Abstract
In many tumor systems, analysis of cells for loss of heterozygosity (LOH) has helped to clarify the role of tumor suppressor genes in oncogenesis. Two important tumor suppressor genes, p53 and the Ink4a/Arf locus, play central roles in the multistep process of Abelson murine leukemia virus (Ab-MLV) transformation. p53 and the p53 regulatory protein, p19Arf, are required for the apoptotic crisis that characterizes the progression of primary transformed pre-B cells to fully malignant cell lines. To search for other tumor suppressor genes which may be involved in the Ab-MLV transformation process, we used endogenous proviral markers and simple-sequence length polymorphism analysis to screen Abelson virus-transformed pre-B cells for evidence of LOH. Our survey reinforces the role of the p53-p19 regulatory pathway in transformation; 6 of 58 cell lines tested had lost sequences on mouse chromosome 4, including the Ink4a/Arf locus. Consistent with this pattern, a high frequency of primary pre-B-cell transformants derived from Ink4a/Arf +/− mice became established cell lines. In addition, half of them retained the single copy of the locus when the transformation process was complete. These data demonstrate that a single copy of the Ink4a/Arf locus is not sufficient to fully mediate the effects of these genes on transformation.
Abelson murine leukemia virus (Ab-MLV) is a rapidly transforming retrovirus that can induce lymphomas in vivo and transform pre-B cells and immortalized fibroblast cell lines in vitro (reviewed in references 39 and 40). The virus carries the v-abl oncogene, and the protein tyrosine kinase it encodes is required for transformation. Despite the strong growth-stimulatory signal provided by the v-Abl protein, Ab-MLV-induced transformation is a multistep process both in vivo and in vitro (16, 17, 59, 60), suggesting that multiple cellular changes are required before a cell becomes fully malignant. Analysis of primary pre-B-cell transformants in vitro has revealed that the p53 tumor suppressor gene and the p19Arf gene, the product of which regulates p53 function (reviewed in references 35 and 46), are intimately involved in the process by which these cells evolve to become fully malignant established cell lines (37, 52, 54). About 50% of all transformants contain mutations affecting p53 (52), and many others express very low levels of the p19Arf protein, a molecule that stabilizes p53, thereby enhancing its function (37).
The factors involved in tumor progression in the Ab-MLV system have received limited attention, and most studies using Ab-MLV or other oncogenic retroviruses have focused on oncogene cooperativity (reviewed in reference 42). However, in many other types of tumors, dominant growth-stimulatory signals generated by oncogenes cooperate with the loss of growth-suppressive signals provided by tumor suppressor genes (26, 45). Alterations in the p53 and NF1 tumor suppressor genes have been reported in some retrovirus-induced tumors (3–6, 53, 56, 61). Other studies have identified chromosomal regions displaying loss of heterozygosity (LOH), a feature associated with the presence of tumor suppressor genes (11, 14, 23). For example, a fraction of Moloney murine leukemia virus (Mo-MLV)-induced thymomas (25) and mammary tumors arising in mouse mammary tumor virus transgenic mice (9) have lost sequences on several chromosomes. Similar changes have also been noted in radiation-induced hematopoietic tumors in mice (8, 30, 32, 47).
The large number of endogenous nonecotropic proviruses carried by inbred mice provide polymorphic markers that facilitate such an analysis (15, 49). Most inbred mice carry between 30 and 60 of these proviruses, and their chromosomal locations have been mapped in many inbred mouse strains (15). In addition, usually more than 60% of the endogenous nonecotropic proviruses carried in the F1 progeny from the cross of two inbred strains will be present in single copy, making LOH detection straightforward. When coupled with PCR-based analysis of simple-sequence length polymorphisms (SSLPs) (10), rapid assessment of the genome for LOH can be achieved.
To search for common deletions in Ab-MLV-transformed pre-B cells, we surveyed transformants derived from the bone marrow of three different F1 crosses, using endogenous proviruses and SSLP genetic markers. These analyses revealed that 7 of 58 cell lines had lost proviral and SSLP markers present on chromosome 4. Although the isolates lost different amounts of information, six of the seven had lost sequences in the vicinity of the Ink4a/Arf locus which maps to this chromosome (36). Loss of chromosome 4 sequences may reflect selection against cells expressing these products; analysis of transformants from Ink4a/Arf +/− mice revealed that a higher frequency of these cells became established more rapidly than cells derived from wild-type littermates. These data suggest that LOH affecting p19Arf can have a significant effect on transformation in the Ab-MLV system and raise the possibility that similar events are important in other types of tumors.
MATERIALS AND METHODS
Cell lines.
Ab-MLV-transformed pre-B-cell lines were maintained in RPMI 1640 medium supplemented to contain 10% fetal calf serum and 50 μM 2-mercaptoethanol. The CXCE (BALB/cByJ × Cast/Ei)F1 transformed pre-B cells were derived by infecting bone marrow with Ab-MLV P160 as described previously (13, 41). The Ab-MLV-transformed CXXB (BALB/Ann.xid × C57BL/10)F1 and CXCB (BALB/cByJ × CBA/Tufts)F1 pre-B cells were derived in a similar fashion and were characterized previously (27). All of these transformants were fully transformed when they were analyzed. The independent origin of all cell lines was confirmed by either the structure of the Igh locus (27) or Southern analysis of the Ab-MLV integration site (data not shown). Ink4a/Arf null mice (44) that had been backcrossed to C57BL/6J mice for five generations were used in some experiments. Primary transformants were plated in 24-well plates in RPMI 1640 supplemented to contain 20% fetal calf serum and 50 μM 2-mercaptoethanol 10 days later. The cells were monitored for growth and viability; when cells filled the well, half of them were transferred to a new well. When viability exceeded 90% and the cells could be subcultured on a regular basis, they were considered to be established (37, 54).
Nucleic acid analysis.
Proviral mapping was carried out as described by Coffin and coworkers (49, 51), with minor modifications. High-molecular-weight DNAs (58) were digested with EcoRI or PvuII (New England Biolabs) and fractionated through 0.8% agarose–0.5× Tris-borate-EDTA gels at 75 V for 22 to 24 h. The ethidium bromide-stained gels were cut in half to facilitate handling and then treated with 1.5 M NaCl–0.5 N NaOH for 30 min and 1.5 M NaCl–1 M Tris-Cl (pH 8) for 30 min. The gels were dried on a slab gel dryer using house vacuum without heat for 1 to 2 h until they were flat and then dried for an additional 30 min at 60°C. The dried gels were treated with 5× SSPE (0.9 M NaCl, 0.05 M NaH2PO4, 0.005 M EDTA [pH 7.4]) and were usually frozen at −20°C for up to 1 month before use. The dried gels were hybridized with radiolabeled JS4, JS5, or JS6/10 oligonucleotide (49) in 5× SSPE–0.1% sodium dodecyl sulfate (SDS)–10 μg of salmon sperm DNA per ml for 20 to 24 h at 62°C. The gels then were washed four times in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate)–0.2% SDS at room temperature for 15 min and twice at 62°C for 30 min in the same solution. The gels were air dried and exposed to Kodak XAR-5 film at −70°C with an intensifier screen for 5 to 14 days. Gels were stripped of oligonucleotide by soaking for 30 min each in 1.5 M NaCl–0.5 N NaOH and 1.5 M NaCl–1 M Tris-HCl (pH 8). For detection of the Ink4a/Arf locus, digested DNAs were fractionated through agarose gels, transferred to nylon membranes, and hybridized with a p16Ink4a exon 1α or p19Arf exon 1β probe (37). Blots were exposed to Kodak XAR-5 film at −70°C with an intensifier screen.
