Abstract
How antiretroviral drug resistance influences human immunodeficiency virus type 1 (HIV-1) evolution is not clear. This study tested the hypothesis that antiretroviral drugs such as 3′-azido-3′-deoxythymidine (AZT) can influence the in vivo mutation rate of HIV-1. It was observed that AZT can increase the rate of HIV-1 mutation by a factor of 7 in a single round of replication. In addition, (−)2′,3′-dideoxy-3′-thiacytidine (3TC) was also found to increase the mutation rate of HIV-1 by a factor of 3. It was also found that HIV-1 drug-resistant reverse transcriptase (RT) variants can influence the in vivo mutation rate. Replication of HIV-1 with AZT-resistant RTs increased the mutation rate by as much as a factor of 3, while replication of HIV-1 with a 3TC-resistant RT (M184V) had no significant effect on the mutation rate. It was observed that only high-level, AZT-resistant RT variants could influence the in vivo mutation rate (i.e., M41L/T215Y and M41L/D67N/K70R/T215Y). In total, these observations indicate that both antiretroviral drugs and drug resistance mutations can influence the in vivo mutation rate of HIV-1.
The emergence of drug-resistant variants of human immunodeficiency virus type 1 (HIV-1) has frustrated efforts to effectively block virus replication at a clinically meaningful level (4, 16). These variants are not only resistant to antiretroviral drugs such as 3′-azido-3′-deoxythymidine (AZT) and (−)2′,3′-dideoxy-3′-thiacytidine (3TC) but can also have altered tropism and virulence properties (29). Reverse transcriptase (RT) is thought to play a role in the generation of retrovirus diversity (38).
Antiretroviral drugs have been previously shown to influence the in vivo mutation rate of spleen necrosis virus (SNV) and murine leukemia virus (MLV). First, 5-azacytidine, which is a nucleoside analog that is incorporated into RNA and inhibits protein synthesis, was found to increase the in vivo SNV mutation rate by a factor of 13 (30). Second, AZT was found to increase the SNV mutant frequency by a factor of 10, while it increased the MLV mutant frequency by a factor of 3 (10). It has been shown that deoxynucleoside triphosphate (dNTP) pool imbalances can influence the SNV and MLV mutation rates but that AZT influences the SNV and MLV rates by a mechanism not involving alterations in dNTP pools (11).
A tractable genetic system has been developed to measure the forward mutation rate of HIV-1 with a vector containing the lacZα peptide gene as a reporter for mutations (21, 23, 24). This system allows for the study of mutations that occur during a single round of HIV-1 replication. The mutation rate of HIV-1 in this system was determined to be 3 × 10−5 mutations per target base pair per cycle in HeLa cells (24) and 4 × 10−5 mutations per target base pair per cycle in a T-lymphoid cell line (20).
Three issues were addressed in the present study. First, we tested the hypothesis that the antiretroviral drugs AZT and 3TC could influence the HIV-1 mutation rate. Second, we tested whether specific mutations in HIV-1 RT that conferred resistance to either AZT or 3TC could influence the rate of HIV-1 mutation. Third, we tested whether there was a correlation between increased AZT drug resistance and the HIV-1 mutation rate. We found that replication of HIV-1 in the presence of either AZT or 3TC can increase the HIV-1 mutation rate. In addition, we found that two AZT-resistant variants, but not a 3TC variant, increased the mutation rate. Finally, we observed that high-level AZT resistance in HIV-1 correlated with significant increases in the in vivo mutation rate.
MATERIALS AND METHODS
Retroviral vectors and expression plasmids.
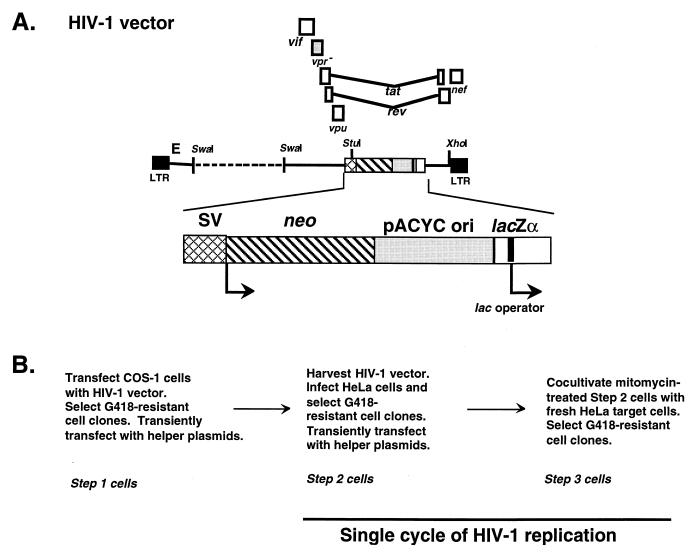
The HIV shuttle vector used in these studies is shown in Fig. 1A and has been described previously (21, 23). Included in the vector is a cassette containing the simian virus 40 (SV40) promoter driving expression of the neomycin phosphotransferase gene (neo), an origin of replication from pACYC 184, and the lacZα peptide gene.
FIG. 1.
(A) HIV-1 vector used in the in vivo mutation rate studies. The vector is shown in the proviral DNA form and has been described previously (21, 23). (B) Protocol for one cycle of HIV-1 vector virus replication. The steps, going from a parental shuttle vector provirus in the step 2 cell to a vector provirus in the step 3 cell, constitute a single cycle of replication.
The HIV-1 gag-pol expression plasmid used was pSVgagpol-rre-r (35), which was a gift from David Rekosh, University of Virginia. This expression plasmid contains the SV40 promoter driving expression of the HIV-1 gag-pol genes. The amphotropic MLV env expression plasmid used, pSV-A-MLV-env (18), was provided by Dan Littman, New York University. The vector used for expression of wild-type Vpr, pCMV-Vpr, has been described previously (21).
