Summary
Despite successful vaccines and updates, constant mutations of SARS-CoV-2 makes necessary the search for new vaccines. We generated a chimeric protein that comprises the receptor-binding domain from spike and the nucleocapsid antigens (SpiN) from SARS-CoV-2. Once SpiN elicits a protective immune response in rodents, here we show that convalescent and previously vaccinated individuals respond to SpiN. CD4+ and CD8+ T cells from these individuals produced greater amounts of IFN−γ when stimulated with SpiN, compared to SARS-CoV-2 antigens. Also, B cells from these individuals were able to secrete antibodies that recognize SpiN. When administered as a boost dose in mice previously immunized with CoronaVac, ChAdOx1-S or BNT162b2, SpiN was able to induce a greater or equivalent immune response to homologous prime/boost. Our data reveal the ability of SpiN to induce cellular and humoral responses in vaccinated human donors, rendering it a promising candidate.
Subject areas: Health sciences, Public health, Immunology, Virology
Graphical abstract

Highlights
-
•
SpiN induced IFN-Υ in CD4 and CD8 T cells of convalescent and vaccinated individuals
-
•
IgG from convalescent and vaccinated individuals recognized SpiN
-
•
Mice respond similarly or better when boosted with SpiN than with commercial vaccines
Health sciences; Public health; Immunology; Virology
Introduction
At the end of 2019, the world was surprised with the emergence of a new virus that generated a true chaos due to its fast spread that led to a pandemic. The genome sequence of the novel beta coronavirus was quickly described,1 the virus was called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the disease named coronavirus disease 19 (COVID-19). The disease comprises a wide spectrum of symptoms, as pneumonia, acute respiratory distress syndrome, liver injury, neurological disease, renal disease, cardiac injury, sepsis and thrombocytosis, that can lead to severe sickness and death.2,3,4 Until now, COVID-19 led to more than 6.8 million of deaths in the world and this number was not higher thanks to the fast manufacturing of vaccines in a record time.
The great legacy that the science and the world took from this pandemic was the effort to rapidly produce vaccines from different platforms and in large scale, with considerable attention to the ones based on nucleic acid. In Brazil two vaccines were firstly approved to be used as massive immunization, CoronaVac (Sinovac, China) and ChAdOx1-S (Oxford, Astrazeneca). Later, the vaccine produced by Pfizer-BioNTech and Janssen (Johnson & Johnson), respectively BNT162b2 and Ad26.COV2.S, were approved. Whereas CoronaVac is a conventional vaccine, made from inactivated virus, ChAdOx1-S and Ad26.COV2.S are vaccines composed of adenoviral vectors and BNT162b2 is a mRNA-based vaccine. It is estimated that vaccines saved about 15 million of lives just in 2021.5 Although vaccines exerted a huge impact in the course of the pandemic, their effectiveness dampens each time a new variant emerges, fact observed mainly with omicron.6,7 The target of most of the COVID-19 vaccines is the Spike (S) protein, the surface protein responsible to bind to human cells receptor ACE-2.8 Receptor binding domain (RBD) of the S protein is the key target of neutralizing antibodies (nAbs)9,10 against SARS-CoV-2 and also a strategical mutation point,11 which lead to the emergence of the variants of concern (VoCs) that evade the immune system. Pfizer-BioNTech and Moderna are developing new strategies targeting both the original virus and the Omicron variant, however, these vaccines require updates for each new variant. This scenario reveals the importance of the production of a new and more effective vaccine targeting different viral proteins and with a mechanism of action that does not rely exclusively on nAbs.
Our group developed a chimeric protein named SpiN composed of the RBD of the canonical S protein and the nucleocapsid (N) protein to be evaluated as a COVID-19 vaccine. In rodents immunized with SpiN and challenged with SARS-CoV-2, we observed a great protection against Wuhan, Delta and Omicron variants. SpiN-mediated protection was primarily based on T lymphocytes, being independent of nAbs.12 Aiming the use of SpiN as boost dose, we investigated if PBMC from vaccinated individuals recognizes and respond to SpiN. A large part of the Brazilian population was immunized with CoronaVac, ChAdOx-1S and BNT126b2, which made it difficult to include individuals vaccinated with Ad26.COV2.S. Here we show that both T and B cells from vaccinated individuals recognize SpiN. The protein induces IFN−γ production by CD4+ and CD8+ T cells from convalescent and vaccinated donors and antibodies produced by B cell from these individuals were able to recognize SpiN. In murine model, we observed that heterologous prime/boost with CoronaVac, ChAdOx1-S and BNT162b2 lead to equal production of IFN−γ in splenocytes and increased anti-S IgG production (ChAdOx1-S), with similar induction of nAbs when compared to homologous prime/boost with the vaccines. The great CD4+ and CD8+ T cell responses induced by SpiN, together with the ability to be recognized by B cell and induce nAbs makes SpiN a good candidate for boost vaccine avoiding the evasion of VoCs from the immune system.
Results
Vaccination induces cellular and humoral responses, but waning immunity is observed after 180 days
First, we evaluated the cellular and humoral immune response elicited by COVID-19 vaccines. To that end, we determined the levels of IFN-γ released by PBMCs stimulated with S or N proteins of SARS-CoV-2 as well as serum IgG levels against these antigens. Two vaccines (CoronaVac and ChAdOx1-S) and two time points (30 days after boost -TP1- and 180 days after the first dose -TP2) were chosen to be compared to control and convalescent groups. Compared to the control group, an increased production of IFN-γ in the convalescent group was observed at the first time point of CoronaVac-vaccinated groups when stimulated with S and N proteins, while no statistically significant difference was observed in ChAdOx1-S-vaccinated group (Figures 1A and 1B). In the humoral branch, we analyzed IgG levels in the serum of individuals. Anti-S IgG production was observed in convalescent and vaccinated groups 30 days after complete vaccination (TP1), with a remarkable decrease 180 days after the first dose (TP2), mainly in CoronaVac group (Figure 1C). As expected, levels of anti-N IgG were observed only in convalescent and CoronaVac-vaccinated groups, in which a dramatic decrease at TP2 was observed (Figure 1D). Reflecting the data of IgG levels, serum neutralization titers were observed in convalescent and ChAdOx1-S-vaccinated group, with significant decrease at TP2. CoronaVac-vaccinated group exhibited minimal neutralizing antibody titers after complete vaccination and even so, it decreases 180 days after the first dose.
Figure 1.
