF00252222 [DOI] [Google Scholar]
- 80. Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745 [DOI] [PubMed] [Google Scholar]
- 81. Wang Y, Wegener G, Hou J, Wang F, Xiao X. 2019. Expanding anaerobic alkane metabolism in the domain of Archaea. Nat Microbiol 4:595–602. doi: 10.1038/s41564-019-0364-2 [DOI] [PubMed] [Google Scholar]
- 82. Liu Y-F, Chen J, Zaramela LS, Wang L-Y, Mbadinga SM, Hou Z-W, Wu X-L, Gu J-D, Zengler K, Mu B-Z. 2020. Genomic and transcriptomic evidence supports methane metabolism in Archaeoglobi. mSystems 5:00651–19. doi: 10.1128/mSystems.00651-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhilina TN, Zavarzin GA. 1990. Extremely halophilic, methylotrophic, anaerobic bacteria. FEMS Microbiol Rev 87:315–322. doi: 10.1111/j.1574-6968.1990.tb04930.x [DOI] [Google Scholar]
- 84. Nobu MK, Narihiro T, Kuroda K, Mei R, Liu WT. 2016. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J 10:2478–2487. doi: 10.1038/ismej.2016.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Garcia PS, Gribaldo S, Borrel G. 2022. Diversity and evolution of methane-related pathways in Archaea. Annu Rev Microbiol 76:727–755. doi: 10.1146/annurev-micro-041020-024935 [DOI] [PubMed] [Google Scholar]
- 86. Wong HL, Visscher PT, White RA, Smith D-L, Patterson MM, Burns BP. 2017. Dynamics of archaea at fine spatial scales in Shark Bay mat microbiomes. Sci Rep 7:46160. doi: 10.1038/srep46160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams AJM. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goh F, Barrow KD, Burns BP, Neilan BA. 2010. Identification and regulation of novel compatible solutes from hypersaline stromatolite-associated cyanobacteria. Arch Microbiol 192:1031–1038. doi: 10.1007/s00203-010-0634-0 [DOI] [PubMed] [Google Scholar]
- 89. López-Sánchez R, Rebollar EA, Gutiérrez-Ríos RM, Garciarrubio A, Juarez K, Segovia L. 2023. Metagenomic analysis of carbohydrate-active enzymes and their contribution to marine sediment biodiversity. Research square. doi: 10.21203/rs.3.rs-3129032/v1 [DOI] [PMC free article] [PubMed]
- 90. Mohapatra M, Yadav R, Rajput V, Dharne MS, Rastogi G. 2021. Metagenomic analysis reveals genetic insights on biogeochemical cycling, xenobiotic degradation, and stress resistance in mudflat microbiome. J Environ Manage 292:112738. doi: 10.1016/j.jenvman.2021.112738 [DOI] [PubMed] [Google Scholar]
- 91. Tartaglia M, Ranauda MA, Falzarano A, Maisto M, Postiglione A, Prigioniero A, Scarano P, Zuzolo D, Sciarrillo R, Guarino C. 2023. Metatranscriptomics of pastures under drought stress show a rhizospheric meta-organism reshape. Rhizosphere 26:100687. doi: 10.1016/j.rhisph.2023.100687 [DOI] [Google Scholar]
- 92. Liu F, Zhang Y, Liang H, Gao D. 2021. Resilience of methane cycle and microbial functional genes to drought and flood in an alkaline wetland: a metagenomic analysis. Chemosphere 265:129034. doi: 10.1016/j.chemosphere.2020.129034 [DOI] [PubMed] [Google Scholar]
- 93. Tabari H, Willems P. 2018. More prolonged droughts by the end of the century in the Middle East. Environ Res Lett 13:104005. doi: 10.1088/1748-9326/aae09c [DOI] [Google Scholar]
- 94. Nashwan MS, Shahid S. 2022. Future precipitation changes in Egypt under the 1.5 and 2.0°C global warming goals using CMIP6 multimodel ensemble. Atmospheric Res 265:105908. doi: 10.1016/j.atmosres.2021.105908 [DOI] [Google Scholar]
- 95. Mazière C, Duran R, Dupuy C, Cravo-Laureau C. 2023. Microbial mats as model to decipher climate change effect on microbial communities through a mesocosm study. Front Microbiol 14:1039658. doi: 10.3389/fmicb.2023.1039658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Verleyen E, Sabbe K, Hodgson D, Grubisic S, Taton A, Cousin S, Wilmotte A, De Wever A, Van der Gucht K, Vyverman W. 2010. Structuring effects of climate-related environmental factors on Antarctic microbial mat communities. Aquat Microb Ecol 59:11–24. doi: 10.3354/ame01378 [DOI] [Google Scholar]
- 97. Mazière C, Bodo M, Perdrau MA, Cravo-Laureau C, Duran R, Dupuy C, Hubas C. 2022. Climate change influences chlorophylls and bacteriochlorophylls metabolism in hypersaline microbial mat. Sci Total Environ 802:149787. doi: 10.1016/j.scitotenv.2021.149787 [DOI] [PubMed] [Google Scholar]
- 98. European Organization For Nuclear Research . 2013. Zenodo. Available from: https://www.zenodo.org/ [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods and Figures S1 to S6.
Solar Lake shallow-water and exposed mat metagenome statistics.
Solar Lake shallow-water and exposed mat metagenome DRAM annotations.
Solar Lake shallow-water and exposed mat MAG summary.
The cyanobacterial MAG relative abundance in the Solar Lake shallow-water and exposed mats.
Nitrogen cycling genes distribution across the Solar Lake shallow-water and exposed archaeal and bacterial MAGs and the relative abundance of nir gene-containing MAGs.
Carbon fixation pathways, Cazyme, and short-chain fatty acids and alcohol conversion genes in Asgardarchaota.
Solar Lake flat mat Myxococcota MAGs annotation using DRAM.
Photooxidative stress gene in Coleofasciculus chthonoplastes MAGs.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The metagenome shotgun sequencing raw data were deposited in the NCBI BioProject database under the accession numbers PRJNA1004913. Metagenome assemblies and metagenome assembled genomes (MAGs) used in this study were deposited in Zenodo repository at https://doi.org/10.5281/zenodo.10521873 (98). The data files names corresponding to each sample are indicated in Table S1.