Abstract
Stathmins are small, unstructured proteins that bind tubulin dimers and are implicated in several human diseases, but whose function remains unknown. We characterized a new stathmin, STMND1 (Stathmin Domain Containing 1) as the human representative of an ancient subfamily. STMND1 features a N-terminal myristoylated and palmitoylated motif which directs it to membranes and a tubulin-binding stathmin-like domain (SLD) that contains an internal nuclear localization signal. Biochemistry and proximity labeling showed that STMND1 binds tubulin, and live imaging showed that tubulin binding inhibits translocation from cellular membranes to the nucleus. STMND1 is highly expressed in multiciliated epithelial cells, where it localizes to motile cilia. Overexpression in a model system increased the length of primary cilia. Our study suggests that the most ancient stathmins have cilium-related functions that involve sensing soluble tubulin.
Stathmins are conserved across most animals and implicated in disease biology, but their evolutionary origin and cellular functions are unclear, especially in the case of STMND1, which has not been the subject of any cellular analysis.
Our phylogenetic analysis argues that STMND1 is the human representative of the most ancient clade. STMND1 is primarily expressed in mutlicilliated epithelial cells, where it localizes to cilia.
Our results indicate that tubulin binding negatively regulates the nuclear transport of STMND1, implying that tubulin binding may also regulate the localization or function of other membrane-bound stathmins.
INTRODUCTION
Stathmins are small, evolutionarily conserved proteins that bind to tubulin dimers through a conserved “Stathmin Like Domain” (SLD). Stathmin 1 (STMN1) was discovered as an abundant phosphoprotein in leukemic cells (Feuerstein and Cooper, 1984). Subsequent research revealed that it binds to two αβ-tubulin heterodimers and negatively regulates microtubule assembly (Belmont and Mitchison, 1996; Jourdain et al., 1997). Vertebrate genomes encode three close homologues, STMN2 (SCG10), STMN3 (SCLIP), and STMN4 (RB3; Chauvin and Sobel, 2015), which feature palmitoylated N-terminal extensions that target them to endomembranes (Chauvin and Sobel, 2015). Structural analysis revealed that the SLD adopts a long α-helix conformation, which binds alongside two αβ-tubulin molecules arranged head-to-tail. This arrangement of stathmin-bound tubulin heterodimers resembles the outwardly-curved protofilaments found at the growing and shrinking ends of microtubules. (Jourdain et al., 1997; Gigant et al., 2000; Ravelli et al., 2004). Additionally, the N-terminal of SLD caps the minus end of one α-tubulin via a β-hairpin (Ravelli et al., 2004).
The cellular functions of the canonical stathmins STMN1-4 are not well understood, and their cellular expression patterns vary (Himi et al., 1994; Ozon et al., 1999; Chauvin and Sobel, 2015). The SLDs of all four canonical stathmins can sequester tubulin (Charbaut et al., 2001), and this activity is negatively regulated by phosphorylation at conserved serine sites of SLDs (Chauvin and Sobel, 2015). Multiple studies proposed that STMN1 serves as a relay between kinase signaling and microtubule assembly (Brattsand et al., 1994; Ulrica Marklund et al., 1996), but other functions have also been proposed (Baldassarre et al., 2005; Belletti and Baldassarre, 2011). More recently, the loss of STMN2 was implicated as a cause of the motor neuron disease, amyotrophic lateral sclerosis (ALS; Klim et al., 2019; Melamed et al., 2019; Krus et al., 2022), reinvigorating interest in the function of the membrane-binding stathmins (STMN2-4). While STMN2-4 could negatively regulate MTs by analogy to STMN1, they may not be expressed in sufficient concentration to sequester a meaningful amount of tubulin in physical environment. An alternative possibility is that they sense soluble tubulin concentration to control some downstream effector function.
We initiated phylogenetic analysis to fill gaps in our understanding of stathmin evolution and diversity, and realized that mammalian genomes encode a fifth small protein with SLD homology, STMND1 (Stathmin Domain Containing 1). STMND1 has been neglected in reviews and lacks publications. It is the closest mammalian homologue of the stathmins encoded in the genomes of choanoflagellates, which resemble the shared metazoan ancestral cell type. We found that STMND1 is dually lipidated at its N-terminus and that it binds tubulin. These features qualify it as a membrane-targeted stathmin. Additionally, a functional NLS (nuclear localization signal) exists in its SLD. STMND1 localizes to cilia, plasma membranes and nuclei. Tubulin binding negatively regulates STMND1 nuclear localization, and we propose that it senses tubulin concentration to regulate biological functions.
RESULTS
Phylogenetic analysis reveals an ancient clade of STMND1-related proteins
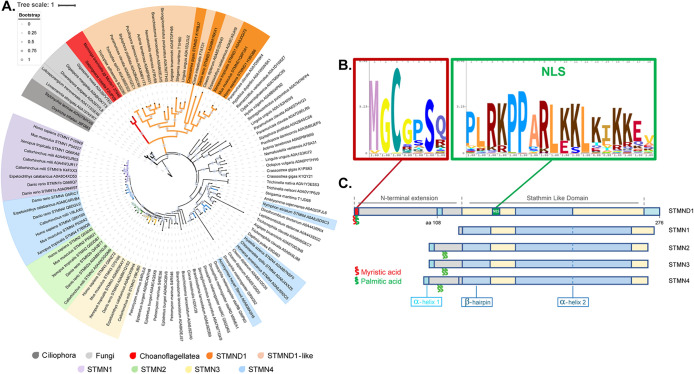
To study the evolutionary history of stathmins, we commenced by identifying protein sequences exhibiting homology to the stathmin-like domain (SLD). We identified 5446 protein sequences with SLD, then focused on proteins where the SLD was the only major domain, with or without an N-terminal extension, similar to STMN1-4. The majority of these SLD-dominated proteins were found in the Opisthokonta supergroup, which comprise Metazoa, fungi, and their unicellular relatives (Steenkamp et al., 2006; Ruiz-Trillo et al., 2008; Shalchian-Tabrizi et al., 2008; Ruggiero et al., 2015). We identified two stathmin homologues from non-Opisthokont species (Stylonychia lemnae A0A078AYX6, Oxytricha trifallax J9F0M3), both of which belong to Ciliophora. We didn’t find SLD-containing proteins from the genome of the model flagellate Chlamydomonas, indicating that no stathmin is part of the highly conserved ciliary axoneme proteome (Figure 1A; Supplemental data). A phylogenetic tree based on SLDs grouped STMN1-4 into distinct clades with a common ancestor (Figure 1A). The tree also revealed a separate clade that emerges adjacent to Choanoflagellate stathmins and includes members from vertebrate and invertebrate genomes that are named STMND1 (orange sector in Figure 1A). STMND1s are well recognized in genomic databases, but we found no publications on them. The branch lengths of the STMND1 clade are longer than those of the canonical STMN1-4, suggesting a longer evolutionary history. STMND1-like proteins are found throughout the animal kingdom (Telford et al., 2008), ranging from primitive comb jelly like Mnemiopsis leidyl (ML16082a) to advanced species like Homo sapiens (H3BQB6).
FIGURE 1:
STMND1 is the orthologue of the ancestral choanoflagellate stathmin. (A) Maximum likelihood phylogenic tree based on the conserved region of SLDs of 106 sequences representing the evolution of stathmins. Branch lengths indicate the expected number of amino acid substitutions per site. Bootstrap values are indicated by circles on the branches. (B) Consensus sequences of the N-terminal dual-lipidation sites (red-outlined) and nuclear localization signal (green-outlined) extracted from 152 annotated STMND1 sequences from UniProt database. (C) Domain arrangement of canonical stathmins and STMND1. The red and green blocks within STMND1 are corresponding to the outline colors of consensus logos in B. The color scheme is corresponding to the color scheme in Supplemental Figure S1C.
Further analysis using full-length alignment of proteins annotated as STMND1 in UniProt unveiled a family of stathmin-like proteins ∼ 280 amino acids in length, characterized by a SLD and an extended N-terminal region. All STMND1 orthologues share a highly conserved N-terminal motif (Consensus: MGCGPSQ). This sequence is predicted to be N-myristoylated at the second position (glycine; Madeo et al., 2022) and S-palmitoylated at the third position (cysteine; Blanc et al., 2015) by standard algorithms. All orthologues examined also contained a predicted nuclear localization signal (Consensus: PLRKPPARLKKLKIKKEV) embedded within the SLD (Nguyen Ba et al., 2009; Figure 1, B and C; Supplemental Figure S1). These features are similarly observed in stathmins from select invertebrate animals which cluster with the STMND1 clade and are probably STMND1 orthologues or close homologues (Figure 1A, light orange).
