Abstract
To derive structural information about the vesicular stomatitis virus (VSV) nucleocapsid (N) protein, the N protein and the VSV phosphoprotein (P protein) were expressed together in Escherichia coli. The N and P proteins formed soluble protein complexes of various molar ratios when coexpressed. The major N/P protein complex was composed of 10 molecules of the N protein, 5 molecules of the P protein, and an RNA. A soluble N protein-RNA oligomer free of the P protein was isolated from the N/P protein-RNA complex using conditions of lowered pH. The molecular weight of the N protein-RNA oligomer, 513,879, as determined by analytical ultracentrifugation, showed that it was composed of 10 molecules of the N protein and an RNA of approximately 90 nucleotides. The N protein-RNA oligomer had the appearance of a disk with outer diameter, inner diameter, and thickness of 148 ± 10 Å, 78 ± 9 Å, and 83 ± 8 Å, respectively, as determined by electron microscopy. RNA in the complexes was protected from RNase digestion and was stable at pH 11. This verified that N/P protein complexes expressed in E. coli were competent for encapsidation. In addition to coexpression with the full-length P protein, the N protein was expressed with the C-terminal 72 amino acids of the P protein. This portion of the P protein was sufficient for binding to the N protein, maintaining it in a soluble state, and for assembly of N protein-RNA oligomers. With the results provided in this report, we propose a model for the assembly of an N/P protein-RNA oligomer.
Vesicular stomatitis virus (VSV) is a nonsegmented, negative-stranded RNA virus belonging to the rhabdovirus family. The 11,161-nucleotide genome of VSV (21) contains five genes and is encapsidated by the nucleocapsid (N) protein to form the ribonucleoprotein (RNP) complex. Associated with the RNP are the phosphoprotein (P protein) and the large polymerase subunit (L protein) (12). These three components are the transcriptionally active unit of the virus (12). The two remaining genes encode the matrix (M) protein and glycoprotein (G). The five genes are arranged in the order 3′ N-P-M-G-L 5′, and the relative abundance of each individual transcript is related to its genomic position. The amounts of the individual mRNAs decrease with distance from the 3′ end of the genome due to a single polymerase entry site at the 3′ end of the genome (14) and localized attenuation at each gene junction (3, 25, 46). Thus, the N and P mRNAs are produced in the greatest abundance. The two proteins encoded by these genes form complexes in virus-infected cells (36, 39) and in vitro when expressed simultaneously (8, 28, 34, 40). In each case, the N/P protein complexes were observed as multiple species with various molar ratios. The optimal molar ratio for efficient viral replication was found to be between 1:1 and 2:1 (N to P protein), while ratios substantially above or below this range had a negative effect on replication (28, 34, 36, 40, 47). The P protein when provided in an appropriate molar ratio to the N protein can prevent the concentration-dependent aggregation of the N protein, which keeps the N protein in a form that can support replication (20).
Previously, the gene sequences encoding the N (1, 15) and P (15) proteins were determined, and the protein sequences were deduced. The N and P proteins have been individually expressed in Escherichia coli with different results. The P protein was expressed and shown to be functional in in vitro transcriptional activity assays (2, 22). The N protein, on the other hand, was successfully expressed in large quantities; however, the protein was produced in an insoluble form which required denaturation and refolding to have measurable activity (7). The refolded N protein required extremely high salt concentrations in order to prevent its aggregation and precipitation, which limited its suitability for further characterization. When the N and P proteins were coexpressed in E. coli, an N/P protein complex was isolated, but the protein was in large aggregates (19).
Since the N and P proteins had previously been observed to interact with each other in cells, many experiments were attempted to determine the regions of each protein that are responsible for their heteromeric interaction (13, 17, 33, 38, 42). The P protein can be divided into three domains. Domain I (residues 1 to 137) is highly acidic and is the site of phosphorylation; domain II is a linker between the first and third domains; the third domain (residues 250 to 274) contains a cluster of basic amino acids and is essential for the interaction with the N protein (13, 17, 33, 38, 42). The P protein binding regions within the N protein are less well defined; nonetheless, Takacs et al. suggested that the extreme C terminus of the N protein was critical for association with the P protein (42). This group showed that removal of the final five amino acids of the N protein abolished binding with the P protein; however, no analyses of the effects on the conformation of the N protein were done.
Numerous experiments have been performed to uncover structural details of rhabdoviral N proteins. Blumberg et al. developed a method to refold N protein in the presence of high-molar salt and viral leader RNA (4). Electron photomicrographs showed that the refolded material bound to the short RNA oligonucleotide was oligomeric and had a toroidal morphology. In a separate set of experiments, Iseni et al. overexpressed the rabies virus N protein in the baculovirus system (23). From this expression system, they recovered nucleocapsids as well as N protein oligomers. The N protein oligomers, although not completely uniform in composition, contained predominantly 10 monomers of the N protein and had a disk-like appearance similar to those observed by Blumberg et al. (4). Perhaps the greatest detail about the structure of the N protein has come from scanning transmission electron microscopy analysis of nucleocapsids isolated from virions (44). The N protein was shown from these studies to have a wedge-shaped, bilobed structure that was elongated down one direction. To date, high-resolution details of the N protein are not available.
Previously published data suggests that the interaction between the N and P proteins is critical to encapsidation of the viral genome. The molar ratio between the N and P proteins determines the optimal condition for VSV replication. To characterize the complex between the N and P proteins or the N protein alone, it would be necessary to produce the individual proteins in a soluble form, which to date has been a major hurdle to structural and biochemical studies. In this report, we show that the N protein can be produced under conditions in which the majority of the protein is in a soluble, encapsidation-competent form in E. coli if it is expressed concomitantly with the P protein. A soluble complex between the N and P proteins and a short RNA has been isolated and characterized. The complex contains 10 molecules of the N protein, five copies of the P protein, and an RNA of ∼90 nucleotides. Removal of the P protein from the complex resulted in a single oligomeric form of the N protein bound to RNA. With the results provided in this report, we propose a model for the assembly of an N/P protein-RNA oligomer. This structure may be representative of the stable RNP of VSV.
MATERIALS AND METHODS
Materials.
Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs. The original plasmids pET-15b, pET-16b, and pET-21b, as well as E. coli DH5α and BL21(DE3), were purchased from Novagen. The TA helper plasmid for direct cloning of PCR products was purchased from Invitrogen. Primers for PCR were purchased from IDT.
Plasmid construction.
The N, P, and Pf (C-terminal 72 amino acids of P) genes were amplified from a cDNA clone of VSV Indiana strain via PCR with the following primers. N-5′ (5′-CCATGGCTTCTGTTACAGTCAAGAGAGAATC-3′) and N-3′ (5′-CCATGGTATATCTCCTTCATTTGTCAAATTCTGAC-3′) each contained the NcoI restriction site. The 3′ primer also contained a ribosomal binding site (RBS), to allow independent translation for the P protein, following the N gene stop codon. P-5′ (5′-GGATTCATATGGATAATCTCACAAAAAGTTC-3′) and P-3′ (5′-GTGATCATATGTTACAGAGAATATTTGACTC-3′) each contained the NdeI restriction site; Pf-5′ (5′-GGAATTCCATATGGCAGTATCAGATGTTTGGTCTC-3′) contained a restriction site for NdeI; and Pf-3′ (5′-CGCGGATCCTTACAGAGAATATTTGACTCTC GC-3′) contained a restriction site for BamHI.
The PCR-amplified N and P genes were ligated individually with T4 DNA ligase into the TA helper vector. The N gene and the vector pET-16b were digested with restriction enzyme NcoI and ligated together; this vector was designated pET-N. The P gene and pET-N were digested with NdeI, and the P gene was then ligated into digested pET-N in frame with the N-terminal polyhistidine tag to create the vector pET-N/P (Fig. 1A).
