Abstract
The genes encoding pestivirus E2 and NS2-3 are separated by a sequence that encodes a small hydrophobic polypeptide with an apparent molecular mass of 6 to 7 kDa (p7). It has been shown that cleavage between E2 and p7 is incomplete, resulting in proteins E2-p7, E2, and p7. We found no precursor-product relationship between E2-p7 and E2, which indicates a stable nature of E2-p7. To study the function of the E2-p7 region of the polyprotein, mutations were introduced into an infectious cDNA of bovine viral diarrhea virus (BVDV). When cleavage between E2 and p7 was abolished, viral RNA replication occurred; however, no infectious virus could be recovered. A corresponding result was obtained with a construct encompassing a large in-frame deletion of p7. To prevent synthesis of E2-p7, a translational stop codon was introduced after the last codon of the E2 gene and an internal ribosome entry site element followed by a signal peptide coding sequence was inserted upstream of the p7 gene. Transfection of RNA transcribed from the bicistronic construct led to the release of infectious virus particles. Thus, synthesis of E2-p7 is not essential for the generation of infectious virions. Cell lines constitutively expressing BVDV p7 and/or E2 were generated for complementation studies. Transfection of BVDV RNAs with point mutations or a deletion in the E2-p7 region into the complementing cell lines led to the generation of infectious virions. According to our studies, p7 as well as E2 can be complemented in trans.
Bovine viral diarrhea virus 1 (BVDV-1) and BVDV-2 are members of the genus Pestivirus, which also comprises Classical swine fever virus (CSFV) and ovine Border disease virus (BDV). Together with the genera Flavivirus and Hepacivirus (Hepatitis C virus), pestiviruses belong to the family Flaviviridae (37, 49). The pestivirus genomic RNA is approximately 12.3 kb in length and consists of one long open reading frame (ORF), which is flanked by untranslated regions at both ends (1, 4, 6, 32, 38, 39). The ORF encodes a polyprotein of about 4,000 amino acids, which is co- and posttranslationally processed by both viral and cellular proteases into at least 11 mature viral proteins: Npro, C, Erns, E1, E2, p7, NS2-3, NS4A, NS4B, NS5A, and NS5B. Npro is an autoprotease which generates its own C terminus (40, 46). C, Erns, E1, and E2 are structural components of the virion (50); Erns has an intrinsic RNase activity (16, 43, 59). The cleavages leading to the release of the envelope proteins are probably mediated by cellular signal peptidases (9, 41). Release of the nonstructural proteins located downstream of NS2-3 is mediated by a serine protease within NS2-3 (47, 60, 61). The latter also exhibits RNA-stimulated nucleoside triphosphatase and helicase activities (12, 13). NS4A acts as a cofactor for the serine protease (61). NS5B has RNA-dependent RNA polymerase activity (17, 26, 63). Recently several infectious cDNA clones of pestiviruses have been constructed (27–29, 31, 39, 55), which allow us to study these viruses by so-called reverse genetics.
The genes encoding pestivirus E2 and NS2-3 are separated by a sequence that encodes a small polypeptide named p7, which has an apparent molecular mass of 6 to 7 kDa and consists mostly of hydrophobic amino acids (9). A comparison of the p7 sequences from different pestiviruses showed that the hydrophilicity profiles and thereby the deduced structures are remarkably conserved. Cleavage between E2 and p7 is apparently mediated by host signal peptidase. In contrast to typical cleavage by signal peptidase, the E2-p7 cleavage occurs inefficiently, resulting in two E2 species, E2 and E2-p7, in pestivirus-infected cells (9, 41).
In the present study, we investigated the processing of the E2-p7 region in the BVDV polyprotein. Furthermore, a mutational analysis based on an infectious cDNA was combined with the use of complementing cell lines to study the role of p7 in replication of BVDV.
MATERIALS AND METHODS
Cells.
MDBK cells were obtained from American Type Culture Collection (Rockville, Md). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum.
Construction of plasmids.
All constructions were verified by restriction analysis and/or sequencing. Numbering of nucleotide and amino acids throughout this study refers to the BVDV CP7 sequence unless otherwise specified. All PCR products were subcloned into pGEM-T (Promega, Mannheim, Germany) and sequenced to confirm the presence of the desired mutation.
(i) SP6/wt.
Silent mutations were introduced into pRN654E2p7NS2 (9), which encompasses the SP6 RNA polymerase promoter sequence, a cDNA encoding the N-terminal domain of preprolactin and BVDV CP7 E2 to NS2 (amino acid positions 693 to 1596). A PCR fragment restricted by AvaI/KpnI (primer, P7+/P7−; template, pRN654E2p7NS2) was substituted for a corresponding fragment of pRN654E2p7NS2, resulting in plasmid SP6/wt, in which new restriction sites AflII (nucleotide [nt] 3543), HaeII (nt 3580), and BssHII (nt 3784) were generated. The introduced mutations do not change the encoded amino acid sequences. All mutations in the study were initially introduced into SP6/wt.
(ii) SP6/p7-NS2.
An NsiI-KpnI PCR fragment (primer, p7+/SM5−; template SP6/wt) was substituted for a corresponding fragment derived from SP6/wt.
(iii) SP6/E2IRESp7.
An AvaI-NcoI PCR fragment (primer, TAGIRES/IRESNcoI; template, pCITE-2a [Novagen, Madison, Wis.]) and an NcoI-HaeII PCR fragment (primer, Sec+/Sec−; template pSecTag2A [Invitrogen, Groningen, Netherlands]) and an HaeII-NotI fragment from SP6/wt were inserted together into SP6/wt digested with AvaI and NotI. The resulting bicistronic construct encompasses the area downstream of the SP6 RNA polymerase promoter, the N-terminal domain of preprolactin, the BVDV E2 gene (nt 2445 to 3566) followed by a stop codon, and the internal ribosome entry site element (IRES) of encephalomyocarditis virus (EMCV). Downstream of the IRES, a cDNA encoding a signal peptide of the mouse immunoglobulin κ chain (derived from pSecTag2A) and BVDV p7 to NS2 (nt 3567 to 5162) are located.
(iv) SP6/p7SVV and pSP6/p7II.
AvaI-PflMI PCR fragment (primer, p7+/SVV− or p7+/II−, respectively; template SP6/wt) were inserted respectively together with a PflMI-NsiI fragment from SP6/wt into SP6/wt digested with AvaI and NsiI.
(v) SP6/Δp715–51.
SP6/wt was digested with HpaI and NsiI and religated after treatment with T4 DNA polymerase. Accordingly, the construct encodes the first 14 and the C-terminal 18 amino acids of p7.
(vi) SP6/E2-p7.
An AflII-BssHII PCR fragment (primer, NR+/p7−; template, SP6/wt) was substituted for a corresponding fragment of SP6/wt.
(vii)CP7/E2p7NS2, CP7/p7-NS2, CP7/E2IRESp7, CP7/p7SVV, CP7/p7II, CP7/Δp715–51, and CP7/E2-p7.
KpnI (nt 2447)-NotI (nt 4905) fragments from the SP6/wt-based plasmids were subsequently substituted for a corresponding fragment of full-length BVDV CP7 clone pA/BVDV (28).
(viii) pcEF-E2IRESp7neo.