STMND1 localization and lipid modification
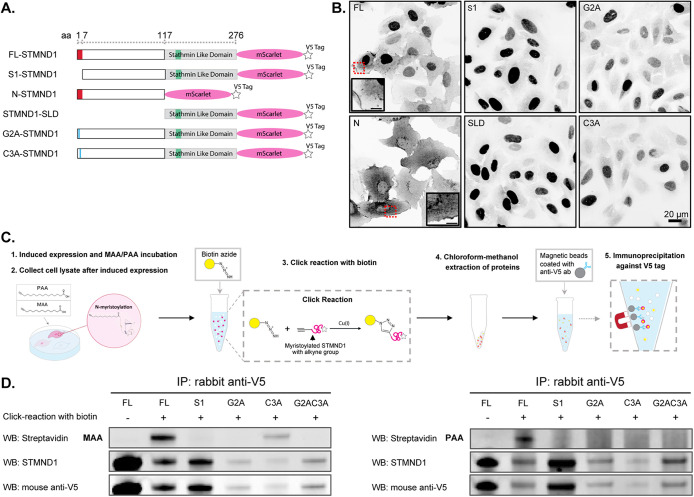
To determine the subcellular localization of STMND1 and test the role of its predicted lipid modification and NLS sites, we expressed full-length, truncated and point mutant versions of human STMND1 fused to a fluorescent protein in proliferating U2OS (human osteosarcoma) cells (Figure 2A). The constructs were doxycycline-inducible, which enabled us to examine the location of STMND1 constructs at different expression levels. High expression levels of constructs that localized to nuclei (after 16 h of induction) caused nuclear morphology defects and cytotoxicity. The data presented are representative of cells with expression at nontoxic levels after 5 h of induction. Full-length STMND1 (FL-STMND1) localized to both plasma membranes and nuclei (Figure 2B). The N-terminal extension of STMND1 (amino acids 1-117), which contains the dual-lipidation motif but lacks the SLD, localized primarily to the plasma membrane and endo-membranes, and was excluded from nuclei (Figure 2B). Truncated STMND1 lacking the first seven amino acids (S1-STMND1) was almost exclusively nuclear, as was STMND1-SLD (Figure 2B). Alanine point mutants in either the conserved second glycine (G2A) or the third cysteine (C3A) abolished membrane location, and nuclear localization predominated (Figure 2B). These results support the predicted function of the first seven amino acids as a plasma membrane localization signal and demonstrate that the NLS within the SLD can promote nuclear localization.
FIGURE 2:
Localization of STMND1 on the plasma membrane and nuclei is driven by the conserved N-myristylation and palmitoylation sites and the NLS within SLD, respectively. (A) STMND1 constructs. (B) Subcellular location of different STMND1 constructs in vivo. Images captured via live imaging following 5 h of induced expression. (C) Workflow for myristic acid/palmitic acid incorporation and IP. (D) Western blots of the IP. Myristic acid (MAA)/palmitic acid (PAA) -incorporated proteins that underwent click-reaction with biotin were detected using streptavidin. The presence of proteins was confirmed by antibodies against STMND1 and V5 tag. Despite efforts to control the gross protein amount at the cell lysate level, precise loading of the same amount of STMND1 proteins proved challenging due to the multiple steps involved in the workflow. Instead, equal volumes of the IP eluate were loaded into each lane. Although the band corresponding to click-reacted FL-STMND1 in the rows of the anti-STMND1 and anti-V5 antibody detection appeared relatively weak, it was the only sample detected by streptavidin in both MAA and PAA treated conditions. Additionally, the C3A mutant showed detectable MAA incorporation.
To investigate whether STMND1 undergoes N-terminal myristoylation and/or palmitoylation, we employed an axide-alkyne click chemistry approach on cell lysates from cell lines expressing FL-STMND1, S1-STMND1, G2A, C3A, or G2A-C3A constructs. Following the induction of expression of these constructs, the cells were treated with 25 μM ω-alkynyl myristic acid or ω-alkynyl palmitic acid to label myristoylated or palmitoylated proteins, respectively (Meldal and Tornøe, 2008). Subsequently, cell lysates were prepared and subjected to azide-alkyne cycloaddition “click chemistry” using azidobiotin to biotinylate the alkyne-tagged lipid and its associated proteins (Meldal and Tornøe, 2008). We then immunoprecipitated total proteins using an antibody against the V5-tag at the C-terminal end of the STMND1 constructs and probed for biotin after SDS–PAGE (Figure 2C). Positive signals in this assay demonstrated that full-length STMND1 underwent myristoylation and/or palmitoylation. Point mutant C3A exhibited clear myristoylation but lacked palmitoylation, whereas G2A showed no acylation (Figure 2D), indicating that the second serine is responsible for myristoylation while the third cysteine is palmitoylated. This observation aligns with the established understanding that palmitoylation necessitates the presence of a proximal myristoyl moiety (Alland et al., 1994; Shenoy-Scaria et al., 1994; Robbins et al., 1995; Godsel and Engman, 1999). In contrast, the S1 construct lacking the first seven amino acids or the mutant with both the second serine and third cysteine replaced did not show these lipid modifications (Figure 2D). These data confirm the lipidations predictions based on protein sequence, but they do not report on what fraction of the protein is lipid modified.
The SLD from STMND1 binds tubulin and inhibits microtubule assembly
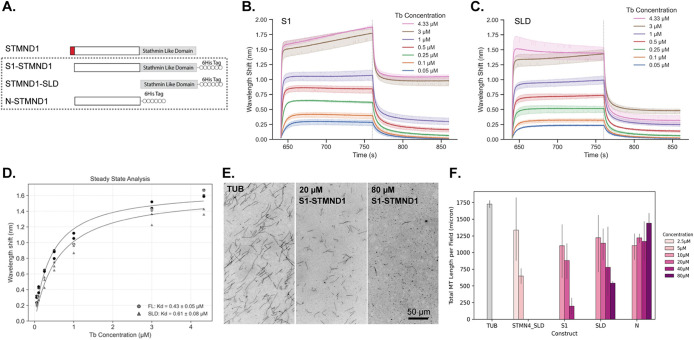
Having confirmed that STMND1 is lapidated on the N-terminus, we were curious whether STMND1 also exhibits the tubulin-binding activity characteristic of other stathmins. The standard method for testing tubulin binding by STMN2,3 and 4 was biochemical analysis of constructs lacking the lipidated N-terminal domain (Gigant et al., 2000; Charbaut et al., 2001). To perform this test with STMND1, we expressed constructs lacking the lipidation motif in bacteria (Figure 3A). We then used biolayer interferometry (BLI) to measure tubulin binding of S1-STMND1 and STMND1-SLD (Figure 3, A, B, and D). Both constructs showed clear evidence of saturable binding to tubulin heterodimers with similar KD values of 0.4 and 0.6 μM, respectively (Figure 3C). We also measured tubulin binding by N-STMND1 and found no evidence of binding activity (Supplemental Figure S3). These results indicate that the SLD of STMND1 binds to tubulin with a KD value similar to other stathmin SLDs (Curmi et al., 1997; Nakao et al., 2004). The total concentration of tubulin in proliferating human cells is ∼10–20 μM (Hiller and Weber, 1978), of which 3–10 μM is soluble dimer (Zhai and Borisy, 1994), so STMND1 likely binds to tubulin in living cells.
FIGURE 3:
STMND1 binds tubulin via SLD, and destabilizes microtubules. (A) The His6-tagged STMND1 constructs without N-terminal lipidation sites for in vitro experiments. (B) Sensorgrams of S1-STMND1 binding to tubulin. (C) Sensorgrams of STMND1-SLD binding to tubulin. (D) Binding curves fitted to the steady-state data based on a 1:1 model, showing the estimated binding constants, Kd, of S1-STMND1 and STMND1-SLD. (E) Rhodamine-labeled tubulin assay to measure the effect of STMND1 proteins on MT dynamics. The images show three typical conditions: tubulin only, tubulin with 20 μM S1-STMND1, and tubulin with 80 μM S1-STMND1. (F) The total length of MTs per imaging field for each protein at different concentrations. The darker color represents higher protein concentration. The SD is shown.
