Abstract
Dendritic spines, the mushroom-shaped extensions along dendritic shafts of excitatory neurons, are critical for synaptic function and are one of the first neuronal structures disrupted in neurodevelopmental and neurodegenerative diseases. Microtubule (MT) polymerization into dendritic spines is an activity-dependent process capable of affecting spine shape and function. Studies have shown that MT polymerization into spines occurs specifically in spines undergoing plastic changes. However, discerning the function of MT invasion of dendritic spines requires the specific inhibition of MT polymerization into spines, while leaving MT dynamics in the dendritic shaft, synaptically connected axons and associated glial cells intact. This is not possible with the unrestricted, bath application of pharmacological compounds. To specifically disrupt MT entry into spines we coupled a MT elimination domain (MTED) from the Efa6 protein to the actin filament-binding peptide LifeAct. LifeAct was chosen because actin filaments are highly concentrated in spines and are necessary for MT invasions. Temporally controlled expression of this LifeAct-MTED construct inhibits MT entry into dendritic spines, while preserving typical MT dynamics in the dendrite shaft. Expression of this construct will allow for the determination of the function of MT invasion of spines and more broadly, to discern how MT-actin interactions affect cellular processes.
It has not been possible to globally inhibit microtubule (MT) polymerization into dendritic spines without affecting MT dynamics throughout the entire neuron as well as any neighboring cells
Transfection with the LifeAct-MTED construct inhibits MT polymerization into dendritic spines in a spatially and temporally specific manor, without affecting native MT dynamics throughout the dendritic shaft
The LifeAct-MTED construct will allow for elucidation of the function of MT entry into dendritic spines as well as other actin-MT interactions in both neuronal and nonneuronal cells.
INTRODUCTION
Microtubule (MT) dynamic instability (Mitchison and Kirschner, 1984; Desai and Mitchison, 1997), the stochastic polymerization and depolymerization of MT polymers, occurs in all cell types, including mammalian neurons. However, as developing neurons mature into highly polarized cells containing elaborate dendrites and a single axon, it has been thought that MTs become stabilized to maintain neuronal architecture. With the discovery and fluorescent labeling of end-binding proteins that track along the tips of polymerizing MTs (Stepanova et al., 2003), it became apparent that a portion of cellular MTs continue to undergo polymerization and depolymerization for the life of the neuron (Hu et al., 2008; Yau et al., 2016). To discover the function of MT dynamics in neurons, researchers have used chemical compounds to inhibit MT dynamicity. At nanomolar concentrations, drugs such as paclitaxel (taxol), which stabilizes MTs, (Schiff and Horwitz, 1980) and nocodazole, which depolymerizes MTs, (Mareel and De Brabander, 1978), both inhibit MT dynamicity, resulting in little polymerization and depolymerization (Vasquez et al., 1997; Mikhailov and Gundersen, 1998; Yvon et al., 1999). Inhibiting MT dynamicity in this way has allowed for the discovery that MT dynamics are essential for many processes in developing neurons, including axon branching (Dent and Kalil, 2001).
More recently, several groups have shown that MTs have robust dynamics in mature cortical, hippocampal and cerebellar neurons, especially in dendrites (Hu et al., 2008; Jaworski et al., 2009; Wagner et al., 2011). These dynamic MTs polymerize throughout the dendritic arbor and can even enter the micron-sized, mushroom-shaped protrusions along dendrites called dendritic spines (Gu et al., 2008; Hu et al., 2008; Jaworski et al., 2009). In excitatory neurons, dendritic spines are the primary site of synaptic contact with presynaptic axons. Dendritic spines are not static structures but undergo morphological and molecular plasticity. They can undergo long-term potentiation (LTP) and enlarge or long-term depression (LTD) and shrink, depending on presynaptic activity (Tada and Sheng, 2006). Interestingly, MT entries into dendritic spines are dependent on neuronal activity (Gu et al., 2008; Hu et al., 2008; Jaworski et al., 2009) and result in long-term spine enlargement (Merriam et al., 2011). Indeed, MT invasion of spines increases during LTP and decreases during LTD (Kapitein et al., 2011; Merriam et al., 2013), showing a direct correlation with neuronal activity.
However, correlation does not indicate causation. To determine the function of MT invasion of dendritic spines requires inhibiting their entry into dendritic spines, while leaving the dynamics of MTs in the dendritic shaft, presynaptic axon and associated glial cells intact. Unfortunately, the primary methodology for inhibiting MT polymerization is the bath application of compounds such as paclitaxel and nocodazole. Addition of nanomolar concentrations of these drugs inhibits MT invasions of spines and results in decreased LTP (Jaworski et al., 2009), abolition of spine growth upon MT entry (Merriam et al., 2011), blockage of BDNF-dependent increase in PSD95 in spines (Hu et al., 2011) and inhibition of transport of a motor/cargo pair (KIF1A/synaptotagmin-IV) into dendritic spines (McVicker et al., 2016). Nevertheless, all these studies used bath applications of nocodazole or paclitaxel, which results in the inhibition of MT dynamics throughout the culture or slice preparation. Even if these pharmacological applications were localized to specific dendrites or spines, they would still affect the MTs present in the presynaptic axon. Therefore, a more spatially restrictive inhibition of MT dynamics is needed to definitively determine the function of MT invasion of dendritic spines.
Recently, others have designed optogenetic tools to depolymerize MTs, including photostatins (Borowiak et al., 2015), a construct based on the depolymerizing activity of kinesin 13 (Lu et al., 2020) or the MT severing enzymes spastin (Liu et al., 2022) or katanin (Meiring et al., 2022). Although these are useful tools to study the function of MTs, they do not lend themselves to the study of MT invasion of dendritic spines because MT invasions are infrequent and transient (Hu et al., 2008; Jaworski et al., 2009). It is also not possible to predict with any certainty which spine along a dendritic arbor will be invaded. Moreover, having a tool that would allow for the specific disruption of MTs into dendritic spines would therefore allow for the functional study of loss of MT entry into dendritic spines in both healthy and disease models (Peris et al., 2022).
Thus, we developed a construct that targets and is maintained in all dendritic spines. This construct contains a potent MT elimination domain (MTED) peptide, derived from the Arf guanine nucleotide exchange factor Efa6 (Qu et al., 2019). We targeted this peptide to dendritic spines by combining it with the LifeAct peptide, which has a high affinity for filamentous actin (f-actin; Riedl et al., 2008), which is concentrated in dendritic spines but is relatively sparse in the dendritic shaft. Moreover, it is well documented that MT invasion of spines is critically dependent on actin filaments in the neck of spines (Merriam et al., 2013; Schatzle et al., 2018). Thus, the depolymerizing peptide (MTED) is positioned on the f-actin that MTs require to enter spines. To control the timing of expression to mature neurons this construct is under the control of a tet-responsive promoter. We show here that expression of the LifeAct-MTED construct for only 16 h robustly inhibits MT invasion of dendritic spines, while preserving typical MT dynamics in the dendritic shaft. Because this construct is expressed in a small percentage of neurons, it does not affect MT dynamics in presynaptic axons or synapse associated glial cells. Thus, transfection with the LifeAct-MTED construct is a much more targeted methodology for inhibiting MT dynamics than application of pharmacological compounds and can be used for studies focused on the interaction of MTs and actin filaments.
