Abstract
Spinal and bulbar muscular atrophy (SBMA) is an X-linked disorder that affects males who inherit the androgen receptor (AR) gene with an abnormal CAG triplet repeat expansion. The resulting protein contains an elongated polyglutamine (polyQ) tract and causes motor neuron degeneration in an androgen-dependent manner. The precise molecular sequelae of SBMA are unclear. To assist with its investigation and the identification of therapeutic options, we report here a new model of SBMA in Drosophila melanogaster. We generated transgenic flies that express the full-length, human AR with a wild-type or pathogenic polyQ repeat. Each transgene is inserted into the same safe harbor site on the third chromosome of the fly as a single copy and in the same orientation. Expression of pathogenic AR, but not of its wild-type variant, in neurons or muscles leads to consistent, progressive defects in longevity and motility that are concomitant with polyQ-expanded AR protein aggregation and reduced complexity in neuromuscular junctions. Additional assays show adult fly eye abnormalities associated with the pathogenic AR species. The detrimental effects of pathogenic AR are accentuated by feeding flies the androgen, dihydrotestosterone. This new, robust SBMA model can be a valuable tool toward future investigations of this incurable disease.
Keywords: genetics, muscle, neuron, polyglutamine, triplet repeat
1 |. INTRODUCTION
The polyglutamine (polyQ) family of disorders comprises nine age-dependent, hereditary neurodegenerative diseases (Johnson et al., 2022; Lieberman et al., 2019). Each is caused by the anomalous expansion of a CAG triplet nucleotide repeat in a disease-specific gene, with subsequent translation resulting in the corresponding protein containing an abnormally expanded polyQ tract (Lieberman et al., 2019). Spinal and bulbar muscular atrophy (SBMA; also known as Kennedy’s disease) is one member of this family of disorders. In SBMA, the CAG triplet repeat expansion resides within the androgen receptor (AR) gene on the X chromosome (La Spada et al., 1991). The other polyQ disease family members are Huntington’s disease, spinocerebellar ataxia types 1, 2, 3, 6, 7, and 17, and dentatorubral-pallidoluysian atrophy (Lieberman et al., 2019). There are currently no effective treatments for any of these diseases.
SBMA is a male-specific disease due to a requirement for circulating testosterone (Beitel et al., 2013; Cortes & La Spada, 2018; Katsuno et al., 2002). Patients experience various symptoms impacting endocrine (Dejager et al., 2002; Rosenbohm et al., 2018), sensory (Manzano et al., 2018), metabolic (Francini-Pesenti et al., 2020; Rosenbohm et al., 2018), and motor functions (Breza & Koutsis, 2019). Of these, the most prominent is motor impairment (Atsuta et al., 2006; Guber et al., 2018; Rhodes et al., 2009). At the cellular level, polyQ-expanded AR has multiple toxic properties, including alterations in transcriptional activity (Badders et al., 2018; Belikov et al., 2015; Irvine et al., 2000; Lieberman et al., 2002; Nakajima et al., 1996; Scaramuzzino et al., 2015; Sheppard et al., 2011) and protein–protein interactions (Pluciennik et al., 2021), increased DNA-binding (Belikov et al., 2015), and impaired nuclear export (Arnold et al., 2019). PolyQ-expanded AR misfolds, forms nuclear aggregates, accumulates in inclusions, and leads to the degeneration of spinal and brainstem motor neurons (Beitel et al., 2013; Sobue et al., 1989). All of these effects culminate in slowly progressing skeletal muscle weakness and atrophy in the proximal limbs, dysarthria, dysphagia, and eventually motor dysfunction (Beitel et al., 2013; Breza & Koutsis, 2019). Although SBMA is classically thought of as a motor neuron disease, there is strong evidence implicating the expression of AR in skeletal muscle as a contributing factor to pathology (Beitel et al., 2013; Cortes et al., 2014; Johansen et al., 2009; Lieberman et al., 2014; Lombardi et al., 2019; Monks et al., 2007; Palazzolo et al., 2009; Yu et al., 2006).
The AR (NR3C4) is a nuclear hormone receptor (Nuclear Receptors Nomenclature Committee, 1999) and transcription factor activated by the binding of androgens, such as testosterone and its derivative, dihydrotestosterone (DHT). When not bound to a ligand, AR resides in a cytoplasmic aporeceptor complex that includes heat shock proteins. Upon ligand binding, AR undergoes conformational changes resulting in its translocation to the nucleus. Within the nucleus, it dimerizes, binds to androgen-response elements (AREs) on DNA, and facilitates the transcription of androgen-responsive genes. Both ligand-binding and subsequent nuclear translocation are implicated in SBMA pathogenesis (Katsuno et al., 2002; Montie et al., 2009; Nedelsky et al., 2010; Sengupta et al., 2022; Takeyama et al., 2002).
Various cell and animal models of SBMA provided valuable insights into disease mechanisms. Still, an additional model that recapitulates both the physiological and biochemical aspects of the disease, while also allowing for rapid investigations of newly emerging theories, would be beneficial to the field. D. melanogaster is a versatile model organism for studying neurodegenerative disease (Ambegaokar et al., 2010; Bolus et al., 2020). Unlike vertebrate models, fruit flies allow for large, lifespan-long experiments to be completed in a short period of time with relatively low cost. Flies also have more complex nervous systems than yeast or cultured cell models. Additionally, because they have high conservation of human disease-associated proteins, the Drosophila models of progressive, age-related neuronal disorders represent a unique opportunity to study disease-induced changes in overall function.
Previous Drosophila models of SBMA (Chan et al., 2002; Funderburk et al., 2009; Jochum et al., 2012; Nedelsky et al., 2010; Pandey et al., 2007; Takeyama et al., 2002) were generated using the modular GAL4/UAS system (Brand & Perrimon, 1993). These models expressed either a truncated (Chan et al., 2002) or full-length form of human polyQ-expanded AR protein (Pandey et al., 2007; Takeyama et al., 2002) in a tissue-specific manner. Tissues targeted for polyQ-expanded AR included the eye (Chan et al., 2002; Nedelsky et al., 2010; Takeyama et al., 2002), salivary gland (Nedelsky et al., 2010), central and peripheral nervous systems (Chan et al., 2002), and motor neurons (Chan et al., 2002; Nedelsky et al., 2010). These models demonstrated ligand-dependent degeneration (Nedelsky et al., 2010), motility defects (Chan et al., 2002; Nedelsky et al., 2010), alterations in larval neuromuscular junction (NMJ) (Nedelsky et al., 2010), and shortened lifespan (Chan et al., 2002; Nedelsky et al., 2010). To align with existing mammalian models and human disease, we developed new Drosophila models of SBMA that express full-length, human AR in both the WT and the pathogenically expanded-CAG repeat form. Unlike prior Drosophila lines, these new models were generated using pHiC31-dependent integration to insert the WT and expanded AR transgenes into a known safe harbor site. This method allows for direct comparisons between the new lines as well as any others generated in the same way, without the potential off-target effects of random insertion or variability in expression due to differences in transgene copy number. Here, we provide the initial characterizations of the new SBMA lines with emphasis on phenotypes particularly relevant to human disease; longevity, motility, and NMJ pathology.
