ABSTRACT
Clonal reproduction of unicellular organisms ensures the stable inheritance of genetic information. However, this means of reproduction lacks an intrinsic basis for genetic variation, other than spontaneous mutation and horizontal gene transfer. To make up for this lack of genetic variation, many unicellular organisms undergo the process of cell differentiation to achieve phenotypic heterogeneity within isogenic populations. Cell differentiation is either an inducible or obligate program. Induced cell differentiation can occur as a response to a stimulus, such as starvation or host cell invasion, or it can be a stochastic process. In contrast, obligate cell differentiation is hardwired into the organism’s life cycle. Whether induced or obligate, bacterial cell differentiation requires the activation of a signal transduction pathway that initiates a global change in gene expression and ultimately results in a morphological change. While cell differentiation is considered a hallmark in the development of multicellular organisms, many unicellular bacteria utilize this process to implement survival strategies. In this review, we describe well-characterized cell differentiation programs to highlight three main survival strategies used by bacteria capable of differentiation: (i) environmental adaptation, (ii) division of labor, and (iii) bet-hedging.
KEYWORDS: cell differentiation, bet-hedging, division of labor, Caulobacter crescentus, Bacillus subtilis, Myxococcus xanthus, signal transduction
INTRODUCTION
Cell differentiation is a biological process used by multiple bacterial species to implement individual and population-level survival strategies. In this review, we discuss how the process of cell differentiation allows bacteria to implement (i) environmental adaptation, (ii) division of labor, and (iii) bet-hedging strategies.
Many bacteria respond to environmental changes by differentiating into new cell types that are better suited to their new conditions. Environmental adaptation is implemented at both the individual level through cell-autonomous signaling pathways and at the population level through cell-cell communication (1, 2).
Division of labor is a population-level survival strategy in which different cell types within a population perform distinct tasks that benefit the population as a whole. This strategy requires individual cells within the population to differentiate into specialized cell types, capable of performing specific tasks. While specialization often comes at a cost to individual cells, this price is outweighed by the increased fitness experienced by the population as a whole (3).
Bet-hedging is a population-level survival strategy that entails the coexistence of multiple cell types, even some maladapted to the current environmental condition. Bet-hedging lowers the population-level risk by spreading it across multiple different cell types. This strategy is particularly useful to bacteria that experience sudden, unpredictable environmental changes. In this instance, a fraction of the population that was maladapted to the previous condition might be well suited for survival under a new, unanticipated condition (4).
The process of cell differentiation can either be induced or obligate. Induced cell differentiation occurs either stochastically or in response to a stimulus, while obligate cell differentiation is hardwired into the bacteria’s life cycle. Bacillus subtilis and Myxococcus xanthus undergo induced cell differentiation from dividing cells to metabolically inactive spores upon sensing nutrient depletion (1, 2), while uropathogenic Escherichia coli differentiate into coccoid and filamentous morphologies upon host cell invasion (5). B. subtilis has been observed to stochastically switch from singlet motile cells to non-motile chaining cells under nutrient-rich conditions (6). In contrast, the aquatic bacterium, Caulobacter crescentus, undergoes obligate cell differentiation from a motile swarmer cell to a stationary stalked cell with each cell cycle. While induced cell differentiation is aptly suited for environmental adaptation, Caulobacter has been shown to respond to environmental cues by modulating the timing of certain cell cycle events (7–10). Bacteria that undergo either induced or obligate cell differentiation can utilize division of labor and/or bet-hedging strategies. This review will mostly focus on cell differentiation programs in relationship to survival strategies. Further details on each differentiation program can be found in the reviews cited throughout each section.
INDUCED CELL DIFFERENTIATION FACILITATES ENVIRONMENTAL ADAPTATION
Cell differentiation has been independently employed as a strategy to withstand environmental adversity by both the gram-positive bacterium, B. subtilis, and the gram-negative bacterium, M. xanthus. Both species form metabolically inactive spores that are protected by a thick coat of proteins and polysaccharides, allowing them to withstand harsh environmental conditions such as starvation, drought, and extreme temperatures (Fig. 1) (2, 11, 12). B. subtilis spore formation requires sequential asymmetric cell division and cell differentiation. This developmental program is initiated by phosphorylation and activation of the master transcription factor, Spo0A (13). Phosphorylation of Spo0A is driven by five histidine kinases, KinA–KinE, which respond to various stimuli (14, 15). Upon compartmentalization, large and small daughter cells initiate distinct gene expression programs, specific to each cell fate (11, 16). At a defined point in this developmental program, the large daughter cell engulfs the small daughter cell, allowing the small cell to fully develop into a spore within the cytoplasm of the large cell (11, 17).
Fig 1.
