Abstract
Polyelectrolyte microparticles (MPs) synthesized on calcium carbonate cores are considered a promising basis for new drug delivery systems. It is known that microparticles entering a physiological environment absorb proteins on their surface, which can change the properties of the microparticles and alter their functional activity. This study aimed to compare the compositions of the adsorbed protein layer formed on microparticles with the core/shell and shell structures obtained by layer-by-layer deposition. The difference in the microparticle structure was associated with changes in their surface topography and ζ-potential. These microparticles were incubated with human serum or plasma at 37°C for 24 h. The adsorbed proteins were eluted and analyzed by means of SDS-PAGE. The protein composition of the eluates was determined by liquid chromatography–tandem mass spectrometry (LC-MS/MS); a total of 357 proteins were identified, and 183 of them were detected in all samples. Our results demonstrate that the relative abundance of proteins of different functional groups (immunoglobulins, complement proteins, and apolipoproteins) varied depending on the structure and surface characteristics of the polyelectrolyte microparticles and the incubation medium. Our findings expand the understanding of the influence of the physicochemical properties of the microparticles on their interaction with proteins, which can help to improve the design of microparticles for drug delivery.
1. Introduction
Core/shell and shell microparticles (MPs) obtained by layer-by-layer deposition are considered to be versatile multifunctional tools for biomedical applications, such as drug delivery and controlled release, gene therapy, sensing, and imaging.1 In the development of drug delivery systems, microparticles offer several advantages, such as the capacities for retaining loaded drugs, controlled drug release, and encapsulation of low-molecular-weight drugs and biomolecules.2,3 Layer-by-layer deposition of the shells allows controlling the resultant characteristics of the MPs, including their size, charge, surface chemistry, structure, and rigidity.4−6 A wide variety of cores, polymers, and surface groups are used to fabricate MPs finely tuned for specific applications.7 During the fabrication of the MPs, different agents can be introduced into the polyelectrolyte shell, such as fluorescent labels,8 quantum dots,9 and magnetic particles,10,11 and their surface can be functionalized with capture molecules,12,13 which extends the use of the MPs in bioimaging and smart drug delivery.
It is known that the physicochemical and functional properties of engineered particles undergo modifications in biological media. The particles interact with biomolecules in blood, which leads to the formation of an additional protein layer on their surface and modification of their biological identity.7,14 This layer on the MP surface can interfere with some biological processes, e.g., cellular uptake of the particles, immune response, and complement activation.15−17 Variations of the protein corona composition have been shown to noticeably affect the association of particles with immune cells.18 Specific association of polymer-coated silica particles carrying affibodies with cancer cells is significantly decreased after their incubation with human serum.19 However, other studies have demonstrated the retention of the targeting capacity and enhancement of targeting specificity of microcapsules coated with a layer of proteins derived from human serum.20,21
At the same time, precoating of particle-based drug delivery systems with specific proteins is widely used to improve their performance in biological media.22 Polymersomes (polymeric vesicles made from synthetic amphiphilic block copolymers) preincubated with proteins (e.g., human IgG) display reduced cytotoxicity.23 Microplastic particles coated with model proteins before incubation in protein-containing media behave differently in protein–cell interactions.24 Precoating with human serum albumin (HSA) reduced the negative effect of plasma protein adsorption on the adhesion of poly(lactic-co-glycolic acid) microparticles to the endothelium.25
Overall, the understanding of factors affecting the adsorption of proteins from biological media on the surface of MPs paves the way to the rational design of safer and more biologically effective particle-based biomedical tools. The size, surface charge, chemical composition, and functionalization of the MPs have been shown to strongly affect the composition of the protein adsorption layer on different types of particles.5,26−28 The roles of other properties, such as structure and surface morphology, are insufficiently understood; therefore, we studied the adsorption of serum and plasma proteins onto MPs with different structures and surface properties.
Here, we obtained two types of polyelectrolyte MPs based on CaCO3 cores, with core/shell and shell structures and various surface charges and surface topologies. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was used to determine differences in the composition of human serum or plasma proteins adsorbed on the surface of these MPs. The results improve the understanding of the relationship between the MP structure and their interaction with proteins and can be used for the optimization of biomedical applications of MPs.
2. Materials and Methods
2.1. Materials
Calcium chloride dihydrate (CaCl2), sodium carbonate (Na2CO3), ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA), poly(allylamine hydrochloride) (PAH, average Mw ≈ 65,000 Da), poly(sodium 4-styrenesulfonate) (PSS, average Mw ≈ 70,000 Da), poly(acrylic acid) (PAA, average Mw ≈ 100,000 Da), Trizma base, sodium dodecyl sulfate (SDS), acrylamide/bis-acrylamide (30% solution), ammonium persulfate ((NH4)2S2O8), sodium phosphate dibasic (Na2HPO4), sodium phosphate monobasic (NaH2PO4), sodium chloride (NaCl), Brilliant Blue R, Sample buffer Laemmli 2× concentrate, N,N,N′,N′-tetramethylethylenediamine (TEMED), and glycerol were purchased from Sigma-Aldrich. Poly(vinyl alcohol) 18–88 (PVA, Emprove) was obtained from Merck KGaA, Germany. Methylcellulose (Metolose SM-100) was purchased from Shin-Etsu Chemical Co., Japan. Hydrochloric acid (HCl) was obtained from PanReac AppliChem (ITW Reagents). All solutions were prepared using ultrapure Milli-Q water with a resistivity of no less than 18.2 mΩ·cm obtained using a Direct-Q water purification system (Millipore, Merck KGaA, Germany) and were additionally filtered through sterile Millex syringe filter units with a pore size of 0.22 μm (Millipore, Merck KGaA, Germany) before use.
2.2. Synthesis of CaCO3 Cores
Calcium carbonate cores were obtained using the coprecipitation reaction in a mixture of equimolar solutions of sodium carbonate and calcium chloride in an aqueous medium containing 1% poly(vinyl alcohol) and 0.05% methylcellulose (PVA/MC) as a thickening agent.29 First, 15 mL of a PVA/MC solution was added to 7.5 mL of 0.33 M CaCl2 and mixed using an UltraTurrax T 25 digital dispersing device (IKA-Werke GmbH & Co. KG, Germany) at 5000 rpm until a homogeneous medium was obtained. After that, 7.5 mL of Na2CO3 was added, and the final mixture was vigorously agitated at 15,000 rpm for 30 s. The resulting suspension was transferred to a 50 mL test tube, Milli-Q water was added to a volume of 50 mL, and the suspension was centrifuged. Then, a fresh portion of Milli-Q water was added, and the suspension was centrifuged at 4000 rpm for 5 min. This step was repeated two times. The resulting pellet of MPs was transferred to a Petri dish and dried in a drying oven at 90 °C overnight.
2.3. Preparation of Core/Shell and Shell Polyelectrolyte Microparticles
The synthesized calcium carbonate cores were used as templates for the fabrication of polyelectrolyte MPs via layer-by-layer deposition as described earlier.12 PAH (Mw ≈ 65,000 Da), PSS (Mw ≈ 70,000 Da), and PAA (Mw ≈ 100,000 Da) were applied onto CaCO3 cores layer by layer to form a multilayer shell of the following structure: PAH/PSS/PAH/PSS/PAH/PSS/PAH/PSS/PAH/PAA.
First, about 1 × 109 CaCO3 cores were suspended in 0.5 mL of ultrapure water and sonicated for 10 min in an ultrasound water bath (Elmasonic P, Elma). Next, 0.5 mL of a 2 mg/mL PAH solution in 0.5 M NaCl was added, and the mixture was sonicated for 60 s and incubated for 20 min at room temperature on an orbital shaker. After the incubation, the excess polyelectrolyte was separated by centrifugation for 3 min at 2604g. The MPs were resuspended in 0.5 mL of ultrapure water. Then, 0.5 mL of a 2 mg/mL PSS solution in 0.5 M NaCl was added, and the mixture was sonicated for 60 s and incubated for 20 min on an orbital shaker. After that, the MP suspension was washed two times with 2.0 mL of ultrapure water by centrifugation for 3 min at 2604g. These steps were repeated to obtain nine layers of polyelectrolytes, PAH forming the upper layer, and then the outer layer of PAA was applied. For this purpose, 0.5 mL of a 2 mg/mL PAA solution in 1 M NaCl was added to the suspension of the MPs. The suspension was sonicated for 60 s and incubated for 20 min with constant stirring on an orbital shaker. After that, the suspension was washed three times with 2.0 mL of ultrapure water by centrifugation and resuspended in 0.5 mL of 50 mM sodium phosphate buffer, pH 8.0. The surface charge of the resultant CaCO3 cores and polyelectrolyte MPs was measured by means of Doppler microelectrophoresis using a Zetasizer Nano ZS instrument (Malvern Instruments Ltd., Worcestershire, U.K.) at each step during the layer-by-layer deposition. To obtain MPs with the shell structure, MPs with the core/shell structure were incubated in an excess of 0.5 M EDTA, pH 8.0, with constant stirring overnight. After the incubation, the resulting shell microparticles were collected by centrifugation for 10 min at 16,873g and washed with ultrapure water three times. At the final step, the MPs were resuspended in 50 mM sodium phosphate buffer, pH 8.0.
