Abstract
Labor is a complex physiological process requiring a well-orchestrated dialogue between the mother and fetus. However, the cellular contributions and communications that facilitate maternal-fetal crosstalk in labor have not been fully elucidated. Herein, single-cell RNA sequencing (scRNA-seq) was applied to decipher maternal-fetal signaling in the human placenta during term labor. First, a single-cell atlas of the human placenta was established, demonstrating that maternal and fetal cell types underwent changes in transcriptomic activity during labor. Cell types most affected by labor were fetal stromal and maternal decidual cells in the chorioamniotic membranes (CAM) and maternal and fetal myeloid cells in the placenta. Cell-cell interaction analyses showed that CAM and placental cell types participated in labor-driven maternal and fetal signaling, including the Collagen, C-X-C motif ligand (CXCL), tumor necrosis factor (TNF), Galectin, and interleukin (IL)-6 pathways. Integration of scRNA-seq data with publicly available bulk transcriptomic data showed that placenta-derived scRNA-seq signatures could be monitored in the maternal circulation throughout gestation and in labor. Moreover, comparative analysis revealed that placenta-derived signatures in term labor were mirrored by those in spontaneous preterm labor and birth. Further, we demonstrated that early in gestation, labor-specific placenta-derived signatures could be detected in the circulation of women destined to undergo spontaneous preterm birth, either with intact or prematurely ruptured membranes. Collectively, our findings provide insight into the maternal-fetal crosstalk of human parturition and suggest that placenta-derived single-cell signatures can aid in the development of non-invasive biomarkers for the prediction of preterm birth.
One Sentence Summary:
Single-cell RNA sequencing of the human placenta reveals the maternal and fetal cell types and communication networks implicated in parturition
INTRODUCTION
Labor is a multi-stage process that involves the coordinated activation of physiological, biochemical, endocrinological, and immunological pathways in the mother and fetus, resulting in successful delivery of the offspring (1, 2). Labor involves extra-uterine and intra-uterine components, of which the latter comprises the increased uterine contractility, the final stage of cervical remodeling (dilation and effacement), and decidual/membrane activation that are considered hallmarks of the common pathway of parturition (3–5). Spontaneous labor at term represents a state of physiological inflammation (6–11) that includes increased leukocyte infiltration and expression of inflammatory mediators in the uterine tissues (12–27), cervix (6, 13–16, 28–31), and chorioamniotic membranes (7, 14, 15, 32–35) as well as in the maternal circulation (16, 36–45). Although previous murine studies proposed that the inflammatory pathways implicated in labor are independently controlled by the mother or the fetus (46–49), recent work has mechanistically proven that such a complex process is driven by both maternal and fetal signaling (50, 51). This concept is supported by human studies showing that, in some cases, the fetus can initiate the onset of preterm parturition (52–54). However, the tightly orchestrated mechanisms whereby maternal-fetal crosstalk can regulate labor are poorly understood, and thus there is a need for deep exploration of the contributions of maternal and fetal cells and their communication. The local sites of contact between maternal and fetal cells are collectively termed the maternal-fetal interface and include the decidual tissues (decidua basalis and decidua parietalis) and the intervillous space of the placenta (55–63). These sites of maternal-fetal interaction are highly heterogeneous and include multiple tissular and immune cell types; therefore, the use of discovery approaches such as single-cell omics is required to decipher the specific cell type signaling pathways implicated in the complex process of labor.
Single-cell RNA sequencing (scRNA-seq) has provided a wealth of information regarding the involved cell types, key cellular interactions, and immune signaling pathways taking place throughout gestation (64). Indeed, we have undertaken investigations using single-cell approaches to unravel the signaling networks altered in the human placenta (65) and myometrium (66) during preterm and/or term labor. However, a comprehensive single-cell atlas of the maternal-fetal interface in human parturition remains to be established, given the proposed role of the decidua as the anatomical site of cellular and molecular processes that dictate the initiation of labor (11, 67–70). Although scRNA-seq continues to provide new insights into pregnancy and its complications, the translation of such findings to a clinical setting remains a challenge. One means of overcoming this limitation has been to integrate single-cell gene signatures derived from the placental tissues with the maternal systemic cellular transcriptome to monitor pregnancy (42, 66) and potentially predict disease. Indeed, this approach has yielded successful models for the non-invasive identification of women with preeclampsia (71, 72) and was proposed to have value for prediction of spontaneous preterm labor (65). However, the potential translational utility of single-cell signatures derived from the placenta, including maternal and fetal cells, for detecting the labor-specific molecular signals in the maternal circulation of women with different subsets of spontaneous preterm birth has not been fully explored.
