ABSTRACT
Recent case reports and epidemiological data suggest that fungal infections represent an underappreciated complication among people with severe COVID-19. However, the frequency of fungal colonization in patients with COVID-19 and associations with specific immune responses in the airways remain incompletely defined. We previously generated a single-cell RNA-sequencing data set characterizing the upper respiratory microenvironment during COVID-19 and mapped the relationship between disease severity and the local behavior of nasal epithelial cells and infiltrating immune cells. Our previous study, in agreement with findings from related human cohorts, demonstrated that a profound deficiency in host immunity, particularly in type I and type III interferon signaling in the upper respiratory tract, is associated with rapid progression to severe disease and worse clinical outcomes. We have now performed further analysis of this cohort and identified a subset of participants with severe COVID-19 and concurrent detection of Candida species-derived transcripts within samples collected from the nasopharynx and trachea. Here, we present the clinical characteristics of these individuals. Using matched single-cell transcriptomic profiles of these individuals’ respiratory mucosa, we identify epithelial immune signatures suggestive of IL17 stimulation and anti-fungal immunity. Further, we observe a significant expression of anti-fungal inflammatory cascades in the nasal and tracheal epithelium of all participants who went on to develop severe COVID-19, even among participants without detectable genetic material from fungal pathogens. Together, our data suggest that IL17 stimulation—in part driven by Candida colonization—and blunted interferon signaling represent a common feature of severe COVID-19 infection.
IMPORTANCE
In this paper, we present an analysis suggesting that symptomatic and asymptomatic fungal coinfections can impact patient disease progression during COVID-19 hospitalization. By looking into the presence of other pathogens and their effect on the host immune response during COVID-19 hospitalizations, we aim to offer insight into an underestimated scenario, furthering our current knowledge of determinants of severity that could be considered for future diagnostic and intervention strategies.
KEYWORDS: SARS-CoV-2, COVID-19, human, nasal mucosa, epithelial immunity, Candida, fungal infection, IL17, cytokine, interferon, anti-viral, scRNA-seq
INTRODUCTION
Infection with SARS-CoV-2, the virus that causes COVID-19, can lead to severe viral pneumonitis and the development of acute respiratory distress syndrome (1, 2). Severe COVID-19 is characterized by peripheral immune dysregulation, and we and others have previously demonstrated blunted interferon responses within the nasal mucosa of patients with severe COVID-19 (3–5). Recent case reports and retrospective cohort studies suggest that secondary infection with fungal pathogens may be a significant contributor to morbidity and mortality in patients with severe COVID-19 (6–11).
The frequency of fungal colonization of the airways in patients with severe COVID-19 and its potential impact on local mucosal immunity remain unknown (9, 12–15). IL17, released by CD4 T cells and innate lymphocytes, is a key effector cytokine that coordinates mucosal anti-fungal immunity among other adaptive and innate leukocytes, granulocytes, and mucosal stroma (16–19). Recent work has uncovered complex interactions between IL17-driven inflammation, type 1 interferon responses, and susceptibility to fungal pathogens; however, the effect of fungal colonization and anti-fungal immune responses during cooccurrent SARS-CoV-2 infection has yet to be explored (20). Here, using a previously published data set derived from a cohort of individuals acutely infected with SARS-CoV-2, we directly assessed cooccurrent fungal colonization in the airways of patients with severe COVID-19 and examined pathways associated with anti-fungal immunity (3).
RESULTS
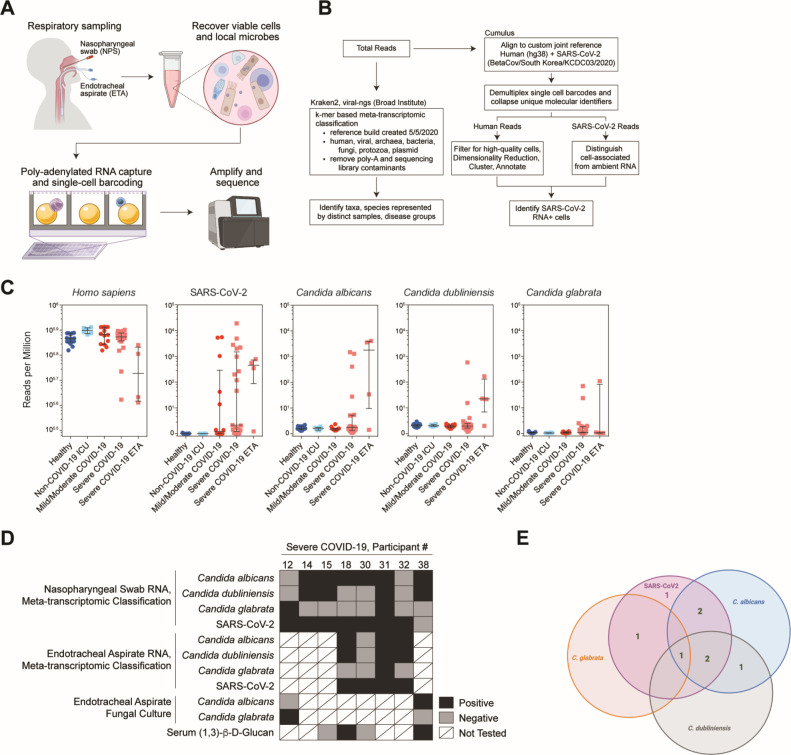
We had previously described a cohort of 58 individuals—56 of which are further characterized here—including 15 healthy participants, 35 individuals diagnosed with acute COVID-19, and 6 intubated patients that were negative for SARS-CoV-2 (3) The two excluded individuals from the original cohort had recovered from COVID-19 (“convalescent COVID-19”) and, therefore, did not represent acute infection. We were assessing phenotypes of respiratory epithelia early following infection. Nasopharyngeal (NP) swabs obtained from these patients were employed in a cross-sectional study of the nasal respiratory cellular composition using single-cell RNA sequencing (scRNA-seq) (Fig. 1A and B). Patients in this cohort with COVID-19 were sampled within 9 days of hospital admission (median: hospital day 2), which we estimated was within 2 weeks of initial respiratory symptoms. Full cohort demographic data and findings relating to the cellular composition, behaviors, and response to SARS-CoV-2 infection between disease groups can be found in our prior manuscript (3). Eight patients from this cohort had detectable Candida species reads from nasopharyngeal swabs or endotracheal aspirate (ETA) samples by meta-transcriptomic analysis. Candida species infection was confirmed in three of these patients by fungal culture and/or serum (1,3)-β-D-glucan. We found no difference in the frequency of antibiotic treatment nor corticosteroid exposure among patients from mild/moderate COVID-19, non-COVID-19 intensive care unit (ICU) intubated controls, severe COVID-19 Candida negative, or severe COVID-19 Candida positive.
Fig 1.
Codetection of host single-cell transcriptome with intracellular and microenvironmental pathogen-derived genomic material. (A) Schematic of biological sample processing pipeline. (B) Schematic of computational pipeline for each sample. (C) Abundance of human, SARS-CoV-2, and Candida spp. by participant and disease group as defined by meta-transcriptomic classification. N = 56 participants. Lines represent median ±interquartile range. (D) Summary of results from sequencing, fungal culture, and serum (1,3)-β-D-glucan assays from eight participants with detectable Candida species reads by nasopharyngeal swab or endotracheal aspirate. (E) Graphical summary of COVID-19 and Candida sp. infection among severe COVID-19 cohort.