Cellular (IFN−γ) and Humoral (IgG and PRNT) responses of controls, convalescents and vaccinated individuals stimulated with SARS-CoV-2 antigens
PBMC and serum from healthy, convalescent or vaccinated individuals were obtained to evaluate IFN-g production (A, B), IgG levels (C, D) and PRNT (E). Samples from vaccinated individuals were obtained 30 days after booster (TP1) and 180 days after the first dose (TP2) with CoronaVac and ChAdOx-1S. IFN-γ and IgG levels were measured by ELISA and PRNT using VERO cells and wild type SARS-CoV-2 from Wuhan. Difference between control and other groups was analyzed by Kruskal–Wallis followed by Dunn’s multiple comparisons test. ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. ∗∗∗∗p < 0.0001. Difference within the groups was analyzed by Mann–Whitney test. #p < 0.05. ##p < 0.01.###p < 0.001. ####p < 0.0001. Data are represented as median with interquartile range. n = 12 for control, 10 for convalescent and 15 for other groups.
Stimulation with SpiN leads to a greater activation of T cells from convalescent and vaccinated individuals
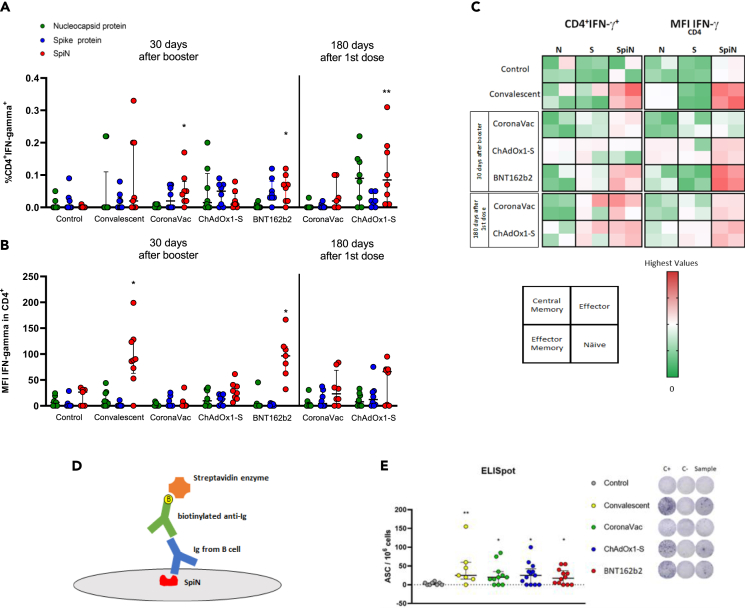
SpiN is a chimeric protein idealized and produced by our research group to be used as the antigen of a new COVID-19 vaccine. SpiN sequence consists in the fusion of the nucleocapsid protein followed by the receptor-binding domain (RBD) as schematically demonstrated in the Figure 2A. Our group published a study validating SpiN as a promising vaccine candidate12 that is now in a phase II clinical trial. Since most of Brazilian population is fully vaccinated, our purpose is to use SpiN as a boost dose. Therefore, we decided to investigate the efficacy of SpiN in activating T cells from convalescent and vaccinated groups.
Figure 2.
SpiN induces IFN−γ production in PBMC from convalescent and vaccinated volunteers and activates CD8+ T cell
Schematic representation of the chimera protein SpiN (A). PBMC from healthy (control), convalescent and vaccinated individuals 30 days after booster were cultured with Nucleocapsid protein, Spike protein or SpiN for 72 h to evaluate IFN−γ production. n = 4 for convalescent and 8 for others groups.
(B). PBMCs from control, convalescent and vaccinated individuals 30 days after booster (TP1) and 180 days after first dose (TP2) were stimulated with recombinant N or S protein from SARS-CoV-2 or with SpiN for 15h in order to evaluate CD8+ T cell activation via IFN−γ production by flow cytometry. The frequencies of IFN−γ producing cells (C) and median fluorescence intensity (MFI) of IFN−γ were evaluated (D). Heatmaps depicting the overall response (CD8+IFN−γ+ frequencies and IFN−γ MFI) by stimuli among compartments of CD8+ T cells are represented: Central Memory (CD45RO+CD27+), Effector (CD45RO−CD27−), Effector Memory (CD45RO+CD27−) and Näive (CD45RO−CD27+) subsets. Major square holds the 4 subsets, as shown in the model below the heatmaps (E). ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. IFN-γ and flow cytometry was analyzed by Kruskal–Wallis followed by Dunn’s multiple comparisons test comparing each stimulus of the groups with their respective control. Data are represented as median with interquartile range. For (C) and (D), n = 9 for convalescent and 8 for other groups.
See also Figure S1.
First, we analyzed IFN-γ production by PBMCs from control, convalescent and individuals vaccinated with CoronaVac, ChAdOx1-S and BNT162b2 at TP1 stimulated with N and S proteins of SARS-CoV-2 or SpiN. The levels of IFN-γ were significantly increased in vaccinated groups stimulated with SpiN, compared to control group. In convalescent group, it is possible to observe an increased production of IFN-γ upon SpiN stimulus compared to N and S protein, but due to the limited number of individuals, we do not observe statistical significance (Figure 2B). Flow cytometry was performed to evaluate the frequencies of IFN-γ responsive T cells stimulated with SARS-CoV-2 antigens or SpiN. Gating strategy is detailed on supplemental information. Briefly, lymphocytes were gated on singlets and live CD3+ cells and then this population was separated in CD4+ and CD8+ cells. To assess memory T cell population, we based on CD45RO and CD27 expression and divided T cells into the following compartments: Central Memory (CD45RO+CD27+), Effector (CD45RO−CD27−), Effector Memory (CD45RO+CD27−) and Naive (CD45RO−CD27+). We assessed IFN-γ expression in both total and compartmentalized T cells populations. In this assay, we compared control and convalescent groups to CoronaVac, ChAdOx1-S and BNT162b2-vacinated groups at TP1 and CoronaVac and ChAdOx1-S-vaccinated group at TP-2.
Observing the CD8+ T cell population, only the convalescent group showed increased frequency of IFN-γ+ cells when stimulated with SpiN, compared to N and S antigens (Figure 2C). We also evaluated the median fluorescence intensity (MFI) of IFN-γ in total CD8+ T cells and observed significant IFN-γ MFI increase only in convalescent and BNT162b2-vaccinated group (Figure 2D). Among CD8+ T cells memory compartment, the stimulation with SpiN leads to higher frequencies of IFN-γ T cells in general, with highlighted attention to effector memory population, as seen in convalescent, BNT162b2 (TP1) and CoronaVac and ChAdOx1-S at TP2 (Figure 2E). When we analyzed the MFI of IFN-γ in CD8+ T subsets, the difference between SpiN and the other antigens becomes remarkable, except for CoronaVac group at TP1 (Figure 2E).