PCR analysis.
For SSLP analyses, genomic DNA was amplified using primers described in the Massachusetts Institute of Technology (MIT) Mouse Genome Database (http://www.genome.wi.mit.edu) and the conditions recommended by the manufacturer (Research Genetics). Briefly, high-molecular-weight DNA was amplified in reactions containing 200 ng of DNA, 200 μM each deoxynucleoside triphosphate, 0.4 μM primers, 1.25 U of Taq, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 0.001% gelatin. After a 3-min incubation at 94°C, the reactions were amplified for 25 cycles of 15 s at 94°C, 3 min at 55°C, and 4 min at 72°C. After the final cycle, the reactions were cooled to 4°C. The products were fractionated through 12% polyacrylamide gels and visualized by ethidium bromide staining. Mice and cell lines derived from the Ink4a/Arf background were genotyped by using PCR with three primers, two Ink4a locus primers (5′-TCCCTCTACTTTTTCTTCTGAC-3′ and 5′-CGGAACGCAAATATCGCAC-3′) and a primer that recognized sequences unique to the targeted allele (5′-CTAGTGAGACGTGCTACTTC-3′). Amplification conditions were similar to those used for the SSLP analysis except that the primers were used at a concentration of 20 μM and 2.5 U of Taq was used. The DNAs were amplified for 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. After a 10-min extension at 72°C, the reactions were cooled to 4°C and the products were analyzed by agarose gel electrophoresis. p19Arf exon 1β sequences were amplified using the primers 5′-CGTGGAGCAAAGATGGGC-3′ and 5′-CCGTCCTGCTTCTACCTCG-3′; exon 2 sequences were amplified using the primers 5′-ACATAGGGCTTCTTTCTTGGGTCC-3′ and 5′-GGACCAACTATGCTCACCTGGGC-3′. DNAs were amplified in reactions containing 200 ng of DNA, 200 μM each deoxynucleoside triphosphate, 25 μM primers, 1.25 U of Taq, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 0.001% gelatin. For exon 1β, the samples were amplified for 29 cycles of 1 min at 94°C, 1.5 min at 60°C, and 1.5 min at 72°C, followed by a 10-min extension at 72°C. The same conditions were used for exon 2 except that the annealing temperature was 63°C. After the final cycle, the reactions were cooled to 4°C. The PCR products were cloned into the TOPO cloning vector (Invitrogen) and sequenced on an ABI 373-Stretch machine (Perkin-Elmer) at the DNA Facility, Department of Physiology, Tufts University. The sequence of exon 1β and exon 2 of the Ink4a/Arf locus in Cast/Ei mice was determined by amplifying these sequences from liver DNA.
Protein analysis.
Cells were lysed in a buffer containing 1% NP-40, 50 mM Tris-HCl (pH 8.0), 0.1 mM NaF, 0.1 mM sodium vanadate, 100 μM phenylmethylsulfonyl fluoride, NaCl (2 μg/ml), and leupeptin (1 μg/ml). The proteins were resolved by electrophoresis through SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). The blots were probed with anti-p19Arf (34) and anti-p16Ink4a and anti-cdk4 (SC1207 and SC260, respectively; Santa Cruz Biotechnologies) antibodies and developed using a chemiluminescence kit (Tropix) according to the manufacturer's instructions.
RESULTS
Several transformants show loss of proviral markers on chromosome 4.
Endogenous nonecotropic proviruses were used as genetic markers to assess the chromosomal constitution of Ab-MLV-transformed pre-B-cell lines. The CXXB panel, derived from (BALB/Ann.xid × C57BL10)F1 mice, and the CXCB panel, from (BALB/cByJ × CBA/Tufts)F1 mice, were screened with oligonucleotide probes that detect the three different classes of endogenous nonecotropic proviruses (49). These crosses provide at least one informative polymorphic marker on all mouse chromosomes except 6 and 17 (Table 1). In both cell panels, chromosomes 6 and 17 carry a single nonpolymorphic provirus, and 33 other nonpolymorphic proviruses are present elsewhere in the genome. Although loss of a single copy of a nonpolymorphic provirus is revealed by a decreased signal, subtle differences in the hybridization of many of the proviruses make it difficult to evaluate the signal intensity accurately. Thus, while no clear cases of loss of one copy of a nonpolymorphic provirus were identified, analyses of the nonpolymorphic proviruses were not included when LOH was assessed.
TABLE 1.
Loss of endogenous proviral markers
| Chromosome | CXXBa
|
CXCBb
|
||
|---|---|---|---|---|
| No. of markersb | Fraction of cell lines with marker loss | No. of markersb | Fraction of cell lines with marker loss | |
| 1 | 3 | 0 | 3 | 0 |
| 2 | 3 | 0 | 1 | 0 |
| 3 | 1 | 0 | 0 | 0 |
| 4 | 3 | 1/27 (Mpmv19) | 5 | 2/16 (Xmv8, Xmv9, Xmv14, Xmv44 |
| 1/27 (Pmv23) | ||||
| 5 | 5 | 0 | 5 | 0 |
| 6 | 0 | 0 | 0 | |
| 7 | 6 | 16/27 (Pmv31) | 5 | 0 |
| 8 | 2 | 0 | 2 | 0 |
| 9 | 1 | 0 | 1 | 0 |
| 10 | 3 | 16/27 (Xmv31) | 2 | 0 |
| 11 | 6 | 27/27 (Mpmv18) | 3 | 1/16 (Xmv20) |
| 12 | 4 | 1/27 (Pmv37) | 0 | |
| 1/27 (Pmv27) | ||||
| 13 | 2 | 0 | 0 | |
| 14 | 2 | 1/27 (Xmv19) | 1 | 0 |
| 15 | 1 | 0 | 0 | |
| 16 | 2 | 0 | 0 | |
| 17 | 0 | 0 | ||
| 18 | 0 | 0 | 1 | 0 |
| 19 | 1 | 0 | 0 | |
| X | 0 | 0 | 1 | 0 |
| Y | 3 | 1/15 (Xmv11) | 1 | 0 |
| 1/15 (Xmv40) | ||||
Twenty-seven cell lines were analyzed for Xmv, Pmv, and Mpmv proviruses on chromosomes 1 to 19; proviruses on the Y chromosome were analyzed in the 15 CXXB cell lines that were derived from male mice. Markers are given in parentheses; only informative, polymorphic proviruses are listed.