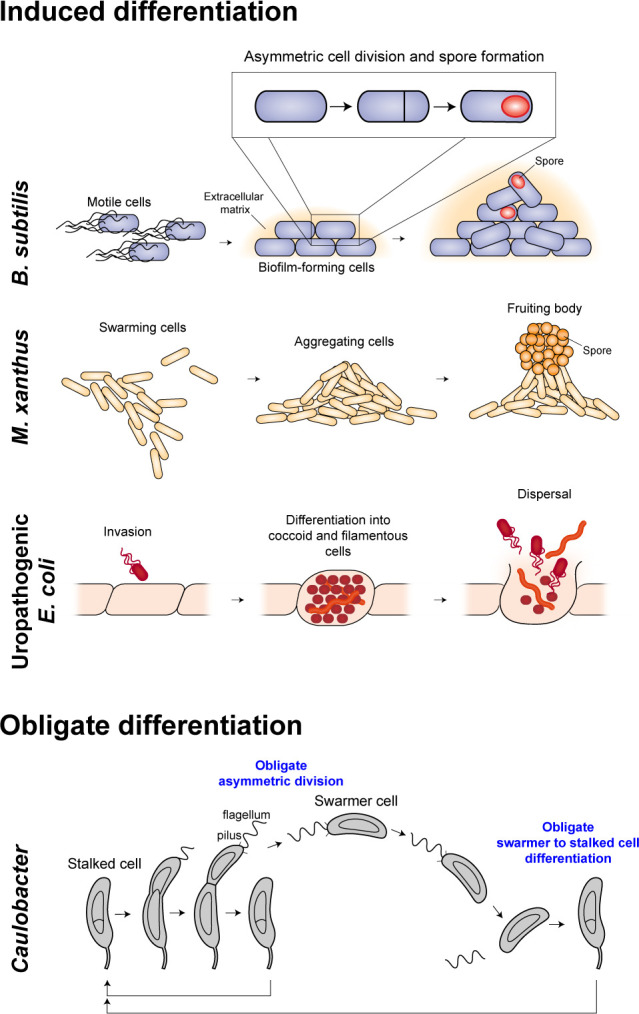
Examples of induced and obligate bacterial cell differentiation. Many bacteria undergo either induced or obligate cell differentiation. Induced cell differentiation is initiated either in response to a stimulus or stochastically. Obligate cell differentiation is hardwired into the organism’s life cycle. Under starvation conditions, B. subtilis cells differentiate into biofilm-forming cells and metabolically inactive spores. Sporulation requires a coordinated process of asymmetric cell division and spore formation. M. xanthus bacteria also form spores upon nutrient depletion. Swarming M. xanthus cells differentiate into aggregating cells that form a fruiting body consisting of spores. M. xanthus sporulation does not require cell division, but instead entails the complete remodeling of the cytoskeleton and cell wall. Uropathogenic E. coli differentiate into coccoid and filamentous morphologies upon host cell invasion. When infected cells rupture, released bacteria are able to invade neighboring cells, resulting in infection persistence. Caulobacter crescentus is an aquatic bacterium that undergoes obligate differentiation from a replication-incompetent motile swarmer cell to a replicating stationary stalked cell, with each cell cycle. Asymmetric division and cell differentiation are both obligate events, hardwired into Caulobacter’s cell cycle.
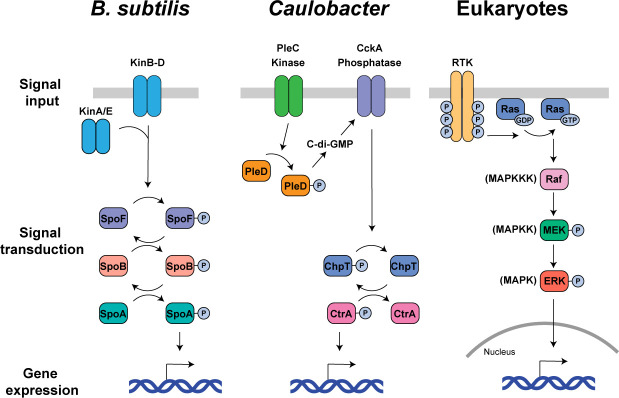
Spo0A phosphorylation is responsible for initiation of both biofilm formation and sporulation survival strategies. Biofilm formation is a strategy used by many bacteria to increase tolerance to chemical and physical stressors as well as to exchange metabolites and other common goods between cells. Moderate levels of phosphorylated Spo0A (Spo0A~P) leads to matrix secretion and biofilm formation, while high levels of Spo0A~P is required for sporulation (13, 18). Activation of one or more of the Kin histidine kinases leads to Spo0A phosphorylation either directly or indirectly by a phosphorelay through the response regulator, Spo0F, and the phosphotransfer protein, Spo0B (Fig. 2) (14). KinA and KinB have been previously identified as the main kinases responsible for sporulation, while KinC and KinD have been linked to biofilm formation (14). Analysis of purified proteins revealed that KinA is a more efficient kinase than either KinC or KinD in vitro (14, 19). Therefore, activation of specific Kin kinases tunes the level of Spo0A~P to initiate differentiation into either biofilm-forming or spore-forming cell fates.
Fig 2.
The basic framework of signal transduction pathways is broadly conserved. Signal transduction is required in cells across all kingdoms in order to integrate external environmental and internal physiological signals into biological outcomes such as cell differentiation, cell cycle progression, and cellular adaptation. B. subtilis and Caulobacter use two-component systems to integrate signals into the decision of whether or not to differentiate. These signaling pathways are built into complex molecular networks that are not depicted in their entirety here. Receptor tyrosine kinase (RTK)/Ras/mitogen-activated protein kinase (MAPK) signaling pathways are an example of eukaryotic signaling pathways that integrate signals into cell fate and cell proliferation decisions. While the specific proteins and their interactions are not conserved between bacterial and eukaryotic signal transduction pathways, the basic framework of these pathways shares many similarities.
While the Kin histidine kinases have been extensively studied, little is presently known about the specific stimuli that activate these kinases. Previous studies have suggested that the sporulation-associated kinases, KinA and KinB, likely respond to changes in growth rate or respiration, rather than to external environmental stimuli (20, 21). In contrast, the biofilm-associated kinases, KinC and KinD, likely respond to B. subtilis-secreted substances such as matrix components and surfactants (2, 22). These studies suggest that activation of the Kin kinases might be secondary to the molecular pathways responsible for directly sensing changes in environmental conditions. Further investigation into the mechanisms involved in perceiving environmental cues and relaying information to the Kin kinases is required to fully understand how B. subtilis differentiation programs are initiated in response to environmental changes. The ability of B. subtilis to differentiate into biofilm-forming and spore-forming cells in response to environmental changes ultimately allows the bacteria to survive under extreme conditions.