Among CD4+ T cells, CoronaVac and BNT162b2-vacinated group at TP1, and ChAdOx1-S-vaccinated people at TP2 responded significantly to stimulation with SpiN (Figure 3A). IFN-γ MFI of CD4+ T cell presented a similar result to that of CD8+ T cells, where only convalescent and BNT162b2-vaccinated group showed significant difference when stimulated with SpiN (Figure 3B). The overall response among CD4+ T cell compartments were similar to those found in CD8+ T cell. Differences to both, frequency of IFN-γ+ cells and MFI of IFN-γ, were observed mainly in convalescent, BNT162b2 (TP1) and CoronaVac and ChAdOx1-S at TP2 stimulated with SpiN (Figure 3C).
Figure 3.
SpiN activates CD4+ T cell and is recognized by clones of B cells from convalescent and vaccinated volunteers
PBMCs from control, convalescent and vaccinated individuals (30 days after booster and 180 days after first dose) were stimulated with recombinant N and S protein from SARS-CoV-2 and with SpiN for 15h in order to evaluate CD4+ T cell activation via IFN−γ production by flow cytometry. Were evaluated frequencies of IFN−γ producing cells (A) and median fluorescence intensity (MFI) of IFN−γ (B) n =. Heatmaps depicting the overall response (CD4+IFN−γ+ frequencies and IFN−γ MFI) by stimuli among compartments of CD4+ T cells are represented: Central Memory (CD45RO+CD27+), Effector (CD45RO−CD27−), Effector Memory (CD45RO+CD27−) and Näive (CD45RO−CD27+) subsets. Major square holds the 4 subsets, as shown in the model below the heatmaps (C). The ability of B cells to recognize SpiN was assessed by ELISpot for detection of igG, which schematic representation is shown (D). PBMC from controls, convalescents and vaccinated individuals (30 days after booster) were stimulated with R848 and human recombinant IL-2 or medium for 72h. After polyclonal stimuli, cells were plated in ELISpot plate coated with anti-IgG (for positive control) or SpiN for 16-24h. Cells without stimulus were used as negative control. After incubation spots representing specific SpiN antigen-secreting cells (ASC) were revealed. n = 7 for convalescent and 12 for other groups (E). ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. Flow cytometry was analyzed by Kruskal–Wallis followed by Dunn’s multiple comparisons test comparing each stimulus of the groups with the respective from control group. ELISpot was analyzed through Mann–Whitney test. C+, positive control; C-, negative control. Data are represented as median with interquartile range. For (A) and (B), n = 9 for convalescent and 8 for other groups.
See also Figure S1.
The activation of both CD8+ and CD4+ T cells memory subsets until 180 days after vaccination indicates that the remaining clones of T cells recognize and are activated by epitopes embedded in SpiN.
In order to find out if also B cells are able to recognize SpiN, we performed an IgG ELISpot assay (Figure 3D), in which the plate was coated with SpiN and incubated with sera from convalescent or vaccinated volunteers. After 72 h of polyclonal stimulation with IL-2 and R848, we found that IgG produced by B cell clones of convalescent and vaccinated volunteers were capable to recognize SpiN (Figure 3E).
Altogether, we found that SpiN strongly stimulates both CD4+ T and CD8+ T cell responses in vaccinated and convalescent individuals toward the production of IFN-γ, still 180 days after the first dose and also that SpiN possesses epitopes recognized by antibodies produced by B cells.
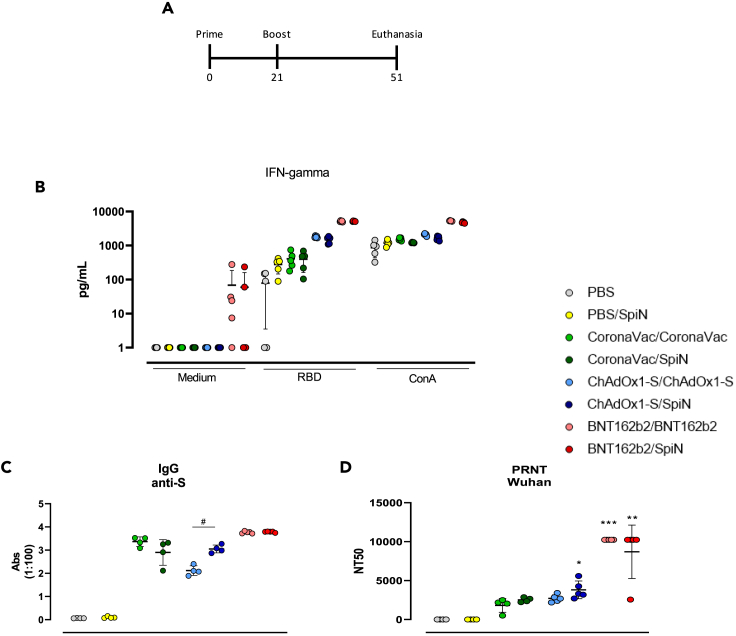
Increased cellular and humoral responses in vaccinated mice boosted with SpiN
Nowadays, virtually all population is vaccinated and the possible use of SpiN would be as a boost. In this way, aiming the clinical use of SpiN, we performed an experiment using a heterologous prime/boost with SpiN as boost dose in mice previously immunized with CoronaVac, ChAdOx1-S and BNT162b2 according to Figure 4A. As control groups, mice were administered with PBS, a single dose of SpiN (PBS/SpiN) and two doses of CoronaVac, ChAdOx1-S and BNT162b2. First, we evaluated the cellular immune response by IFN-γ production after in vitro stimulation of splenocytes from immunized mice. As depicted on Figure 4B, splenocytes from mice that received both homologous or heterologous prime/boost of each vaccine present an increase in IFN-γ production after incubation with RBD in comparison with PBS and a single dose of SpiN. Importantly, stimulation with RBD led to similar levels of IFN-γ in splenocytes from mice administered with heterologous prime/boost protocol when compared with two doses of each vaccine. We also investigated the humoral response through anti-S IgG antibodies (Figure 4C), and it was observed that animals vaccinated with ChAdOx1-S/SpiN presented higher levels of antibodies in comparison with two doses of ChAdOx1-S. Heterologous prime/boost with CoronaVac and BNT162b2 led to an equivalent level of anti-S IgG compared to receiving two doses of either vaccine alone. Consistently with these results, nAbs are also elevated in mice that received ChAdOx1-S/SpiN, while there is no difference between control or two doses of ChAdOx1-S. Similar neutralizing titer was found in homologous and heterologous prime/boost with BNT162b2 (Figure 4D). In conclusion, our data suggests that the boost with SpiN is able to induce a cellular immune response as efficient as the homologous prime/boost with CoronaVac, ChAdOx1-S, and BNT162b2 and, more importantly, increases the robustness of the humoral response anti-SARS-CoV-2 in comparison with two doses of the vaccines.