A KpnI (nt 2447)-Ecl136II (nt 5868) fragment from pA/BVDV was ligated to pCITE2a digested with KpnI and PmlI. The plasmid was digested with XhoI and KpnI and then ligated with an XhoI-KpnI-cohesive-ends adapter encoding an CSFV Erns signal sequence, resulting in pCITEsigE2-NS3. The adapter was made by annealing two complementary oligonucleotides, Erns signal+ and Erns signal−. pCITEsigE2-NS3 digested with AvaI and KpnI was ligated with an AvaI-NotI fragment from pSP6/E2IRESp7, resulting in plasmid pCITEsigE2IRESp7-NS3. An XhoI-EcoRV fragment from pCITEsigE2IRESp7-NS3 was made blunt with the Klenow fragment of DNA polymerase I (Klenow) and introduced into cdEF321swxneo (15) digested with SwaI. Then the cos and the spacer region were deleted from the plasmid by EcoRV, resulting in plasmid pcEF-E2IRESp7neo. The bicistronic construct encompasses a human elongation factor 1α promoter (EF-1α) sequence and a cDNA encoding a signal peptide sequence of Erns derived from CSFV Alfort Tübingen (nt 1120 to 1170) and E2 of BVDV CP7 (nt 2445 to 3566) with a translational stop codon at the end of the E2 gene followed by the EMCV IRES, and a cDNA encoding a pSecTag2A-derived signal sequence (see above) and p7 to the N-terminal half of NS2 (nt 3567 to 4597) of BVDV CP7.
(ix) pcEF-p7neo.
pCITEsigE2IRESp7-NS3 digested with NcoI was treated with Klenow, and an XbaI linker (CTCTAGAG; New England Biolabs, Schwalbach, Germany) was ligated to generate an optimal translation initiation sequence following the consensus sequence of Kozak (20). An XbaI-EcoRV fragment from the construct treated with Klenow was ligated into cdEF321swxneo in the same way. The resulting construct encompasses the EF-1α promoter sequence and a cDNA encoding a signal peptide sequence of Igκ and p7 to the N-terminal half of NS2 (nt 3567 to 4597) of BVDV CP7.
The sequences of the oligonucleotides primers are as follows: (the polarity of the oligonucleotides is indicated by + [sense orientation] or − [antisense orientation]): p7+, 5′-GGCCTCGGGTGTCCAGTATGCGCCGGTGAAATAGTGATGAT-3′; p7−, 5′-TAGGTACCCCTGCGCGCCTGGTTCAGCCTTTGCCAT-3′; p7AflII−, 5′-GACACCCGAGGCCATCTGTTCGCTTAAGATCATGTA-3′; E2+, 5′-CAAGGGTACCCAGAC-3′; SM5−, 5′-TCTAGGTACCCCTGGGCGCCTGGTTCATTCTTTGCCATC-3′; NR+, 5′-TTAAGCGAACAGATGAACTCGAGGGTCCAGTATGGC-3′; SVV−, 5′-TGACCCATTTTTTGGTGTTTACCACGCTTAAGAGTAGGTATAGTAG-3′; II−, 5′-CCCATATTA TGGTGTTTTCCTCTCTTAAGAGTAGGTATAG-3′ (the underlined sequences represent exchanges of the BVDV-derived cDNA sequence); Ernssignal+, 5′-TCGAGATCCACCATGGCCCTGTTGGCTTGGGCGGTGATAACAATCTTGCTGTACCAGCCTGTAGCAGGGTAC-3′; Ernssignal-, 5′-CCTGCTACAGGCTGGTACAGCAAGATTGTTATCACCGCCCAAGCCAACAGGGCCATGGTGGATC-3′; TAGIRES+, 5′-GCCTCGGGTTAGCGAATTAATTCCGGTTAT-3′; IRESNcoI−, 5′-TGGCCATGGTATTATCATCGTGTT-3′; Sec+, 5′-AATGGGAGTTTGTTTTGG-3′; Sec−, 5′-ACCGGCGCCATACTGGACACCAGTGGAACCTGGAAC-3′.
In vitro translation.
Uncapped RNA transcripts were synthesized by SP6 RNA polymerase (NatuTec, Frankfurt, Germany) from the SP6/wt-based plasmids. The RNA transcripts were translated in nuclease-treated rabbit reticulocyte lysates (Promega, Mannheim, Germany) in the presence of canine pancreatic microsomal membranes (Promega), which were added to the translation reaction mixtures at a concentration of 1.6 equivalents per 10 μl. Translation occurred at 30°C for 1 h essentially as specified by the manufacturer in the presence of 5 μCi [35S]methionine (Amersham Pharmacia Biotech, Freiburg, Germany). The translated products were resuspended in ice-cold phosphate-buffered saline (PBS) containing 50 mM EDTA, and microsomal membrane fractions were collected by centrifugation at 10,000 × g for 20 min. The samples were denatured in 2× Laemmli sample buffer with 2-mercaptoethanol at 95°C for 2 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a Tris-Tricine buffer system (44). The gels were processed for fluorography by using Enlightning (NEN Life Science, Zavantem, Belgium).
Amino (N)-terminal sequence analysis.
Metabolically labeled NS2 was synthesized in the presence of 300 μCi of [3H]leucine by in vitro translation from pRN653E2p7NS2. The radiolabeled NS2 protein was separated by SDS-PAGE, transferred to an Immobilon polyvinylidine difluoride membrane, and localized by radioautography. The partial amino acid sequence of the NS2 protein was determined by the method of Edman degradation as described previously (47).
RNA transcription and transfection.
Full-length BVDV CP7 cDNA constructs were linearized with SmaI and purified by phenol extraction and ethanol precipitation. Runoff transcripts were synthesized using T7 RNA polymerase (Stratagene Europe, Amsterdam, The Netherlands). The RNA transcripts were transfected into MDBK cells using electroporation as described previously (48).
IF analysis.
Viral RNA replication in MDBK cells was monitored by indirect immunofluorescence (IF) analysis with monoclonal antibody (MAb) 8.12.7 directed against pestivirus NS3 (5) as described previously (48). The presence of infectious virions was monitored at 72 h after transfection of viral RNA. The cell culture supernatant was clarified by centrifugation at 10,000 × g in a microcentrifuge for 5 min, filtered through a 0.45-μm-pore-size membrane, and subsequently used for infection of MDBK cells. At 48 h post infection (p.i.), the cells were tested by IF analysis with MAb directed against NS3 for the presence of BVDV.
RT-PCR.
Total cellular RNA in virus infected cells was extracted with RNeasy (Qiagen, Hilden, Germany). A cDNA was synthesized from the RNA template with thermostable reverse transcriptase (Display Thermo-RT; Appligene Oncor, Heidelberg, Germany) and the CP7 genome-specific primer. The synthesized cDNAs were amplified with ExTaq polymerase (BioWhittaker Europe). The primers correspond to the CP7 sequence: a, nt 3450 to 3483 (sense); b, nt 3765 to 3800 (antisense). The reverse transcription-PCR (RT-PCR) products were cloned into pGEM-T (Promega) and used for sequence analysis.
Virus growth analysis.
MDBK cells were infected at a multiplicity of infection of about 0.05. Virus titers were determined by a method which was essentially described previously (21) as log 50% tissue culture infective dose (TCID50) per milliliter.