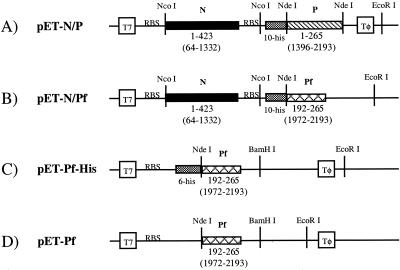
FIG. 1.
Design of plasmids pET-N/P (A), pET-N/Pf (B), pET-Pf-His (C), and pET-Pf (D). Plasmids A and B allowed for the coexpression of N protein with P and Pf proteins, respectively. The parent vector for both A and B was pET-16b (Novagen). In contruct A, the P gene was inserted into the NdeI restriction site, while in B, the Pf gene was inserted into the NdeI and EcoRI restriction sites. In each case, the genes are in frame with an N-terminal 10-histidine tag. The N gene (A and B) was cloned into the NcoI restriction site with the addition of an RBS following the stop codon for the N gene and prior to the P (or Pf) gene. In each of the coexpression plasmids (A and B), one T7 promoter (T7) controls the transcription of both N and P (or Pf) genes. The individual RBSs preceding each gene allow each protein to be translated independently from this single transcript. Plasmids C and D are for single expression of the Pf protein. In both cases, the Pf gene was inserted into the NdeI and BamHI sites of the parent vector, pET-15b (Novagen) (C) or pET-21b (Novagen) (D). In construct C, the Pf gene is in frame with an N-terminal six-histidine tag. In A to D, numbers below the boxes for the N, P, and Pf genes represent the amino acids of the proteins encoded by the corresponding genes, with nucleotide numbers according to GenBank entry g335873 in parentheses. Tφ, T7 terminator.
The vectors pET-15b and pET-21b and the PCR product corresponding to the C-terminal 72-amino-acid gene segment of P, which we have designated Pf, were digested with restriction enzymes NdeI and BamHI. The digested Pf gene was ligated into both the pET-15b and pET-21b vectors, which resulted in the vectors pET-Pf-His, which adds a polyhistidine tag on the N terminus of Pf, and pET-Pf, respectively (Fig. 1C and 1D).
To generate a plasmid containing the full-length N gene and the P gene fragment Pf, plasmid pET-Pf was digested with restriction enzymes NdeI and EcoRI to liberate the Pf gene. The vector pET-N/P was digested with NdeI and EcoRI, which resulted in the elimination of the P gene. The Pf gene was then ligated to the digested pET-N/P vector, resulting in the vector pET-N/Pf, which again provides an N-terminal polyhistidine tag on Pf (Fig. 1B).
Protein expression and purification.
In all cases, cDNA clones were transformed into E. coli strain BL21(DE3) and proteins were expressed according to standard protocols (Novagen). Briefly, E. coli was grown at 37°C in LB broth (Difco) in the presence of ampicillin, when an A600 equal to 0.5 was reached, protein expression was induced at 30°C with isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 1 mM) for 5 h. The cells harboring pET-N/P, pET-N/Pf, and pET-Pf-His were individually harvested by centrifugation, resuspended in 50 mM Tris buffer (pH 7.9) containing 500 mM NaCl, sonicated, and centrifuged at 15,000 × g for 30 min. Soluble His-tagged N/P, N/Pf, and Pf produced from pET-Pf-His were purified over an Ni affinity column (Pharmacia) according to the Novagen protocol. N/P and N/Pf were chromatographed on a Sephacryl S-300 gel filtration column in 50 mM Tris buffer (pH 7.5) containing 300 mM NaCl. Cells containing pET-PF were harvested and resuspended in 50 mM HEPES buffer (pH 7.5) containing 50 mM NaCl. The cells were then sonicated and centrifuged at 15,000 × g for 30 min, and soluble fractions were loaded onto a HiTrap SP ion-exchange column (Pharmacia). Native Pf was eluted with a linear NaCl gradient. Native and His-tagged Pf proteins were further purified on a Superdex S-75 (Pharmacia) gel filtration column with an elution buffer consisting of 50 mM Tris buffer (pH 7.5) containing 300 mM NaCl.
Western blot analysis of the N/P and N/Pf complexes and the Pf protein.
N/P and N/Pf protein complexes and His-tagged Pf protein were expressed, isolated by Ni column chromatography, and electrophoresed as described in this report. Proteins were transferred from the gels to polyvinylidene difluoride membranes by electroblotting. Monospecific polyclonal rabbit antibodies made to bacterially expressed N protein were used as the primary antibody for identification of the N protein. Likewise, monospecific polyclonal rabbit antibodies made to bacterially expressed P protein were used as the primary antibody for identification of the P protein. The N and P antibodies were shown to immunoprecipitate authentic VSV N and P proteins, respectively, synthesized in virally infected cells (data not shown). Primary antibodies were incubated with membranes in a blocking buffer consisting of 100 mM Tris (pH 7.5) containing 1.5 mM NaCl and 0.1% Tween 20 (TTBS). Membranes were washed with TTBS followed by incubation in TTBS with anti-rabbit immunoglobulin G antibodies conjugated with alkaline phosphatase (Sigma). Membranes were again washed with TTBS, then transferred to 100 mM Tris buffer (pH 9.5) containing 100 mM NaCl and 5 mM MgCl2, and developed with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (Gibco).
Densitometric analysis of the N/P protein complexes.
Densitometric analysis of the N/P protein complexes was performed as follows. The N protein was purified as described by low-pH treatment of Ni affinity-purified N/P protein complexes followed by gel filtration chromatography. To isolate the P protein, Ni affinity-purified N/P protein complexes were denatured in 8 M urea, and the P protein was purified by Ni affinity chromatography under denaturing conditions, with all buffers containing 8 M urea. Standard curves for the staining profiles of the N and P proteins with Coomassie blue stain were made as follows. Concentrations of the purified N protein and purified P protein solutions were quantitated by Bradford assay, and known amounts of 0.26, 0.47, 0.943, and 1.8 μg for the N protein and 0.06, 0.12, 0.24, 0.48, 0.95, and 1.9 μg for the P protein were loaded onto a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, electrophoresed, and stained with Coomassie brilliant blue R250 stain. Following destaining, the gels were photographed with an AlphaImager 2000 system (Alpha Innotech Corporation). Protein bands were selected on the images, and the optical densities of the bands were integrated with the aid of the Alphasease program, version 3.2 (Alpha Innotech). Protein amounts were plotted versus optical densities and curves were fit to the data by the least squares method. These curves served as the standard curves for each proteins staining profile with Coomassie blue stain. N/P protein complexes, isolated by gel filtration chromatography (see Fig. 3A, peaks 2 and 4), were electrophoresed and photographed, and protein quantities were determined by comparison with the appropriate standard curve.
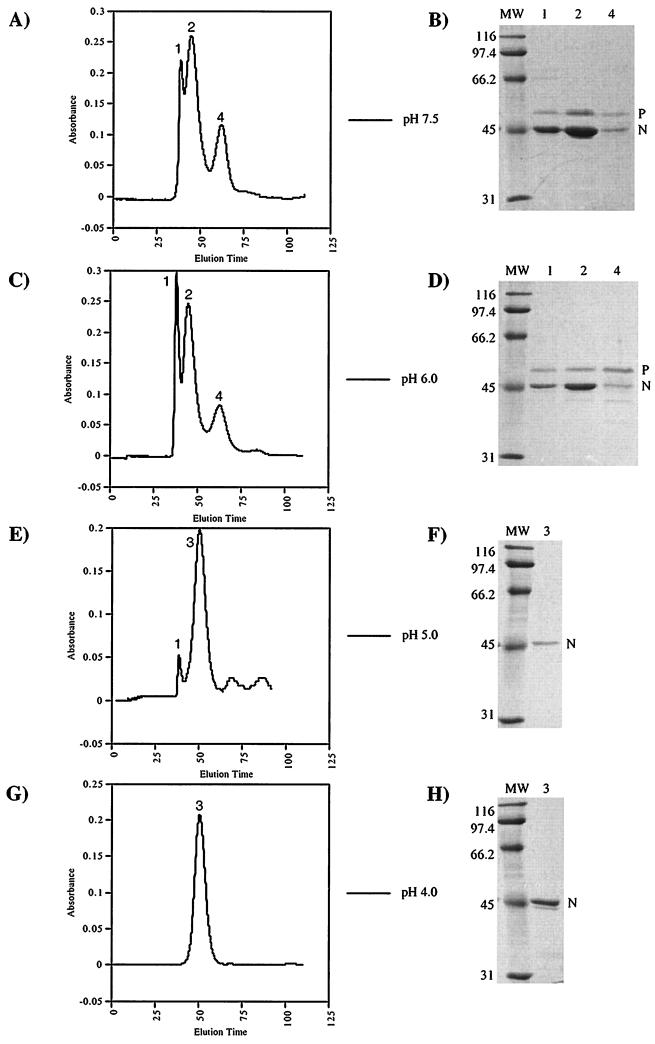
FIG. 3.
