Abstract
Rous sarcoma virus (RSV), a simple retrovirus, needs to export unspliced viral RNA from the nucleus to the cytoplasm, circumventing the host cell restriction on cytoplasmic expression of intron-containing RNA. The cytoplasmic accumulation of full-length viral RNA is promoted by two cis-acting direct repeat (DR) elements that flank the src gene; at least one copy of the DR sequence is necessary for viral replication. We show here that the DR mediates export of a reporter construct from the nucleus, suggesting it is a constitutive transport element (CTE). In contrast, human immunodeficiency virus type 1 (HIV-1) and other complex retroviruses encode accessory proteins, Rev or Rex, which promote export of incompletely spliced viral transcripts. This RNA export pathway is CRM1 dependent and can be blocked by the cytotoxic agent leptomycin B. We show here that DR-mediated export is CRM1 independent, suggesting that RSV uses a different export pathway from that of HIV-1 and other complex retroviruses. The simian retroviruses have a CTE which interacts with the cellular Tap export protein. However, we were unable to detect binding of the RSV DR RNA to Tap, suggesting it may use a different export pathway from that of the simian retroviruses. These data suggest that the RSV DR element uses a novel nucleocytoplasmic export pathway.
A defining feature of eukaryotic cells is their division into nucleoplasm and cytoplasm. One consequence of the nuclear membrane is the partitioning of unspliced pre-mRNA away from the cytoplasm where spliced mRNA is translated. Cellular mRNAs are exported from the nucleus only after processing is completed (10, 26, 27). Retroviruses use cellular factors for RNA transcription, capping, splicing, and polyadenylation; however, they require unspliced RNA to be present in the cytoplasm both for translation of gag and pol genes and for packaging into virions (11). The export of these incompletely processed RNAs is dependent on cis-acting elements within the RNA (reviewed in references 3, 12, and 21).
The complex retroviruses encode accessory proteins that interact with viral cis-acting RNA elements. The human immunodeficiency virus type 1 (HIV-1) protein Rev binds directly to the Rev response element (RRE) within the env gene to accumulate a cytoplasmic population of unspliced and partially spliced RNA species (reviewed in reference 12). Xenopus oocyte injection experiments have shown that Rev mediates export of RRE-containing RNA and that it can inhibit export of cellular 5S rRNA and spliceosomal U snRNA (15). Rev contains a nuclear export signal (15) that binds the cellular protein, chromosomal region maintenance protein 1 (CRM1), or exportin-1, which is linked to the GTP-Ran transport cofactor (16). CRM1 binds to nucleoporins, including Nup214/Can, thus targeting the complex to the nuclear pore (17). The cytotoxin leptomycin B (LMB) inhibits Rev-mediated export by binding CRM1 and preventing assembly of the Rev-CRM1-Ran-GTP complex (16, 25, 44). Other complex retroviruses, including human T-cell leukemia virus type 1 (HTLV-1), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), and the human endogenous retrovirus HTDV/HERV-K also encode nuclear export signal (NES)-containing proteins that interact with CRM1, and their export is blocked by LMB (7, 28, 35).
In contrast, simple retroviruses do not encode accessory proteins; they rely upon the interaction of cellular factors with their cis-acting elements for export of their unspliced RNA species (reviewed in references 3, 12, and 21). The simian type D retroviruses Mason-Pfizer monkey virus (MPMV) and SRV-1 contain a cis-acting constitutive transport element (CTE) that is necessary for unspliced RNA export (9, 14, 46). The CTE appears to form a stem-loop structure which is highly conserved (13, 42). CTE-containing intron lariats are exported from Xenopus oocyte nuclei and can compete for export with some cellular mRNAs (36, 37). The export of MPMV RNA is LMB independent (35), suggesting it does not use the CRM1 export pathway. Recently, the hepatitis B virus posttranscriptional regulatory element has also been shown to be resistant to LMB treatment (35); however, its cofactors have not been identified.
Several different cellular cofactors have been proposed for the closely related MPMV and SRV-1 CTEs. These CTEs bind the cellular protein Tap specifically in vitro, and this interaction promotes CTE-mediated export in Xenopus oocytes and in transfected quail cells (6, 20, 24). The C-terminal domain of Tap interacts with the nuclear pore complex (2). Interestingly, Tap is homologous to Mex67p, an mRNA export factor in yeast (38). RNA helicase A (RHA) has also been reported to bind specifically to the MPMV CTE and to colocalize with it in infected cells (43).
The Rous sarcoma virus (RSV) direct repeat (DR) sequences each contain approximately 100 nucleotides (nt) and flank the src gene in RSV. In the Prague C (PrC) strain of RSV, these two sequences are located at nt 6897 and nt 8791 and have been termed DR1 and DR2, respectively (33). One copy of either of these DR sequences has been shown to be sufficient for cytoplasmic RNA accumulation (1, 33). The closely related avian leukosis virus (ALV) has a single copy of this sequence in the 3′ untranslated region of the genome (45). Mutations within this element have been shown in our laboratory to suppress viral replication and to markedly reduce the levels of full-length viral RNA in the cytoplasm (32, 33). We have also observed a slightly decreased half-life of unspliced viral RNAs lacking both DR elements (33). However, others have seen a smaller decrease in cytoplasmic RNA levels with their DR mutants and have proposed that a major function of the DR element involves packaging of virion RNA (1, 41). Impairment of Gag protein processing was also observed with DR mutants, leading to the proposal that the DR has a role in virion assembly, possibly by affecting the cytoplasmic localization of the RNA (31, 39, 40).