2.4. Physicochemical Characterization of the CaCO3 Cores and Microparticles
The size and shape of the obtained CaCO3 cores and polyelectrolyte MPs were determined using an Axio Observer 3 optical microscope (Zeiss, Jena, Germany). Microphotographs were taken using Zeiss ZEN version 2.5 software (blue edition) (Zeiss, Jena, Germany) and processed using Fiji (ImageJ) version 1.8.0 software. In addition, the MPs were examined by using scanning electron microscopy (SEM). To obtain SEM samples, the suspension of MPs was washed with ultrapure water 10 times, and a drop of a diluted suspension containing 103–104 MPs per 1 μL was placed onto a preliminarily purified silicon substrate and dried at room temperature. SEM was performed by using a JSM-7001F scanning electron microscope (JEOL, Japan) equipped with a Schottky cathode. To achieve better contrast, the MPs were additionally coated with 5 nm of tungsten during scanning by means of magnetron sputtering (Torr MagSput-DC-RF). The resultant samples were scanned at a beam current of 100 pA and an accelerating voltage of 15 kV.
2.5. AFM Measurements
The size and morphology of the MPs were characterized with a Bioscope Resolve atomic force microscope (Bruker) combined with an Axio Observer inverted optical microscope (Carl Zeiss, Germany) both in air and in liquid (PBS). For imaging in air, the 10 μL drop of a diluted suspension containing 103–104 MPs per 1 μL was dried on glass coverslips and then scanned in PeakForce Tapping mode by using ScanAsyst Air cantilevers (Bruker) with a nominal spring constant of 0.4 N/m and a nominal tip radius of 2 nm. For stable imaging, the MPs were attached to the glass coverslips via EDC (1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide hydrochloride, Thermo Scientific) coupling. Briefly, 10 μL of a diluted suspension containing 103–104 MPs per 1 μL was placed on a glass coverslip, and after 1 h of incubation, the coverslip was immersed in a solution of EDC (10 mg/mL) in PBS for 2 h at room temperature. Imaging was performed in PBS in PeakForce Tapping mode using ScanAsyst Fluid cantilevers (Bruker) with a nominal spring constant of 0.7 N/m and a nominal tip radius of 20 nm. 3 × 3 μm2 scans with 256 × 256 points were acquired at 0.8 Hz scan rate, 1 kHz cantilever oscillation frequency, and 100 nm amplitude with a set point of 5 nN. The acquired topography images were processed (flattened) with Gwyddion open software.30 The height of at least 55 MPs of each type was measured. The root-mean-squared (rms) surface roughness (Rq) was calculated in 1 × 1 μm2 areas in the middle of MPs for samples in PBS, 5 particles per sample were analyzed. Additionally, the effective Young’s modulus of the particles was estimated by means of Force Volume mode in PBS. Before that, the values of cantilever spring constant and tip radius were determined by the thermal tune method and by scanning the titanium roughness sample (Bruker), respectively. To calculate Young’s modulus, the extend parts of force curves recorded over the top part of MPs were processed by the previously developed MATLAB (the MathWorks, Natick, MA) routine.31 Briefly, the contact point was determined automatically, and the Hertz’s model for spherical indentation was fitted to the contact region of the extend curves. The force curve acquisition speed was 30 Hz, and maps with a resolution of 80 × 80 points and a size of 20 × 20 μm2 were obtained.
2.6. Human Serum and Plasma Preparation
Human blood samples were obtained with informed consent from healthy volunteers at the N.N. Blokhin National Medical Research Center of Oncology and performed in accordance with ethical standards. Blood was collected in Vacutest collection tubes (Vacutest KIMA, Arzergrande, Italy) containing a clot activator for obtaining serum or lithium heparin for obtaining plasma. After 30 min at room temperature, the collection tubes were centrifuged at 1300g for 10 min. The obtained serum and plasma samples were aliquoted and stored at a temperature of −20 °C before use.
2.7. Incubation of Polyelectrolyte Core/Shell and Shell Microparticles with Human Serum or Plasma
Equivalent amounts of MPs with different structures (4 × 107 particles) were dispersed in 50 μL of 50 mM sodium phosphate buffer (pH 8.0). After addition of 250 μL of human serum, human plasma, or phosphate-buffered saline (PBS) (control samples), the samples were incubated with constant shaking at 300 rpm in an Eppendorf ThermoMixerC device (Eppendorf, Germany) at 37 °C for 24 h. After the incubation, the MPs were separated by centrifugation for 20 min at 16,873g and washed with 0.5 mL of PBS five times by centrifugation (2 min, 16,873g). Then, 50 μL of an elution buffer (Sample Buffer (Sigma-Aldrich) for SDS-PAGE, 62.5 mM Tris-HCl (pH 6.8), 10% glycerol, 1% SDS, and 5% 2-mercaptoethanol for MS analysis) were added to the pellets, which was followed by incubation for 20 min at 95 °C to elute proteins from the surface of the MPs. Then, the MPs were separated by centrifugation for 5 min at 16,873g, and the supernatants were collected for further analysis.
2.8. SDS-Polyacrylamide Gel Electrophoresis
For SDS-PAGE analysis, we used a 5% concentrating polyacrylamide gel and a 12% resolving polyacrylamide gel. Samples containing eluted proteins in Sample Buffer were applied onto the gel and separated in Tris-glycine buffer at 70 V for 15 min and then at 150 V for 75 min. The gels were stained in a 0.25% Coomassie Brilliant Blue R-250 solution overnight and washed with ultrapure water by heating. Each gel contained one lane of prestained protein molecular weight markers (PageRuler Plus Prestained Protein Ladder, Thermo Fisher Scientific). To quantify the amount of absorbed proteins, we performed SDS-PAGE of the obtained samples and reference samples containing different amounts of bovine serum albumin (BSA) (5–0.312 μg) as described below. The images of gels were captured using a ChemiDoc XRS+ imaging system (Bio-Rad Laboratories Ltd., Hertfordshire, U.K.), and their densitometric analysis was performed using Image Lab version 5.0 software (Bio-Rad Laboratories Ltd., Hertfordshire, U.K.).
2.9. Sample Preparation for Liquid Chromatography–Tandem Mass Spectrometry
Samples for LC-MS/MS were obtained as described above (Section 2.7). The number of samples was increased to reach the amount of protein required for the MS analysis, and supernatants obtained from the same type of MPs and incubation media were pooled. Before the analysis, the samples were concentrated with Microcon-10, Ultracel PL-10 centrifugal filters (Merck Millipore, Ireland). Lysis buffer containing 10% SDS in 50 mM triethylammonium bicarbonate (TEAB), pH 7.55 (200 μL) was added to the samples. Then, the samples were treated with an ultrasonic probe three times for 30 s at 0 °C and centrifuged for 10 min at 13,000g. For reduction and alkylation, 5 μL of a solution containing 0.5 M Tris (2-carboxyethyl)phosphine hydrochloride (TCEP), and 10 μL of 0.4 M chloroacetamide (CAA) in 50 mM TEAB was added to the supernatant liquid. The resulting solutions were stirred, incubated for 30 min at 80 °C, and cooled to room temperature. Next, a 12% aqueous solution of phosphoric acid was added to the samples in a volume equal to 10% of the sample volume and mixed. Then, a 6-fold volume of binding buffer (90% methanol solution containing 100 mM TEAB, pH 7.5) was added, and the mixture was stirred well. After that, the samples were transferred to an S-trap filter (ProtiFi), 170 μL per filter, and centrifuged for 3 min at 4000g. The application was repeated until the samples were completely transferred to the filter. Next, the samples were washed (four to six times) by adding 150 μL of a 90% methanol solution in 100 mM TEAB to the filter and centrifuged for 4 min at 4000g. After the final centrifugation, the filter was transferred to a clean 1.5 mL test tube for further hydrolysis. For enzymatic hydrolysis, 40 μL of hydrolysis buffer in 50 mM TEAB containing trypsin at a ratio to protein of 1:50 was added. The samples were incubated for 2 h at 47 °C. After the incubation, 40 μL of a solution containing 0.2% formic acid in 50 mM TEAB was added to the filter and the samples were centrifuged for 4 min at 4000g. Then, peptides were eluted from the filter by adding 40 μL of a solution containing 50% acetonitrile in 0.1% formic acid and centrifuging the mixture for 4 min at 4000g. The eluate was transferred from the collection tube to a glass vial and dried in a rotary evaporator at 45 °C. After complete drying, the samples were redissolved in water for subsequent measurement of the amounts of peptides using a Pierce Quantitative Colorimetric Peptide Assay kit (Thermo Scientific, Waltham, MA). The data on the mean concentrations were used to equalize the amount of protein in all samples before analysis. Finally, the samples were dried in a rotary evaporator and diluted with 0.1% formic acid to 2 μg/μL for subsequent analysis.