In contrast to B. subtilis, spore formation in M. xanthus does not involve cell division. Instead, individual rod-shaped cells completely remodel their cytoskeleton and secrete coat components in order to differentiate into spherical spores (12). Under nutrient-rich, vegetative conditions, cells swarm radially outward, collectively feeding on prey bacteria (1). Upon nutrient depletion, cells undergo a highly coordinated process of aggregation, fruiting body formation, and sporulation that ultimately allows the species to persist through different environmental conditions (Fig. 1) (1, 3, 23). It has been shown that the developmental program responsible for fruiting body formation initiates in reaction to an intracellular accumulation of the stringent response alarmones (p)ppgpp (24, 25). Therefore, similar to B. subtilis sporulation, the molecular pathway responsible for M. xanthus differentiation senses a starvation-induced change in cell physiology rather than nutrient limitation itself. Subsequent to (p)ppgpp synthesis, cells synthesize and release a quorum-sensing A-signal required for cell aggregation (26). Once the A-signal surpasses a threshold level, cells re-direct their motility, aggregating into a protruding mound that develops into a large (~0.08 mm) structure that is the fruiting body (Fig. 1) (27). Within the fruiting body, only ~1%–15% of cells differentiate into spores, while ~80% undergo programmed cell death, and ~5% remain as non-aggregating peripheral rod cells (28, 29). It has been speculated that this large-scale cell death is an example of bacterial altruism (28). Transition from a population of vegetative, predatory cells to a fruiting body consisting of spores requires initiation of an intricate molecular program acting at the individual and population levels. This coordinated program of cell aggregation, fruiting body formation, and sporulation ultimately allows M. xanthus to survive through periods of harsh environmental conditions.
Pathogenic bacteria use cell differentiation to facilitate their survival inside infected hosts (5). Upon invasion of bladder epithelial cells, uropathogenic Escherichia coli (UPEC) transition from rod to coccoid morphology (5). In addition, a small subset of UPEC cells adopt a long, filamentous morphology that hinders phagocytosis, allowing these cells to evade the host immune system (Fig. 1) (30, 31). Infected epithelial cells eventually rupture, releasing bacteria into the extracellular space. The released bacteria are able to invade neighboring epithelial cells, leading to persistence of the infection (Fig. 1) (5). Many other pathogenic bacteria differentiate into filamentous cell morphologies upon host infection, including Mycobacterium tuberculosis and Salmonella typhimurium (32, 33). Understanding the signaling pathways associated with these differentiation programs can provide putative targets for decreasing the virulence of these bacterial pathogens.
PHENOTYPIC HETEROGENEITY ENABLES DIVISION OF LABOR AND BET-HEDGING STRATEGIES
While the process of induced cell differentiation is especially suited for environmental adaptation, species that employ this type of cell differentiation are not precluded from utilizing division of labor or bet-hedging strategies. It has been proposed that B. subtilis and M. xanthus use both of these population-level survival strategies (3, 4). Division of labor involves distinct cell types performing specialized tasks that benefit the population as a whole. These tasks are divided among individual cells in a way that bears a population-level fitness advantage. Therefore, bacteria are able to bypass the evolutionary pressure put on individual cells by prioritizing the success of the population. In a system in which individual cells expend resources to produce public goods or services, one can imagine that cells that do not act cooperatively can exploit the efforts of the surrounding cells. The emergence of exploitative cells may be less prevalent in monocultures where cells are considered to have a “common goal.” While this review focuses on isogenic populations, cooperation has been documented in multi-species bacterial communities. Therefore, division of labor might be a survival strategy that is employed by both clonal populations and multi-species communities (34, 35).
B. subtilis biofilms comprise a consortium of matrix-secreting cells, exoprotease-producing cells, sporulating cells, motile cells, and competent cells, capable of acquiring new DNA (2). Matrix-secreting cells provide a service to the population by producing and secreting matrix components that are required for biofilm integrity. Exoprotease-producing cells secrete proteases that degrade proteins into peptides which can be taken up by cells within the biofilm (36). This type of specialization comes at a cost as specialized cells must expend energy to provide these services. This energetic cost is outweighed by the emergent benefits that cooperation provides to all the cells within the population. In isogenic populations, spores also provide a service as their presence helps to ensure the long-term success of the population. M. xanthus spores and non-sporulating cells have been observed to participate in a seemingly one-sided cooperation. In fruiting bodies, aggregating cells form a stalk-like structure that props up the spores. Placement of spores at the end of this cell-made stalk structure has been proposed to aid in spore dispersal (37). Therefore, seemingly altruistic cooperation between various cell types brings forth an increased probability of success for the entire population.
In addition to division of labor, bacteria utilize bet-hedging as another population-level survival strategy. Bet-hedging is a strategy that lowers the population-level risk by spreading it across multiple cell types, even some that are maladapted to the current environmental condition (4). While sporulation is initiated in response to starvation conditions, only a small fraction of B. subtilis cells enter the spore-forming cell fate (38, 39). To deter neighboring cells from differentiating, sporulating cells release signaling factors that not only prevent nearby cells from initiating sporulation, but also cause cell lysis. Metabolites from lysed cells are used by their siblings to continue to grow and divide, in a process described as “cannibalism” (40). These dividing cells are poised to initiate rapid growth in case of nutrient influx (41). Conversely, dormant spores are able to persist through prolonged harsh conditions, continuing the genetic legacy of the population. In this way, the population sets itself up for optimal success in the event of any unforeseen changes in nutrient availability.