Establishment of MDBK-p7 and MDBK-E2IRESp7 cells.
pcEF-p7neo and pcEF-E2IRESp7neo were used for establishment of MDBK-p7 and MDBK-E2IRESp7 cells. First, 2 μg of the respective plasmids was linearized with EcoRV in the vector backbone and electroporated into MDBK cells at 960 μF and 1,100 V/cm with a Gene Pulser II (Bio-Rad, Munich, Germany). At 3 days posttransfection (p.t.), cell culture medium was exchanged with DMEM containing 1 mg of G418 (Calbiochem, Frankfurt, Germany) per ml. G418-resistant colonies were isolated and replated twice for purification.
Metabolic labeling and radioimmunoprecipitation analysis.
For metabolic labeling of BVDV CP7-infected MDBK cells, 1.5 × 106 MDBK cells were infected with CP7 virus at a multiplicity of infection of 0.5. At 18 h post infection (p.i.), cell culture medium was replaced with DMEM without methionine and cysteine (Sigma Aldrich, Steinheim, Germany) and incubated for 90 min. The cells were then labeled with 100 μCi of 35S-protein-labeling mixture (Amersham Pharmacia Biotech) for 15 min, washed with PBS and incubated with DMEM containing 1.5 mg of methionine per ml, 0.26 mg of cysteine per ml, and 100 μg of cycloheximide (Sigma Aldrich, Steinheim, Germany) per ml for the indicated periods at 37°C. After labeling, the cells were washed twice with PBS and lysed with a solution of 1% NP-40, 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA, and 100 μM of PefablocSC (Merck, Darmstadt, Germany). The lysates were clarified by centrifugation at 10,000 × g for 20 min and incubated with Sepharose 4B conjugated with MAb D5 directed against BVDV E2 (58). Immunoprecipitated proteins were eluted from the Sepharose beads at 95°C for 10 min in 50 mM sodium phosphate (pH 7.5) containing 0.5% SDS and 1% 2-mercaptoethanol. For deglycosylation, peptide N-glycosidase F (PNGaseF: New England Biolabs, Schwalbach, Germany) was used essentially as specified by the manufacturer. Digested and undigested samples were mixed with 2× Laemmli sample buffer and analyzed by SDS-PAGE as described above.
For metabolic labeling of MDBK-p7 and MDBK-E2IRESp7 cells, 1.5 × 106 cells were labeled with 20 μCi of 35S-protein cell-labeling mixture at 37°C for 3 h and subjected to radioimmunoprecipitation analysis.
Western blot analysis.
BVDV p7 expressed in MDBK-p7 cells and MDBK-E2IRESp7 cells was monitored by Western blotting as described previously (9) with some minor modifications. After electrotransfer of protein, the nitrocellulose membrane was blocked with PBS containing 2% skim milk and 0.05% Tween 20 (TPBS-skim milk) for 2 h. Rabbit serum against BVDV p7 was used at a dilution of 1:3,000 in the TPBS-skim milk. As a secondary antibody, peroxidase-conjugated anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany) was used at a dilution of 1:20,000 in TPBS. Antigen-antibody complexes were visualized with the Supersignal chemiluminescence kit (Kmf Laborchemie, St. Augustin, Germany).
Complementation experiments.
MDBK-p7 cells and MDBK-E2IRESp7 cells were transfected with BVDV RNAs carrying point mutations or a deletion in the p7 gene. At 72 h p.t., cell culture supernatants were clarified by centrifugation at 10,000 × g in a microcentrifuge for 5 min, filtered through a 0.45-μm-pore-size membrane, and used for infection of MDBK cells. At 48 h p.i., the replication of BVDV in the infected cells was monitored by IF analysis with a MAb directed against NS3.
For neutralization of the rescued viruses, bovine anti-BVDV serum (019), which was collected from cattle experimentally infected with BVDV-1 New York strain (P. Becher, unpublished data) was used. The culture supernatant was incubated with a 1:100 dilution of the immune serum at 37°C for 3 h and subsequently used for infection of MDBK cells.
The infectious titer of the complemented virus was calculated essentially as described by Khromykh et al. (18): titer per 1.2 × 106 initially transfected cells = N × (SW/SIA) × (V/VI), where N is the average number of NS3-positive cells in the image area, calculated from five different image areas; SW is the surface of the well in a 24-well plate (177 mm2); SIA is the surface of the image area (1.96 mm2, using defined magnification parameters of the reflected-light Axiovert 35 microscope [Zeiss, Oberkochen, Germany]); V is the total volume of the cell culture supernatant (usually 2.5 ml per 35-mm-diameter dish) collected from the population of 1.2 × 106 initially transfected cells; and VI is the volume used for inoculation (usually 500 μl).
RESULTS
N terminus of pestivirus NS2-3.
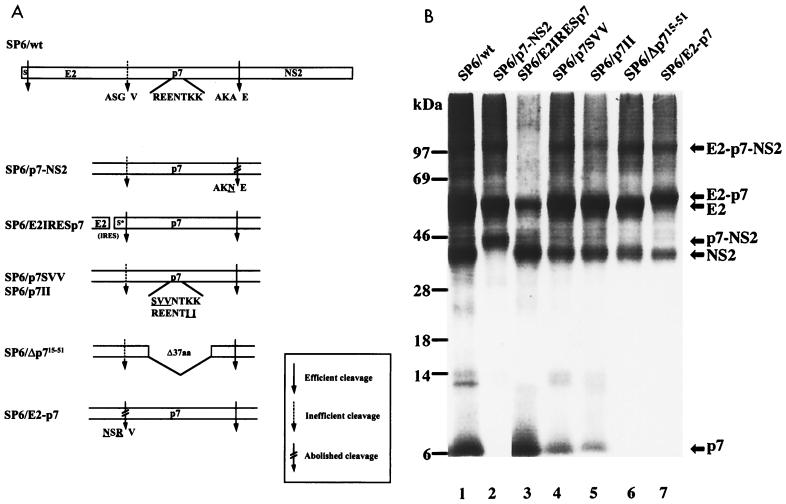
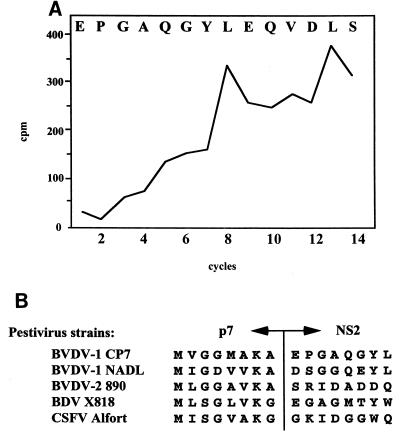
The polyprotein of noncytopathogenic pestiviruses is cleaved into at least 11 proteins. The exact positions of almost all cleavage sites in the pestivirus polyprotein have been determined (9, 41, 46, 47, 61), but the one at the p7-NS2 site has not. For studies on the expression and function of the E2-p7 region, knowledge about the authentic termini of the respective proteins is helpful. Transient-expression experiments suggested that the cleavage depends on the presence of microsomal membranes and that the N terminus of NS2 is located in the region between amino acids 1110 and 1150 (9). The SignalSeq program of the GCG DNA analysis package (7) predicted a high probability for signal peptidase cleavage between Ala1136 and Glu1137 (data not shown). The upstream sequence of the putative cleavage site fulfilled the algorithm of von Heijne (57), which predicts small amino acids in the −1 (Ala1136) and −3 (Ala1134) positions. We decided to study processing between p7 and NS2 by in vitro translation. SP6/wt contains downstream of the SP6 RNA polymerase promoter sequence a cDNA encoding the N-terminal domain of preprolactin which acts as a signal peptide and BVDV E2 to NS2 (Fig. 1A). In vitro translation in the presence of microsomal membranes led to generation of E2, E2-p7, p7, and NS2 (Fig. 1B, lane 1). Next, the Ala codon in the −1 position of the putative p7-NS2 cleavage site was changed for an Asn codon, resulting in the construct SP6/p7-NS2 (Fig. 1A). After in vitro translation, a p7-NS2 fusion protein, but neither p7 nor NS2, could be detected; cleavage at the E2-p7 site was unaffected (Fig. 1B, lane 2). Moreover, substitution at the putative −3 position also strongly interfered with the cleavage between p7 and NS2 (data not shown). To directly determine the N terminus of the pestivirus NS2 protein, the latter was synthesized by in vitro translation in the presence of [3H]leucine, purified by SDS-PAGE, electroblotted onto an Immobilon polyvinylidine difluoride membrane, and subjected to Edman degradation analysis. The release of radioactivity during the course of the cyclic Edman degradation peaked in cycles 8 and 13. The spacing of the two peaks fitted with leucine residues at positions 1144 and 1149 of the polyprotein and identified glutamic acid 1137 as the N terminus of NS2 (Fig. 2A). Accordingly, p7 encompasses 70 amino acids. An alignment of the amino acid sequences of representatives of all four pestivirus species showed that the character of the amino acids at −3 and −1 positions of the putative p7-NS2 cleavage site in all cases was in line with the requirements for a signal peptidase cleavage. It is suggested that the determined cleavage site is representative of all pestiviruses (Fig. 2B).