Gel filtration profiles of the N/P protein complexes at pH 7.5 (A), 6.0 (C), 5.0 (E), and 4.0 (G). The N and P proteins were coexpressed in E. coli harboring plasmid pET-N/P. Following protein expression, the N/P protein complexes were isolated by Ni affinity chromatography. These partially purified protein complexes were dialyzed in either 0.05 M Tris (pH 7.5) containing 300 mM NaCl or 0.1 M citrate (pH 6.0, 5.0, or 4.0) containing 0.25 M NaCl. Following dialysis, the complexes were chromatographed on a Sephacryl S-300 size exclusion column (16/60; Pharmacia) at a flow rate of 1 ml/min. In each case, the elution buffer was the same as the dialysis buffer. The horizontal axis shows the elution time in minutes; the vertical axis shows A280. The proteins contained in the peaks from each purification were analyzed by SDS-polyacrylamide gel electrophoresis (gel B for panel A, gel D for panel C, gel F for panel E, and gel H for panel G). (B, D, F, and H). The proteins were electrophoresed on 10% gels and stained with Coomassie brilliant blue R-250. Lane numbers correspond to the peak numbers on the corresponding chromatogram to the left. Lanes containing molecular weight standards are labeled MW; positions are indicated in kilodaltons.
Isolation of uncomplexed N protein and characterization of the N/P protein complex.
Previous work has shown that VSV replication is affected by pH and that the reason for this is that the amount of P protein bound to N protein is decreased with a decrease in pH (6, 28). Given this, we used pH as a method for separating the N and P proteins. The N and full-length P proteins were coexpressed and purified with a Ni column as described above. The protein complexes were then dialyzed in 0.1 M citrate buffer (pH 4) containing 250 mM NaCl. This resulted in dissociation of the complex and precipitation of P protein from the solution while N protein remained in solution. Uncomplexed N protein was dialyzed against 50 mM Tris buffer (pH 7.5) containing 50 mM NaCl and purified on a Sephacryl S-300 gel filtration column. N protein was verified by N-terminal amino acid sequencing and Western blotting. All subsequent characterization of the uncomplexed N protein was performed with N protein prepared in this manner from N/full-length P protein complexes unless stated otherwise.
To study the dissociation of the N/P protein complex, Ni column-purified N/P protein complexes were dialyzed against 0.1 M citrate buffer containing 0.25 M NaCl at pH 6.0, 5.0, or 4.0. Post dialysis, the resulting complexes were filtered through a 0.22-μm-pore-size syringe filter to remove precipitated protein and then chromatographed on a Sephacryl S-300 gel filtration column in citrate buffer with the pH of the dialysis buffer. The proteins present in the peaks were analyzed by electrophoresis on an SDS–10% polyacrylamide gel containing an acrylamide-to-bisacrylamide ratio of 75:1.
The ability of Pf protein to bind with purified N protein was assessed by a binding assay. Purified His-tagged Pf protein was bound to charged chelating Sepharose beads, and the beads were washed with the binding buffer. Purified N protein was then incubated with the Pf-bound beads. The beads were washed and eluted. In addition, N protein was incubated with the beads in the absence of the Pf protein, washed, and then eluted with same procedure. Eluted fractions were analyzed by electrophoresis on SDS-polyacrylamide gels as above.
Analytical ultracentrifugation of purified N protein.
Sedimentation velocity measurements were made in a Beckman XL-A analytical ultracentrifuge using band centrifugation (45). For analytical band centrifugation, the macromolecular solution was transferred onto a denser bulk solution from a small sample well upon the application of a centrifugal field, a density gradient immediately formed by diffusion of small solute molecules from the bulk solution into the macromolecular lamella. Band centerpieces were either purchased from Beckman-Coulter Instruments (part no. 331359) or fabricated from standard double-sector centerpieces obtained from that company. For the band experiments, the sample well was filled, while the cell was partially assembled, with 30 μl of N protein at a concentration of approximately 0.75 mg/ml. After sample filling, the cell was fully assembled using the same procedure as for a conventional boundary experiment. The sample and reference sectors were then filled with 340 and 380 μl of buffered bulk solution described below. A four-cell An-60 Ti rotor was used for the band centrifugation velocity analysis at 58,000 rpm at 20°C, with scanning at 230 nm. Sedimentation coefficients were determined from the band data using the software program MLAB (Civilized Software, Bethesda, Md.) with algorithms developed by Peter Schuck (National Institutes of Health).
For N band centrifugation, we used either 100 or 90% D2O:H2O (by volume) containing 0.1 M NaCl and 0.05 M phosphate buffer for density gradient stabilization of bands. The density and relative viscosity values at 20°C for the 100% buffered D2O solution are 1.1145 and 1.2685, respectively; for the 90% buffered D2O solution, they are 1.1040 and 1.2455. Values for the density and viscosity of D2O were obtained from Kirschenbaum (27); those for 0.1 M NaCl and 0.05 M phosphate buffer were obtained from Laue et al. (29). These values were rapidly calculated using the software program Sednterp, developed by D. Hayes, J. Philo, and T. Laue. The standard correction equation was used to convert measured s values to s20,w values using the density and relative viscosity values given above. The partial specific volume of the N protein (0.735) and the sequence molecular weight (47,257) were also calculated using the Sednterp software program.
Sedimentation equilibrium analysis was also performed in a model XL-A analytical ultracentrifuge using a four-cell An-60 Ti rotor. A double-sector cell was loaded with 120 μl of N protein in 50 mM NaCl–50 mM Tris (pH 7.5). A density of 1.00163 was evaluated for the above buffered solution using the Sednterp software program. Sedimentation equilibrium data were obtained at 5,000 and 8,000 rpm at 20°C. Equilibrium was considered reached when two consecutive sets of data taken 2 h apart were completely superimposable with small root mean squares difference and the difference spectra showed no systematic deviations. Sedimentation equilibrium was achieved in 72 h at 5,000 rpm, and the angular velocity was changed to 8,000 rpm. We allowed 48 h to obtain equilibrium at the higher centrifugal velocity. The data analysis software package of the XL-A (version 4.0 and Microcal Origin version 4.1) allows for a global nonlinear regression of the sedimentation equilibrium data to determine the molecular weight of a single component or for the modeling of self-associating systems if appropriate.
Electron microscopy of the VSV N protein.
Purified N protein at a concentration of 0.05 mg/ml was placed on carbon-coated grids, fixed briefly with glutaraldehyde, and stained with 0.1% NH4-phosphotungstic acid. Images were taken with a Philips CM-10 electron microscope at a magnification of ×105,000.
Isolation of RNA from purified N protein and N/P protein complexes.