Bet-hedging is also observed under nutrient-rich conditions. Exponentially growing B. subtilis populations consist of singlet motile cells and chains of non-motile cells joined at the poles by extracellular matrix. Motile cells are thought of as foragers, able to seek new nutrient sources, while non-motile chains take full advantage of the current niche (6). Non-motile chains also serve as starting points for biofilm formation, should environmental conditions change (42). Entry into the chaining cell fate is a stochastic process, dictated at its core by just two proteins, the SinR transcriptional repressor and its antagonist, SinI. When free and active, SinR binds to and represses the transcription of genes related to the chaining phenotype. SinR repressor activity is inhibited by direct binding by SinI. Small fluctuations in the relative abundance of SinR and SinI proteins are enough to drive stochastic entry into a SinI dominant state, corresponding with the chaining phenotype (43, 44). Interestingly, the reverse switch from chaining to motile state is not stochastic and relies on a molecular feedback loop that encodes a generational memory, committing cells to the chaining phenotype for a stereotypical number of generations before disassembly into singlet motile cells (45). Constant switching between these two cell types allows B. subtilis populations to simultaneously explore and colonize new habitats while being poised to initiate biofilm formation in the event of any unforeseen environmental changes.
How are different gene expression programs initiated in a population of cells that are experiencing relatively the same environment? While heterogeneous micro-environments caused by nutrient and oxygen gradients can explain some level of phenotypic heterogeneity, it has been proposed that many bacteria utilize noise to generate mutually exclusive phenotypic states (46). Positive and negative feedback loops enable small amounts of noise to amplify into large changes in magnitude, resulting in activation or repression of a bistable switch. Such feedback loops within differentiation programs have been proposed to give rise to multiple cell types within isogenic populations (46, 47). As discussed previously, activation of one or many of the KinA–E kinases leads to phosphorylation of the master regulator, Spo0A, responsible for sporulation. In addition, Spo0A~P itself participates in feedback loops that breed further Spo0A synthesis and phosphorylation. For example, Spo0A~P stimulates transcription of both its own gene and that of the upstream response regulator, Spo0F (Fig. 2) (48, 49). While starvation induces Spo0A phosphorylation, it curiously also leads to the synthesis of phosphatases that act on Spo0A~P and Spo0F~P. It has been shown that dephosphorylation of Spo0A~P and Spo0F~P is critical for garnering phenotypic bistability (38, 50, 51). In addition to activating developmental programs associated with biofilm formation and spore formation, Spo0A~P inhibits transcription of the fla/che operon, required for the motile cell fate (52). In this way, small amounts of noise in the Spo0A signaling pathway give rise to motile, biofilm-forming, and spore-forming cells, within the same population.
OBLIGATE DIFFERENTIATION ALLOWS FOR DIVISION OF LABOR AND BET-HEDGING STRATEGIES
Unlike induced cell differentiation, obligate differentiation is hardwired into an organism’s life cycle. The aquatic bacterium, Caulobacter crescentus, is studied as a model bacterium that undergoes obligate cell differentiation and asymmetric cell division with each cell cycle (53). Motile, replication-incompetent swarmer cells undergo differentiation into sessile, replicating stalked cells (Fig. 1). This developmental program involves shedding of the polar flagellum, pili retraction, degradation of the chemotaxis machinery, initiation of genome replication, and biogenesis of the stalk appendage (54). These developmental events result from an intricate signaling cascade, initiated by activation of the PleC histidine kinase (Fig. 2) (55–57). PleC phosphorylates the diguanylate cyclase, PleD, allowing for rapid synthesis of the small molecule, cyclic di-GMP (c-di-GMP) (58). This rapid increase in intracellular c-di-GMP concentration leads to dephosphorylation and degradation of the master transcription factor, CtrA (Fig. 2) (59, 60). As phosphorylated CtrA binds to the origin of replication, thereby inhibiting replication initiation, CtrA dephosphorylation and degradation is required to initiate chromosome replication (61). c-di-GMP synthesis additionally triggers the histidine kinase, ShkA, prompting the onset of the stalk biogenesis program (62). Therefore, activation of the PleC kinase initiates an intricate signaling cascade that ultimately results in swarmer to stalked cell differentiation (55, 57).
Conversely, Caulobacter asymmetric cell division involves phosphorylation and activation of CtrA in pre-divisional cells (59, 63, 64). Once phosphorylated and active, CtrA promotes the transcription of over 90 developmental genes, many involved in the development of the new flagellar pole (65). Distinct biomolecular condensates positioned at the stalked and flagellar poles allow asymmetric localization of cellular components prior to cell division (66–69). Caulobacter’s bi-phasic cell cycle gives rise to two distinct sub-populations: planktonic swarmer cells and stationary stalked and pre-divisional cells. This bi-phasic cell cycle allows for both division of labor and bet-hedging strategies. Division of labor is achieved by stationary cells performing the tasks of proliferation and biofilm formation, while swarmer cells are responsible for dispersal. While this specialization may seem trivial as many other species of bacteria are able to simultaneously disperse and replicate, this division of labor brings forth an emergent advantage for Caulobacter. In low-nutrient environments, bacteria must be prudent in their resource allocation. Halting replication allows swarmer cells to persist through various conditions and disperse to new habitats prior to committing to genome replication and matrix secretion, which is required for biofilm formation. In addition, biofilm formation and proliferation exclusively by stalked and pre-divisional cells maintains the integrity of already established communities. These established communities serve as proliferation centers whereby newly born swarmer cells disperse.
Bet-hedging is built into the Caulobacter cell cycle as the two cell types are specifically suited to withstand distinct challenges. For example, stalked cells are particularly suited to withstand phage infection as they lack flagella and pili, known sites of phage attachment (70, 71). Additionally, they possess an outer lipopolysaccharide capsule that covers and obscures the proteinaceous S-layer, another phage attachment site (72). In contrast, only swarmer cells are able to enter the persister cell fate, in which cells experience increased antibiotic resistance (73). Caulobacter’s continual cycles of colonization and dispersal provide an additional form of bet-hedging. The constant establishment of new communities diminishes the population-level impact associated with the destruction of previously established communities. Utilization of these two strategies, division of labor and bet-hedging, enables Caulobacter to thrive in fluctuating, low-nutrient environments such as fresh-water lakes and streams (74). This bi-phasic lifestyle is not only employed by bacteria. Some eukaryotic organisms have convergently evolved life cycles with obligate free-living and sessile phases, including sea jellies and colonial tunicates. Interestingly, these metazoans live in ocean environments where nutrient availability is often low and inconsistent (75, 76).