Next, we tested whether STMND1 can inhibit microtubule assembly as reported for STMN1-4. STMND1 constructs were mixed with bovine brain tubulin and GTP. After polymerization and fixation, microtubules were visualized by fluorescence. S1-STMND1 caused a concentration-dependent inhibition of microtubule assembly, but the concentrations required were ∼2-fold higher than for the SLD from STMN4 (Figure 3, E and F). The SLD of STMND1 alone also inhibited microtubule assembly, but less potently than S1-STMND1 did (Figure 3F). The N-terminal domain did not inhibit microtubule assembly (Figure 3F). Nevertheless, given that the STMND1:Tubulin ratio in this biochemical assay is predicted to be higher than in cells, we conclude that negative regulation of microtubules is unlikely to be the main function of STMND1.
Cellular Binding partners of STMND1
We next employed proximity labeling (Rhee et al., 2013; Hung et al., 2014) to investigate the molecular environment and binding partners of STMND1 in cells. We constructed APEX2-fused versions of FL-STMND1, S1-STMND1, N-STMND1, and STMND1-SLD using an inducible expression system and made stably transfected cell lines. The cells were induced for STMND1 expression for a total of 4 h, with the last hour including biotin-phenol treatment, followed by a 1-min pulse of H2O2. APEX2 oxidizes the probe into short-lived reactive radicals that react with nearby amino acids (Rhee et al., 2013; Hung et al., 2014). Biotinylated proteins were then isolated and analyzed by SDS–PAGE and mass spectrometry. To our knowledge, this experiment was the first application of proximity labeling to identify stathmin binding partners in living cells.
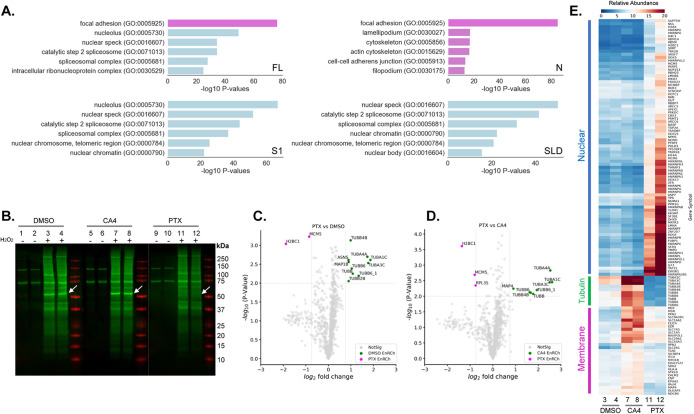
Figure 4A shows ontology analysis of proximity-labeled proteins, reporting the six most significantly enriched GO terms. GO terms dominated by cytoplasmic proteins are colored magenta, while those dominated by nuclear proteins are colored blue. FL-STMND1 caused proximity labeling of both plasma membrane-related and nuclear proteins (Figure 4A). In contrast, N-STMND1 predominantly labeled membrane and cytoskeleton proteins, while S1-STMND1 and STMND1-SLD exhibited exclusive labeling of nuclear proteins (Figure 4A; Supplemental Figure S4). These data are consistent with the cellular locations of the same constructs by imaging (Figure 2). Proteins related to cell junctions were statistically enriched after labeling with the cytoplasmic constructs (FL- and N-STMND1), while nucleolus-associated proteins were enriched for the nuclear constructs (S1-STMND1 and STMND1-SLD; Figure 4A; Supplemental Figure S4).
FIGURE 4:
Proximity labeling reveals the cellular locations of STMND1 and the correlation of STMND1’s tubulin dissociation and nuclear localization. For all samples, the expression of STMND1 constructs in U2OS cells were induced for total of 4 h. For drug-treated samples, MT drugs or DMSO were introduced into the media after 2 h of induction, and cell lysates were collected after an additional 2 h of induction. (A) Gene ontology annotation of the enriched genes of STMND1 constructs. (Magenta, membrane related genes; blue, nuclear genes) (B) Western blot analysis was conducted on APEX2-labeled samples pulled down by streptavidin magnetic beads from cells treated with either PTX (1 μM) or CA4 (200 nM), with or without H2O2 activation. The experiments were performed in duplicate. The membrane is blotted by streptavidin (Green). White arrows point to the ∼50 KDa bands. The remaining samples were subsequently labeled with TMT and then subjected to proteomics. (C and D) Volcano plots showing statistical significance versus protein enrichment in cells expressing FL-STMND1-APEX2 and treated with PTX versus DMSO (C), or PTX versus CA4 (D). The P values (statistical significance of enrichment calculated from unpaired Student’s t tests) for 864 quantified proteins were plotted against the TMT ratios of the comparison pair. Magenta and green dots indicate proteins that are increased or decreased in PTX-treated samples, respectively, while grey dots represent proteins with no significant changes. (E) Hierarchical two-way cluster analysis of an FL-STMND1-APEX2/TMT experiment treated with microtubule-binding drugs. Ward’s minimum variance method was used to cluster the relative abundances of each identified protein (rows) in the individual samples (columns). The relative abundance of a given protein was normalized by the sum of TMT signals in each sample. The color scheme of relative abundances is shown at the bottom of panel.
Tubulin exhibited strong labeling in close proximity to FL-STMND1, indicating their close association within a physiological environment. To more critically test if STMND1 binds tubulin heterodimers in living cells, we performed proximity labeling after manipulating the concentration of soluble tubulin using microtubule polymerizing or depolymerizing drugs (Mitchison and Kirschner, 1984). We subjected FL-STMND1 expressing cells to treatment with 1 μM paclitaxel (PTX) to deplete soluble tubulin or 200 nM combretastatin A4 (CA4) to increase it. PTX forces most tubulin to polymerize, where it cannot binds to a SLD. CA4 binding causes most tubulin to depolymerize, and SLDs are known to bind tubulin-CA4 complexes (Gaspari et al., 2017; PDB: 5LYJ). Immunoblot analysis of the isolated biotinylated proteins showed that PTX and CA4 had opposite effects on proximity labeling of a prominent 50 KDa band, which significantly weakened upon PTX treatment and intensified upon CA4 treatment (Figure 4B). Proteomic analysis indicated that tubulin isomers were the only proteins with molecular weight of 50 KDa, whose enrichment dramatically changed in response to PTX or CA4 treatment (Figure 4, C–E). These data validate the proximity between STMND1 and tubulin/microtubule in cells and show that the degree of proximity responds to changes in tubulin concentration over a physiologically plausible range. PTX treatment, which decreases soluble tubulin and increases tubulin-free STMND1, caused an increase in the enrichment of nuclear proteins such as H2BC1 in the proximity interactome (Figure 4, C–E). These data suggested a negative correlation between tubulin binding and nuclear localization of STMND1.
STMND1 translocates to the nucleus when the soluble tubulin concentration decreases in cytoplasm
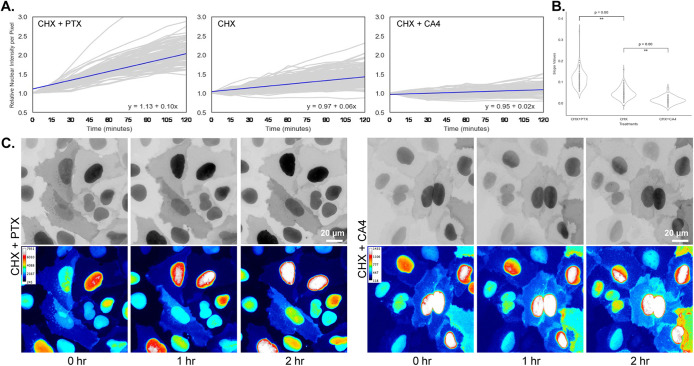
To directly test whether soluble tubulin regulates STMND1’s localization, we performed live-cell imaging of FL-STMND1-mScarlet expressing cells. After inducing sufficient expression of FL-STMND1-mScarlet with doxycycline for 5 h, once the fluorescent intensity reached a suitable level for imaging, we replaced the media containing doxycycline with media containing the protein synthesis inhibitor, cycloheximide. This step ensured that our observations were focused on translocation of existing proteins rather than translation of new proteins. At the same time, we added PTX or CA4 to the media and started imaging. We quantified nuclear intensity throughout the imaging period using an automated algorithm. We refrained from attempting to quantify the nucleus/cytoplasm ratio due to the unreliable segmentation and measurement of the plasma membrane-associated signal. With no microtubule drug, we observed a small increase in nuclear STMND1 over 2 h (Figure 5B). This must be due to mScarlet maturation and translocation, because translation was blocked. PTX, which decreases the soluble tubulin concentration in cytoplasm, significantly increased nuclear translocation (Figure 5A), while CA4, which increases soluble tubulin, suppressed it (Figure 5C). These data confirm the proximity labeling results and suggest that STMND1 senses soluble tubulin concentration, with decreased level of soluble tubulin leading to enhanced nuclear accumulation of STMND1.