We had previously applied a meta-transcriptomic taxonomic classification analysis to each sample to assign both cell-associated and ambient/extracellular sequencing reads to a reference database of human and microbial genomes (generated on 5 May 2020 from the NCBI Reference Sequence Database including archaeal, bacterial, viral, protozoan, and fungal genomes) (21, 22). This approach, in addition to direct reference-based alignment, enabled us to quantify respiratory abundances of SARS-CoV-2 and connect viral abundances to the cellular sources of viral replication, as well as concurrent epithelial host responses (Fig. 1B). Here, we describe further analysis of the data generated from these nasopharyngeal samples, as well as additional data generated from matched ETAs obtained from four of the individuals in the original cohort with severe COVID-19.
Candida species coinfection was limited to severe COVID-19 patients
Using meta-transcriptomic alignment of scRNA-seq data, we identified additional high-abundance microbial taxa across healthy participants and those with COVID-19 including common commensal microbes such as Cutibacterium acnes, Malassezia restricta, and Staphylococcus aureus (Fig. S1A; Table S1). After SARS-CoV-2, the second-most abundant microbe detected was Candida albicans, which was detected on six NP samples obtained from patients with severe COVID-19 (Fig. 1C through E; Fig. S1A). For a subset of patients with PCR-positive SARS-CoV-2 and clinical evidence of COVID-19 (both requirements for inclusion into our “COVID-19” group), there are no reads that align with SARS-CoV-2. However, SARS-CoV-2 RNA was detected from scRNA-seq libraries in 80% (28/35) of participants diagnosed with COVID-19. This proportion is in accordance with larger studies that assess the likelihood of a positive Real Time Polymerase Chain Reaction (RT-PCR) result by nasopharyngeal sampling. In one meta-analysis by Mallett et al., 89% of RT-PCR results were positive within 4 days of symptom onset for patients with eventual confirmed COVID-19, and 81% of RT-PCR tests remained positive within the first 4 days of hospital admission (23). Further, we have assessed for concordance between Kraken2-based meta-transcriptomic capture as well as direct alignment against a SARS-CoV-2 reference sequence (Fig. S1C).
We also identified high levels of Candida glabrata in NP samples from two patients with severe COVID-19 and Candida dubliniensis in four samples. All samples that were positive for C. glabrata or C. dubliniensis were also positive for C. albicans with the exception of the NP sample obtained from COVID-19 participant 12 (Fig. 1D and E). Candida albicans was also detected in three of four ETA samples obtained from patients with severe COVID-19. For one patient (COVID-19 participant 32), C. albicans was detected via ETA but was not detected on their matched NP swab. All three ETA samples that were positive for C. albicans were also positive for C. dubliniensis, and one of these was also positive for C. glabrata (Fig. 1C through E; Fig. S1A and B). Notably, no Candida spp. or other fungal pathogens were detected within samples obtained from healthy individuals, those obtained from individuals with mild/moderate COVID-19, or those obtained from SARS-CoV-2 negative intubated patients in the intensive care unit with severe respiratory failure due to alternative causes. Thus, all Candida spp. reads were detected among patients who developed severe COVID-19 requiring intubation and mechanical ventilation (WHO severity score of 6–8). Nearly all NP or ETA samples that were positive for Candida spp. were collected within 1 week of hospital admission (Fig. S2A). The majority (6/8) had been intubated for at least 1 day, and 5/8 had received at least 1 day of corticosteroid treatment prior to sample collection (Fig. S2A). Clinical evaluations during hospitalization were performed to search for possible fungal infection for three of the eight patients with high abundances of Candida spp. by meta-transcriptomic classification. COVID-19 participant 12, who had a high abundance of Candida glabrata RNA sampled by NP swab on hospital day 2, was intubated on hospital day 1 (Fig. S2A), and ETA fungal cultures sampled on hospital day 2 revealed growth of Candida glabrata (Fig. 1D). For two participants, detection of Candida spp.-derived RNA via NP/ETA sampling significantly preceded clinical diagnostic testing for fungal pathogens. COVID-19 participant 38, whose NP swab revealed both Candida albicans and Candida dubliniensis RNA on hospital day 8, tested positive for serum (1,3)-β-D-glucan on hospital day 14 and had Candida albicans growth from ETA culture on hospital day 16 (Fig. S2A). Both the NP and ETA samples from COVID-19 participant 18 obtained on hospital day 6 contained high abundances of reads classified as C. albicans, and C. dubliniensis was detected in the ETA sample. On hospital day 12, (1,3)-β-D-glucan was detected in this individual’s serum (Fig. S2A), prompting treatment with micafungin (an echinocandin anti-fungal) (24). Four out of 21 patients with severe COVID-19 had Candida reads on our meta-transcriptomic analysis, but only two of those were positive by serum (1,3)-β-D-glucan testing. Together, this demonstrates that for a subset of patients, there was significant clinical concern during their hospitalization to prompt additional testing to evaluate for fungal respiratory infection or fungemia.
Among those individuals, we find concordance between the detection of Candida-derived RNA from scRNA-seq libraries and clinical assays during their hospitalization. Additionally, we compared the proportion of patients in each subgroup in our data set receiving corticosteroids in advance or at the time of sample collection. Thirty-three percent (2/6) of patients in the intubated non-COVID-19 ICU group were receiving corticosteroids, and 0% (0/6) had detectable Candida reads by our analysis. Among patients with severe COVID-19 and no Candida species detected via meta-transcriptomic analyses, 61% (8/13) were receiving corticosteroids. Finally, 75% (6/8) of patients with severe COVID-19 and Candida species detected were receiving corticosteroids. Additionally, among COVID-19 patients not in the ICU (our mild/moderate COVID-19 group), 42.9% (6/14) were receiving corticosteroids, and 0% (0/14) had detectable Candida reads. We did not identify any significant differences in the proportion of corticosteroid treatment across any of these four groups by the chi-squared test. Although limited by sample size, our data do not suggest that corticosteroid treatment represented a major confounder in our detection of Candida.
The majority of COVID-19 patients with severe disease included in our analysis were treated with antibiotics during their hospitalization. The most common antibiotic combination used was azithromycin and ceftriaxone. Among patients with severe COVID-19 and no Candida detected, 77% (10/13) were treated with antibiotics for at least 1 day prior to sampling. Among patients with severe COVID-19 and Candida species detected, 88% (7/8) were treated with antibiotics for at least 1 day in advance of nasal swab.
We did not detect a difference in demographics or clinical characteristics—apart from the severity of COVID-19—between patients whose samples did or did not contain Candida-specific reads (Table 1; Fig. S2B through G). Twenty-eight-day mortality rates among individuals with severe COVID-19 were similar between Candida spp. positive vs negative groups: 62.5% (5/8) among participants with Candida spp. detected, compared to 84.6% (11/13) among participants without Candida spp. detected. Nearly all (7/8) participants with COVID-19 whose samples contained Candida spp.-aligning RNA were previously diagnosed with type 2 diabetes mellitus (T2DM), and 8/8 were diagnosed with chronic hypertension. Notably, although T2DM represents an independent risk factor for mucosal Candida colonization, we did not find Candida or other fungal species among individuals with T2DM within the healthy non-COVID-19 intubated or COVID-19 mild/moderate groups (25). Additionally, for individuals with recently measured HbA1c, we did not identify significant differences in the degree of glycemic control between individuals with different COVID-19 severity or by detection of Candida-specific reads (Fig. S2G) (26).