N/P protein complexes were expressed and isolated by Ni affinity chromatography. These N/P protein complexes were further purified by gel filtration chromatography as described above. N/P protein complexes from peak 2 (see Fig. 3A) were used for RNA extraction. Also, N protein was isolated by low-pH treatment of either the N/P or N/Pf protein complexes which were isolated by Ni affinity chromatography. Then 0.5 mg of either total protein of purified N/P protein complexes or purified N protein from either N/P or N/Pf complexes was mixed with an equal volume of phenol chloroform, vortexed, and centrifuged. The nucleic acid-containing aqueous layer was removed and added to an equivalent volume of chloroform, vortexed, and centrifuged. The aqueous layer was again removed, and the RNA was precipitated with 2.5 volumes of ethanol with the addition of NaCl at −70°C. RNA was analyzed by electrophoresis on an 8 M urea–polyacrylamide gel.
RESULTS
Cloning and expression of the N/P protein complex in E. coli.
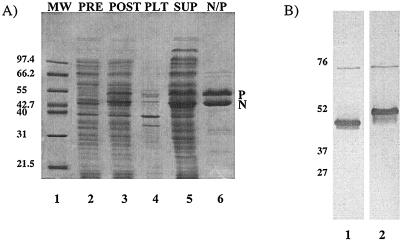
Previous work expressing the N protein in E. coli resulted in the majority of the protein being insoluble. Since the P protein has been shown to function to maintain the N protein in a soluble form in infected cells (20, 40), we designed a single vector to express the N and P proteins concomitantly in E. coli that would result in the production of the proteins in approximately the same molar ratios as found in infected cells. The genes encoding the N and P proteins were placed in the Novagen vector pET-16b to generate plasmid pET-N/P (Fig. 1A). The N gene was placed in front of the P gene, and the transcription of both genes was controlled by a single T7 transcription promoter situated upstream of the N gene so that the T7 polymerase transcribed the N gene first and continued directly into the P gene. In order for the translation of each protein to remain independent, the plasmids were designed so that each gene was preceded by its own bacterial RBS. E. coli harboring pET-N/P was grown at 37°C (Fig. 2A, lane 2) and induced to express the N and P proteins at 30°C (lane 3). Following induction, cells were harvested and lysed. The whole-cell extracts were then centrifuged to separate the insoluble portion of the cell extract from the soluble portion. Both the soluble and insoluble fractions were electrophoresed on an SDS-polyacrylamide gel. The results showed that the N and P proteins were predominantly in the soluble fraction (lane 5), while both proteins were barely detectable in the insoluble fraction (lane 4). Initial purification of the N and P proteins was performed on a Ni affinity column, by taking advantage of the N-terminal histidine tag on the P protein. Ni column-purified proteins were analyzed by electrophoresis on SDS-polyacrylamide gels, which showed that the N protein copurified along with histidine-tagged P protein during this single affinity chromatography step (lane 6). This indicated that the N and P proteins were in a complexed form. The N protein migrated to its characteristic position of ∼50 kDa, while the P protein migrated much slower than would be expected from its calculated molecular mass of 29 kDa on an SDS-polyacrylamide gel, as has been reported previously (31), due to the numerous acidic amino acid residues within the N-terminal half of the protein (lane 6). The mobility of the P protein was also retarded due to the high acrylamide-to-bis-acrylamide ratio used in preparation of the gels. The identity of each protein was confirmed by Western blotting with monospecific polyclonal antibodies to either the N or P protein, as shown in Fig. 2B. Approximately 35 mg of purified soluble complexes was obtained from 1 liter of culture.
FIG. 2.
Expression, purification, and detection of the N and P proteins. The N and P proteins were coexpressed in E. coli harboring plasmid pET-N/P. These cells were harvested and sonicated. The insoluble proteins were then pelleted from the total-cell extracts by centrifugation. Soluble proteins were applied to a Ni affinity column, and the N and P proteins were purified. (A) Coomassie brilliant blue R-250-stained SDS–10% polyacrylamide gels (containing an acrylamide-to-bisacrylamide ratio of 75:1) with molecular weight markers (MW), total-cell extract prior to N/P protein induction with IPTG (PRE), total-cell extract after induction with IPTG (POST), insoluble fraction from the total postinduction cell extract (PLT), supernatant fraction from the total postinduction cell extract (SUP), and Ni affinity column-purified N/P protein complexes (N/P). (B) Western blot of proteins isolated by Ni column chromatography as in panel A (lane 6). The N/P protein complexes were electrophoresed on an SDS–10% polyacrylamide gel (containing an acrylamide-to-bisacrylamide ratio of 75:1). Lanes 1, N protein detected using a monospecific polyclonal antibody for the N protein; 2, P protein detected using a monospecific polyclonal antibody for the P protein. Sizes are indicated in kilodaltons.
Isolation of N/P protein oligomers.
The Ni column-purified N/P protein complexes were chromatographed on a Sephacryl S-300 (Pharmacia) gel filtration column for further purification. Multiple peaks that contained both N and P proteins eluted from the column with time (Fig. 3A, peaks 1, 2, and 4). The proteins within each peak were electrophoresed on an SDS-polyacrylamide gel and detected with Coomassie brilliant blue blue R-250 stain (Fig. 3B, lanes 1, 2, and 4). Peak 1 was a high-molecular-weight species that eluted at the void volume of the gel filtration column, indicating a size of greater than 1,500 kDa, the limit of the column. It was not possible to determine if this peak corresponded to a single molar complex or contained more than one species of the N/P protein complex, because all protein complexes equal to or larger than 1,500 kDa would elute at this position. Peak 2 contained the most abundant species of the N/P protein complex. The size of this complex was determined to be ∼669 kDa, based on its elution time in relation to known molecular weight standards. The molar ratio of N to P protein in this species was determined by densitometry to be 2:1. The size of the last species (peak 4) was estimated by size exclusion chromatography on three separate columns (Sephacryl S-300, Sephacryl S-200, and Superose 6) and determined to be between 133 and 151 kDa. The composition of peak 4 is unclear, as densitometric measurements were not consistent. We believe that this peak could correspond to oligomers of P protein, possibly a pentamer.
Low-pH treatment of N/P protein complexes and purification of the N protein.
To study the N protein individually, we needed to separate the N/P protein complexes. pH often affects the conformation and/or charge of proteins, and as a result, it is possible to use pH as the driving force to dissociate protein-protein complexes (48). When the soluble N/P protein complexes were dialyzed to low pH, the changes of the complexes at different pH points between 7.5 and 4 were monitored by size exclusion chromatography (Fig. 3). The retention times of the protein complexes were not changed from pH 7.5 (Fig. 3A, peaks 1, 2, and 4) to pH 6.0 (Fig. 3C, peaks 1, 2, and 4). As the pH was lowered to 5.0 and below, the P protein was observed to precipitate. The precipitates were removed by filtering the protein solutions through a 0.22-μm-pore-size filter prior to analysis by chromatography. The multiple species present at higher pH were reduced to a single major species (peak 3) of molecular mass estimated to be 487 kDa (Fig. 3E, peak 3). Gel electrophoresis showed that this single species corresponded to a soluble N protein complex in the absence of the P protein (Fig. 3F, lane 3). At pH 4 (Fig. 3G), all detectable P protein had dissociated from the N protein and fallen out of solution (Fig. 3H, lane 3). The only remaining peak (3) corresponded to the uniform N protein complex which was soluble in the absence of salt (data not shown). The size of the N protein complex was unchanged from pH 5.0 to 4.0. The identity of the purified N protein was confirmed by N-terminal amino acid sequencing as SVTVK. This evidence along with mass spectrometry results (data not shown) suggested that the N-terminal methionine was absent. The change in the elution profiles of N/P protein complexes with respect to pH was apparently due to the dissociation of the P protein from the N protein. By this procedure, 6 to 10 mg of the soluble N protein complex could be purified from 1 liter of culture.
Cloning and production of the P protein C-terminal fragment that interacts with N protein.