2.10. Liquid Chromatography–Tandem Mass Spectrometry
Proteomic analysis was performed using an Ultimate 3000 RSLCnano HPLC system (Thermo Scientific) connected to a Q-Exactive HFX mass spectrometer (Thermo Scientific), which was equipped with a NESI ion source (Thermo Scientific). The peptide mixture was loaded onto an Acclaim μ-Precolumn enrichment column (0.5 mm × 3 mm; particle size, 5 μm; Thermo Scientific) at a flow rate of 10 μL/min for 4 min in the isographic mode using a buffer solution containing 2% acetonitrile and 0.1% formic acid in deionized water as a mobile phase. Next, the peptides were separated on a Peaky Efficiency FE HPLC column (100 μm × 20 cm; particle size, 1.9 μm; Molecta, Russia) in the gradient elution mode. The gradient was formed with mobile phase A (0.1% formic acid) and mobile phase B (80% acetonitrile and 0.1% aqueous formic acid) at a flow rate of 0.3 μL/min. The column was washed with 2% mobile phase B for 10 min, after which the concentration of mobile phase B was linearly increased first to 35% during 68 min and then to 99% during 2 min. After 2 min of washing with 99% buffer B, the concentration of this buffer was linearly reduced to the original 2% during 3 min. The total duration of the analysis was 90 min. The peptides were analyzed in the positive ionization mode at an emitter voltage of 2.1 kV and a capillary temperature of 240 °C. Panoramic scanning was performed in a mass range from 300 to 1500 m/z at a resolution of 120 000. In tandem scanning, the resolution was set to 15,000 in a mass range from 100 m/z to the upper limit, which was determined automatically on the basis of the precursor mass but did not exceed 2000 m/z. Isolation of precursor ions was carried out in a window of ±1 Da. The maximum number of ions isolated in the MS2 mode was set to 40, while the cutoff for selecting the precursor for tandem analysis was set to 50,000 units and the normalized collision energy (NCE) was 29. For tandem scanning, only ions with z values from 2+ to 6+ were taken into account. The maximum accumulation time for precursor ions was 50 ms; for fragment ions, it was 110 ms. The automatic gain control (AGC) values for the precursors and fragment ions were set to 1 × 106 and 2 × 105, respectively. All of the measured precursors were dynamically excluded from the tandem MS/MS analysis for 90 s. Each sample was analyzed in a technical duplicate.
2.11. Analysis of Liquid Chromatography–Tandem Mass Spectrometry Data
Proteins were identified by means of the MaxQuant v.2.0.3.0 software using the Andromeda search algorithm.32 The UniProt sequence database UP000005640 (Homo sapiens proteome) was used to identify proteins with the following search parameters: trypsin as the cleaving enzyme, the accuracy of mass determination of monoisotopic peptides of ±4.5 ppm, the accuracy of mass determination in the MS/MS spectra of ±20 ppm, and the possibility of skipping two trypsin cleavage sites. Oxidation of methionines and acetylation of the N terminus of the protein molecule were selected as possible modifications of the peptide, and carbamidomethylation of cysteine was selected as mandatory modification. For validation of comparisons (pairing) of the Peptide-Spectrum Matches (PSMs) and identification of peptides and proteins, the False Discovery Rate (FDR) value was set to no more than 1.0%. Proteins were considered to be reliably identified if at least two peptides were found for them. Label-free quantification (LFQ) of proteins was based on the mean LFQ intensities. Relative mean LFQ intensities were used for a comparison of protein amounts in the samples. A heatmap was generated by the Heatmapper software (http://www.heatmapper.ca/) using Log 2 values of the mean LFQ intensities calculated from analytical duplicates. The physicochemical parameters of proteins, including molecular weight (Mw), isoelectric point (pI), and grand average of hydropathicity (GRAVY), were calculated from the primary sequences using the ProtParam software (www.expasy.org/resources/protparam).
2.12. Statistical Analysis
The MS Office Excel 2013 and GraphPad Prism 8 software were used for statistical analysis of the data. The results are presented as the means and standard deviations for three independent experiments, unless otherwise specified. The data on the surface charge were compared using Student’s t test. The data on the protein amounts were compared using one-way analysis of variance (ANOVA) followed by Tukey’s test for multiple comparison between samples by means of the GraphPad Prism 8 software. The differences were considered significant at p < 0.05.
3. Results and Discussion
3.1. Engineering of Core/Shell and Shell Microparticles
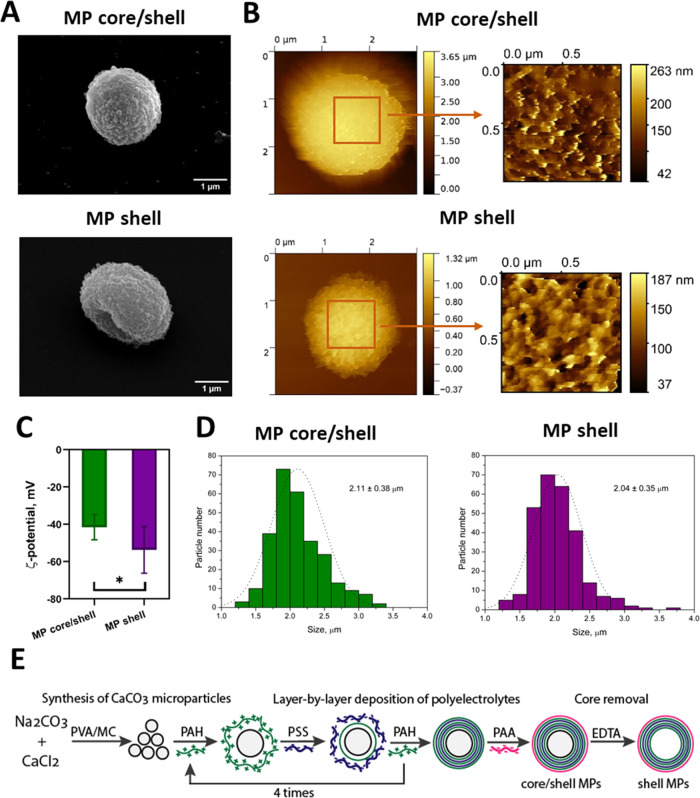
The common approach to controlled synthesis of CaCO3 MPs using additives allows obtaining MPs of various sizes. In this study, we used an aqueous solution of poly(vinyl alcohol) and methylcellulose as a thickening agent to obtain CaCO3 cores with a mean size of ∼2 μm (Figure S1A). SEM images show that the cores had a spherical shape and highly porous surface, which is typical for the vaterite crystalline form of calcium carbonate33 (Figure S1B). Optical microscopy also confirmed the spherical shape of the CaCO3 particles and the absence of the calcite polymorph (data not shown). Formation of the vaterite polymorph of calcium carbonate is critical for further use in biomedical applications and can be provided by controlling the synthesis conditions. In our study, the conditions of synthesis (temperature, salt concentrations, and presence of a thickening agent) yielded CaCO3 particles of the vaterite polymorph with a narrow size distribution.
Using CaCO3 cores as templates, we obtained core–shell and shell MPs via layer-by-layer deposition. The polyelectrolyte shell was formed by alternating polycation (PAH) and polyanion (PSS) layers, with an outer layer of polyacrylamide acid (PAA). Changes in the surface charge during multilayer shell formation were determined by means of Doppler microelectrophoresis (Figure S1C). The initial CaCO3 MPs had a small negative charge, which allowed us to apply PAH as the first polyelectrolyte layer of the shell. Applying PAA in the final step of polyelectrolyte shell formation ensured a large negative surface charge. These results demonstrate the successful deposition of polyelectrolyte layers accompanied by alternation of positive and negative ζ-potentials of the MPs, which yielded a polyelectrolyte shell consisting of the following layers: (PAH/PSS)4/PAH/PAA. Core/shell MPs were incubated with excess EDTA to remove the CaCO3 cores and obtain shell MPs.