Herein, we utilized scRNA-seq to generate a comprehensive single-cell landscape of the human placenta during parturition by examining the placenta [including placental villous tree attached to the basal plate (the decidua basalis)] and extraplacental tissues (chorioamniotic membranes including the decidua parietalis) from women who underwent term delivery with or without spontaneous labor. We described the cellular composition of each compartment and identified those maternal and fetal cell types most impacted by the labor process. The analysis of cell type transcriptomic profiles highlighted labor-associated enrichment of specific pathways. Cell-cell interaction analyses were used to infer communication networks among cell types in each tissue, including key signaling pathways implicated in labor. We then leveraged existing bulk transcriptomic datasets of the maternal peripheral blood to demonstrate that placenta-derived scRNA-seq signatures could be non-invasively monitored throughout gestation, and that a fraction of these signatures is enriched during labor. Furthermore, we found that scRNA-seq signatures modulated by term labor in the chorioamniotic membranes were mirrored in those tissues from women who underwent preterm labor and birth; yet the inflammatory and microbiological status of the amniotic cavity drove specific transcriptomic activity in maternal and fetal cell types. Lastly, we demonstrated enrichment of labor-specific placenta-derived scRNA-seq signatures in the maternal circulation during the second and third trimesters that can distinguish women who ultimately undergo spontaneous preterm labor with intact membranes or preterm prelabor rupture of membranes, which could potentially allow for the non-invasive prediction of spontaneous preterm birth.
RESULTS
Establishment of a single-cell atlas of the human placenta during labor
To explore the cellular interactions between the mother and fetus during parturition, we first established a comprehensive single-cell atlas of the human placenta by using placental and extraplacental tissues from women who underwent spontaneous labor (n = 24) or who were delivered in the absence of spontaneous labor (n = 18) at term (tables S1 and S2). The placentas were harvested immediately after delivery and included samples of the chorioamniotic membranes (CAM, comprising the amnion, chorion, and decidua parietalis) as well as a biopsy that included basal plate (decidua basalis) and placental villi (BPPV) (Fig. 1A). Collected CAM and BPPV were used to prepare single-cell suspensions followed by single-cell RNA-sequencing (scRNA-seq) and for maternal and fetal genotyping to assign cell type origin during single-cell analyses, as previously described (73) (Fig. 1A). Unsupervised clustering analysis and cell type annotation resulted in the identification of 33 distinct clusters corresponding to immune and non-immune cell types across the CAM and BPPV (Fig. 1, B and C). Non-immune cells included stromal cells (7 clusters), decidual cells (3 clusters), syncytiotrophoblast (STB), cytotrophoblast (CTB, 4 clusters), extravillous trophoblast (EVT, 2 clusters), npiCTB (non-proliferative interstitial cytotrophoblast), a CTB subset within the EVT trajectory (CTB/EVT), Endothelial cells, and Endometrial cells (Fig. 1, B and C). Identified immune cell types were Monocytes, macrophages (4 clusters), innate lymphoid cells (ILCs), lymphatic endothelial decidual (LED) cells, NK cells, T cells (3 clusters), and B cells (Fig. 1, B and C). From this point, capitalized cell names are used to refer to the individual cell clusters that we identified using scRNA-seq. A tally of cell counts for each cluster indicated differences in cell type composition between tissues, with endothelial cells (clusters 14 and 23), stromal cells (clusters 0, 2, 8, 15, and 22), and decidual cell types (clusters 10, 12 and 17) being more abundant in the CAM compared with the BPPV (data file S1). Lymphoid cell composition did not markedly differ between compartments (Fig. 1, B and C and data file S1). By contrast, myeloid cells were more predominant in the BPPV (Fig. 1, B and C and data file S1). Stromal-4 and Stromal-6 (cluster 20 and 27) were primarily present in the BPPV (Fig. 1, B and C and data file S1). As expected, a variety of trophoblast types was more evident in the BPPV except for EVT-1 (cluster 1), which was more abundant in CAM (Fig. 1, B and C and data file S1).