Sixteen cell lines were analyzed for Xmv and Mpmv proviruses, and 13 were analyzed for Pmv proviruses. All but two of these cell lines were derived from female mice.
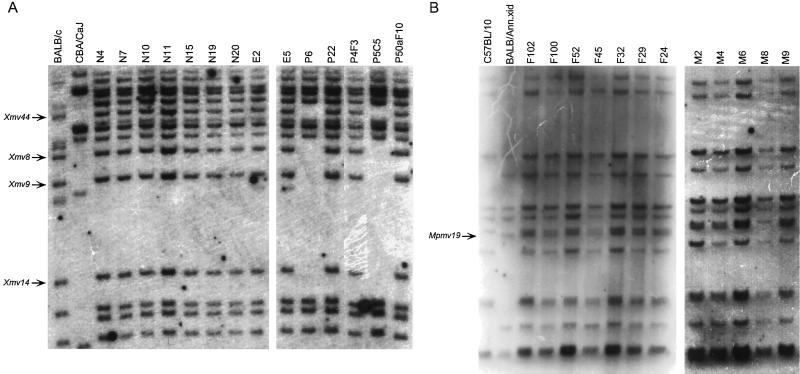
Analysis of polymorphic proviruses revealed that 7 of 58 cell lines had lost proviral markers present on chromosome 4. Two of sixteen CXCB cell lines, P6 and P5C5, had lost the four polymorphic chromosome 4 proviruses that are inherited from the BALB/c parent, Xmv8, Xmv9, Xmv14, and Xmv44 (Fig. 1A). Loss of chromosome 4 markers was also observed in the CXXB panel (data not shown); M8 was missing Mpmv19, the only polymorphic chromosome 4 marker inherited from the BALB/c parent in this cross (Fig. 1B), and F18 was missing Pmv23, a marker inherited from the B10 parent (data not shown). Similarly, 3 of 14 cell lines from the CXCE panel [derived from BALB/cByJ × Cast/Ei)F1 mice], 511-5, 511-13, and 511-32, had lost Xmv8, Xmv9, and Xmv44, the three informative chromosome 4 proviruses inherited from the BALB/c parent (data not shown). The presence of the fourth polymorphic chromosome 4 Xmv provirus lost in the CXCE cross could not be scored in these cells because proviral bands from the Cast/Ei parent comigrate with these fragments in both EcoRI- and PvuII-digested DNAs. Additional proviral analysis was not done on this panel because the Cast/Ei proviruses have not been mapped.
FIG. 1.
Loss of chromosome 4 proviruses. DNAs from CXCB cells, digested with EcoRI (A), and those from CXXB cells, digested with PvuII (B), were fractionated through agarose gels. The dried gels were probed with JS6/10 (A) or JS4 (B) (49) and exposed to XAR film at −70°C for 2 weeks. The fragments corresponding to Xmv8, Xmv9, Xmv14, and Xmv44 (A) and Mpmv19 (B) are indicated by the arrows. Control DNAs were prepared from BALB/c, CBA/CaJ, and C57BL/10 liver.
In contrast to the recurrent pattern of chromosome 4 sequence loss, most other proviral markers were retained by the cells. However, all of the CXXB cell lines had lost Mpmv18, the single BALB/c-derived polymorphic provirus found on chromosome 11. This provirus was retained by all of the CXCB cell lines (Table 1). These data suggest that deletions in the immediate vicinity of Mpmv18 are not common in all transformants. Only one other cell line, P29, had lost a provirus mapping to chromosome 11. This cell line was missing Xmv20, inherited from the CBA parent which maps about 44 centimorgans (cM) distal to Mpmv18. Over half of the CXXB cell lines lost both Xmv31 and Pmv31. These BALB/c-inherited proviruses are located on chromosomes 10 and 7, respectively. However, neither of these proviruses nor any of the other 12 proviruses located on these chromosomes were lost in any of the other cell lines. Individual CXXB cell lines also lost proviruses found on chromosomes 12 and 14 and the Y chromosome. Because these patterns of loss were not found in both cell panels or occurred only once, loss of regions in the immediate vicinity of these proviruses is not required for transformation of cells from all strains of mice.
SSLP analysis reveals loss of all or most of one copy of chromosome 4 in some transformants.
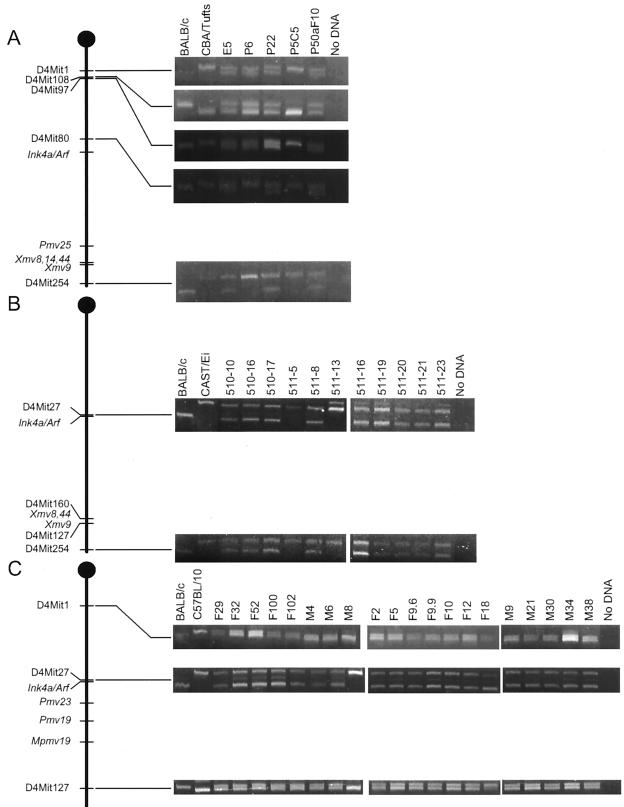
To obtain a clearer picture of types of deletions involved in the loss of the endogenous proviral markers, primers that detect SSLPs located near the different proviruses were used to analyze DNAs from the cell panels (10). None of the cell lines that retained all of the proviral markers found on chromosome 4 showed evidence of deletion. However, six of seven cell lines that had lost chromosome 4 proviral markers had lost SSLP markers mapping to the region occupied by the proviral markers. P5C5, 511-5, 511-13, 511-32, and M8 cells, which had lost proviral markers from the BALB/c-inherited copy, had also lost all of the SSLP markers tested from this copy of chromosome 4 (Fig. 2), suggesting that this chromosome was probably completely deleted. P6 cells retained three SSLP markers located on proximal chromosome 4 but had lost two others which map to the central and distal regions (Fig. 2), demonstrating that a minimum of 16 cM and a maximum of 31 cM of chromosome 4 sequences are retained. F18 cells retained all 15 SSLP markers tested. When considered with information from the proviral mapping, these data suggest that F18 cells may have lost a maximum of about 2 cM from the central portion of the chromosome. No evidence of marker loss affecting the second copy of chromosome 4 was observed in any of these cells. Interestingly, loss of markers affecting large regions of this chromosome, including those involved in the Ab-MLV transformants, was also observed in studies of Mo-MLV- and radiation-induced thymomas (8, 25, 30, 32).