RESULTS
Development of a methodology for localized disruption of MT dynamics
Use of pharmacological compounds, such as paclitaxel or nocodazole, to inhibit MT polymerization into dendritic spines is a nonspecific approach that introduces global MT disruption and a myriad of off-target effects. To address and mitigate confounding variables, we have developed a plasmid construct to specifically inhibit MT polymerization into dendritic spines, while preserving native dynamics within the dendritic shaft (Figure 1A). The primary component of this construct is a 20-amino-acid peptide referred to as the MTED from the protein Efa6, which has been shown to potently depolymerize MTs (Qu et al., 2019). A scrambled version of the MTED peptide serves as a control in a separate plasmid (Figure 1A). To selectively target the MTED or scrambled peptide to dendritic spines, we combined it with LifeAct, a 17-amino-acid peptide that readily localizes to f-actin (Riedl et al., 2008), which is highly enriched in the head and neck regions of spines. The fluorescent protein mScarlet (Bindels et al., 2017) was incorporated between the LifeAct and MTED or scrambled peptide to allow for construct visualization. To control the temporal expression of the constructs we utilized the Tet-On system, in which a doxycycline-inducible tet promoter is used in conjunction with a constitutively active hPGK promoter driving expression of the rTTA gene (David Root, RRID: Addgene_41393). The LifeAct-mScarlet-MTED or the LifeAct-mScarlet-scramble fusion protein sequence was inserted after the tet promoter, but before the hPGK promoter (Figure 1A).
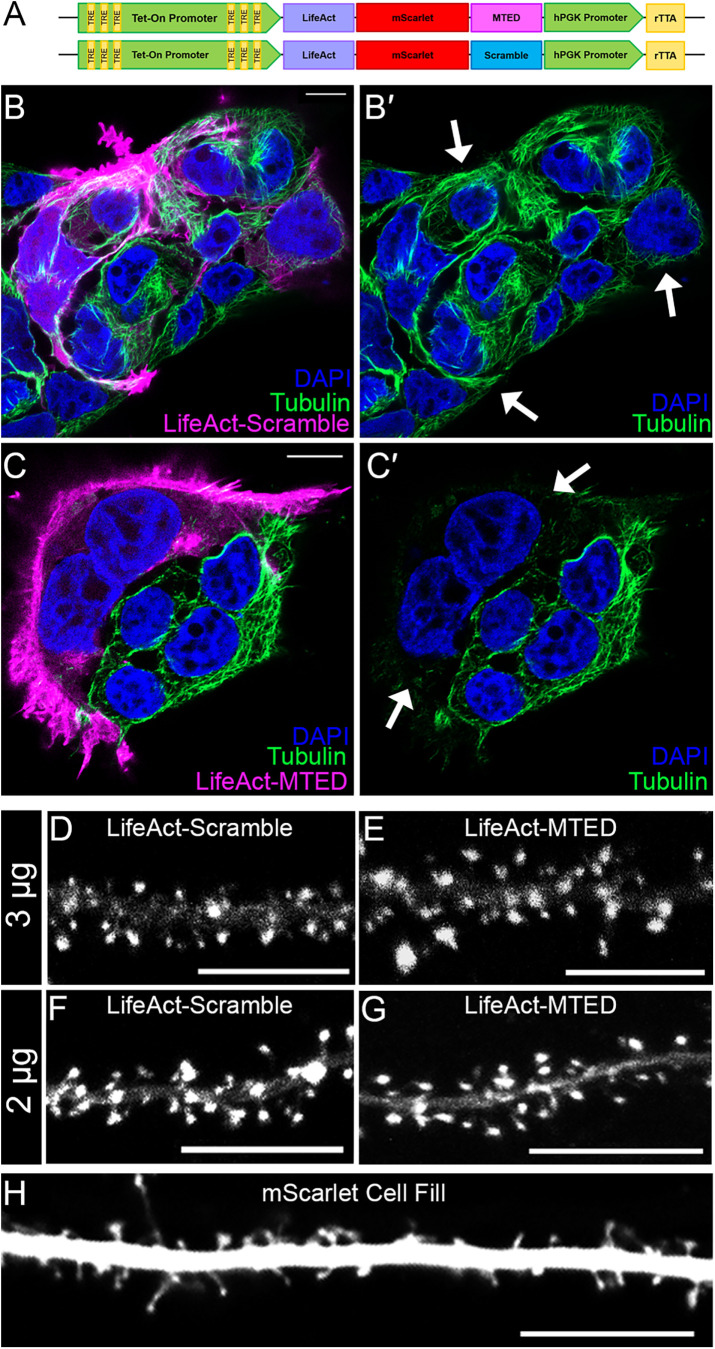
FIGURE 1:
Design of the MTMTED plasmid construct. (A) Schematic of MTED and scramble plasmid constructs containing Tet-On doxycycline inducible promoter (green) with tet-response elements (yellow), LifeAct (purple)-mScarlet (red)-MTED/scramble (magenta/blue) fusion protein sequence, separate hPGK promoter (green) and constitutively active rTTa gene sequence (yellow). (B–C’) Confocal images of fixed HEK293T cells transfected with the LifeAct-scramble (B and B’) or LifeAct-MTED (C and C’) plasmid construct and immunostained for beta-tubulin (green) with cell nuclei stained blue (DAPI). The leftmost panels (B and C) show transfected cells in magenta while the corresponding panels on the right (B’ and C’) show the same image, without the magenta overlay. Arrows in B’ and C’ point to transfected cells that contain MTs (B’) or lack MTs (C’). (D–H) Confocal images of living primary hippocampal neurons (DIV 21–25) transfected with 3 μg LifeAct-scramble (D), 3 μg LifeAct-MTED (E), 2 μg LifeAct-scramble (F), 2 μg LifeAct-MTED (G) or mScarlet fluorescent cell fill (H). All scale bars are 10 µm.
For initial validation of the LifeAct-mScarlet-MTED and LifeAct-mScarlet-scramble plasmid constructs, which we will henceforth refer to as LifeAct-MTED or LifeAct-scramble, we utilized immortalized HEK293T cells, due to their rapid and robust culturing when compared with primary neuronal cultures. HEK293T cells were transfected with either LifeAct-MTED or LifeAct-scramble, then fixed and stained for tubulin and cell nuclei (Figure 1, B–C′). Tubulin content within cells transfected with LifeAct-scramble appears similar to the tubulin content of adjacent, untransfected cells (depicted by white arrows) (Figure 1, B and B′). Conversely, cells transfected with LifeAct-MTED have a visible reduction of tubulin staining when compared with nearby untransfected cells (Figure 1, C and C′), illustrating the MT-depolymerizing power of the MTED peptide. It should be noted that most of the MTs were depolymerized throughout the transfected cell in Figure 1, C and C’. This nonspecific, extensive MT depolymerization is likely due to high expression levels of the constructs in the HEK293T cells, resulting in their localization to f-actin, but with excess LifeAct-MTED dispersed throughout the cytoplasm.
While the LifeAct-MTED and LifeAct-scramble constructs appeared to be localizing to f-actin appropriately in nonneuronal cells, it was important to assess their localization in neuronal cultures. Hippocampal neurons were transfected with either LifeAct-MTED or LifeAct-scramble at two different plasmid concentrations (2 or 3 µg), allowed to mature in culture (21–25 DIV), and induced to express overnight (16 h). We chose low concentrations of plasmid and short expression times to minimize mislocalization and disruption of MT dynamics throughout the neuron. Both concentrations of the LifeAct-scramble (Figure 1, D and F) and LifeAct-MTED (Figure 1, E and G) show distinct enrichment in dendritic spines, when compared with cells transfected with a fluorescent cell fill (Figure 1H). These results suggest that the plasmid constructs are localizing as expected in mature hippocampal neurons following short-term expression. Although a recent publication indicated LifeAct was less concentrated in spines than other actin labels (Ignacz et al., 2023), we detected very low levels of LifeAct label in the dendrite shaft and significant enrichment in dendritic spines. This discrepancy may be due to differing levels and timing of expression.
