Abstract
Several studies have indicated that the genetic diversity of human T-cell leukemia virus type 1 (HTLV-1), a virus associated with adult T-cell leukemia, is significantly lower than that of other retroviruses, including that of human immunodeficiency virus type 1 (HIV-1). To test whether HTLV-1 variation is lower than other retroviruses, a tractable vector system has been developed to measure reverse transcription accuracy in one round of HTLV-1 replication. This system consists of a HTLV-1 vector that contains a cassette with the neomycin phosphotransferase (neo) gene, a bacterial origin of DNA replication, and the lacZα peptide gene region (the mutational target). The vector was replicated by trans-complementation with helper plasmids. The in vivo mutation rate for HTLV-1 was determined to be 7 × 10−6 mutations per target base pair per replication cycle. The majority of the mutations identified were base substitution mutations, namely, G-to-A and C-to-T transitions, frameshift mutations, and deletion mutations. Mutation of the methionine residue in the conserved YMDD motif of the HTLV-1 reverse transcriptase to either alanine or valine (i.e., M188A or M188V) led to a factor of two increase in the rate of mutation, indicating the role of this motif in enzyme accuracy. The HTLV-1 in vivo mutation rate is comparable to that of bovine leukemia virus (BLV), another member of the HTLV/BLV genus of retroviruses, and is about fourfold lower than that of HIV-1. These observations indicate that while the mutation rate of HTLV-1 is significantly lower than HIV-1, this lower rate alone would not explain the low diversity in HTLV-1 isolates, supporting the hypothesis that HTLV-1 replicates primarily as a provirus during cellular DNA replication rather than as a virus via reverse transcription.
Human T-cell leukemia virus type 1 (HTLV-1) is a member of the human T-cell leukemia virus/bovine leukemia virus (HTLV/BLV) genus of the Retroviridae family. HTLV-1 has been shown by epidemiology to be associated with adult T-cell leukemia, HTLV-1-associated myelopathy-tropical spastic paraparesis, and polymyositis (5). Infection of HTLV-1 is endemic in Melanesia, Japan, the Caribbean, and sub-Saharan Africa. There is a remarkable amount of homogeneity among HTLV-1 isolates (12, 13, 15, 25, 35, 41, 43–45, 52). For example, isolates from Japan have close to 99% homology, and isolates from Japan, the Caribbean, and Africa can also share as much as 99% homology. It has been suggested that HTLV-1 isolates endemic in different races may be of utility in studying the movement of ancient human populations or in anthropologic studies (12). HTLV-1 isolates from Melanesia would not be as useful, since there is not as much sequence homology to the original Japanese isolate (11). This suggests that HTLV-1 may have originated in the Pacific Rim rather than in Africa. Genetic diversity among isolates of HTLV-2 is equally low (48).
The low level of genetic diversity in HTLV-1 has been speculated to be due to oligoclonal expansion of infected cells and very low levels of virus replication in infected individuals (6, 7, 54). The low levels of virus replication observed in cell culture has been used to support this hypothesis. Replication of the viral genome primarily as a provirus during cellular DNA replication would provide a higher-fidelity mode of virus replication than viral nucleic acid replication via reverse transcription. An advantage for the virus in doing this is that HTLV may be able to escape immune selection. It has been shown that the HTLV envelope protein becomes nonfunctional with a limited number of mutations, which is in contrast to the human immunodeficiency virus type 1 (HIV-1) envelope (38).
To help dissect the basis of genetic variation of retroviruses, the mutation rate per replication cycle has been studied extensively. This work was initiated by designing systems to determine the mutation rate per base pair per replication cycle for spleen necrosis virus (SNV), an avian C-type retrovirus similar to the murine type C retroviruses, using an amber codon reversion assay with an SNV vector (9, 10). A similar reversion assay was used to determine the mutation rate of murine leukemia virus (MLV) (53). The in vivo forward mutation rates for various types of mutations were calculated with the lacZα peptide gene as a reporter gene for mutations and the blue-white colony color selection method for identifying mutant proviruses in Escherichia coli (36, 37). The major types of mutations found were base-pair substitutions, frameshifts, simple deletions, and deletions with insertions. The overall in vivo mutation rate of SNV in this system was determined to be 10−5 mutations/target base pair/replication cycle.
These studies have been extended to BLV (32). BLV was found to have a mutation rate of 4 × 10−6, which is 2.5 times less than that for SNV. A similar distribution of mutation types was found with BLV relative to that of SNV, indicating that a common property of reverse transcriptase (RT) is responsible for all of these error processes. Temin speculated that this common property was the strand-transfer process (50). The mutation rate of HIV-1 was determined with a vector containing the lacZα peptide gene (26, 31). The mutation rate of HIV-1 in this system was determined to be 3 × 10−5 mutations per target base pair per cycle. The Vpr protein of HIV-1 influences the mutation rate and involves the interaction of Vpr with the cellular DNA repair enzyme uracil DNA glycosylase (27, 30).