FIG. 1.
(A) Schematic representation of SP6/wt and mutant plasmids which encode the BVDV E2 to NS2 proteins. The wt is shown at the top: the N-terminal domain of preprolactin (S) acts as a signal peptide (9) and is fused to E2. Amino acid sequences around the cleavage sites and the charged stretch of p7 are shown below. Amino acids which were substituted in this study are underlined. S*, signal peptide of the mouse immunoglobulin κ chain; aa, amino acids. The degree of cleavage at different sites after the introduction of mutations is indicated (see the box). (B) In vitro translation analysis of RNAs transcribed from SP6/wt and mutant plasmids. Microsomal membrane fractions of the translated products were analyzed by SDS-PAGE (9% polyacrylamide). The positions of BVDV proteins are indicated on the right. Sizes (in kilodaltons) of marker proteins are indicated on the left. The lanes combined in the figure are derived from the same gel.
FIG. 2.
Processing at the p7-NS2 cleavage site of pestiviruses. (A) N terminus of BVDV NS2, strain CP7. [3H]leucine-labeled NS2 was synthesized from pRN653E2p7NS2 (9) by in vitro translation and used for N-terminal sequence analysis. The graph shows the radioactivity (in counts per minute [cpm]) released during each cycle. The N-terminal amino acid sequence of CP7 NS2 is shown at the top of the graph. (B) Alignment of the amino acid sequences in the region of the p7-NS2 cleavage site of representative strains from the four pestivirus species, BVDV-1 CP7, BVDV-1 NADL, BVDV-2 890, BDV X818, and CSFV Alfort.
Processing at the E2-p7 site.
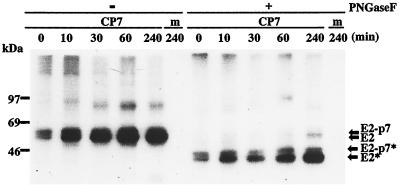
Cleavage at the E2-p7 site of the pestivirus polyprotein appears to be incomplete, since not only E2 and p7 but also the fusion protein E2-p7 can be detected (9, 41). However, it is so far not known whether E2-p7 represents a precursor or remains stable in the infected cells. To this end, we looked for any precursor-product relationship between E2-p7 and its processing products. The relative amounts of E2-p7 and E2 were investigated in BVDV CP7-infected MDBK cells by a pulse-chase experiment. As shown in Fig. 3, E2-p7 and E2 could be detected after a 15-min pulse. Moreover, while the amount of E2 increased gradually during this period, the E2-p7 molecule did not appear to be subject to further cleavage at least during a 4-h chase period. This conclusion was supported by the result obtained after digestion of the same precipitates with PNGase F (Fig. 3). There is thus no clear precursor-product relationship between E2-p7 and E2.
FIG. 3.
Pulse-chase analysis of the processing of E2-p7. MDBK cells infected with BVDV CP7 (CP7) or mock infected (m) are shown. At 19.5 h p.i., the cells were labeled with 35S-protein-labeling mixture for 15 min and chased for the indicated times. Cell extracts were immunoprecipitated with a MAb directed against BVDV E2 and then incubated in the absence (−) or presence (+) of PNGase F. The precipitates were analyzed under reducing conditions by SDS-PAGE (11% polyacrylamide). The positions of E2-p7 and E2 before and after PNGase F digestion (indicated by *) are indicated by arrows on the right. Sizes (in kilodaltons) of marker proteins are indicated on the left.
It should be noted that the level of E2 appeared to increase during the chase even though cycloheximide had been added. In contrast, such an increase was not observed during the same chase for NS2 (data not shown). One possible explanation for this unexpected observation is that the BVDV E2-specific MAb used for the immunoprecipitation preferentially recognizes the slowly forming mature conformation of E2. Alternatively, a precursor containing E2 and p7 might not be recognized by the E2-specific MAb. To address this point directly, we tried to establish whether the amount of p7 is also increasing during the chase. Unfortunately, p7 could not be detected under the chosen conditions.
E2-p7 is not required for generation of infectious virions.
The result from the pulse-chase experiment described above indicated that E2-p7 represents a stable protein in BVDV-infected cells. Based on this observation, we wanted to address whether E2-p7 is essential for the viral life cycle. To abolish the synthesis of E2-p7, a bicistronic construct with a translational stop codon at the end of the E2 gene, an IRES, and a sequence encoding a foreign signal peptide upstream of the p7 gene (SP6/E2IRESp7; Fig. 1A) was generated. In vitro translation analysis showed that an RNA transcribed from SP6/E2IRESp7 led to the synthesis of E2, p7, and NS2 but not E2-p7 (Fig. 1B, lane 3). Subsequently, the same genetic element was inserted into the full-length CP7 cDNA clone, resulting in plasmid CP7/E2IRESp7. To monitor viral RNA replication and generation of infectious virions, an RNA transcribed from CP7/E2IRESp7 was transfected into MDBK cells. At 24 h p.t., NS3 expression was taken as a measure of viral replication and monitored by IF analysis. As a control, BVDV CP7 RNA encompassing a deletion in the active site of the RNA polymerase gene was transfected into MDBK cells. NS3 could not be detected by IF analysis in the MDBK cells transfected with this replication-defective RNA (data not shown). Corresponding results were observed in our previous study (48). At 72 h p.t., cell culture supernatants were collected and used for infection of MDBK cells. These cell cultures were analyzed by IF at 48 h p.i. The IF analysis revealed that the RNA derived from CP7/E2IRESp7 led to viral RNA replication and generation of infectious virions (data not shown). The results indicated that E2-p7 is dispensable for both viral RNA replication and generation of infectious virions.