The RT variants analyzed in these experiments were constructed by introducing mutations coding for RT amino acid substitutions into pSVgagpol-rre-r by a primary/combinatorial two-step PCR protocol (9, 21).
Transfections, infections, and cocultivations.
The HeLa and COS-1 cell lines used were obtained from the American Type Culture Collection (Manassas, Va.) and were maintained in Dulbecco's modified Eagle's medium containing 10% calf serum or 10% fetal bovine serum, respectively. HIV-1 vectors and expression plasmids were transfected into HeLa cells by use of dimethyl sulfoxide (DMSO)–Polybrene (12) or with Superfect (Qiagen). HeLa cells were infected in the presence of Polybrene (7). Infection of HeLa target cells was also done by cocultivation of virus-producing cells with target cells (22, 25). Briefly, virus-producing cells (typically 2.5 × 105 cells in a 60-mm petri dish) were treated with mitomycin C (10 μg/ml), an inhibitor of host cell DNA synthesis, for 2 h at 37°C. The cells were then washed three times with fresh medium, and 2.5 × 105 HeLa target cells were added. Two days after cocultivation, selective medium containing G418 was added. Control experiments were done with each cocultivation experiment to ensure that mitomycin C-treated, virus-producing cells did not proliferate and no longer adhered to the surfaces of culture dishes.
The influence of the antiretroviral drugs AZT and 3TC on the HIV-1 mutation rate was determined by either pretreatment, posttreatment, or both pre- and posttreatment of cells with drug. Pretreatment refers to maintaining the virus-producing cells in medium supplemented with either AZT or 3TC for 24 h before cocultivation. AZT or 3TC pretreatment could influence the HIV-1 mutation rate by altering the accuracy of the transcription of the provirus into RNA by cellular RNA polymerase II. Posttreatment refers to maintaining HeLa target cells in medium supplemented with various concentrations of AZT or 3TC for 2 h before cocultivation and continuing until 24 h after cocultivation. Posttreatment with AZT or 3TC may influence the HIV-1 mutation rate only during reverse transcription.
Experimental protocol for a single cycle of HIV-1 replication.
The experimental protocol developed to obtain a single cycle of HIV-1 shuttle vector replication is shown in Fig. 1. The protocol contains three steps. In step 1, the HIV-1 shuttle vector was introduced into COS-1 cells by transfection and placed under G418 selection. Cell clones were then transiently transfected with the helper plasmids pSVgagpol-rre-r, pSV-A-MLV-env, and pCMV-Vpr. In step 2, vector virus was harvested from step 1 cells at 48 h posttransfection and was used to infect fresh HeLa cells. Step 2 clones were tested by Southern analysis to ensure that only a single vector proviral DNA was present. The lacZα peptide gene in the vector proviral DNA of step 2 clones was sequenced to confirm that no mutations had been introduced. G418-resistant cell clones were transiently transfected with the helper plasmids (step 2 cells). The step 2 clones used in these studies have been used previously in mutation rate analyses (21, 23). In step 3, vector virus was transferred to fresh HeLa target cells by cocultivation; cells were then placed under G418 selection (step 3 cells). Cocultivation was used to produce step 3 cells because it was desirable to obtain the largest number of step 3 cells for analysis of mutant frequency.
Proviral DNA recovery.
Purified genomic DNA (34) from pools of step 3 clones was digested with the restriction enzymes StuI and XhoI to release the neo, pACYC origin of replication, and lacZα peptide gene sequences from the HIV-1 shuttle vector proviral DNA (Fig. 1). Proviral DNA was purified with the Lac repressor protein as previously described (25). The Lac repressor protein was purified from Escherichia coli strain HB101/lac pIQ (kindly supplied by Tom Record, University of Wisconsin, Madison) as previously described (17). The purified proviral DNA was filled in using the Klenow fragment of DNA polymerase and was ligated and used to electroporate competent E. coli XLI Blue cells (Stratagene). Kanamycin-resistant bacterial colonies were selected in the presence of the isopropyl-β-d-thiogalactoside (IPTG) inducer. The ratio of white plus light-blue bacterial colonies to total bacterial colonies observed provided a forward mutation rate for a single retroviral replication cycle. Plasmid DNA was purified (34) and sequenced in the lacZα peptide gene region.
RESULTS
HIV-1 vector replication.
The experimental protocol developed to assay a single cycle of HIV-1 shuttle vector replication is shown in Fig. 1. The step 1 cells and the cell clones transfected with helper plasmids to make step 2 cells have been described previously (21). The HIV-1 shuttle vector used was a previously used HIV-1 vector (Fig. 1), which contains a deletion in the gag-pol and env genes with an insertion, in the env gene, of a cassette containing the neo gene, the pACYC origin of replication, and the lacZα peptide gene. In addition, a mutation which prevents expression of Vpr was introduced (21). This vector can replicate in mammalian cells as a virus and can be selected with the neomycin analog G418. The vector can replicate in E. coli as a plasmid and is selected by using the drug kanamycin. To be packaged into a virus particle, the vector is complemented in trans by transient transfection of cells with an HIV-1 gag-pol expression plasmid, the amphotropic MLV env expression plasmid, and the wild-type vpr expression plasmid.
Vector virus produced from either COS-1 or HeLa cells was used to infect fresh HeLa target cells (Fig. 1). Cocultivation was used to produce step 3 cells from step 2 cells because it was desirable to obtain the largest number of step 3 cells for analysis of the mutant frequency. Cocultivation of mitomycin C-treated step 2 cells (typically 2.5 × 105 cells per 60-mm petri dish, 5 × 105 cells per 100-mm petri dish, or 7.5 × 105 cells per 150-mm petri dish) with fresh HeLa target cells led to 8 × 102 to 3 × 103 CFU/2.5 × 105 HeLa target cells. The steps going from a parental shuttle vector provirus in the step 2 cells to a vector provirus in the step 3 cells constitute a single cycle of replication (Fig. 1). These steps include transcription of the proviral DNA by the cellular transcription machinery, packaging of the viral RNA, release of viral particles, infection of target cells, reverse transcription, and integration of newly synthesized viral DNA to generate a vector provirus.