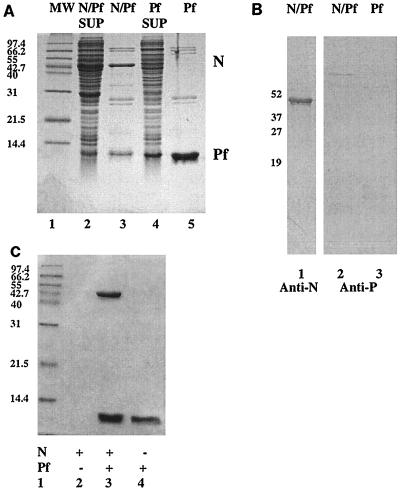
The P protein was found to be digested to a 72-amino-acid fragment (Pf) corresponding to its C terminus when the N/P complex was stored over an extended period of time at 4°C in the absence of protease inhibitors. We therefore cloned and expressed His-tagged (pET-Pf-His [Fig. 1C]) and untagged protein (pET-Pf [Fig. 1D]) corresponding to the C-terminal 72-amino-acid fragment of the P protein in E. coli. His-tagged Pf protein was purified by affinity chromatography on a nickel affinity column (Fig. 4A, lane 5), while untagged Pf protein was purified by ion-exchange chromatography (data not shown). In both cases, gel filtration profiles indicated the Pf protein eluted in the position that corresponded to a monomeric size. Transcriptionally active P protein is a multimer (16). Thus, the C-terminal portion of the Pf protein is not responsible for P protein multimerization. To assess if the bacterially expressed Pf had the ability to bind our N protein oligomer that was purified away from coexpressed P protein by prior pH treatment, His-tagged Pf protein was bound to Ni affinity beads and then incubated with N protein. The complexes were then washed and eluted (Fig. 4C). Purified Pf was shown to bind the low-pH-purified N protein. This indicated that the low-pH treatment of N protein did not impair its ability to bind to the P protein and that the N binding site of the P protein was localized to the last 72 amino acids of the P protein.
FIG. 4.
Expression and purification of the N/Pf protein complex and Pf protein. E. coli containing either pET-N/Pf or pET-Pf-His was grown to the appropriate density and induced with IPTG for production of the N and Pf proteins or the Pf protein alone, respectively. In each case, the total-cell extracts from postinduced cells were separated into soluble and insoluble fractions. Proteins in the soluble fractions were then purified by Ni affinity columns, and either the N/Pf protein complex or the Pf protein was purified. (A) Coomassie brilliant blue R-250-stained SDS–13.5% polyacrylamide gels of N/Pf or Pf protein expression and purification. Lanes: 1, molecular weight (MW) markers (positions are indicated in kilodaltons); 2, soluble fraction of N/Pf protein expression; 3, Ni column-purified soluble N/Pf protein complexes; 4, soluble fraction of Pf protein expression; 5, Ni column-purified soluble His-tagged Pf protein. (B) Western blot of proteins purified by Ni column chromatography. Partially purified N/Pf protein complexes (lane 1 and 2) and Pf protein (lane 3) were electrophoresed on SDS–13.5% polyacrylamide gels. The N protein was detected using a monospecific polyclonal antibody for the N protein (lane 1), while the monospecific polyclonal antibody for P protein did not recognize Pf (lanes 2 and 3). Identities of the Pf proteins were determined by N-terminal amino acid sequencing and mass spectrometry. (C) Coomassie brilliant blue R-250-stained gel of the N Pf protein interaction. To show that purified N protein complex was able to bind the C-terminal domain of the P protein, a binding assay was performed. His-tagged Pf protein was bound to Ni affinity beads in Eppendorf tubes, and purified N protein was then incubated with these beads. The beads were washed and eluted. Lanes: 1, size markers; 2, N protein alone; 3, N protein with Pf protein; 4, Pf protein alone.
Having shown that the Pf protein had the ability to bind the N protein, we tested whether this domain of the P protein was sufficient to allow the soluble expression of the N protein in E. coli. The full-length P gene in plasmid pET-N/P was replaced with the Pf gene to create plasmid pET-N/Pf, which we used to express both the N and Pf proteins in E. coli. The expressed N/Pf complex also copurified in a single affinity chromatography procedure by utilizing the N-terminal histidine tag on Pf (Fig. 4A, lanes 2 and 3). The identity of the N protein was confirmed by Western blotting (Fig. 4B, lane 1); however, the monospecific polyclonal antibodies (from our lab) made to the full-length P protein did not recognize the Pf protein (Fig. 4B, lanes 2 and 3). This result could indicate that this domain of the P protein is buried by the rest of the native full-length P; therefore, the epitope for our antibodies was not found on the Pf protein. The majority of the N/Pf protein complexes aggregated upon elution from the nickel column, as determined from the chromatographic profile in which a significant portion of the protein eluted at the void volume of the S-300 size exclusion column (data not shown). However, a peak containing the N/Pf protein complex was observed within the volume of the column. It is possible that the whole N/P protein complex is better stabilized by the P protein N-terminal domains in high-salt conditions. These domains are missing from the Pf protein, which may be why the N/Pf protein complexes aggregate upon elution from the Ni column.
Bacterially expressed N protein binds RNA.
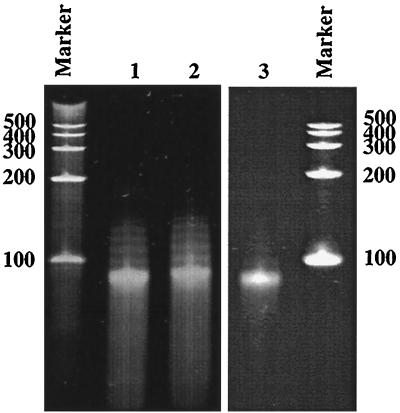
The A260/280 ratio for the purified N protein complex containing no P protein was determined to be 0.9744. This indicated that the protein was possibly associated with nucleic acid; moreover, this ratio suggested that there was 3.84% nucleic acid bound to the protein (18). This was surprising since authentic VSV genomic RNA, which is usually the template for encapsidation, was not present in our expression system and because the N protein does not bind mRNA during a viral infection (37). In addition, it has been reported previously that in the presence of the P protein, the N protein would bind only VSV-specific RNA (32). To confirm the presence of nucleic acid, purified N/P protein complexes and N protein oligomers, isolated from N/P or N/Pf protein complexes, were phenol extracted. Nucleic acid was recovered from all three of these complexes and analyzed by electrophoresis on a denaturing polyacrylamide gel, which showed that it migrated as one major band at the position of about 90 bases (Fig. 5, lanes 1 to 3). This length is consistent with the estimation that 1,200 copies of the N protein encapsidate the 11-kb genome of VSV, which results in each monomer of the N protein covering approximately nine bases of the genome (44). The presence of RNA in the N/P protein complexes confirmed that the N/P protein complex binds the RNA. Unbound RNA was susceptible to digestion by RNase but was RNase resistant when bound to the N protein or the N/P protein complex (data not shown). Since VSV leader or genomic RNA was not made available in our E. coli expression system, the sequence of the RNA present in the oligomer complex was expected to be random. Preliminary experiments indicated the RNA was mRNA for the N and P genes (data not shown). The sequence of specificity of binding is being addressed in further work.
FIG. 5.
Bacterially coexpressed N and P proteins bind RNA. The N and P proteins or the N and Pf proteins were coexpressed and purified, separately, as described in the text. The N protein was isolated by low-pH dialysis of either the N/P protein complexes or the N/Pf protein complexes, separately. N/P protein complexes were purified by gel filtration chromatography (Fig. 3A, peak 2). RNA was then phenol extracted from these purified N/P protein complexes and N protein complexes (lacking the P or Pf protein) and electrophoresed on a 10% 8 M urea–polyacrylamide gel. Shown on the gel is RNA isolated from N protein oligomers originally coexpressed with the full-length P protein (lane 1) or with the Pf protein (lane 2) and RNA from intact N/P protein complexes (lane 3). The size of the RNA oligomer in each case is approximately 90 bases by comparison with the RNA molecular weight marker (Century Marker; Ambion).