While obligate cell differentiation is encoded in the Caulobacter cell cycle, the process of cell differentiation can be either stimulated or stalled, depending on environmental cues. Multiple studies have reported that the process of swarmer to stalked cell differentiation can be stimulated by surface attachment. In this instance, signal transduction through either the polar pili or flagellum initiates cell differentiation (7, 8, 10). It has also been reported that under starvation conditions, differentiating swarmer cells initiate stalk biogenesis but halt stalk elongation, resulting in a stunted stalk appendage. These cells fail to initiate chromosome replication, not fully committing to the stalked cell fate (9, 77). Therefore, while cell differentiation is obligate to the Caulobacter cell cycle, timing of this differentiation event is modulated in response to various environmental cues. In this way, Caulobacter is capable of environmental adaption by tuning the duration of specific cell cycle events.
Conservation among cell differentiation signaling pathways
Both induced and obligate cell differentiation processes enable employment of population-level survival strategies. While activation of these two types of differentiation programs differ, all cell differentiation programs require initiation of a signal transduction pathway. Two-component systems are responsible for most signal transduction in prokaryotes. They consist of a histidine kinase that either phosphorylates or dephosphorylates a response regulator, that then either directly or indirectly enacts a change in gene expression (78, 79). In B. subtilis, differentiation into biofilm-forming or spore-forming cells occurs as a result of activation of one or more of the Kin histidine kinases (Fig. 2). Increase in phosphorylation of the master regulator, Spo0A, either directly or indirectly by the Kin kinases, enacts changes in gene expression that result in cell differentiation (14, 15). Obligate swarmer to stalked cell differentiation in Caulobacter results from activation of the histidine kinase, PleC, which phosphorylates the response regulator, PleD (55–57). Phosphorylated PleD synthesizes c-di-GMP, indirectly leading to dephosphorylation and degradation of the master regulator, CtrA (Fig. 2) (59, 60). This signaling pathway ultimately results in a change in gene expression, initiation of chromosome replication, and stalk biogenesis.
While two-component systems are found rarely in eukaryotes (although there are reported examples in plants [80]), the basic framework of many eukaryotic signaling pathways shares similarities with those of bacteria. For example, once activated, receptor tyrosine kinase (RTK)/Ras/mitogen-activated protein kinase (MAPK) pathways transmit a signal through a series of protein-protein interactions and post-translational modifications, ultimately resulting in a change in gene expression (81, 82). Typically, when an RTK is bound to a ligand, it undergoes transphosphorylation, in which each dimer subunit phosphorylates tyrosine residues on the other subunit. The resulting multi-phosphorylated protein domain enables docking of signaling proteins which leads to conversion of Ras-GDP to Ras-GTP. Active Ras-GTP stimulates a cascade of MAPK activation and phosphorylation events that results in a change in gene expression (Fig. 2) (83). RTK/Ras/MAPK signaling pathways are used by many eukaryotic cell types to control cell proliferation and cell differentiation (84). While the individual proteins involved in eukaryotic signaling pathways are not homologous to those found in bacterial signaling pathways, the basic framework of many prokaryotic and eukaryotic signaling pathways is quite similar.
CELL CYCLE-DEPENDENT ACTIVATION OF A SIGNAL PATHWAY INITIATES OBLIGATE DIFFERENTIATION
Signal transduction pathways are designed to initiate in response to a signal. So, how is obligate Caulobacter cell differentiation initiated in the absence of an external stimulus? The signal transduction pathway responsible for swarmer to stalked cell differentiation is initiated by PleC kinase activation (55). Therefore, PleC kinase must be activated in a cell cycle-dependent manner to ensure proper timing of swarmer to stalked cell differentiation. PleC kinase has been shown to be activated through three distinct but coordinated cell cycle-dependent events: (i) dissociation and degradation of the PodJ polar complex (57), (ii) subcellular localization of the DivK signaling protein (55), and (iii) retraction of the polar pili (56). When present, the polar scaffold protein, PodJ, localizes to the flagellar pole and inhibits PleC kinase activity (85, 86). Upon PodJ proteolysis and dissociation from the cell pole, PleC is released from the pole and freed from PodJ inhibition (57). At this time, the signaling protein, DivK, is localized to the same pole, where it allosterically binds to PleC, stimulating PleC kinase activity (55). Pili retraction at the same pole increases the PilA concentration in the inner membrane, which binds to PleC and further stimulates PleC activity (56). Therefore, at least three distinct cell cycle-dependent events coordinate the molecular signaling pathway responsible for swarmer to stalked cell differentiation with the rest of the Caulobacter cell cycle.
Progression through the Caulobacter cell cycle is dictated by seven master regulators, whose peak abundance oscillates out of phase with each other. These master regulators are responsible for initiating specific gene expression programs associated with cell cycle events such as flagellum assembly or chromosome segregation (87). Acting in concert with these core master regulators are spatially resolved signaling circuits that, once initiated, execute cell cycle events at the correct time and location. Each signaling circuit acts as a functional module that communicates with other modules both to indicate their completion and to signal for the initiation of the next module (88). The three signaling events that contribute to swarmer to stalked cell differentiation provide an example of how seemingly independent modules coordinate cell cycle progression. Prior to swarmer to stalked cell differentiation, the PodJ polar complex is dissociated from the cell pole through displacement by another polar scaffold protein, SpmX (66). At this time, PodJ is also degraded by the proteases PerP and MmpA (89). Once PodJ is removed from the cell pole, PleC is freed from kinase inhibition. Stimulation of PleC kinase activity through allosteric binding of the phosphotransfer protein, DivK, requires a high concentration of DivK at the cell pole. While DivK is abundant in swarmer cells, it is not localized to the cell pole until it is recruited by SpmX through interaction with the histidine kinase, DivJ (90, 91). In addition to PleC, DivJ also phosphorylates PleD, further ramping up the intracellular concentration of c-di-GMP (55). Therefore, PleC release from kinase inhibition and kinase stimulation depends on PodJ removal and SpmX localization at the incipient stalked pole. Lastly, the PleC kinase is further stimulated by direct binding of PilA to PleC’s N-terminal transmembrane domain upon retraction of the polar pili. This additional stimulation coordinates PleC kinase activity with the deconstruction of a swarmer cell-specific polar appendage (56). Therefore, multiple signaling pathways feed into PleC kinase activation to initiate swarmer to stalked cell differentiation. Input from these three signaling pathways allows for modulation at multiple levels and coordination of swarmer to stalked cell differentiation with the rest of the cell cycle.