In an attempt to resolve the controversy over whether or not the DR element functions as a CTE, we chose to assay it in a nonviral context. This removes complications due to different rates of viral replication, as well as possible effects of the DR element on packaging of RNA into virions or on Gag protein processing. We also wanted to be able to assay the DR activities in various cell types, irrespective of the ability of RSV to replicate in them. Several different laboratories have used different methods of cell fractionation, perhaps contributing to differences in determination of the relative levels of cytoplasmic viral RNA (1, 33, 40, 41). To resolve this, we wanted to test the possible role of the DR as a CTE, by using a method that did not require cell fractionation. We chose to use the pCMV128 chloramphenicol acetyltransferase (CAT) reporter construct, because it has been widely used to assay RNA export in other systems (22, 23, 30, 43). Using this reporter construct, we have observed that the DR can mediate HIV-1-CAT unspliced mRNA export. Furthermore, DR mutations that inhibit viral replication (32) similarly inhibited export of unspliced CAT mRNA. These experiments suggest that at least one function of the DR element is that of a CTE.
We next asked whether the RSV DR element uses the same export pathway as either the HIV-1 Rev-RRE complex or the SRV CTE. We report that LMB, an inhibitor of the CRM1 pathway for RNA export used by complex retroviruses, did not inhibit DR-mediated export of a CAT reporter construct. Furthermore, we were unable to detect binding of Tap to the RSV DR RNA in a gel shift assay, suggesting its export pathway may differ from that of the SRV CTE. Finally, we observed some activity of the DR element in mammalian as well as in avian cells.
MATERIALS AND METHODS
Plasmids.
pCMV128 and pCMVRev were obtained from Tom Hope. pCMV128 was derived from pDM128 (22) by replacement of the simian virus 40 (SV40) promoter with the CMV promoter (30). The DR elements were inserted at a unique BamHI site directly downstream of the CAT gene in pCMV128 (Fig. 1A). The DR fragments inserted into pCMV128 were generated by PCR methods. The primers used for the generation of DR2 and the DR2 point mutants were 5′ 8770 GAGTGAGGATCCGCGTCCTGCGTTGCTCCG and 3′ 8925 GAGTGAGGATCCCAGGGAAGACGCCATCAT. These primers were used with viral DNA templates pPrC (wild-type Pr-C RSV) (33), pPrC-G8844C, and pPrC-G8863C (32). The DR1 construct was generated with primers 5′ 6863 GAGTGAGGATCCTAGAGCTCAGTTATAATA and 3′ 7037 AGTGAGGATCCATATTAAGACTACATTTTT and the pPrC template.
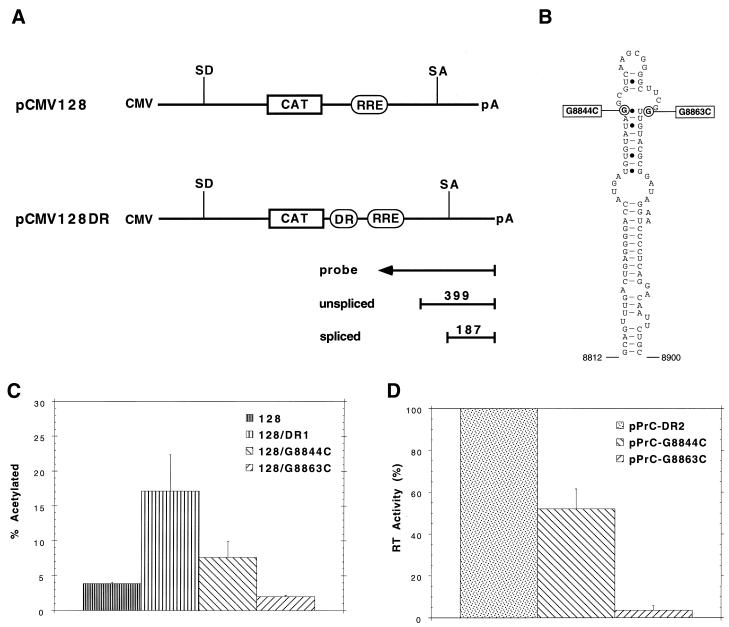
FIG. 1.
RSV DR is a CTE. (A) Schematic diagrams of the pCMV128 CAT reporter constructs, depicting the location of the DR element insertions. These constructs are derived from the HIV-1 env gene (22, 30). Since cat expression is dependent on unspliced RNA translation in the cytoplasm, the inserted elements are tested for their ability to promote RNA export. Because this construct contains the HIV-1 RRE, a construct expressing the HIV-1 Rev protein can be cotransfected with pCMV128 as a positive control. An antisense riboprobe used in RNase protection assays spans the 3′ splice site as shown. This probe protects 399 nt of the unspliced RNA and 187 nt of the spliced RNA. SD, splice donor; SA, splice acceptor. (B) The predicted secondary structure of the RSV DR2 element. This structure was generated by using the MFOLD computer program (47) with the constraint that nt 8852 be single stranded, as demonstrated by nuclease probing. The locations of DR2 point mutations used in this study are shown. (C) Point mutations in the DR element decrease RNA export and viral replication. pCMV128 constructs bearing either wild-type or mutated DR elements were transfected into CEFs, and CAT activity was measured 48 h later. Note that the parental pCMV128 construct reproducibly yields more CAT activity than the G8863C mutant. (D) RT activity of full-length viral constructs bearing a single copy of wild-type or mutant DR elements was determined 48 h after transfection of CEFs.