2 |. RESULTS
2.1 |. Generation of WT and polyQ-expanded AR Drosophila lines
To generate the new SBMA lines, cDNA sequences encoding the full-length human AR with either wild-type (Q20) or expanded (Q112) repeats (Figure 1a) were inserted into a safe harbor locus using pWalium.10.moe as a targeting vector (Figure 1b). Our models contain a mixed CAG/CAA repeat sequence. We have used mixed CAG/CAA tract for all models of polyQ diseases that we prepared recently, including SCAs 1, 2, 3, 6, 7, 17, HD, and DRPLA. The new SBMA transgenics belong to this compendium of the polyQ family of disease models. Critical to this investigatory toolset is our focus on the disease protein without potential confounding effects from mRNA toxicity or nontraditional protein translation (Figura et al., 2015; Shieh & Bonini, 2011; Sobczak et al., 2003; Sobczak & Krzyzosiak, 2005; Zu et al., 2018). We recognize that in the future the SBMA model would benefit from additional transgenic lines that express AR with pure CAG repeats. In fact, a primary strength of the new model that we present here is its ability to be compared side by side to any other transgenic lines with a similar design.
FIGURE 1.

New transgenic fly lines for SBMA. (a) Amino acid sequence of the human AR protein used. The polyQ is encoded by a CAG/CAA doublet repeat to circumvent the possibility of mRNA- and non-AUG translation-dependent toxicity. Bolded: HA epitope tag. (b) Insertion strategy into site VK00033 on chromosome 3 of the fly. (c) Western blotting from adult flies expressing the noted transgenes. Whole fly lysates. elav-Gal4: pan-neuronal expression. Mef2- Gal4: muscle cell expression. Black arrows: main AR bands; solid red bracketed line: SDS-resistant, polyQ-expanded AR trapped in the stacking gel; dotted, red bracketed line, polyQ-expanded AR trapped in resolving gel; gray bracketed line: likely proteolytic products of AR. Each lane is from an independent repeat.
The presence of AR protein was visualized via Western blotting (WB) (Figure 1c). The main wild-type (WT) and polyQ-expanded (SBMA) AR bands (black arrows) migrate above the 100 kDa marker, with the expanded variant running higher due to the longer polyQ (Figure 1c). Also of note is the presence of SDS-resistant, polyQ-expanded species in the stacking (solid red bracket) and resolving (dotted-red bracket) portions of the gel, as well as lower molecular weight bands that are likely proteolytic products of AR (gray bracket).
2.2 |. Expression of polyQ-expanded AR in neurons or muscles reduces lifespan and impairs mobility in adult Drosophila
To characterize the phenotypes of these lines, we examined if expression of WT or SBMA AR causes deficits in the absence of ligand. Because AR toxicity affects both motor neurons and skeletal muscle (Beitel et al., 2013; Breza & Koutsis, 2019), we utilized the Gal4/UAS system to drive AR in the nervous system or muscles in flies. This modular system utilizes the exogenous yeast transcriptional activator, Gal4 and its binding sequence, UAS (Upstream Activating Sequence) to manipulate expression of target transgenes in specific tissues within the fly throughout development and adulthood (Brand & Perrimon, 1993; Caygill & Brand, 2016). By mating one fly containing Gal4 under the control of a tissue-specific promotor (driver) and one fly containing the UAS (responder) sequence upstream of the target gene, transcription can be targeted to the tissue of interest. Myocyte enhancer factor-2 (Mef2) is required for Drosophila muscle differentiation and development and is expressed in both cardiac and skeletal muscle (Lilly et al., 1995; Ranganayakulu et al., 1995, 1998). Expressing Gal4 under the control of the Mef2 promoter (Mef2-Gal4) allows for muscle-specific expression of target transgenes. Embryonic lethal abnormal vision (elav) is expressed pan-neuronally and is required for developing and maintaining the Drosophila nervous system (Campos et al., 1987; Robinow & White, 1991).
We found that in the absence of ligand, SBMA AR dramatically reduced longevity with both neuronal and muscle tissue expression (Figure 2a; additional statistical information for figure 2 is in Table S1). Expressing WT AR in neuronal cells also resulted in modest reductions to longevity (Figure 2a). However, the reduction in longevity with WT AR appears to be inconsistent, as it is absent in other repetitions (please refer to the “EtOH” group in Figure 4a). In muscles, WT AR expression slightly, but significantly, increased longevity (Figure 2a; the same pattern was independently confirmed in Figure 5a); the cause of this modest increase is unclear and will be a focus of future investigations. In assessments of climbing ability, SBMA flies became impaired significantly more quickly than control flies (Figure 2b).
FIGURE 2.
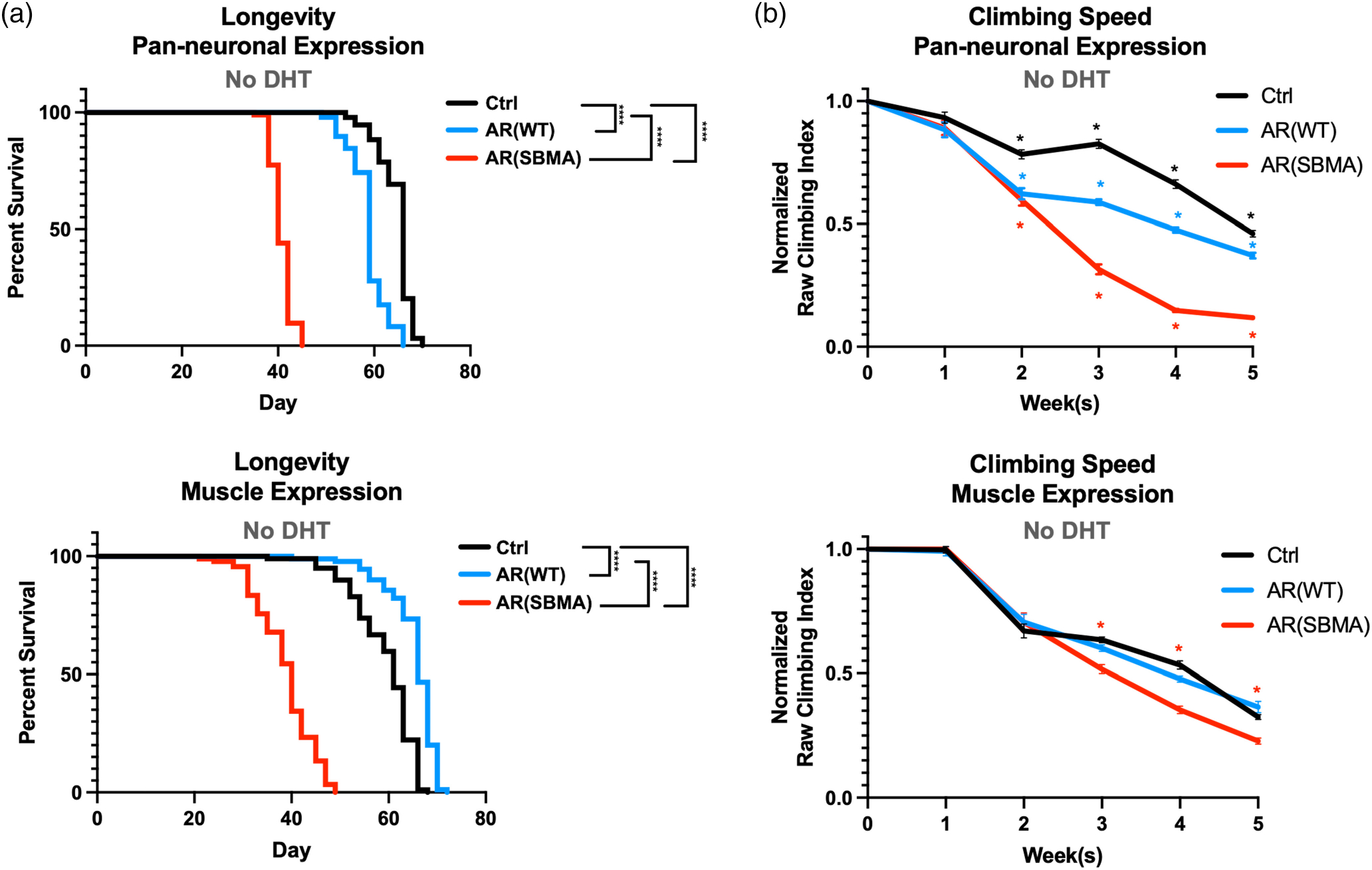
Longevity and motility outcomes from expression of W T or polyQ-expanded AR in neuronal or muscle cells. (a) Longevity assays of female flies expressing the noted transgenes. No DHT was introduced at any point of their lives. N ≧ 100 flies per group. ****p < .0 0 01, log-rank tests. (b) Normalized motility assays of female flies expressing the noted transgenes in neurons or muscles. N ≧ 100 flies per group *p < .05. Statistical analyses used: two-way ANOVA with Bonferroni post hoc correction. For (a) and (b), Ctrl denotes the background line used to generate the transgenic lines, in trans with the respective driver. Details about statistical analyses and outcomes are in Table S1.