Figure 4.
Cellular and humoral responses of mice boosted with SpiN
C57BL/6 mice were immunized in a regimen involving homologous and heterologous (with SpiN) prime/boost with CoronaVac, ChAdOx1-S or BNT162b2. Mice immunized with PBS or only SpiN served as the control group, following the specified protocol (A). Levels of IFN−γ was measured on culture supernatant of splenocytes stimulated with RBD or concanavalin A for 72 h n = 5 (B). The serum was used to assess levels of IgG anti-S by ELISA (C) or neutralizing titer to the wild type SARS-CoV-2 from Wuhan by PRNT (D). Difference between control (PBS) and other groups was analyzed by Kruskal–Wallis followed by Dunn’s multiple comparisons test. ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. Difference within the groups was analyzed by Mann–Whitney test. #p < 0.05. Difference within the groups was analyzed by Mann–Whitney test. #p < 0.05. Data are represented as median with interquartile range. For (C) and (D), n = 5 for BNT162b2 groups and 4 for others.
Discussion
The COVID-19 pandemic compelled researchers from around the world to develop vaccination strategies to fight against the new common enemy, the SARS-CoV-2. Despite the success of the vaccines, due to the emerging variants of SARS-CoV-2, the pandemic remains among us still causing some damage and leaving the need of a vaccine that efficiently blocks the transmission and dampens the disease caused by SARS-CoV-2. In the present study we present data indicating that SpiN, a chimeric protein, can be used as a new vaccine boost candidate. First, we show that cellular and humoral response of vaccines drops 180 days after vaccination with the conventional vaccines, data that is reinforced by a range of studies. Afterward, we demonstrate that SpiN activates T cells from convalescent and vaccinated people, leading to cell activation by IFN-γ production. Moreover, B cells from convalescent and vaccinated individuals produced antibodies that recognized SpiN. Lastly, we show that heterologous prime/boost protocol with ChAdOx1-S and SpiN in mice was able to induce substantial cellular and humoral responses, demonstrated by the production of IFN-γ, anti-S IgG and nAbs, proving that SpiN is a great boost candidate.
Since the controversial immunization with cowpox made by Janner, which led to the protection of a child against smallpox, vaccines changed the history of humanity preventing much more than 10 million of deaths between the 60’s and 2015, just with viral vaccines.13,14 Vaccines acts essentially on the adaptive immune system, activating both cellular and humoral effector and memory mechanisms of defense. Some vaccines, such as those against yellow fever or smallpox, can confer long lasting memory and are considered the most successful vaccines, whereas other vaccines prompt to a shorter protection and require booster vaccine to maintain prophylactic immunity.15,16 The pandemic triggered by the spread of SARS-CoV-2 gathered the world science in a common effort to achieve vaccines able to mitigate the devastation caused by COVID-19. Several types of vaccines were rapidly approved to be used in all continents and they were, without any doubt, responsible to save millions of lives. Despite the success of the vaccines, after more than two years since the beginning of vaccination campaigns, we are still into a pandemic due to the emergence of SARS-CoV-2 variants, that use mutations to escape from the immune system. These circumstances lead to a constant search for a more efficient vaccine.
Brazil started to vaccinate its population at the beginning of 2021 and two vaccines were initially approved for use: CoronaVac, from Sinovac, and the Oxford-AstraZeneca, ChAdOx1-S. Corroborating previously published data, we found that both cellular and humoral response of these vaccines drop after six months.17,18 Interestingly and apparently controversial, we found levels of antibodies anti-N in individuals vaccinated with ChAdOx1-S. Once we excluded COVID-19 diagnosed individuals, these data suggest that these individuals were previously exposed to SARS-CoV-2 without presenting the disease. In this work, we assessed mainly CoronaVac and ChAdOx1-S due to the Brazilian National Immunization Program, which included the third boost dose before the volunteers complete 180 days from the first dose, but it is known that other vaccines follow the same fate, with immunity decay after 180 days.19
Protection elicited by vaccines against SARS-CoV-2 are strongly dependent on nAbs,20,21,22,23 but it is now well known that T cell response is as important as nAbs.24,25,26 A positive clinical outcome was associated with an effective T cell response, like the viral control associated with CD4+ T cells and expression of high levels of effector molecules by CD8+ T cells.27,28,29 The target of most, if not all, COVID-19 vaccines is the Spike protein, the main point of mutation of SARS-CoV-2, which turned it into a great evader of the immune system, showing ability to escape antibody neutralization.30,31,32 An ideal vaccine should aim to elicit robust cellular and humoral immune responses. To enhance effectiveness and reduce the risk of viral evasion, it is advisable to target not only the highly mutable Spike protein but also other conserved parts of the virus. By broadening the immune response to encompass multiple viral components, vaccines can potentially provide better protection and reduce the likelihood of viral escape mechanisms. It has been well described that nucleocapsid (N) protein from SARS-CoV-2 possesses several epitopes recognized by CD4+ and CD8+ T cells, including memory subsets.33,34,35,36 Taus et al. showed that CD8+ T cells recognize peptides from nucleocapsid, spike and matrix protein from SARS-CoV-2, but the findings pointes that nucleocapsid is immunodominant.36 Moreover, N protein is highly conserved in VoCs.12,33,37 These data and a broad of other studies38,39,40 point N protein as a great and relevant target to be part of an effective vaccine. This strategy has been proposed by other authors as well, including the utilization of a chimera molecule that comprises part of N along with RBD.41,42,43,44,45,46,47
In a recently published work of our group, it was shown that SpiN adjuvanted with Poly ICLC (SpiN-Tec) protects mice against infection with SARS-CoV-2 Wuhan isolate and also Delta and Omicron variants. Interestingly, the elicited protection is T-cell-dependent, but nAbs-independent. The potency of T cell response to a specific antigen is often assessed by IFN-γ production. CD4+ and CD8+ T cells from convalescent and vaccinated individuals express IFN-γ in response to predicted epitopes of SARS-CoV-2.33,36,48 Our findings show that in response to SpiN, CD4+ and CD8+ T cells, including memory compartments, of convalescent and vaccinated individuals produce IFN-γ in higher or similar levels to stimulation with N and S proteins, similar to previously reported frequencies.49,50,51 These results evidence that SpiN possesses epitopes that are recognized by CD4+ and CD8+ T cells, which makes SpiN a great inductor of cellular response when used as boost dose. Recently, Yang et al. described the mechanism that shapes the production of antibodies against VoCs after a third dose of a vaccine. In short, this study shows that the subsequent doses of vaccines select B cells specific to subdominant epitopes that are less mutated in Omicron, therefore the third dose of vaccine will expand these clones and enhance nAbs against variants.52 We showed that convalescent and vaccinated individuals possess B cell clones that recognize SpiN, thus the use of SpiN as a boost could lead to the production of nAbs against variants, once in Brazil most of the population has received at least two doses of COVID-19 vaccine. In our study, we found that using SpiN as a booster in mice previously immunized with CoronaVac, ChAdOx1-S, and BNT162b2 resulted in similar levels of IFN-γ when splenocytes were stimulated with RBD. Interestingly, compared to homologous prime/boost, the heterologous prime/boost using ChAdOx1-S and SpiN led to the production of increased levels of IgG anti-S and similar, but slightly higher, neutralizing titers against the canonical Wuhan SARS-CoV-2. Mice immunized with CoronaVac and BNT162b2 exhibited comparable levels of anti-S IgG and neutralizing titer for both homologous and heterologous regimens. There was no difference between homologous and heterologous prime/boost in neutralizing titer against Omicron variant. These data show that SpiN has the ability to increase the production of, not only ligand antibodies, but also nAbs, confirming what we discussed before.