The N protein was stable in a wide spectrum of pH from 4 to 11. The RNA was also stable at these various pH conditions. At basic pH, naked RNA ordinarily undergoes base-catalyzed hydrolysis during which the 2′-OH on the nucleotides is deprotonated, and the adjacent 3′ phosphate undergoes nucleophilic attack by this deprotonated 2′-O−. One might expect that the RNA bound to the N protein oligomer would undergo such hydrolysis at pH 11, but this did not happen. When incubated at pH 11 for several days, RNA was still bound to the N protein and was intact, and there was no change in the size of the protein oligomer as determined by gel filtration chromatography. This suggested a tight association between the N protein and the 2′-OH of the nucleotides, thus preventing base-catalyzed hydrolysis of the RNA. This idea is consistent with the previous observations that RNA encapsidated by the N protein was not susceptible to RNase (11, 24, 26). Taken together, these data suggest that the phosphate backbone and the ribose moiety are not accessible or are possibly buried away from the solvent, yet the bases are exposed to the solvent as was shown by Iseni et al. (24) and Keene et al. (26).
Characterization of the N protein-RNA oligomer by analytical ultracentrifugation.
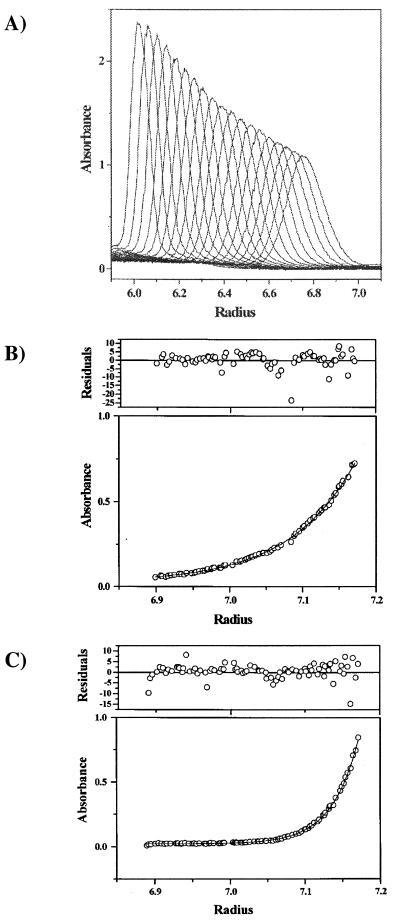
To determine the sedimentation coefficient and molecular weight of the N protein-RNA complex in the absence of the P protein, we performed analytical band and analytical equilibrium centrifugation, respectively. Analytical band centrifugation experiments were performed as described by Lebowitz et al. (30). The N protein-RNA complex used in this set of experiments was derived from the coexpression in E. coli with full-length P protein. The P protein was then dissociated from N/P protein-RNA complexes by the low-pH dialysis method and purified as described previously. Figure 6A shows an overlay of 19 band scans at 230 nm taken at different time points during a single experiment. This shows that the N protein-RNA complex sediments as a single oligomer, with no evidence of larger or smaller components. The mean sedimentation coefficient for five band experiments in 0.1 M NaCl and 0.05 M phosphate-buffered D2O was 9.75S ± 0.36S (R2 values of ln r versus ω2 t plots were 0.999 to 1.000 for all s determinations). Correction for the density and viscosity of D2O and buffer components (see Materials and Methods) gave an s20,w for N of 18.17S ± 0.67S. By combining the Svedberg and Stokes equations according to the method of Teller et al. (43), a molecular mass of 482,800 kDa was predicted from this sedimentation velocity coefficient. Because this estimate of molecular mass assumes that the N oligomer is spherical, which is not the case (as was determined from electron microscopic photographs of this protein described next), sedimentation equilibrium analysis, which measures mass independent of the shape of the protein or protein complex, was also used to determine a molecular weight for the N protein oligomer. Figures 6B and C show the sedimentation equilibrium data for the N oligomer at 5,000 and 8,000 rpm, respectively. Global nonlinear regression fitting of both data sets was performed using an ideal single-component model. The residuals for each fit (expressed in terms of standard deviations, i.e., the average absorbance collected at each radial position) are shown as an upper panel above the absorbance-versus-radial distribution profiles. The distribution of error was essentially random about a zero mean. A molecular weight of 513,879, with 95% confidence limits of 501,226 and 526,558, was obtained from this global fit. Using the Teller et al. (43) empirical relationship cited above, we calculate an s20,w of 18.59 for the N protein oligomer. From the analytical centrifugation characterization of this N protein oligomer, there is good agreement between the s20,w calculated from the molecular weight obtained from sedimentation equilibrium and the experimental s20,w from band sedimentation. A stoichiometry of 10.9 monomer subunits is obtained for the N oligomer complex using the molecular weight of 513,879 obtained from sedimentation equilibrium. This value is reduced to 10.3 monomer subunits with a consideration of the estimated molecular weight of a 90-base RNA found to be bound. Also, evaluation of the molecular weight of the N protein complex assumes that the partial specific volume of the N protein remains unchanged upon formation of the oligomer. A release of water upon oligomerization would decrease the partial specific volume. Allowing a 2% decrease in partial specific volume upon complex formation would decrease the molecular weight to 486,710 and yield a stoichiometry of 10.3 subunits, or 9.7 subunits with consideration of RNA. The data from the sedimentation velocity and equilibrium characterization are in agreement and suggest that N protein purified as described earlier is an oligomer composed of 10 monomer subunits plus RNA.
FIG. 6.
Analytical ultracentrifugation of the N protein oligomer. (A) Analytical band centrifugation of the VSV N protein oligomeric complex. Overlapping absorption-versus-radial distance band sedimentation scans of the N protein complex are shown. The sedimentation velocity experiment was performed at 58,000 rpm at 20°C. The bulk solution used for stabilizing band transport was 100% D2O containing 0.1 M NaCl and 0.05 M phosphate buffer. A sedimentation coefficient of 9.75S ± 0.36S (mean of five different band experiments) was calculated. Correction for the density and viscosity of D2O and buffer components gave an s20,w for N of 18.17S ± 0.67S. (B and C) Sedimentation equilibrium profiles of the absorbance as function of radial position for the N protein at 20°C. The sedimentation equilibrium data were obtained at both 5,000 (B) and 8,000 (C) rpm. All data shown were globally fit using a single ideal component as the model. The solid lines denote the fitted curves for the two data sets. The residual at each radial position has been defined as the ratio of the difference of the measured absorbance and the corresponding fitted value to the standard deviation of the measurement. A molecular weight of 513,879, with 95% confidence limits of 501,226 and 526,558, was obtained from this global fit.
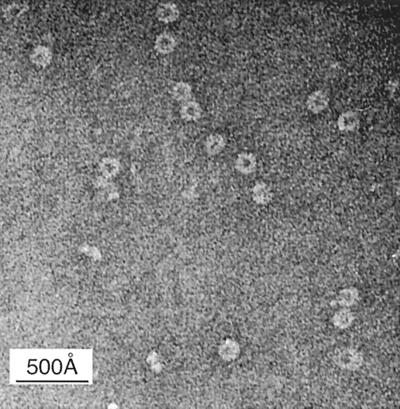
Recombinant N protein-RNA oligomers have a disk-like appearance.
Electron photomicrographs of purified N protein-RNA oligomers taken at a magnification of ×105,000 showed a ring-like overall morphology (Fig. 7). N protein oligomers used in this experiment were isolated from complexes with full-length P protein, as described earlier. Dimensions of the N protein disks were measured from photographs enlarged 2.56× and are as follows. The outer diameter of the disk measures 148 ± 10 Å, and the inner diameter of the disk measures 78 ± 9 Å. The thickness of the disk measures 83 ± 8 Å. The numbers of measurements were 60, 60 (the same disks were used for both the outer and inner diameter measurements), and 30, respectively. The outer radius minus the inner radius gives an N protein height of 35 ± 7 Å.
FIG. 7.
Electron micrographs of the N protein-RNA oligomer negatively stained with 0.1% NH4-phosphotungstic acid (magnification, ×105,000). The N protein-RNA oligomer has a disk-like morphology with the following dimensions: outer and inner diameters, 148 ± 10 Å and 78 ± 9 å, respectively; thickness, 83 ± 7 Å.