Effect of LifeAct-MTED in mature hippocampal neurons
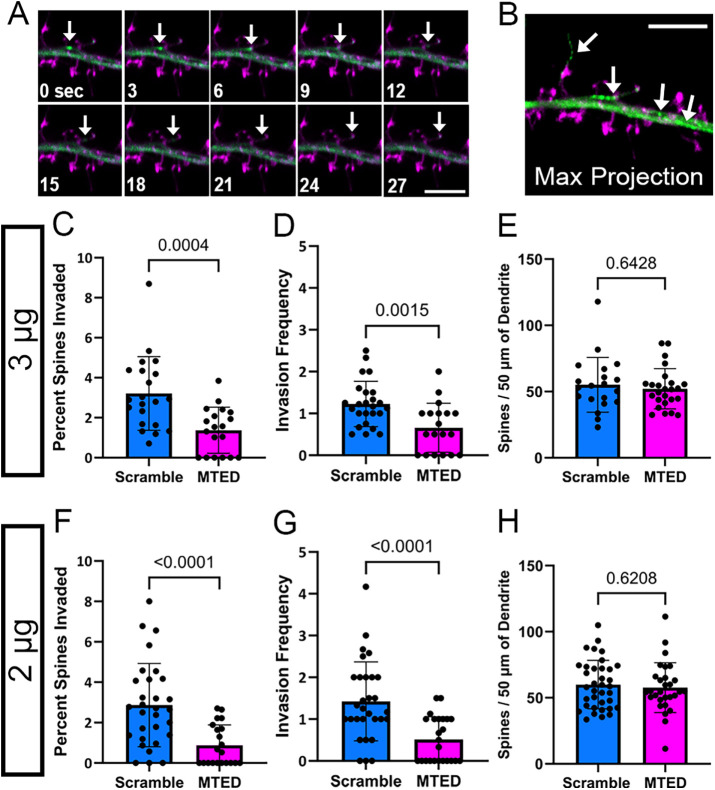
Once we confirmed that the LifeAct-MTED and LifeAct-scramble constructs are selectively enriched in dendritic spines (Figure 1, D–G), we next investigated whether the LifeAct-MTED could effectively reduce rates of MT polymerization into spines. This was done using neurons cotransfected with a fluorescent MT end binding protein, EB3-mNeon, and varying concentrations (2 or 3 µg) of either LifeAct-MTED or LifeAct-scramble. Fluorescent EB3 binds to polymerizing ends of MTs, allowing us to visualize MT “comets” moving within the dendritic shaft and into dendritic spines in time-lapse (Figure 2A). Maximum projection of the collected time series shows in a single frame where MTs were polymerizing within the dendrite (Figure 2B). MT invasion rates were assessed by the percentage of spines invaded (invaded spines/total number of spines) and invasion frequency (total invasions/number of invaded spines) as we have done previously (Hu et al., 2008, 2011; Merriam et al., 2011; Merriam et al., 2013; McVicker et al., 2016). There is a significant reduction in the percentage of spines invaded in neurons transfected with both the higher (Figure 2C) and lower concentrations of LifeAct-MTED (Figure 2F), compared with LifeAct-scramble controls. This suggests that the MTED effectively and specifically reduces the percentage of dendritic spines being targeted for invasion by polymerizing MTs. Additionally, there is a significant reduction in invasion frequency in neurons transfected with both the higher (Figure 2D) and lower concentration of LifeAct-MTED (Figure 2G). To assess whether expression of our construct influenced the density of dendritic spines along dendrites, we also analyzed the total number of dendritic spines per 50 μm segment of dendrite of neurons transfected with the higher (Figure 2E) and lower (Figure 2H) concentration of LifeAct-MTED. In both instances, there was no significant difference in dendritic spine density when compared with neurons transfected with a corresponding concentration of LifeAct-scramble. These data illustrate that there is both a reduction in the percentage of spines being targeted for invasion and the total number of invasions. However, these changes in MT invasion parameters did not affect total dendritic spine density. Thus, it appears that the LifeAct-MTED plasmid construct is affecting invading MTs as intended and restricting MTs from polymerizing into spines.
FIGURE 2:
Reduction of MT invasions into dendritic spines within LifeAct-MTED transfected hippocampal neurons. (A) Confocal time-series of living mature hippocampal neuron (DIV 23) taken at 3-s intervals. The neuron was transfected with LifeAct-scramble (magenta) and mNeon EB3 (green) plasmid constructs. Invading EB3 comet is shown with a white arrow in each frame. (B) Corresponding maximum intensity projection of the time-series shown in A. Traces of an invading EB3 comet, as well as EB3 comets within the dendritic shaft, are shown in green and are identified by white arrows. Dendritic spines of the transfected neuron are shown in magenta. Scale bars are 5 µm (A and B). (C–H) Bar graphs show mean ± SD and black dots are individual dendritic segments. (C) Quantification of percent of dendritic spines invaded (number of invaded dendritic spines/total number of dendritic spines within field of view) for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED plasmid (n = 21 scramble, n = 19 MTED from five separate biological replicates for both C and D). (D) Quantification of invasion frequency (total number of invasions throughout the course of a time-series containing 100 frames/invaded dendritic spines within field of view) for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED plasmid. (E) Quantification of dendritic spine density normalized to 50 µm dendritic segments for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED (n = 20 scramble, n = 25 MTED from six separate biological replicates). (F) Quantification of percent of dendritic spines invaded for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED (n = 30 scramble, n = 21 MTED from five separate biological replicates for both F and G). (G) Quantification of invasion frequency for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED plasmid. (H) Quantification of dendritic spine density normalized to 50 µm dendritic segments for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED (n = 36 scramble, n = 29 MTED from six separate biological replicates). P values in (C–H) shown above bars are calculated with two-tailed Student’s t test or Mann-Whitney depending on normality of data distribution.
Examination of potential off-target effects of LifeAct-MTED in mature hippocampal neurons
Although LifeAct-MTED markedly inhibits MT polymerization into dendritic spines (Figure 2, C–H), it remains critical to determine whether the construct has any off-target effects within dendrites. Thus, we examined MT dynamics within the dendritic shaft of transfected neurons to assess whether the typical MT dynamicity was being perturbed in the presence of LifeAct-MTED and LifeAct-scramble. Kymographs, graphical representations of EB3 comet spatial position over time, were created to visualize motion of EB3 comets within dendritic shafts of transfected neurons (Figure 3A). Mature hippocampal neurons were again cotransfected with EB3-mNeon and either 2 or 3 µg of LifeAct-MTED or LifeAct-scramble. Each EB3 comet, representative of a polymerizing MT, can be visualized as a line in the kymograph, with the slope of a line representing EB3 comet velocity. The (x, y) coordinates of the beginning (x1,y1 in Figure 3A) and end of a line (x2,y2 in Figure 3A) represent the movement of EB3 comets over time. Such positions can, therefore, be used to calculate the distance traveled by an individual EB3 comet (x2-x1) as well as how long the comet was visualized or the “lifetime” of a comet (y2-y1) within a dendritic shaft segment.