FIGURE 4.

Effect of DHT on SBMA model motility and longevity when expressed in neurons. (a) Longevity and (b) normalized motility outcomes from expression of normal or polyQ-expanded AR in neuronal cells, without or with DHT. N ≧ 100 flies per group for (a) and (b). Statistical analyses used: Log-rank tests, ****p < .0 0 01 (longevity), two-way ANOVA with Bonferroni post hoc correction. Red asterisks compare Ctrl to SBMA. Additional details about statistical analyses and outcomes are in Table S1.
FIGURE 5.

Effect of DHT on SBMA model motility and longevity when expressed in muscles. (a) Longevity and (b) normalized motility outcomes from expression of normal or polyQ-expanded AR in muscle cells, without or with DHT. N ≧ 100 flies per group for (a) and (b). Statistical analyses used: Log-rank tests, *p < .05, ***p < .001, ****p < .0001 (longevity), two-way ANOVA with Bonferroni post hoc correction. Red asterisks compare Ctrl to SBMA. Additional details about statistical analyses and outcomes are in Table S1.
Interestingly, although the effect was milder than with SBMA AR, pan-neuronal expression of WT AR also reduced climbing ability compared to controls lacking AR. This decline-of-motility phenotype was absent in flies with muscle expression of AR (Figure 2b). At first, these data showing negative phenotypes in the absence of DHT were surprising since polyQ-expanded AR toxicity requires ligand activation. However, our findings are in agreement with more recent reports of some androgen-independent pathology in disease conditions (Feng et al., 2022).
The experiments in Figures 1 and 2 were conducted using female flies. Male flies also showed marked reductions in longevity with neuronal and muscle expression of polyQ-expanded AR, while robust motor deficits were seen only with neuronal expression (Figure S1). For additional characterizations, we elected to move forward focusing only on female flies. Even though SBMA is male-specific in humans, AR is exogenous to both male and female flies; therefore, the choice to examine female flies in this context rather than males does not lessen its relevance to the human disease. Moreover, the use of male mice is necessary in mammalian models of this disease, because they produce androgens; flies do not produce testosterone or DHT and since SBMA is hormone-dependent, the flies in the study do not need to be male. Additional rationale for this choice is as follows: using one sex reduces the overall amount of DHT required for the study, thus minimizing the use of a controlled substance; since both sexes showed strong phenotypes with expression of polyQ-expanded AR, utilizing one sex still provides pertinent information that can be extrapolated to both; lastly, for the purposes of histological and biochemical preparations, the larger size of female flies compared to males yields modestly higher amounts of material for downstream applications and eases anatomical dissections and observations.
To visualize the relationship between physiological outputs and AR protein levels and aggregation, we next conducted WBs. We selected two time points for each expression pattern: day 1 and week 3 for neurons, and day 1 and week 5 for muscle.
We selected the 3-week time point for neuronal expression as it coincides with sustained, significant reduction in mobility for this tissue type; we chose the 5-week mark for muscle expression for the same reason. As shown in Figure 3, expression of AR was consistent during this time frame, in each tissue. With the disease-causing version, we observed the accumulation of SDS-resistant smears over time, concomitant with a reduction in signal of the primary AR band. This effect was especially pronounced in neuronal tissue.
FIGURE 3.

Expression of AR protein in neurons and muscles. Western blots from whole flies expressing the noted transgenes in neurons (a) or muscles (b), for the amounts of time indicated. Images in top part of panel (a) are from the same blot and exposure, cropped and rearranged to ease visualization. Blots in bottom part of panel (a) are from independent lysates. In both panels, each lane is an independent repeat, that is, for (a) N = 3 for top panel, except for the first lane without AR and the last lane, with muscle expression, where N = 1; N = 4 for day 1 and N = 6 for week 3 on the bottom panel; for (b): N = 3 for all lanes, except first and last lanes, where N = 1. The last lanes on the top blot (a) and in blot (b) shows the relative expression of muscle (a) and neuronal (b) AR for comparative purposes. A note for panel (a): soluble monomeric AR is nearly undetectable at week 3 in flies with neuronal expression. This is likely due to the enhanced aggregation of polyQ AR in these flies, as evidenced by substantial high molecular weight, aggregated AR.
From these results (Figures 1–3), we conclude that expression of polyQ-expanded AR is toxic when expressed in fly neuronal and muscle tissue and that its toxicity coincides with increased aggregation of the insulting protein.
2.3 |. DHT reduces longevity and exacerbates motor impairment in SBMA model flies
Because SBMA is an androgen-dependent disorder (Beitel et al., 2013; Cortes & La Spada, 2018), we next evaluated the role of DHT in the longevity and motility of WT and SBMA AR flies. With pan-neuronal expression, DHT supplementation had no significant effect on the longevity of either the background control or WT AR flies (Figure 4a; additional statistical information for figure 4 is in Table S1). In agreement with non-DHT studies, polyQ-expanded AR expression in neurons resulted in severe reduction to lifespan, further enhanced by DHT supplementation (Figure 4a). These toxic effects carried over to mobility, where SBMA lines rapidly declined over time, a phenotype that was again exacerbated by DHT (Figure 4b, Table S1).