It had to be verified that the infectious virions obtained after transfection of the bicistronic RNA were not derived from revertant RNA molecules which had lost the inserted sequences. At 72 h p.t., cell culture supernatants of MDBK cells transfected with RNA derived from CP7/wt or CP7/E2IRESp7 were harvested and were subsequently used for infection of MDBK cells. Following five serial passages of both viruses on MDBK cells, total cellular RNA was extracted from infected cells and analyzed by RT-PCR with BVDV-specific primers (Fig. 4A). DNA fragments of the predicted size were obtained from both viral RNAs (Fig. 4B, lanes 2 and 3). The nature of the RT-PCR products was further confirmed by the use of six different restriction enzymes (data not shown). Accordingly, the analyses revealed no evidence for the generation of wild-type revertants after transfection of CP7/E2IRESp7 RNA and five passages.
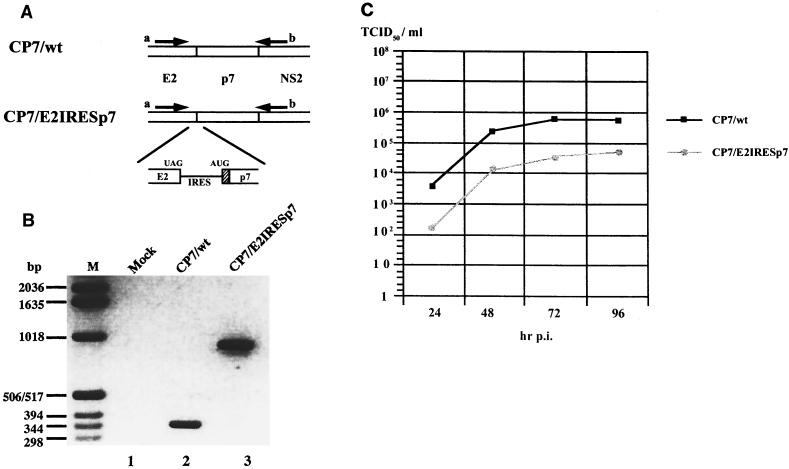
FIG. 4.
Characterization of viruses derived from CP7/wt and CP7/E2IRESp7. (A) Schematic representation of the RT-PCR analysis. Viral RNAs were transcribed from CP7/wt and CP7/E2IRESp7. UAG, translational stop codon; AUG, translational initiation codon; hatched box, signal sequence from the mouse immunoglobulin κ chain; arrows a and b, locations of primers used for the RT-PCR analysis. (B) RT-PCR analysis. cDNAs were synthesized from total RNAs of MDBK cells infected with viruses derived from CP7/wt (lane 2) and CP7/E2IRESp7 (lane 3). Mock-infected MDBK cells served as a control (lane 1). The amplified DNAs were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. M, marker. (C) Growth analysis of viruses derived from CP7/E2IRESp7 and CP7/wt. MDBK cells were infected with the CP7/wt virus or CP7/E2IRESp7 virus at a multiplicity of infection of 0.05. Cell culture supernatants were collected at the indicated time points p.i.; virus titers were determined as log TCID50 per milliliter.
The growth properties of viruses derived from constructs CP7/E2IRESp7 and CP7/wt in MDBK cells were evaluated by infection with both viruses at a multiplicity of infection of 0.05. Cell culture supernatants were collected at the indicated times to determine the titers of infectious viruses. CP7/wt reached a titer of 105.8 TCID50/ml (Fig. 4C), while CP7/E2IRESp7 reached a titer of 104.7 TCID50/ml. The fact that the CP7/E2IRESp7 RNA leads to the generation of significant amounts of infectious virions excludes an essential role of E2-p7 in this process.
p7 is required for generation of infectious virions.
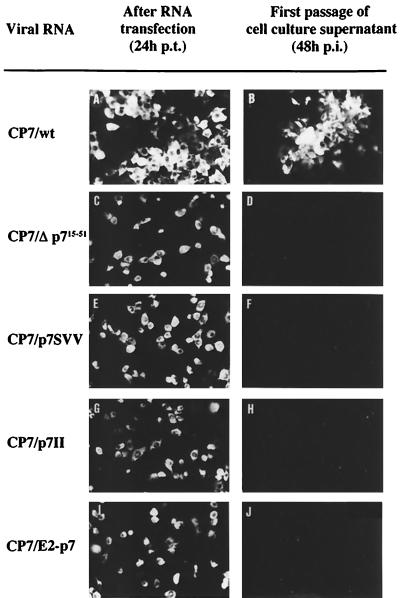
We next wanted to determine whether p7 itself is required for the viral life cycle. Accordingly, we generated a plasmid which encompasses an in-frame deletion from amino acid residues 15 to 51 of p7 (SP6/Δp715–51; Fig. 1A). This plasmid encodes the complete E2 but is not capable of expressing authentic p7 and E2-p7. As expected, in vitro translation of an RNA transcribed from SP6/Δp715–51 did not lead to the generation of p7 (Fig. 1B, lane 6). Since E2 and NS2 translated from the same RNA were indistinguishable from the respective wild-type proteins, cleavages at the E2-p7 site and the p7-NS2 site were apparently not affected. The corresponding deletion was subsequently introduced into the full-length CP7 cDNA, resulting in plasmid CP7/Δp715–51. RNAs transcribed from CP7/wt and CP7/Δp715–51 were transfected into MDBK cells, and viral RNA replication and generation of infectious virions were monitored as described above. CP7/Δp715–51 RNA replicated in MDBK cells, but infectious virions could not be detected in the culture supernatant (Fig. 5C and D); furthermore, no infectious virus could be recovered if the cell extract was prepared by freezing and thawing (data not shown). MDBK cells transfected with this RNA showed a single-cell fluorescence, which also indicated that this RNA is not capable of producing infectious progeny virions. The RNA was capable of inducing a cytopathic effect in the absence of plaque formation, which is typically observed in CP7-infected cells. On the other hand, the formation of foci of infected cells as well as severe cytopathic effect was observed in cells transfected with RNA transcript of CP7/wt (Fig. 5A and B). These results demonstrated that p7 is required for the generation of infectious virions.
FIG. 5.
IF analysis of MDBK cells transfected with RNAs transcribed from CP7/wt, CP7/Δp715–51, CP7/p7SVV, CP7/p7II, and CP7/E2-p7. At 24 h p.t., NS3 expression was analyzed by IF (A, C, E, G, and I). At 72 h p.t., cell culture supernatants were collected and subsequently used for incubation of MDBK cells. At 48 h p.i., the replication of BVDV in the cells was monitored by IF analysis (B, D, F, H, and J). Only after transfection of CP7/wt RNA were infectious virions released to the supernatant.
The pestivirus p7 protein is composed mainly of hydrophobic amino acids interrupted by a stretch of charged amino acid residues (Fig. 1A) which is highly conserved among pestiviruses. By analogy, p7 of HCV also encompasses a stretch of charged residues. The conservation of this element was taken as an indication of functional importance. To test this directly, mutations were introduced into the charged stretch of p7. In the polyprotein encoded by SP6/p7SVV, three amino acids were exchanged: Arg1100 to Ser, Glu1101 to Val, and Glu1102 to Val (Fig. 1A). Moreover, we constructed SP6/p7II, which encodes an exchange of two amino acids: Lys1105 to Ile and Lys1106 to Ile (Fig. 1A). RNAs transcribed from both constructs were used for in vitro translation. Cleavages at both the E2-p7 site and the p7-NS2 site were apparently not affected by the respective mutations (Fig. 1B, lane 4 and 5). To test whether the mutations affect viral replication and/or generation of infectious virions, full-length CP7 cDNAs containing the mutations present in SP6/p7SVV and SP6/p7II were constructed, resulting in plasmids CP7/p7SVV and CP7/p7II. Viral RNAs transcribed from both mutated full-length cDNA constructs were transfected into MDBK cells, and viral replication and the generation of infectious virions were monitored as described above. RNAs derived from CP7/p7SVV and CP7/p7II replicated, but infectious virions could not be demonstrated in the respective cell culture supernatant (Fig. 5E to H). In addition, MDBK cells transfected with the RNAs were passaged five times but infectious virus was not observed. These results showed that the charged stretch of BVDV p7 is essential for the generation of infectious virions.