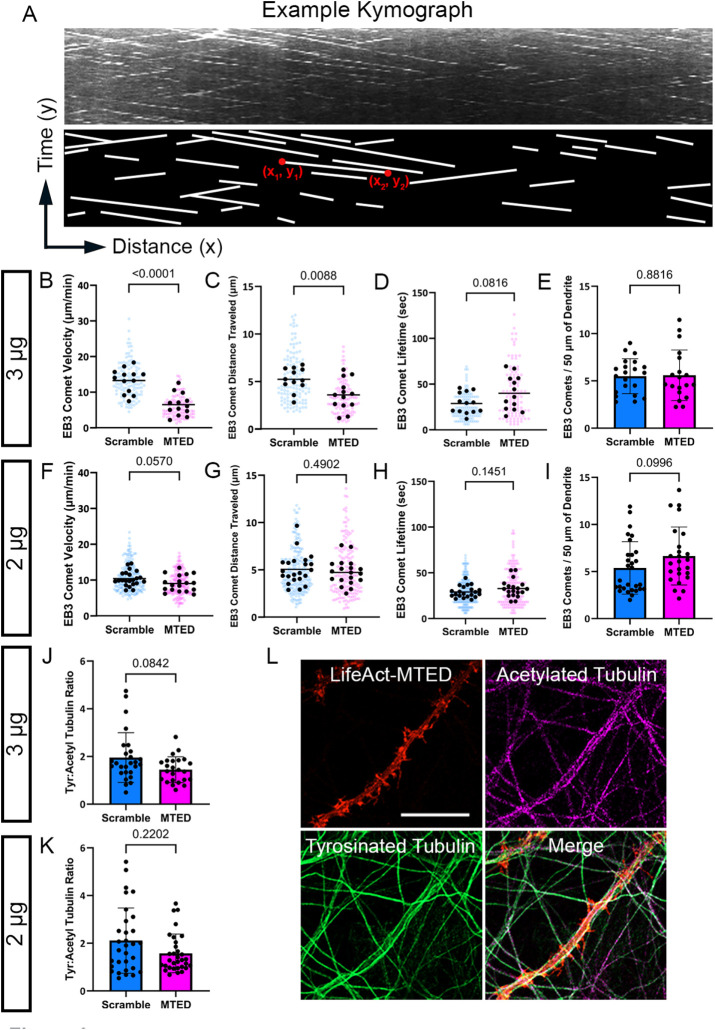
FIGURE 3:
Limited off-target effects following transfection of LifeAct-MTED in mature hippocampal neurons. (A) Representative kymograph, a graphical representation of EB3 comet spatial position over time, with corresponding illustration below for simplified visualization. Each white line is representative of a single, moving EB3 comet. Coordinates of the beginning (x1,y1) and end (x2,y2) of an example line are used for calculation of distance traveled (difference in x value) and how long the comet was visualized, also referred to as the comet “lifetime” (difference in y value). (B–D) Scatter plots displaying EB3 comet velocity (B), EB3 comet distance traveled (C), and EB3 comet lifetime (D) obtained from neurons transfected with 3 μg of LifeAct-MTED or LifeAct-scramble. Mean is shown by a black bar. Each black dot is representative of the average measurement per neuron ((B) n = 13 scramble, n = 13 MTED; (C) n = 12 scramble, n = 13 MTED; (D) n = 13 scramble, n = 13 MTED from four separate biological replicates), while the blue or magenta dots in the background represent individual EB3 comet measurements ((B) n = 125 scramble, n = 108 MTED; (C) n = 121 scramble, n = 105 MTED; (D) n = 118 scramble, n = 107 MTED). (E) Quantification of EB3 comet abundance (number of comets per 50 μm of dendrite) within the dendritic shaft of neurons transfected with 3 μg of LifeAct-MTED or LifeAct-scramble. Bar graph shows mean ± SD and black dots are individual dendritic segments (n = 21 scramble, n = 19 MTED from five separate biological replicates). (F–H) Scatter plots displaying EB3 comet velocity (F), EB3 comet distance traveled (G), and EB3 comet lifetime (H) obtained from neurons transfected with 2 μg of LifeAct-MTED or LifeAct-scramble. Mean is shown by a black bar. Each black dot is representative of the average measurement per neuron ((F) n = 22 scramble, n = 18 MTED; (G) n = 22 scramble, n = 18 MTED; (H) n = 21 scramble, n = 18 MTED from five separate biological replicates), while the blue or magenta dots in the background represent individual EB3 comet measurements ((F) n = 195 scramble, n = 164 MTED; (G) n = 192 scramble, n = 165 MTED; (H) n = 187 scramble, n = 161 MTED). (I) Quantification of EB3 comet abundance within the dendritic shaft of neurons transfected with 2 μg of LifeAct-MTED or LifeAct-scramble. Bar graph shows mean ± SD and black dots are individual dendritic segments (n = 29 scramble, n = 25 MTED from five separate biological replicates). (J and K) Ratio of tyrosinated tubulin:acetylated tubulin within the dendritic shaft of neurons transfected with either 3 μg (J) (n = 27 scramble, n = 24 MTED) or 2 μg (K) (n = 31 scramble, n = 32 MTED) of LifeAct-MTED or LifeAct-scramble from three separate biological replicates. Bar graph shows mean ± SD and black dots are individual dendritic segments. (L) Representative confocal images of fixed, hippocampal neurons transfected with the Life-Act MTED (red) and stained for acetylated tubulin (magenta) and tyrosinated tubulin (green). Such images were used for quantification of tyrosinated tubulin:acetylated tubulin ratios (J and K). P values in (B–K) shown above bars are calculated with two-tailed Student’s t test or Mann-Whitney depending on normality of data distribution.
We observed a significant slowing of MT polymerization following transfection with the higher concentration (3 µg) of LifeAct-MTED compared with the LifeAct-scramble control (Figure 3B). This decrease in velocity was negated when neurons were transfected with a lower concentration (2 µg) of plasmid (Figure 3F), suggesting that there may be expression level-dependent off-target effects. Thus, the LifeAct-MTED construct must be titrated to avoid disruption of MT polymerization within the dendritic shaft. It is possible this disruption of MT velocity in the dendrite is due, in part, to the presence of low, but not negligible, levels of f-actin within the dendritic shaft (Figure 1, D–G). To assess the mechanism by which EB3 comet velocity was decreasing with higher concentrations of LifeAct-MTED, we measured both the distance and lifetime of each polymerizing MT. Interestingly, in neurons transfected with 3 µg of LifeAct-MTED there is a significant reduction in the distance traveled by EB3 comets (Figure 3C). EB3 comet lifetime, however, is not significantly different compared with LifeAct-scramble control, although it does trend longer (Figure 3D). Thus, in neurons transfected with 3 µg of LifeAct-MTED, MTs polymerize for the same amount of time but over shorter distances, resulting in decreased comet velocity in the dendritic shaft. Together, these data suggest that MT dynamics can be significantly altered in the dendritic shaft at higher concentrations of LifeAct-MTED expression.
To decrease these off-target effects detected with 3 µg of plasmid, we transfected neurons with a lower concentration (2 µg) of LifeAct-MTED or LifeAct-scramble. We discovered with 2 µg of plasmid there is no significant difference in EB3 comet velocity (Figure 3F), distance traveled (Figure 3G), or comet lifetime (Figure 3H) between LifeAct-MTED and LifeAct-scramble. To examine whether the reduction in MT invasion rates (Figure 2, B, C, F, and G) resulted from a reduction in the number of polymerizing MTs, we also measured comet abundance as a proxy for the number of polymerizing MTs in the dendrite shaft but found no significant difference between LifeAct-MTED and LifeAct-scramble control (Figure 3, E and I). For additional rigor, we also compared the populations of posttranslationally modified tubulin. Tyrosinated tubulin is traditionally interpreted as being incorporated into short-lived, dynamic MTs while acetylated tubulin is associated with long-lived, stable MTs (Tas et al., 2017).
This balance of various posttranslational modifications of tubulin is critical for neuronal health as illustrated by disruption of MT tyrosination/detyrosination cycles resulting in synaptic dysfunction characteristic of disease as well as deficits in axonal transport (Peris et al., 2022; Konietzny et al., 2024). To assess whether we were preferentially disrupting one specific population of MTs we fixed and stained for tyrosinated and acetylated tubulin in neurons transfected with either 3 or 2 µg of LifeAct-MTED (Figure 3L) or LifeAct-scramble (unpublished data). However, we found no significant difference between the tyrosinated:acetylated tubulin ratio in neurons transfected with either concentration of LifeAct-MTED when compared with LifeAct-scramble controls (Figure 3, J and K). Taken together, these data suggest that transfection with a low concentration of LifeAct-MTED is sufficient for markedly inhibiting MT polymerization into dendritic spines, without disrupting MT dynamics and stability in the dendrite shaft.