TABLE 1.
Demographics and medical comorbidities of patients with severe COVID-19a
| COVID-19 severe (WHO score 6–8) Candida spp. negative | COVID-19 severe (WHO score 6–8) Candida spp. positive | |
|---|---|---|
| Case number | 23.2% (13/56) | 14.3% (8/56) |
| Age (years) | ||
| Minimum | 28 | 38 |
| Median (IQR) | 63 (49) | 57 (54.3) |
| Maximum | 79 | 84 |
| Sex | ||
| Female | 38.5% (5/13) | 62.5% (5/8) |
| Male | 61.5% (8/13) | 37.5% (3/8) |
| Ethnicity | ||
| Hispanic | 7.7% (1/13) | 0% (0/13) |
| Not Hispanic | 92.3% (12/13) | 100% (13/13) |
| Race | ||
| Black/African American | 53.8% (7/13) | 75% (6/8) |
| White | 23.1% (3/13) | 25% (2/8) |
| American Indian | 23.1% (3/13) | 0% (0/8) |
| BMI | ||
| Median (IQR) | 29.9 (27.8) | 37.0 (33.6) |
| Pre-existing conditions | ||
| Diabetes | 61.5% (8/13) | 87.5% (7/8) |
| Chronic kidney disease | 15.4% (2/13) | 25% (2/8) |
| Congestive heart failure | 7.7% (1/13) | 0% (0/8) |
| Lung disorder | 30.1% (4/13) | 50% (4/8) |
| Hypertensionb | 69.2% (9/13) | 100% (8/8) |
| IBD | 0% (0/13) | 0% (0/8) |
| Treatment | ||
| Corticosteroids | 61.5% (8/13) | 75% (6/8) |
| Remdesivir | 7.7% (1/13) | 0% (0/8) |
| 28-day mortalityc | 84.6% (11/13) | 62.5% (5/8) |
IQR, interquartile range; BMI, body mass index; IBD, inflammatory bowel disease.
*p = 0.023.
***p < 0.0001.
In summary, we identified 0% (0/14) of patients with mild/moderate COVID-19 with Candida reads despite equivalent library quality characteristics. This contrasts with the 38% (8/21) of patients with severe COVID-19 with associated Candida-mapping reads, corresponding to P = 0.0118 by the Fisher exact test. Additionally, we found Candida reads in 0% (0/6) of patients who were intubated in the intensive care unit. Critically, this analysis is based on a limited sample size and merits further investigation with adequately powered cohorts.
The nasal epithelium of patients with severe COVID-19 presented increased expression of IL17A-induced genes
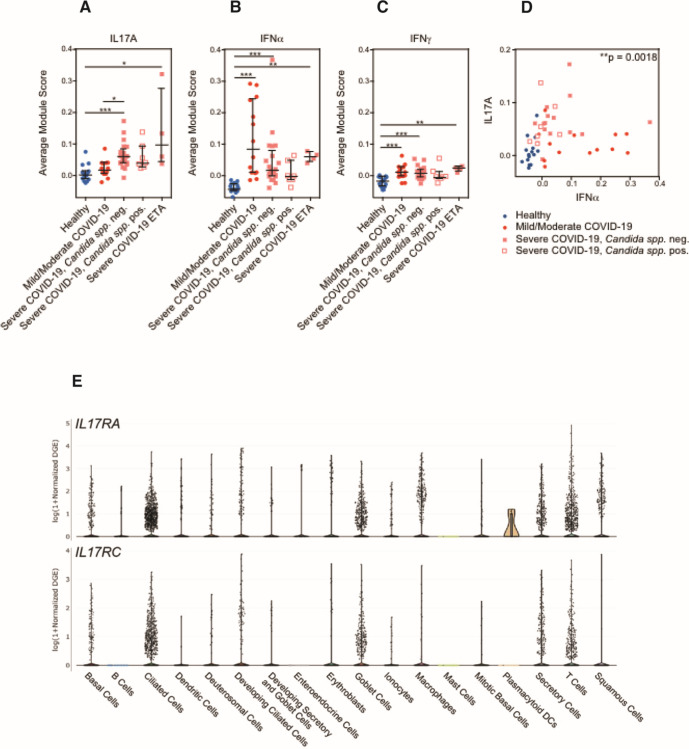
Previous findings have reported reductions in type I interferon (27–31) and an increase in IL17 signaling (32–35) in severe COVID-19 patients. To address the impact in the mucosal environment upon exposure to these cytokines, we directly scored epithelial cells for expression of gene signatures indicative of IL17A, IFNα, or IFNγ (Fig. 2A through C). Individuals who developed severe COVID-19 expressed consistently higher abundances of IL17A-induced genes compared to healthy participants or those with mild/moderate COVID-19 (Fig. 2A). Interestingly, we did not detect higher abundances of genes in the IL17A gene module among participants with severe COVID-19 and detectable Candida spp. compared to participants with severe COVID-19 without detectable fungal pathogens. Likewise, we did not detect significant differences in levels of interferon-induced signatures between individuals with and without Candida spp. detected (Fig. 2B and C). On an individual level, participants whose nasal epithelial cells expressed higher abundances of IFNα-induced genes did not express IL17A-induced genes, and vice versa (Fig. 2D). Together, this suggests that factors beyond Candida spp. colonization may be responsible for the induction of IL17A-stimulated genes in the nasal epithelium. However, this underscores that an IL17A-induced epithelial cell state represents a shared feature of individuals who develop severe COVID-19 and is correlated with the absence of robust interferon-induced anti-viral responses.
Fig 2.
Average module scores in different disease groups in all epithelial cells. (A–D) Average gene module scores calculated for each participant, separated by disease group. Module score expression was computed over all epithelial cells. Input module genes derived from in vitro stimulation with each labeled cytokine: IL17A (A), IFNα (B), and IFNγ (C). Statistical testing by Kruskal–Wallis test with Dunn’s post hoc testing. ***P < 0.001, **P < 0.01, and *P < 0.05. Lines represent the mean ± SEM. (E) Top: expression of IL17RA by cell type. Bottom: expression of IL17RC by cell type. Data accessed from https://singlecell.broadinstitute.org/single_cell/study/SCP1289.
As expected, the majority of IL17 sources are derived from T cells (Fig. S4A). To further evaluate the relationship between severe COVID-19 and response to IL17 stimulation, we have analyzed our data for the expression of IL17 receptor on epithelial cells. The expression of IL17RA and IL17RC (which form the heterodimer receptor complex for IL17A) was indeed expressed among nasopharyngeal epithelial cells (Fig. 2E). Ciliated cells, goblet cells, secretory cells, and, to a lesser extent, basal cells were the major epithelial cell types expressing IL17RA and IL17RC.