The viral constructs pPrC-DR2 and mutants pPrC-G8863C and pPrC-G8844C were described previously (32, 33). They contain the full-length nonpermuted genome of PrC RSV, but lack src and the upstream DR1 sequence. For in vitro binding studies, DR2 (nt 8770 to 8925) was cloned into pBluescript KS+ (Stratagene) at the 5′ SalI and 3′ BamHI sites. The SRV-1 CTE and mutant CTE were described previously (37).
Cell culture and DNA transfection.
Secondary chicken embryo fibroblasts (CEFs) were cultured in medium 199 containing 2% tryptose phosphate, 1% calf serum, 1% heat-inactivated chick serum, and 1% antibiotic-antimycotic (all from Life Technologies). For CAT assays, a 6-cm-diameter dish of cells was cotransfected with 0.5 μg of the pCMV128 reporter construct and 0.25 μg of pGL3-control luciferase reporter construct (Promega) in medium 199 containing 200 μg of DEAE-dextran per ml. After 4 h, cells were subjected to a 10% dimethyl sulfoxide shock for 2 min; they were harvested 48 h later. For the LMB experiments, medium was removed 24 h after transfection and replaced with medium containing either 5 or 10 nM LMB (25), a kind gift from Minoru Yoshida. Cells were harvested 18 h after LMB addition. For preparation of nuclear and cytoplasmic RNA, 15-cm-diameter plates were transfected with 10 μg of pCMV128 constructs.
HeLa cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal calf serum and 1% antibiotic-antimycotic (all from Life Technologies). Cells were transfected with 7.0 μg of plasmid DNA per 10-cm-diameter dish by using Lipofectamine (Life Technologies) for 5 h. This mixture was removed and overlaid with fresh medium, and cells were harvested 48 h later.
CAT assays.
CAT activity was measured by the method of Gorman et al. (19). Acetylated and nonacetylated [14C]chloramphenicol (Amersham) products were resolved by thin-layer chromatography and quantified with an InstantImager (Packard). Luciferase assays to measure expression of the cotransfected pGL3-control plasmid were carried out on a Berthold luminometer. All CAT data shown are the average of at least three separate transfections and are normalized relative to the luciferase control.
RT assay.
A reverse transcriptase (RT) assay was used to monitor viral replication as described previously (33). Cells were transfected with full-length viral constructs. Forty-eight hours later, medium was harvested from the cells. Viral particles were concentrated from 2 ml of medium by centrifugation for 2 h at 14,000 × g in a refrigerated centrifuge (Savant). The pellet was resuspended in RT buffer for assay.
RNA isolation and RNase protection assays.
Total cellular RNA was harvested by using RNAzol B (Tel-Test) 48 h after transfection. Transcription of antisense riboprobes and RNase protection analysis were performed as previously described (4). The 128 riboprobe construct, which spans the 3′ splice site, was a gift from Gordon Carmichael (23). The DNA template was linearized with EcoRI and transcribed with T3 RNA polymerase (Life Technologies). The riboprobe was hybridized to nuclear or cytoplasmic RNA fractions for 16 h at 55°C. RNase digestions utilized 10 μg of RNase A and 10 U of RNase T1 per ml (both from Calbiochem) and were carried out at 30°C for 1 h. Electrophoresis of protected RNA was on a 6% polyacrylamide gel containing 8 M urea, and quantitation of protected bands was carried out on an InstantImager (Packard).
Cell fractionation.
Cytoplasmic and nuclear RNAs were isolated by a citric acid cell fractionation protocol described previously (5, 33). This is a stringent cell fractionation method that strips the outer nuclear membrane. As a control for the fractionation procedure, an aliquot of RNA from each fraction was electrophoresed on a 1.3% agarose gel in MOPS (morpholinepropanesulfonic acid) buffer (0.04 M MOPS, 10 mM sodium acetate, 1 mM EDTA [pH 7.0]) for 30 min at 100 V. Prior to loading, RNA samples were precipitated, resuspended in 2 μl of water, and mixed with 10 μl of loading buffer (1× MOPS, 50% formamide, 1/5 volume of 37% formaldehyde, and 5 μg of ethidium bromide). The samples were then heated for 10 min at 65°C and placed on ice prior to loading. This method allowed visualization of cellular rRNA and pre-rRNA, as well as tRNA and snRNA, in the subcellular fractions.
Electrophoretic mobility shift assay.
Tap binding assays were performed with in vitro-transcribed 32P-labeled RSV DR2 RNA and SRV-1 CTE RNA. Tap protein was synthesized in a rabbit reticulocyte lysate, and the gel shift assays were carried out as described by Braun et al. (8).
RESULTS
The RSV DR sequence promotes nucleocytoplasmic RNA export.