3.2. Characterization of the Structure and Surface Properties of the Core/Shell and Shell Microparticles
The morphology of the obtained core/shell and shell MPs was estimated by SEM (Figure 1A). Analysis of SEM images showed that the core/shell MPs had a spherical shape similar to that of the CaCO3 cores, whereas the shell MPs were somewhat elongated, probably due to the absence of the core in their structure. The size and surface topographies of MPs with different structures were analyzed by means of atomic force microscopy both in air (Figure S2A) and in liquid (PBS) (Figure 1B). In PBS, the core/shell and shell MPs had similar heights of 2.2 ± 0.4 and 2.2 ± 0.7 μm, respectively, while dried shell MPs had a substantially lower height of 1.2 ± 0.3 μm, versus 1.9 ± 0.3 μm for core/shell MPs (Figure S2B), which confirmed the dissolution of the core in the shell MPs. Both core/shell and shell MPs had a rough surface with similar root-mean-squared surface roughness of 32 ± 3 and 31 ± 11 nm, respectively. The MPs also had similar Young’s modulus of 3.5 ± 1.3 MPa for core/shell MPs and 3.3 ± 1.3 MPa for shell MPs (nonsignificant difference, Studentʼs t test), as measured by AFM in PBS. The close values of Young’s moduli could be due to the large number of polyelectrolyte layers in the shell, which provides mechanical strength in the absence of the core, as indicated earlier.34
Figure 1.
Physicochemical and structural characteristics of the microparticles (MPs) with different structures. (A) Scanning electron microscopy images of core/shell polyelectrolyte MPs obtained by layer-by-layer deposition and shell MPs obtained after core removal. (B) Atomic force microscopy images of core/shell and shell MPs in PBS; the size of the zoomed regions is 1.0 × 1.0 μm2. (C) Surface charges (ζ-potentials) of core/shell polyelectrolyte MPs obtained by layer-by-layer deposition and shell MPs obtained after core removal, comparison using Studentʼs t test, N = 8 (* p < 0.05). (D) Size distribution of core/shell and shell MPs obtained by means of optical microscopy. (E) Schema of the preparation of core/shell and shell MPs based on CaCO3 cores. PVA, poly(vinyl alcohol); MC, methylcellulose; PAH, poly(allylamine hydrochloride); PSS, poly(sodium 4-styrenesulfonate); PAA, poly(acrylic acid).
The ζ-potential measurements showed that both types of MPs had a negative surface charge. However, the shell MPs had a significantly larger negative surface charge (p < 0.05, Studentʼs t test), which also indicated changes in the surface characteristics caused by removal of the core (Figure 1C). Both types of MPs were characterized by a narrow size distribution with similar mean sizes, 2.11 ± 0.38 and 2.04 ± 0.35 μm for core/shell and shell MPs, respectively (Figure 1D). As a result, we obtained two types of MPs with similar sizes and identical compositions of the polyelectrolyte shell but different structures (Figure 1E). Thus, our results clearly indicate that modification of the MP structure (removal of the core) leads to changes in the shape, surface charge, and morphology.
3.3. Characterization of Protein Adsorption on the Microparticles with Different Structures by SDS-Polyacrylamide Gel Electrophoresis
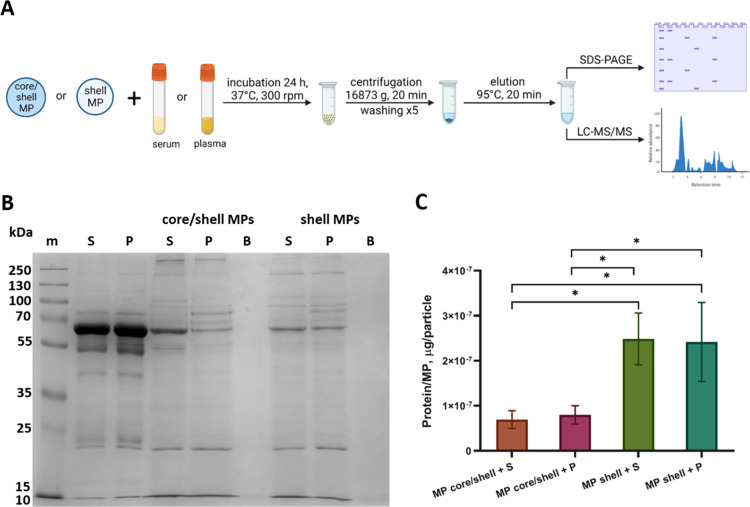
Study of protein adsorption on the surface of engineered particles in biological fluids is a topical subject in biomedicine. Understanding the factors affecting the composition of absorbed proteins and their influence on subsequent interaction of MPs with cells and tissues is crucial for fabricating safer and more effective particle-based biomedical tools. In this study, we analyzed the influence of the structure of polyelectrolyte MPs on protein adsorption in human serum and plasma. Figure 2A shows the design of experiments for estimating the quantitative and qualitative differences in protein adsorption on different types of MPs.
Figure 2.
(A) Schematics of the preparation of samples eluted from the surface of core/shell and shell MPs after their incubation with human serum or plasma. Created with BioRender.com. (B) Results of the SDS-PAGE of the samples eluted from the surface of core/shell and shell MPs after incubation at 37 °C for 24 h with human serum (S) or plasma (P). Lane m contains protein molecular weight markers (kDa). (C) Amount of protein per particle adsorbed on the core/shell and shell MPs after their incubation with serum or plasma as estimated by means of densitometry; comparison of samples using one-way ANOVA with Tukey’s test for multiple comparison, N = 3 (* p < 0.05).
Samples containing proteins eluted from the surface of core/shell and shell MPs after their incubation with serum or plasma were analyzed by using SDS-PAGE in a 12% resolving gel (Figure 2B). Analysis of the number and location of protein bands in each lane shows clear differences between the samples eluted from core/shell and shell MPs. The electrophoregrams of most samples have intense bands that are absent in those of the control samples of diluted serum and plasma (Figure 2B, lanes S and P, respectively), which indicates accumulation of specific proteins on the MP surface. We also detected some differences in the band pattern between the samples obtained using the same types of MPs incubated with different blood derivatives (serum and plasma).
Semiquantitative densitometric analysis of experimental samples and reference samples containing known amounts of BSA was used to estimate the total amount of protein adsorbed on the surface of MPs (Figure 2C). This amount was significantly higher for shell MPs than for core/shell MPs incubated with serum or plasma (one-way ANOVA with Tukey’s test, p < 0.05) (Figure 2B). This phenomenon can be explained by differences in the surface charge and topology of MPs with different structures. Incubation of MPs with serum or plasma did not significantly influence the amount of proteins bound to the same type of MPs. In addition, it has been shown that PAH/PSS microcapsules effectively adsorb proteins (BSA) due to their interaction with charged groups of the internal polyelectrolyte complex,35 which explains the high adsorption capacity of the shell MPs.
3.4. Identification of Proteins Adsorbed on Microparticles with Different Structures by Means of Liquid Chromatography–Tandem Mass Spectrometry
We used mass spectrometry analysis of eluates from the surface of MPs to detect qualitative and quantitative differences in the composition of proteins adsorbed on the two types of particles. LC-MS/MS analysis identified 357 individual proteins in samples obtained from the surface of core/shell and shell MPs after their incubation with serum and plasma. A complete list of the proteins is presented in the Supporting Information (Table S1). Among these proteins, 183 were found in all samples. In samples of core/shell MPs incubated with serum or plasma, five unique proteins were detected; in samples of shell MPs incubated with serum or plasma, two unique proteins. In samples obtained after incubation of both types of MPs with serum, nine unique proteins were detected; in samples obtained after incubation of both types of MPs with plasma, eight unique proteins.
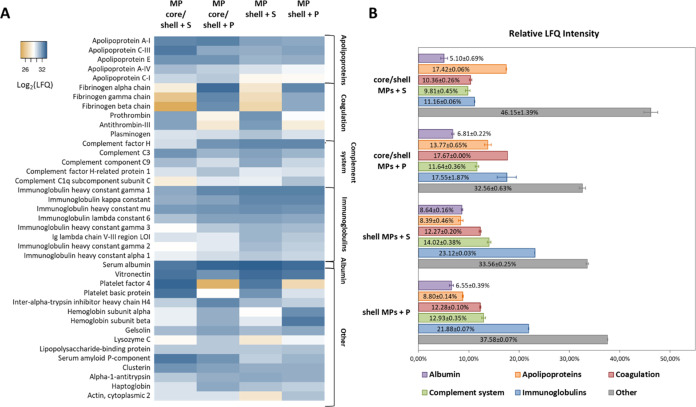
The amounts of proteins in different samples were estimated from the mean LFQ intensities calculated from two replicates (Figure 3A). All proteins were divided into groups according to their physiological functions: apolipoproteins, coagulation system proteins, complement system, immunoglobulins, and others (Figure 3B). The Other group includes a wide range of proteins, such as cytokines, acute phase proteins, hormones, and cell membrane and intracellular proteins. Albumin formed a separate group because of its high content in all samples (5.10–8.64%). The amount of apolipoproteins was noticeably higher in samples of core/shell MPs (17.42 and 13.77% for samples incubated with serum and plasma, respectively) than in samples of shell MPs (8.39 and 8.80%, respectively). On the other hand, proteins of the complement system were more abundant in the samples obtained from shell MPs (14.02 and 12.93%) than in samples of core/shell MPs (9.81 and 11.64%) incubated with both serum and plasma, respectively. Similarly, immunoglobulins were more abundant in samples of shell MPs incubated with serum and plasma (23.12 and 21.88%, respectively) than in those of core/shell MPs (11.16 and 17.55%, respectively). The highest relative amount of coagulation proteins was found in the sample of core/shell MPs incubated with plasma (17.67%), their amounts in other samples being close to one another.