FIGURE 5:
Cytosolic tubulin concentration affects nuclear-translocation of STMND1. (A) Graphs depict the relative nuclear intensity per pixel compared with initial points over a 2-h imaging period. Cells were induced to express STMND1-mScarlet for 5 h. And then the media used for induction was replaced with media containing 20 μg/ml cycloheximide (CHX) along with either 1 μM paclitaxel (PTX) or 200 nM combretastatin A4 (CA4) for an additional 2 h. Imaging commenced immediately after the introduction of the CHX and MT drugs cocktail. Each plot is generated from ∼80 nuclei and features a trend line, accompanied by its equation located in the bottom right corner. (B) Plots of the slopes of each nucleus’ intensity change in Figure A. (C) Examples showing time points from live imaging of cells expressing STMND1-mScarlet. Top row: the gray intensity represents the fluorescence of STMND1-mScarlet. Bottom row: Pseudocolor representation illustrates the change in fluorescence intensity over time, with the color scale for each field displayed in the upper left corner. It is observed that the intensity in nuclei and the intensity in cytoplasm change in reverse directions, particularly noticeable in CHX+PTX treated samples. Scale bar: 20 μm.
STMND1 is highly enriched in motile cilia
We next investigated the tissue and cell type expression of STMND1 in public databases. Bulk mRNA data showed high expression in multiciliated epithelial tissues, such as the airway and fallopian tube. Single-cell transcriptome data from the Tabula Sapiens database (Consortium* et al., 2022; Supplemental Figure S4) and immunohistochemistry (IHC) data in the Human Protein Atlas (HPA; Uhlén et al., 2015) showed strong expression in multiciliated epithelial cells, as well as immunolocalization to cilia in the HPA. Consistent with these data, STMND1 mRNA was strongly induced upon differentiation of multiciliated epithelial cells, along with other known cilium proteins (He et al., 2020).
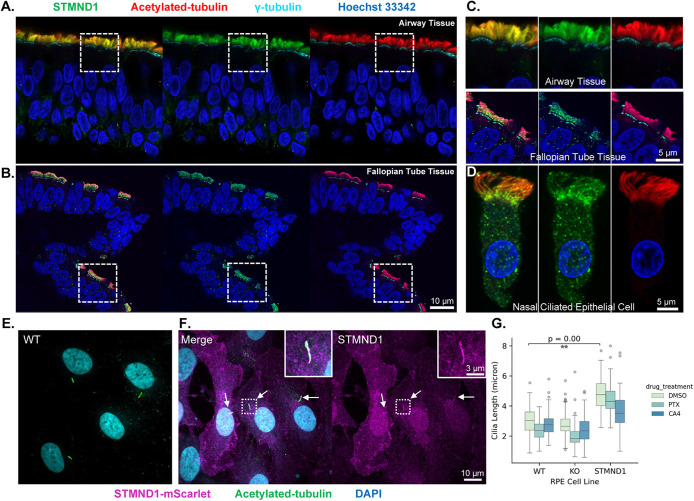
To characterize the subcellular localization of STMND1 in multiciliated tissues, we imaged human tissue using super resolution immunofluorescence microscopy. We first validated the specificity of the anti-STMND1 antibody (Atlas Antibodies, HPA067970) by western blotting the U2OS cell lines which expressed FL-STMND1, N-STMND1 or STMND1-SLD following doxycycline induction (Supplemental Figure S2). IHC on formalin-fixed, paraffin-embedded airway and fallopian tube tissue confirmed that STMND1 proteins localizes to motile cilia (Figure 6, A, B, and C). We then costained with antibodies to γ-tubulin to mark basal bodies and acetylated-tubulin to mark the full length of cilia. We observed that STMND1 was predominantly present in small puncta, primarily enriched within the cilium’s basal and middle regions, alongside distribution in the cytoplasm and nuclei of multiciliated epithelial cells. (Figure 6; Supplemental Videos S1 and S2).
FIGURE 6:
STMND1 is highly enriched on cilia. (A) Immunofluorescence of 20 μm thick sinonasal epithelium tissue. The composite image on the left shows enrichment of STMND1 (green) in the motile cilia marked by acetylated tubulin (red). 𝛾-tubulin (cyan) marks the root of cilia and Hoechst 33342 (blue) stains the nuclei. (B) Immunofluorescence of 20 μm thick fallopian tube tissue. Scale bars for A. and B. is 10 μm. (C) 4X enlargement view of the areas marked by white squares in panel A. and B. with a scale bar of 5 μm. (D) Single nasal epithelial cell immunofluorescence that shows the puncta pattern and distribution of STMND1 throughout the motile cilia. Scale bar is 5 μm. (E) Immunofluorescence of RPE cells with induced primary cilia after 48 h serum-starvation. (F) RPE1 cells expressing STMND1 serum-starved to generate primary cilia. Left, the composite. Right, the channel of STMND1 only. Three examples of STMND1-positive cilia are indicated by arrows. Scale bars for (E) and (F) is 10 μm. 3X enlargement of one of the cilia is shown in squares with a scale bar of 3 μm. STMND1-mScarlet (magenta), acetylated-tubulin (green), DAPI (cyan). (G) Statistical analysis was conducted on the primary cilia length in RPE1 cell lines treated with or without MT drugs. Cilia lengths were measured after a 2-h treatment of MT drugs or DMSO. For wild type (WT) RPE1 cells and STMND1-knock-out (KO) RPE1 cells, DMSO or the MT drugs were directly added to the media. For cells expressing STMND1-mScarlet, serum starvation was performed for 48 h to generate primary cilia, followed by induction of STMND1-mScarlet expression for 5 h. Subsequently, the induction media was replaced by fresh media, and the MT drugs were introduced. WT, wild type RPE1; KO, STMND1-knock-out RPE1; STMND1, RPE1 expressing STMND1-mScarlet.
Movie S1.
Fallopian Tube Cilia Staining
Movie S2.
Airway Cilia Staining
STMND1 localized to primary cilia and elongated them
Genetic perturbation of multiciliated cells poses technical challenges, so we utilized primary cilia as a model system to investigate the effects of STMND1 expression on cilia. The RPE1 cell line, derived from immortalized retinal pigment epithelium (RPE), develops primary cilia when subjected to serum starvation and has been extensively used as a model (Pugacheva et al., 2007). We detected primary cilia in serum-starved RPE1 cells using antibodies to acetylated tubulin, but were unable to detect STMND1 signals using the anti-STMND1 antibody employed for tissue staining (Figure 6E; Supplemental Figure S6). Furthermore, knock out of the STMND1 gene did not impact primary ciliogenesis or cilium length (Figure 6G; Supplemental Figures S6 and S7). However, when we induced exogenous expression of FL-STMND1-mScarlet in RPE1 cells after ciliogenesis, we observed robust STMND1 enrichment within primary cilia (Figure 6F). Interestingly, the cilia in STMND1-positive cells exhibited a significant increase in length compared with control cells or cells with the STMND1 gene knocked out (Figure 6G; Supplemental Figure S7).
Previous studies showed that microtubule-targeting drugs can influence ciliary length, presumably by modulating the levels of soluble tubulin (Sharma et al., 2011). Treatment of ciliated WT or STMND1-KO RPE cells with microtubule drugs revealed that cilia length decreased in cells treated with PTX but showed no significant change in cells treated with CA4, consistent with previous findings (Sharma et al., 2011). However, when we treated STMND1-expressing ciliated RPE1 cells with CA4, the length of STMND1-positive cilia decreased by ∼30% (Figure 6G). These findings suggest that STMND1 is not necessary for primary cilia formation, at least in the RPE1 model. However, the overexpression of STMND1 in cilia was associated with an increase in ciliary length and made length more susceptible to changes in tubulin concentration.