FIG. 2.
Loss of chromosome 4 markers in CXCE, CXCB, and CXXB cells. DNAs from CXXB, CXCE, and CXCB cells were amplified with the primers indicated; the products were fractionated on agarose gels and visualized by ethidium bromide staining. Controls included DNAs from BALB/c, CBA, Cast/Ei, and C57Bl/10 liver and reactions that did not contain DNA. Representative analyses are shown on the right; the drawing on the left depicts the relative locations of all informative markers tested. Marker location is based on information in the MIT Center for Genome Research and Mouse Genome databases (http://www.genome.wi.mit.edu and http://www.jax.org).
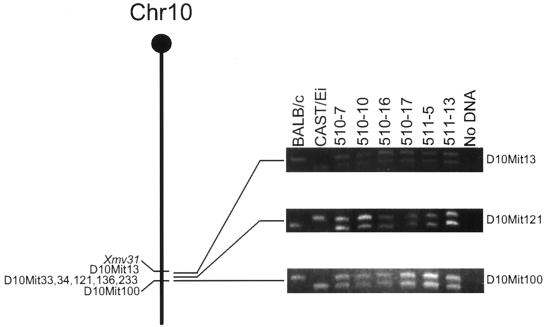
In contrast to the pattern obtained with chromosome 4, analysis of different markers located in the vicinity of Xmv31 on chromosome 10, including the representatives shown in Fig. 3, revealed that none of the seven markers analyzed in the CXCE panel were deleted. In addition, analysis exploiting a restriction fragment length polymorphism (RFLP) revealed that these cells retain both copies of the Mdm2 gene which maps about 6 cM distal to Xmv31 (data not shown). These data, and those from the proviral mapping of the CXCB panel, reinforce the idea that loss of sequences in the vicinity of Xmv31 is not a general feature of the transformants. In a similar vein, although analyses of the CXXB cell lines that had lost Pmv31 revealed that they were missing D7Mit62, an SSLP marker which maps proximal to the provirus, none of the cell lines in the CXCB or CXCE panels were missing this marker, or a second marker, D7Mit211, which maps distal to Pmv23. Eleven cell lines from the CXXB panel were examined with four SSLP markers within 6 cM proximal and distal of Mpmv18, the BALB/c-derived chromosome 11 provirus lost in these cells. Among this group, 2 of 11 cell lines were missing D11Mit162, D11Mit80, and D11Mit152 but retained D11Mit83, a marker approximately 6 cM distal to the provirus. This last marker was retained in all CXXB cell lines (data not shown).
FIG. 3.
SSLP markers are retained on chromosome 10. DNAs from CXCE cell lines were amplified with primer D10Mit121 (A) or D10Mit100 (B). The products were fractionated on an agarose gel and visualized by ethidium bromide staining. Controls included DNAs from BALB/c, CBA, Cast/Ei, and C57Bl/10 liver and reactions that did not contain DNA. Representative analyses are shown on the right; the drawing on the left depicts the relative locations of all informative markers tested. Marker location is based on information in the MIT Center for Genome Research and Mouse Genome databases (http://www.genome.wi.mit.edu and http://www.jax.org).
Loss of Ink4a/Arf expression in the Ab-MLV transformants.
The p19Arf protein, encoded by the Ink4a/Arf locus, is a tumor suppressor already known to play an important role in the Ab-MLV transformation process (37). The Ink4a/Arf locus is located on chromosome 4, approximately 42 cM from the centromere in the same region as Pmv23 (Fig. 2) (36), and sequences lost in six of the seven cell lines missing chromosome 4 markers should include the locus. Although the F18 cell line is missing Pmv23, two SSLP markers which map between Pmv23 and the Ink4a/Arf locus are retained in this cell line (data not shown). Two patterns of Ink4a/Arf expression have been identified in Ab-MLV-transformed pre-B cells. Transformants which express wild-type p53 have very low to undetectable levels of Ink4a/Arf locus products; consistent with the negative effect of p53 on the locus (38, 48), transformants which have acquired p53 mutations usually express abundant p16Ink4a and p19Arf (37).
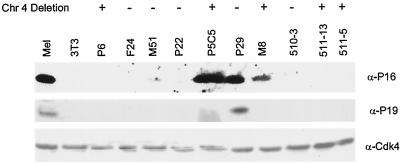
To determine if the pattern of Ink4a/Arf expression could be correlated to LOH involving chromosome 4, the presence of p16Ink4a and p19Arf was analyzed by using Western blotting with antibodies directed against p16Ink4a and p19Arf. Because expression of these proteins usually correlates with p53 status, the cells were screened for sensitivity of γ-irradiation-induced apoptosis, a response that depends on the presence of wild-type p53 in Ab-MLV-transformed pre-B cells (37, 52). Transformants from all of the cell panels, including those which displayed LOH involving chromosome 4, as well as lysates from NIH 3T3 cells, which do not express these proteins, and the erythroleukemia cell line MEL, which expresses readily detectable Ink4a/Arf locus products (36), were examined. Consistent with earlier studies, cell lines which expressed mutant forms of p53, as represented by the P29 cell line, expressed readily detectable Ink4a/Arf locus products (Fig. 4). All of the cell lines which had lost chromosome 4 sequences expected to include the Ink4a/Arf locus failed to express p19Arf, and all of these expressed wild-type p53 (data not shown). However, two of these cell lines, P5C5 and M8, expressed p16Ink4a. Expression of p16Ink4a in the absence of p19Arf has been observed in about 5% of Ab-MLV-transformed pre-B-cell lines (37).
FIG. 4.
Expression of p16Ink4a and p19Arf in representative transformants. Cell lysates were fractionated through SDS-polyacrylamide gels, and the proteins were transferred to a membrane which was probed with anti-p16Ink4a, anti-p19Arf, and anti-Cdk4 antibodies. NIH 3T3 cells, which do not express Ink4a/Arf locus products, and MEL cells, which express readily detectable p16Ink4a and p19Arf, were used as controls. Samples were from cells in which chromosome (Chr) 4 sequences have been lost (+) and from cells that do not have a detectable chromosome 4 deletion (−). The P29 cell line expresses mutant p53; all of the other pre-B-cell transformants shown express wild-type p53. The pattern of Ink4a/Arf product expression observed in 511-32, another cell line missing chromosome 4 sequences, was identical to that observed in 511-13 and 511-5 (data not shown).
At least one copy of the Ink4a/Arf locus is retained in all CXCE cell lines.