Figure 3.
(A) Heatmap of label-free quantification intensities of major proteins (>1% of the total protein content) adsorbed on core/shell and shell microparticles after incubation with serum or plasma. Created using the Heatmapper (http://www.heatmapper.ca/). (B) Relative amounts of proteins divided into groups according to their biological functions in samples obtained from core/shell and shell microparticles after their incubation with serum or plasma. The Apolipoproteins group includes 15 proteins; Coagulation, 33 proteins; Complement system, 27 proteins; Immunoglobulins, 34 proteins; Other, 244 proteins. Albumin forms a separate group because of its large amounts in all samples. The data are presented as the mean and standard deviation calculated from two replicates.
Among immunoglobulins, IGHG1, IGKC, and IGHM were the most abundant. In the group of complement proteins, both activators and inhibitors were present, with the largest amounts (>1%) of complement components C1q, C3, C9, complement factor H, and CFH-related protein 1. The group of apolipoproteins with relative LFQ intensities in samples >1% consisted of ApoA1, C3, E, A4, and C1 proteins. Among other proteins, HSA, vitronectin, and platelet factor 4 were the most abundant in all of the samples analyzed (Figure 3A). Thus, LC-MS/MS analysis revealed differences in protein composition between samples obtained after incubation of core/shell and shell MPs with serum and plasma in terms of the presence/absence and relative amounts of individual proteins, as well as distribution of proteins into groups.
3.5. Biological Significance of the Adsorption of Specific Proteins on the Microparticles
It is known that adsorption of proteins on the surface of particles may affect their targeting, biodistribution, and elimination by the immune system. HSA is the most abundant protein in serum and is a common component of the protein corona of various particles. HSA is frequently used to block nonspecific interaction of cells with biomaterials, but, at the same time, HSA adsorption has been shown to be associated with platelet activation.36 In our study, albumin was the most abundant protein in the adsorption layer of both core/shell and shell MPs. Another protein with a high relative amount in all samples, vitronectin, promotes monocyte adhesion.37 Interestingly, spontaneous coating of drug delivery particles with vitronectin after their incubation with human plasma resulted in efficient uptake of particles by cancer cells.38 Platelet factor 4, whose content in the adsorption layer on core/shell MPs was relatively high, increases particle uptake by endothelial cells.39 Therefore, differences in the adsorption of specific proteins on MPs of different types may affect their interaction with cells and elimination by the immune system.
3.6. Possible Effects of the Adsorption of Immunoglobulins and Complement Proteins on the Microparticles
Immunoglobulins and complement proteins enhance macrophage recognition and uptake of particles, thus reducing their blood circulation time.40 Complement component C3 is a key activator of the complement system that regulates both classical and alternative pathways. Binding of C3 to biomaterial and particle surface has been found to be related with monocyte and B-cell adhesion.18,41 Complement component C1q has been shown to induce complement activation and enhance opsonization of particles with C3.42 The presence of immunoglobulins in the protein adsorption layer on nanoparticles triggers their opsonization with complement proteins.17 In our study, shell MPs adsorbed higher amounts of both complement proteins (C1q and C3) and immunoglobulins, which can be explained by the aforementioned mechanisms. On the contrary, the presence of complement factor H on the surface of different foreign materials is known to significantly inhibit complement activation via an alternative pathway.43,44 Note that the shell MPs also adsorbed larger amounts of complement factor H and CFH-related protein 1.
3.7. Possible Effects of the Adsorption of Apolipoproteins on the Microparticles
Apolipoproteins constituted another group of proteins whose content noticeably differed between core/shell and shell MPs. Apolipoproteins are involved in the transport of lipids and maintenance of the structure of lipoprotein particles, as well as function as cofactors of lipid metabolism enzymes.45 In the context of biomedical applications of particles, apolipoproteins can affect the blood circulation time and cell uptake. Apolipoproteins B and E enhance the transport of drug-loaded nanoparticles through the blood–brain barrier.46 A study of the protein corona formed on polymeric nanoparticles has demonstrated different effects of adsorbed proteins: apolipoproteins A4 and C3 significantly inhibit cellular uptake, whereas apolipoprotein H enhances it.47 The presence of apolipoprotein A-I in the protein coronas of different nanoparticles is negatively correlated with macrophage uptake and clearance rates.48 ApoA1 has been shown to be the key protein modulating the blood circulation time of particles depending on their elasticity.49 In our study, apolipoproteins modulating the in vivo behavior of particles (ApoA1, ApoE, ApoA4, and ApoC3) were detected in all samples, with a higher content in core/shell MPs.
3.8. Differences in the Adsorption of Proteins Related to Blood Coagulation
Among proteins related to blood coagulation, fibrinogen (α, β, and γ chains) prevails in samples incubated with plasma, which is explained by the differences between plasma and serum compositions. However, fibrinogen was also found in samples of MPs incubated with serum, because some fibrinogen is known to remain in serum even after most of it has been removed in the form of the fibrin clot during serum preparation.50 Fibrinogen adsorption onto the surface of biomaterials is considered to be an important mechanism promoting cell adhesion and ensuring their biocompatibility. However, the total amount of adsorbed fibrinogen is poorly correlated with platelet adhesion, whereas its conformation, associated with the availability of platelet-binding peptides, exhibits a strong correlation with this process.51 In contrast, prothrombin and antithrombin were more abundant in the samples of MPs incubated with serum than in those incubated with plasma. Prothrombin is the inactive form of thrombin, a serine protease that catalyzes the conversion of fibrinogen into fibrin and activation of platelets.52 The presence of thrombin on the surface of nanoparticles is correlated with fibrinogen activation.53 Antithrombin III is a member of the serpin superfamily, which regulates the proteolytic activity of coagulation factors.54 Binding of antithrombin III into a complex with heparin is used for enhancing the anticoagulation properties of biomaterial surfaces.55 Plasminogen, the precursor of plasmin, a protease involved in fibrin clot degradation, was present in comparable amounts in all of the analyzed samples. Development of affinity surfaces with affinity for plasminogen is a useful approach to obtaining fibrinolytic surfaces.56 The proportions of procoagulation and anticoagulation factors in the adsorption layer of MPs seem to determine their effect on blood clotting.
In general, protein adsorption onto particles in a biological environment strongly modulates their subsequent biological behavior, including the interaction with cells, distribution, and targeting as well as the immune response to them. For example, a study on the protein adsorption on amphiphilic nanoparticles has shown that the presence of HSA, complement proteins, apolipoproteins, immunoglobulins, and proteins involved in the coagulation cascade (fibrinogen and prothrombin) weakens nanoparticle–cell interactions and reduces the cytotoxic and hemolytic effects.57 Thus, differences in the content of adsorbed proteins between core/shell and shell MPs used in this study may cause differences in the biological response to these MPs. However, prediction of the biological response based on protein identification alone is difficult due to the complexity of this process.58 Our study can continue to investigate protein adsorption on MPs with different structures and surface properties in connection with the biological responses to these MPs.