We showed previously that the mutation or deletion of both copies of the DR sequence in full-length viral constructs results in the inhibition of viral replication and a decrease in unspliced viral RNA accumulation in the cytoplasm (32, 33). However, we did not see a buildup of unspliced RNA in the nucleus with these viral mutants, so it was not clear whether the DR sequence promoted RNA export or stabilization (nuclear or cytoplasmic) or both. Our working hypothesis is that the DR mutants inhibit export of the viral unspliced RNA, leading to its splicing or degradation in the nucleus and reduced cytoplasmic accumulation. However, other investigators have proposed that the major role of the DR in viral replication involves packaging of virion RNA (1, 41) or virion assembly (39, 40). Since the assays for RNA export were based on quantitation of RNA levels in the nucleus and the cytoplasm after cell fractionation, it is possible that the differences observed are due to different methods of cell fractionation.
Thus, in this study, we used an assay that was independent of viral replication and also did not require cell fractionation to ask whether the block in viral replication observed with DR point mutations (32) correlates with a block in RNA export. The pCMV128 reporter construct was developed by Hope and colleagues to study nucleocytoplasmic RNA export (30). This construct differs from pDM128 (22) by replacement of the SV40 promoter with the CMV promoter (Fig. 1A). Like pDM128, it contains the CAT gene in the tat-rev intron of the HIV-1 env gene. We left the RRE intact because we found that its removal increased background levels of CAT expression in some cell types (data not shown). Since the tat-rev intron is inefficiently spliced due to suboptimal HIV-1 splice sites (34), both unspliced and spliced RNAs are generated in the nucleus. CAT protein synthesis requires the unspliced cat mRNA to be exported to the cytoplasm. This assay has been used to measure Rev-RRE-mediated RNA export (22), as well as MPMV CTE export (43). The DR1 and DR2 wild-type sequences and flanking sequences (RSV nt 6863 to 7037 and 8770 to 8925, respectively) and two DR2 point mutants were cloned into the pCMV128 construct downstream of the CAT gene as shown in Fig. 1A.
A computer-predicted secondary structure model of the Prague subgroup C (Pr-C) strain RSV DR2 element is shown in Fig. 1B. Nuclease probing data (not shown) were consistent with this structure after nt 8852G in the hairpin loop was constrained to be single stranded. The upper region of the lower stem is highly conserved between different avian retroviruses (33) and is similar to that predicted for the Pr-C DR1 structure (1) and the ALV CTE (45). This lower stem structure is also supported by phylogenetic variation. For example, a G-C base pair between PrC nt 8823 and 8887 is conserved in most DR sequences, but is replaced by an A-U base pair in the Schmidt-Ruppin (SR) A strain DR1 element and by a G-U base pair in SR-A DR2 (according to the alignment of Ogert et al. [33]). Nevertheless, substitution mutations in the lower stem had relatively minor effects on viral replication; substitution of 4 G's beginning at nt 8825 with 4 U's resulted in only about a 25% decrease in viral replication (32).
Although DR1 and DR2 are functionally interchangeable in viral constructs and have 82% sequence identity (33), it has proven difficult to predict a common structure for the central region (DR2 nt 8830 to 8880) located above the lower stem. In fact, the predicted structures for DR1 (1), DR2 (Fig. 1B), and the ALV CTE (45) exhibit significant differences in this region. Thus, if there is a common structure, it may have significant non-Watson-Crick base pairing. Nevertheless, the hairpin loop determined for the ALV CTE (45) is similar to that of our DR2 model, but contains 3 additional nt on the 3′ side. One of the most critical functional regions of all of the DRs appears to be in the nt 8861 to 8863 region, because single point mutations are very inhibitory to DR function and viral replication (1, 32, 39, 45). In this report, we have analyzed DR2 point mutations at nt 8844 and 8863 in a pCMV128 vector and in a viral construct. The position of these mutations on opposite sides of the upper stem of the predicted secondary structure is shown in Fig. 1B.
To ask whether the RSV DR can function as a CTE, the pCMV128DR constructs were transfected into CEFs, and CAT activity was assayed 48 h later. pGL3-control luciferase constructs were cotransfected and assayed as a control for transfection efficiency. We observed a fourfold increase in CAT activity with the CMV128 construct containing the DR over that with the related construct lacking the DR (Fig. 1C). pCMV128/DR1 and pCMV128/DR2 gave identical results in this assay (data not shown). When the DR was inserted in the antisense orientation, less CAT activity was detected than with the parental pCMV128 construct alone (data not shown). Furthermore, DR mutant G8863C resulted in 12% ± 0.2% of wild-type DR activity, while mutant G8844C resulted in an intermediate level (40% ± 2.3% of wild-type activity). These results are consistent with the DR element being a CTE.
The proportional decrease in CAT activity caused by these point mutations was compared to their effects on viral replication. This activity was measured by RT assays of virus in the medium 48 h after transfection with full-length viral constructs bearing the same mutations (Fig. 1D). The pPrC-G8863C mutant yielded 4% of wild-type pPrC-DR2 levels of RT activity, whereas pPrC-G8847C resulted in 52% of wild-type levels. Thus, we observed a strong correlation between the effects of these mutations on viral RT activity and pCMV128 CAT activity. This suggested that the decrease in viral replication we observed could be explained by a decrease in cytoplasmic accumulation of RNA, presumably due to a decrease in RNA export. However, this does not rule out the possibility that the DR has additional functions in an infected cell.
DR sequence promotes cytoplasmic accumulation of unspliced CAT mRNA.