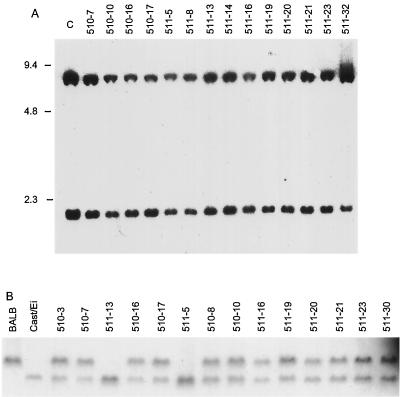
The protein analysis revealed that many of the cell lines, including those which had lost chromosome 4 sequences, failed to express Ink4a/Arf locus products. This phenotype is common in Ab-MLV-transformed cells and could suggest that these sequences have been deleted from both copies of chromosome 4 in the transformants. Small deletions could easily have escaped detection in the mapping study because the markers closest to the locus are still several centimorgans proximal and distal, respectively. Homozygous deletion of Ink4a/Arf has been observed in a number of human tumors (22, 31, 33). To examine this possibility, DNAs from the CXCE panel were examined by Southern blotting (Fig. 5A). All of the cell lines retained at least one copy of the locus, suggesting that other mechanisms are responsible for the pattern of expression.
FIG. 5.
The CXCE cell lines retain at least one copy of the Ink4a/Arf locus. (A) DNAs were digested with PstI, fractionated through an agarose gel, and analyzed by Southern blotting with a full-length p16Ink4a cDNA probe (36). C, control DNA prepared from Cast/Ei liver. (B) DNAs were digested with HindIII, fractionated through an agarose gel, and analyzed by Southern blotting with a p16Ink4a exon 1α probe.
The digestion strategy illustrated in Fig. 5A does not effectively monitor LOH. To determine if cell lines in addition to those detected in the mapping screen displayed LOH involving these sequences, DNAs from BALB/cJ and Cast/Ei mice and their F1 progeny were screened with several restriction enzymes for polymorphisms that distinguished the alleles inherited from each parent. Digestion with HindIII revealed the presence of two easily separable fragments that could be detected with a p16Ink4a exon 1α probe (Fig. 5B), and digestion with PstI revealed an RFLP that could be detected with a p19Arf exon 1β probe (data not shown). Analysis of the CXCE panel revealed that each of the cell lines which had lost chromosome 4 sequences had lost the BALB/c-derived copies of both p16Ink4a and p19Arf. None of the other cell lines from this panel had lost these sequences. These data suggest that LOH involving Ink4a/Arf sequences occurs in about 10% of the transformants. However, because about 50% of all transformants down-modulate expression of the Ink4a/Arf locus (37), deletion does not appear to be the major pathway by which expression of these sequences is controlled.
In many instances, loss of one copy of a tumor suppressor gene is accompanied by mutation of the second copy. To determine if mutations affecting p19Arf sequences were common in transformants retaining a single copy of these sequences, PCR was used to amplify exon 1β and exon 2 sequences from 511-5, 511-13, and 511-32, three of the CXCE cell lines that retained the Cast/Ei copy of the Ink4a/Arf locus. The sequences of exon 1β and exon 2 were also amplified from Cast/Ei liver DNA. Comparison of the Cast/Ei exon 1β sequence to that of DBA mice, considered to represent the wild-type sequence of laboratory mouse strains (62), revealed two nucleotide substitutions in noncoding sequence. One of these, G39C, was in the leader region of the mRNA; the second was in the intron 3′ of exon 1β. Analysis of exon 2 sequences revealed the presence of two nucleotide substitutions within the coding region of exon 2 in the Cast/Ei gene. One of these, G292C, does not affect the sequence of p16Ink4a but results in substitution of a glutamine for a glutamic acid in p19Arf. The second, C487G, results in the substitution of a tryptophan for a cysteine in p19Arf and the substitution of a valine for a leucine in p16Ink4a. The sequences obtained from all three cell lines were identical to those obtained from the Cast/Ei liver DNA. These data indicate that mutations which affect the coding sequence of p19Arf do not commonly account for the absence of protein expression.
Loss of a single copy of the Ink4a/Arf locus predisposes to Ab-MLV transformation.
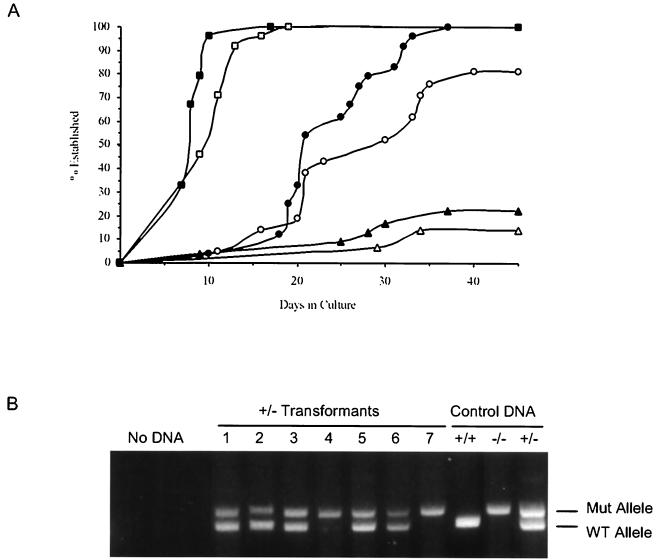
Analyses of the transformed cell lines suggest that the presence of a single copy of the Ink4a/Arf locus could be sufficient to influence the transformation process. Complete loss of the locus allows primary transformants to bypass the apoptotic crisis characteristic of Ab-MLV-induced pre-B-cell transformation (37). To determine if loss of one copy of the locus affects transformation, bone marrow from Ink4a/Arf −/−, +/−, and +/+ littermates was infected with Ab-MLV and plated in soft agar (41). When primary transformants were scored 10 days later, consistent differences in colony frequency that correlated with genotype were not observed (data not shown). Primary transformants represent the first stage in the Ab-MLV transformation process (37, 54). Expansion in liquid medium, the second stage in the process, is marked by an apoptotic crisis; only a fraction of primary transformants from normal mice survive this phase and become fully established cell lines. As expected (37), when primary transformants from Ink4a/Arf null animals were plated in liquid medium, all of them established rapidly; only 15 to 20% of those from +/+ litter mates became established. This pattern is similar to that obtained with primary transformants derived from other strains of normal mice (54). Primary transformants derived from Ink4a/Arf +/− mice displayed an intermediate pattern. All 24 primary transformants became established in one experiment, and 21 of 24 were established in a second experiment. In addition, the apoptotic crisis characteristic of the transformation process and the time required for the cells to become established was less than that required for cells derived from normal mice. Analyses of 16 established transformants derived from heterozygous animals revealed that eight, including the representatives shown (Fig. 6B), still retained the single, nontargeted copy of the locus. These data demonstrate that heterozygosity at the Ink4a/Arf locus confers a selective advantage to Ab-MLV transformants.
FIG. 6.