E2-p7 cannot substitute for E2 and p7.
Next we tested whether the two proteins E2 and p7 can be replaced by fusion protein E2-p7. To do this, cleavage at the E2-p7 site was prevented by the exchange of two codons (those encoding Ala1064 to Asn and Gly1066 to Arg) at the E2-p7 cleavage site in SP6/wt, resulting in construct SP6/E2-p7 (Fig. 1A). In vitro translation of the corresponding RNA transcript revealed that E2-p7, but not the final products E2 and p7, was generated (Fig. 1B, lane 7). On the basis of this plasmid, we generated a full-length BVDV CP7 cDNA construct containing the mutations described above, resulting in plasmid CP7/E2-p7. After transfection, the corresponding RNA replicated but infectious virions were not generated (Fig. 5 I and J). Thus, E2-p7 cannot substitute for E2 and p7.
Functional complementation of E2 and p7 in trans.
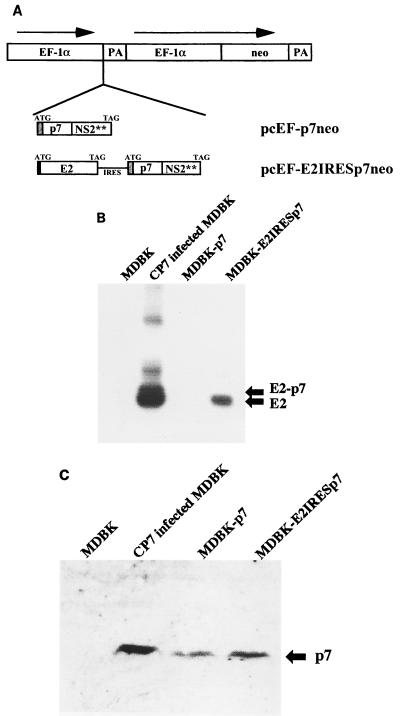
The results described above suggest that p7 as well as E2 is essential for the generation of infectious virions. To directly address this point, a trans-complementation study was started by the construction of two expression plasmids (Fig. 6A). Plasmid pcEF-p7neo is designed to express p7 in mammalian cells, whereas pcEF-E2IRESp7neo is aimed at the expression of E2 as well as p7, but not E2-p7. pcEF-p7neo encompasses the human EF-1α promoter sequence (19), a cDNA encoding a foreign signal sequence, p7, and the N-terminal half of NS2 of BVDV CP7. pcEF-E2IRESp7neo encompasses a cDNA encoding a CSFV Erns signal sequence followed by E2, a translational stop codon and the EMCV IRES. Downstream of the IRES, the same gene fragment of pcEF-p7neo was inserted (Fig. 6A). Following transfection of linearized pcEF-p7neo or pcEF-E2IRESp7neo into MDBK cells, carrying the plasmids were selected in medium containing G418. After isolation of G418-resistant colonies, expression of pestivirus proteins in each clone was analyzed by immunoprecipitation and Western blotting. Two cell lines were established and were named MDBK-p7 (carrying pcEF-p7neo) and MDBK-E2IRESp7 (carrying pcEF-E2IRESp7neo). Analyses of BVDV protein expression in both cell lines are shown in Fig. 6B and C. These cell lines were subsequently tested for their ability to rescue functionally defective viral genomes.
FIG. 6.
(A) Schematic representation of pcEF-p7neo and pcEF-E2IRESp7neo, which were used to establish the cell lines MDBK-p7 and MDBK-E2IRESp7. The expression vector is schematically shown at the top: the promoter region of human EF-1α, the polyadenylation signal from the simian virus 40 late region (PA), and a gene encoding aminoglycoside 3′-phosphotransferase (neo). The inserted fragments in the expression vector are shown below: the translational initiation codon (ATG); the IRES; genes encoding BVDV CP7 E2, p7, and the N-terminal half of NS2 (E2, p7, and NS2**); and the translational stop codon (TAG). The black box in pcEF-E2IRESp7neo indicates the signal peptide sequence derived from CSFV Erns. The shaded boxes indicate the signal sequence of the mouse immunoglobulin κ chain. (B and C) Immunoprecipitation analysis with MAb directed against BVDV E2 (B) and Western blot analysis with rabbit serum directed against BVDV p7 (C) in MDBK, BVDV CP7-infected MDBK, MDBK-p7, and MDBK-E2IRESp7 cells.
Transfection of RNA from CP7/Δp715–51 into MDBK cells did not lead to the generation of infectious virions (see above). At 72 h after transfection of the same RNA into MDBK-p7 cells, cell culture supernatants were used for infection of MDBK cells. At 48 h p.i., these cells tested positive for the presence of BVDV antigen by IF analysis. Thus, transfection of CP7/Δp715–51 RNA into MDBK-p7 cells led to the generation of infectious virions. This result demonstrated that p7 provided from the cell line was capable of a functional complementation of defective CP7/Δp715–51 RNA which expresses E2 but, due to the deletion within p7, neither authentic E2-p7 nor p7 (Fig. 7, second row). This result also confirmed that E2-p7 is not required for the generation of infectious virions. The rescued CP7/Δp715–51 RNA led to single-cell fluorescence upon infection of MDBK cells, which indicated that the recovered viruses were not capable of producing infectious virions in the absence of p7 (data not shown). The release of infectious virions upon transfection of CP7/Δp715–51 RNA into MDBK-p7 cells was therefore not due to RNA recombination. Furthermore, RNA extracted from MDBK cells infected with the rescued CP7/Δp715–51 virus was amplified by RT-PCR with BVDV-specific primers and three independent subclones of the RT-PCR products were sequenced. The sequences of the RT-PCR products exactly corresponded to the sequence of the primary cDNA construct, demonstrating directly the absence of genetic recombination (data not shown). Incubation with a bovine serum against BVDV (see Materials and Methods) led to complete neutralization of the rescued CP7/Δp715–51 virus, which proved that the transmission of the CP7/Δp715–51 RNA was mediated by infectious virions. The infectious titer of the rescued virus at 72 h p.i. was calculated as described by Khromykh et al. (18). Transfection of CP7/Δp715–51 RNA in MDBK-p7 cells resulted in ∼7.5 × 103 infectious particles per 106 transfected cells, while transfection of CP7/wt RNA yielded ∼3.2 × 105 infectious particles per 106 transfected cells.
FIG. 7.
Schematic illustration of complementation experiments. Viral RNAs were transfected into MDBK, MDBK-p7, and MDBK-E2IRESp7 cells. Cell culture supernatants were used for incubation of MDBK cells. Replication of BVDV in the cells was monitored by IF analysis. The CP7-encoded polyprotein is shown at the top. Individual BVDV proteins encoded by the different viral RNAs are shown: E2-p7 (white box); E2 (shaded box); p7 (dotted box). BVDV proteins expressed in the MDBK-p7 and MDBK-E2IRESp7 cell lines are illustrated below their names in the same way. Rescue of viral RNA is indicated in columns at the right: +, positive; −, negative.