The influence of AZT or 3TC on the rate of HIV-1 mutation was determined by maintaining the HeLa target cells in medium supplemented with various concentrations of AZT or 3TC. The target cells were treated for 2 h before infection as well as 24 h after infection. The AZT or 3TC treatments were initiated 2 h before infection to ensure that dNTP pools were altered by the drugs at the time of infection, when the process of reverse transcription initiates. The target cells were maintained in medium with the drug for 24 h after infection to ensure that the dNTP pool imbalance was present throughout HIV-1 replication and establishment as a provirus (∼8 h after infection). The choice of drug concentrations was based both on physiological relevance and on the experimental requirement to avoid greatly diminishing the level of infection of target cells.
At increased concentrations of AZT and 3TC, it was found that virus transfer to target cells was reduced to 20 and 9% of that in the absence of drug treatment, respectively. Mutant frequencies increased in a concentration-dependent manner for both AZT and 3TC. AZT increased mutant frequencies by a factor of 7, whereas 3TC increased mutant frequencies by a factor of 3. These observations indicated that concentrations of AZT and 3TC that significantly influenced HIV-1 virus transfer also led to statistically significant increases in mutant frequencies (see below).
Trypan blue exclusion was used to assess whether decreased virus transfer was due to killing of target cells by drug treatment. For each drug, 5 × 105 HeLa cells were plated on three 100-mm dishes per group treated. After 24 h, the cell culture medium was replaced with a drug-containing medium for another 24 h. Numbers of viable cells were then determined. The results indicated that treatment with AZT or 3TC decreased the fraction of viable cells by 10 or 20% relative to that for untreated controls, respectively. This indicates that the cytotoxic effects observed with AZT and 3TC treatment do not account for the reductions in virus transfer.
AZT treatment of cells increases HIV-1 mutant frequencies.
To determine whether treatment of cells with AZT would influence the mutant frequency of HIV-1, the effects of postinfection AZT treatment of infected target cells were determined. In parallel experiments, target cells infected with the HIV-1 vector were grown in the presence of AZT, ranging from 0 to 0.4 μM (Table 1). In the absence of AZT, the average mutant frequency was 0.005 (35 of 6,427) mutation/cycle. Posttreatment of cells with 0.1 μM AZT resulted in a mutant frequency of 0.007 (60 of 8,150) mutation/cycle, which was comparable (chi square = 2; P > 0.1) to the control mutant frequency. In contrast, posttreatment of cells with either 0.2 or 0.4 μM AZT resulted in a mutant frequency of 0.019 (127 of 6,579) or 0.038 (217 of 5,712) mutation/cycle, respectively. Therefore, posttreatment with 0.02 or 0.04 μM AZT significantly increased the mutant frequency to either 3 (chi square = 50; P < 0.001) or 7 (chi square = 151; P < 0.001) times that of the control, respectively. These observations indicate that AZT treatment leads to an increase in the mutant frequency of HIV-1 in a dose-dependent manner. The relative amount of virus transfer to target cells compared to that for controls was reduced to 0.8 with 0.1 μM AZT, and to 0.4 and 0.2 with 0.2 and 0.4 μM AZT, respectively. This indicates that concentrations of AZT that reduced titers to 60 and 80% significantly increased the mutant frequencies to 3 and 7 times that of the control, respectively.
TABLE 1.
Effects of postinfection AZT treatmenta on the mutant frequency in recovered HIV-1 proviruses
| AZT concn (μM) | Relative amt of virus production | No. of independent mutants/total no. of bacterial colonies | Mutant frequencyb (no. of mutations/cycle) | Relative mutation frequency |
|---|---|---|---|---|
| 0.0 | 1 | 8/1,879 | 0.004 | 1 |
| 12/2,207 | 0.005 | |||
| 15/2,341 | 0.006 | |||
| 0.1 | 0.8 | 14/2,452 | 0.006 | 1.4 |
| 24/2,978 | 0.008 | |||
| 22/2,720 | 0.008 | |||
| 0.2 | 0.4 | 34/1,985 | 0.017 | 3.8 |
| 44/2,341 | 0.019 | |||
| 49/2,253 | 0.022 | |||
| 0.4 | 0.2 | 63/1,726 | 0.037 | 7.6 |
| 75/2,068 | 0.036 | |||
| 79/1,918 | 0.041 |
Referred to as posttreatment. Target cells were treated with the indicated concentration of AZT for 2 h prior to cocultivation, and AZT was continued for 24 h after cocultivation with mitomycin C-treated virus-producing cells. Proviral DNA from target cells was then purified and screened for mutations in E. coli.
Standard deviations are 0.001, 0.002, 0.004, and 0.004 mutant/cycle for mutant frequencies at AZT concentrations of 0, 0.1, 0.2, and 0.4 μM, respectively.
To determine if both pre- and posttreatment with AZT would lead to a significant increase in mutant frequency, both the virus-producing cells and the virus target cells were treated with 0.2 μM AZT (Table 2). The mutant frequency with AZT pretreatment was 0.012 (47 of 3,960) mutant/cycle, while AZT posttreatment led to a mutant frequency of 0.023 (83 of 3,595) mutant/cycle. Pre- plus posttreatment with 0.2 μM AZT led to a mutant frequency of 0.029 (110/3,788) mutant/cycle, which is not significantly different from that with AZT posttreatment alone (chi square = 2.4; P > 0.1). These data indicate that AZT pretreatment does not significantly increase the mutant frequency and that the majority of the mutations identified occurred during reverse transcription. The significant increase in mutant frequency observed with AZT posttreatment was interpreted as due to increases in the error rate of reverse transcription.
TABLE 2.