Loss of a single copy of the Ink4a/Arf locus confers a selective advantage to Ab-MLV transformants. (A) Primary transformants from Ink4a/Arf −/− (squares), +/− (circles), and +/+ (triangles) mice were plated in liquid medium, and their ability to develop into established transformants was monitored. A transformant is considered established when levels of apoptosis are less than 10% and the cells can be subcultured at regular intervals (37, 54). Each point represents the frequency of primary transformants that were established at the day shown. Filled and open symbols illustrate results obtained with two different litters. (B) DNAs from established transformants derived from Ink4a/Arf +/− mice were amplified by PCR to detect the presence of the wild-type (WT) and mutant (Mut) alleles. Control DNAs were prepared from mice from our Ink4a/Arf colony.
DISCUSSION
Our analysis of chromosomal markers in three panels of Ab-MLV-transformed pre-B cells suggests that loss of sequences on chromosome 4 is an important but not obligate step in the transformation process. Deletions affecting this chromosome were the only consistent feature detected in the 58 cell lines studied. The cell panels provided at least one polymorphic proviral marker on all chromosomes except 6 and 17, and the majority of cell lines retained most of the markers. Although our survey did not screen a large number of markers on all chromosomes, LOH does not appear to be a common feature of Ab-MLV transformation. A similar conclusion was reached in a study of Mo-MLV-induced thymomas (25). A number of other mouse tumors, including hepatocellular carcinomas (29), lung carcinomas (18), insulinomas (11), and chemically induced thymomas (63), also show similar low frequencies of allelic loss at different chromosomal sites. However, radiation-induced lymphomas display much higher frequencies of allelic loss (8, 30, 32, 47), suggesting that both the tumor type and the way in which the tumor is induced affect LOH.
Other investigators have examined Ab-MLV transformants and tumor cells for the presence of chromosomal abnormalities that can be correlated with the development of a fully malignant phenotype (1, 7, 24). One study noted a deletion affecting a minimum of about 10 cM involving the region 15 and 25 cM from the centromere of chromosome 13 in three tumorigenic clones (7). This region is proximal to Xmv13 and Pmv9, the two chromosome 13 proviral markers analyzed in the CXXB panel, and neither of these proviruses was deleted in any of the cell lines. Indeed, the region lost in two of the three clones (7) should include the Xmv13 provirus. Other analyses of Ab-MLV transformants, Ab-MLV-induced thymic tumors, and BCR/ABL-induced murine tumors have noted the presence of trisomy 5, X-chromosome breaks, and amplification of sequences on chromosomes 12, 14, and 17 (1, 7, 57). Although the proviral mapping technique used here is not designed to readily detect amplified sequences, none of the proviral fragments appeared to be dramatically overrepresented, suggesting that amplifications may not be a prominent feature of in vitro-derived transformants. Indeed, in our study and those of others (1, 7, 24), the pattern that emerges is consistent with the idea that many of the transformants retain a diploid karyotype.
Cells in the CXXB panel have lost several proviral markers that are not affected in the CXCB and CXCE cell lines. For example, even though about 60% of the CXXB cells were missing Xmv31, a chromosome 10 provirus, no chromosome 10 losses were observed in the 30 CXCB and CXCE cell lines analyzed. In addition, SSLP mapping failed to reveal evidence of marker loss in the vicinity of Xmv31 in the CXCE cell lines. Similarly, even though all of the CXXB cell lines had lost Mpmv18, this provirus was not lost in the CXCB panel. All of the proviral markers affected in the CXXB cross except those mapping to chromosome 4 and the Xmv14 provirus on chromosome 14 are inherited from the BALB/Ann.xid parent. Perhaps these losses reflect segregation of these proviruses in the BALB/Ann.xid background. Unfortunately, DNA from parents used to generate the F1 mice from which the CXXB panel was derived is not available. Thus, even though the proviruses affected in the cell lines are retained by BALB/Ann.xid mice (data not shown), the status of these loci in the particular female mouse used in the CXXB mating from which the cell lines were derived cannot be determined. While changes that are absolutely required for transformation should be present in samples from independent sources, changes unique to cell lines from a particular cross could reflect effects of the genetic background of the strains. Further analyses would be required to determine if different sets of genes are activated or inactivated in particular strain combinations.
Chromosome 4 has been identified as a target of LOH in several murine lymphoma models (8, 25, 30, 32). However, because the deletions are very large in all these cases, implicating a particular gene has not been possible. In a similar vein, most of the deletions identified here encompass very large amounts of chromosome 4. An exception is the F18 cell line. However, this deletion is based on the loss of a single proviral marker and may not reflect a deletion of other chromosomal sequence; loss of proviral sequences via homologous long terminal repeat-based recombination has been observed at a low frequency (43, 50, 55), raising the possibility that cellular sequences may not be missing. SSLP mapping data failed to reveal additional deletions and demonstrated that sequences lying between Pmv31 and the Ink4a/Arf locus are retained in F18 cells. Nonetheless, most of the chromosome 4 deletions in all the lymphoma systems suggest that loss of sequences in the vicinity of the Ink4a/Arf locus (8, 30, 32) and loss of the p73 gene which maps to distal chromosome 4 may be important (20).
Although p73 expression and function have not been studied in Ab-MLV-transformed cells, the Ink4a/Arf locus is known to be important in Ab-MLV transformation (37). p19Arf can trigger apoptosis in Ab-MLV transformants that retain wild-type p53, and Ink4a/Arf locus deletion allows primary transformants to bypass the apoptotic crisis characteristic of the transformation process (37). Indeed, one pathway leading to full transformation involves down-regulation of p19Arf expression. Analyses of transformation using cells from Ink4a/Arf +/− mice, coupled with the mapping studies, suggest that LOH involving the Ink4a/Arf locus can facilitate this process. Accelerated tumor development has also been observed in Eμ-Myc transgenic mice that are hemizygous for the Arf locus (12), raising the possibility that the Arf locus may be regulated in this fashion in a variety of tumor systems.
LOH can reveal the presence of recessive mutations affecting tumor suppressor genes. However, sequence analyses of p19Arf sequences in three of the cell lines that retain a single copy of the locus failed to reveal mutations. In addition, analysis of seven transformants derived from Msh2 null mice and from two transformants derived from p53 null mice revealed that p19Arf sequences are not mutated in these cells, even though the transformants from Msh2 null animals are unable to mediate DNA mismatch repair and display a high frequency of p53 mutations (54; J. Jenab-Wolcott and N. Rosenberg, unpublished data). Thus, mutations affecting coding sequence do not appear to be a common mechanism by which p19Arf expression and function is modulated in Ab-MLV transformants.
Cells which must inactivate only a single copy of p19Arf display a selective growth advantage and have a higher probability of becoming dominant within a population. Neither mutation nor large deletions which remove both copies of the locus appear to be the principal way by which this is achieved in the Ab-MLV system, and DNase sensitivity experiments suggest that differences in chromatin structure are not involved (A. Halgren and N. Rosenberg, unpublished data). Methylation of sequences upstream of the gene could be important; methylation correlates with decreased expression of Ink4a, Arf, or the closely linked p15Ink4b gene in some cases (2, 19, 21, 28, 31, 33, 38). Because all stages in the transformation process can be followed closely in the Ab-MLV model, the way in which altered expression of p19Arf is orchestrated in these cells and the possible role played by de novo methylation can be readily assessed. In addition, because p19Arf expression is altered in a wide range of tumors, such analyses are likely to shed light on a general mechanism of oncogenesis.