DISCUSSION
Phylogenetic analysis revealed an ancient stathmin clade represented by STMND1 in mammals. STMND1 meets the criteria to be called a stathmin based on its sequence homology and ability to bind soluble tubulin with a physiologically relevant affinity. While it shares the capability to inhibit microtubule assembly in biochemical assays with the canonical stathmins STMN1-4, this effect required a high concentration of STMND1. High expression level of STMND1 caused nuclear morphology defects in transfected cells so we think it is unlikely that the cellular function of STMND1 is to regulate cytoplasmic microtubules. Rather, our data suggests STMND1 regulates some aspect of motile cilium biology in multiciliated epithelial cells and perhaps mediates cilium-to-nucleus communication. STMND1 likely evolved to serve this unknown function in early eukaryotes that resembled chaonoflagellates, and its expression pattern in human tissues suggested it may serve a parallel function in modern multicilliated epithelial. However, it is not part of the core cilium proteome, because it was undetectable in primary cilia in RPE1 cells, no homologue was found in Chlamydomonas. In mouse tracheal epithelial cells, the expression level of STMND1 only becomes detectable in mature ciliated cells, suggesting that STMND1 is not required for ciliogenesis (He et al., 2020). Gene expression analysis showed that STMND1 is significantly downregulated in nasal mucosa after ammonia exposure, consistent with a sensory role (Wang et al., 2020). We further speculate that the concept that a membrane-targeting stathmin senses tubulin concentration to regulate some nonmicrotubule-focused function may extend to STMN2 and be relevant to its recently-discovered neuroprotective function (Klim et al., 2019; Melamed et al., 2019; Guerra San Juan et al., 2022; Krus et al., 2022; Thornburg-Suresh et al., 2023).
Conserved features of the STMND1 subfamily revealed by our work include a N-terminal dual-lipidation motif and a predicted NLS within the tubulin-binding SLD. N-myristoylation is usually cotranslational and irreversible (Towler et al., 1987; Farazi et al., 2001), while S-palmitoylation is posttranslational and reversible (Blaskovic et al., 2013). The presence of both modifications near the N-terminus is essential for cilium targeting of some proteins (Godsel and Engman, 1999; Bijlmakers and Marsh, 2003), so the N-terminal motif in STMND1 is probably a ciliary membrane targeting sequence. The predicted NLS located within the SLD is functional, because all constructs containing it exhibited nuclear localization. The SLD is also functional in tubulin binding, as evidenced by biochemical and proximity labeling assays. The position of the NLS within the SLD suggests that tubulin binding sterically competes with recognition of STMND1 by nuclear import factors. Consistent with this hypothesis, addition of paclitaxel, which lowers soluble tubulin concentration by forcing polymerization, caused STMND1 to translocate more into nuclei in both proximity labeling and live cell imaging assays albeit in nonciliated U2OS cells.
A full understanding of STMND1 function will require experiments in mutliciliated cells, and we need to be careful interpreting data from a primary cilium model that does not normally express STMND1. With that important qualification, our data hint at interesting possible functions. We currently favor models in which STMND1 acts as a sensor of soluble tubulin, rather than a negative regulator of microtubule polymerization. Our evidence that tubulin binding and nuclear transport are mutually exclusive points to a model in which STMND1 has some nuclear function which is negatively regulated by tubulin binding. In the nucleus, STMND1 appeared to associate with spliceosomal proteins in nucleolar speckles, based on proximity labeling and live imaging data. This suggests a role in regulating pre-mRNA processing. Alternatively or coordinately, STMND1 might function in cilia, for example, to participate in length regulation. The lipid modification of STMND1 resembles that of the important cilium assembly factor ARL13b, and both increased primary cilia length when overexpressed (Lu et al., 2015). These proteins have different binding partners, but might share the ability to facilitate transport of vesicles to the base of the cilium to promote membrane for growth. Once concentrated in cilia, STMND1 might use its tubulin binding activity to regulate the growth of axonemal microtubules or to sense the local concentration of soluble tubulin. Two important questions we have not yet addressed are related to interpreting possible functions in cilia: can STMND1 bind membranes and tubulin at the same, and is its targeting to membranes reversible? Addressing possible functions of STMND1 in cilia and nuclei will require phenotypic analysis in multciliated cells combined with deeper biochemical analysis to answer these questions.
MATERIAL AND METHODS
Request a protocol through Bio-protocol.
Bioinformatics
We searched InterPro 92.0 (Paysan-Lafosse et al., 2023) and identified 5446 protein sequences with homology to stathmin that contain a SLD; InterPro Entry: PF00836). We also performed BLASTp against JGI-Monosiga brevicollis MX1 v1.0 (King et al., 2008; Nordberg et al., 2014) using the Salphigoeca rosetta stathmin as the query sequence (accession #: F2UPV8), to identified candidate Monosiga brevicollis stathmin sequences. Additionally, we searched Ensembl (Yates et al., 2022) and KEGG (Acropora digitifera; Shinzato et al., 2011) databases for more information on nonvertebrate genomes. From this sequence pool, we selected 106 stathmin amino acid sequences from reference species that represent the general evolutionary history of life for phylogenetic analysis. The representative species of Metazoa were chosen according to Borowiec et al. (2015) and Laumer et al. (2019). The selected species include major model animals and those representing the major diverging events in evolutionary history (Supplemental data). For each selected species, all detected stathmin variants of are included. Besides the representative metazoan stathmins, the 106 sequences also include all the nonmetazoan stathmins identified from Fungi, Ciliophora, and Choanoflagellatea. We excluded several SLD-containing proteins identified by InterProt from our analysis, for example Rhodosorus marinus (A0A7S3EF07, A0A7S2ZSJ6) that differ largely from the typical stathmins, containing an SLD at the N-terminus and a testis-specific protein PBS13 T-complex 11 (TCP11) at the C-terminus (unpublished data).
We aligned the amino acid sequences using Clustal OMEGA (Sievers and Higgins, 2021), removed sites with less than 70% coverage, to create a final dataset for estimating phylogenetic tree that comprised 145 amino acids in the conserved region of SLDs. To infer the evolutionary history, we conducted Maximum Likelihood tree inference in IQ-Tree 2.0 (Minh et al., 2020). Branch support was calculated with 1000 ultrafast bootstrap replicates (Minh et al., 2013; Hoang et al., 2018) and Shimodaira–Hasegawa approximate likelihood ratio tests (Guindon et al., 2010). The best model for the data was selected using ModelFinder method (Kalyaanamoorthy et al., 2017). The LG+G4 model (Yang, 1994; Le and Gascuel, 2008) was chosen as the best-fit model according to the Bayesian information criterion scores and weights. The phylogenetic tree was annotated and presented though iTOL (Letunic and Bork, 2021).
We used SVMyr (Madeo et al., 2022) for myristoylation site prediction, SwissPalm (Blanc et al., 2015) for palmitoylation site prediction, and NLStradamus (Nguyen Ba et al., 2009) for nuclear location sequence prediction. To generate the consensus motifs of STMND1, we collected 152 explicitly-annotated STMND1 sequences from Uniprot (The UniProt Consortium, 2023), aligned with Clustal OMEGA, and used Skylign (Wheeler et al., 2014) with hidden Markov models to generate consensus logos for the N-myristoylation and S-palmitoylation motif, and nuclear location sequences.
Molecular biology and construct of cell lines
All STMND1 constructs are based on human STMND1 (Uniprot H3BQB6). All primers and DNA blocks were ordered from Integrated DNA Technologies. The doxycycline-inducible plasmids were based upon a piggyBac vector, PB-TRE-EGFP-EF1a-rtTA (Addgene #104454), and synthesized DNA cassettes were cloned between NcoI and XhoI. Each construct was transfected together with a transposase vector at ratio of 4:1, using Lipofectamine 3000 (Thermo Fisher Scientific, L3000008). Transfected cells were further selected with 1 μg/ml puromycin (Thermo Fisher Scientific, A1113803).
We generated STMND1 knock-out cell lines using CRISPR (Hsu et al., 2013) system. The sgRNA targeting the exon4 and its flanking regions were designed according to CRISPick (Doench et al., 2016) and CHOPCHOP (Labun et al., 2019). We inserted a pair of sgRNAs into two expression vectors for sgRNAs and for expression of Cas9 linked to mCherry (Addgene #64324) or GFP (Addgene #48138). The two vectors were transfected into cells simultaneously using Lipofectamine 3000. Transfected cell lines were initially selected by flow cytometry on the expression of both mCherry and GFP, followed by screening of purified genomic DNA (Invitrogen, K182001) using PCR, and finally targeted fragments were sequenced.
To purify soluble proteins from bacteria, we excluded the first seven amino acids that include conserved lipidation sites from all STMND1 constructs used in the in vivo experiments. We cloned the full-length STMND1 (amino acids 8-276), SLD (amino acids 118-276), and the N-terminal region of STMND1 (amino acids 8-117) from human into the pHAT5 vector (Addgene #112586) and subsequently transformed them into BL21 Rosetta cells.