Phylogenetic analysis of RTs has indicated several conserved domains and structures that are important for RT function (34). For example, the fingers, palm, and thumb subdomains of the HIV-1 RT catalytic domain are thought to be well conserved among other RTs because of the importance of this region of the enzyme for contacting the primer-template, and binding the incoming deoxynucleoside triphosphate (20). The conserved YXDD motif is important because it has been associated with resistance to nucleoside analogs, decreases in enzymatic activity and viral infectivity, and changes in the positioning of the primer in the template-primer complex (39).
The objectives of this study were to determine the accuracy of HTLV-1 reverse transcription and to see if amino acid substitutions in the conserved YMDD motif of HTLV-1 RT would influence the rate of HTLV-1 mutation. To do this, a tractable genetic system was developed to measure the forward rate of mutation using the lacZα peptide gene as a reporter for mutations. The mutation rate of HTLV-1 was determined to be 7 × 10−6 mutations/target base pair/replication cycle. This rate is comparable to that of BLV, but is about fourfold lower than the mutation rate of HIV-1. Mutation of the YMDD motif doubled the mutation rate of HTLV-1.
MATERIALS AND METHODS
HTLV-1 vector construction.
The HTLV-1 shuttle vector pH1sh (Fig. 1) was constructed from pHTLV-CMVNEO (kindly provided by David Derse, National Cancer Institute) (8) in a two-step process. First, a deletion of the pol region was done by deletion of a HindIII fragment. Second, a cassette containing the simian virus 40 (SV40) promoter driving expression of the neomycin phosphotransferase (neo) gene, a bacterial origin of DNA replication, and the lacZα peptide gene region from a previously described HIV-1 vector (31) was inserted in place of the CMVNEO cassette to create pH1sh. The plasmid pCMV-HT1 (kindly provided by David Derse) was used as a helper plasmid to trans-complement the HTLV-1 shuttle vector with gag, pol, tax, and rex (8). The plasmid pSV-A-MLV-env was also used as a helper plasmid and has been previously described (24). The HTLV-1 RT variants M188A and M188V were made in pCMV-HT1 by a primary-combinatorial two-step PCR protocol (17).
FIG. 1.
HTLV-1 vector used for reverse transcription accuracy studies. (A) HTLV-1 vector. The vector is shown in the proviral DNA form and has a cassette containing the SV40 promoter driving expression of the neomycin phosphotransferase gene (neo), a bacterial plasmid origin of DNA replication (pACYC ori), and the lacZα peptide gene region (lacZα) that includes the lac operator sequence. (B) Protocol for one cycle of HTLV-1 vector virus replication. The steps going from a parental shuttle vector provirus in the step 2 cell to a vector provirus in the step 3 cell constitute a single cycle of replication.
Transfections, infections, and cocultivations.
The CEM-A cell line used was obtained from the NIH AIDS Reagent Program and was maintained in RPMI 1640 supplemented with 2 mM l-glutamine containing 10% calf serum. The HTLV-1 vector and expression plasmids were transfected into CEM-A cells by use of either dimethyl sulfoxide-Polybrene (18) or Superfect (Qiagen). CEM-A cells were infected in the presence of Polybrene (28). Cocultivation of CEM-A target cells with virus-producing cells was also done as previously described (29, 32). Briefly, virus-producing cells (typically, 2.5 × 105 cells per 60-mm petri dish or 5 × 105 cells per 100-mm petri dish or 7.5 × 105 cells per 150-mm petri dish were treated with mitomycin C (8 μg/ml), an inhibitor of host cell DNA synthesis, for 1.5 h at 37°C. The cells were then washed three times with fresh medium, and CEM-A target cells equivalent to the number of treated virus-producing cells were added. Two days after cocultivation, selective medium containing G418 was added. Control experiments were done with each cocultivation experiment to ensure that mitomycin C-treated, virus-producing cells did not proliferate and no longer adhered to the surfaces of culture dishes. Cells expressing the neo gene were selected with the neomycin phosphotransferase analog, G418, until the formation of colonies (typically about 3 weeks).
Analysis of HTLV-1 reverse transcription accuracy in a single replication cycle.