3.9. Physicochemical Characterization of the Proteins Adsorbed on the Microparticles with Different Structures
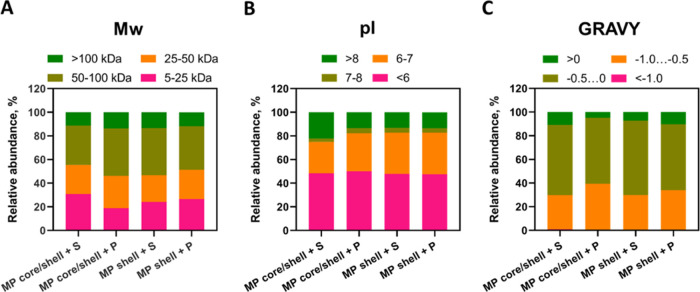
We compared three physicochemical characteristics of the proteins identified in the samples, Mw, pI, and GRAVY, which is considered a suitable parameter for assessing the hydrophilicity/hydrophobicity of proteins.59 The proportions of proteins grouped according to their Mw, pI, and GRAVY were almost identical in all samples of the MPs. Proteins with Mw between 50 and 100 kDa constituted the major part of all adsorbed proteins (33.03–40.02%), and proteins with Mw > 100 kDa were the least abundant (Figure 4A). The ratio between the proteins with molecular weights of 5–25 and 25–50 kDa varied in different samples: in core/shell MPs incubated with serum and shell MPs incubated with either serum or plasma, the relative amount of 5–25 kDa proteins was higher, whereas in core/shell MPs incubated with plasma, 25–50 kDa proteins were more abundant. Regarding pI values (Figure 4B), negatively charged proteins were prevailing in all samples: the relative abundance of proteins with pI < 6 varied between 47.58 and 50.03% and those with a pI of 6–7, between 26.54 and 35.18%. The proportion of proteins with a pI of 7–8 was the lowest in all samples (2.90–4.42%). The amount of proteins with pI > 8 was almost the same in core/shell MPs incubated with plasma and shell MPs incubated with either serum or plasma (13.30–13.53%), and it was larger in core/shell MPs incubated with serum (22.19%). Because MPs used in this study had a negative ζ-potential, we assume that their preferential interaction with negatively charged proteins is determined by nonelectrostatic forces, e.g., hydrogen bonds. We also suppose that the outer layer of the polyelectrolyte shell consists of a mixture of PAH and PAA molecules, which can explain the high content of negatively charged proteins (albumin and apolipoproteins). In addition, heterogeneity of the protein surface charge distribution can lead to the formation of complexes of acidic proteins with negatively charged polyelectrolytes.60 The GRAVY score distribution (Figure 4C) demonstrated that most adsorbed proteins were hydrophilic (GRAVY < 0), but hydrophobic proteins were also found in all samples (4.75–10.70%). More than half of all identified proteins had slightly negative GRAVY scores (between −0.5 and 0), and less than 1% had GRAVY scores lower than −1.0. Thus, most proteins in all samples had molecular weights between 50 and 100 kDa, negative charges in the physiological environment (pI < 6), and GRAVY scores between −0.5 and 0 (i.e., they were hydrophilic). In general, the physicochemical characteristics of the proteins adsorbed on the core/shell and shell MPs did not differ noticeably from each other.
Figure 4.
Relative label-free quantification intensities of proteins grouped according to their (A) molecular weights (Mw), (B) isoelectric points (pI), and (C) grand average of hydropathicity (GRAVY) scores adsorbed on core/shell and shell MPs upon their incubation with serum or plasma. The Mw, pI, and GRAVY were calculated on the basis of the primary sequences of the proteins using ProtParam software.
3.10. Influence of Biological Media Used for Incubation on the Protein Adsorption Pattern
Most studies used only one blood derivative as a source of proteins. In contrast, we incubated MPs with both serum and plasma as biologically relevant media. The results showed differences in the composition of adsorbed proteins between MPs of the same type incubated with serum and plasma. Our results agree with the data on protein adsorption from serum and plasma onto methacrylic acid copolymer beads.58 These results should be taken into account in planning studies on the interaction of engineered particles with components of the biological environment.
4. Conclusions
Polyelectrolyte MPs and microcapsules are promising tools with great potential in biomedical applications. However, the inevitable interaction of engineered particles with constituents of biological media and their possible effects on the functioning and safety limit their wide use. The key factors influencing this interaction are the physicochemical parameters of the particles, including their size and surface charge as well as their functionalization. Here, we have compared the interaction of human serum and plasma proteins with polyelectrolyte MPs of two types differing in structure: core/shell MPs and shell MPs with a dissolved core. SDS-PAGE and LC-MS/MS have been used to identify qualitative and quantitative differences in the protein adsorption pattern between core/shell and shell MPs, as well as between MPs incubated with blood serum and plasma. We have demonstrated that some protein groups, such as lipoproteins, are predominantly adsorbed onto core/shell MPs, and others, such as immunoglobulins and complement proteins, are adsorbed onto shell MPs. Adsorbed lipoproteins are known to affect the blood circulation time and cell adhesion of particles. Immunoglobulins and lipoproteins can also modify the interaction of particles with cells, complement activation, and the uptake of the particles on which they are adsorbed. Because protein adsorption is crucial for biological response to engineered particles and biomaterials, these differences should be considered in the design of MPs to improve their biomedical performance. The observed differences in the composition of proteins adsorbed on the surface of MPs with different structures can be used for the selective adsorption of various proteins. The results of our study add to the understanding of factors affecting protein adsorption on polyelectrolyte MPs and can be used in the development of particle-based applications.
Acknowledgments
A.S. and I.N. acknowledge the support of the French Ministry of Higher Education, Research and Innovation, and the University of Reims Champagne-Ardenne. The authors thank Galina Nifontova for technical assistance and Vladimir Ushakov for proofreading the manuscript.
Glossary
Abbreviations
- AFM
atomic force microscopy
- AGC
automatic gain control
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- EDTA
ethylenediaminetetraacetic acid
- FDR
false discovery rate
- GRAVY
grand average of hydropathicity
- HSA
human serum albumin
- Ig
immunoglobulin
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- LFQ
label-free quantification
- MC
methylcellulose
- MP
microparticle
- NCE
normalized collision energy
- PAA
poly(acrylic acid)
- PAGE
polyacrylamide gel electrophoresis
- PAH
poly(allylamine hydrochloride)
- PBS
phosphate-buffered saline
- PSMs
peptide-spectrum matches
- PSS
poly(sodium 4-styrenesulfonate)
- PVA
poly(vinyl alcohol)
- SDS
sodium dodecyl sulfate
- SEM
scanning electron microscopy
- TCEP
tris (2-carboxyethyl)phosphine hydrochloride
- TEAB
triethylammonium bicarbonate
- TEMED
N,N,N′,N′-tetramethylethylenediamine
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c03307.
Results of scanning electron microscopy analysis of CaCO3 cores (Figure S1A); size distribution of the CaCO3 cores estimated by means of optical microscopy (Figure S1B); surface charge changes during the fabrication of polyelectrolyte core/shell microparticles (Figure S1C); AFM images of core/shell MPS, shell MPs, and CaCO3 cores in air (Figure S2A); heights of core/shell MPS, shell MPs, and CaCO3 cores in PBS and in air (Figure S2B); data on the effective Young’s modulus of the core/shell MPS and shell MPs in PBS (Figure S2C); and list of proteins identified by means of liquid chromatography–tandem mass spectrometry in samples obtained after incubation of core/shell and shell microparticles with serum or plasma (Table S1) (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The ITMO Cancer of Aviesan within the framework of the 2021–2030 Cancer Control Strategy, on funds administered by the French National Institute of Health and Medical Research, grant no. 22CP174–00 “Smart-Nano”. The Russian Science Foundation (RSF) grant no. 22–75–10103 in the part of the work related to the synthesis of microcapsules and the mass spectrometry studies, and grant no. 23–75–30016 in the part of the work related to the microcapsules functionalization and bioadaptation.
The authors declare no competing financial interest.