In vitro STMND1 expression and purification
Transformed BL21 Rosetta bacteria were grown in Luria-Bertani medium containing carbenicillin (27 μg/ml) and chloramphenicol (50 μg/ml) at 37°C. For protein expression, the bacteria were grown until reaching an O.D.600 of ∼0.5, and then induced by IPTG for ∼4 h. After pelleting the cells by centrifuge at 4°C, the pellet was resuspended in 10 ml of Lysis Buffer (50 mM Tris-HCl [p.H. 7.4], 150 mM NaCl, 1 mM BME, 25 mM Imidazole, 10% glycerol) supplemented with protease inhibitor cocktail per 1 g of cell pellet. The cells were lysed by French Press and the cell lysate was clarified by centrifugation at 14,500 g (11 krpm on Sorval SS34 rotor) at 4°C for 25 min. The protein was purified from the centrifuged supernatant by nickel-charged affinity chromatography (Ni-NTA-resin, Qiagen), concentrated by centrifugal filter with size cutoff, and stored at –80°C until use. The protein concentration was measured by Bradford test.
Binding affinity measurements
Binding Affinity Measurements were performed through biolayer interferometry (BLI) using an Octet Red 384 instrument (ForteBio) housed in the Center for Molecular Interactions (CMI) at Harvard Medical School. C-terminally His6-tagged STMND1 constructs (FL-STMND1 [amino acids 8-276] and STMND1-SLD [amino acids 118-276]) were diluted to a concentration of 0.22 μM in BLI reaction buffer (50 mM Tris, 50 mM NaCl, 0.5 mM GTP, 1 mM MgCl2, 1 mM DTT, pH 7.4) supplemented with 0.02% Tween. Bovine brain tubulin purified from calf brains was used to make tubulin dilutions (0 μM, 0.05 μM, 0.1 μM, 0.25 μM, 0.5 μM, 1 μM, 3 μM, and 4.33 μM) using the BLI reaction buffer supplemented with 0.02% Tween. Tubulin dilutions were prepared immediately after thawing on ice and kept at room temperature. The Octet 384-Well Tilted-Bottom Microplate (Sartorius; Item No.: 18-5080) and Ni-NTA Octet Biosensors (Sartorius; Item No.: 18-5101) were used according to the manufacturer’s instructions for the Octet BLI instrument.
We defined the experimental wells and sensor type, and established the assay timing using the Octet BLI Discovery Software for the experimental setup. Our assays involved a 600-s delay to ensure appropriate sensor hydration, followed by an equilibration step (180 s), a loading phase (400 s), a baseline step (60 s), an association phase (120 s), and a dissociation phase (100 s). To determine the KD values, we used the Octet Data Analysis HT 11.0 Suite by assessing the average response of each sample once the sensor reached steady state.
For the FL-STMND1 (amino acids 8-276) comparison with N-STMND1 (amino acids 8-117), we used the same protocol as above but only four tubulin dilutions were used (0 μM, 0.05 μM, 0.5 μM, and 4.33 μM). The assay was performed in triplicate in independent runs on different days.
The raw data were analyzed using Jupyter Lab environment. The data of steady status were used to fit parameters for binding curves based on 1:1 binding model, and we calculated the KD from the fitter parameters. The binding curves and KD estimations were plotted using matplotlib and seaborn. The Jupyter notebooks containing the code are available.
Microtubule polymerization assays
Tubulin and tubulin labeled with Biotium CF 543 NHS ester were prepared as described previously Hyman et al. (1991) and mixed at a ratio of 20:1 unlabeled to labeled together with diluted STMND1 proteins to a final concentration of 3.5 mg/ml tubulin and 1 mM GTP in BRB80 buffer (80 mM KPIPES (pH 6.8), 1 mM MgCl2, 1 mM EGTA) at 0°C – 4°C. This mix (10 μl) was incubated at 37°C for 10 min, then diluted 1:1000 into fixative (60% glycerol, 0.2% glutaraldehyde, 80 mM KPIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA) and mixed by gently pipetting up and down three times. The mix (4 μl) was placed on a glass slide, covered with a 18 × 18-mm coverslip, and imaged by fluorescence microscopy.
Images were acquired with a Nikon Ti 2 motorized inverted microscope, using a 40x/1.30 NA objective lens. The images were processed in ImageJ FUJI for presentation. Images of microtubule fibers were then analyzed in a Jupyter Lab environment using pandas, SciPy, NumPy and plotted with matplotlib and seaborn. The pipelines, and Jupyter notebooks are available.
Cells and antibodies
Antibodies against the following antigens were used: polyclonal anti-STMND1 antibody (Atlas Antibodies, HPA067970); V5-Tag (D3H8Q) rabbit mAb (Cell Signaling Technology, #1320); V5 epitope tag mouse antibody (E10/V4RR; Novus Biologicals, NBP2-37825); anti-α-tubulin antibody (DM1A; Sigma-Aldrich, 05-829); antiacetylated tubulin antibody, mouse monoclonal, (Sigma-Aldrich, T7451); GAPDH (D16H11) XP rabbit mAb (Cell Signaling Technology, #5174); acetylated alpha tubulin antibody (6-11B-1) Alexa Fluor 488 (Santa Cruz, sc-23950 AF488); Alexa Fluor 647 anti-gamma tubulin antibody (TU-30)-C-terminal (Abcam, ab19114).
Cell culture and inducible expression of STMND1 in mammalian cells
U2OS and retinal pigmented epithelial cells (RPE1-hTERT) were cultured in DMEM (Corning, 10-013-CV) supplemented with 10% FBS (Life Technologies, 26140-079). The STMND1 transgene cells were maintained with a final concentration of 1 μg/ml puromycin (Thermo Fisher Scientific, A1113803). The expression of STMND1 constructs in mammalian cells was induced by treatment with 2 μg/ml doxycycline (Sigma D3072) for 2–4 h. The impact of prolonged induced expression of STMND1 is demonstrated in Supplemental Figure 2. To halt further synthesis of STMND1 following induced expression, the induction media was replaced with media containing cycloheximide at a concentration of 20 μg/ml. To induce ciliation, RPE cells were starved in DMEM without FBS for 48 h.
Immunofluorescence and imaging
Cells plated on #1.5 coverslips were washed with PBS and crosslinked with freshly made 4% paraformaldehyde and 0.2% glutaraldehyde in PBS for 15 min at room temperature, and washed three times with PBS afterwards. Permeabilization was carried out with 0.1% Triton-X in PBS, 5 min at RT. Cells were blocked with 5% BSA (constituted from powder BSA, Roche Fraction V, sold by Sigma Catalogue Number: 10735078001) in PBS for 1 h at RT. Primary antibodies were diluted in PBS, and cells were incubated with diluted primary antibodies for ∼16 h at 4°C in a humidified chamber. Cells were then washed three times with in PBS and incubated with fluorescently labeled secondary antibodies, diluted 1:500 in PBS for 1 h at RT, and washed three times with PBS. The coverslips were mounted on glass slides using VECTASHIELD plus antifade mountaining medium with DAPI (Vector Laboratories, H-2000) and left in a dark chamber overnight before imaging.
The following antibodies were used for immunofluorescence: polyclonal anti-STMND1 antibody (Atlas Antibodies, HPA067970); V5-Tag (D3H8Q) rabbit mAb (Cell Signaling Technology, #1320); V5 epitope tag mouse antibody (E10/V4RR; Novus Biologicals, NBP2-37825); anti-α-tubulin antibody (DM1A; Sigma-Aldrich, 05-829); antiacetylated tubulin antibody, mouse monoclonal, (Sigma-Aldrich, T7451).
Images were acquired with a Nikon Ti motorized inverted microscope equipped with Yokogawa CSU-W1 spinning disk confocal at Harvard Nikon Imaging Center, using a 63x/1.40 NA Oil objective lens. The images were processed in ImageJ FUJI for presentation. Images of cilia were then analyzed in a Jupyter Lab environment using pandas, SciPy, NumPy and plotted with matplotlib and seaborn. The pipelines and Jupyter notebooks are available on GitHub.