Effects of AZT pretreatment, posttreatment, and combined pre- and posttreatment on the mutant frequency in recovered HIV-1 proviruses
| Treatmenta | Relative amt of virus production | No. of independent mutants/total no. of bacterial colonies | Mutant frequencyb (no. of mutations/cycle) | Relative mutation frequency |
|---|---|---|---|---|
| No treatment | 1.0 | 17/2,509 | 0.007 | 1 |
| 10/2,371 | 0.004 | |||
| Pretreatment | 0.7 | 12/1,137 | 0.011 | 2 |
| 14/1,080 | 0.013 | |||
| 21/1,743 | 0.012 | |||
| Posttreatment | 0.5 | 37/1,589 | 0.023 | 3.8 |
| 21/1,037 | 0.020 | |||
| 25/969 | 0.026 | |||
| Pre- and posttreatment | 0.3 | 37/1,362 | 0.027 | 4.8 |
| 39/1,239 | 0.031 | |||
| 34/1,187 | 0.029 |
Pretreatment, cells producing the parental HIV-1 shuttle vector were treated with AZT (0.2 μM) for 24 h prior to cocultivation with target cells. Posttreatment, target cells were treated with AZT (0.2 μM) for 2 h prior to cocultivation, and AZT was continued for 24 h after cocultivation with mitomycin C-treated virus-producing cells. Proviral DNA from target cells was then purified after AZT treatments and screened for mutations in E. coli.
Standard deviations are 0.001, 0.004, and 0.003 mutant/cycle for mutant frequencies with AZT (0.2 μM) pretreatment, posttreatment, and combined pre- and posttreatment, respectively.
3TC treatment of cells increases the mutation frequency of HIV-1 in a dose-dependent manner.
To determine whether treatment of cells with 3TC would increase the mutant frequency of HIV-1, the effects of postinfection 3TC treatment of infected target cells were determined. In parallel experiments, target cells infected with the HIV-1 vector were grown in the presence of 3TC, ranging from 0 to 0.6 μM (Table 3). In the absence of 3TC, the average mutant frequency was 0.006 (37 of 6,551) mutation/cycle. Posttreatment of cells with 0.1 μM 3TC resulted in a mutant frequency of 0.007 (46 of 7,013) mutation/cycle, which was comparable (chi square = 0.46; P > 0.1) to the control mutant frequency. However, posttreatment of cells with either 0.3 or 0.6 μM 3TC resulted in a mutant frequency of 0.014 (96 of 6,746) or 0.017 (81 of 4,909) mutant/cycle, respectively. Therefore, posttreatment with 0.3 or 0.6 μM 3TC increased the mutant frequency to either 2 (chi square = 24; P < 0.001) or 3 (chi square = 32; P < 0.001) times that of the control. These observations indicate that 3TC treatment can increase the mutant frequency of HIV-1 in a dose-dependent manner. The relative amount of virus transfer to target cells compared to that for controls was reduced to 0.9 with 0.1 μM 3TC, and to 0.5 and 0.09 with 0.3 and 0.6 μM 3TC, respectively. This indicates that concentrations of 3TC that reduced titers to 50 and 91% significantly increased the mutant frequencies to 2 and 3 times that of the control, respectively.
TABLE 3.
Effects of postinfection 3TC treatmenta on the mutant frequency in recovered HIV-1 proviruses
| 3TC concn (μM) | Relative amt of virus production | No. of independent mutants/total no. of bacterial colonies | Mutant frequencyb (no. of mutations/cycle) | Relative mutation frequency |
|---|---|---|---|---|
| 0.0 | 1 | 13/2,267 | 0.006 | 1 |
| 6/1,570 | 0.004 | |||
| 18/2,714 | 0.007 | |||
| 0.1 | 0.9 | 11/2,043 | 0.005 | 1.2 |
| 19/2,592 | 0.007 | |||
| 16/2,378 | 0.007 | |||
| 0.3 | 0.5 | 26/1,956 | 0.013 | 2.3 |
| 33/2,102 | 0.016 | |||
| 37/2,688 | 0.014 | |||
| 0.6 | 0.09 | 38/2,111 | 0.018 | 2.8 |
| 27/1,778 | 0.015 | |||
| 16/1,020 | 0.016 |
Referred to as posttreatment. Target cells were treated with the indicated concentration of 3TC for 2 h prior to cocultivation, and 3TC was continued for 24 h after cocultivation with mitomycin C-treated virus-producing cells. Proviral DNA from target cells was then purified and screened for mutations in E. coli.
Standard deviations are 0.002, 0.001, 0.002, and 0.003 mutant/cycle for mutant frequencies at 3TC concentrations of 0, 0.1, 0.3, and 0.6 μM, respectively.
To determine if both pre- and posttreatment with 3TC would lead to a significant increase in mutant frequency, both the virus-producing cells and the virus target cells were treated with 0.3 μM 3TC (Table 4). The mutant frequency with 3TC pretreatment was 0.007 (39 of 5,379) mutant/cycle, while 3TC posttreatment led to a mutant frequency of 0.013 (69 of 5,450) mutant/cycle. Pre- plus posttreatment with 0.3 μM 3TC led to a mutant frequency of 0.017 (81/4,907) mutant/cycle, which is not significantly different from that with 3TC posttreatment alone (chi square = 2.6; P > 0.1). As was observed with AZT pretreatment, these data indicate that 3TC pretreatment does not significantly increase the mutant frequency and that the majority of the mutations identified occurred during reverse transcription. The increase in mutant frequency observed with 3TC posttreatment was interpreted to be due to an increase in the error rate of reverse transcription.
TABLE 4.