Figure 1. Single-cell atlas of the maternal-fetal interface in parturition.
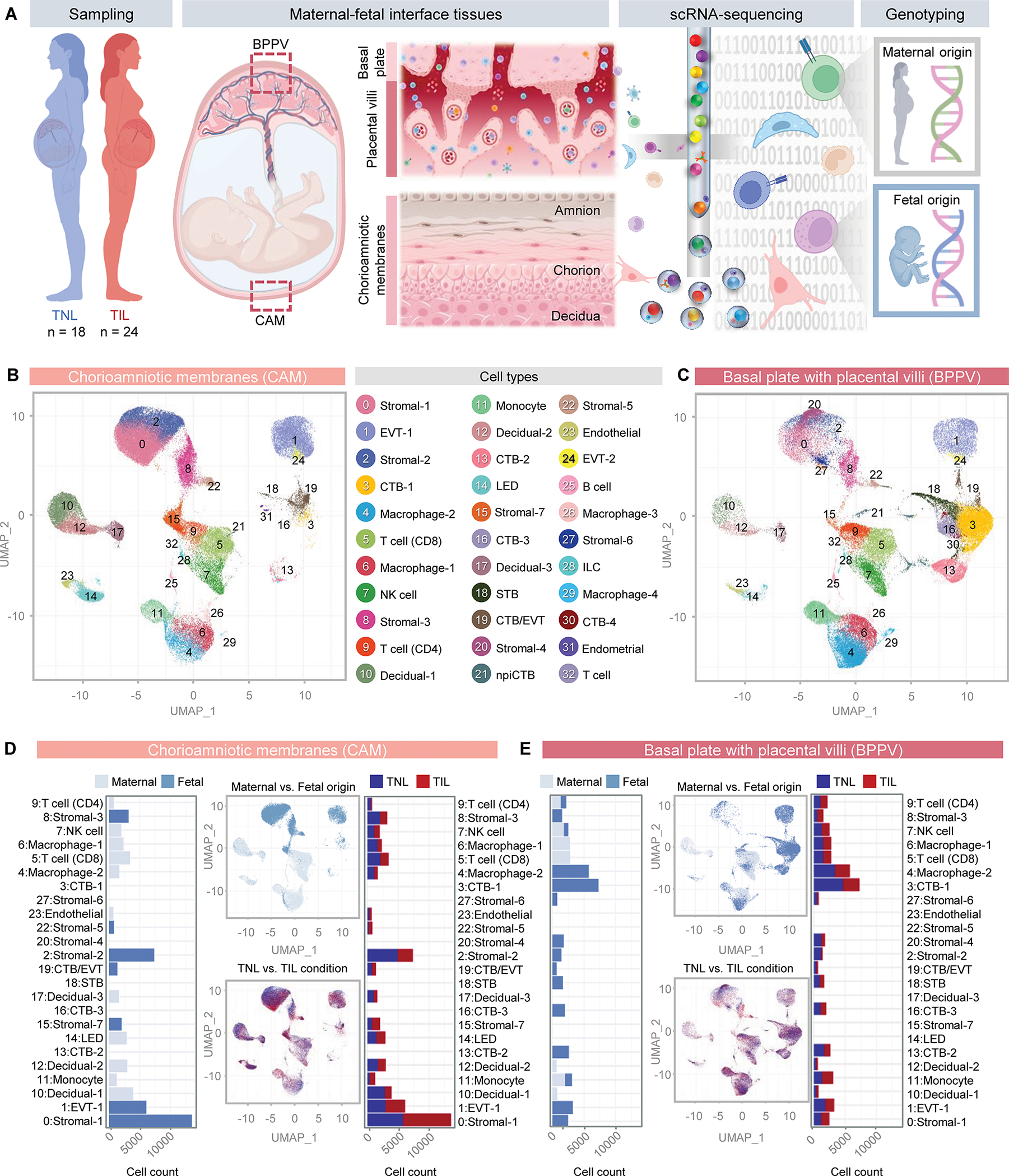
(A) Study design showing the collection of placental tissues from women who delivered at term with (TIL, n = 24) or without (TNL, n = 18) labor. Briefly, chorioamniotic membranes (CAM) and basal plate with placental villi (BPPV) were sampled to generate single-cell suspensions for single-cell RNA-sequencing (scRNA-seq). Genotyping of maternal and neonatal tissues was also performed to allow assignment of maternal (M) or fetal (F) origin to cells. (B and C) Uniform manifold approximation and projection (UMAP) plots show all cell types identified in the (B) CAM and (C) BPPV. (D and E) Bar plots represent the total numbers of each maternal (light blue) or fetal (dark blue) cell type in the (D) CAM and (E) BPPV and the numbers of each cell type with labor (red bars) and without labor (blue bars). Corresponding UMAP plots show the cell clusters in the CAM and BPPV according to maternal (light blue) or fetal (dark blue) origin and how each changes with labor (red clusters) and without labor (blue clusters). Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; ILC, innate lymphoid cell; npiCTB, non-proliferative interstitial cytotrophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell; STB, syncytiotrophoblast.
After identifying the changes in the cell composition of the CAM and BPPV, we then used genotyping information to determine the maternal and/or fetal origin of all identified cells (Fig. 1, D and E). As expected, trophoblast cells in the CAM and BPPV were of fetal origin (Fig. 1, D and E). The majority of stromal cells in the CAM were also fetal (Fig. 1D). Decidual cells were of maternal origin, as expected, whereas endothelial, lymphoid, and myeloid cell types showed mixed maternal and fetal origin across the CAM and BPPV (Fig. 1, D and E). There were no major