Since the CAT assay is a sensitive measure of enzymatic activity that can be affected by factors other than RNA export, we also analyzed the levels of nuclear and cytoplasmic RNAs resulting from transfection with the pCMV128 constructs. Cells were fractionated by a stringent citric acid fractionation method that strips the outer nuclear membrane (5, 33). As a control for the fractionation procedure, we electrophoresed an aliquot of the RNA from each subcellular fraction on an agarose gel and stained it with ethidium bromide (Fig. 2B). We detected 45S pre-rRNA only in the nuclear fractions and 28S and 18S rRNA only in the cytoplasmic fraction. We also detected a large amount of tRNA in the cytoplasm. Thus, the fractionation appeared to give good separation of the nuclear and cytoplasmic RNA species.
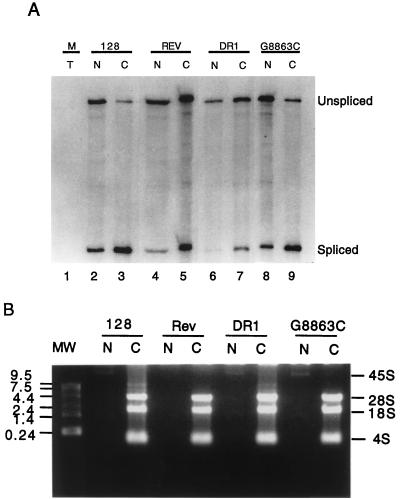
FIG. 2.
The DR element increases cytoplasmic levels of unspliced CAT RNA. CEFs were either mock transfected (M) or transfected with pCMV128 (128), pCMV128 and pCMVRev (REV), pCMV128/DR1 (DR1), and pCMV128/G8863C (G8863C). After 48 h, cells were fractionated into nuclear (N) and cytoplasmic (C) fractions by using a citric acid protocol. (A) RNase protection assays were carried out with a riboprobe complementary to the 3′ splice site as shown in Fig. 1A. The percentage of the total unspliced and spliced RNA was determined for each construct after InstantImager analysis. In this experiment, we observed the following percentage of total RNA as unspliced RNA in the cytoplasm: 128, 9%; Rev, 30%; DR1, 47%; and G8863C, 12%. (B) Control for fractionation efficiency. An aliquot of each RNA fraction was electrophoresed on an agarose gel and stained with ethidium bromide as described in Materials and Methods. 45S pre-rRNA was observed in the nucleus, and 28S and 18S rRNA and 4S tRNA were observed in the cytoplasm.
Levels of spliced and unspliced CAT RNAs were determined by an RNase protection assay, using a riboprobe spanning the 3′ splice site of pCMV128 as depicted in Fig. 1A. The results of the RNase protection assays, carried out on nuclear and cytoplasmic RNA fractions from transfected CEFs, are shown in Fig. 2A. The protected bands were quantified in an instantImager (Packard). For each construct, the relative amount of each RNA species was calculated as a percentage of the total spliced and unspliced RNAs protected, correcting for the difference in probe size protected for the two RNA species.
In this assay, we consistently saw a low level of the parental pCMV128 unspliced RNA in the cytoplasm (Fig. 2A, lane 3), as did Huang and Carmichael (23). However, there was a marked increase (four- to fivefold) in the relative amount of the unspliced cytoplasmic RNA after cotransfection with Rev (lane 5) or after insertion of the wild-type DR1 sequence (lane 7). In contrast, DR2 mutant G8863C (lane 9) showed a level of cytoplasmic unspliced RNA similar to that of the parental pCMV128 construct alone (lane 3). This increase in the levels of unspliced RNA in the cytoplasm correlated very well with the increase in CAT activity observed with the DR constructs (Fig. 1C). Thus, the quantitative RNA analysis is consistent with the DR and Rev being mediators of unspliced RNA export and/or stability.
Importantly, the RNase protection assay (Fig. 2A) showed that the majority of unspliced CAT RNA was in the nucleus with constructs lacking the DR or Rev (lanes 2 and 8). These constructs also showed much more spliced RNAs than did the DR construct. Thus, this result is supportive of the idea that the DR element is promoting export of the RNA. Furthermore, the total amounts of spliced and unspliced RNA recovered were similar for constructs with and without an export element, suggesting that differences in RNA stability may not be the cause of the increase in unspliced cytoplasmic RNA.
RSV DR is active in mammalian cells.
We had originally identified the RSV DR elements as having Rev-like activity by using an HIV-1 gag-pol-RRE expression construct to measure Gag protein expression in the absence of Rev (33). Using a Western blot assay, we detected Gag protein expression in CEFs, but not in COS cells. Conversely, we detected MPMV CTE activity in COS cells, but not in CEFs, by this assay. Since the CMV128 CAT assay is more sensitive than the HIV-1 Gag protein Western assay, we reexamined the activity of the DR in mammalian cells (Fig. 3).
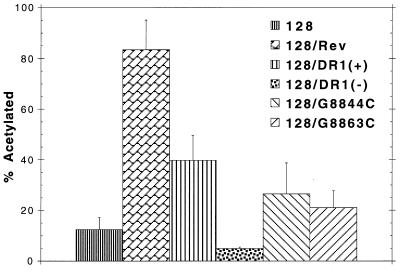
FIG. 3.
DR element promotes RNA export in HeLa cells. The following constructs were transfected into HeLa cells, and CAT activity was assayed 48 h later: pCMV128 alone (128) and cotransfected with Rev (128/Rev) and pCMV128 constructs containing either the wild-type DR1 element in the sense (+) or antisense (−) orientation or DR point mutations (128/G8844C and 128/G8863C).