Effects of 3TC pretreatment, posttreatment, and combined pre- and posttreatment on the mutation frequency in recovered HIV-1 proviruses
| Treatmenta | Relative amt of virus production | No. of independent mutants/total no. of bacterial colonies | Mutant frequencyb (no. of mutations/cycle) | Relative mutation frequency |
|---|---|---|---|---|
| No treatment | 1 | 7/1,064 | 0.007 | 1 |
| 14/2,830 | 0.005 | |||
| Pretreatment | 0.8 | 19/2,736 | 0.007 | 1.4 |
| 8/1,053 | 0.008 | |||
| 12/1,590 | 0.008 | |||
| Posttreatment | 0.5 | 32/2,342 | 0.014 | 2.6 |
| 23/1,720 | 0.013 | |||
| 14/1,388 | 0.010 | |||
| Pre- and posttreatment | 0.3 | 24/1553 | 0.015 | 3.4 |
| 33/2,078 | 0.016 | |||
| 24/1,276 | 0.019 |
Pretreatment, cells producing the parental HIV-1 shuttle vector were treated with 3TC (0.3 μM) for 24 h prior to cocultivation with target cells. Posttreatment, target cells were treated with 3TC (0.3 μM) for 2 h prior to cocultivation, and 3TC was continued for 24 h after cocultivation with mitomycin C-treated virus-producing cells. Proviral DNA from target cells was then purified after 3TC treatments and screened for mutations in E. coli.
Standard deviations are 0.001, 0.003, and 0.004 mutant/cycle for mutant frequencies with 3TC (0.3 μM) pretreatment, posttreatment, and combined pre- and posttreatment, respectively.
Sequence analysis of HIV-1 vector mutants following AZT or 3TC pretreatment, posttreatment, or pre- and posttreatment.
To determine whether AZT or 3TC treatment of cells led to significant changes in the spectrum of mutations in HIV-1 from that for HIV-1 replicating in the absence of drug, the lacZα peptide gene region was sequenced in a sampling of mutant clones. Table 5 shows that following either AZT pretreatment, posttreatment, or combined pre- and posttreatment, the general spectrum of mutations observed was base pair substitution mutations, frameshift mutations, deletion mutations, and mutants containing multiple mutations (hypermutants). The predominant types of base pair substitution mutations observed were G-to-A and C-to-T transition mutations. In addition, frameshift mutations were observed as primarily −1 frameshifts in runs of T's and in A's. Deletion mutations included simple deletion mutations and two deletions with an insertion of a nucleotide sequence of unknown origin. The hypermutants characterized were predominantly G-to-A and C-to-T hypermutants (Table 5). Interestingly, one hypermutant was identified with two frameshift mutations in runs of A's.
TABLE 5.
Sampling of characterized mutations in HIV-1 proviruses following treatment with AZT
| Nucleotide change(s) | No. of independent mutants recovereda with the indicated treatment
|
|||
|---|---|---|---|---|
| Pretreatment | Posttreatment | Pre- and posttreatment | Control | |
| G to A | 8 | 9 | 11 | 7 |
| C to T | 5 | 6 | 6 | 2 |
| T to C | 1 | 2 | 1 | |
| T to G | 2 | 1 | 1 | |
| G to A hypermutants | 2 | 3 | 1 | 1 |
| C to T hypermutants | 1 | |||
| TTTT to TTT | 3 | 2 | 3 | 1 |
| TTTT to TTTTT | 1 | 1 | 1 | |
| AAAA to AAA | 3 | 3 | 1 | 2 |
| AAAA to AAAAA | 1 | 1 | 1 | |
| CCC to CC | 1 | 1 | 1 | 1 |
| CCCCC to CCCC | 1 | 1 | ||
| AAA to AA, AAAA to AAA | 1 | |||
| Δ37 | 1 | |||
| Δ41 | 1 | |||
| Δ39 | 1 | |||
| Δ23, +6 | 1 | |||
| Δ15 | 1 | |||
| Δ7 | 1 | |||
| Δ16, +23 | 1 | |||
| Δ19 | 1 | |||
| Δ33 | 1 | |||
A general comparison of the characterized mutants from AZT pretreatment, posttreatment, or combined pre- and posttreatment with mutants of the HIV-1 vector in the absence of AZT revealed that the spectrum and general types of mutations characterized were similar (Table 5). In addition, the general locations of mutations within the lacZα peptide gene region were similar to those identified in the absence of the drug (data not shown). This indicates that the increase in the overall rate of mutations observed with AZT posttreatment and with AZT pre- and posttreatment was due to a general increase in substitution, frameshift, and deletion mutations. These results suggest that AZT increased the propensity to mutations, but not the specific types of mutations or locations within the lacZα peptide gene region.
Analysis of the spectrum of mutations identified from a sampling of mutants following 3TC pretreatment, posttreatment, or combined pre- and posttreatment revealed base pair substitutions, frameshifts, and deletion mutations (Table 6). The general trends indicated that the mutations identified for 3TC treatment were similar in type and rate to those observed when the HIV-1 vector was replicated in the absence of 3TC. The characterized substitution mutations were predominantly G-to-A and C-to-T mutations, while the frameshift mutations were mostly −1 frameshifts. Deletion mutations were observed and included both simple deletions and deletions with an insertion. Several G-to-A hypermutants were identified (Table 6). The general location of mutations after 3TC treatment was similar to that seen in the absence of drug (data not shown). This suggests that 3TC increased the frequency of mutations but not the mutation type or location in the lacZα peptide gene region.
TABLE 6.