In conclusion, this study reveals that SpiN can be used as boost vaccine candidate against COVID-19. Our previous study demonstrated significant protection of rodents immunized with SpiN and then challenged with SARS-CoV-2. SpiN elicited protection independent of nAbs, acting by T cell responses. Here we show that in convalescent and vaccinated individuals, not only CD4+ and CD8+ T cells recognize SpiN epitopes, but also B cells. Recently, SpiN was approved by the Brazilian Health Regulatory Agency for clinical trial phase II, which is ongoing. The solid cellular response triggered by SpiN associated with the possible induction of nAbs makes SpiN a formidable vaccine candidate.
Limitations of the study
Due to Brazilian National Immunization Program, which offered the third vaccine dose before BNT162b2-vacinated volunteers complete 180 days from the first dose, we could not obtain samples of this time point of BNT162b2-vacinated volunteers. Furthermore, the Ad26.COV2.S vaccine was included in the Brazilian National Immunization Program after the majority of individuals had already received other vaccines, which made it difficult to find volunteers who had been vaccinated with Ad26.COV2.S.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgG-HRP antibody | Fapon | BEEIGGI201 |
| anti-CD4 -BV605 | BD | RPA-T4; RRID:AB_2744420 |
| anti-CD8 - AlexaFluor700 | Biolegend | SK1; RRID: AB_2562789 |
| anti-CD45RO - BV786 | BD | UCHL1; RRID:AB_2738733 |
| anti-CD27 - APC-Cy7 | Biolegend | O323; RRID: AB_571976 |
| anti-IFN-g - PE-Cy7 | eBioscience | 4 S.B3; RRID: AB_469682 |
| anti-CD3 - FITC | BD | UCHT1; RRID:AB_395739 |
| anti-IgG mouse | SOUTHERN BIOTECH | 1030-05; RRID: AB_2794348 |
| Bacterial and virus strains | ||
| Wuhan - SARS-CoV-2 | Rede Vírus | Ministry of Science, Technology and Innovation |
| Omicron - SARS-CoV-2 | Rede Vírus | Ministry of Science, Technology and Innovation |
| E. coli SHuffle T7 Express | New England biolabs (NEB) | C3029J |
| E. coli BL21 (DE3) | New England biolabs (NEB) | C2527H |
| Chemicals, peptides, and recombinant proteins | ||
| Nucleocapside | In house | |
| Spike | In house | |
| SpiN | In house | |
| RBD | In house | |
| Critical commercial assays | ||
| Human IgG ELISpotbasic kit | Mabtech | 3850-2A |
| OptEIA Human IFN-gamma | BD | 555142 |
| Mouse IFN-gamma DuoSet ELISA | R&D Systems | DY485 |
| Experimental models: Cell lines | ||
| VERO cells | ATCC | CCL-81 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | Center for Laboratory Animal Facilities of the Federal University of Minas Gerais (CEBIO-UFMG) | |
| Software and algorithms | ||
| GraphPad Prism 6.0 | GraphPad Inc, USA | |
| FlowJo v10.5.3 | BD Biosciences | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bruno Valiate (valiate.bvs@gmail.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data produced in this paper is available upon request to the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Ethics statement
Ethical Committees on Human Experimentation from Fundação Hospitalar do Estado de Minas Gerais (FHEMIG) approved this study performed with human donors (CAAE: 43335821.4.0000.5119). All individuals provided an informed consent to participate in the research. Experiments with mice were conducted according to institutional guidelines for animal ethics and approved by the Institutional Ethics Committees from Oswaldo Cruz Foundation and Ethics, Commission on Animal Use (CEUA) LW 25/20.
Blood donors
Human blood samples were collected from vaccinated individuals (n = 33), convalescent patients (n = 13), and healthy controls (n = 9). All individuals were between 18 and 70 years old (36 ± 11, female:male ratio = 3.5). Detailed data regarding age and sex of the participants can be found in Table S1. Vaccinated individuals received two doses of CoronaVac (Sinovac, China), ChAdOx1-S (Oxford, Astrazeneca) or BNT162b2 (Pfizer-BioNTech) and were sampled 27-54 days after the second dose (Time Point 1 – TP1) and 180-195 days after the first dose just for CoronaVac and ChAdOx1-S (Time Point 2 – TP2). Convalescent individuals reported having mild COVID-19 between 24–196 days before sampling, confirmed by PCR. All individuals were briefly interviewed before sampling and consent forms were signed. We excluded vaccinated individual who were previously diagnosed with COVID-19. There was no participant compensation.
PBMC culture
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by Ficoll gradient. Briefly, blood layered on Ficoll-Paque plus (GE-Healthcare) were centrifuged 410 x g, 40 minutes, RT. One-million cells per well were distributed in 96-well flat-bottom plates and incubated in complete media (RPMI 1640, 10% FBS, 100 mg/ml streptomycin, 100 U/ml penicillin) with 5 μg/mL of N, S or SpiN recombinant antigen or anti-CD3 (1 μg/mL) and anti-CD28 (0.5 μg/mL) as positive controls. Unstimulated cells were used to assess the background production of cytokines. Culture supernatants were harvested after 72 h and frozen at −80°C until analysis.