Unlike with neuronal expression, DHT supplementation with muscle expression of AR negatively impacted the lifespan of both WT AR and SBMA AR flies (Figure 5a; additional statistical information for figure 5 is in Table S1), although this phenotype was more severe with SBMA AR. The reduction in longevity observed in flies not expressing AR in the presence of DHT was not a consistent outcome, as we did not always observe a significant reduction in lifespan in these flies (Figure S2). Muscle expression of polyQ-expanded AR consistently reduced lifespan both with and without the addition of DHT. However, this effect was worsened by DHT supplementation (Figure 5a). In assessments of mobility, as observed in Figure 2b, SBMA lines exhibited significantly reduced climbing ability starting in week 2 and continuing through week 5 (Figure 5b). Interestingly, DHT did not impact the rate of decline in climbing ability for any of the lines tested (Figure 5b, Table S1).
We next examined the effect of DHT on WT and SBMA AR using WBs. We focused again on week 3 for neurons and week 5 for muscles, similar to the experiments conducted in the absence of DHT (Figure 3). As reported in other models (Kemppainen et al., 1992; Zhou et al., 1995), DHT led to increased levels of AR protein in both WT and SBMA models, in both neurons and muscles (Figure 6; additional statistical information for figure 6 is in Table S1). With polyQ-expanded AR, we again observed the presence of SDS-resistant species regardless of the treatment with vehicle control or DHT, but the types of smears seen were different, indicating differences in the types of aggregates formed in the presence of ligand. We also noticed trends that did not reach statistical significance with the levels of SDS-resistant species in the absence versus presence of DHT: higher levels with SBMA AR in neurons and muscles (Figure 6), and reduced (neurons; Figure 6a) or similar levels (muscle; Figure 6b) with WT AR.
FIGURE 6.

AR protein in the absence and presence of DHT. Western blots from whole flies expressing the noted AR proteins. Black arrows: main AR bands; red bracketed lines: SDS-resistant, polyQ-expanded AR; blue bracketed arrow, SDS-resistant, WT AR. Quantifications are from the images on top, where each lane represents an independent repeat. *p < .05; **p < .01 based on Student’s t-tests. Each lane is an independent repeat.
Altogether, these data indicate that DHT worsens phenotypes caused by polyQ-expanded AR when expressed in neurons or in muscles, in agreement with reports from other models of SBMA (Montie et al., 2009; Nedelsky et al., 2010; Takeyama et al., 2002; Walcott & Merry, 2002).
2.4 |. The effect of SBMA AR on neuromuscular junction complexity
In mouse and previous fly models of SBMA, the structure and function of the neuromuscular junction (NMJ) is pathologically altered (Badders et al., 2018; Molotsky et al., 2022; Nedelsky et al., 2010; Poort et al., 2016; Xu et al., 2016, 2018). Here, we examined NMJ complexity in the dorsal longitudinal flight muscles to assess the effect of polyQ-expanded AR expression in our fly models (Figures 7 and S3; additional statistical information for figure 7 is in Table S1). On day 1, no significant differences in NMJ complexity were observed with pan-neuronal or muscle expression of either AR transgene (Figure 7). At 3 weeks of age, pan-neuronal expression of SBMA AR significantly reduced NMJ complexity, with or without DHT supplementation; DHT led to a trend of further reduction in complexity, but this did not reach statistical significance (Figure 8a,b; additional statistical information for figure 8 is in Table S1). Interestingly, the NMJ complexity of WT AR flies also trended lower with the addition of DHT (Figure 8a,b); however, this also did not reach statistical significance. We did not observe significant differences in NMJ complexity at 5 weeks of age in flies with muscle-specific expression of W T or polyQ-expanded AR (Figure 8c,d).
FIGURE 7.

Effects of AR on neuronal-muscle structural relation in young flies. (a) Representative images from adult female flies whose flight muscles were dissected and stained as outlined in methods. White bracketed lines: muscle fibers; open arrows: motor neuron branches that are adjacent to muscle fibers; yellow boxes: neuronal branching. (b) Quantification of axonal branching complexity categorized by expressing tissue; (N), neuron, (M) muscle. Statistics: not significant by one-way ANOVA. N = 12 muscles per condition.
FIGURE 8.

Effects of AR expression on neuronal-muscle structural relation in aged adult flies with or without ligand. (a) Representative images from adult female flies whose flight muscles were dissected and stained as noted. Pan-neuronal expression of WT or SBMA AR with DHT or EtOH. White bracketed lines: muscle fibers; open arrows: motor neuron branches that are adjacent to muscle fibers; yellow boxes: neuronal branching. (b) Quantification of axonal branching with pan-neuronal expression of AR. N = 12 muscles per condition. Statistics: two-way ANOVA with Bonferroni post hoc correction. *p < .05, ***p < .001, ****p < .0 0 01. (c) Representative images from adult female flies whose flight muscles were dissected and stained as noted. Muscle expression of W T or SBMA AR with DHT or EtOH. White bracketed lines: muscle fibers; open arrows: motor neuron branches that are adjacent to muscle fibers; yellow boxes: neuronal branching. (d) Quantification of axonal branching with pan-neuronal expression of AR. Statistics: not significant by two-way ANOVA. N = 12 muscles per condition.
2.5 |. ARQ112 causes deterioration in Drosophila eyes
Eye-specific expression of mutant genes in Drosophila is a well-established tool in the study of neurodegenerative diseases—it is useful for conducting large genetic screens due to the ease of phenotype evaluation. Moreover, mutations that would be lethal in other cell types (e.g., neurons or muscles) can be assessed in fly eyes without impacting viability. Previous work established a useful scoring system (illustrated in Figure 9a) for tracking eye deterioration over time (Johnson et al., 2020, 2021). We utilized this scoring system by crossing each of our lines to the eye-restricted driver, GMR-Gal4. The scoring was blinded.
FIGURE 9.

Effects of AR expression in fly eyes. (a) Scoring system. (b, d) Impact of AR expression in fly eyes for the noted days, in the presence or absence of DHT. WT: wild-type AR. SBMA: polyQ-expanded AR. Means. +/−SD. Statistics: two-tailed Wilcoxon tests, where *p < .05, **p < .01, ***p < .001, ****p < .0001. N ≧ 42 per group per time point in (b) and N ≧ 8 (d). For (d), we kept the “N” small to highlight the significance of the phenotype. (c) Western blots from dissected fly heads expressing the noted forms of AR in eyes in the absence of ligand. Each lane represents an independent repeat. Black arrows: main AR bands; red bracketed line: SDS-resistant AR; gray bracketed line: likely proteolytic fragments of AR protein.
In the absence of DHT, SBMA AR expression worsened eye phenotypes beginning in week 3, while WT AR and non-AR expressing controls remained unaffected throughout the timeline of investigations (Figure 9b; additional statistical information for figure 9 is in Table S1). Examination via WBs showed a pattern of migration for WT and SBMA AR similar to what we observed in other tissues, with SBMA AR migrating as a main band as well as SDS-resistant species (Figure 9c). The SBMA AR-dependent phenotype in fly eyes worsened with the addition of DHT, being exacerbated as early as the second week and continuing through week 4 (Figure 9d). Thus, as with other tissues that we examined, SBMA AR also causes pathology in the fly eye.