In vitro exposure to IL17 in nasal epithelial cells enhances pro-inflammatory responses
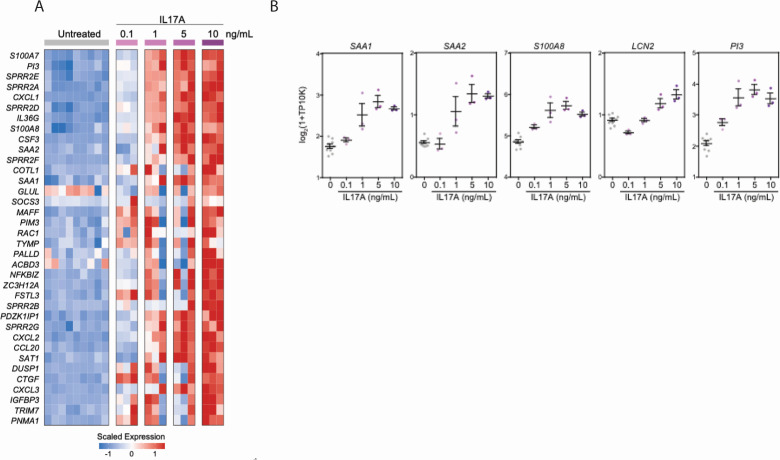
Given that Candida spp. colonization became a clinically relevant infection among some individuals who developed severe COVID-19, we wondered whether the nasal mucosa of these individuals exhibits evidence for reactive or aberrant IL17 responses. To better define the response of the human nasal epithelium to IL17, we reanalyzed a subset of 15 healthy and 8 severe COVID-19 donors from our previously published population RNA-seq data that reflect gene expression in human nasal epithelial cells following in vitro exposure to a range of doses of IL17A (Fig. 3A and B) (36, 37). Across multiple human donors, IL17A exposure led to the upregulation of genes involved in keratinization (SPRR2E, SPRR2F, and SPRR2G); chemoattractant cytokines for lymphocytes, monocytes, and neutrophils (CCL20, CXCL1, CXCL2, and CXCL3); and pro-inflammatory factors such as S100A7 and S100A8 (Fig. 3A and B) (38–40). IL17A additionally resulted in the dose-dependent induction of serum amyloid A genes SAA1 and SAA2 from nasal epithelial cells, which has previously been linked to pathogenic Th17 responses at barrier tissues (41). Using RNA-seq data, we generated consensus gene sets for each tested cytokine that were robust across distinct donors, thereby giving us cell type-specific gene expression modules for IL17A, IFNα, IFNγ, IL1β, and IL4 (Fig. S3A; Table S2).
Fig 3.
Respiratory epithelial transcriptional signatures following in vitro IL17A stimulation and in vivo fungal colonization in a severe COVID-19 cohort. (A) Heatmap of population RNA-seq data comparing untreated nasal epithelial cells to those treated with increasing concentrations of IL17A as indicated across columns. Genes (rows) with significant differential expression between untreated and IL17A-treated conditions False Discovery Rate [(FDR)-corrected P < 0.05)]. (B) Expression of select genes following 12-hour stimulation with increasing doses of IL17A. Each gene significantly upregulated following IL17A exposure by likelihood ratio test, FDR-adjusted P-value <0.001. Lines represent the mean ± SEM.
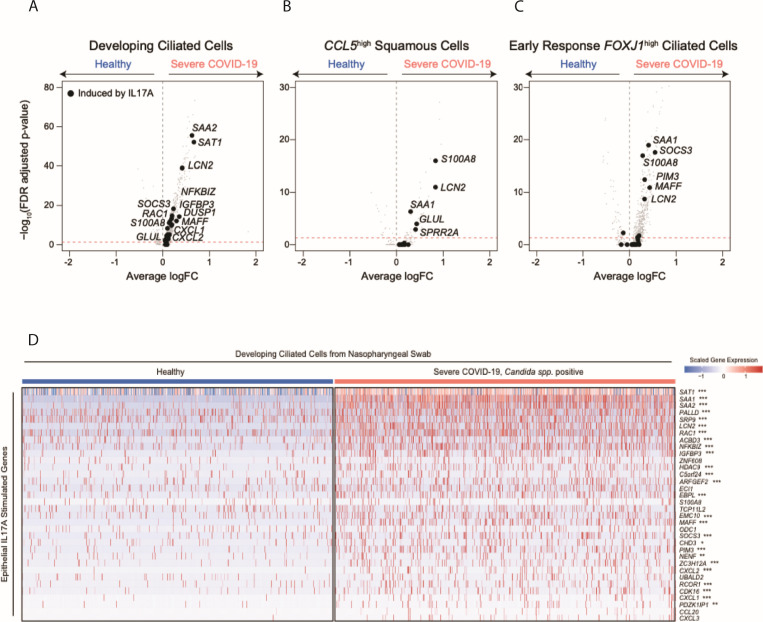
Next, we returned to the scRNA-seq data from our human COVID-19 cohort and evaluated epithelial cells for transcriptomic signatures consistent with exposure to each cytokine. Compared to epithelial cells isolated from 15 healthy controls, epithelial cells isolated from eight individuals with severe COVID-19 expressed significantly higher levels of genes that were also induced by IL17A exposure in vitro (Fig. 4). In particular, IL17A-induced genes SAA1, SAA2, SAT1, LCN2, S100A8, and GLUL were repeatedly significantly upregulated among diverse epithelial cell subsets in severe COVID-19 (Fig. 4A through C) but were not found to be significantly induced within the nasal epithelia of patients with milder COVID-19 (Fig. S3B). We confirmed that treatment of nasal epithelial cells with other inflammatory signals potentially found within the respiratory epithelium, including IL4 and IL1β, does not appreciably induce these factors, suggesting that the induction of this gene module is a specific downstream effect of IL17 sensing (Fig. S3A).
Fig 4.
Transcriptional responses of respiratory epithelial cells following in vitro exposure to various cytokines (A–C). Volcano plots of differentially expressed genes between select epithelial cell types from healthy participants vs those with severe COVID-19: developing ciliated cells (A), CCL5high squamous cells (B), and early response FOXJ1high ciliated cells (C). Gray points, all genes; black points, genes induced in human nasal epithelial cells following IL17A treatment (as in Fig. 3A). (D) Heatmap of IL17A-induced genes among developing ciliated cells from NP swabs. Heatmap colors reflect scaled gene expression: red, higher expression; blue, lower expression. Left columns (blue bar), developing ciliated cells from n = 15 healthy participants; right (pink bar), developing ciliated cells from n = 8 participants with severe COVID-19 and Candida spp. detected. Genes (rows) selected represent genes significantly upregulated among cultured human nasal epithelia cells following in vitro exposure to IL17A. Statistical significance comparing healthy-derived vs severe COVID-19, Candida spp. positive-derived single cells by likelihood ratio test assuming an underlying negative binomial distribution. ***FDR-corrected P < 0.001; **FDR < 0.01; *FDR < 0.05.
DISCUSSION
Our data demonstrate Candida spp. colonization of the upper respiratory tract in a significant proportion (38%) of individuals hospitalized with severe COVID-19 in a cohort from the University of Mississippi Medical Center sampled during the summer 2020 COVID-19 peak. Notably, no fungal pathogens were identified among individuals with mild or moderate COVID-19, non-COVID-19 ICU patients, or healthy controls. Fungal reads were detected by NP swabs at early timepoints following hospital admission and within the acute phase of patients’ COVID-19 disease trajectory, suggesting that in some patients, fungal colonization and infection likely occurred prior to hospital admission and in advance of nosocomial exposures. Testing for fungal infection among patients with prolonged intubation and clinical concern for infection is a common occurrence in the intensive care unit. These details provide clinical context and note interesting examples of agreement between the two approaches, but we agree that it does not reflect orthogonal validation.