ACKNOWLEDGMENTS
This work represents equal contributions of the first two authors.
We are grateful to Peter Brodeur for supplying the CXXB and CXCB DNAs and to John Coffin, Jonathan Stoye, and Wayne Frankel for assistance with the proviral mapping and for useful discussions.
This work was supported by grant CA 33771 from the NIH.
REFERENCES
- 1.Aubert D, Heuze F, Diez E, Jotereau F, Berger R. Cytogenetics of Abelson virus-induced malignant lymphoma cell lines in the mouse. Cancer Genet Cytogenet. 1987;28:119–125. doi: 10.1016/0165-4608(87)90361-x. [DOI] [PubMed] [Google Scholar]
- 2.Balova A, Diccianni M B, Yu J C, Nobori T, Link M P, Pullen J, Yu A L. Frequent and selective methylation of p15 and deletion of both p15 and p16 in T-cell acute lymphoblastic leukemia. Cancer Res. 1997;57:832–836. [PubMed] [Google Scholar]
- 3.Baxter E W, Blyth K, Donehower L A, Cameron E R, Onions D E, Neil J C. Moloney murine leukemia virus-induced lymphomas in p53-deficient mice: overlapping pathways in tumor development? J Virol. 1996;70:2095–2100. doi: 10.1128/jvi.70.4.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-David Y, Bernstein A. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron D, Houde J, Polquin L, Barbeau B, Rassart E. Analysis of proviruses integrated in the Fli-1 and Evi-1 regions in Cas-Br-E MuLV-induced non-T, non B-cell leukemias. Virology. 1993;191:661–669. doi: 10.1016/0042-6822(92)90241-g. [DOI] [PubMed] [Google Scholar]
- 6.Cawthon R M, Anderson L B, Buchberg A M, Xu G, O'Connell P, Viskochil D, Weiss R B, Wallace M R, Marchuk D A, Culver M, Stevens J, Jenkins N A, Copeland N G, Collins F S, White R. cDNA sequences and genomic structure of EVI-2B, a gene lying within an intron of the neurofibromatosis type 1 gene. Genomics. 1991;9:446–460. doi: 10.1016/0888-7543(91)90410-g. [DOI] [PubMed] [Google Scholar]
- 7.Clark S S, Liang Y, Reedstrom C K, Wu S Q. Nonrandom cytogenetic changes accompany malignant progression in clonal lines of Abelson virus-infected lymphocytes. Blood. 1994;84:4301–4309. [PubMed] [Google Scholar]
- 8.Cleary H J, Wright E, Plumb M. Specificity of loss of heterozygosity in radiation-induced mouse myeloid and lymphoid leukemias. Int J Radiat Biol. 1999;75:1223–1230. doi: 10.1080/095530099139377. [DOI] [PubMed] [Google Scholar]
- 9.Cool M, Jolicoeur P. Elevated frequency of loss of heterozygosity in mammary tumors arising in mouse mammary tumor virus/neu transgenic mice. Cancer Res. 1999;59:2438–2444. [PubMed] [Google Scholar]
- 10.Dietrich W F, Miller J, Steen R, Mercahnt M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O'Connor T J, Evans C A, DeAngelis M M, Levinson D M, Kruglyak L, Goodman N, Copeland N G, Jenkins N A, Hawkins T L, Stein L, Page D C, Lander E S. A comprehensive genetic map of the mouse genome. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich W F, Randy E H, Smith J S, Bishop J M, Hanahan D, Lander E S. Genome-wide search for loss of heterozygosity in transgenic mouse tumors reveals candidate tumor suppressor genes on chromosomes 9 and 16. Proc Natl Acad Sci USA. 1994;91:9451–9455. doi: 10.1073/pnas.91.20.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman A, Rosenberg N. Temperature-sensitive mutants of Abelson murine leukemia virus deficient in protein tyrosine kinase activity. J Virol. 1990;64:4242–4251. doi: 10.1128/jvi.64.9.4242-4251.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearon E R, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 15.Frankel W N, Stoye J P, Taylor B A, Coffin J M. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green P L, Kaehler D A, Bennett L M, Risser R. Multiple steps are required for the induction of tumors by Abelson murine leukemia virus. J Virol. 1989;63:1989–1994. doi: 10.1128/jvi.63.5.1989-1994.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green P L, Kaehler D A, Risser R. Clonal dominance and progression in Abelson murine leukemia virus lymphomagenesis. J Virol. 1987;61:2192–2197. doi: 10.1128/jvi.61.7.2192-2197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegi M E, Devereux T R, Dietrich W F, Cochran C J, Lander E S, Foley J F, Maronpot R R, Anderson M W, Wiseman R W. Allelotype analysis of mouse lung carcinomas reveal frequent allelic losses on chromosome 4 and an association between allelic imbalances on chromosome 6 and K-ras activation. Cancer Res. 1994;54:6257–6264. [PubMed] [Google Scholar]
- 19.Herman J G, Civin C I, Issa J-P J, Collector M I, Sharkis S J, Baylin S B. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 20.Herranz M, Santos J, Salido E, Fernandez-Piqueras J, Serrano M. Mouse p73 gene maps to the distal part of chromosome 4 and might be involved in the progression of γ-irradiation-induced T-cell lymphomas. Cancer Res. 1999;59:2068–2071. [PubMed] [Google Scholar]
- 21.Hirama T, Koeffler H P. Role of the cyclin-dependent kinase inhibitors in the development of cancer. Blood. 1995;86:841–854. [PubMed] [Google Scholar]
- 22.Jen J, Harper W, Bigner S H, Bigner D D, Papadopoulas N, Markowitz S, Willson J K V, Kinzler K W, Vogelstein B. Detection of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6355–6358. [PubMed] [Google Scholar]
- 23.Kinzler K W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 24.Klein G, Ohno S, Rosenberg N, Wiener F, Spira J, Baltimore D. Cytogenetic studies on Abelson-virus-induced mouse leukemias. Int J Cancer. 1980;25:805–811. doi: 10.1002/ijc.2910250617. [DOI] [PubMed] [Google Scholar]
- 25.Lander J K, Fan H. Low-frequency loss of heterozygosity in Moloney murine leukemia virus-induced tumors in BRAKF1/J mice. J Virol. 1997;71:3940–3952. doi: 10.1128/jvi.71.5.3940-3952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Mainville C A, Sheehan K M, Klaman L D, Giorgetti C A, Press J L, Brodeur P H. Deletional mapping of fifteen mouse VH gene families reveals a common organization for three Igh haplotypes. J Immunol. 1996;156:1038–1046. [PubMed] [Google Scholar]
- 28.Malumbres M, Perez de Castro I, Santos J, Melendez B, Mangues R, Serrano M, Pellicer A, Fernandez-Piqueras J. Inactivation of the cyclin-dependent kinase inhibitor p15INK4b by deletion and de novo methylation with independence of p16INK4a alterations in murine primary T-cell lymphomas. Oncogene. 1997;14:1361–1370. doi: 10.1038/sj.onc.1200969. [DOI] [PubMed] [Google Scholar]