Transfection of RNA derived from CP7/E2-p7 (no E2-p7 cleavage) into MDBK cells did not lead to the generation of infectious virions (see above). Upon transfection of this RNA into MDBK-p7 cells, no infectious virions could be detected in the cell culture supernatants (Fig. 7, third row). This indicated that p7 supplemented by the cell line was not sufficient to rescue the defect in RNA CP7/E2-p7. In contrast, rescued virus was detected in the culture supernatant after transfection of the same RNA into MDBK-E2IRESp7 cells (Fig. 7, fourth row). Thus, E2 provided together with p7 in trans was capable of a functional complementation of defective RNA CP7/E2-p7. Taken together, these experiments demonstrated that E2 is essential for the generation of infectious virions.
It should be noted that MDBK-E2IRESp7 cells and MDBK-p7 cells are not permissive for infections with either BVDV CP7/wt or the complemented viruses CP7/Δp715–51 and CP7/E2-p7. The reason for the nonpermissiveness is not known and is the subject of ongoing studies.
To test whether E2-p7 and E2 are sufficient for production of infectious virus in the absence of free p7, we attempted to complement uncleaved E2-p7 expressed from viral RNA CP7/E2-p7 to CP7/Δp715–51 virus. Accordingly, we cotransfected both RNAs into MDBK cells. No infectious virions could be detected in the supernatant of the respective cell cultures (Fig. 7, fifth row). Thus, E2-p7 and E2 in the absence of p7 are not sufficient for the generation of infectious virions.
DISCUSSION
Reverse genetics on an infectious BVDV cDNA was used for functional studies on the E2-p7 region of the BVDV polyprotein. Furthermore, a system for trans complementation of the respective proteins was established and applied. It could be demonstrated that both p7 and E2 are essential for the generation of infectious virions while the fusion protein E2-p7 is dispensable for this process.
Previous transient-expression experiments (9), our in vitro translation study, and the determined cleavage site strongly suggest that the cleavage between p7 and NS2 is catalyzed by host signal peptidase. Thus, the C-terminal domain of p7 most probably acts as a signal sequence for translocation of NS2-3, which is also supported by computer-assisted sequence analysis. The significance of the cleavage at the p7-NS2 site for replication of BVDV has not been addressed so far, and therefore mutations abolishing this cleavage were introduced into the infectious BVDV cDNA. No viral RNA replication was detected in the cells transfected with the corresponding RNA (data not shown). This indicates that cleavage at the p7-NS2 site and thus the generation of mature NS2-3 is required for RNA replication. In this context, it is interesting that an RNA transcript from an infectious CSFV cDNA, in which most of the p7 gene was removed, did not replicate (33). Taking into account the p7-NS2 cleavage site determined in our study, the respective construct still encodes two C-terminal amino acid residues of p7. Unfortunately, processing of the respective mutant polyprotein was not investigated, and it therefore remains uncertain whether authentic NS2-3 was released from the polyprotein. These data suggest that authentic processing at the N terminus of NS2-3 is essential for pestivirus RNA replication.
A pulse-chase experiment demonstrated that there is no precursor-product relationship between E2-p7 and E2. This situation is reminiscent of the HCV system, where a stable E2-p7 molecule has been observed during the chase period (24). The occurrence of stable E2-p7 in pestiviruses and HCV suggested an important function for this protein. However, the results from transfection of the bicistronic full-length BVDV RNA and the trans-complementation experiments demonstrated that E2-p7 is dispensable not only for RNA replication but also for the formation of infectious virions. Even though E2-p7 is not essential for the life cycle of pestiviruses in cell culture, it remains to be determined whether it plays a role in fitness in the natural host.
Our complementation experiments demonstrated that p7 supplied by the MDBK-p7 cells is not sufficient to rescue the defect in RNA transcribed from CP7/E2-p7 (no E2-p7 cleavage). In addition, cotransfection of RNAs transcribed from CP7/E2-p7 and CP7/Δp715–51 (supplying E2) into MDBK cells did not lead to the generation of infectious virions. Taken together, the two experiments indicated that E2 and p7 are both required for the generation of infectious virions. Pestivirus E2 represents a main structural component of the virion (50) and was found to be the major target for virus-neutralizing antibodies (8, 14, 21, 34, 54, 58). The function of p7, however, is not known. It is certainly not a major structural component of the virion (9). According to a model proposed for the topology of HCV p7, the molecule comprises two transmembrane segments which are separated by a few charged residues localized on a short cytoplasmic loop (24, 30). A corresponding topology can be predicted for pestivirus p7 (9). The conservation of the structure was taken as an indicator of the functional importance of the charged cytoplasmic loop. In line with this hypothesis, the RNAs transcribed from CP7/p7SVV and CP7/p7II, in which the preserved charged residues of p7 are mutated, did not lead to the generation of infectious virions. According to the rules of Gafvelin et al. (11), it is conceivable that the mutated p7 polypeptides might be perturbed in their membrane topology and may subsequently lose their function. Taken together, our data suggest that p7 is essential for the production of progeny virus.
Proteins with certain properties similar to ones of p7 have been described in other virus systems. The small hydrophobic 6-kDa polypeptide of alphaviruses (the 6K polypeptide) has been demonstrated to correspond in its topology on the endoplasmic reticulum membrane to the one described above for p7 of Hepatitis C virus and pestiviruses (22). The physiological role of 6K has been investigated by a variety of reverse genetic approaches. These studies suggested that the molecule is important in the final assembly of virions and facilitates virus release from the plasma membrane of mammalian cells (23, 25, 62). The small membrane protein E of the coronavirus Mouse hepatitis virus (MHV) also appears to be analogous to p7 and 6K in some of its properties, including hydrophobicity, a cluster of charged amino acids, and a critical role in formation and release of virions from cells (36, 56). A mutation introduced into the charged amino acid cluster of the MHV E protein yielded viruses which appeared to have aberrant morphology (10). Furthermore, the E protein of Equine arteritis virus, an arterivirus, has a structure which corresponds to that of MHV E protein, and this protein also appears to be essential for the production of infectious virions (45). Recently, several viral proteins capable of modulating membrane permeability have been shown to play a crucial role in the release of virions. The new family of viral proteins are called viroporins (2). Viroporins are rather small polypeptides, which form a hydrophilic pore on the membrane by oligomerization and subsequently cause membrane destabilization. It has been proposed that the following proteins belong to the viroporin family: 2B of Coxsackievirus (53), NSP4 of Rotavirus (51, 52), ion channel M2 of Influenza virus (35), NS2B of Japanese encephalitis virus (3), and 6K of alphaviruses (42). Some viroporins encompass a short stretch of basic amino acids flanked by membrane-interacting domains, comparable to p7, and are thought to participate in membrane permeabilization (2). Therefore, it is tempting to speculate that p7 represents a further member of the viroporins with a function in the release of infectious progeny virus.
ACKNOWLEDGMENTS
We thank Tillman Rümenapf, Robert Stark, Matthias König, and Gabriele Rinck for helpful discussions and Astrid Kaiser for excellent technical assistance. We are grateful to Knut Elbers for his contribution to the determination of the N terminus of NS2.