3 |. DISCUSSION
We sought to generate robustly phenotypic Drosophila models of SBMA that can help the field with expedited investigations of the biology of this disease and the identification of therapeutic options. The experiments summarized here demonstrate the value of the new SBMA models. In agreement with data from mammalian models of this disease, the new Drosophila SBMA lines have reduced lifespan, motility, and NMJ complexity, coinciding with aggregation of polyQ-expanded AR protein. These phenotypes are generally worsened by the addition of DHT. In line with our data, DHT-dependent lethality and motor impairment have also been reported in previous adult fly and larval models expressing human full-length polyQ-expanded AR (Badders et al., 2018; Bott et al., 2016; Nedelsky et al., 2010). When expressed in neurons, SBMA AR consistently and rapidly reduced each of the physiological outputs that we measured and led to the deterioration of NMJ complexity. DHT-dependent reduction in NMJ branching complexity was previously reported in fly larvae expressing human polyQ-expanded AR in a motor neuron-specific manner (Nedelsky et al., 2010). In our model, we did not observe this difference at the time points we examined. We chose histology timepoints based on the strong phenotypic deficits that we observed, rationalizing that they would concur with NMJ alterations; however, it is possible that there are significant NMJ changes bet ween the treatment s that were not captured by this specific choice of timeline. When expressed in muscles, SBMA AR again led to general reductions in physiological outputs; however, these did not coincide with structural deteriorations in the NMJ. Lastly, it is formally possible that motility and longevity defects that resulted from expression of SBMA AR in all neurons and associated NMJ morphological anomalies were due not solely to toxicity in motorneurons, but also in other neuronal cell populations. This is a point that we will keep in mind in future investigations. Alongside reports from studies in mammals, these data support important roles for both neuronal and muscle tissue in future investigations of SBMA biology of disease.
A key feature of SBMA pathogenesis is the ligand-enhanced translocation of polyQ-expanded AR to the nucleus. Contrary to other Drosophila models (Nedelsky et al., 2010; Pandey et al., 2007; Takeyama et al., 2002), expressing human polyQ-expanded AR in our fly models resulted in marked phenotypes in the absence of ligand. In our ongoing work, we noticed that both WT and SBMA AR indeed localize to the nucleus in fly tissues in the absence of DHT; presence in the nucleus is enhanced by the supplementation of DHT for SBMA AR, whereas WT AR shows a trend of increased nuclear fractionation, but does not reach statistical significance (Figure S4). This is distinct from earlier studies in PC12 cells in which polyQ-expanded AR aggregation and toxicity is completely ligand dependent (Montie et al., 2009; Walcott & Merry, 2002).
Moreover, forced localization of the expanded AR to the nucleus did not result in its aggregation in the absence of ligand (Montie et al., 2009). Only when the AR was activated (conformationally altered) by ligand binding, did its toxic effects ensue. However, our observations of ligand-independent phenotypes with SBMA AR are congruent with recent studies in transgenic mice, which suggested that early events in SBMA can be androgen independent (Xu et al., 2018). Additionally, it is known that AR can be imported into the nucleus in the absence of androgens (Gregory et al., 2001), (although even in this location, it is largely inactive in the absence of ligand.)
Ligand-free, AR-induced toxicity at the level of the mitochondria was recently reported in patient-derived cells (Feng et al., 2022); ligand-binding did exacerbate toxicity (Feng et al., 2022). It is of interest that mixed or alternating CAG/CAA repeats were used in the development of these cell lines (Feng et al., 2022), similar to the design of our models. Pure versus mixed polyQ-encoding repeats lead to different types of mRNA structures: pure CAG repeats result in relatively stable hairpins, while mixed CAG/CAA repeats result in unstable hairpins (Figura et al., 2015). One possibility is that the different mRNA structures formed by polyQ-encoding repeats lead to mRNA-based toxicity at the motor unit. mRNA toxicity has been implicated in various triplet repeat diseases, although that toxicity usually has been associated with pure CAG repeats, rather than mixed ones (Malik et al., 2021). Whether such unstable, or reduced, hairpin formation enhances mixed CAG/CAA repeat translation efficiency, leading to its enhanced nuclear localization and aggregation, or an RNA-mediated mechanism of toxicity, is the focus of ongoing investigations.
An additional point of interest is the negative impact of WT AR overexpression in previous Drosophila models of SBMA (Nedelsky et al., 2010; Scaramuzzino et al., 2015). These findings are consistent with studies in mice, where substantial overexpression of WT AR in muscle resulted in severe phenotypes (Monks et al., 2007). Because there appear to be expression-level dependent influences on phenotype severity, it may be that higher expression of WT AR protein amplifies native AR functions and contributes to toxicity (Nedelsky et al., 2010). In our model, we saw minor or no phenotypes with WT AR expression in the tissues we examined. For example, WT AR expression in neurons negatively impacted climbing ability when flies were given no treatment (Figure 2b; reduced compared to Ctrl), but not so when treated with DHT/EtOH (Figure 4b; no difference compared to Ctrl). Perhaps, treatment with EtOH alone or mixed with DHT benefited the climbing ability of flies and accounts for this discrepancy. However, lack of improvement in background control lines supplemented with DHT/EtOH does not support this possibility. Conversely, in some cases, the phenotypes seen with WT AR were improved outcomes, compared with controls (Figure 2a; longevity with muscle expression). However, these differences were unreliable and varied with age. Collectively, we conclude that overexpression of WT AR in our fly models is non-toxic.
In conclusion, we introduce a new model of SBMA in D. melanogaster. This model recapitulates key phenotypic manifestations of the disease in neuronal and muscle tissues. Because of its conceptual design, the model enables the generation of additional, isogenic fly lines with specific AR mutations and various polyQ repeat lengths that can be compared side by side for hypothesis-based examinations or screening efforts to help further our understanding of SBMA and the discovery of therapeutic interventions for it.
4 |. MATERIALS AND METHODS
4.1 |. Fly stocks and maintenance
All flies were aged at 25°C under controlled 50% humidity and 12-h light/dark cycle. Virgin females and males used for crosses and experiments were collected under light CO2 within 2 h of eclosion over a 72-h period. All flies used in experiments were age-matched and heterozygous for driver and responder. Stocks used for experiments were obtained as follows: GMR-Gal4 from BDSC (Bloomington Drosophila Stock Center) (Stock #8605), elav-Gal4 was generously gifted by Dr. Daniel Eberl, University of Iowa, and Mef2-Gal4 from Dr. Rolf Bodmer, Sanford-Burnham Medical Research Institute, California.
4.2 |. New fly line generation
ARQ20 and ARQ112 cDNA sequences were based on the NCBI human AR reference sequence NM_000044.2. Mutagenesis was carried out by Genscript (genscript.com) and codon-optimized for expression in Drosophila. The transgenes were subcloned into pWalium10-moe and fly lines were generated using pHiC31-dependent integration into VK00033 on chromosome 3. All resulting transformants were migrated into the w1118 background.