Mice immunization and splenocyte culture
Female C57BL/6 mice (6-10 weeks) were immunized using a heterologous prime/boost protocol. Mice received PBS, CoronaVac (180 SU/animal), ChAdOx1-S (1010 PFU/animal) or BNT162b2 (100 μL/animal). Twenty-one days after the first dose, the animals received a booster dose of 10 μg SpiN-Tec (recombinant SpiN plus adjuvant). Both doses were administered intramuscularly in tibial muscle. Thirty days after administration of the second dose, the animals were euthanized and splenocytes were isolated by spleen maceration using a 100 μm pore cell strainer (Cell Strainer, BD Falcon). Then, erythrocytes were lysed with ammonium-chloride potassium (ACK) buffer and the cell number was adjusted to 1x106 cells/well and stimulated with 10 μg/mL of RBD. As positive control, Concanavalin A at 5 μg/mL (Sigma) was used.
Method details
Plasmid constructions and recombinant antigens production
Plasmids containing sequences encoding the full-length N, the RBD of Spike and the chimeric SpiN protein with codons optimized for expression in E. coli were purchased from Genscript. Competent E. coli Star (DE3) were transformed with the pET24 vector with N or the RBD sequences and E. coli SHuffle with the pET24_with SpiN. Transformed bacteria were grown in LB medium with kanamycin (50 μg/ml) at 37°C until OD600 0.6 was reached. At this point, protein expression was induced by adding IPTG to the culture at a final concentration of 0.5 mM. The induction of expression of the three proteins was done at 37°C for 3 h for N and RBB and for 18 h for SpiN. The N and RBD proteins contained a histidine tag and were purified through affinity chromatography step with the Histrap HP (GE HealthCare) column following the manufacturer’s instructions. After bacterial lysis, N protein was purified from the soluble fraction and RBD protein from the insoluble fraction by adding 8 M urea in the buffers for solubilization. The SpiN protein, expressed without histidine tag, was purified from the soluble and insoluble fractions of the bacteria cell lisate after the addition of 8 M urea and through two steps of chromatography. A cation exchange chromatography with the Hitrap SP HP column (GE HealthCare) was followed by molecular exclusion with the column HiPrep 26/60 Sephacryl S-100 HR (GE HealthCare), following the manufacturer’s instructions. The S protein expressed in mammalian cells was kindly provided by Dr. Leda Castilho from Universidade Federal do Rio de Janeiro.
IFN-γ measurement
Levels of IFN−γ from human serum were measured by ELISA following the manufacturer’s protocol (BD, OptEIA Human IFN−γ, Cat 555142). The supernatant of splenocytes culture was collected for determination of IFN-γ levels by ELISA (R&D Systems) following manufacturer’s instructions.
Detection of human and mouse antigen-specific antibodies
Plates were coated overnight with 0.4 μg/well of N or S recombinant proteins and blocked for 2 h with PBS containing 2% bovine serum albumin (PBS-2% BSA) at 37°C. Serum was diluted at 1:100 in PBS-2% BSA and incubated for 1 h at 37°C, and then incubated with anti-human IgG-HRP antibody (Fapon, BEEIGGI201) or with anti-mouse IgG-HRP antibody (Southern Biotech, 1030-05), all diluted 1:5000. After 5 washes, plates were revealed with 1-Step ultra TMB substrate solution (Biolegend) for 15 minutes in the dark, and reaction was stopped by adding 2 N H2SO4 (Sigma). Plates were read in 450 nm and results were expressed as raw optical density (OD) for murine IgG and as ELISA index (division of the OD450 of each sample by the cut-off value) for human IgG.
Flow cytometry
PBMCs from healthy donors, convalescents or vaccinated donors were thawed in RPMI 1640 (Sigma-Aldrich) with benzonase nuclease (20 U/mL, Sigma). Cells were plated in complete media (RPMI 1640, 10% FBS, 100 mg/ml streptomycin, 100 U/ ml penicillin), rested for 5 h and then incubated with positive controls (anti-CD3, 1 μg/mL and anti-CD28, 0.5 μg/mL, BD), negative controls (SpiN Vehicle) or 20 μg/mL of SpiN in 5% CO2 at 37°C. After 12 h incubation, 2.5 μg/mL each of Brefeldin A and Monensin was added for additional 6 h. After total 18 h incubation cells were harvested and stained for viability (Fixable Aqua Dead Cell Stain Kit, TermoFischer) and surface antigens anti-CD4 (BV605, RPA-T4, BD), anti-CD8 (AlexaFluor700, SK1, Biolegend), anti-CD45RO (BV786, UCHL1, BD) and anti-CD27 (APC-Cy7, O323, Biolegend). After washing, cells were fixed and permeabilized according to the manufacturer’s instructions (FoxP3 staining buffer set, eBioscience). Cells were then stained for the intracellular antigens anti-IFN−γ (PE-Cy7, 4 S.B3, eBioscience) and anti-CD3 (FITC, UCHT1, BD). Samples were acquired on a CytoFlex (Beckman Coulter) and analyzed on FlowJo. T-cell subpopulations were gated on viable CD3+ CD4+ and CD3+ CD8+ events. More detailed analysis of naive and memory sub-populations were done based on CD27/CD45RO expression and intracellular IFN−γ: effector memory (EM, CD45RO+CD27-), central memory (CM, CD45RO+CD27+), effector (Eff, CD45RO-CD27-), and naïve (Nv, CD45RO-CD27+) cells.
Detection of SpiN-Specific antibody-secreting cells (ASC)
ELISpot to detect ASCs was performed with the use of the Human IgG ELISpotbasic kit (Mabtech, AB, Nacka, Sweden). In brief, PBMCs were incubated for three days in RPMI-1640 medium with 10% FBS, supplemented with the TLR7 and TLR8 agonist imidazoquinoline resiquimod (R848, 1 mg/ml), and recombinant human IL-2 (10 ng/ml) for stimulation of memory B cells. The ELISpot plates (MAHAS4510, Millipore, Bedford, MA, USA) pre-coated with 50 μg/mL of SpiN or capturing monoclonal anti-human IgG antibodies were incubated with a total of 5x105 or 5x104 pre-stimulated cells per well for detection of SpiN-specific IgG and total IgG (positive control) secreting cells, respectively. PBMC cultured without R848 and IL-2 were used as negative control. Spots detection were made using biotinylated monoclonal antibody (MT91/145), streptavidin-ALP and substrate BCIP/ NBT-plus (Mabtech, AB, Nacka, Sweden). The number of B cells secreting IgG antibodies specific for SpiN and cells secreting IgG (total IgG) were measured using the CTL ImmunoSpot® Reader.