T.H. was supported by a Japan Science and Technology Overseas Research Fellowship. This study was supported by the SFB 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Becher P, Shannon A D, Tautz N, Thiel H-J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 2.Carrasco L. Modification of membrane permeability by animal viruses. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y S, Liao C L, Tsao C H, Chen M C, Liu C I, Chen L K, Liu Y L. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J Virol. 1999;73:6257–6264. doi: 10.1128/jvi.73.8.6257-6264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 5.Corapi W V, Donis R O, Dubovi E J. Characterization of a panel of monoclonal antibodies and their use in the study of the antigenic diversity of bovine viral diarrhea virus. Am J Vet Res. 1990;51:1388–1394. [PubMed] [Google Scholar]
- 6.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–869. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donis R O, Corapi W, Dubovi E J. Neutralizing monoclonal antibodies to bovine viral diarrhoea virus bind to the 56K to 58K glycoprotein. J Gen Virol. 1988;69:77–86. doi: 10.1099/0022-1317-69-1-77. [DOI] [PubMed] [Google Scholar]
- 9.Elbers K, Tautz N, Becher P, Stoll D, Rümenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gafvelin G, Sakaguchi M, Andersson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- 12.Grassmann C W, Isken O, Behrens S E. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol. 1999;73:9196–9205. doi: 10.1128/jvi.73.11.9196-9205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu B, Liu C, Lin-Goerke J, Maley D R, Gutshall L L, Feltenberger C A, Del Vecchio A M. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J Virol. 2000;74:1794–1800. doi: 10.1128/jvi.74.4.1794-1800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond J M, McCoy R J, Jansen E S, Morrissy C J, Hodgson A L, Johnson M A. Vaccination with a single dose of a recombinant porcine adenovirus expressing the classical swine fever virus gp55 (E2) gene protects pigs against classical swine fever. Vaccine. 2000;18:1040–1050. doi: 10.1016/s0264-410x(99)00347-3. [DOI] [PubMed] [Google Scholar]
- 15.Harada T, Kim D W, Sagawa K, Suzuki T, Takahashi K, Saito I, Miyamura T. Establishment and characterization of hepatoma cell line expressing HCV proteins by transfection of viral cDNA. J Gen Virol. 1995;76:1215–1221. doi: 10.1099/0022-1317-76-5-1215. [DOI] [PubMed] [Google Scholar]
- 16.Hulst M M, Himes G, Newbigin E, Moormann R J. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology. 1994;200:558–565. doi: 10.1006/viro.1994.1218. [DOI] [PubMed] [Google Scholar]
- 17.Kao C C, Del Vecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 18.Khromykh A A, Varnavski A N, Westaway E G. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D W, Harada T, Saito I, Miyamura T. An efficient expression vector for stable expression in human liver cells. Gene. 1993;134:307–308. doi: 10.1016/0378-1119(93)90115-j. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 21.König M, Lengsfeld T, Pauly T, Stark R, Thiel H-J. Classical swine fever virus: independent induction of protective immunity by two structural glycoproteins. J Virol. 1995;69:6479–6486. doi: 10.1128/jvi.69.10.6479-6486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liljestrom P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liljestrom P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, Lindenbach B D, Pragai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewy A, Smyth J, von Bonsdorff C H, Liljestrom P, Schlesinger M J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann V, Overton H, Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- 27.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyers G, Thiel H-J, Rümenapf T. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol. 1996;70:1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima H, Hijikata M, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moormann R J, van Gennip H G, Miedema G K, Hulst M M, van Rijn P A. Infectious RNA transcribed from an engineered full-length cDNA template of the genome of a pestivirus. J Virol. 1996;70:763–770. doi: 10.1128/jvi.70.2.763-770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moormann R J, Warmerdam P A, van der Meer B, Schaaper W M, Wensvoort G, Hulst M M. Molecular cloning and nucleotide sequence of hog cholera virus strain Brescia and mapping of the genomic region encoding envelope protein E1. Virology. 1990;177:184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- 33.Moser C, Stettler P, Tratschin J D, Hofmann M A. Cytopathogenic and noncytopathogenic RNA replicons of classical swine fever virus. J Virol. 1999;73:7787–7794. doi: 10.1128/jvi.73.9.7787-7794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton D J, Lowings J P, Barrett A D. Epitope mapping of the gp53 envelope protein of bovine viral diarrhea virus. Virology. 1992;190:763–772. doi: 10.1016/0042-6822(92)90914-b. [DOI] [PubMed] [Google Scholar]
- 35.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 36.Raamsman M J, Locker J K, de Hooge A, de Vries A A, Griffiths G, Vennema H, Rottier P J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–959. [Google Scholar]
- 38.Ridpath J F, Bolin S R. The genomic sequence of a virulent bovine viral diarrhea virus (BVDV) from the type 2 genotype: detection of a large genomic insertion in a noncytopathic BVDV. Virology. 1995;212:39–46. doi: 10.1006/viro.1995.1451. [DOI] [PubMed] [Google Scholar]
- 39.Ruggli N, Tratschin J D, Mittelholzer C, Hofmann M A. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rümenapf T, Stark R, Heimann M, Thiel H-J. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J Virol. 1998;72:2544–2547. doi: 10.1128/jvi.72.3.2544-2547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rümenapf T, Unger G, Strauss J H, Thiel H-J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67:3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanz M A, Perez L, Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J Biol Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- 43.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H-J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 44.Schägger H, Jagow G V. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 45.Snijder E J, van Tol H, Pedersen K W, Raamsman M J, de Vries A A. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stark R, Meyers G, Rümenapf T, Thiel H-J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tautz N, Harada T, Kaiser A, Rinck G, Behrens S, Thiel H-J. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J Virol. 1999;73:9422–9432. doi: 10.1128/jvi.73.11.9422-9432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel H-J, Plagemann P G, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 50.Thiel H-J, Stark R, Weiland E, Rümenapf T, Meyers G. Hog cholera virus: molecular composition of virions from a pestivirus. J Virol. 1991;65:4705–4712. doi: 10.1128/jvi.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian P, Ball J M, Zeng C Q, Estes M K. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 1996;70:6973–6981. doi: 10.1128/jvi.70.10.6973-6981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian P, Ball J M, Zeng C Q, Estes M K. Rotavirus protein expression is important for virus assembly and pathogenesis. Arch Virol Suppl. 1996;12:69–77. doi: 10.1007/978-3-7091-6553-9_8. [DOI] [PubMed] [Google Scholar]
- 53.van Kuppeveld F J, Hoenderop J G, Smeets R L, Willems P H, Dijkman H B, Galama J M, Melchers W J. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Rijn P A, van Gennip H G, de Meijer E J, Moormann R J. Epitope mapping of envelope glycoprotein E1 of hog cholera virus strain Brescia. J Gen Virol. 1993;74:2053–2060. doi: 10.1099/0022-1317-74-10-2053. [DOI] [PubMed] [Google Scholar]
- 55.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vennema H, Godeke G J, Rossen J W, Voorhout W F, Horzinek M C, Opstelten D J, Rottier P J. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiland E, Stark R, Haas B, Rümenapf T, Meyers G, Thiel H-J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol. 1990;64:3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Windisch J M, Schneider R, Stark R, Weiland E, Meyers G, Thiel H-J. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J Virol. 1996;70:352–358. doi: 10.1128/jvi.70.1.352-358.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 61.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao J S, Strauss E G, Strauss J H. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]