4.3 |. DHT treatment
Flies used for ligand experiments were collected and then divided at random into cohorts of DHT- or ethanol (EtOH) (vehicle)-treated. After being separated, DHT flies were transferred onto vials containing 80 μL of 20 mM DHT dissolved in 10 0% EtOH, while EtOH controls were transferred into vials containing 80 μL of 10 0% EtOH. DHT or EtOH was pipetted directly on top of the food and allowed to absorb for 1 h prior to use.
Fresh vials were provided three times each week for the duration of the study. The first day of DHT treatment was considered “Day One” in all DHT experiments. DHT was procured from Cayman Chemicals (cat. # 15874).
4.4 |. Rapid iterative negative geotaxis speed assay
Climbing speed was assessed in Rapid Iterative Negative Geotaxis (RING) assays using groups of 100 flies per cohort. Five vials containing 20 flies each were set up in a RING apparatus and negative geotaxis stimulus was initiated by tapping the apparatus down on the countertop twice firmly and rapidly. The height of the flies in the vials was captured by photo image 2 s after the start of climbing. Flies were longitudinally tested twice per week for 5 weeks to assess climbing ability at each time point. Raw image files obtained for each cohort were analyzed by semi-automated methods using ImageJ software to eliminate examiner bias. Briefly, a single raw image was used to develop a macro code with parameters able to binarize and assign a pixel height value for each individual fly pictured in the image. These same macro parameters were then applied to batch-process all images in the dataset under the exact same conditions, and to generate the average values. Longitudinal climbing speed is normalized to day-one climbing data within each genotype and treatment. For descriptions in further detail refer to Damschroder et al. (2018) and Sujkowski et al. (2022).
4.5 |. Longevity
Differences in survival were measured using cohorts of 100 sex- and age-matched flies per condition. Vials were inspected and changed every other day to count and remove any dead flies. Flies that escaped the vial or died for reasons other than natural aging were excluded from analysis.
4.6 |. NMJ isolation and staining
NMJ dissections and staining were modified from Sidisky and Babcock (2020). Briefly, whole flies were anesthetized in light CO2 then submerged for 30 s in 70% EtOH to remove wax coating from cuticle before being transferred to a gel-bottomed dissecting dish. The wings, legs, head, and abdomen were removed, and thoraxes were transferred to 4% paraformaldehyde for 60 min. Fixed thoraces were submerged in liquid nitrogen for 10 s, bisected with a sharp razor blade under a dissecting microscope, and transferred to ice-cold PBS. Hemithoraces were then blocked for 2 h before staining overnight with anti-22c10 primary antibody (mouse a-22c10, 1:100, DSHB). The following day, tissues were washed, and stained with fluorophore-conjugated secondary antibody and heptapeptide (a-mouse AF594, 1:20 0, ThermoFisher and phalloidin AF488, 1:500, ThermoFisher, respectively), and imaged using a confocal microscope. Each of the data points used for quantifications of NMJ complexity were provided as the average number of branch points of four individual nodes within a single muscle prep for each fly analyzed (Figure S3). Images were blinded prior to analysis.
4.7 |. Western blots
Western blotting was performed with five whole adult flies (neuronal or muscle expression) or 10 dissected fly heads (eye expression) per sample. Each sample was physically homogenized in 95°C lysis buffer (50 mM Tris pH 6.8, 2% SDS, 10% glycerol, 10 0 mM dithiothreitol), sonicated, boiled for 10 min, and then centrifuged for 10 min at room temperature at 13,300 rpm. Homogenized samples were electrophoresed through 4%–20% Tris/Glycine gels (Bio-Rad). ChemiDoc (Bio-Rad) was used to image Western blots, which were then quantified with ImageLab (Bio-Rad).
Direct Blue staining of the total protein signal was used as loading control. Direct blue staining was performed as follows: PVDF membranes were incubated for 10 min in .008% Direct Blue 71 (Sigma-Aldrich) in 40% EtOH and 10% acetic acid, rinsed with 40% EtOH/10% acetic acid, air dried, and then imaged.
4.8 |. Eye scoring
Eye scores were represented in a scoring system, with higher numbers indicating worsening phenotypes characterized as follows:
(1) Normal (wild-type-looking) eye with a clear pseudopupil; (2) faded pseudopupil that has begun to lose its clarity; (3) no visible pseudopupil; (4) color variegation/depigmentation at the edge of the eye in addition to pseudopupil loss; and (5) depigmentation throughout the eye in addition to pseudopupil loss.
4.9 |. Statistics
Prism 9 (GraphPad) was used for graphics and statistical analyses.
Statistical analyses used are noted in the figure legends.
Supplementary Material
Statistical information related to main figures and supplemental data.
Significance.
We describe the generation and characterization of a new fruit fly (Drosophila melanogaster) model of the incurable polyglutamine-dependent disease, spinal and bulbar muscular atrophy (SBMA), also known as Kennedy’s disease. SBMA is caused by abnormal expansion in the polyglutamine repeat of the androgen receptor (AR). These new flies express in an inducible manner the full-length, human AR protein with a normal or pathogenic polyglutamine; they will be useful to the SBMA research community to further model this disease and to find therapeutic options for it.
FUNDING INFORMATION
This study was supported by a Thomas Rumble Fellowship from Wayne State University Graduate School (KR), T32 HL120822 (KR), T34 GM140932 (ALH), Waite-Griffin Memorial Fellowship (AS), R01 NS108114 (DEM), R01 AG059683 (R JW), R21 NS121276 (R JW, SV T), and R01 NS086778 (SV T).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
Edited by Cristina Antonella Ghiani and Barrington Burnett. Reviewed by Maria Pennuto, Carlo Rinaldi and Christopher Grunseich.
DATA AVAILABILITY STATEMENT
Data are available upon request to the corresponding author.