DISCUSSION
Previous work expressing the N protein from E. coli resulted in the production of N protein that was insoluble except in high salt (7). Since prior experiments (20, 36, 39) showed that the P protein could maintain the N protein in a soluble state, functional for replication, we developed a system for N and P protein coexpression which allowed the production of soluble N and P proteins in large quantities from bacterial cells. This permitted us to reinvestigate the role of the VSV P protein as a solubility factor for the N protein and as a potential component in the assembly pathway of the VSV RNP. We found the C-terminal domain of the P protein was important for the interaction between the N and P proteins, which is in agreement with previous descriptions (17, 41, 42). Takacs et al. demonstrated that as few as the final 10 C-terminal residues of the P protein are critical for interaction with the N protein. Furthermore, we showed that the C-terminal domain of the P protein was sufficient to maintain the N protein soluble in bacterial cells. This domain was, however, not responsible for P-to-P protein oligomerization, as this domain of the P protein, in the absence of the N protein, existed as a monomer in solution. From N/P protein complexes we isolated a stable oligomer of the N protein that remained soluble upon removal of the P protein. This suggests that the role of the P protein in maintaining N protein solubility is transient. Finally, we demonstrated that bacterially expressed N protein was functional, as deduced from its ability to bind RNA. From the studies presented here, we believe that the P protein may control the concentration-dependent aggregation of the N protein by regulating the assembly of the N protein during N/P protein coexpression.
Multiple species of the complex between the N and P proteins were observed in VSV-infected cells (8, 34). Our data showed the N and P proteins also form species of different molar ratios when expressed concomitantly in a bacterial host. An N-to-P protein stoichiometry of between 1:1 and 2:1 has been shown to be optimal for supporting efficient VSV replication and encapsidation (20, 28, 36, 40, 47). Our experiments are consistent with these observations, since the major N/P protein complex that we isolated from bacteria had a 10:5 molecular composition (molar ratio of 2:1) and was bound to RNA. Our data suggest that the 2:1 N/P ratio is optimal for N/P protein-RNA assembly; however, it does not rule out the 1:1 molar complex between the N and P proteins as optimal for an RNA encapsidation precursor. The ability to achieve the right stoichiometry in E. coli is dependent on the expression levels of the T7 transcript in the bacteria. Gupta et al. (19) coexpressed the N and P proteins in E. coli by using another approach in which each gene, N and P, was transcribed and translated independently from a single plasmid. Soluble proteins were recovered, but only a large set of aggregates was observed when the soluble protein was purified by size exclusion chromatography. The large aggregates produced in that experiment are consistent with the first of three complexes purified from our expression system on the same column.
Using the expression approach described here, the majority of the complexes of N and P protein isolated were soluble and in addition bound RNA. This difference suggests that our expression design, a single transcript encoding both genes, with separate RBSs for their independent translation, may allow the level of each protein to more closely approach the ratio of N to P protein that is critical for not only the solubility of N protein but also the encapsidation of RNA and assembly.
Having established that N protein solubility is dependent on the presence of P protein during expression, we tested whether P protein was constantly required for N protein solubility. If so, upon their separation, we would expect N protein to aggregate. We monitored the aggregation states of the N/P protein complexes at different pHs (Fig. 3). Following the complete dissociation of the P protein from the N/P protein complex at pH lower than 5, the N protein-RNA oligomer remained stable and soluble in solution. This oligomer had a longer retention time than the 10:5 N/P protein-RNA complex on the size exclusion column, which meant that the N protein-RNA oligomer had a lower molecular weight. This change in molecular weight was attributed to the loss of the five P molecules.
The molecular weight of the N protein-RNA oligomer was determined by analytical centrifugation experiments to be 513,879, consistent with an oligomer comprised of 10 N protein molecules bound with an RNA oligonucleotide of ∼90 bases. Electron photomicrographs showed that the N protein-RNA oligomer has a disk-like appearance (Fig. 7). This morphology is consistent with that observed previously for the N protein bound to short VSV leader RNA (4) and for rabies virus N protein bound to RNA (23).
Model for VSV nucleocapsid assembly.
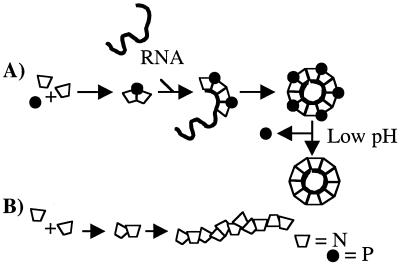
In order to form the helical nucleocapsid characteristic of VSV, individual molecules of the N protein have to interact. Cross-linking studies by Chatterjee et al. (5) demonstrated that the N protein in mature nucleocapsids, isolated from viruses, existed as dimers. The N/N dimer, prior to incorporation into the nucleocapsid, becomes associated with the P protein. At this point, the N protein is blocked from the aggregation seen when the N protein was expressed in the absence of the P protein (7). We propose a model for the assembly of the N/P protein-RNA oligomer in our bacterial system (Fig. 8). In this model, two molecules of the N protein come together with one molecule of the P protein, which may impose an overall curvature on the complex. The association of the P protein with the N protein dimer is through the C terminus (Pf). This 2:1 molecular complex binds the RNA and is followed by the addition of subsequent 2:1 complexes. When five 2:1 N/P complexes become associated with the RNA, a disk-like oligomer, which is consistent with one turn of the RNP helix (10, 44), is formed.
FIG. 8.
Proposed model of assembly of the N/P protein-RNA oligomer. We propose that the P protein allows for the correct assembly of the N protein by possibly inducing curvature on the N/N dimers. The dimers bound by a single P protein bind RNA. This is followed by the binding of subsequent 2:1 (N:P protein) complexes to the RNA. Following the addition of five of these complexes to the RNA, a disk, equivalent to one turn of the RNP helix, is completed. If the P protein is not present, the N protein may form random aggregates (B).
A key component in this assembly process is the RNA, which we believe plays a major role in stability of the N protein oligomer. Since the RNAs isolated from the N protein disks have a finite length of ∼90 bases, one of the interesting questions is why the oligomerization of N molecules stops at 10 in this system. This could allude to an encapsidation control function by the P protein, a specific sequence in authentic VSV genomic RNA that promotes further encapsidation, or a viral or host factor that is responsible for continuous encapsidation. The latter two are not present in the current bacterial expression system. An alternative explanation could be that a predominance of short RNA in the cell determines the size of the oligomer. This hypothesis is offered by Iseni et al., who recently reported the isolation of RNA from rabies virus N protein oligomers expressed in the baculovirus system (23). The authors inferred from their data that the cellular RNA in uninfected cells was predominantly 87 nucleotides long, the length of the RNA encapsidated by their N protein oligomers. We do not believe that this is true for the bacterial milieu, as the T7 expression system in our case results in overproduction of the mRNA transcript encoding the N and P genes, which is greater than 2,000 nucleotides in length. Therefore, more relevant to our case, we believe, are the findings of Moyer et al., who showed that the N protein could encapsidate authentic VSV RNA in vitro, but the limit for encapsidation was ∼90 nucleotides even if the RNA transcripts being encapsidated were longer (35). Consequently, the likely explanation for this assembly product is perhaps that a viral or host factor required for continuous nucleocapsid assembly is not available in our system. Upon finishing the first turn of the RNP helix, monomer 1 and monomer 10 of the oligomer come into contact to form the disk, a state similar to that seen for tobacco mosaic virus (9). However, unlike the tobacco mosaic virus disk, the VSV disk is bound to RNA. After being assembled in this manner, oligomeric N protein will not aggregate further upon loss of the P protein, and oligomer N protein can remain in a soluble form at a multitude of pH conditions and with no salt requirement.