CONCLUSIONS
Many species of bacteria have evolved cell differentiation programs. Whether to survive the onset of harsh environmental conditions, to adapt to life inside a host, or to employ division of labor or bet-hedging strategies, cell differentiation has been implemented by a multitude of bacterial species to excel in various niches. Both induced and obligate cell differentiation programs have come about by organizing seemingly simple two-component systems into intricate molecular networks. Cell differentiation in eukaryotes is achieved through signal transduction pathways that are divergent from those in bacteria but accomplish the same goal of coordinating signal input with differential gene expression and morphological change. In this review, we described well-characterized examples of bacterial cell differentiation. Beyond these examples, there is undoubtedly a wide array of bacterial differentiation programs that result in various cell fates, contributing to the widespread success of bacteria over billions of years.
Contributor Information
Lucy Shapiro, Email: shapiro@stanford.edu.
Marcio Rodrigues, Instituto Carlos Chagas, Curitiba, Brazil.
REFERENCES
- 1. Kaiser D, Robinson M, Kroos L. 2010. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb Perspect Biol 2:a000380. doi: 10.1101/cshperspect.a000380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis . FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- 3. van Gestel J, Vlamakis H, Kolter R. 2015. Division of labor in biofilms: the ecology of cell differentiation. Microbiol Spectr 3:MB–0002. doi: 10.1128/microbiolspec.MB-0002-2014 [DOI] [PubMed] [Google Scholar]
- 4. Morawska LP, Hernandez-Valdes JA, Kuipers OP. 2022. Diversity of bet-hedging strategies in microbial communities—recent cases and insights. WIREs Mech Dis 14:e1544. doi: 10.1002/wsbm.1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Justice SS, Harrison A, Becknell B, Mason KM. 2014. Bacterial differentiation, development, and disease: mechanisms for survival. FEMS Microbiol Lett 360:1–8. doi: 10.1111/1574-6968.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Losick R, Desplan C. 2008. Stochasticity and cell fate. Science 320:65–68. doi: 10.1126/science.1147888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hershey DM, Fiebig A, Crosson S. 2021. Flagellar perturbations activate adhesion through two distinct pathways in Caulobacter crescentus. mBio 12:1–17. doi: 10.1128/mBio.03266-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Britos L, Abeliuk E, Taverner T, Lipton M, McAdams H, Shapiro L. 2011. Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One 6:e18179. doi: 10.1371/journal.pone.0018179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. 2017. Second messenger–mediated tactile response by a bacterial rotary motor. Science 358:531–534. doi: 10.1126/science.aan5353 [DOI] [PubMed] [Google Scholar]
- 11. Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586. doi: 10.1016/j.mib.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 12. Müller FD, Schink CW, Hoiczyk E, Cserti E, Higgs PI. 2012. Spore formation in Myxococcus xanthus is tied to cytoskeleton functions and polysaccharide spore coat deposition. Mol Microbiol 83:486–505. doi: 10.1111/j.1365-2958.2011.07944.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Strauch M, Webbt V, Spiegelmant G, Hoch JA. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A 87:1801–1805. doi: 10.1073/pnas.87.5.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LeDeaux JR, Yu N, Grossman AD. 1995. Different roles for kinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol 177:861–863. doi: 10.1128/jb.177.3.861-863.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x [DOI] [PubMed] [Google Scholar]
- 16. Chareyre S, Li X, Anjuwon-Foster BR, Updegrove TB, Clifford S, Brogan AP, Su Y, Zhang L, Chen J, Shroff H, Ramamurthi KS. 2024. Cell division machinery drives cell-specific gene activation during differentiation in Bacillus subtilis. Proc Natl Acad Sci U S A 121:e2400584121. doi: 10.1073/pnas.2400584121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS. 2014. A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett 358:129–136. doi: 10.1111/1574-6968.12554 [DOI] [PubMed] [Google Scholar]
- 18. Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. 1999. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol 33:389–395. doi: 10.1046/j.1365-2958.1999.01481.x [DOI] [PubMed] [Google Scholar]
- 20. Kolodkin-Gal I, Elsholz AKW, Muth C, Girguis PR, Kolter R, Losick R. 2013. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev 27:887–899. doi: 10.1101/gad.215244.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiehler B, Haggett L, Fujita M. 2017. The PAS domains of the major sporulation kinase in Bacillus subtilis play a role in tetramer formation that is essential for the autokinase activity. Microbiologyopen 6:1–12. doi: 10.1002/mbo3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. 2010. Kind is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis Biofilms. mBio 1:e00035-10. doi: 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murphy P, Comstock J, Khan T, Zhang J, Welch R, Igoshin OA. 2023. Cell behaviors underlying Myxococcus xanthus aggregate dispersal . mSystems 8:mSystems doi: 10.1128/msystems.00425-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manoil C, Kaiser D. 1980. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol 141:297–304. doi: 10.1128/jb.141.1.297-304.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harris BZ, Kaiser D, Singer M. 1998. The guanosine nucleotide (P)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev 12:1022–1035. doi: 10.1101/gad.12.7.1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaplan HB, Plamann L. 1996. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett 139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x [DOI] [PubMed] [Google Scholar]