Sampling of characterized mutations in HIV-1 proviruses following treatment with 3TC
| Nucleotide change(s) | No. of independent mutants recovereda with the indicated treatment
|
|||
|---|---|---|---|---|
| Pretreatment | Posttreatment | Pre- and posttreatment | Control | |
| G to A | 10 | 8 | 11 | 6 |
| C to T | 6 | 3 | 4 | 3 |
| T to C | 1 | 1 | 1 | |
| T to G | 1 | 2 | 1 | 1 |
| T to A | 1 | |||
| G-to-A hypermutants | 3 | 2 | 2 | 2 |
| C-to-T hypermutants | 1 | |||
| TTT to TT | 1 | 1 | ||
| TTTT to TTT | 2 | 3 | 1 | 1 |
| TTTT to TTTTT | 1 | 1 | 2 | |
| AAAA to AAA | 1 | 3 | 1 | 2 |
| AAAA to AAAAA | 1 | 1 | ||
| CCC to CC | 1 | 2 | 2 | 1 |
| CCCCC to CCCC | 1 | 1 | 1 | |
| Δ52 | 1 | |||
| Δ23 | 1 | |||
| Δ7 | 1 | |||
| Δ15 | 1 | |||
| Δ12, +7 | 1 | |||
| Δ24 | 1 | |||
| Δ43 | 1 | |||
| Δ8 | 1 | |||
| Δ19 | 1 | |||
A total of 30 mutants were characterized for 3TC pretreatment, posttreatment, and pre- and posttreatment. Ten mutants from each pool (see Table 4) were randomly selected for sequencing analysis, except for the 3TC pretreatment, where 11 mutants were randomly sequenced from two pools and 8 mutants were sequenced from the third pool. Twenty mutants total (7 from 1 pool, 13 from the other) were characterized from the control (see Table 4).
An AZT-resistant HIV-1 RT variant can increase mutant frequencies and the in vivo HIV-1 mutation rate.
A series of HIV-1 RT variants that confer AZT resistance was tested for their ability to influence the HIV-1 mutant frequency. The RT variants selected for analysis correspond to AZT-resistant RT variants that appear during the course of AZT therapy. Specifically, three different variants, corresponding to RT variants with increasing levels of AZT resistance, were tested: T215Y, M41L/T215Y, and M41L/D67N/K70R/T215Y. The HIV-1 vector was replicated in one round of replication with each RT variant, and then proviral DNA from pools of infected target cells was purified and introduced into E. coli in order to determine mutant frequencies. The mutant frequency of HIV-1 replicated with the T215Y RT variant led to an average mutant frequency of 0.005 (35 of 7,087) mutant/cycle (Table 7). The mutant frequency of HIV-1 replicated with T215Y was not significantly different (chi square = 0.33; P > 0.5) from that of HIV-1 replicated with wild-type RT, 0.004 (33 of 7,690) mutant/cycle (Table 7). Replication of HIV-1 with the M41L/T215Y RT variant resulted in an average mutant frequency of 0.013 (63 of 5,005) mutant/cycle, which is 2 times higher (chi square = 27; P < 0.001) than that for virus replication with wild-type RT. When HIV-1 was replicated with the M41L/D67N/K70R/T215Y variant, the resulting average mutant frequency was 0.017 (84 of 4,943) mutant/cycle, which is 3 times higher (chi square = 52; P < 0.001) than the mutant frequency of HIV-1 replicated with wild-type HIV-1 RT (Table 7). HIV-1 was also replicated with M184V RT, which confers resistance to 3TC. The average mutant frequency was found to be 0.003 (14 of 4,846) mutant/cycle. This frequency is not significantly different (chi square = 1.6; P > 0.1) from that obtained when HIV-1 was replicated with wild-type RT (Table 7).
TABLE 7.
Mutant frequencies in recovered HIV-1 proviruses replicated by AZT- or 3TC-resistant HIV-1 RT variants
| RT | Relative amt of virus production | No. of independent mutants/total no. of bacterial colonies | Mutant frequencya (no. of mutations/cycle | Relative mutant frequency |
|---|---|---|---|---|
| Wild type | 1 | 7/2,029 | 0.003 | 1 |
| 10/1,726 | 0.006 | |||
| 7/2,043 | 0.003 | |||
| 9/1,892 | 0.005 | |||
| T215Y | 0.9 | 12/2,385 | 0.005 | 1.3 |
| 10/1,979 | 0.005 | |||
| 13/2,723 | 0.004 | |||
| M41L/T215Y | 0.6 | 27/2,417 | 0.011 | 3.3 |
| 19/1,150 | 0.017 | |||
| 17/1,438 | 0.012 | |||
| M41L/D67N/K70R/ T215Y | 0.4 | 23/1,522 | 0.015 | 4.3 |
| 44/2,378 | 0.019 | |||
| 17/1,043 | 0.016 | |||
| M184V | 1 | 7/1,904 | 0.004 | 0.8 |
| 3/1,709 | 0.002 | |||
| 4/1,233 | 0.003 |
Standard deviations are 0.003, 0.001, 0.005, 0.004, and 0.001 mutant/cycle for mutant frequencies with wild-type, T215Y, M41L/T215Y, M41L/D67N/K70R/T215Y, and M184V RT.
Sequence analysis from a sampling of mutants was done to characterize the types of mutations that had occurred during replication. The spectrum of mutations observed subsequent to the replication of HIV-1 containing the T215Y alteration was found to be similar to that seen when HIV-1 was replicated with wild-type RT (Table 8). The mutations characterized from proviruses that had been replicated with either the M41L/T215Y or the M41L/D67N/K70R/T215Y alteration in RT also revealed trends in mutations that were comparable to that identified by replication with T215Y or with wild-type RT. The locations of the mutations identified when HIV-1 was replicated in the presence of AZT were similar to those obtained when HIV-1 was replicated in the absence of the drug (data not shown). This indicates that the AZT resistance mutations did not alter the type or location of mutations but increased their frequency.
TABLE 8.