In HeLa cells, we observed approximately an eightfold activation by cotransfection of pCMV128 with pCMVRev. The pCMV128/DR1 construct showed about a fourfold activation in the same cells. The DR was only active in the sense orientation. The DR point mutations at nt 8844 and 8863 both led to a decrease in CAT activity over that with the wild-type DR construct. Similar to the result in CEFs, the G8844C mutant again yielded about 50% of the activity of the wild-type construct. One difference between the behavior of these constructs in CEFs and HeLa cells is that the G8863C mutant's CAT activity was reduced relative to those of the wild type and G8844C, but it was still above background levels observed with pCMV128 alone. In contrast, this mutant was reproducibly below background levels in CEFs.
RSV DR uses a different export pathway from that of HIV-1.
Rev acts to export RRE-containing RNA through the interaction of its nuclear export signal (NES) with CRM1. CRM1 can be inhibited by LMB, rendering it unable to bind the NES of Rev (18). LMB has been shown to inhibit Rev-mediated export (16, 44). We asked whether the RSV DR used the same export pathway as Rev. To this end, the pCMV128 constructs described above were transfected into CEFs. After 24 h, various doses of LMB were added, and the cells were harvested 18 h later. We compared the effects of LMB on pCMV128 constructs cotransfected with Rev (Fig. 4A) and on the pCMV128DR constructs in the absence of Rev (Fig. 4B).
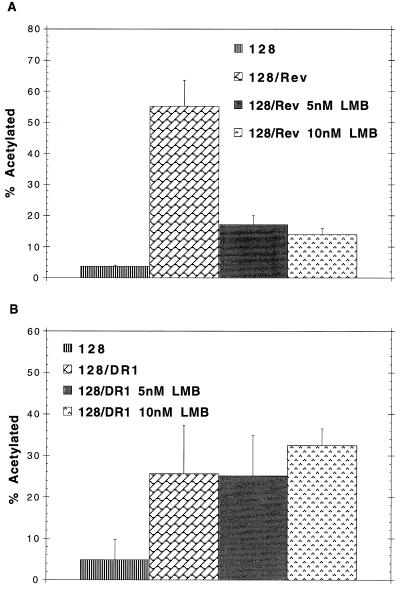
FIG. 4.
LMB does not inhibit export mediated by the DR element. (A) LMB inhibits Rev-mediated export in transfected CEFs. pCMV128 was cotransfected with pCMV Rev and incubated either without LMB or with 5 or 10 nM LMB for 18 h. (B) Effect of LMB on DR-mediated CAT activity. The pCMV128/DR1 construct was transfected into CEFs and incubated either without LMB or with 5 or 10 nM LMB for 18 h.
While the Rev-mediated CAT expression was inhibited by 5 nM LMB in CEFs (Fig. 4A), we did not see any inhibition of the DR constructs at either 5 or 10 nM LMB (Fig. 4B). In fact, a slight increase in CAT activity was observed with the higher concentration of LMB. Luciferase constructs (pGL3-control) were cotransfected and assayed as a measure of transfection efficiency and toxicity. We found that LMB was very toxic to cell lines such as 293 or 3T3 and had increased toxicity on subconfluent plates of CEF cells. Thus, the experiments shown were performed by transfection of approximately 80% confluent CEFs, where toxicity was minimal. This experiment showed that Rev-mediated export in CEFs requires interaction with CRM1 and is sensitive to LMB, just as has been seen in other cell types (25, 35). The resistance of the DR construct to LMB suggested that the DR does not use the CRM1 export pathway.
RSV DR RNA does not bind Tap directly.
As we have seen for RSV DR-mediated export (see Fig. 4B), SRV-1 or MPMPV CTE-mediated export is resistant to LMB, suggesting it does not bind CRM1 (25, 35). Since the SRV CTE has been shown to bind the cellular protein Tap and this interaction appears to be important for CTE function (6, 8, 20, 24), we asked whether the DR RNA might also bind the human Tap protein. To this end, we carried out gel shift assays comparing interactions between Tap and either the SRV-1 CTE RNA or the RSV DR RNA.
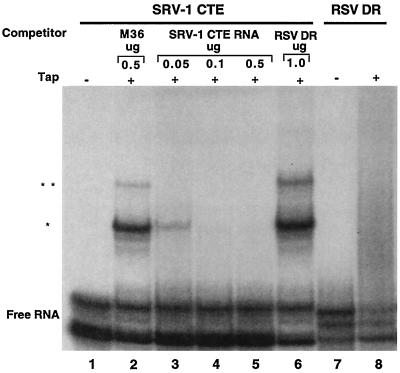
Figure 5 shows the results of an electrophoretic mobility shift assay using human Tap protein synthesized in a rabbit reticulocyte lysate. As a negative control, 32P-labeled SRV CTE RNA was incubated with reticulocyte lysate lacking Tap; this did not result in a gel shift (Fig. 5, lane 1). As a positive control, the SRV-1 CTE RNA was incubated with reticulocyte lysate containing Tap and competed with either the M36 mutant CTE RNA (lane 2) or with various amounts of wild-type SRV-1 CTE RNA (lanes 3 to 5). Two shifted bands were observed, which may be due to the binding of one or two copies of Tap per CTE RNA. While binding of 32P-labeled SRV CTE RNA to Tap was not inhibited by cold M36 RNA (lane 2), increasing amounts of wild-type SRV-1 CTE RNA effectively competed for Tap binding (lanes 3 to 5). In contrast, incubation with 1 μg of RSV DR2 RNA did not affect Tap binding (lane 6), so no competition was seen between the SRV-1 CTE and the RSV DR RNA. As a further test of a possible interaction between Tap and the DR RNA, labeled DR2 RNA was used in a gel shift assay (lane 8). No gel shift bands were observed, suggesting there is no detectable interaction between Tap and the DR RNA under these stringent conditions. This result suggests that the DR sequence may use a different export pathway from that of the SRV CTE. However, this experiment does not rule out the possibility that Tap might interact with the DR RNA under less stringent conditions or in the presence of a putative bridging factor.