The experimental protocol developed to assay a single cycle of HTLV-1 vector replication is shown in Fig. 1. The protocol contains three steps. In step 1, the HTLV-1 vector was introduced into CEM-A cells by transfection and placed under G418 selection. Cell clones were then transiently transfected with the helper plasmids. In step 2, vector virus was harvested 48 h posttransfection from step 1 cells and used to infect fresh CEM-A cells. G418-resistant cell clones were transiently transfected with the helper plasmids (step 2 cells). Step 2 clones were tested by Southern analysis to ensure that only a single vector proviral DNA was present. The lacZα peptide gene in the vector proviral DNA of step 2 clones was sequenced to confirm that no mutations were introduced. In step 3, vector virus was transferred to fresh CEM-A target cells by cocultivation for 24 to 48 h after transient transfection of helper plasmids; cells were then placed under G418 selection (step 3 cells). Cocultivation was used to produce step 3 cells to maximize the number of step 3 cells for analysis of the mutant frequency.
Recovery of shuttle vector proviral DNA and DNA sequencing of the lacZα peptide region.
Purified genomic DNA (42) from pools of step 3 clones was digested with the restriction enzyme SalI to release the neo, pACYC origin of replication, and lacZα peptide gene sequences from the HTLV-1 shuttle vector proviral DNA (Fig. 1). Proviral DNA was purified with the Lac repressor protein as previously described (32). The Lac repressor protein was purified from E. coli HB101/lac pIQ (kindly supplied from Tom Record, University of Wisconsin-Madison) as previously described (23). The purified proviral DNA was ligated and used to electroporate competent E. coli XL1 Blue cells (Stratagene). Kanamycin-resistant bacterial colonies were selected in the presence of the isopropyl-β-d-thiogalactoside (IPTG) inducer. The ratio of white plus light-blue bacterial colonies to total bacterial colonies observed provided a forward mutant frequency for a single retroviral replication cycle. Plasmid DNA was purified (42) and sequenced in the lacZα peptide gene region in order to determine the mutation rate.
RESULTS
A HTLV-1 single cycle replication assay for mutation detection.
In order to measure HTLV-1 reverse transcription accuracy, a replication assay was developed to measure the in vivo mutation rate in a single cycle of replication. An HTLV-1 shuttle vector was constructed in order to identify mutations that had occurred during HTLV-1 replication. The vector contained a cassette which included the neo gene under control of the SV40 promoter, a bacterial origin of plasmid DNA replication, and the lacZα peptide gene region as a mutational target. This cassette has been previously used for analysis of reverse transcription accuracy of HIV-1 and BLV (31, 32). This vector, pH1sh, can replicate in both mammalian cells as a virus and in bacterial cells as a plasmid.
The assay developed is shown in Fig. 1. CEM-A cells were used in these studies because they are adherent T-lymphoid cells that are permissive for HTLV-1 replication (51). The utility of adherent cells is that they could be placed under drug selection when vector virus was introduced either by transfection or infection and form drug-resistant colonies. The HTLV-1 shuttle vector, pH1sh, was first introduced into fresh CEM-A cells by transfection and placed under G418 selection (step 1 cells). G418-resistant colonies were pooled and then transfected with helper plasmids. The supernatant from these cells was used to infect fresh CEM-A cells. Titers of the vector virus were very low (∼10 CFU/ml) but generated several infected cell clones for further studies. Prior to use in the single cycle replication assay, a few selected clones were analyzed for the presence of a single integrated provirus. Data indicated that the selected clones contained single integrated copies of the HTLV-1 vector (data not shown). Transfection of helper plasmids into these cells (step 2 cells) was done to allow for virus production of the vector. These virus-producing cells were treated with mitomycin C and cocultured with fresh CEM-A cells. Cocultivation was used to produce step 3 cells to maximize the number of step 3 cells for analysis of the mutant frequency. Following cocultivation, cells were placed under G418 selection. Approximately 1,000 to 1,800 G418-resistant colonies were obtained per 7.5 × 105 target cells cocultivated with step 2 cells. The G418 resistant cells (step 3 cells) were then pooled from over 40,000 colonies, and the total DNA was purified. The purified DNA was digested with SalI, and the cassette containing the lacZα peptide gene region mutational target containing the Lac operator sequence was purified using the Lac repressor protein.
Mutant frequency, type, and location in HTLV-1 replicated with wild-type RT.
The mutant frequency from several parallel experiments indicated that the average mutant frequency was 0.0009 (33/36,561) mutation per target base pair per replication cycle (Table 1). Nucleotide sequence analysis of the lacZα peptide gene region was done to determine the types of mutations responsible for the mutant colony color phenotype (Table 2). Of the 33 mutants recovered, 20 (61%) had base substitution mutations. Of the 20, 14 (70%) were G-to-A and C-to-T transition mutations. One C-to-T hypermutant was identified, which contained two C-to-T transition mutations within the mutational target. Of the 33 mutants, 7 (21%) had frameshift mutations. Six of the seven frameshift mutations were +1 frameshifts in runs of A's, C's, or T's. Finally, 6 of the 33 mutants (18%) had deletion mutations. The deletion mutations were either simple deletions (four of six) that contained base pair homology at the deletion junctions or were more complex deletions with insertion mutations (inserted sequences of unknown origin).