Candida is not typically present in the nares of healthy people, being more readily detected in the oropharynx; thus, the identification of Candida from NP swabs of a subset of patients with severe COVID-19 would suggest either that severe COVID-19 predisposes to ectopic colonization in some hosts or alternatively that our methodology is detecting increased fungal abundances derived from the oral mucosa (42). Further direct comparisons of colonization in the mouth and nose of patients with severe COVID-19 will be necessary to clarify this issue (43).
Our analysis additionally unites the use of single-cell transcriptomic technologies in human clinical cohorts with emerging computational approaches for meta-genomic pathogen classification of human samples, all derived from limited cellular material captured on a single NP swab (21, 22). By linking unbiased pathogen detection with single-cell nasal epithelial and immune transcriptional profiles, we have identified specific host behaviors indicative of response to a fungal pathogen and IL17 signaling. For a subset of patients, early detection of Candida spp. in the upper respiratory mucosa corresponded to the development of more extensive colonization and clinical concern for secondary fungal pneumonia and/or candidemia. Mounting evidence across various clinical cohorts, including our prior work, suggests that severe COVID-19 arises in individuals with impaired anti-viral immunity (3, 31, 44–50). While there is a paucity of evidence to suggest that type I/III interferon signaling directly restricts fungal colonization, prior in vitro studies indicate that IL17 signaling among airway epithelial cells may attenuate cellular responses to type I/III interferon (20, 51–54). Additionally, enhanced virally induced epithelial injury resulting from impaired IFN signaling could facilitate Candida colonization (29, 55). Surprisingly, we observe that IL17A-induced gene sets are elevated among epithelial cells from all individuals with severe COVID-19 in our cohort, even those patients without genomic or clinical evidence for coincident fungal infection.
Crucially, our data do not allow us to determine whether our observation of elevated IL17 responses in patients with severe COVID-19 without overt evidence for fungal colonization (i) is the result of colonization below levels of detection in these assays or (ii) suggests that IL17 elevation represents a general phenotype of epithelial cells of patients with severe COVID-19, independent of fungal colonization. Future experiments encompassing longitudinal sampling of patients with COVID-19 could shed additional light on whether variability in the dynamics of IL17 and interferon signaling may underlie Candida colonization in the upper airways.
Together, our data suggest that upper respiratory Candida colonization and infection represent an underappreciated phenomenon among patients with severe COVID-19. We acknowledge the limited sample number as a limitation to this study, and further research with larger cohorts is warranted to understand the frequency and timing of cooccurring infection with Candida spp. and other fungal pathogens following SARS-CoV-2 infection. Our data suggest that dedicated, multi-institutional studies are required to disentangle how clinical and subclinical fungal infections impact patient outcomes during hospitalization for COVID-19 and may hold key insights into determinants of severe respiratory failure for these patients and new strategies for diagnostic or therapeutic intervention.
MATERIALS AND METHODS
Participant recruitment and respiratory sampling
Full participant characteristics are provided in a previously published study (3). The UMMC Institutional Review Board approved the study under IRB#2020-0065. All participants or their legally authorized representatives provided written informed consent. Briefly, participants were eligible for inclusion in the COVID-19 group if they were at least 18 years old; had a positive nasopharyngeal swab for SARS-CoV-2 by PCR; had COVID-19-related symptoms including fever, chills, cough, shortness of breath, and sore throat; and weighed more than 110 lb. Participants were eligible for inclusion in the healthy group if they were at least 18 years old, had a current negative SARS-CoV-2 test (PCR or rapid antigen test), and weighed more than 110 lb. COVID-19 participants were classified according to the eight-level ordinal scale proposed by the WHO representing their peak clinical severity and level of respiratory support required. Nasopharyngeal samples and endotracheal aspirate samples were collected by a trained healthcare provider; all processing and handling were carried out as previously described (3, 36).
scRNA-seq data generation and alignment
Annotated scRNA-seq data were recovered from the Single Cell Portal (see Data Availability), and single-cell annotations were used as described in reference (3). Briefly, data represent aligned scRNA-seq libraries generated using Seq-Well S3; libraries were generated using Illumina Nextera XT Library Prep Kits and sequenced on NextSeq 500/550 High Output 75 cycle v2.5 kits to an average depth of 180 million aligned reads per array: read 1: 21 (cell barcode, UMI), read 2: 50 (digital gene expression), and index 1: 8 (N700 barcode) (56). Libraries were aligned using STAR within the Drop-Seq Computational Protocol (https://github.com/broadinstitute/Drop-seq) and implemented on Cumulus (https://cumulus.readthedocs.io/en/latest/drop_seq.html, snapshot 9, default parameters) (57, 58). As previously described, data were aligned using a custom reference, which combined human GRCh38 (from CellRanger version 3.0.0, Ensembl 93) and SARS-CoV-2 RNA genomes (3, 59).
Meta-transcriptomic pathogen classification
To identify codetected microbial taxa present in the cell-associated or ambient RNA of nasopharyngeal swabs, we used the Kraken2 software implemented using the Broad Institute viral-ngs pipelines on Terra (https://github.com/broadinstitute/viral-pipelines/tree/master) (21, 22). A previously published reference database included human, archaea, bacteria, plasmid, viral, fungi, and protozoa species and was constructed on 5 May 2020, therefore including sequences belonging to the novel SARS-CoV-2 virus. Inputs to Kraken2 were the following: kraken2_db_tgz = “gs://pathogen-public-dbs/v1/kraken2-broad-20200505.tar.zst,” krona_taxonomy_db_kraken2_tgz = “gs://pathogen-public-dbs/v1/krona.taxonomy-20200505.tab.zst,” ncbi_taxdump_tgz = “gs://pathogen-public-dbs/v1/taxdump-20200505.tar.gz,” trim_clip_db = “gs://pathogen-public-dbs/v0/contaminants.clip_db.fasta,” and spikein_db = “gs://pathogen-public-dbs/v0/ERCC_96_nopolyA.fasta.” Results were collected using the merge_metagenomics tool (https://viral-pipelines.readthedocs.io/en/latest/merge_metagenomics.html), and analysis and visualization of each sample’s metagenomic alignments were implemented in Prism (v6) or R [v4.0.2; packages ggplot2 (v3.3.2), Seurat (v3.2.2), and ComplexHeatmap (v2.7.3)]. All classification data are included in Table S1.
Human nasal epithelial cell response to in vitro cytokine exposure
Gene lists representing human nasal epithelial cell responses to various exogenous cytokines in vitro are derived from previously published population RNA-seq data (36, 37). Briefly, human nasal epithelial basal cells from two donors were stimulated in vitro with 0.1–10 ng/mL IFNα, IL17A, IFNγ, IL1β, or IL4 for 12 hours. Following stimulation, cells were lysed, and bulk RNA-seq libraries were generated using the SMART-Seq2 protocol (60). We identified epithelial gene sets induced by each cytokine independently by testing for differentially expressed genes compared to matched, untreated nasal epithelial samples (n = 10). Differential expression testing was carried out using a likelihood ratio test assuming a negative binomial distribution, implemented with the Seurat (v3.1.5) FindAllMarkers function (test.use = “negbinom”). We considered genes as differentially expressed with an FDR-adjusted P-value <0.05 (61).