Supplementary Material
References
- Li J.; Parakhonskiy B. V.; Skirtach A. G. A Decade of Developing Applications Exploiting the Properties of Polyelectrolyte Multilayer Capsules. Chem. Commun. 2023, 59, 807–835. 10.1039/D2CC04806J. [DOI] [PubMed] [Google Scholar]
- Galogahi F. M.; Zhu Y.; An H.; Nguyen N. T. Core-Shell Microparticles: Generation Approaches and Applications. J. Sci.: Adv. Mater. Devices 2020, 5, 417–435. 10.1016/j.jsamd.2020.09.001. [DOI] [Google Scholar]
- Kruk T.; Chojnacka-Górka K.; Kolasińska-Sojka M.; Zapotoczny S. Stimuli-Responsive Polyelectrolyte Multilayer Films and Microcapsules. Adv. Colloid Interface Sci. 2022, 310, 102773 10.1016/j.cis.2022.102773. [DOI] [PubMed] [Google Scholar]
- Zan X.; Garapaty A.; Champion J. A. Engineering Polyelectrolyte Capsules with Independently Controlled Size and Shape. Langmuir 2015, 31, 7601–7608. 10.1021/acs.langmuir.5b01578. [DOI] [PubMed] [Google Scholar]
- Barrantes A.; Wengenroth J.; Arnebrant T.; Haugen H. J. Poly-L-Lysine/Heparin Multilayer Coatings Prevent Blood Protein Adsorption. J. Colloid Interface Sci. 2017, 485, 288–295. 10.1016/j.jcis.2016.09.046. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Guo J.; Yan Y.; Ang C. S.; Bertleff-Zieschang N.; Caruso F. Cell-Conditioned Protein Coronas on Engineered Particles Influence Immune Responses. Biomacromolecules 2017, 18, 431–439. 10.1021/acs.biomac.6b01545. [DOI] [PubMed] [Google Scholar]
- Nifontova G.; Tsoi T.; Karaulov A.; Nabiev I.; Sukhanova A. Structure-Function Relationships in Polymeric Multilayer Capsules Designed for Cancer Drug Delivery. Biomater. Sci. 2022, 10, 5092–5115. 10.1039/D2BM00829G. [DOI] [PubMed] [Google Scholar]
- Song X.; Li H.; Tong W.; Gao C. Fabrication of Triple-Labeled Polyelectrolyte Microcapsules for Localized Ratiometric PH Sensing. J. Colloid Interface Sci. 2014, 416, 252–257. 10.1016/j.jcis.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Nifontova G.; Krivenkov V.; Zvaigzne M.; Efimov A.; Korostylev E.; Zarubin S.; Karaulov A.; Nabiev I.; Sukhanova A. Nanoparticle-Doped Hybrid Polyelectrolyte Microcapsules with Controlled Photoluminescence for Potential Bioimaging Applications. Polymers 2021, 13, 4076 10.3390/polym13234076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdepérez D.; del Pino P.; Sánchez L.; Parak W. J.; Pelaz B. Highly Active Antibody-Modified Magnetic Polyelectrolyte Capsules. J. Colloid Interface Sci. 2016, 474, 1–8. 10.1016/j.jcis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Wang H.; Xu S.; Fan D.; Geng X.; Zhi G.; Wu D.; Shen H.; Yang F.; Zhou X.; Wang X. Multifunctional Microcapsules: A Theranostic Agent for US/MR/PAT Multi-Modality Imaging and Synergistic Chemo-Photothermal Osteosarcoma Therapy. Bioact. Mater. 2022, 7, 453–465. 10.1016/j.bioactmat.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nifontova G.; Kalenichenko D.; Baryshnikova M.; Gomes F. R.; Alves F.; Karaulov A.; Nabiev I.; Sukhanova A. Biofunctionalized Polyelectrolyte Microcapsules Encoded with Fluorescent Semiconductor Nanocrystals for Highly Specific Targeting and Imaging of Cancer Cells. Photonics 2019, 6, 117 10.3390/photonics6040117. [DOI] [Google Scholar]
- Ferrari P. F.; Zattera E.; Pastorino L.; Perego P.; Palombo D. Dextran/Poly-L-Arginine Multi-Layered CaCO3-Based Nanosystem for Vascular Drug Delivery. Int. J. Biol. Macromol. 2021, 177, 548–558. 10.1016/j.ijbiomac.2021.02.058. [DOI] [PubMed] [Google Scholar]
- Monopoli M. P.; Åberg C.; Salvati A.; Dawson K. A. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol. 2012, 7, 779–786. 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Abdallah M. N.; Tran S. D.; Abughanam G.; Laurenti M.; Zuanazzi D.; Mezour M. A.; Xiao Y.; Cerruti M.; Siqueira W. L.; Tamimi F. Biomaterial Surface Proteomic Signature Determines Interaction with Epithelial Cells. Acta Biomater. 2017, 54, 150–163. 10.1016/j.actbio.2017.02.044. [DOI] [PubMed] [Google Scholar]
- Coron A. E.; Fonseca D. M.; Sharma A.; Slupphaug G.; Strand B. L.; Rokstad A. M. A. MS-Proteomics Provides Insight into the Host Responses towards Alginate Microspheres. Mater. Today Bio 2022, 17, 100490 10.1016/j.mtbio.2022.100490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu V. P.; Gifford G. B.; Chen F.; Benasutti H.; Wang G.; Groman E. V.; Scheinman R.; Saba L.; Moghimi S. M.; Simberg D. Immunoglobulin Deposition on Biomolecule Corona Determines Complement Opsonisation Efficiency of Preclinical and Clinical Nanoparticles. Nat. Nanotechnol. 2019, 14, 260–268. 10.1038/s41565-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y.; Kelly H. G.; Dagley L. F.; Reynaldi A.; Schlub T. E.; Spall S. K.; Bell C. A.; Cui J.; Mitchell A. J.; Lin Z.; et al. Person-Specific Biomolecular Coronas Modulate Nanoparticle Interactions with Immune Cells in Human Blood. ACS Nano 2020, 14, 15723–15737. 10.1021/acsnano.0c06679. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Yan Y.; Guo J.; Björnmalm M.; Cui J.; Sun H.; Caruso F. Targeting Ability of Affibody-Functionalized Particles Is Enhanced by Albumin but Inhibited by Serum Coronas. ACS Macro Lett. 2015, 4, 1259–1263. 10.1021/acsmacrolett.5b00627. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Yan Y.; Ang C. S.; Kempe K.; Kamphuis M. M. J.; Dodds S. J.; Caruso F. Monoclonal Antibody-Functionalized Multilayered Particles: Targeting Cancer Cells in the Presence of Protein Coronas. ACS Nano 2015, 9, 2876–2885. 10.1021/nn506929e. [DOI] [PubMed] [Google Scholar]
- Ju Y.; Dai Q.; Cui J.; Dai Y.; Suma T.; Richardson J. J.; Caruso F. Improving Targeting of Metal-Phenolic Capsules by the Presence of Protein Coronas. ACS Appl. Mater. Interfaces 2016, 8, 22914–22922. 10.1021/acsami.6b07613. [DOI] [PubMed] [Google Scholar]
- Peng Q.; Zhang S.; Yang Q.; Zhang T.; Wei X. Q.; Jiang L.; Zhang C. L.; Chen Q. M.; Zhang Z. R.; Lin Y. F. Preformed Albumin Corona, a Protective Coating for Nanoparticles Based Drug Delivery System. Biomaterials 2013, 34, 8521–8530. 10.1016/j.biomaterials.2013.07.102. [DOI] [PubMed] [Google Scholar]
- de Oliveira F. A.; Albuquerque L. J. C.; Castro C. E.; Riske K. A.; Bellettini I. C.; Giacomelli F. C. Reduced Cytotoxicity of Nanomaterials Driven by Nano-Bio Interactions: Case Study of Single Protein Coronas Enveloping Polymersomes. Colloids Surf., B 2022, 213, 112387 10.1016/j.colsurfb.2022.112387. [DOI] [PubMed] [Google Scholar]
- Jasinski J.; Wilde M. V.; Voelkl M.; Jérôme V.; Fröhlich T.; Freitag R.; Scheibel T. Tailor-Made Protein Corona Formation on Polystyrene Microparticles and Its Effect on Epithelial Cell Uptake. ACS Appl. Mater. Interfaces 2022, 14, 47277–47287. 10.1021/acsami.2c13987. [DOI] [PubMed] [Google Scholar]
- Lopez-Cazares G.; Eniola-Adefeso O. Dual Coating of Chitosan and Albumin Negates the Protein Corona-Induced Reduced Vascular Adhesion of Targeted PLGA Microparticles in Human Blood. Pharmaceutics 2022, 14, 1018 10.3390/pharmaceutics14051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia J.; Yazdimamaghani M.; Moghaddam S. P. H.; Ghandehari H. Differential Protein Adsorption and Cellular Uptake of Silica Nanoparticles Based on Size and Porosity. ACS Appl. Mater. Interfaces 2016, 8, 34820–34832. 10.1021/acsami.6b09950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzer P.; Svec F.; Sekot G.; Jungbauer A. Protein Adsorption onto Nanoparticles Induces Conformational Changes: Particle Size Dependency, Kinetics, and Mechanisms. Eng. Life Sci. 2016, 16, 238–246. 10.1002/elsc.201500059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasampanthan T.; Beyer C. D.; Yu W.; Karthäuser J. F.; Wanka R.; Spöllmann S.; Becker H. W.; Aldred N.; Clare A. S.; Rosenhahn A. Effect of Multilayer Termination on Nonspecific Protein Adsorption and Antifouling Activity of Alginate-Based Layer-by-Layer Coatings. Langmuir 2021, 37, 5950–5963. 10.1021/acs.langmuir.1c00491. [DOI] [PubMed] [Google Scholar]
- Nifontova G.; Kalenichenko D.; Kriukova I.; Terryn C.; Audonnet S.; Karaulov A.; Nabiev I.; Sukhanova A. Impact of Macrophages on the Interaction of Cetuximab-Functionalized Polyelectrolyte Capsules with EGFR-Expressing Cancer Cells. ACS Appl. Mater. Interfaces 2023, 15, 52137–52149. 10.1021/acsami.3c10864. [DOI] [PubMed] [Google Scholar]