TABLE 1.
Analysis of HTLV-1 reverse transcription accuracy by recovery of mutant proviruses following replication with either wild-type HTLV-1 RT or YXDD variants
| RT | Relative amt of virus production | Step 2 clone | No. of independent mutants/total no. of bacterial colonies [total] | Mutant frequency (mutant/cycle) [avg] |
|---|---|---|---|---|
| Wild type | 1.0 | 1a | 3/3,217 | 0.0009 |
| 2a | 3/4,592 | 0.0007 | ||
| 3a | 2/3,648 | 0.0005 | ||
| 1b | 3/2,843 | 0.0011 | ||
| 2b | 4/4,331 | 0.0009 | ||
| 3b | 2/2,720 | 0.0007 | ||
| 1a | 6/5,035 | 0.0012 | ||
| 2a | 4/4,792 | 0.0008 | ||
| 3c | 6/5,383 | 0.0011 | ||
| [33/36,561] | [0.0009]a | |||
| M188A | 0.7 | 1a | 10/4,126 | 0.0024 |
| 2a | 8/3,790 | 0.0021 | ||
| [18/7,916] | [0.0023] | |||
| M188V | 0.6 | 1a | 12/5,264 | 0.0023 |
| 1b | 8/4,477 | 0.0018 | ||
| [20/9,741] | [0.0021] |
Standard deviation of 0.0005 mutant/cycle.
TABLE 2.
Spectrum of mutations in the lacZα peptide gene region of recovered HTLV-1 vector proviruses after replication with either wild-type HTLV-1 RT or by YXDD variants
| Nucleotide change(s) | No. of independent mutants recovered
|
||
|---|---|---|---|
| Wild type | M188A | M188V | |
| G-to-A | 8 | 3 | 3 |
| C-to-T | 6 | 2 | 3 |
| T-to-C | 2 | 1 | |
| T-to-A | 1 | 1 | |
| G-to-T | 1 | 1 | |
| T-to-G | 1 | ||
| C-to-T hypermutant | 1 | 1 | |
| G-to-A hypermutant | 1 | ||
| TTTT to TTTTT | 1 | 1 | 2 |
| TTT to TTTT | 2 | 1 | |
| TTTT to TTT | 1 | 1 | |
| AAA to AAAA | 1 | 1 | 1 |
| AAAA to AAAAA | 2 | 1 | 1 |
| AAAA to AAA | 1 | 2 | |
| CCCCC to CCCCCC | 1 | 1 | |
| CCC to CCCC | 1 | 1 | 1 |
| Δ53 | 1 | ||
| Δ93 | 1 | ||
| Δ13 | 1 | ||
| Δ56 | 1 | ||
| Δ45, +20 | 1 | ||
| Δ86, +65 | 1 | ||
| Δ34 | 1 | ||
| Δ7 | 1 | ||
| Δ12 | 1 | ||
| Δ47 | 1 | ||
| Δ43, +88 | 1 | ||
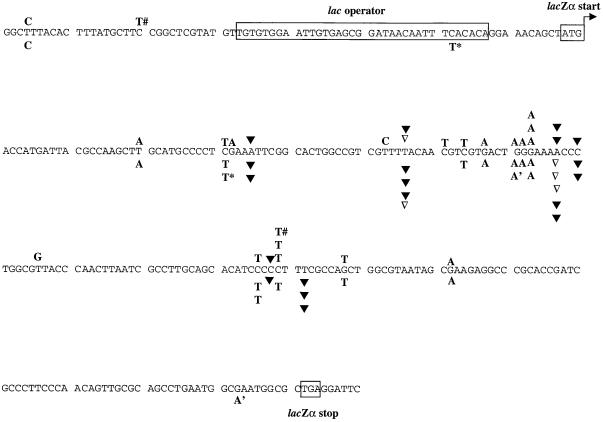
Analysis of the location of mutations indicated that the base substitutions and frameshift mutations occurred at locations in the lacZα peptide gene region where substitutions and frameshift mutations had been previously characterized for both BLV and HIV-1 (Fig. 2). This suggests that there are mutational hotspots within the lacZα peptide gene region that are recognized as such by many RTs. The deletion mutations were located throughout the lacZα peptide gene region but in most instances had deletion junctions near locations that may be mutational hotspots. The deletion junctions for the simple deletions had either 3- or 4-bp homology (Fig. 3). Two deletions with insertion mutants were identified that did not have base pair homology at the deletion junctions.
FIG. 2.