The methods described here will be applied toward further study of the N and P proteins from VSV and likewise may be used in the study of other members of the Rhabdoviridae family of viruses. The knowledge of how to produce and purify these proteins may also be applied to the studies of their homologues in other viruses. In addition, the ability to produce sufficient protein for structural characterization could lead to the discovery of new VSV protein structures. Coexpression of the N and P proteins led to the production of suitable amounts of protein for setting up crystallization trials. The purified N oligomer and the N oligomer complexed with Pf have now been crystallized (unpublished data).
ACKNOWLEDGMENT
We thank L. R. Melson for work involving electron microscopy.
REFERENCES
- 1.Banerjee A K, Rhodes D P, Gill D S. Complete nucleotide sequence of the mRNA coding for the N protein of vesicular stomatitis virus (New Jersey serotype) Virology. 1984;137:432–438. doi: 10.1016/0042-6822(84)90237-x. [DOI] [PubMed] [Google Scholar]
- 2.Barik S, Banerjee A K. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J Virol. 1991;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee P K, Cervera M M, Penman S. Formation of vesicular stomatitis virus nucleocapsid framework-bound N protein: possible model for structure assembly. Mol Cell Biol. 1984;4:2231–2234. doi: 10.1128/mcb.4.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinton G M, Burge B W, Huang A S. Effects of phosphorylation and pH on the association of NS protein with vesicular stomatitis virus cores. J Virol. 1978;27:340–346. doi: 10.1128/jvi.27.2.340-346.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das T, Banerjee A K. Expression of the vesicular stomatitis virus nucleocapsid protein gene in Escherichia coli: analysis of its biological activity in vitro. Virology. 1993;193:340–347. doi: 10.1006/viro.1993.1130. [DOI] [PubMed] [Google Scholar]
- 8.Davis N L, Arnheiter H, Wertz G W. Vesicular stomatitis virus N and NS proteins form multiple complexes. J Virol. 1986;59:751–754. doi: 10.1128/jvi.59.3.751-754.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durham A C H, Finch J T, Klug A. Polymerization of tobacco mosaic virus protein and its control. Nat New Biol. 1971;229:47–50. doi: 10.1038/newbio229042a0. [DOI] [PubMed] [Google Scholar]
- 10.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson S. Transcription of vesicular stomatitis virus. In: Wagner R, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 245–269. [Google Scholar]
- 12.Emerson S U, Wagner R R. Dissociation and reconstitution of the transcriptase activities of the vesicular stomatitis B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson S U, Schubert M. Localization of the binding domains for the RNA polymerase L and the riboucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1987;84:5655–5659. doi: 10.1073/pnas.84.16.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson S U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 15.Gallione C J, Greene J R, Iverson L E, Rose J K. Nucleotide sequences of the mRNA's encoding the vesicular stomatitis virus N and NS proteins. J Virol. 1981;39:529–535. doi: 10.1128/jvi.39.2.529-535.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill D S, Chattopadhyay D, Banerjee A K. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc Natl Acad Sci USA. 1986;83:8873–8877. doi: 10.1073/pnas.83.23.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasel J A. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. BioTechniques. 1995;18:62–63. [PubMed] [Google Scholar]
- 19.Gupta A K, Banerjee A K. Expression and purification of vesicular stomatitis virus N-P complex from Escherichia coli: role in genome RNA transcription and replication in vitro. J Virol. 1997;71:4264–4271. doi: 10.1128/jvi.71.6.4264-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard M, Wertz G. Vesicular stomatitis virus RNA replication: a role for the NS protein. J Gen Virol. 1989;70:2683–2694. doi: 10.1099/0022-1317-70-10-2683. [DOI] [PubMed] [Google Scholar]
- 21.Huang A S, Wagner R R. Comparative sedimentation coefficients of RNA extracted from plaque-forming and defective particles of vesicular stomatitis virus. J Mol Biol. 1966;22:381–384. doi: 10.1016/0022-2836(66)90143-4. [DOI] [PubMed] [Google Scholar]
- 22.Hudson L D, Condra C, Lazzarini R A. Cloning and expression of a viral phosphoprotein: structure suggests vesicular stomatitis virus NS may function by mimicking an RNA template. J Gen Virol. 1986;67:1571–1579. doi: 10.1099/0022-1317-67-8-1571. [DOI] [PubMed] [Google Scholar]
- 23.Iseni F, Barge A, Baudin F, Blonbel D, Ruigrok R W H. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol. 1998;79:2909–2919. doi: 10.1099/0022-1317-79-12-2909. [DOI] [PubMed] [Google Scholar]
- 24.Iseni F, Florence B, Blondel D, Riogrok R W H. Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA. 2000;6:270–281. doi: 10.1017/s135583820099109x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 26.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirschenbaum I. Physical properties and analysis of heavy water. New York, N.Y: McGraw-Hill; 1951. pp. 1–41. [Google Scholar]
- 28.La Ferla F M, Peluso R W. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. Computer aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, United Kingdom: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 30.Lebowitz J, Teale M, Schuck P W. Analytical band centrifugation of proteins and protein complexes. Biochem Soc Trans. 1998;26:745–749. doi: 10.1042/bst0260745. [DOI] [PubMed] [Google Scholar]
- 31.Marnell L L, Summers D F. Characterization of the phosphorylated small enzyme subunit, NS, of the vesicular stomatitis virus RNA polymerase. J Biol Chem. 1984;259:13518–13524. [PubMed] [Google Scholar]
- 32.Masters P S, Banerjee A K. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J Virol. 1988;62:2658–2664. doi: 10.1128/jvi.62.8.2658-2664.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masters P S, Banerjee A K. Efficient interaction of the vesicular stomatitis virus P protein with the L protein or the N protein in cells expressing the recombinant proteins. Virology. 1995;208:821–826. doi: 10.1006/viro.1995.1219. [DOI] [PubMed] [Google Scholar]
- 34.Masters P S, Banerjee A K. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J Virol. 1988;62:2651–2657. doi: 10.1128/jvi.62.8.2651-2657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer S A, Smallwood-Kentro S, Haddad A, Prevec L. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol. 1991;65:2170–2178. doi: 10.1128/jvi.65.5.2170-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattnaik A K, Wertz G W. Replication and amplification of defective interferring particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNA. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patton J T, Davis N L, Wertz G W. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul P R, Chattopadhyay D, Banerjee A K. The functional domains of the phosphoprotein of vesicular stomatitis virus (Indiana serotype) Virology. 1988;166:350–357. doi: 10.1016/0042-6822(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 39.Peluso R W. Kinetic, quantitative, and functional analysis of multiple forms of the vesicular stomatitis virus nucleocapsid protein in infected cells. J Virol. 1988;62:2799–2807. doi: 10.1128/jvi.62.8.2799-2807.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peluso R W, Moyer S A. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology. 1988;162:369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- 41.Takacs A M, Banerjee A K. Efficient interaction of the vesicular stomatitis virus P protein with the L protein or the N protein in cells expressing the recombinant proteins. Virology. 1995;208:821–826. doi: 10.1006/viro.1995.1219. [DOI] [PubMed] [Google Scholar]
- 42.Takacs A M, Das T, Banerjee A K. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teller D C, Swanson E, Dehaen C. The translation friction coefficient of proteins. Methods Enzymol. 1979;61:103–124. doi: 10.1016/0076-6879(79)61010-8. [DOI] [PubMed] [Google Scholar]
- 44.Thomas D, Newcomb W W, Brown J C, Hall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinograd J, Bruner R, Kent R, Weigle J. Band centrifugation of macromolecules and viruses in self-generating gradients. Proc Natl Acad Sci USA. 1963;49:902–910. doi: 10.1073/pnas.49.6.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertz G W, Perepelitsa V, Ball A. Gene rearrangment attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci USA. 1998;95:3501–3501. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wertz G W, Howard M B, Davis N, Patton J. The switch from transcription to replication of a negative-strand RNA virus. Cold Spring Harbor Symp Quant Biol. 1987;52:367–371. doi: 10.1101/sqb.1987.052.01.042. [DOI] [PubMed] [Google Scholar]
- 48.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]