REFERENCES
- Ambegaokar SS, Roy B, & Jackson GR (2010). Neurodegenerative models in Drosophila: Polyglutamine disorders, Parkinson disease, and amyotrophic lateral sclerosis. Neurobiology of Disease, 40(1), 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold FJ, Pluciennik A, & Merry DE (2019). Impaired nuclear export of polyglutamine-expanded androgen receptor in spinal and bulbar muscular atrophy. Scientific Reports, 9(1), 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsuta N, Watanabe H, Ito M, Banno H, Suzuki K, Katsuno M, Tanaka F, Tamakoshi A, & Sobue G (2006). Natural history of spinal and bulbar muscular atrophy (SBMA): A study of 223 Japanese patients. Brain, 129(Pt 6), 1446–1455. [DOI] [PubMed] [Google Scholar]
- Badders NM, Korff A, Miranda HC, Vuppala PK, Smith RB, Winborn BJ, Quemin ER, Sopher BL, Dearman J, Messing J, Kim NC, Moore J, Freibaum BD, Kanagaraj AP, Fan B, Tillman H, Chen PC, Wang Y, Freeman BB III, … Taylor JP (2018). Selective modulation of the androgen receptor AF2 domain rescues degeneration in spinal bulbar muscular atrophy. Nature Medicine, 24(4), 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel LK, Alvarado C, Mokhtar S, Paliouras M, & Trifiro M (2013). Mechanisms mediating spinal and bulbar muscular atrophy: Investigations into polyglutamine-expanded androgen receptor function and dysfunction. Frontiers in Neurology, 4, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Bott LC, Fischbeck KH, & Wrange Ö (2015). The polyglutamine-expanded androgen receptor has increased DNA binding and reduced transcriptional activity. Biochemistry and Biophysics Reports, 3, 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolus H, Crocker K, Boekhoff-Falk G, & Chtarbanova S (2020). Modeling neurodegenerative disorders in Drosophila melanogaster. International Journal of Molecular Sciences, 21(9), 3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott LC, Badders NM, Chen KL, Harmison GG, Bautista E, Shih CCY, Katsuno M, Sobue G, Taylor JP, Dantuma NP, Fischbeck KH, & Rinaldi C (2016). A small-molecule Nrf1 and Nrf2 activator mitigates polyglutamine toxicity in spinal and bulbar muscular atrophy. Human Molecular Genetics, 25(10), 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, & Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118(2), 401–415. [DOI] [PubMed] [Google Scholar]
- Breza M, & Koutsis G (2019). Kennedy’s disease (spinal and bulbar muscular atrophy): A clinically oriented review of a rare disease. Journal of Neurology, 266(3), 565–573. [DOI] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, & White K (1987). Molecular analysis of the locus elav in Drosophila melanogaster: A gene whose embryonic expression is neural specific. The EMBO Journal, 6(2), 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, & Brand AH (2016). The GAL4 system: A versatile system for the manipulation and analysis of gene expression. Methods in Molecular Biology, 1478, 33–52. [DOI] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Andriola I, Merry D, & Bonini NM (2002). Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Human Molecular Genetics, 11(23), 2895–2904. [DOI] [PubMed] [Google Scholar]
- Cortes CJ, & La Spada AR (2018). X-linked spinal and bulbar muscular atrophy: From clinical genetic features and molecular pathology to mechanisms underlying disease toxicity. Advances in Experimental Medicine and Biology, 1049, 103–133. [DOI] [PubMed] [Google Scholar]
- Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, Ly L, Tokunaga S, Lopez E, Sopher BL, Bennett CF, Shelton GD, Cleveland DW, & la Spada AR (2014). Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron, 82(2), 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damschroder D, Cobb T, Sujkowski A, & Wessells R (2018). Drosophila endurance training and assessment of its effects on systemic adaptations. Bio-Protocol, 8(19), e3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejager S, Bry-Gauillard H, Bruckert E, Eymard B, Salachas F, LeGuern E, Tardieu S, Chadarevian R, Giral P, & Turpin G (2002). A comprehensive endocrine description of Kennedy’s disease revealing androgen insensitivity linked to CAG repeat length. The Journal of Clinical Endocrinology and Metabolism, 87(8), 3893–3901. [DOI] [PubMed] [Google Scholar]
- Feng X, Cheng XT, Zheng P, Li Y, Hakim J, Zhang SQ, Anderson SM, Linask K, Prestil R, Zou J, Sheng ZH, & Blackstone C (2022). Ligand-free mitochondria-localized mutant AR-induced cytotoxicity in spinal bulbar muscular atrophy. Brain, 146, 278–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figura G, Koscianska E, & Krzyzosiak WJ (2015). In vitro expansion of CAG, CAA, and mixed CAG/CAA repeats. International Journal of Molecular Sciences, 16(8), 18741–18751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francini-Pesenti F, Vitturi N, Tresso S, & Sorarù G (2020). Metabolic alterations in spinal and bulbar muscular atrophy. Revue Neurologique (Paris), 176(10), 780–787. [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Shatkina L, Mink S, Weis Q, Weg-Remers S, & Cato ACB (2009). Specific N-terminal mutations in the human androgen receptor induce cytotoxicity. Neurobiology of Aging, 30(11), 1851–1864. [DOI] [PubMed] [Google Scholar]
- Gregory CW, Johnson RT Jr., Mohler JL, French FS, & Wilson EM (2001). Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Research, 61(7), 2892–2898. [PubMed] [Google Scholar]
- Guber RD, Kokkinis AD, Schindler AB, Bendixen RM, Heatwole CR, Fischbeck KH, & Grunseich C (2018). Patient-identified impact of symptoms in spinal and bulbar muscular atrophy. Muscle & Nerve, 57(1), 40–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, & Coetzee GA (2000). Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Human Molecular Genetics, 9(2), 267–274. [DOI] [PubMed] [Google Scholar]
- Jochum T, Ritz ME, Schuster C, Funderburk SF, Jehle K, Schmitz K, Brinkmann F, Hirtz M, Moss D, & Cato ACB (2012). Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochimica et Biophysica Acta, 1822(6), 1070–1078. [DOI] [PubMed] [Google Scholar]
- Johansen JA, Yu Z, Mo K, Monks DA, Lieberman AP, Breedlove SM, & Jordan CL (2009). Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiology of Disease, 34(1), 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Libohova K, Blount JR, Sujkowski AL, Prifti MV, Tsou WL, & Todi SV (2021). Targeting the VCP-binding motif of ataxin-3 improves phenotypes in Drosophila models of spinocerebellar ataxia type 3. Neurobiology of Disease, 160, 105516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Ranxhi B, Libohova K, Tsou WL, & Todi SV (2020). Ubiquitin-interacting motifs of ataxin-3 regulate its polyglutamine toxicity through Hsc70–4-dependent aggregation. eLife, 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Tsou WL, Prifti MV, Harris AL, & Todi SV (2022). A survey of protein interactions and posttranslational modifications that influence the polyglutamine diseases. Frontiers in Molecular Neuroscience, 15, 974167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, & Sobue G (2002). Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron, 35(5), 843–854. [DOI] [PubMed] [Google Scholar]
- Kemppainen JA, Lane MV, Sar M, & Wilson EM (1992). Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. The Journal of Biological Chemistry, 267(2), 968–974. [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, & Fischbeck KH (1991). Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature, 352(6330), 77–79. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Harmison G, Strand AD, Olson JM, & Fischbeck KH (2002). Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Human Molecular Genetics, 11(17), 1967–1976. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Shakkottai VG, & Albin RL (2019). Polyglutamine repeats in neurodegenerative diseases. Annual Review of Pathology, 14, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, Yu XX, Cortes CJ, Bennett CF, Monia BP, la Spada AR, & Hung G (2014). Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Reports, 7(3), 774–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, & Olson EN (1995). Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science, 267(5198), 688–693. [DOI] [PubMed] [Google Scholar]