Plus strand nucleotide sequence of the lacZα gene region in the HTLV-1 vector provirus. The start and stop codons of the lacZα open reading frame (small boxed sequences) and the lac operator sequence (large boxed sequence) are shown. Nucleotide positions of base pair substitutions (letters above the sequence), +1 frameshifts (letters with ▾ above the sequence), −1 frameshifts (▿ above the sequence) are indicated. G-to-A and C-to-T hypermutants are indicated with a number sign (#), asterisk, or apostrophe adjacent to the letter above or below the sequence. The locations of base substitution and frameshift mutations in the parental HTLV-1 vector provirus replicated with wild-type HTLV-1 RT are indicated above the sequence, while the mutations in the HTLV-1 provirus replicated with the M188A and M188A RT variants are indicated below the nucleotide sequence.
FIG. 3.
Nucleotide sequence analysis of deletions and deletions with insertions. (A) Deletions and deletions with insertions in HTLV-1 proviruses replicated with wild-type RT. Short direct repeats at the deletion junctions are shown in boxes. The numbers of nucleotides deleted are indicated between the deletion junctions and are preceded by a minus sign; the number of inserted nucleotides are preceded by a plus sign. (B) Deletions and deletions with insertions in HTLV-1 proviruses replicated with M188A and M188V HTLV-1 RTs. Short direct repeats and numbers of nucleotides deleted and inserted are as described in panel A.
Mutant frequency and characterization of HTLV-1 replicated with RT variants in one round of replication.
To determine whether amino acid substitutions in the HTLV-1 RT could influence the accuracy of the reverse transcription process, the methionine residue at position 188 was changed to either alanine (M188A) or valine (M188V). M188 lies in the highly conserved YXDD motif. The HTLV-1 vector was then replicated in parallel with RT containing either the M188A mutation or the M188V mutation or with wild-type RT. Following recovery of proviruses from infected CEM-A target cells, the mutant frequency observed after replication with the M188A RT was found to be 0.0023 (18/7,916) mutant/cycle (Table 1). This is significantly higher (χ2 = 11; P < 0.005) than the mutant frequency found by replication with wild-type HTLV-1 RT (33/36,561) (Table 1). The mutant frequency observed following replication with M188V RT was 0.0021 (20/9,741) mutant/cycle (Table 1). The mutant frequency for M188V is also significantly higher (χ2 = 9; P < 0.005) than the mutant frequency obtained after replication with wild-type HTLV-1.
To determine whether the types of mutations that occurred when the HTLV-1 vector was replicated with either M188A or M188V were different than when the vector was replicated with wild-type RT, the 38 mutants isolated were sequenced. Table 2 summarizes the types of mutants identified. A total of 39 and 35% of the mutations identified for both M188A and M188V were substitution mutations (7 of 18 and 7 of 20), respectively. For both M188A and M188V, G-to-A and C-to-T transition mutations were the most common substitutions, occurring either 71% (5 of 7) or 86% (6 of 7) of the time, respectively. Frameshift mutations were observed for both M188A and M188V and represented 44% (8 of 18) or 45% (9 of 20) of the total mutants, respectively. This trend indicates a doubling of frameshift mutations compared to that with wild-type HTLV-1 RT (i.e., 7 of 33, 21%) (Table 2). M188A and M188V led to deletion mutations either 11% (2 of 18) or 15% (3 of 20) of the total, respectively. The deletion mutations included both simple deletions and deletions with insertions. A G-to-A hypermutant was recovered with M188A that had two G-to-A mutations, and a C-to-T hypermutant was recovered with M188V that had two C-to-T mutations in the lacZα peptide gene region.
The locations of the base substitution and frameshift mutations were compared to that observed with wild-type RT (Fig. 2). In general, the locations of mutations identified when HTLV-1 was replicated with either M188A or M188V were similar to that seen with wild-type RT.
The deletion mutations were located throughout the lacZα peptide gene region but in most instances had deletion junctions near locations that may be mutational hotspots. The deletion junctions for the simple deletions had either 3- or 1-bp homology (Fig. 3). Interestingly, one simple deletion had no base-pair homology at the deletion junction. One deletion with an insertion mutant was also identified that did not have base pair homology at the deletion junctions. The observation that the simple deletions had fewer homologous base pairs at the deletion junctions suggests that M188A and M188V may be less processive than wild-type HTLV-1 RT.
Relative rate of mutation for HTLV-1 to that of BLV, HIV-1, and SNV using the lacZα peptide gene region as a mutational target.
The characterized mutants from replicating HTLV-1 with wild-type RT allowed for calculation of the in vivo mutation rate for HTLV-1. The mutation rate was calculated to be 7 × 10−6 mutation/target base pair/replication cycle (Table 3). Target nucleotides in the mutational target have been previously described (1, 3, 36). The mutant frequency for HTLV-1 is not significantly different (χ2 = 1.2; P > 0.1) than that of BLV, but is fourfold lower than that of HIV-1 (χ2 = 77; P < 0.001) and is twofold lower than the mutant frequency of SNV (χ2 = 15; P < 0.005). This indicates that the HTLV-1 and BLV mutation rates are comparable, whereas the HTLV-1 mutation rate is significantly different from the HIV-1 and SNV mutation rates (Table 3).