To score for cytokine-specific gene expression among COVID-19 or healthy scRNA-seq samples, we first subsetted our scRNA-seq data to only epithelial cells using “coarse” cell types, as defined by a cell typing procedure carried out in prior publication (3). Coarse cell type groups that were included in the analysis include “ciliated cells,” “developing ciliated cells,” “secretory cells,” “goblet cells,” “ionocytes,” “deuterosomal cells,” “squamous cells,” “basal cells,” “mitotic basal cells,” and “developing secretory and goblet cells.” We calculated module scores over all epithelial cells using the Seurat function AddModuleScore with default inputs. The average module score for each NP or ETA sample was utilized as a representative measure of epithelial behavior for each participant, as represented in Fig. 2A through D.
scRNA-seq analysis of differential expression
To compare gene expression between cells from distinct disease groups (e.g., healthy vs severe COVID-19, Candida spp.), we employed a likelihood ratio test assuming a negative binomial distribution as described above [using Seurat FindAllMarkers function (test.use = “negbinom”)] (61). We considered genes as differentially expressed with an FDR-adjusted P-value <0.001 and log fold change >0.25. Results from select “detailed” cell types, as defined and previously reported by Ziegler et al., are represented in Fig. 4A through C (3). Full results of differential expression as represented in Fig. 4A through C can be found in supplementary tables.
Statistical testing
All statistical tests were implemented in R (v4.0.2) or Prism (v6) software. Specific statistical tests, P-values, n, and all summary statistics are provided in Results, figure legends, and/or figure panels.
ACKNOWLEDGMENTS
We thank the study participants and their families for enabling this research, the clinical support staff at UMMC for their assistance in sample collection, and the members of the Shalek, Ordovas-Montanes, Horwitz, and Glover labs for the thoughtful discussion and comments on the project. This project has been made possible in part by grant number 2020-216949 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation to A.K.S. and J.O.-M., and by CZI grant DAF2021-237665 and grant DOI https://doi.org/10.37921/881723wbyqpp from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation (funder DOI 10.13039/100014989) to J.O.-M., B.H.H., S.C.G., and A.K.S. C.G.K.Z. was supported by award numbers T32GM007753 and T32GM144273 from the National Institute of General Medical Sciences. J.O.-M. is a New York Stem Cell Foundation–Robertson Investigator. J.O.-M. was supported by the AbbVie-Harvard Medical School Alliance, the Richard and Susan Smith Family Foundation, the AGA Research Foundation’s AGA-Takeda Pharmaceuticals Research Scholar Award in IBD (AGA2020-13-01), the Leona M. and Harry B. Helmsley Charitable Trust, the Pew Charitable Trusts Biomedical Scholars, the Broad Next Generation Award, the Mathers Foundation, the New York Stem Cell Foundation, NIH R01 HL162642, NIH R01 DE031928, and the Cell Discovery Network at Boston Children’s Hospital supported by the Manton Foundation and Warren Alpert Foundation. B.H.H. was supported by DK122532-01A1, NIH; 12019PG-CD002, the Leona M. and Harry B. Helmsley Charitable Trust; SRA #54518; and Crohn’s and Colitis Foundation. A.K.S. was supported by the Bill and Melinda Gates Foundation, Sloan Fellowship in Chemistry, the NIH (5U24AI118672), and the Ragon Institute of MGH, MIT, and Harvard. This work was supported in part by the Division of Intramural Research of the NIAID.
Conceptualization was performed by J.O.-M., A.K.S., S.C.G., and B.H.H. Methodology was performed by C.G.K.Z., A.H.O., S.C.G., J.O.-M., A.K.S., and B.H.H. Formal analysis was performed by C.G.K.Z. and A.H.O. Investigation was performed by C.G.K.Z. and A.H.O. Resources were secured by S.C.G., A.H.O., M.Sl., H.L., M.Se., and G.E.A. Data curation was performed by T.O.R., Y.T., M.Sl., A.H.O., H.L., H.B.W., and C.G.K.Z. Writing (original draft) was performed by C.G.K.Z., A.H.O., J.O.-M., A.K.S., S.C.G., and B.H.H. Writing (review and editing) was performed by C.G.K.Z., M.G.-P., G.D., S.K., V.N.M., A.H.O., A.W.N., Y.T., J.D.B., P.L., M.S.L., M.Sl., H.L., H.B.W., M.G., R.S.D., Y.P., G.E.A., M.Se., T.O.R., A.K.S., S.C.G., B.H.H., and J.O.-M. Visualization was performed by C.G.K.Z. Supervision was performed by J.O.-M., A.K.S., S.C.G., B.H.H., and T.O.R. Project administration was performed by Y.P. and T.O.R. Funding acquisition was performed by J.O.-M., A.K.S., S.C.G., and B.H.H.
Contributor Information
Alex K. Shalek, Email: shalek@mit.edu.
Jose Ordovas-Montanes, Email: jose.ordovas-montanes@childrens.harvard.edu.
Bruce H. Horwitz, Email: bruce.horwitz@childrens.harvard.edu.
Sarah C. Glover, Email: sglover3@tulane.edu.
Joshua J. Obar, Geisel School of Medicine at Dartmouth, Lebanon, New Hampshire, USA
DATA AVAILABILITY
All scRNA-seq and RNA-seq data analyzed in this study are publicly available from prior manuscripts. Aligned and annotated scRNA-seq data can be downloaded via the Single Cell Portal, study SCP1289. Aligned TPM-normalized RNA-seq data can be downloaded via the Single Cell Portal, study SCP822. Results from Kraken2 metagenomic classification are reported in Table S1.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03516-23.
Fig. S1 to S4.
Metatranscriptomic data output from kraken2.
Gene sets.