Sampling of characterized mutations in HIV-1 proviruses replicated by AZT- or 3TC-resistant HIV-1 RT
| Nucleotide change(s) | No. of independent mutants recovereda by use of the indicated RT
|
||||
|---|---|---|---|---|---|
| T215Y | M41L/ T215Y | M41L/D67N/ K70R/T215Y | M184V | Control | |
| G to A | 11 | 11 | 9 | 3 | 8 |
| C to T | 5 | 6 | 5 | 2 | 7 |
| T to C | 4 | 2 | 3 | 1 | 4 |
| T to G | 1 | 1 | |||
| G to T | 2 | ||||
| G-to-A hypermutants | 1 | 2 | 2 | 1 | 3 |
| TTTT to TTT | 1 | 2 | 2 | 2 | 1 |
| TTTT to TTTTT | 1 | 1 | 1 | ||
| AAAA to AAA | 2 | 2 | 1 | 2 | 1 |
| AAAA to AAAAA | 1 | 1 | 1 | ||
| CCC to CC | 1 | 1 | 1 | ||
| CCCCC to CCCC | 1 | ||||
| TTTT to TTT, AAAA to AAA | 1 | ||||
| Δ34 | 1 | ||||
| Δ31 | 1 | ||||
| Δ11, +32 | 1 | ||||
| Δ6, +14 | 1 | ||||
| Δ37 | 1 | ||||
| Δ33 | 1 | ||||
| Δ14 | 1 | ||||
| Δ27, +19 | 1 | ||||
| Δ43, +11 | 1 | ||||
| Δ8 | 1 | ||||
| Δ41, +25 | 1 | ||||
Thirty mutants total (10 mutants per pool [see Table 7]) were randomly selected and characterized by sequencing for HIV-1 replicated with the following RT variants: T215Y, M41L/T215Y, and M41L/D67N/K70R/T215Y. All 14 mutants isolated following replication of HIV-1 with the M184V RT variant were sequenced. Twenty-eight mutants total (7 mutants per pool [see Table 7]) were characterized from the control.
Replication of HIV-1 with M184V RT resulted in a pattern of mutations that suggests a trend different from that for wild-type RT. Fifty percent of the mutations (7 of 14) characterized from vectors replicated with M184V RT were substitution mutations, while 50% (7 of 14) were frameshift mutations (Table 8). No deletion mutations were observed in vectors replicated with M184V RT. For the control, 88% (24 of 28) of the characterized mutants had substitutions, 7% (2 of 28) had frameshifts, and 7% (2 of 28) had deletion mutations.
DISCUSSION
AZT can increase the HIV-1 in vivo mutation rate.
The data presented here indicate that the antiretroviral drug AZT can increase the rate of HIV-1 mutation. Mutant frequencies were determined in a single cycle of replication with an HIV-1 vector containing the lacZα peptide gene as a mutational target. Postinfection treatment of target cells revealed that the mutant frequency increased by a factor of 3 or 7 with an AZT concentration of 0.2 or 0.4 μM, respectively. AZT pretreatment led to a mutant frequency that was not significantly different from that of HIV-1 replicated in the absence of the drug, and AZT pre- and posttreatment led to a mutant frequency that was not significantly different from the mutant frequency with AZT posttreatment alone. These data indicate that AZT treatment significantly influences reverse transcription.
Potential mechanisms for the AZT-mediated increase in the HIV-1 mutation rate.
Sequencing analysis of a sampling of HIV-1 vector mutants recovered after AZT treatment indicated that the general location and spectrum of substitution, frameshift, and deletion mutations observed with AZT pretreatment, posttreatment, and combined pre- and posttreatment were comparable to the spectrum and location of mutants observed in the absence of drug. This indicates that the mechanisms by which mutations occurred were similar but that the rate had increased.
The HIV-1 vector used in these studies cannot readily be used to determine whether mutations occurred during minus-strand or plus-strand DNA synthesis, but the locations of the mutations suggest particular mechanisms for their creation. The majority of the G-to-A transitions characterized, including those from the G-to-A hypermutants, were in GpA dinucleotides. This has been previously observed with HIV-1 hypermutants (6, 39). Transition mutations adjacent to runs of a single nucleotide have been suggested to occur by the mechanism of dislocation mutagenesis (1, 14). According to this model, dislocation of the primer with respect to the template produces an unpaired nucleotide base; realignment occurs between the primer and the template, resulting in a mismatch, followed by elongation beyond the mismatch. Most of the G-to-A transition mutations occurred at sites adjacent to a run of nucleotides, suggesting that these mutations may have resulted from dislocation mutagenesis.
The frameshift mutations characterized were mainly −1 frameshifts in runs of T's and A's. These general trends are in agreement with those found for frameshift mutations made by purified HIV-1 RT (1). Plus-one frameshift mutations in runs of T's and A's occurred with SNV in vivo (3, 32). The frameshift mutations in homo-oligomeric runs suggest that these result from template primer slippage (2, 13, 36, 37). The +1 frameshift mutations may have occurred during minus-strand DNA synthesis (3), while the −1 frameshift mutations could have occurred during either minus- or plus-strand DNA synthesis. Simple deletion mutants and deletion-with-insertion mutants have been previously identified, and the mechanisms by which they could have occurred have been proposed (31, 33).
Increases in the SNV and MLV mutation rate have been observed previously when these viruses were replicated in the presence of AZT (11). Several potential mechanisms have been proposed for this effect on the mutation rate. One proposed mechanism is direct interaction of AZT with RT in a noncatalytic manner, which could induce a conformational change that alters enzyme fidelity. Although the influence of AZT on the SNV and MLV mutation rates has been shown to be due to a mechanism not involving alterations in dNTP pools (11), it is possible that this is not the case with HIV-1 (15, 26, 28, 40).
3TC increases the in vivo mutation rate of HIV-1.
It was found that another antiretroviral drug, 3TC, could also increase the HIV-1 mutation rate. The mutant frequency was observed to increase by postinfection treatment of target cells with 0.3 and 0.6 μM 3TC by factors of 2 and 3, respectively. Pretreatment of virus-producing cells with 0.3 μM 3TC did not significantly influence the mutant frequency, and pre- and posttreatment together (a factor-of-2 increase) did not significantly increase the mutant frequency over that of just posttreatment alone (a factor-of-3 increase). This indicates, as was observed with AZT, that 3TC influences the reverse transcription step of the HIV-1 life cycle. Sequencing analysis of a sampling of mutants recovered after 3TC treatment indicated that the spectrum and location of mutations observed were similar to those seen in the absence of the drug.
Increased AZT drug resistance correlates with an increase in the HIV-1 in vivo mutation rate.