Plaque reduction neutralization test
To access the neutralizing antibodies, serum samples from human and mice were tested in Biosafety level 3 facilities in an in-house plaque reduction neutralization test (PRNT). The assay was performed in duplicates, using 24-well tissue culture plates (TPP- Technno Plastic Products AG, Trasadingen, Switzerland). For this purpose, CCL-81 Vero cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin-streptomycin. The sera were diluted and incubated with SARS-CoV-2. After 1 hour, the mix was added to Vero cells (2x105 cells) plated in a 24 wells plate and incubated for 1 hour, then the mix was removed and was added DMEM supplemented with 2% carboxymethylcellulose (CMC) and incubates for 72 hours at 37°C and 5% CO2 to allow the viral the viral plaque formation. PRNT50 was defined as the highest sample dilution that showed 50% reduction in number of plaques formed compared with possible control who consist in the number of plaques in wells inoculated with SARS-CoV-2 alone.53
Quantification and statistical analysis
Statistical analysis was conducted using GraphPad Prism 6.0 for Mac (GraphPad Inc, USA). First, outliers were detected with Grubbs’s test and then D’Agostino-Pearson was run to verify data normality. The tests used on each data analysis are explained on figure legends. In general, comparison between the groups was performed through unpaired t-test or Mann–Whitney U test, according to data distribution. Statistical differences were considered significant when p values ≤0.05.
Acknowledgments
Rede Virus from the Ministry of Science, Technology and Innovation, (Finep 01.20.0010 and 01.20.0005.00; CNPq 403514/2020-7 and 403701/2020-1 R.T.G.); Science and Technology of Vaccines (FAPEMIG APQ-03608-17/CNPq 465293/2014-0 R.T.G.); FAPESP (2020/05527-0 R.T.G.); Fundação Hospitalar do Estado de Minas Gerais (FHEMIG). We thank Leda Castilho and Aline Magalhães for providing the S protein and for the English checking, respectively, our project analyst Ms. Cristiane Gomes, our secretary Elizabeth Araújo, and the technicians Raquel Caldeira Horsth e Débora Ribeiro Soares.
Author contributions
F.F.B., N.S. and S.R.T were responsible for protein expression. B.V.S.V., T.G.M., G.G.A., L.I.O., G.B.F.M., M.A.A.R and L.P.F. performed blood samples processing and PBMC culture. B.V.S.V., T.G.M., G.G.A. performed IgG and IFN-γ ELISAS of human samples. B.V.S.V. and G.G.A. performed flow cytometry assays. B.V.S.V. and J.T.C. performed ELISpot assay. B.V.S.V., F.F.B., L.P.F., J.T.C. and N.S.H.S. performed mice immunization, IgG and IFN-γ ELISAS of mice samples. B.V.S.V. and L.A.F.A. performed PRNT assay. B.V.S.V., G.G.A., and R.T.G. contributed to study’s conception and design. B.S.V. wrote the manuscript.
Declaration of interests
R.T.G., S.R.T., N.S., J.T.C. and N.S.H-S. are co-inventors of the potential COVID-19 vaccine evaluated in this study. The patent is under evaluation process, application number BR1020210095733. The other authors declare no competing interests.
Published: June 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110177.
Contributor Information
Bruno Vinicius Santos Valiate, Email: valiate.bvs@gmail.com.
Gregório Guilherme Almeida, Email: vet.greg@gmail.com.
Supplemental information
References
- 1.Tan W., Zhao X., Ma X., Wang W., Niu P., Xu W., Gao G.F., Wu G. Notes from the Field A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases-Wuhan, China 2019−2020. China CDC Wkly. 2020;2:61–62. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao R., Liang J., Shen J., Ghosh S., Zhu L.R., Yang H., Wu K.C., Chen M.H., Chinese Society of IBD, Chinese Elite IBD Union. Chinese IBD Quality Care Evaluation Center Committee Implications of COVID-19 for patients with pre-existing digestive diseases. Lancet Gastroenterol. Hepatol. 2020;5:425–427. doi: 10.1016/S2468-1253(20)30076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. doi: 10.1001/JAMA.2020.12839. [DOI] [PubMed] [Google Scholar]
- 4.Merad M., Blish C.A., Sallusto F., Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/SCIENCE.ABM8108. [DOI] [PubMed] [Google Scholar]
- 5.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMOA2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/S41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 10.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/SCIENCE.ABC5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormick K.D., Jacobs J.L., Mellors J.W. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/SCIENCE.ABG4493. [DOI] [PubMed] [Google Scholar]
- 12.Castro J.T., Azevedo P., Fumagalli M.J., Hojo-Souza N.S., Salazar N., Almeida G.G., Oliveira L.I., Faustino L., Antonelli L.R., Marçal T.G., et al. Promotion of neutralizing antibody-independent immunity to wild-type and SARS-CoV-2 variants of concern using an RBD-Nucleocapsid fusion protein. Nat. Commun. 2022;13:4831. doi: 10.1038/s41467-022-32547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orenstein W.A., Ahmed R. Simply put: Vaccination saves lives. Proc. Natl. Acad. Sci. USA. 2017;114:4031–4033. doi: 10.1073/PNAS.1704507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olshansky S.J., Hayflick L. The Role of the WI-38 Cell Strain in Saving Lives and Reducing Morbidity. AIMS Public Health. 2017;4:127–138. doi: 10.3934/PUBLICHEALTH.2017.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/NM917. [DOI] [PubMed] [Google Scholar]
- 16.Pulendran B., Ahmed R. Immunological Mechanisms of Vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Q., Zhang H., Yao P., Xu N., Sun Y., Lu H., Xu F., Liao Y., Yang J., Mao H., et al. Immunogenicity and immune-persistence of the CoronaVac or Covilo inactivated COVID-19 Vaccine: a 6-month population-based cohort study. Front. Immunol. 2022;13 doi: 10.3389/FIMMU.2022.939311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marking U., Havervall S., Greilert-Norin N., Ng H., Blom K., Nilsson P., Phillipson M., Hober S., Nilsson C., Mangsbo S., et al. Duration of SARS-CoV-2 Immune Responses Up to Six Months Following Homologous or Heterologous Primary Immunization with ChAdOx1 nCoV-19 and BNT162b2 mRNA Vaccines. Vaccines (Basel) 2022;10:359. doi: 10.3390/vaccines10030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menni C., May A., Polidori L., Louca P., Wolf J., Capdevila J., Hu C., Ourselin S., Steves C.J., Valdes A.M., Spector T.D. COVID-19 vaccine waning and effectiveness and side-effects of boosters: a prospective community study from the ZOE COVID Study. Lancet Infect. Dis. 2022;22:1002–1010. doi: 10.1016/S1473-3099(22)00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 21.