TABLE 3.
Relative rates of mutation for HTLV-1, BLV, HIV-1, and SNV using the lacZα peptide gene region as a mutational target
| Virus | No. of independent mutant vector proviruses recovered
|
Overall mutant frequency (no. of mutants/total no. of colonies) | Mutation ratea | |||
|---|---|---|---|---|---|---|
| Base pair substitution | Frameshift | Deletion | Deletion with insertion | |||
| HTLV-1 | 20 | 7 | 4 | 2 | 33/36,561 (0.0009) | 7 × 10−6 |
| BLVb | 3 | 4 | 2 | 2 | 11/18,009 (0.0006) | 4 × 10−6 |
| HIV-1c | 57 | 26 | 7 | 3 | 93/20,696 (0.004) | 3 × 10−5 |
| SNVd | 11 | 5 | 12 | 7 | 37/16,867 (0.002) | 1 × 10−5 |
Rates are in units of mutations/target base pair/replication cycle. The rates of mutation were calculated as the sums of the rates of base pair substitution, frameshift, and deletion mutations per number of independent vector proviruses recovered per 114 target nucleotides for substitutions, per 150 target nucleotides for frameshifts, or per 280 target nucleotides for deletion mutations. Target nucleotides have been previously described (1, 3, 36).
Data from Mansky and Temin (32).
Data from Pathak and Temin (37).
DISCUSSION
Determination of the HTLV-1 in vivo mutation rate.
The accuracy of HTLV-1 reverse transcription has been determined. Using an HTLV-1 vector containing the lacZα peptide gene region as a mutational target, the mutant frequency was determined to be 0.0009 mutant/replication cycle. Sequence analysis of the recovered mutants indicated substitution, frameshift, and deletion mutations had occurred. The predominant type of mutations to occur were G-to-A and C-to-T transition mutations. Deletion mutations were found to represent about one-quarter of the mutants recovered. The calculated in vivo mutation rate for HTLV-1, 7 × 10−6 mutation per target base pair per replication cycle, is comparable to that previously reported for BLV but is significantly different than that of HIV-1 and SNV.
Possible mechanisms responsible for the creation of mutations.
The HTLV-1 vector used in these studies does not allow the determination of whether mutations occurred during minus-strand or plus-strand DNA synthesis, but the locations of the mutations suggest particular mechanisms for their creation. The majority of the G-to-A transitions characterized were in GpA dinucleotides. Transition mutations adjacent to runs of a single nucleotide appear to occur by the mechanism of dislocation mutagenesis (1, 22). In this model, dislocation of the primer to the template produces an unpaired nucleotide base; realignment occurs between the primer and the template resulting in a mismatch, followed by elongation beyond the mismatch. Most of the G-to-A transition mutations occurred at sites adjacent to a run of nucleotides, which suggests that these mutations could have occurred by dislocation mutagenesis.
The frameshift mutations characterized were mainly +1 frameshifts in runs of A's and T's. Plus-one frameshift mutations in runs of T's and A's occurred with SNV in vivo (4, 37). The frameshift mutations in homo-oligomeric runs suggest that these result from template-primer slippage (2, 21, 46, 47). The +1 frameshift mutations may have occurred during minus-strand DNA synthesis (4), while the −1 frameshift mutations could have occurred during either minus- or plus-strand DNA synthesis. Simple deletion and deletion with insertion mutants have been previously identified and the mechanisms by which they could have occurred have been proposed (36, 40).
Mutation of M188 in HTLV-1 RT decreases reverse transcription accuracy.
Two HTLV-1 RT variants, M188A and M188V, were found to significantly increase the rate of HTLV-1 mutation by a factor of 2 and therefore decrease the accuracy of HTLV-1 reverse transcription by twofold. The types of mutations that occurred during replication with these variants indicated a spectrum similar to what was observed with wild-type RT. However, limited number of mutants characterized suggests that the frequency of frameshift mutations doubled. Mutation of the YXDD motif may influence the template-primer affinity and could potentially influence frameshift fidelity.
Deletion rates.
It has been observed that defective proviruses represent about 25 to 40% of all HTLV-1 genomes present in lymphocytes from infected individuals (many of which are defective due to deletion mutations in gag, pol, and/or env) (16, 19, 33, 49). A large number of deleted HTLV-1 proviruses have also been observed in cell lines infected with a variety of HTLV-1 isolates (14). These observations could indicate that these deletion mutations are created during the reverse transcription process at a higher rate than that observed for other retroviruses. However, comparison of the frequency of deletion mutations among retroviruses in which in vivo mutation rates have been determined using the lacZα peptide gene as a mutational target does not support this (Table 3). Rather, it indicates that HTLV-1 is no more prone to deletion mutations than BLV, HIV-1, or SNV.