FIG. 5.
CTE-Tap interactions tested by electrophoretic mobility shift assays. A gel mobility retardation assay was performed with reticulocyte lysate programmed with cDNAs encoding Tap. Lanes 1 to 6 contain the labeled SRV-1 CTE RNA, and lanes 7 and 8 contain labeled RSV DR RNA. The reaction mixtures contained the following unlabeled competitor RNAs at the concentrations shown above the lanes: M36 (lane 2), SRV-1 CTE (lanes 3 to 5), and RSV DR (lane 6). Lanes 1 and 7 are controls containing reticulocyte lysate lacking Tap protein. Lane 8 shows the RSV DR2 RNA incubated with Tap. The Tap-CTE RNA complexes are indicated on the left. The upper complex may represent two molecules of Tap bound to the CTE RNA.
Since the SRV-1 CTE mediates RNA export in Xenopus oocytes (36, 37), we have carried out similar assays with the DR element. Although the DR RNA by itself is readily exported from Xenopus nuclei after injection, it does not promote export of heterologous RNA (U. Fischer and K. L. Beemon, unpublished data). Furthermore, the DR does not compete for export with tRNA or dihydrofolate reductase mRNA (E. Izaurralde and K. L. Beemon, unpublished data). Thus, the RSV DR element appears to use a different export pathway from that of either the HIV-1 Rev-RRE complex or the SRV-1 CTE. Conversely, the MPMV or SRV-1 CTE is inactive in quail cells in the absence of exogenous Tap (24), whereas the ALV or RSV DR element is active in quail cells (45; data not shown). This also suggests that the avian DR export activity may not require Tap.
RHA has also been shown to bind to the MPMV CTE and has been proposed to play a role in the nuclear export of viral RNA (43). To ask whether RHA might be involved in DR-mediated export, we cotransfected the pCMV128DR construct with an expression plasmid for RHA. We did not observe any upregulation of CAT activity in CEFs with RHA (data not shown). However, this experiment is inconclusive, because RHA may be present in saturating amounts in these cells.
DISCUSSION
Simple retrovirus dependence on cellular factors for the unorthodox transport of incompletely spliced mRNAs makes these viruses a useful tool for the study of nucleocytoplasmic export of RNA. Previous work on retroviral RNA export has shown that overexpression of the HIV-1 Rev NES domain inhibits export of both cellular 5S rRNA and U snRNA, suggesting that these pathways share common factors (15). Furthermore, the export factor CRM1 is the cellular mediator between Rev and the nuclear pore (16, 17). Similarly, the MPMV or SRV-1 CTE utilizes an mRNA export pathway and interacts with the cellular export protein Tap (2, 6, 8, 20, 24, 36, 37). RHA has also been implicated in the SRV CTE export pathway (43).
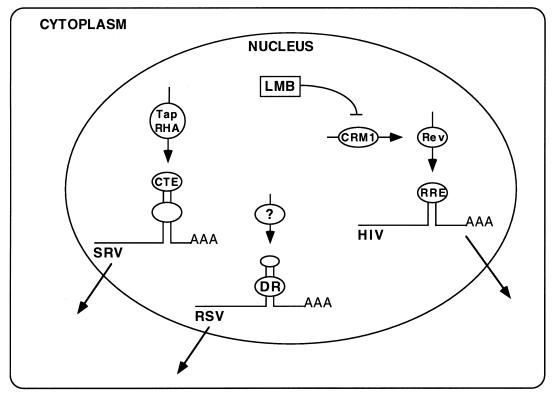
In this paper, we propose that the avian retroviral DR uses a third viral export pathway. We have found that DR-mediated export is resistant to LMB, suggesting it does not require CRM1. Thus, RSV appears to use a different RNA export pathway from that of HIV-1 and the other complex retroviruses, HTLV-1, EIAV, FIV, and HERV-K (7, 28, 35). Furthermore, we have been unable to demonstrate binding of the DR RNA to Tap in vitro. While we have not ruled out the possibility of weak or indirect interactions between Tap and the DR RNA, our data suggest that the RSV RNA export pathway differs from that of the simian type D retroviruses. We conclude that the avian DR element uses a novel export pathway, as depicted in the model in Fig. 6. It will be of great interest to determine factors involved in the export of the DR system, because this may teach us more about cellular nucleocytoplasmic export mechanisms.
FIG. 6.