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/J.CELL.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Mao Q., Wu X., He Q., Bian L., Bai Y., Wang Z., Wang Q., Zhang J., Liang Z., Xu M. Considerations for the Feasibility of Neutralizing Antibodies as a Surrogate Endpoint for COVID-19 Vaccines. Front. Immunol. 2022;13:1897. doi: 10.3389/FIMMU.2022.814365/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 25.Cox R.J., Brokstad K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020;20:581–582. doi: 10.1038/s41577-020-00436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadarangani M., Marchant A., Kollmann T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021;21:475–484. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular Immune Responses to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection in Senescent BALB/c Mice: CD4 + T Cells Are Important in Control of SARS-CoV Infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Ploeg K., Kirosingh A.S., Mori D.A.M., Chakraborty S., Hu Z., Sievers B.L., Jacobson K.B., Bonilla H., Parsonnet J., Andrews J.R., et al. TNF-α+ CD4+ T cells dominate the SARS-CoV-2 specific T cell response in COVID-19 outpatients and are associated with durable antibodies. Cell Rep. Med. 2022;3 doi: 10.1016/J.XCRM.2022.100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell. 2020;183:1479–1495.e20. doi: 10.1016/J.CELL.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubio-Casillas A., Redwan E.M., Uversky V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines. 2022;10 doi: 10.3390/BIOMEDICINES10061339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/J.CELL.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2020;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 34.Nelson R.W., Chen Y., Venezia O.L., Majerus R.M., Shin D.S., Carrington M.N., Yu X.G., Wesemann D.R., Moon J.J., Luster A.D., et al. SARS-CoV-2 epitope–specific CD4+ memory T cell responses across COVID-19 disease severity and antibody durability. Sci. Immunol. 2022;7:9464. doi: 10.1126/SCIIMMUNOL.ABL9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. SARS-CoV-2–specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131 doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taus E., Hofmann C., Ibarrondo F.J., Hausner M.A., Fulcher J.A., Krogstad P., Ferbas K.G., Tobin N.H., Rimoin A.W., Aldrovandi G.M., Yang O.O. Dominant CD8+ T Cell Nucleocapsid Targeting in SARS-CoV-2 Infection and Broad Spike Targeting From Vaccination. Front. Immunol. 2022;13:506. doi: 10.3389/FIMMU.2022.835830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilocca B., Soggiu A., Sanguinetti M., Musella V., Britti D., Bonizzi L., Urbani A., Roncada P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 2020;22:188–194. doi: 10.1016/J.MICINF.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhagen J., van der Meijden E.D., Lang V., Kremer A.E., Völkl S., Mackensen A., Aigner M., Kremer A.N. Human CD4+ T cells specific for dominant epitopes of SARS-CoV-2 Spike and Nucleocapsid proteins with therapeutic potential. Clin. Exp. Immunol. 2021;205:363–378. doi: 10.1111/CEI.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/SCIENCE.ABF4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee E., Sandgren K., Duette G., Stylianou V.V., Khanna R., Eden J.-S., Blyth E., Gottlieb D., Cunningham A.L., Palmer S. Identification of SARS-CoV-2 Nucleocapsid and Spike T-Cell Epitopes for Assessing T-Cell Immunity. J. Virol. 2021;95 doi: 10.1128/JVI.02002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hevesi Z., Gerges D.A., Kapps S., Freire R., Schmidt S., Pollak D.D., Schmetterer K., Frey T., Lang R., Winnicki W., et al. Preclinical Establishment of a Divalent Vaccine against SARS-CoV-2. Vaccines (Basel) 2022;10 doi: 10.3390/VACCINES10040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mambelli F., Marinho F.V., Andrade J.M., De Araujo A.C.V.S.C., Abuna R.P.F., Fabri V.M.R., Santos B.P.O., Da Silva J.S., De Magalhães M.T.Q., Homan E.J., et al. Recombinant Bacillus CalmetteŸGuérin Expressing SARS-CoV-2 Chimeric Protein Protects K18-hACE2 Mice against Viral Challenge. J. Immunol. 2023;210:1925–1937. doi: 10.4049/jimmunol.2200731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T., Wang L., Wang H., Li X., Zhang S., Xu Y., Wei W. Serum SARS-COV-2 Nucleocapsid Protein: A Sensitivity and Specificity Early Diagnostic Marker for SARS-COV-2 Infection. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/FCIMB.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahlén G., Frelin L., Nikouyan N., Weber F., Höglund U., Larsson O., Westman M., Tuvesson O., Gidlund E.K., Cadossi M., et al. The SARS-CoV-2 N Protein Is a Good Component in a Vaccine. J. Virol. 2020;94:10. doi: 10.1128/JVI.01279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutta N.K., Mazumdar K., Gordy J.T. The Nucleocapsid Protein of SARS-CoV-2: a Target for Vaccine Development. J. Virol. 2020;94 doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matchett W.E., Joag V., Stolley J.M., Shepherd F.K., Quarnstrom C.F., Mickelson C.K., Wijeyesinghe S., Soerens A.G., Becker S., Thiede J.M., et al. Nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. bioRxiv. 2021 doi: 10.1101/2021.04.26.441518. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira S.C., de Magalhães M.T.Q., Homan E.J. Immunoinformatic Analysis of SARS-CoV-2 Nucleocapsid Protein and Identification of COVID-19 Vaccine Targets. Front. Immunol. 2020;11:2758. doi: 10.3389/FIMMU.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moderbacher C.R., Ramirez S.I., Dan J.M., Smith D.M., Sette A., Crotty S. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/J.CELL.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L., Van Beek M., Wang Z., Muecksch F., Canis M., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., Chakraborty A.K. Antigen presentation dynamics shape the response to emergent variants like SARS-CoV-2 Omicron strain after multiple vaccinations with wild type strain. bioRxiv. 2022 doi: 10.1101/2022.08.24.505127. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bagno F.F., Andrade L.A.F., Sérgio S.A.R., Parise P.L., Toledo-Teixeira D.A., Gazzinelli R.T., Fernandes A.P.S.M., Teixeira S.M.R., Granja F., Proença-Módena J.L., et al. Previous Infection with SARS-CoV-2 Correlates with Increased Protective Humoral Responses after a Single Dose of an Inactivated COVID-19 Vaccine. Viruses. 2022;14:510. doi: 10.3390/V14030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data produced in this paper is available upon request to the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.