ACKNOWLEDGMENTS
I thank L. Bernard, P. Pandya, M. Reinhardt, and A. Waggoner for outstanding technical assistance. I also thank M. Williams for comments on the manuscript.
This work was supported by the Public Health Service (GM56615), the American Cancer Society, and the Ohio Cancer Research Associates.
REFERENCES
- 1.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 2.Bebenek K, Kunkel T A. The fidelity of retroviral reverse transcriptases. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 3.Boyer J C, Bebenek K, Kunkel T A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci USA. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns D P W, Temin H M. High rates of frameshift mutations within homo-oligomeric runs during a single cycle of retroviral replication. J Virol. 1994;68:4196–4203. doi: 10.1128/jvi.68.7.4196-4203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cann A J, Chen I S Y. Human T-cell leukemia virus types I and II. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1849–1880. [Google Scholar]
- 6.Cavrois M, Leclercq I, Gout O, Gessain A, Wain-Hobson S, Wattel E. Persistent oligoclonal expansion of human T-cell leukemia virus type 1-infected circulating cells in patients with tropical spastic paraparesis/HTLV-1 associated myelopathy. Oncogene. 1998;17:77–82. doi: 10.1038/sj.onc.1201906. [DOI] [PubMed] [Google Scholar]
- 7.Cavrois M, Wain-Hobson S, Gessain A, Plumelle Y, Wattel E. Adult T-cell leukemia/lymphoma on a background of clonally expanding human T-cell leukemia virus type 1-positive cells. Blood. 1996;88:4646–4650. [PubMed] [Google Scholar]
- 8.Copeland K F T, Haaksma A G M, Derse D, Heeney J L. Detection of human T-cell leukemia virus 1 permissive cells using cell lines producing selectable recombinant virions. J Virol Methods. 1994;50:219–226. doi: 10.1016/0166-0934(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 9.Dougherty J P, Temin H M. Determination of the rate of base-pair substitution and insertion mutation in retrovirus replication. J Virol. 1988;62:2817–2822. doi: 10.1128/jvi.62.8.2817-2822.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dougherty J P, Temin H M. High mutation rate of a spleen necrosis virus-based retrovirus vector. Mol Cell Biol. 1986;68:4387–4395. doi: 10.1128/mcb.6.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessain A, Boeri E, Yanagihara R, Gallo R C, Franchini G. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type I (HTLV-I) variant from Melanesia: genetic and phylogenetic relationship to HTLV-I strains from other geographical regions. J Virol. 1993;67:1015–1023. doi: 10.1128/jvi.67.2.1015-1023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gessain A, Gallo R C, Franchini G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol. 1992;66:2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray G S, White M, Bartman T, Mann D. Envelope gene sequence of HTLV-I isolate MT-2 and its comparison with other HTLV-I isolates. Virology. 1990;177:391–395. doi: 10.1016/0042-6822(90)90498-g. [DOI] [PubMed] [Google Scholar]
- 14.Hill S A, Shuh M, Derse D. Comparisons of defective HTLV-1 proviruses predict the mode of origin and coding potential of internally deleted genomes. Virology. 1999;263:273–281. doi: 10.1006/viro.1999.9922. [DOI] [PubMed] [Google Scholar]
- 15.Hinuma Y. Seroepidemiology of adult T-cell leukemia virus (HTLV-1/ATLV): origin of virus carriers in Japan. AIDS Res. 1986;2:517–522. [PubMed] [Google Scholar]
- 16.Hiramatsu K, Yoshikura H. Frequent partial deletion of human T-cell leukemia virus type 1 proviruses in experimental transmission: pattern and possible implication. J Virol. 1986;58:508–512. doi: 10.1128/jvi.58.2.508-512.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 18.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi N, Konishi H, Sabe H, Shigesada K, Noma T, Honjo T, Hatanaka M. Genomic structure of HTLV (human T-cell leukemia virus): detection of defective genome and its amplification in MT-2 cells. EMBO J. 1984;3:1339–1343. doi: 10.1002/j.1460-2075.1984.tb01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohlstaedt L A, Wang J, Rice P A, Friedman J M. The structure of HIV-1 reverse transcriptase. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 223–249. [Google Scholar]
- 21.Kunkel T A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A, Alexander P S. The base substitution fidelity of eukaryotic DNA polymerases-mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986;261:160–166. [PubMed] [Google Scholar]