Avian retroviral RNA uses a novel export pathway. A model of the nucleocytoplasmic transport pathways used by different retroviruses to export incompletely spliced viral RNAs is shown. The simian D type retroviruses, MPMV and SRV-1, have a CTE that interacts with the cellular protein Tap to transport unspliced RNA to the nuclear pore. RHA has also been implicated in this export pathway. Complex retroviruses, including HIV-1, HTLV-1, EIAV, FIV, and HERV-K, encode a viral Rev or Rev-like protein. The Rev nuclear export signal interacts with the CRM1 export protein to facilitate export of incompletely spliced RNA. The CRM1 pathway can be inhibited with LMB. Whereas RSV appears to use a different pathway from that of either HIV or SRV for the export of its unspliced RNA, the export protein or proteins for RSV remain to be identified.
We have shown here that the DR elements are active in the pCMV128 reporter system, commonly used as an assay for RNA export (22, 23, 45). Furthermore, point mutations that impair RSV replication were found to impair CAT activity to similar extents. In the reporter system, we observed an accumulation of unspliced RNA in the nucleus with DR mutants, further supporting its involvement in RNA export. This suggests that one function of the DR element is the promotion of unspliced RNA export. However, we have previously shown that full-length RNAs lacking both DR elements have about a twofold shorter half-life than wild-type RNAs (34). Thus, a role for stabilization of RSV unspliced RNA by the presence of the DR elements is also likely. The instability observed in the absence of both DR elements may be a result of a block in export, because similar results were observed with HIV-1 when Rev was mutated (29). In addition, others have proposed a role for the DR in RNA packaging (1, 41) and cytoplasmic localization of the RNA, which may be related to virion assembly (31, 39, 40). Thus, it seems likely that this element may play more than one role in the viral life cycle. In fact, if there is a coupling between RNA nuclear export and its cytoplasmic localization, stabilization, and translation, it would make sense for one cis-acting element to be involved in all of these functions. However, different cellular factors may interact with the DR to promote these different functions.
In this study, we have observed DR-induced CAT activity in HeLa cells, demonstrating its ability to function in a mammalian cell line. In contrast, we previously failed to detect DR-mediated export in COS cells using a less sensitive Western assay (33). Constructs bearing mutations in the DR2 element were less active than the wild type in HeLa cells, but to a lesser degree than in avian cells. This could be due to the presence of additional binding factors in HeLa cells. The DR elements have also been shown to be inactive in Xenopus oocytes where Rev-RRE and the MPMV CTE are both active. Conversely, the RSV DR is active in quail cells, where the MPMV CTE has been reported to be inactive (24).
After the manuscript for this article had been initially submitted, Yang and Cullen (45) reported the presence of a CTE in the single DR sequence of RAV-2 ALV. This sequence is closely related to that of RSV DR1 and DR2 (33). Using the pDM128 assay, CAT activity was observed in both quail and mammalian cell lines (45). This ALV CTE activity was shown to be CRM1 independent with truncated CAN nucleoporin as a competitive inhibitor of CRM1 (45), thus confirming our experiments with LMB and the RSV DR element.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant RO1CA48746 from the National Cancer Institute. R.E.P. and R.A.O. were supported in part by NIH predoctoral training grant 52T32G07231.
We thank Minoru Yoshida, Tom Hope, Barbara Felber, Gordon Carmichael, and Flossie Wong-Staal for reagents and Jennifer Rahmandar and Utz Fischer for help with experiments.
REFERENCES
- 1.Aschoff J M, Foster D, Coffin J M. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J Virol. 1999;73:7421–7429. doi: 10.1128/jvi.73.9.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks J, Beemon K, Linial M. RNA regulatory elements in the genomes of simple retroviruses. Semin Virol. 1997;8:194–204. [Google Scholar]
- 4.Barker G F, Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11:2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker G F, Beemon K. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol. 1994;14:1986–1996. doi: 10.1128/mcb.14.3.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 11.Coffin J, Hughes S, Varmus H. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 12.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 13.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst R K, Bray M, Rekosh D, Hammarskjöld M L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component, Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 19.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 21.Hammarskjold M L. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 22.Hope T J, Huang X J, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Carmichael G G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 26.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 27.Luo M-J, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magin C, Löwer R, Löwer J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 1999;73:9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald D, Hope T J, Parslow T G. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J Virol. 1992;66:7232–7238. doi: 10.1128/jvi.66.12.7232-7238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasioulas G, Hughes S H, Felber B K, Whitcomb J M. Production of avian leukosis virus particles in mammalian cells can be mediated by the interaction of the human immunodeficiency virus protein Rev and the Rev-responsive element. Proc Natl Acad Sci USA. 1995;92:11940–11944. doi: 10.1073/pnas.92.25.11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogert R A, Beemon K L. Mutational analysis of the Rous sarcoma virus DR posttranscriptional control element. J Virol. 1998;72:3407–3411. doi: 10.1128/jvi.72.4.3407-3411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Reilly M M, McNally M T, Beemon K L. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 35.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 38.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson S B, Guo W, Winistorfer S C, Craven R C, Stoltzfus C M. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology. 1998;247:86–96. doi: 10.1006/viro.1998.9233. [DOI] [PubMed] [Google Scholar]
- 40.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorge J, Ricci W, Hughes S H. cis-Acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983;48:667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 44.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Cullen B. Structural and functional analysis of the avian leukemia virus constitutive transport element. RNA. 1999;5:1645–1655. doi: 10.1017/s1355838299991616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]