Assessment of correlation between various effects of LifeAct-MTED
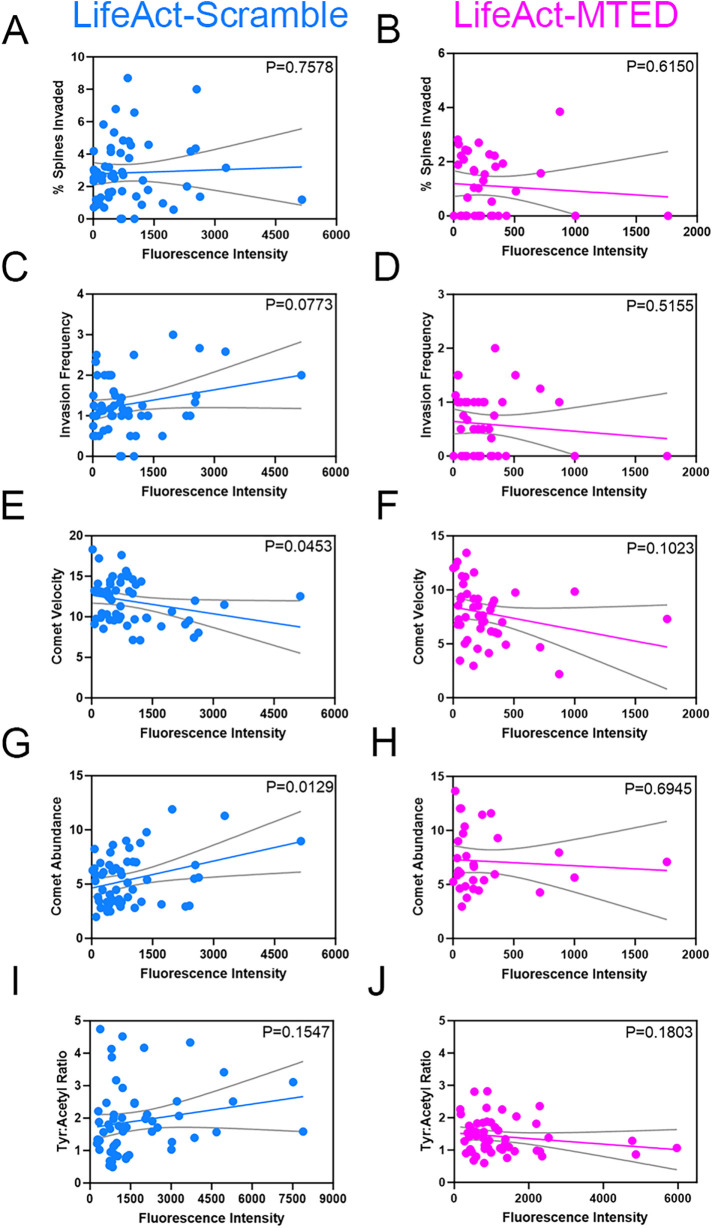
The results of our study show that transfecting hippocampal neurons with low (2 µg) levels of the LifeAct-MTED plasmid specifically inhibits MT polymerization into dendritic spines, without affecting MT dynamics and stability in the dendrite shaft (Figure 3, F–I). However, expressing slightly more LifeAct-MTED plasmid (3 µg) is sufficient to significantly decrease MT polymerization velocity by decreasing the distance MTs polymerize in the dendritic shaft (Figure 3, B and C). One would expect that expression of either 2 or 3 µg of plasmid would result in a distribution of expression levels in individual neurons. Therefore, we measured expression levels of LifeAct-MTED or LifeAct-scramble in individual neurons and constructed scatter plots, fitted with linear regression lines against the MT metrics in spines and dendrite shafts that we measured above. We found a weak negative correlation in both the percentage of spines invaded and the invasion frequency plotted against fluorescence intensity of dendrites for the LifeAct-MTED transfected neurons compared with a weak positive correlation for the LifeAct-scramble transfected neurons (Figure 4, A–D). However, the slopes of the linear regression lines are not significantly different from zero, with p values ranging from 0.0773 to 0.7578. Moreover, for neurons expressing the LifeAct-MTED plasmid we found a weak negative correlation between fluorescence intensity of dendrites and comet velocity (Figure 4F), comet abundance (Figure 4H) and tyrosinated:acetylated tubulin ratios (Figure 4J). However, neurons transfected with LifeAct-scramble have a significant (p = 0.0453) negative correlation between comet velocity and fluorescence intensity (Figure 4E), while having a significant (p = 0.0129) positive correlation between comet abundance and fluorescence intensity (Figure 4G). Nevertheless, it should be noted that the fluorescent intensity values for the LifeAct-scramble construct include average fluorescence intensity values (0-6000) much higher than for the LifeAct-MTED construct (<2000). The technical reason why higher fluorescent values (2000–6000) of the LifeAct-MTED construct are not included is because neurons expressing higher values of the construct lacked discernable EB3 comets that could be used to quantify MT dynamics. Thus, at higher concentrations LifeAct-MTED is likely present throughout the cytoplasm and negatively affects all MT polymerization events.
FIGURE 4:
Correlation between LifeAct-MTED/scramble expression levels and MT dynamics. (A–J) Scatter plots of expression levels of LifeAct-scramble (A, C, E, G, and I) or LifeAct-MTED (B, D, F, H, and J) within hippocampal neurons, as measured by fluorescent intensity of the cell dendrite. Linear regression lines (colored) and 95% confidence intervals (gray) were plotted to compare percent spines invaded (A and B), invasion frequency (C and D), EB3 comet velocity (E and F), EB3 comet abundance (G and H) and tyrosinated tubulin:acetylated tubulin ratios (I and J) to the fluorescent intensity values. The slope of each linear regression line was analyzed for whether it significantly differed from zero, with the p value being displayed in the upper right corner of each graph. N = 57 (A), n = 40 (B), n = 54 (C), n = 42 (D), n = 57 (E), n = 44 (F), n = 56 (G), n = 31 (H), n = 57 (I), n = 52 (J) from 16 separate biological replicates.
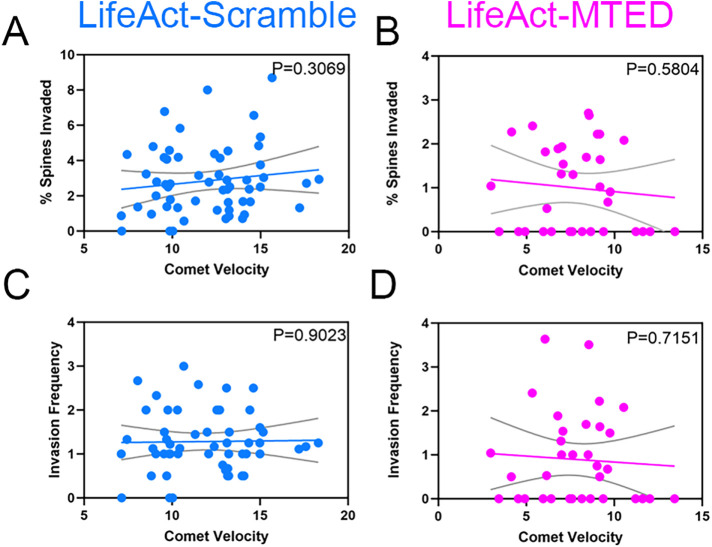
With EB3 comet velocity appearing to be the most sensitive to variations in LifeAct-MTED expression, we sought to assess whether the magnitude of comet slowing was correlated with metrics of dendritic spine invasion. Thus, we constructed scatter plots of average EB3 comet velocity in neurons transfected with either LifeAct-MTED or LifeAct-scramble and fit linear regression lines against the percentage of spines invaded (Figure 5, A and B) and invasion frequency (Figure 5, C and D). We observed a slight, positive correlation of both the percentage of spines invaded and invasion frequency for LifeAct-scramble transfected neurons (Figure 5, A and C), while there was a slight, negative correlation of both in LifeAct-MTED transfected neurons (Figure 5, B and D). While the slopes of the linear regression lines did not significantly differ from zero, we believe this importantly shows that the reduction in MT invasion rates observed with the LifeAct-MTED construct (Figure 2, B, C, F, and G) is not dependent on a reduction in EB3 comet velocity. Additionally, this weak correlation between comet velocity and measurements of MT polymerization into spines demonstrates a range of usability for our construct in that expression levels can be titrated, abolishing off-target effects, without reducing the desired effect on MT invasion rates.