- 23.Laiken S L, Gross C A, von Hippel P H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972;66:143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- 24.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik K T A, Eveir J, Karpas A. Molecular cloning and complete nucleotide sequence of an adult T-cell leukemia virus/HTLV-I (ATLV/HTLV-I) isolate of Caribbean origin: relationship to other members of the ATLV/HTLV-I subgroup. J Gen Virol. 1985;69:1695–1710. doi: 10.1099/0022-1317-69-7-1695. [DOI] [PubMed] [Google Scholar]
- 26.Mansky L M. Forward mutation rate of human immunodeficiency virus type 1 in a T-lymphoid cell line. AIDS Res Hum Retrovir. 1996;12:307–314. doi: 10.1089/aid.1996.12.307. [DOI] [PubMed] [Google Scholar]
- 27.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 28.Mansky L M. Retroviral-vector-mediated gene transfer. In: Griffiths J B, Doyle A, Newell D G, editors. Cell and tissue culture: laboratory procedures, update. 6th ed. New York, N.Y: John Wiley & Sons; 1994. pp. 27B:5.1–27B:5.10. [Google Scholar]
- 29.Mansky L M, Krueger A E, Temin H M. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J Virol. 1995;69:3282–3289. doi: 10.1128/jvi.69.6.3282-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansky L M, Preveral S, Selig L, Benarous R, Benichou S. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansky L M, Temin H M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994;68:494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka M, Tamiya S, Takemoto S, Yamaguchi K, Takatsuki K. HTLV-1 provirus in the clinical subtypes of ATL. Leukemia. 1997;11(Suppl. 3):67–69. [PubMed] [Google Scholar]
- 34.McClure M A. Evolutionary history of reverse transcriptase. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 425–444. [Google Scholar]
- 35.Paine E, Garcia J, Philpott T C, Shaw G, Ratner L. Limited sequence variation in human T-lymphotropic virus type 1 isolates from North American and African patients. Virology. 1991;182:111–123. doi: 10.1016/0042-6822(91)90654-t. [DOI] [PubMed] [Google Scholar]
- 36.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990b;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990a;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pique C, Tursz T, Dokhelar M C. Mutations introduced along the HTLV-I envelope gene result in a non-functional protein: a basis for envelope conservation? EMBO J. 1990;9:4243–4248. doi: 10.1002/j.1460-2075.1990.tb07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad V R. Genetic analysis of retroviral reverse transcriptase structure and function. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 135–162. [Google Scholar]
- 40.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salemi M, Desmyter J, Vandamme A-M. Tempo and mode of human and simian T-lymphotropic virus (HTLV/STLV) evolution revealed by analyses of full-genome sequences. Mol Biol Evol. 2000;17:374–386. doi: 10.1093/oxfordjournals.molbev.a026317. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Seiki M, Hatton S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman M P, Saksena N K, Dube D K, Yanagihara R, Poiesz B J. Evolutionary insights on the origin of human T-cell lymphoma/leukemia virus type I (HTLV-I) derived from sequence analysis of a new HTLV-I variant from Papua New Guinea. J Virol. 1992;66:2556–2563. doi: 10.1128/jvi.66.4.2556-2563.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slattery J P, Franchini G, Gessain A. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 1999;9:525–540. [PubMed] [Google Scholar]
- 46.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Streisinger G, Owen J. Mechanism of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Switzer W M, Pieniazek D, Swanson P, Samdal H H, Soriano V, Khabbaz R F, Kaplan J E, Lal R B, Heneine W. Phylogenetic relationship and geographic distribution of multiple human T-cell lymphotropic virus type II subtypes. J Virol. 1995;69:621–632. doi: 10.1128/jvi.69.2.621-632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamiya S, Matsuoka M, Etoh K, Watanabe T, Kamihira S, Yamaguchi K, Takasuki K. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood. 1996;88:3065–3073. [PubMed] [Google Scholar]
- 50.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tremblay M, Sullivan A K, Rooke R, Geleziunas R, Tsoukas C, Shematek G, Gilmore N, Wainberg M A. New CD4(+) cell line susceptible to infection by HIV-1. J Med Virol. 1989;28:243–249. doi: 10.1002/jmv.1890280408. [DOI] [PubMed] [Google Scholar]
- 52.Tsujimoto H, Terunchi T, Imamura J, Shimotohno K, Miyoshi I, Miwa M. Nucleotide sequence analysis of a provirus derived from HTLV-I-associated myelopathy (HAM) Mol Biol Med. 1988;5:29–42. [PubMed] [Google Scholar]
- 53.Varela-Echavarria A, Garvey N, Preston B D, Dougherty J D. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992;267:24681–24688. [PubMed] [Google Scholar]
- 54.Wattel E, Vartanian J P, Pannetier C, Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]