- 29.Manenti G, DeGregorio L, Gariboldi M, Dragani T A, Pierotti M A. Analysis of loss of heterozygosity in murine hepatocellular tumors. Mol Carcinog. 1995;13:191–200. doi: 10.1002/mc.2940130309. [DOI] [PubMed] [Google Scholar]
- 30.Melendez B, Santos J, Fernandez-Piqueras J. Loss of heterozygosity at the proximal-mid part of mouse chromosome 4 defines two novel tumor suppressor gene loci in T-cell lymphomas. Oncogene. 1999;18:4166–4169. doi: 10.1038/sj.onc.1202826. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa S, Hirano N, Sato N, Takahashi T, Hangaishi A, Tanaka K, Kurokawa M, Tanaka T, Mitani K, Yazaki Y, Hirai H. Homozygous loss of cyclin-dependent kinase 4-inhibitor (p16) gene in human leukemias. Blood. 1994;84:2431–2435. [PubMed] [Google Scholar]
- 32.Okumoto M, Park Y G, Song C W, Mori N. Frequent loss of heterozygosity on chromosomes 4, 12, and 19 in radiation-induced lymphomas in mice. Cancer Lett. 1999;135:223–228. doi: 10.1016/s0304-3835(98)00305-x. [DOI] [PubMed] [Google Scholar]
- 33.Pinyol M, Hernandez L, Cazorla M, Balbin M, Jares P, Fernandez P L, Montserrat E, Cardesa A, Lopez-Otin C, Campo E. Deletions and loss of expression of P16INK4a and P21Waf1 genes are associated with aggressive variants of mantle cell lymphomas. Blood. 1997;89:272–280. [PubMed] [Google Scholar]
- 34.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 35.Prives C. Signaling to p53: breaking the MDM2p-53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 36.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter K H, Walker C, Beach D, Sherr C J, Serrano M. Cloning and characterization of murine p16INK4a and p15INK4b. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 37.Radfar A, Unnikrishnan I, Lee H-W, DePinho R A, Rosenberg N. P19Arf induces p53-dependent apoptosis during Abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg N. abl oncogenes. Semin Virol. 1991;2:365–374. [Google Scholar]
- 40.Rosenberg N. abl-mediated transformation, immunoglobulin gene rearrangements and arrest of B lymphocyte differentiation. Semin Cancer Biol. 1994;5:95–102. [PubMed] [Google Scholar]
- 41.Rosenberg N, Baltimore D. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J Exp Med. 1976;143:1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg N, Jolicoeur P. Retrovirus pathogenesis. In: Coffin J M, Hughes S, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–586. [PubMed] [Google Scholar]
- 43.Seperack P K, Strobel M C, Corrow D J, Jenkins N A, Copeland N G. Somatic and germ-line reverse mutations rates of the retrovirus-induced dilute coat-color mutation of DBA mice. Proc Natl Acad Sci USA. 1988;85:189–192. doi: 10.1073/pnas.85.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 45.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 46.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2981. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 47.Shinbo T, Matsuki A, Matsumoto Y, Kosugi S, Takahashi Y, Niwa O, Kominami R. Allelic loss mapping and physical delineation of a region harboring a putative thymic lymphoma suppressor gene on mouse chromosome 12. Oncogene. 1999;18:4131–4136. doi: 10.1038/sj.onc.1202767. [DOI] [PubMed] [Google Scholar]
- 48.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoye J P, Coffin J M. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J Virol. 1988;62:168–175. doi: 10.1128/jvi.62.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoye J P, Fenner S, Greenoak G E, Moran C, Coffin J M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 51.Stoye J P, Frankel W N, Coffin J M. DNA hybridization in dried gels with fragmented probes: an improvement over blotting techniques. Technique. 1991;3:123–128. [Google Scholar]
- 52.Thome K, Radfar A, Rosenberg N. Mutation of Tp53 contributes to the malignant phenotype of Abelson virus-transformed lymphoid cells. J Virol. 1997;77:8149–8156. doi: 10.1128/jvi.71.11.8149-8156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulrich E, Boehmelt G, Bird A, Beug H. Immortalization of conditionally transformed chicken cells: loss of normal p53 expression is an early step that is independent of cell transformation. Genes Dev. 1992;86:876–887. doi: 10.1101/gad.6.5.876. [DOI] [PubMed] [Google Scholar]
- 54.Unnikrishnan I, Radfar A, Jenab-Wolcott J, Rosenberg N. p53 mediates apoptotic crisis in primary Abelson virus-transformed pre-B cells. Mol Cell Biol. 1999;19:4825–4831. doi: 10.1128/mcb.19.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varmus H E, Quintrell N E, Ortiz S. Retroviruses as mutagens: insertion and excision of a non-transforming provirus alters expression of a resident transforming provirus. Cell. 1981;25:23–26. doi: 10.1016/0092-8674(81)90228-2. [DOI] [PubMed] [Google Scholar]
- 56.Viskochil D, Buchberg A M, Xu G, Cawthon R M, Stevens J, Wolff R K, Culver M, Carey J C, Copeland N G, Jenkins N, White R, O'Connell P. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 57.Voncken J W, Morris C, Pattengale P, Dennert G, Kikly C, Groffen J, Heisterkamp N. Clonal development and karyotype evolution during leukemogenesis of BCR/ABL transgenic mice. Blood. 1992;79:1029–1036. [PubMed] [Google Scholar]
- 58.Wang L C, Rosenberg N. RAG-1 and RAG-2 are not sufficient to direct all phase of immunoglobulin rearrangement in pre-B cell lines. Mol Cell Biol. 1993;13:3890–3899. doi: 10.1128/mcb.13.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitlock C A, Witte O N. Abelson virus-infected cells exhibit restricted in vitro growth and low oncogenic potential. J Virol. 1981;40:577–584. doi: 10.1128/jvi.40.2.577-584.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitlock C A, Ziegler S F, Witte O N. Progression of the transformed phenotype in clonal lines of Abelson virus-infected lymphocytes. Mol Cell Biol. 1983;3:596–604. doi: 10.1128/mcb.3.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf D, Rotter V. Inactivation of p53 gene expression by an insertion of Moloney murine leukemia virus-like DNA sequences. Mol Cell Biol. 1984;4:1402–1410. doi: 10.1128/mcb.4.7.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S, Ramsay E S, Mock B A. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci USA. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhuang S, Eklund L K, Cochran C, Rao G N, Wiseman R W, Soderkvist P. Allelotype analysis of 2′,3′-dideoxycytidine- and 1,3-butadiene-induced lymphomas in B6C3F1 mice. Cancer Res. 1996;56:3338–3343. [PubMed] [Google Scholar]