FIGURE 5:
Lack of correlation between EB3 comet velocity and MT invasion of dendritic spines. (A–D) Scatter plots of average EB3 comet velocity within hippocampal neurons transfected with LifeAct-scramble (A and C) or LifeAct-MTED (B and D). Linear regression lines (colored) and 95% confidence intervals (gray) were plotted against metrics measuring likelihood of MT polymerization into dendritic spines, including percent spines invaded (A and B) and invasion frequency (C and D). The slope of each linear regression line was analyzed for whether it significantly differed from zero, with the p value being displayed in the upper right corner of each graph. N = 57 (A), n = 34 (B), n = 54 (C), n = 34 (D) from 11 separate biological replicates.
DISCUSSION
Despite previous assumptions that MTs are largely stable polymers in mature neurons, it is increasingly appreciated that they remain dynamic throughout the life of the cell (Gu et al., 2008; Hu et al., 2008; Jaworski et al., 2009). Moreover, our group, as well as others, have observed MTs transiently polymerizing into, or invading, dendritic spines of hippocampal and cortical neurons (Gu et al., 2008; Hu et al., 2008, 2011; Jaworski et al., 2009; Kapitein et al., 2011; Merriam et al., 2011; Merriam et al., 2013; McVicker et al., 2016; Schatzle et al., 2018). This phenomenon appears to be highly correlated with mechanisms of learning and memory, such as LTP and LTD, in which disruption of MT dynamicity impairs learning and memory as assessed by electrophysiology or behavioral paradigms (Shumyatsky et al., 2005; Jaworski et al., 2009; Barnes et al., 2010; Fanara et al., 2010; Uchida et al., 2014). A recent study has also shown that MT invasions of spines is inhibited by application of a soluble amyloid-β peptide, suggesting a role for MT invasion of spines in Alzheimer’s disease (Peris et al., 2022). Many of the aforementioned studies, however, relied on pharmacological disruption of MT dynamics. General, bath application of drugs makes it difficult to discern the origin of observed outcomes, for it effects MTs in both presynaptic axonal compartments and postsynaptic dendritic compartments, as well as any glia present within the culture. Thus, drug treatment induces many confounding variables that may contribute to, or be responsible for, the impairment of MT polymerization into dendritic spines.
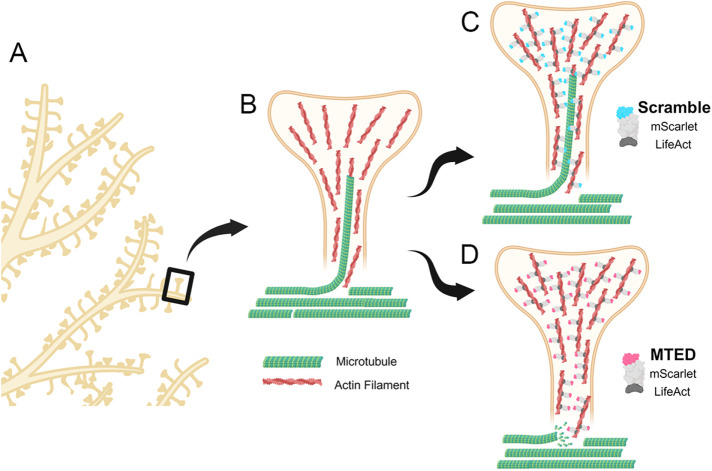
To overcome the limitations associated with generalized bath application of pharmacological compounds such as paclitaxel or nocodazole, we have developed a novel plasmid, termed LifeAct-MTED, containing the discrete MTED of the Efa6 protein (Figure 6). A scrambled version of the MTED is used as a control in a separate plasmid construct termed LifeAct-scramble. The MTED binds directly to tubulin, preventing polymerization, and is effective at abolishing exploratory MTs through mechanisms that remain to be fully elucidated (Qu et al., 2019). The MTED, coupled with LifeAct, a small peptide that readily associates with f-actin, allows for specific enrichment of the MTED to f-actin rich dendritic spine heads and necks (Merriam et al., 2013; Schatzle et al., 2018). LifeAct-MTED effectively inhibits not only the percentage of dendritic spines being targeted for invasion by polymerizing MTs, but also the total number of invasions occurring across the dendritic field without apparent impact on total dendritic spine density (Figure 2). In addition to decreasing the percentage of spines targeted and the invasion frequency of MTs polymerizing into dendritic spines, LifeAct-MTED appears to have limited off-target effects on MT dynamics within the dendritic shaft (Figure 3). Of the examined metrics, only EB3 comet velocity was significantly different within neurons transfected with 3 µg of LifeAct-MTED when compared with LifeAct-scramble controls. Additionally, this effect was negated upon reducing the plasmid concentration being used to transfect the neurons from 3 to 2 µg.
FIGURE 6:
Schematic of LifeAct-MTED/scramble effects on MT invasions of dendritic spines. (A) Section of a representative dendritic arbor with one spine boxed. (B) Representative dendritic spine (yellow) rich with f-actin (red filaments) being invaded by a polymerizing MT (green). Other noninvading MTs are shown in the dendrite shaft (unpublished data). (C) Dendritic spine (yellow) of neuron transfected with LifeAct-scramble (blue and gray fusion protein) that is appropriately localizing to actin filaments (red) but does not affect the likelihood of a MT (green) directly polymerizing into the dendritic spine. (D) Dendritic spine (yellow) of neuron transfected with LifeAct-MTED (pink and gray fusion protein) that has localized to actin filaments (red) in the spine head and neck. LifeAct-MTED is shown depolymerizing a MT (green) before its entry into the dendritic spine. Created with BioRender.com.
The need to titrate expression levels was largely anticipated due to the apparent potency of the MTED as well as the low, but nonnegligible levels of f-actin present within dendritic shafts. Additionally, it has been observed that the MTED can be found within the cytosol when high expression levels result in saturation of the target to which it is localized (Qu et al., 2019). It is intriguing, however, that plasmid expression levels, measured by fluorescent intensity of transfected neurons, are not highly correlated with metrics assessed in dendritic spines and shafts (Figure 4). We believe this illustrates the flexibility of our tool in that there is a working range of expression that can be used to induce the desired effect on invading MTs. Nonetheless, the slowing of MT polymerization observed within neurons transfected with 3 µg of LifeAct-MTED remains an important consideration when applying this tool to other lines of research. Nevertheless, we found that the velocity of MT polymerization in dendrites was not associated with the percentage of spines invaded by MTs or the MT invasion frequency for both LifeAct-MTED and LifeAct-scramble (Figure 5). This result suggests that even though the LifeAct-MTED can decrease MT polymerization velocity at higher concentrations, this decrease is not correlated with a decrease in MT polymerization into dendritic spines. Another interesting observation was that cells transfected with LifeAct-MTED were consistently at lower levels of fluorescence in comparison to those transfected with LifeAct-scramble. We believe this was due to the difficultly of assessing MT dynamics within neurons highly expressing LifeAct-MTED, in which we could no longer visualize fluorescent EB3 comets, suggesting the LifeAct-MTED construct was appreciatively depleting MT polymerization throughout the dendrite.
Taken together, these experiments serve to illustrate that the LifeAct-MTED tool is a specific and effective means for inhibiting transiently polymerizing MTs from entering dendritic spines (Figure 6). The magnitude of inhibition of MT invasions induced by LifeAct-MTED as compared with other MT-targeting agents remains unknown at this time. However, considering the likelihood of pharmacological compounds affecting cellular processes well beyond MT polymerization into dendritic spines upon their application, we would hypothesize equal or subdued effects with transfection of LifeAct-MTED. Thus, implementation of this construct could shed light on off-target effects of various MT-stabilizing and destabilizing drugs by comparing global disruption of MT dynamics to focal disruption only into dendritic spines. An important consideration is that such proposed studies would require implementation of labeled tubulin as opposed to EB3 as used here, because even nanomolar concentrations of compounds such as nocodazole (Mimori-Kiyosue et al., 2000) and paclitaxel (Pagano et al., 2012) induce EB protein dissociation from the polymerizing plus ends of MTs, preventing tracking of MT invasions into spines.