- 27. Jelsbak L, Søgaard-Andersen L. 2000. Pattern formation: fruiting body morphogenesis in Myxococcus xanthus. Curr Opin Microbiol 3:637–642. doi: 10.1016/s1369-5274(00)00153-3 [DOI] [PubMed] [Google Scholar]
- 28. Nariya H, Inouye M. 2008. MazF, an mRNA Interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55–66. doi: 10.1016/j.cell.2007.11.044 [DOI] [PubMed] [Google Scholar]
- 29. O’Connor KA, Zusman DR. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol 173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Möller J, Luehmann T, Hall H, Vogel V. 2012. The race to the pole: how high-aspect ratio shape and heterogeneous environments limit phagocytosis of filamentous Escherichia coli bacteria by macrophages. Nano Lett 12:2901–2905. doi: 10.1021/nl3004896 [DOI] [PubMed] [Google Scholar]
- 31. Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol 6:162–168. doi: 10.1038/nrmicro1820 [DOI] [PubMed] [Google Scholar]
- 32. Chauhan A, Madiraju M, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. 2006. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol 188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenberger CM, Gallo RL, Finlay BB. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc Natl Acad Sci U S A 101:2422–2427. doi: 10.1073/pnas.0304455101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oliveira NM, Niehus R, Foster KR. 2014. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci U S A 111:17941–17946. doi: 10.1073/pnas.1412673111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun X, Cai P, Sørensen SJ, Mortimer M, Gao C, Huang Q, Wang Y, Lin X, Wu Y, Zhu D, Chen R. 2020. Interspecific interactions in dual-species biofilms of soil bacteria: effects of fertilization practices. J Soils Sediments 20:1494–1501. doi: 10.1007/s11368-019-02500-6 [DOI] [Google Scholar]
- 36. Veening JW, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, Kuipers OP. 2008. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol Syst Biol 4:184. doi: 10.1038/msb.2008.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zusman DR. 1984. Cell-cell interactions and development in Myxococcus xanthus. Q Rev Biol 59:119–138. doi: 10.1086/413780 [DOI] [Google Scholar]
- 38. Veening JW, Hamoen LW, Kuipers OP. 2005. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol Microbiol 56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x [DOI] [PubMed] [Google Scholar]
- 39. Chung JD, Stephanopoulos G, Ireton K, Grossman AD. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol 176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Losick RM. 2020. Bacillus subtilis: a bacterium for all seasons. Curr Biol 30:R1146–R1150. doi: 10.1016/j.cub.2020.06.083 [DOI] [PubMed] [Google Scholar]
- 41. Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A 105:4393–4398. doi: 10.1073/pnas.0700463105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lord ND, Norman TM, Yuan R, Bakshi S, Losick R, Paulsson J. 2019. Stochastic antagonism between two proteins governs a bacterial cell fate switch. Science 366:116–120. doi: 10.1126/science.aaw4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dannenberg S, Penning J, Simm A, Klumpp S. 2024. The motility-matrix production switch in Bacillus subtilis —a modeling perspective. J Bacteriol 206:e0004723. doi: 10.1128/jb.00047-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Norman TM, Lord ND, Paulsson J, Losick R. 2013. Memory and modularity in cell-fate decision making. Nature 503:481–486. doi: 10.1038/nature12804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x [DOI] [PubMed] [Google Scholar]
- 47. Smits WK, Kuipers OP, Veening JW. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271. doi: 10.1038/nrmicro1381 [DOI] [PubMed] [Google Scholar]
- 48. Chastanet A, Vitkup D, Yuan GC, Norman TM, Liu JS, Losick RM. 2010. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 107:8486–8491. doi: 10.1073/pnas.1002499107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strauch MA, Wu JJ, Jonas RH, Hoch JA. 1993. A positive feedback loop controls transcription of the Spo0F gene, a component of the sporulation phosphorelay in Bacillus subtilis. Mol Microbiol 7:967–974. doi: 10.1111/j.1365-2958.1993.tb01188.x [DOI] [PubMed] [Google Scholar]
- 50. Phillips ZEV, Strauch MA. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci 59:392–402. doi: 10.1007/s00018-002-8431-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gallegos-Monterrosa R, Kovács ÁT. 2023. Phenotypic plasticity: the role of a phosphatase family rap in the genetic regulation of Bacilli. Mol Microbiol 120:20–31. doi: 10.1111/mmi.15060 [DOI] [PubMed] [Google Scholar]
- 52. Fujita M, González-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barrows JM, Goley ED. 2023. Synchronized swarmers and sticky stalks: Caulobacter crescentus as a model for bacterial cell biology. J Bacteriol 205:e0038422. doi: 10.1128/jb.00384-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newton A, Ohta N. 1990. Regulation of the cell division cycle and differentiation in bacteria. Annu Rev Microbiol 44:689–719. doi: 10.1146/annurev.mi.44.100190.003353 [DOI] [PubMed] [Google Scholar]
- 55. Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–461. doi: 10.1016/j.cell.2008.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Del Medico L, Cerletti D, Schächle P, Christen M, Christen B. 2020. The type IV pilin PilA couples surface attachment and cell-cycle initiation in Caulobacter crescentus. Proc Natl Acad Sci U S A 117:9546–9553. doi: 10.1073/pnas.1920143117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chong TN, Panjalingam M, Saurabh S, Shapiro L. 2024. Phosphatase to kinase switch of a critical enzyme contributes to timing of cell differentiation. mBio 15:e0212523. doi: 10.1128/mbio.02125-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mann TH, Seth Childers W, Blair JA, Eckart MR, Shapiro L. 2016. A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat Commun 7:11454. doi: 10.1038/ncomms11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joshi KK, Bergé M, Radhakrishnan SK, Viollier PH, Chien P. 2015. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell 163:419–431. doi: 10.1016/j.cell.2015.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A 95:120–125. doi: 10.1073/pnas.95.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaczmarczyk A, Hempel AM, von Arx C, Böhm R, Dubey BN, Nesper J, Schirmer T, Hiller S, Jenal U. 2020. Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in caulobacter. Nat Commun 11:816. doi: 10.1038/s41467-020-14585-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20:329–341. doi: 10.1016/j.devcel.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Childers WS, Xu Q, Mann TH, Mathews II, Blair JA, Deacon AM, Shapiro L. 2014. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol 12:e1001979. doi: 10.1371/journal.pbio.1001979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the caulobacter cell cycle. Proc Natl Acad Sci U S A 99:4632–4637. doi: 10.1073/pnas.062065699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tan W, Cheng S, Li Y, Li XY, Lu N, Sun J, Tang G, Yang Y, Cai K, Li X, Ou X, Gao X, Zhao GP, Childers WS, Zhao W. 2022. Phase separation modulates the assembly and dynamics of a polarity-related scaffold-signaling hub. Nat Commun 13:7181. doi: 10.1038/s41467-022-35000-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lasker K, von Diezmann L, Zhou X, Ahrens DG, Mann TH, Moerner WE, Shapiro L. 2020. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat Microbiol 5:418–429. doi: 10.1038/s41564-019-0647-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saurabh S, Chong TN, Bayas C, Dahlberg PD, Cartwright HN, Moerner WE, Shapiro L. 2022. ATP-responsive biomolecular condensates tune bacterial kinase signaling. Sci Adv 8:eabm6570. doi: 10.1126/sciadv.abm6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu N, Duvall SW, Zhao G, Kowallis KA, Zhang C, Tan W, Sun J, Petitjean HN, Tomares DT, Zhao GP, Childers WS, Zhao W. 2023. Scaffold-scaffold interaction facilitates cell polarity development in Caulobacter crescentus. mBio 14:MBio doi: 10.1128/mbio.03218-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Skerker JM, Shapiro L. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19:3223–3234. doi: 10.1093/emboj/19.13.3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmidt JM. 1966. Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J Gen Microbiol 45:347–353. doi: 10.1099/00221287-45-2-347 [DOI] [PubMed] [Google Scholar]
- 72. Ardissone S, Fumeaux C, Bergé M, Beaussart A, Théraulaz L, Radhakrishnan SK, Dufrêne YF, Viollier PH. 2014. Cell cycle constraints on capsulation and bacteriophage susceptibility. Elife 3:1–30. doi: 10.7554/eLife.03587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou X, Eckart MR, Shapiro L. 2021. A bacterial toxin perturbs intracellular amino acid balance to induce persistence. mBio 12:1–17. doi: 10.1128/mBio.03020-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. doi: 10.1128/br.28.3.231-295.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Helm RR. 2018. Evolution and development of scyphozoan jellyfish. Biol Rev Camb Philos Soc 93:1228–1250. doi: 10.1111/brv.12393 [DOI] [PubMed] [Google Scholar]
- 76. Gasparini F, Manni L, Cima F, Zaniolo G, Burighel P, Caicci F, Franchi N, Schiavon F, Rigon F, Campagna D, Ballarin L. 2015. Sexual and asexual reproduction in the colonial ascidian Botryllus schlosseri. Genesis 53:105–120. doi: 10.1002/dvg.22802 [DOI] [PubMed] [Google Scholar]
- 77. Hallgren J, Jonas K. 2024. Nutritional control of bacterial DNA replication. Curr Opin Microbiol 77:102403. doi: 10.1016/j.mib.2023.102403 [DOI] [PubMed] [Google Scholar]
- 78. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 79. Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cashin P, Goldsack L, Hall D, O’Toole R. 2006. Contrasting signal transduction mechanisms in bacterial and eukaryotic gene transcription. FEMS Microbiol Lett 261:155–164. doi: 10.1111/j.1574-6968.2006.00295.x [DOI] [PubMed] [Google Scholar]
- 81. Heppner DE, Eck MJ. 2021. A structural perspective on targeting the RTK/Ras/MAP kinase pathway in cancer. Protein Sci 30:1535–1553. doi: 10.1002/pro.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roberts PJ, Der CJ. 2007. Targeting the RAF-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26:3291–3310. doi: 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- 83. Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. doi: 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rauch N, Rukhlenko OS, Kolch W, Kholodenko BN. 2016. MAPK kinase signalling dynamics regulate cell fate decisions and drug resistance. Curr Opin Struct Biol 41:151–158. doi: 10.1016/j.sbi.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 85. Hinz AJ, Larson DE, Smith CS, Brun YV. 2003. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol 47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x [DOI] [PubMed] [Google Scholar]
- 86. Zhang C, Zhao W, Duvall SW, Kowallis KA, Childers WS. 2022. Regulation of the activity of the bacterial histidine kinase PleC by the scaffolding protein PodJ. J Biol Chem 298:101683. doi: 10.1016/j.jbc.2022.101683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lasker K, Mann TH, Shapiro L. 2016. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr Opin Microbiol 33:131–139. doi: 10.1016/j.mib.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. 2006. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J 25:377–386. doi: 10.1038/sj.emboj.7600935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Radhakrishnan SK, Thanbichler M, Viollier PH. 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 22:212–225. doi: 10.1101/gad.1601808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Perez AM, Mann TH, Lasker K, Ahrens DG, Eckart MR, Shapiro L. 2017. A localized complex of two protein oligomers controls the orientation of cell polarity. mBio 8:1–16. doi: 10.1128/mBio.02238-16 [DOI] [PMC free article] [PubMed] [Google Scholar]