- Lombardi V, Querin G, Ziff OJ, Zampedri L, Martinelli I, Heller C, Foiani M, Bertolin C, Lu CH, Malik B, Allen K, Rinaldi C, Zetterberg H, Heslegrave A, Greensmith L, Hanna M, Soraru G, Malaspina A, & Fratta P (2019). Muscle and not neuronal biomarkers correlate with severity in spinal and bulbar muscular atrophy. Neurology, 92(11), e1205–e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik I, Kelley CP, Wang ET, & Todd PK (2021). Molecular mechanisms underlying nucleotide repeat expansion disorders. Nature Reviews. Molecular Cell Biology, 22(9), 589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano R, Sorarú G, Grunseich C, Fratta P, Zuccaro E, Pennuto M, & Rinaldi C (2018). Beyond motor neurons: Expanding the clinical spectrum in Kennedy’s disease. Journal of Neurology, Neurosurgery, and Psychiatry, 89(8), 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotsky E, Liu Y, Lieberman AP, & Merry DE (2022). Neuromuscular junction pathology is correlated with differential motor unit vulnerability in spinal and bulbar muscular atrophy. Acta Neuropathologica Communications, 10(1), 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, Yu Z, Lieberman AP, Breedlove SM, & Jordan CL (2007). Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proceedings of the National Academy of Sciences of the United States of America, 104(46), 18259–18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montie HL, Cho MS, Holder L, Liu Y, Tsvetkov AS, Finkbeiner S, & Merry DE (2009). Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Human Molecular Genetics, 18(11), 1937–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Kimura F, Nakagawa T, Furutama D, Shinoda K, Shimizu A, & Ohsawa N (1996). Transcriptional activation by the androgen receptor in X-linked spinal and bulbar muscular atrophy. Journal of the Neurological Sciences, 142(1–2), 12–16. [DOI] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, & Taylor JP (2010). Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron, 67(6), 936–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. (1999). A unified nomenclature system for the nuclear receptor superfamily. Cell, 97(2), 161–163. [DOI] [PubMed] [Google Scholar]
- Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, Sobue G, Taylor JP, Sumner CJ, Fischbeck KH, & Pennuto M (2009). Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron, 63(3), 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, & Taylor JP (2007). HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature, 447(7146), 859–863. [DOI] [PubMed] [Google Scholar]
- Pluciennik A, Liu Y, Molotsky E, Marsh GB, Ranxhi B, Arnold FJ, St.-Cyr S, Davidson B, Pourshafie N, Lieberman AP, Gu W, Todi SV, & Merry DE (2021). Deubiquitinase USP7 contributes to the pathogenicity of spinal and bulbar muscular atrophy. The Journal of Clinical Investigation, 131(1), e134565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poort JE, Rheuben MB, Breedlove SM, & Jordan CL (2016). Neuromuscular junctions are pathological but not denervated in two mouse models of spinal bulbar muscular atrophy. Human Molecular Genetics, 25(17), 3768–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Elliott DA, Harvey RP, & Olson EN (1998). Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development, 125(16), 3037–3048. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, & Schulz RA (1995). A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Developmental Biology, 171(1), 169–181. [DOI] [PubMed] [Google Scholar]
- Rhodes LE, Freeman BK, Auh S, Kokkinis AD, la Pean A, Chen C, Lehky TJ, Shrader JA, Levy EW, Harris-Love M, di Prospero NA, & Fischbeck KH (2009). Clinical features of spinal and bulbar muscular atrophy. Brain, 132(Pt 12), 3242–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, & White K (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. Journal of Neurobiology, 22(5), 443–461. [DOI] [PubMed] [Google Scholar]
- Rosenbohm A, Hirsch S, Volk AE, Grehl T, Grosskreutz J, Hanisch F, Herrmann A, Kollewe K, Kress W, Meyer T, Petri S, Prudlo J, Wessig C, Müller HP, Dreyhaupt J, Weishaupt J, Kubisch C, Kassubek J, Weydt P, & Ludolph AC (2018). The metabolic and endocrine characteristics in spinal and bulbar muscular atrophy. Journal of Neurology, 265(5), 1026–1036. [DOI] [PubMed] [Google Scholar]
- Scaramuzzino C, Casci I, Parodi S, Lievens PMJ, Polanco MJ, Milioto C, Chivet M, Monaghan J, Mishra A, Badders N, Aggarwal T, Grunseich C, Sambataro F, Basso M, Fackelmayer FO, Taylor JP, Pandey UB, & Pennuto M (2015). Protein arginine methyltransferase 6 enhances polyglutamine- expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron, 85(1), 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M, Pluciennik A, & Merry DE (2022). The role of ubiquitination in spinal and bulbar muscular atrophy. Frontiers in Molecular Neuroscience, 15, 1020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard RL, Spangenburg EE, Chin ER, & Roth SM (2011). Androgen receptor polyglutamine repeat length affects receptor activity and C2C12 cell development. Physiological Genomics, 43(20), 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, & Bonini NM (2011). Genes and pathways affected by CAG-repeat RNA-based toxicity in Drosophila. Human Molecular Genetics, 20(24), 4810–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidisky JM, & Babcock DT (2020). Visualizing synaptic degeneration in adult Drosophila in association with neurodegeneration. Journal of Visualized Experiments, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, de Mezer M, Michlewski G, Krol J, & Krzyzosiak WJ (2003). RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Research, 31(19), 5469–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, & Krzyzosiak WJ (2005). CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. The Journal of Biological Chemistry, 280(5), 3898–3910. [DOI] [PubMed] [Google Scholar]
- Sobue G, Hashizume Y, Mukai E, Hirayama M, Mitsuma T, & Takahashi A (1989). X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain, 112(Pt 1), 209–232. [DOI] [PubMed] [Google Scholar]
- Sujkowski A, Richardson K, Prifti MV, Wessells R, & Todi SV (2022). Endurance exercise ameliorates phenotypes in Drosophila models of spinocerebellar ataxias. eLife, 11, e75389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, & Kato S (2002). Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron, 35(5), 855–864. [DOI] [PubMed] [Google Scholar]
- Walcott JL, & Merry DE (2002). Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. The Journal of Biological Chemistry, 277(52), 50855–50859. [DOI] [PubMed] [Google Scholar]
- Xu Y, Halievski K, Henley C, Atchison WD, Katsuno M, Adachi H, Sobue G, Breedlove SM, & Jordan CL (2016). Defects in neuromuscular transmission may underlie motor dysfunction in spinal and bulbar muscular atrophy. The Journal of Neuroscience, 36(18), 5094–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Halievski K, Katsuno M, Adachi H, Sobue G, Breedlove SM, & Jordan CL (2018). Pre-clinical symptoms of SBMA may not be androgen-dependent: Implications from two SBMA mouse models. Human Molecular Genetics, 27(14), 2425–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, Robins DM, & Lieberman AP (2006). Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. The Journal of Clinical Investigation, 116(10), 2663–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Lane MV, Kemppainen JA, French FS, & Wilson EM (1995). Specificity of ligand-dependent androgen receptor stabilization: Receptor domain interactions influence ligand dissociation and receptor stability. Molecular Endocrinology, 9(2), 208–218. [DOI] [PubMed] [Google Scholar]
- Zu T, Pattamatta A, & Ranum LPW (2018). Repeat-associated non-ATG translation in neurological diseases. Cold Spring Harbor Perspectives in Biology, 10(12), a033019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical information related to main figures and supplemental data.
Data Availability Statement
Data are available upon request to the corresponding author.