Future studies could also apply this methodology to investigate the role of MT invasion in maintenance and/or stabilization of dendritic spine morphology. Others have reported that acute treatment (3–4 h) with nanomolar concentrations of nocodazole (Jaworski et al., 2009) or paclitaxel (Qu et al., 2017) can cause changes in spine morphology toward more immature spines or a decrease in dendritic spine density, respectively. We do not detect a change in dendritic spine density after a 16hr induction of LifeAct-MTED at either concentration used here. Future work is focused on determining if longer expression of LifeAct-MTED affects spine number and morphology and how it may or may not differ from pharmacological treatments. Additionally, the LifeAct-MTED construct should allow for the determination of the postsynaptic components of dendritic spines that are dependent on MT polymerization into dendritic spines. Importantly, the use of this construct is not limited to neurons and should prove useful to dissect MT/f-actin interactions in any type of cell.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Plasmid design and construction
MTED and scramble plasmids were assembled using standard molecular cloning techniques including PCR amplification, restriction digest and Gibson assembly. From 5′ to 3′ the constructs consist of a Tet-On doxycycline inducible promoter (a gift from David Root (Addgene plasmid # 41393; http://n2t.net/addgene:41393; RRID:Addgene_41393), a Kozak consensus sequence and a LifeAct – mScarlet (Bindels et al., 2017) – MTED/scramble fusion protein sequence followed by a separate hPGK promoter and a constitutively produced rTTA gene. MTED (gcgccgcgctttgaagcgtatatgatgaccggcgatctgattctgaacctgagccgcacc; APRFEAYMMTGDLILNLSRT) and scramble (atgattaccgcgccgcgcgaatttgattatctgaacctgcgcgcgggcctgagcatgacc; MITAPREFDYLNLRAGLSMT) sequences (Qu et al., 2019) were ordered from IDT as single-stranded ultramers with overhangs complementary to the destination vector. These, along with PCR amplified mScarlet, were inserted into Not1 and Mlu1 digested backbone by Gibson assembly reaction to create an intermediate plasmid. LifeAct peptide sequence was ordered from IDT as an ultramer with complementary overhangs and was inserted via Gibson assembly into the Ale1 and Not1 digested intermediate vector. Sequences were confirmed with Sanger sequencing (QuintaraBio). Complete maps may be found on Addgene for each respective plasmid, or are available by reasonable request.
HEK293T cell culture, transfection, and immunostaining
HEK293T cells (Sigma, 12022001) were maintained at 37°C, 5% CO2 in standard HEK media comprised of DMEM High Glucose (Life Technologies), sodium pyruvate (Life Technologies) and 10% FBS. Cells were passaged at ∼80% confluency and plated onto 0.1% polyethylenimine (PEI; Sigma) polymer-coated glass coverslips. HEK cells were transfected 24 h after plating with Lipofectamine 3000 (Thermo Fisher Scientifc) transfection reagent according to the manufacturer provided protocol. Tet-On plasmid constructs were induced with 1 µg/ml doxycycline and allowed to express overnight. HEK cells were then fixed with 4% paraformaldehyde-KREB-sucrose (PKS), blocked in 10% bovine serum albumin (BSA) overnight at 4°C and incubated with primary antibody overnight at 4°C followed by secondary antibody with DAPI (1:250) overnight at 4°C. Antibodies used were mouse alpha tubulin (1:500, Sigma T9026) and goat anti-mouse AlexaFluor 488 (1:500, Thermo Fisher Scientifc A11029). Glass coverslips were mounted onto frosted microscope slides with Fluoromount-G (SouthernBiotech).
Primary neuron cell culture and transfection
Primary hippocampal neurons were prepared from Sprague Dawley rats (Envigo) at embryonic day 18.5 (E18.5). Rat hippocampi were dissected and trypsinized. Dissociated neurons were resuspended in nucleofector solution (Mirus) and transfected using an Amaxa/Lonza Nucleofector II. Transfected neurons were plated at a density of 5 × 104 neurons per cm2 on 0.1% PEI-coated glass bottom dishes (35 mm) with 14-mm microwells. Neurons were plated with plating media (PM; neurobasal media with 5% defined fetal bovine serum (dFBS), B27 supplement, 2 mM Glutamax, 0.3% glucose and 37.5 mM NaCl) for 2 h at 5.0% CO2 and 37°C after which the chambers were flooded with 2 ml of serum-free media (PM with no added dFBS). Neurons used within experiments ranged from DIV 21–25. All procedures were approved by the University of Wisconsin Committee on Animal Care and were in accordance with the NIH guidelines.
Tyrosinated:acetylated tubulin immunocytochemistry
Primary hippocampal neurons were allowed to develop to DIV 21–25, and then induced with 1 µg/ml doxycycline for overnight expression. After ∼16 h, neurons were fixed with 4% PKS for 20 min (Dent and Meiri, 1992). Blocking solution of 10% BSA was applied and allowed to incubate overnight at 4°C. Primary antibody was incubated at 4°C, as was secondary antibody. Antibodies used were rat tyrosinated alpha-tubulin (1:1000, Millipore MAB1864-I), mouse acetylated alpha-tubulin (1:2000, Thermo Fisher Scientifc 32-2700), goat antirat AlexaFluor 488 (1:500, Thermo Fisher Scientifc A11006) and donkey antimouse AlexaFluor 647 (1:200, Thermo Fisher Scientifc A32787).
Confocal imaging
Images were acquired on a Zeiss LSM 800 confocal microscope with a 63x/1.4NA Plan Apochromat oil objective. For spine invasion and comet velocity/abundance data, quick successive time-lapse images were acquired in a single channel (EB3-mNeon) at a rate of 1 frame/3 s for a duration of 100 frames. During live, time-lapse microscopy, neurons were kept at 37°C in a warmed chamber enclosing the microscope and with a glass ring sealed with silicone grease and a glass coverslip to maintain appropriate CO2 levels. Fixed cell imaging for collection of acetylated:tyrosinated tubulin data utilized z-stacks that were acquired in red, green, and far-red channels with slice increments of 0.24 µm.
Image analysis
Images were processed using ImageJ software (NIH). Spine invasions were defined as distinct EB3 puncta or “comets” at least two times brighter than the background fluorescence of the dendritic shaft moving into a dendritic spine and persisting there for at least two frames. The percentage of spines invaded was determined by dividing the number of invaded spines by the total number of spines in the dendritic field. Invasion frequency was defined by the total number of invasions divided by the number of spines invaded. EB3 comet velocities were calculated by the slope of the line produced by the kymograph tool in ImageJ and did not include stationary or paused events. EB3 comet distance traveled was derived from the difference in x values of the coordinates at the beginning and end of the kymograph line and EB3 comet lifetime was derived from the difference in y values. Fluorescence intensity measurements were collected from confocal microscopy images. The average background fluorescence was subtracted from the entire image. Normalized fluorescence was measured in five regions of interest along each dendritic field. Fluorescence intensities were averaged between three dendritic sections per neuron.
Graphing and statistical analysis
All statistical tests and graphing were performed in Prism 8 (GraphPad). Outliers were identified by the ROUT method in which Q = 1% and subsequently excluded. Data were tested for normality using the Kolmorgorov-Smirnov test of normality. If data were normal, then a two-tailed t test was performed. If data were not normal, then a Mann-Whitney U test was performed. P values for scatter plots were determined by simple linear regression analysis in Prism. Data with P values less than 0.05 were considered statistically significant. Complete data are available upon request.
Acknowledgments
This work was supported by National Institute of Health (NIH) grants R01-NS098372 and R01-NS115400 to E.D.. E.H. was supported, in part, by NIH training grant T32-NS105602. We thank Innes Hahn (University of York) and Andreas Prokop (University of Manchester) for providing reagents and advice at the beginning of this project. We thank Daniel Bolt at the Waisman Center for constructive statistical advice.