High-resolution tissue imaging
The BOND RX Automated Stainer was used to bake formalin-fixed, paraffin-embedded (FFPE) specimen slides at 56°C for 55 min, dewax using Bond Dewax solution at 72°C for 30 min., and perform antigen retrieval using Epitope Retrieval 1 (LeicaTM) solution at 100°C for 20 min. Hoechst 33342 and all antibodies were diluted in SuperBlock Blocking Buffer (Thermo Fisher) and incubated overnight at 4°C in the dark. 24 × 50 coverslips were wet mounted using 150 μL of 50% Glycerol in PBS before imaging.
Antibodies used for histology staining are: polyclonal anti-STMND1 antibody (Atlas Antibodies, HPA067970); acetylated alpha tubulin antibody (6-11B-1) Alexa Fluor 488 (Santa Cruz, sc-23950 AF488); Alexa Fluor 647 antigamma tubulin antibody (TU-30)-C-terminal (Abcam, ab19114).
Images were taken using a 63x/1.4 NA objective lens with Airyscan 2 super-resolution on a Zeiss LSM980 microscope using ZEN software (v.3.4.91) at Harvard Microscopy Resource On the North Quad (MicRoN) core, and three-dimensional rendering was completed in Imaris (v.9.9.1).
Immunoblotting, immunoprecipitation (IP) of alkyne-myristate/palmitate proteins from cells
The cells were subjected to treatment with 25 μM ω-alkynyl myristic acid (MAA) or alkynyl palmitic acid (PAA; Click Chemistry Tools, #1164, or #1165)while simultaneously inducing STMND1 protein expression with doxycycline overnight. To collect the cell lysate, the cells were washed with cold PBS, harvested, and then lysed using cold EDTA-free NP-40 buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1% NP-40 substitute, EDTA-free complete protease inhibitor [Roche, 11836170001]), while rocking for 15 min at 4°C. The protein concentrations of the cell lysate were determined using a BCA assay (Thermo Fisher Scientific, #23225), following the manufacturer’s instructions. Click chemistry (Click Chemistry Tools, #1001) was then used to react the resulting cell lysates with 10 μM azidobiotin (Click Chemistry Tools, #1488), which labeled the MAA/PAA-tagged proteins with biotin. The resulting protein was precipitated by chloroform-methanol extraction and reconstituted according to the manufacturer’s manual (Click Chemistry Tools, #1001). For IP, ∼ 200 μg of biotin-labeled protein for each condition was incubated with Dynabeads Protein G (Invitrogen, #10004D) that had been coated with mouse anti-V5 antibody (Novus Biologicals, NBP2-37825) for 1 h, and then extensively washed with NP-40 buffer. Electrophoresis samples were prepared by adding 4 × loading buffer, boiling for 5 min, and then loading onto 12.5% acrylamide gels. After electrophoresis, the gels were transferred onto 0.2 μM nitrocellulose membrane (Bio-Rad) and probed with the following antibodies: polyclonal anti-STMND1 antibody (Atlas Antibodies, HPA067970); V5-Tag (D3H8Q) rabbit mAb (Cell Signaling Technology, #1320); anti-α-tubulin antibody (DM1A; Sigma-Aldrich, 05-829); GAPDH (D16H11) XP rabbit mAb (Cell Signaling Technology, #5174).
Proximity proteomics
Proximity labeling was performed on cells expressing APEX2-fused STMND1 constructs. After 4 h of doxycycline-induced expression, biotinyl tyramide (Toronto Research Chemicals, B397770) was added to the media at a final concentration of 0.5 μM, and cells were incubated in the labeling media for 1 h. To initiate labeling, H2O2 (Sigma-Aldrich, H1009) was added to a final concentration of 1 mM. Exactly 1 min after H2O2 treatment, the labeling solution was decanted, and cells were washed three times with cold quenching solution containing 10 mM sodium ascorbate (VWR 95035-692), 5 mM Trolox (Sigma-Aldrich, 238813), and 10 mM sodium azide (Sigma-Aldrich, S2002). The cell lysate was harvested by scraping and cleared by centrifugation at 16,000 × g for 20 min, and the resulting pellet was flash-frozen and stored at −80°C until streptavidin pull-down. The cell pellet was lysed using a cell lysis solution (8 M Urea, 100 mM sodium phosphate pH 8, 1% SDS (wt/vol), 100 mM NH4HCO3, 10 mM TCEP, sterile-filtered). Protein was extracted by adding 55% TCA (Sigma-Aldrich, 91228) at a 1:1 ratio to the lysate and precipitated by centrifugation. The protein pellet was then washed three times with −20°C cold acetone. Protein was subjected to cysteine alkylation using 20 mM iodoacetamide (Sigma-Aldrich, I6125), quenched by 50 mM DTT (Sigma-Aldrich, 43815), and pulled down using streptavidin magnetic beads (VWR, PI88817). A detailed procedure for the protein processing procedure can be found in Kalocsay, 2019.
Subsequent protein processing procedures and MS analysis were carried out as described. The digested peptides were labeled with TMTpro 16-plex (Thermo Fisher Scientific, A44520) for 1 h. Data collection followed a MultiNotch MS3 TMT method using an Orbitrap Lumos mass spectrometer coupled to a Proxeon EASY-nLC 1200 liquid chromatography system (both Thermo Fisher Scientific). Peptides were searched against a size-sorted forward and reverse database of the Homo sapiens reference proteome (Uniprot 03/2021) using SEQUEST (v.28, rev. 12)-based software. Spectra were first converted to mzXML. For the searches, a mass tolerance of 20 p.p.m. for precursors and a fragment ion tolerance of 0.9 Da were used. The search allowed for a maximum of two missed cleavages per peptide.
Carboxyamidomethylation on cysteine was set as a static modification (+57.0214 Da), and oxidized methionine residues (+15.9949 Da) were searched for dynamically. A target decoy database strategy was applied, and a false discovery rate (FDR) of 1% was set for peptide-spectrum matches after filtering by linear discriminant analysis. The FDR for final collapsed proteins was 1%. MS1 data were calibrated postsearch, and searches performed again. Quantitative information on peptides was derived from MS3 scans. Quantitative tables were generated requiring an MS2 isolation specificity of >70% for each peptide and a sum of TMT (tandem mass tags) signal:noise ratio (s:n) of >200 over all channels for any given peptide, and then exported to Excel and further processed therein. Proteomics raw data and search results were deposited in the PRIDE archive. The relative summed TMT s:n for proteins between two experimental conditions was calculated from the sum of TMT s:n for all peptides of a given protein quantified.
Supplementary Material
Acknowledgments
The authors want to thank Simon Rayner, Hannah Mitchison, and Peter G. Czarnecki for helpful discussions. We acknowledge the Nikon Imaging Center at Harvard Medical School (NIC) for providing technical support of imaging. This study was supported by the National Institute of General Medical Sciences, grant R35GM131753 to Timothy Mitchison, BWH President’s Scholar Award no. U54-CA225088 to Sandro Santagata. This work was conducted with support from the Cancer Prevention and Research Institute of Texas, grant RR220032, to M.K. who is a CPRIT Scholar in Cancer Research. The authors declare no competing interests.
Abbreviations used:
- CA4
combretastatin A4
- CHX
cycloheximide
- MAA
myristic acid
- MT
microtubule
- NLS
nuclear localization signal
- PAA
palmitic acid
- PTX
paclitaxel
- RPE
retinal pigment epithelium
- STMND1
Stathmin Domain Containing 1
- STMN
stathmin
- SLD
stathmin-like domain
- TB
tubulin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-12-0514) on April 17, 2024.