Control vs COVID in epithelial cells.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, Liu L, Shan H, Lei C-L, Hui DSC, et al. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ziegler CGK, Miao VN, Owings AH, Navia AW, Tang Y, Bromley JD, Lotfy P, Sloan M, Laird H, Williams HB, George M, Drake RS, Christian T, Parker A, Sindel CB, Burger MW, Pride Y, Hasan M, Abraham GE, Senitko M, Robinson TO, Shalek AK, Glover SC, Horwitz BH, Ordovas-Montanes J. 2021. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 184:4713–4733. doi: 10.1016/j.cell.2021.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida M, Worlock KB, Huang N, Lindeboom RGH, Butler CR, Kumasaka N, Dominguez Conde C, Mamanova L, Bolt L, Richardson L, et al. 2022. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature 602:321–327. doi: 10.1038/s41586-021-04345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, et al. 2020. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat Biotechnol 38:970–979. doi: 10.1038/s41587-020-0602-4 [DOI] [PubMed] [Google Scholar]
- 6. Hassan I, Powell G, Sidhu M, Hart WM, Denning DW. 2009. Excess mortality, length of stay and cost attributable to candidaemia. J Infect 59:360–365. doi: 10.1016/j.jinf.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 7. Kayaaslan B, Eser F, Kaya Kalem A, Bilgic Z, Asilturk D, Hasanoglu I, Ayhan M, Tezer Tekce Y, Erdem D, Turan S, Mumcuoglu I, Guner R. 2021. Characteristics of candidemia in COVID-19 patients; increased incidence, earlier occurrence and higher mortality rates compared to non-COVID-19 patients. Mycoses 64:1083–1091. doi: 10.1111/myc.13332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chumbita M, Puerta-Alcalde P, Garcia-Pouton N, García-Vidal C. 2021. COVID-19 and fungal infections: etiopathogenesis and therapeutic implications. Rev Esp Quimioter 34:72–75. doi: 10.37201/req/s01.21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol M, Pitart C, Inciarte A, Bodro M, Morata L, Ambrosioni J, Grafia I, Meira F, Macaya I, Cardozo C, Casals C, Tellez A, Castro P, Marco F, García F, Mensa J, Martínez JA, Soriano A, COVID-19 Researchers Group . 2021. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect 27:83–88. doi: 10.1016/j.cmi.2020.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arastehfar A, Carvalho A, Nguyen MH, Hedayati MT, Netea MG, Perlin DS, Hoenigl M. 2020. COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J Fungi (Basel) 6:211. doi: 10.3390/jof6040211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. White PL, Dhillon R, Cordey A, Hughes H, Faggian F, Soni S, Pandey M, Whitaker H, May A, Morgan M, Wise MP, Healy B, Blyth I, Price JS, Vale L, Posso R, Kronda J, Blackwood A, Rafferty H, Moffitt A, Tsitsopoulou A, Gaur S, Holmes T, Backx M. 2021. A national strategy to diagnose coronavirus disease 2019-associated invasive fungal disease in the intensive care unit. Clin Infect Dis 73:e1634–e1644. doi: 10.1093/cid/ciaa1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mastrangelo A, Germinario BN, Ferrante M, Frangi C, Li Voti R, Muccini C, Ripa M, COVID-BioB Study Group . 2021. Candidemia in coronavirus disease 2019 (COVID-19) patients: incidence and characteristics in a prospective cohort compared with historical non-COVID-19 controls. Clin Infect Dis 73:e2838–e2839. doi: 10.1093/cid/ciaa1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nucci M, Barreiros G, Guimarães LF, Deriquehem VAS, Castiñeiras AC, Nouér SA. 2021. Increased incidence of candidemia in a tertiary care hospital with the COVID-19 pandemic. Mycoses 64:152–156. doi: 10.1111/myc.13225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Omrani AS, Koleri J, Ben Abid F, Daghfel J, Odaippurath T, Peediyakkal MZ, Baiou A, Sarsak E, Elayana M, Kaleeckal A, Almaslamani MA. 2021. Clinical characteristics and risk factors for COVID-19-associated Candidemia. Med Mycol 59:1262–1266. doi: 10.1093/mmy/myab056 [DOI] [PubMed] [Google Scholar]
- 15. Bretagne S, Sitbon K, Botterel F, Dellière S, Letscher-Bru V, Chouaki T, Bellanger A-P, Bonnal C, Fekkar A, Persat F, et al. 2021. COVID-19-associated pulmonary Aspergillosis, Fungemia, and Pneumocystosis in the intensive care unit: a retrospective multicenter observational cohort during the first French pandemic wave. Microbiol Spectr 9:e0113821. doi: 10.1128/Spectrum.01138-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sparber F, LeibundGut-Landmann S. 2019. Interleukin-17 in antifungal immunity. Pathogens 8:54. doi: 10.3390/pathogens8020054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mengesha BG, Conti HR. 2017. The role of IL-17 in protection against mucosal Candida infections. J Fungi (Basel) 3:52. doi: 10.3390/jof3040052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Puel A, Cypowyj S, Maródi L, Abel L, Picard C, Casanova J-L. 2012. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol 12:616–622. doi: 10.1097/ACI.0b013e328358cc0b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trautwein-Weidner K, Gladiator A, Nur S, Diethelm P, LeibundGut-Landmann S. 2015. IL-17-mediated antifungal defense in the oral mucosa is independent of neutrophils. Mucosal Immunol 8:221–231. doi: 10.1038/mi.2014.57 [DOI] [PubMed] [Google Scholar]
- 20. Break TJ, Oikonomou V, Dutzan N, Desai JV, Swidergall M, Freiwald T, Chauss D, Harrison OJ, Alejo J, Williams DW, et al. 2021. Aberrant type 1 immunity drives susceptibility to mucosal fungal infections. Science 371:eaay5731. doi: 10.1126/science.aay5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemieux JE, Siddle KJ, Shaw BM, Loreth C, Schaffner SF, Gladden-Young A, Adams G, Fink T, Tomkins-Tinch CH, Krasilnikova LA, et al. 2021. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science 371:eabe3261. doi: 10.1126/science.abe3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mallett S, Allen AJ, Graziadio S, Taylor SA, Sakai NS, Green K, Suklan J, Hyde C, Shinkins B, Zhelev Z, Peters J, Turner PJ, Roberts NW, di Ruffano LF, Wolff R, Whiting P, Winter A, Bhatnagar G, Nicholson BD, Halligan S. 2020. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med 18:346. doi: 10.1186/s12916-020-01810-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiederhold NP, Cota JM, Frei CR. 2008. Micafungin in the treatment of invasive candidiasis and invasive aspergillosis. Infect Drug Resist 1:63–77. doi: 10.2147/IDR.S3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bader MS, Lai SM, Kumar V, Hinthorn D. 2004. Candidemia in patients with diabetes mellitus: epidemiology and predictors of mortality. Scand J Infect Dis 36:860–864. doi: 10.1080/00365540410021126 [DOI] [PubMed] [Google Scholar]
- 26. Rodrigues CF, Rodrigues ME, Henriques M. 2019. Candida sp. infections in patients with diabetes mellitus. J Clin Med 8:76. doi: 10.3390/jcm8010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, Choi B, Nam SK, Sa M, Kwon J-S, Jeong SJ, Lee HK, Park SH, Park S-H, Choi JY, Kim S-H, Jung I, Shin E-C. 2020. Immunophenotyping of covid-19 and influenza highlights the role of type i interferons in development of severe covid-19. Sci Immunol 5:1554. doi: 10.1126/sciimmunol.abd1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galani I-E, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, Koukaki E, Fragkou PC, Panou V, Rapti V, Koltsida O, Mentis A, Koulouris N, Tsiodras S, Koutsoukou A, Andreakos E. 2021. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 22:32–40. doi: 10.1038/s41590-020-00840-x [DOI] [PubMed] [Google Scholar]