Abbreviations used:
- BSA
bovine serum albumin
- f-actin
filamentous actin
- FBS
fetal bovine serum
- LTP
long-term potentiation
- LTD
long-term depression
- MT
microtubule
- MTED
microtubule elimination domain
- PKS
paraformaldehyde/Krebs/Sucrose
- Tet
tetracycline.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E24-02-0093) on April 17, 2024.
REFERENCES
- Barnes SJ, Opitz T, Merkens M, Kelly T, von der Brelie C, Krueppel R, Beck H (2010). Stable mossy fiber long-term potentiation requires calcium influx at the granule cell soma, protein synthesis, and microtubule-dependent axonal transport. J Neurosci 30, 12996–13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TW, Jr. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- Borowiak M, Nahaboo W, Reynders M, Nekolla K, Jalinot P, Hasserodt J, Rehberg M, Delattre M, Zahler S, Vollmar A, et al. (2015). Photoswitchable Inhibitors of Microtubule Dynamics Optically Control Mitosis and Cell Death. Cell 162, 403–411. [DOI] [PubMed] [Google Scholar]
- Dent EW, Kalil K. (2001). Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci 21, 9757–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Meiri KF (1992). GAP-43 phosphorylation is dynamically regulated in individual growth cones. J Neurobiol 23, 1037–1053. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ (1997). Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Fanara P, Husted KH, Selle K, Wong PY, Banerjee J, Brandt R, Hellerstein MK (2010). Changes in microtubule turnover accompany synaptic plasticity and memory formation in response to contextual fear conditioning in mice. Neuroscience 168, 167–178. [DOI] [PubMed] [Google Scholar]
- Gu J, Firestein BL, Zheng JQ (2008). Microtubules in dendritic spine development. J Neurosci 28, 12120–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Viesselmann C, Nam S, Merriam E, Dent EW (2008). Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci 28, 13094–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Ballo L, Pietila L, Viesselmann C, Ballweg J, Lumbard D, Stevenson M, Merriam E, Dent EW (2011). BDNF-induced increase of PSD-95 in dendritic spines requires dynamic microtubule invasions. J Neurosci 31, 15597–15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignacz A, Nagy-Herczeg D, Hausser A, Schlett K (2023). Dendritic effects of genetically encoded actin-labeling probes in cultured hippocampal neurons. Mol Biol Cell 34, br8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. (2009). Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61, 85–100. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Yau KW, Gouveia SM, van der Zwan WA, Wulf PS, Keijzer N, Demmers J, Jaworski J, Akhmanova A, Hoogenraad CC (2011). NMDA receptor activation suppresses microtubule growth and spine entry. J Neurosci 31, 8194–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konietzny A, Han Y, Popp Y, van Bommel B, Sharma A, Delagrange P, Arbez N, Moutin MJ, Peris L, Mikhaylova M (2024). Efficient axonal transport of endolysosomes relies on the balanced ratio of microtubule tyrosination and detyrosination. J Cell Sci 137, jcs261737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, Chen SC, Lee GH, Shaiv K, Chen PY, Cheng H, Hong SR, Yang WT, Huang SH, Chang YC, et al. (2022). Precise control of microtubule disassembly in living cells. EMBO J 41, e110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lakonishok M, Liu R, Billington N, Rich A, Glotzer M, Sellers JR, Gelfand VI (2020). Competition between kinesin-1 and myosin-V defines Drosophila posterior determination. eLife 9, e54216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareel MM, De Brabander MJ (1978). Effect of microtubule inhibitors on malignant invasion in vitro. J Natl Cancer Inst 61, 787–792. [PubMed] [Google Scholar]
- McVicker DP, Awe AM, Richters KE, Wilson RL, Cowdrey DA, Hu X, Chapman ER, Dent EW (2016). Transport of a kinesin-cargo pair along microtubules into dendritic spines undergoing synaptic plasticity. Nat Commun 7, 12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring JCM, Grigoriev I, Nijenhuis W, Kapitein LC, Akhmanova A (2022). Opto-katanin, an optogenetic tool for localized, microtubule disassembly. Curr Biol 32, 4660–4674. [DOI] [PubMed] [Google Scholar]
- Merriam EB, Lumbard DC, Viesselmann C, Ballweg J, Stevenson M, Pietila L, Hu X, Dent EW (2011). Dynamic microtubules promote synaptic NMDA receptor-dependent spine enlargement. PLoS One 6, e27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam EB, Millette M, Lumbard DC, Saengsawang W, Fothergill T, Hu X, Ferhat L, Dent EW (2013). Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin. J Neurosci 33, 16471–16482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A, Gundersen GG (1998). Relationship between microtubule dynamics and lamellipodium formation revealed by direct imaging of microtubules in cells treated with nocodazole or taxol. Cell Motil Cytoskeleton 41, 325–340. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S (2000). The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol 10, 865–868. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Pagano A, Honore S, Mohan R, Berges R, Akhmanova A, Braguer D. (2012). Epothilone B inhibits migration of glioblastoma cells by inducing microtubule catastrophes and affecting EB1 accumulation at microtubule plus ends. Biochem Pharmacol 84, 432–443. [DOI] [PubMed] [Google Scholar]
- Peris L, Parato J, Qu X, Soleilhac JM, Lante F, Kumar A, Pero ME, Martinez-Hernandez J, Corrao C, Falivelli G, et al. (2022). Tubulin tyrosination regulates synaptic function and is disrupted in Alzheimer’s disease. Brain 145, 2486–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Hahn I, Lees M, Parkin J, Voelzmann A, Dorey K, Rathbone A, Friel CT, Allan VJ, Okenve-Ramos P, et al. (2019). Efa6 protects axons and regulates their growth and branching by inhibiting microtubule polymerisation at the cortex. Elife 8, e50319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yuan FN, Corona C, Pasini S, Pero ME, Gundersen GG, Shelanski ML, Bartolini F (2017). Stabilization of dynamic microtubules by mDia1 drives Tau-dependent Abeta(1-42) synaptotoxicity. J Cell Biol 216, 3161–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzle P, Esteves da Silva M, Tas RP, Katrukha EA, Hu HY, Wierenga CJ, Kapitein LC, Hoogenraad CC (2018). Activity-Dependent Actin Remodeling at the Base of Dendritic Spines Promotes Microtubule Entry. Curr Biol 28, 2081–2093. [DOI] [PubMed] [Google Scholar]
- Schiff PB, Horwitz SB (1980). Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci USA 77, 1561–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumyatsky GP, Malleret G, Shin RM, Takizawa S, Tully K, Tsvetkov E, Zakharenko SS, Joseph J, Vronskaya S, Yin D, et al. (2005). Stathmin, a gene enriched in the amygdala, controls both learned and innate fear. Cell 123, 697–709. [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J Neurosci 23, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M (2006). Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol 16, 95–101. [DOI] [PubMed] [Google Scholar]
- Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC (2017). Differentiation between Oppositely Oriented Microtubules Controls Polarized Neuronal Transport. Neuron 96, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Martel G, Pavlowsky A, Takizawa S, Hevi C, Watanabe Y, Kandel ER, Alarcon JM, Shumyatsky GP (2014). Learning-induced and stathmin-dependent changes in microtubule stability are critical for memory and disrupted in ageing. Nat Commun 5, 4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L (1997). Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell 8, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W, Brenowitz SD, Hammer JA, 3rd. (2011). Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol 13, 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Schatzle P, Tortosa E, Pages S, Holtmaat A, Kapitein LC, Hoogenraad CC (2016). Dendrites In Vitro and In Vivo Contain Microtubules of Opposite Polarity and Axon Formation Correlates with Uniform Plus-End-Out Microtubule Orientation. J Neurosci 36, 1071–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon AM, Wadsworth P, Jordan MA. (1999). Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol Biol Cell 10, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]