The mutant frequencies for several AZT-resistant RTs (i.e., T215Y, M41L/T215Y, and M41L/D67N/K70R/T215Y) were characterized in order to determine if increasing levels of AZT drug resistance were associated with higher mutation rates. T215Y did not influence the mutant frequency compared to that of wild-type RT, but M41L/T215Y and M41L/D67N/K70R/T215Y resulted in mutant frequencies that were increased by factors of 2 and 3, respectively. This indicates that there is a correlation between increased drug resistance and the ability to influence mutant frequency. However, this relationship is not linear, as the drug resistance levels of T215Y, M41L/T215Y, and M41L/D67N/K70R/T215Y RT are about 16, 70, times, and 180 times that of wild-type RT, respectively (19). Sequencing analysis of a sampling of mutants indicated that the observed mutations were similar in type and location to those observed with wild-type RT, indicating that the AZT-resistant RTs made errors more frequently, but by the same mechanisms as wild-type RT.
A mechanism of AZT resistance has been recently reported in a cell-free reaction (27). An AZT-resistant RT (containing the D67N, K70R, T215F, and K219Q amino acid substitutions) was found to have an increased ability compared with wild-type RT to extend the primer past several potential termination sites in the presence of AZTTP when ATP was added to the cell-free reaction mixture. It was also observed that transfer of the AZTMP residue from the primer terminus to ATP to form dinucleoside polyphosphate and unblocked primer was enhanced in the mutant enzyme. The inhibition of this activity by the next complementary dNTP was found to be reduced compared to that for the wild type. The crystal structure of a covalently trapped catalytic complex containing HIV-1 RT, chain-terminated primer and template, and dTTP has been determined (8). The majority of the AZT resistance mutations are in the neighborhood of the incoming nucleotide, which appears to provide structural support for this mechanism of AZT resistance.
A 3TC-resistant RT variant does not influence the mutation rate.
Replication of the HIV-1 vector with an M184V RT variant did not lead to a significant change in the mutant frequency. This indicates that the threefold increase in fidelity by M184V in nucleotide insertion assays (41) does not have a phenotype that can be distinguished from wild-type RT in the in vivo mutation rate assay. It has previously been shown that M184V does not affect the overall cell-free error rate of HIV-1 RT by use of the lacZα peptide gene as a mutational target (5). Sequencing analysis of the mutations in recovered mutants indicated a pattern in which there were an approximately equal number of base substitution and frameshift mutations recovered. This trend is different from that for wild-type RT, where there are about twice as many substitution mutations observed as frameshift mutations.
ACKNOWLEDGMENTS
We thank R. Cherian, M. Mauck, S. Uchida, S. Webb, and A. Waggoner for outstanding technical assistance. We also thank M. Williams for comments on the manuscript.
This work was supported by the Public Health Service (GM56615), the American Cancer Society, and the Ohio Cancer Research Associates.
REFERENCES
- 1.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 2.Bebenek K, Kunkel T A. The fidelity of retroviral reverse transcriptases. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 3.Burns D P W, Temin H M. High rates of frameshift mutations within homo-oligomeric runs during a single cycle of retroviral replication. J Virol. 1994;68:4196–4203. doi: 10.1128/jvi.68.7.4196-4203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 5.Drosopoulos W C, Prasad V R. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J Virol. 1998;72:4224–4230. doi: 10.1128/jvi.72.5.4224-4230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgibbon J E, Mazar S, Dubin D T. A new type of G-A hypermutation affecting human immunodeficiency virus. AIDS Res Hum Retrovir. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- 7.Hu W-S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 9.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 10.Julias J G, Kim T, Arnold G, Pathak V K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J Virol. 1997;71:4254–4263. doi: 10.1128/jvi.71.6.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julias J G, Pathak V K. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel T A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T A, Alexander P S. The base substitution fidelity of eukaryotic DNA polymerases—mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986;261:160–166. [PubMed] [Google Scholar]
- 15.Kunz B A, Kohalmi S E. Modulation of mutagenesis by deoxyribonucleotide levels. Annu Rev Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 16.Kuritzkes D R. Clinical significance of drug resistance in HIV-1 infection. AIDS. 1996;10(Suppl. 5):S27–S33. doi: 10.1097/00002030-199612005-00005. [DOI] [PubMed] [Google Scholar]
- 17.Laiken S L, Gross C A, von Hippel P H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972;66:143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- 18.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larder B A. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J Gen Virol. 1994;75:951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 20.Mansky L M. Forward mutation rate of human immunodeficiency virus type 1 in a T-lymphoid cell line. AIDS Res Hum Retrovir. 1996;12:307–314. doi: 10.1089/aid.1996.12.307. [DOI] [PubMed] [Google Scholar]
- 21.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 22.Mansky L M. Retroviral-vector-mediated gene transfer. In: Griffiths J B, Doyle A, Newell D G, editors. Cell and tissue culture: laboratory procedures. 6th ed. New York, N.Y: John Wiley & Sons; 1994. pp. 27B:5.1–27B:5.10. [Google Scholar]
- 23.Mansky L M, Preveral S, Selig L, Benarous R, Benichou S. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansky L M, Temin H M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994;68:494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews C K, Ji J. DNA precursor asymmetries, replication fidelity, and variable genome evolution. Bioessays. 1992;14:295–301. doi: 10.1002/bies.950140502. [DOI] [PubMed] [Google Scholar]
- 27.Meyer P R, Matsuura S E, Mian A M, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 28.Meyerhans A, Vartanian J-P, Hultgren C, Plikat U, Karlsson A, Wang L, Eriksson S, Wain-Hobson S. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak V K, Temin H M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Smith A J, Cho M-L, Hammarskjold M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Streisinger G, Owen J. Mechanism of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vartanian J-P, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G-to-A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vartanian J-P, Meyerhans A, Sala M, Wain-Hobson S. G-A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci USA. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M A, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]