- 29. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. doi: 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meffre E, Iwasaki A. 2020. Interferon deficiency can lead to severe COVID. Nature 587:374–376. doi: 10.1038/d41586-020-03070-1 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. doi: 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, Gozzi L, Iannone A, Lo Tartaro D, Mattioli M, et al. 2020. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun 11:3434. doi: 10.1038/s41467-020-17292-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pacha O, Sallman MA, Evans SE. 2020. COVID-19: a case for inhibiting IL-17? Nat Rev Immunol 20:345–346. doi: 10.1038/s41577-020-0328-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ponde NO, Shoger KE, Khatun MS, Sarkar MK, Dey I, Taylor TC, Cisney RN, Arunkumar SP, Gudjonsson JE, Kolls JK, Gottschalk RA, Gaffen SL. 2023. SARS-CoV-2 ORF8 mediates signals in macrophages and monocytes through MyD88 independently of the IL-17 receptor. J Immunol 211:252–260. doi: 10.4049/jimmunol.2300110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hasan MZ, Islam S, Matsumoto K, Kawai T. 2021. SARS-CoV-2 infection initiates interleukin-17-enriched transcriptional response in different cells from multiple organs. Sci Rep 11:16814. doi: 10.1038/s41598-021-96110-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, Wadsworth MH, Hughes TK, Kazer SW, Yoshimoto E, Cahill KN, Bhattacharyya N, Katz HR, Berger B, Laidlaw TM, Boyce JA, Barrett NA, Shalek AK. 2018. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560:649–654. doi: 10.1038/s41586-018-0449-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, et al. 2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181:1016–1035. doi: 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cabral A, Voskamp P, Cleton-Jansen AM, South A, Nizetic D, Backendorf C. 2001. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J Biol Chem 276:19231–19237. doi: 10.1074/jbc.M100336200 [DOI] [PubMed] [Google Scholar]
- 39. Christmann C, Zenker S, Martens L, Hübner J, Loser K, Vogl T, Roth J. 2020. Interleukin 17 promotes expression of alarmins S100A8 and S100A9 during the inflammatory response of keratinocytes. Front Immunol 11:599947. doi: 10.3389/fimmu.2020.599947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. 2011. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat Immunol 12:853–860. doi: 10.1038/ni.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J-Y, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin W-Y, Yeung ST, Silva HM, Li D, Hine A, Loke P, Hudesman D, Martin JC, Kenigsberg E, Merad M, Khanna KM, Littman DR. 2020. Serum amyloid A proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell 180:79–91. doi: 10.1016/j.cell.2019.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salehi M, Ahmadikia K, Mahmoudi S, Kalantari S, Jamalimoghadamsiahkali S, Izadi A, Kord M, Dehghan Manshadi SA, Seifi A, Ghiasvand F, Khajavirad N, Ebrahimi S, Koohfar A, Boekhout T, Khodavaisy S. 2020. Oropharyngeal candidiasis in hospitalised COVID-19 patients from Iran: species identification and antifungal susceptibility pattern. Mycoses 63:771–778. doi: 10.1111/myc.13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, NIH Intramural Sequencing Center Comparative Sequencing Program . 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Galani I-E, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, Koukaki E, Fragkou PC, Panou V, Rapti V, Koltsida O, Mentis A, Koulouris N, Tsiodras S, Koutsoukou A, Andreakos E. 2021. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 22:32–40. doi: 10.1038/s41590-020-00840-x [DOI] [PubMed] [Google Scholar]
- 45. Kusnadi A, Ramírez-Suástegui C, Fajardo V, Chee SJ, Meckiff BJ, Simon H, Pelosi E, Seumois G, Ay F, Vijayanand P, Ottensmeier CH. 2021. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8+ T cells. Sci Immunol 6:eabe4782. doi: 10.1126/sciimmunol.abe4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu C, Martins AJ, Lau WW, Rachmaninoff N, Chen J, Imberti L, Mostaghimi D, Fink DL, Burbelo PD, Dobbs K, et al. 2021. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell 184:1836–1857. doi: 10.1016/j.cell.2021.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bastard P, Orlova E, Sozaeva L, Lévy R, James A, Schmitt MM, Ochoa S, Kareva M, Rodina Y, Gervais A, et al. 2021. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med 218:e20210554. doi: 10.1084/jem.20210554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Q, Matuozzo D, Le Pen J, Lee D, Moens L, Asano T, Bohlen J, Liu Z, Moncada-Velez M, Kendir-Demirkol Y, et al. 2022. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med 219:e20220131. doi: 10.1084/jem.20220131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370:eabd4585. doi: 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Combes AJ, Courau T, Kuhn NF, Hu KH, Ray A, Chen WS, Chew NW, Cleary SJ, Kushnoor D, Reeder GC, et al. 2021. Global absence and targeting of protective immune states in severe COVID-19. Nature 591:124–130. doi: 10.1038/s41586-021-03234-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Espinosa V, Dutta O, McElrath C, Du P, Chang Y-J, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, Kotenko SV, Rivera A. 2017. Type III interferon is a critical regulator of innate antifungal immunity. Sci Immunol 2:eaan5357. doi: 10.1126/sciimmunol.aan5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smeekens SP, Ng A, Kumar V, Johnson MD, Plantinga TS, van Diemen C, Arts P, Verwiel ETP, Gresnigt MS, Fransen K, van Sommeren S, Oosting M, Cheng S-C, Joosten LAB, Hoischen A, Kullberg B-J, Scott WK, Perfect JR, van der Meer JWM, Wijmenga C, Netea MG, Xavier RJ. 2013. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat Commun 4:1342. doi: 10.1038/ncomms2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marié IJ, Brambilla L, Azzouz D, Chen Z, Baracho GV, Arnett A, Li HS, Liu W, Cimmino L, Chattopadhyay P, Silverman G, Watowich SS, Khor B, Levy DE. 2021. Tonic interferon restricts pathogenic IL-17-driven inflammatory disease via balancing the microbiome. Elife 10:e68371. doi: 10.7554/eLife.68371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niwa M, Fujisawa T, Mori K, Yamanaka K, Yasui H, Suzuki Y, Karayama M, Hozumi H, Furuhashi K, Enomoto N, Nakamura Y, Inui N, Suzuki T, Maekawa M, Suda T. 2018. IL-17A attenuates IFN-λ expression by inducing suppressor of cytokine signaling expression in airway epithelium. J Immunol 201:2392–2402. doi: 10.4049/jimmunol.1800147 [DOI] [PubMed] [Google Scholar]
- 55. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, et al. 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584:463–469. doi: 10.1038/s41586-020-2588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gierahn TM, Wadsworth II MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. 2017. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14:395–398. doi: 10.1038/nmeth.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li B, Gould J, Yang Y, Sarkizova S, Tabaka M, Ashenberg O, Rosen Y, Slyper M, Kowalczyk MS, Villani A-C, Tickle T, Hacohen N, Rozenblatt-Rosen O, Regev A. 2020. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat Methods 17:793–798. doi: 10.1038/s41592-020-0905-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, et al. 2021. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595:107–113. doi: 10.1038/s41586-021-03570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A. 2014. Preparation of single-cell RNA-Seq libraries for next generation sequencing. Curr Protoc Mol Biol 107:4. doi: 10.1002/0471142727.mb0422s107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4.
Metatranscriptomic data output from kraken2.
Gene sets.
Control vs COVID in epithelial cells.
Data Availability Statement
All scRNA-seq and RNA-seq data analyzed in this study are publicly available from prior manuscripts. Aligned and annotated scRNA-seq data can be downloaded via the Single Cell Portal, study SCP1289. Aligned TPM-normalized RNA-seq data can be downloaded via the Single Cell Portal, study SCP822. Results from Kraken2 metagenomic classification are reported in Table S1.