REFERENCES
- Alland L, Peseckis SM, Atherton RE, Berthiaume L, Resh MD (1994). Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J Biol Chem 269, 16701–16705. [PubMed] [Google Scholar]
- Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A (2005). p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell 7, 51–63. [DOI] [PubMed] [Google Scholar]
- Belletti B, Baldassarre G (2011). Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets 15, 1249–1266. [DOI] [PubMed] [Google Scholar]
- Belmont L, Mitchison TJ (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623–631. [DOI] [PubMed] [Google Scholar]
- Bijlmakers M-J, Marsh M (2003). The on–off story of protein palmitoylation. Trends Cell Biol 13, 32–42. [DOI] [PubMed] [Google Scholar]
- Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, Goot FG van der (2015). SwissPalm: Protein Palmitoylation database. F1000Res 4, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovic S, Blanc M, van der Goot FG (2013). What does S-palmitoylation do to membrane proteins? The FEBS Journal 280, 2766–2774. [DOI] [PubMed] [Google Scholar]
- Borowiec ML, Lee EK, Chiu JC, Plachetzki DC (2015). Extracting phylogenetic signal and accounting for bias in whole-genome data sets supports the Ctenophora as sister to remaining Metazoa. BMC Genomics 16, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattsand G, Marklund U, Nylander K, Roos G, Gullberg M (1994). Cell-cycle-regulated phosphorylation of oncoprotein 18 on Ser16, Ser25 and Ser38. Eur J Biochem 220, 359–368. [DOI] [PubMed] [Google Scholar]
- Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A (2001). Stathmin Family Proteins Display Specific Molecular and Tubulin Binding Properties*. J Biol Chem 276, 16146–16154. [DOI] [PubMed] [Google Scholar]
- Chauvin S, Sobel A (2015). Neuronal stathmins: A family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Prog Neurobiol 126, 1–18. [DOI] [PubMed] [Google Scholar]
- Consortium* TTS, Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, et al. (2022). The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curmi PA, Andersen SS, Lachkar S, Gavet O, Karsenti E, Knossow M, Sobel A (1997). The stathmin/tubulin interaction in vitro. J Biol Chem 272, 25029–25036. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI (2001). The Biology and Enzymology of ProteinN-Myristoylation. J Biol Chem 276, 39501–39504. [DOI] [PubMed] [Google Scholar]
- Feuerstein N, Cooper HL (1984). Rapid phosphorylation-dephosphorylation of specific proteins induced by phorbol ester in HL-60 cells. Further characterization of the phosphorylation of 17-kilodalton and 27-kilodalton proteins in myeloid leukemic cells and human monocytes. J Biol Chem 259, 2782–2788. [PubMed] [Google Scholar]
- Gaspari R, Prota AE, Bargsten K, Cavalli A, Steinmetz MO (2017). Structural Basis of cis- and trans-Combretastatin Binding to. Tubulin. Chem 2, 102–113. [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M (2000). The 4 Å X-Ray Structure of a Tubulin:Stathmin-like Domain Complex. Cell 102, 809–816. [DOI] [PubMed] [Google Scholar]
- Godsel LM, Engman DM (1999). Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J 18, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra San Juan I, Nash LA, Smith KS, Leyton-Jaimes MF, Qian M, Klim JR, Limone F, Dorr AB, Couto A, Pintacuda G, et al. (2022). Loss of mouse Stmn2 function causes motor neuropathy. Neuron 110, 1671–1688.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- He M, Wu B, Ye W, Le DD, Sinclair AW, Padovano V, Chen Y, Li K-X, Sit R, Tan M, et al. (2020). Chloride channels regulate differentiation and barrier functions of the mammalian airway. eLife 9, e53085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller G, Weber K (1978). Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell 14, 795–804. [DOI] [PubMed] [Google Scholar]
- Himi T, Okazaki T, Wang H, McNeill TH, Mori N (1994). Differential localization of SCG10 and p19/stathmin messenger RNAs in adult rat brain indicates distinct roles for these growth-associated proteins. Neuroscience 60, 907–926. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018). UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol 35, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee H-W, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY (2014). Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol Cell 55, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T (1991). [39]Preparation of modified tubulins. In: Methods in Enzymology, Academic Press, 478–485. [DOI] [PubMed] [Google Scholar]
- Jourdain L, Curmi P, Sobel A, Pantaloni D, Carlier M-F (1997). Stathmin: A Tubulin-Sequestering Protein Which Forms a Ternary T 2 S Complex with Two Tubulin Molecules. Biochemistry 36, 10817–10821. [DOI] [PubMed] [Google Scholar]
- Kalocsay M (2019). APEX Peroxidase-Catalyzed Proximity Labeling and Multiplexed Quantitative Proteomics. In: Proximity Labeling: Methods and Protocols, ed. Sunbul M, Jäschke A, New York, NY: Springer, 41–55. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R, et al. (2019). ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krus KL, Strickland A, Yamada Y, Devault L, Schmidt RE, Bloom AJ, Milbrandt J, DiAntonio A (2022). Loss of Stathmin-2, a hallmark of TDP-43-associated ALS, causes motor neuropathy. Cell Rep 39, 111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47, W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumer CE, Fernández R, Lemer S, Combosch D, Kocot KM, Riesgo A, Andrade SCS, Sterrer W, Sørensen MV, Giribet G (2019). Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc R Soc B 286, 20190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008). An Improved General Amino Acid Replacement Matrix. Mol Biol Evol 25, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49, W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Toh MT, Narasimhan V, Thamilselvam SK, Choksi SP, Roy S (2015). A function for the Joubert syndrome protein Arl13b in ciliary membrane extension and ciliary length regulation. Dev Biol 397, 225–236. [DOI] [PubMed] [Google Scholar]
- Madeo G, Savojardo C, Martelli PL, Casadio R (2022). SVMyr: A Web Server Detecting Co- and Post-translational Myristoylation in Proteins. J Mol Biol 434, 167605. [DOI] [PubMed] [Google Scholar]
- Melamed Z, López-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, Freyermuth F, McMahon MA, Beccari MS, Artates JW, et al. (2019). Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci 22, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal M, Tornøe CW (2008). Cu-Catalyzed Azide−Alkyne Cycloaddition. Chem Rev 108, 2952–3015. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A (2013). Ultrafast Approximation for Phylogenetic Bootstrap. Mol Biol Evol 30, 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020). IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner M (1984). Dynamic InstabIlity of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Nakao C, Itoh TJ, Hotani H, Mori N (2004). Modulation of the Stathmin-like Microtubule Destabilizing Activity of RB3, a Neuron-specific Member of the SCG10 Family, by Its N-terminal Domain. J Biol Chem 279, 23014–23021. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM (2009). NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I (2014). The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42, D26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozon S, Mestikawy SE, Sobel A (1999). Differential, regional, and cellular expression of the stathmin family transcripts in the adult rat brain. J Neurosci Res 56, 553–564. [DOI] [PubMed] [Google Scholar]
- Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, et al. (2023). InterPro in 2022. Nucleic Acids Res 51, D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA (2007). HEF1-Dependent Aurora A Activation Induces Disassembly of the Primary Cilium. Cell 129, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M (2004). Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202. [DOI] [PubMed] [Google Scholar]
- Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY (2013). Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 339, 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins SM, Quintrell NA, Bishop JM (1995). Myristoylation and Differential Palmitoylation of the HCK Protein-Tyrosine Kinases Govern Their Attachment to Membranes and Association with Caveolae. Mol Cell Biol 15, 3507–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero MA, Gordon DP, Orrell TM, Bailly N, Bourgoin T, Brusca RC, Cavalier-Smith T, Guiry MD, Kirk PM (2015). A Higher Level Classification of All Living Organisms. PLOS ONE 10, e0119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF (2008). A Phylogenomic Investigation into the Origin of Metazoa. Mol Biol Evol 25, 664–672. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T (2008). Multigene Phylogeny of Choanozoa and the Origin of Animals. PLOS ONE 3, e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Kosan ZA, Stallworth JE, Berbari NF, Yoder BK (2011). Soluble levels of cytosolic tubulin regulate ciliary length control. MBoC 22, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM (1994). Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J Cell Biol 126, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG (2021). The Clustal Omega Multiple Alignment Package. Methods Mol Biol 2231, 3–16. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL (2006). The protistan origins of animals and fungi. Mol Biol Evol 23, 93–106. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Bourlat SJ, Economou A, Papillon D, Rota-Stabelli O (2008). The evolution of the Ecdysozoa. Philos Trans R Soc B: Biol Sci 363, 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2023). UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res 51, D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg-Suresh EJC, Richardson JE, Summers DW (2023). The Stathmin-2 membrane-targeting domain is required for axon protection and regulated degradation by DLK signaling. J Biol Chem 299, 104861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, Gordon JI (1987). Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc Natl Acad Sci USA 84, 2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Marklund U, Larsson N, Gradin HM, Brattsand G, Gullberg M (1996). Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J 15, 5290–5298. [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang M, Liu C, Huang L, Gao Y, Yu M, Zhao S, Li X (2020). Ammonia Exposure Induced Cilia Dysfunction of Nasal Mucosa in the Piglets. BioMed Res Int 2020, e1705387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Clements J, Finn RD (2014). Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics 15, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (1994). Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol 39, 306–314. [DOI] [PubMed] [Google Scholar]
- Yates AD, Allen J, Amode RM, Azov AG, Barba M, Becerra A, Bhai J, Campbell LI, Carbajo Martinez M, Chakiachvili M, et al. (2022). Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Research 50, D996–D1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Borisy GG (1994). Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J Cell Sci 107, 881–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.