- Nečas D.; Klapetek P. Gwyddion: An open-source software for SPM data analysis. Open Phys. 2012, 10 (1), 181–188. 10.2478/s11534-011-0096-2. [DOI] [Google Scholar]
- Efremov Yu. M.; Shpichka A. I.; Kotova S. L.; Timashev P. S. Viscoelastic mapping of cells based on fast force volume and PeakForce Tapping. Soft Matter 2019, 15, 5455–5463. 10.1039/C9SM00711C. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Cox J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Trushina D. B.; Bukreeva T. V.; Kovalchuk M. V.; Antipina M. N. CaCO3 Vaterite Microparticles for Biomedical and Personal Care Applications. Mater. Sci. Eng. C 2014, 45, 644–658. 10.1016/j.msec.2014.04.050. [DOI] [PubMed] [Google Scholar]
- Palankar R.; Pinchasik B.-E.; Schmidt S.; De Geest B. G.; Fery A.; Möhwald H.; Skirtach A. G.; Delcea M. Mechanical strength and intracellular uptake of CaCO3-templated LbL capsules composed of biodegradable polyelectrolytes: The influence of the number of layers. J. Mater. Chem. B 2013, 1, 1175–1181. 10.1039/c2tb00319h. [DOI] [PubMed] [Google Scholar]
- Volodkin D. V.; Petrov A. I.; Prevot M.; Sukhorukov G. B. Matrix Polyelectrolyte Microcapsules: New System for Macromolecule Encapsulation. Langmuir 2004, 20 (8), 3398–3406. 10.1021/la036177z. [DOI] [PubMed] [Google Scholar]
- Sivaraman B.; Latour R. A. Delineating the Roles of the GPIIb/IIIa and GP-Ib-IX-V Platelet Receptors in Mediating Platelet Adhesion to Adsorbed Fibrinogen and Albumin. Biomaterials. 2011, 32, 5365–5370. 10.1016/j.biomaterials.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalakshan R. M.; Bright R.; Burzava A. L. S.; Barker A. J.; Simon J.; Ninan N.; Palms D.; Wood J.; Martínez-Negro M.; Morsbach S.; et al. Antibacterial Nanostructured Surfaces Modulate Protein Adsorption, Inflammatory Responses, and Fibrous Capsule Formation. ACS Appl. Mater. Interfaces 2023, 15, 220–235. 10.1021/acsami.2c13415. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Cardarelli F.; Pozzi D.; Salomone F.; Maccari G.; Bardi G.; Capriotti A. L.; Cavaliere C.; Papi M.; Laganà A. Selective Targeting Capability Acquired with a Protein Corona Adsorbed on the Surface of 1,2-Dioleoyl-3-Trimethylammonium Propane/Dna Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 13171–13179. 10.1021/am404171h. [DOI] [PubMed] [Google Scholar]
- Lin Z. P.; Ngo W.; Mladjenovic S. M.; Wu J. L. Y.; Chan W. C. W. Nanoparticles Bind to Endothelial Cells in Injured Blood Vessels via a Transient Protein Corona. Nano Lett. 2023, 23, 1003–1009. 10.1021/acs.nanolett.2c04501. [DOI] [PubMed] [Google Scholar]
- Barui A. K.; Oh J. Y.; Jana B.; Kim C.; Ryu J. Cancer-Targeted Nanomedicine: Overcoming the Barrier of the Protein Corona. Adv. Ther. 2020, 3, 1900124 10.1002/adtp.201900124. [DOI] [Google Scholar]
- Wang X.; Schmidt D. R.; Joyce E. J.; Kao W. J. Application of MS-Based Proteomics to Study Serum Protein Adsorption/Absorption and Complement C3 Activation on Poly(Ethylene Glycol) Hydrogels. J. Biomater. Sci., Polym. Ed. 2011, 22, 1343–1362. 10.1163/092050610X508400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano R.; Gabrielli L.; Lubian E.; Fedeli C.; Visentin S.; Laureto P.; De; Arrigoni G.; Geffner-smith A.; Chen F.; Simberg D.; et al. C1q-Mediated Complement Activation and C3 Opsonization Trigger Recognition of Stealth Poly(2-Methyl-2-Oxazoline)-Coated Silica Nanoparticles by Human Phagocytes. ACS Nano 2019, 12, 5834–5847. 10.1021/acsnano.8b01806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belling J. N.; Jackman J. A.; Avsar S. Y.; Park J. H.; Wang Y.; Potroz M. G.; Ferhan A. R.; Weiss P. S.; Cho N. J. Stealth Immune Properties of Graphene Oxide Enabled by Surface-Bound Complement Factor H. ACS Nano 2016, 10, 10161–10172. 10.1021/acsnano.6b05409. [DOI] [PubMed] [Google Scholar]
- Wu Y.-Q.; Qu H.; Sfyroera G.; Tzekou A.; Kay B. K.; Nilsson B.; Ekdahl K. N.; Ricklin D.; Lambris J. D. Protection of Nonself Surfaces from Complement Attack by Factor H-Binding Peptides: Implications for Therapeutic Medicine. J. Immunol. 2011, 186, 4269–4277. 10.4049/jimmunol.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.; Shapiro M. D. Apolipoproteins in Vascular Biology and Atherosclerotic Disease. Nat. Rev. Cardiol. 2022, 19, 168–179. 10.1038/s41569-021-00613-5. [DOI] [PubMed] [Google Scholar]
- La Barbera L.; Mauri E.; D’Amelio M.; Gori M. Functionalization Strategies of Polymeric Nanoparticles for Drug Delivery in Alzheimer’s Disease: Current Trends and Future Perspectives. Front. Neurosci. 2022, 16, 939855 10.3389/fnins.2022.939855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz S.; Schöttler S.; Kotman N.; Baier G.; Musyanovych A.; Kuharev J.; Landfester K.; Schild H.; Jahn O.; Tenzer S.; Mailänder V. Protein Corona of Nanoparticles: Distinct Proteins Regulate the Cellular Uptake. Biomacromolecules 2015, 16, 1311–1321. 10.1021/acs.biomac.5b00108. [DOI] [PubMed] [Google Scholar]
- Assali A.; Razzazan S.; Akhavan O.; Mottaghitalab F.; Adeli M.; Atyabi F. The Bio-Interface between Functionalized Au NR@GO Nanoplatforms with Protein Corona and Their Impact on Delivery and Release System. Colloids Surf., B 2019, 173, 891–898. 10.1016/j.colsurfb.2018.10.042. [DOI] [PubMed] [Google Scholar]
- Li M.; Jin X.; Liu T.; Fan F.; Gao F.; Chai S.; Yang L. Nanoparticle Elasticity Affects Systemic Circulation Lifetime by Modulating Adsorption of Apolipoprotein A-I in Corona Formation. Nat. Commun. 2022, 13, 4137 10.1038/s41467-022-31882-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaq H. J.; Xiao Z.; Veenstra T. D. Serum and Plasma Proteomics. Chem. Rev. 2007, 107, 3601–3620. 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- Horbett T. A. Fibrinogen Adsorption to Biomaterials. J. Biomed. Mater. Res., Part A 2018, 106, 2777–2788. 10.1002/jbm.a.36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi N.; Di Cera E. Prothrombin Structure: Unanticipated Features and Opportunities. Expert Rev. Proteomics 2014, 11, 653–655. 10.1586/14789450.2014.971763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahniuk M. S.; Alshememry A. K.; Unsworth L. D. Human Plasma Protein Adsorption to Elastin-like Polypeptide Nanoparticles. Biointerphases 2020, 15, 021007 10.1116/6.0000027. [DOI] [PubMed] [Google Scholar]
- Rezaie A. R.; Giri H. Anticoagulant and Signaling Functions of Antithrombin Alireza. J. Thromb. Haemostasis 2020, 18, 3142–3153. 10.1111/jth.15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J. M.; Berry L. R.; Atkinson H. M.; Cornelius R. M.; Sandejas D.; Rochow N.; Selvaganapathy P. R.; Fusch C.; Chan A. K. C.; Brash J. L. Surface Modification of Poly(Dimethylsiloxane) with a Covalent Antithrombin-Heparin Complex for the Prevention of Thrombosis: Use of Polydopamine as Bonding Agent. J. Mater. Chem. B 2015, 3, 6032–6036. 10.1039/C5TB00808E. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Li D.; Zhan W.; Luan Y.; Du H.; Liu X.; Brash J. L.; Chen H. Surfaces Having Dual Affinity for Plasminogen and Tissue Plasminogen Activator: In Situ Plasmin Generation and Clot Lysis. J. Mater. Chem. B 2015, 3, 6939–6944. 10.1039/C5TB01308A. [DOI] [PubMed] [Google Scholar]
- de Castro C. E.; Panico K.; Stangherlin L. M.; Ribeiro C. A. S.; da Silva M. C. C.; Carneiro-Ramos M. S.; Dal-Bó A. G.; Giacomelli F. C. The Protein Corona Conundrum: Exploring the Advantages and Drawbacks of Its Presence around Amphiphilic Nanoparticles. Bioconjugate Chem. 2020, 31, 2638–2647. 10.1021/acs.bioconjchem.0c00564. [DOI] [PubMed] [Google Scholar]
- Wells L. A.; Guo H.; Emili A.; Sefton M. V. The Profile of Adsorbed Plasma and Serum Proteins on Methacrylic Acid Copolymer Beads: Effect on Complement Activation. Biomaterials 2017, 118, 74–83. 10.1016/j.biomaterials.2016.11.036. [DOI] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- de Vries R.; Weinbreck F.; de Kruif C. G. Theory of polyelectrolyte adsorption on heterogeneously charged surfaces applied to soluble protein-polyelectrolyte complexes. J. Chem. Phys. 2003, 118 (10), 4649–4659. 10.1063/1.1543981. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.