shifts in cell type abundance with labor, with the only change occurring in the Stromal-5 subset from the BPPV (data file S1 and S2). Together, these data provide a comprehensive single-cell landscape of the human placenta in labor, showing that, in abundance, the most influenced cell types in the CAM are fetal stromal cells, whereas those in the BPPV are the CTB and fetal macrophages.
Maternal and fetal cell types display labor-driven transcriptomic activity
Next, we explored the cell type-specific transcriptomic changes driven by labor in the CAM and BPPV (Fig. 2A). In the CAM, fetal Stromal-1, Stromal-2, and Stromal-3 showed the greatest labor-driven changes in differential gene expression (Fig. 2A). Maternal Decidua-1 cells also showed a considerable number of differentially expressed genes (DEGs); yet this change was moderate compared to those shown by fetal stromal cells (Fig. 2A). Among BPPV cell types, the largest numbers of DEGs were predominantly in the maternal Monocyte cell type followed by fetal Macrophage-2, likely corresponding to infiltrating inflammatory monocytes and Hofbauer cells, respectively (Fig. 2A). Quantile-quantile (Q-Q) plots show the most affected cell types of fetal and maternal origin in the CAM and BPPV (Fig. 2B), which were largely consistent with the cell types displaying the greatest differential gene expression. Notably, fetal stromal cell types were found to be affected by labor in the CAM, as were maternal decidual cells (Fig. 2B). Similarly, maternal Monocyte and fetal Macrophage-2 were represented as most affected by labor in the BPPV (Fig. 2B). Correlation analysis of the effects of labor on gene expression between cell types indicated similarities among maternal myeloid cells, decidual cells, and fetal stromal cells, among others (Fig. 2C).
Figure 2. Parturition induces transcriptomic changes in maternal and fetal cells in the CAM and BPPV.
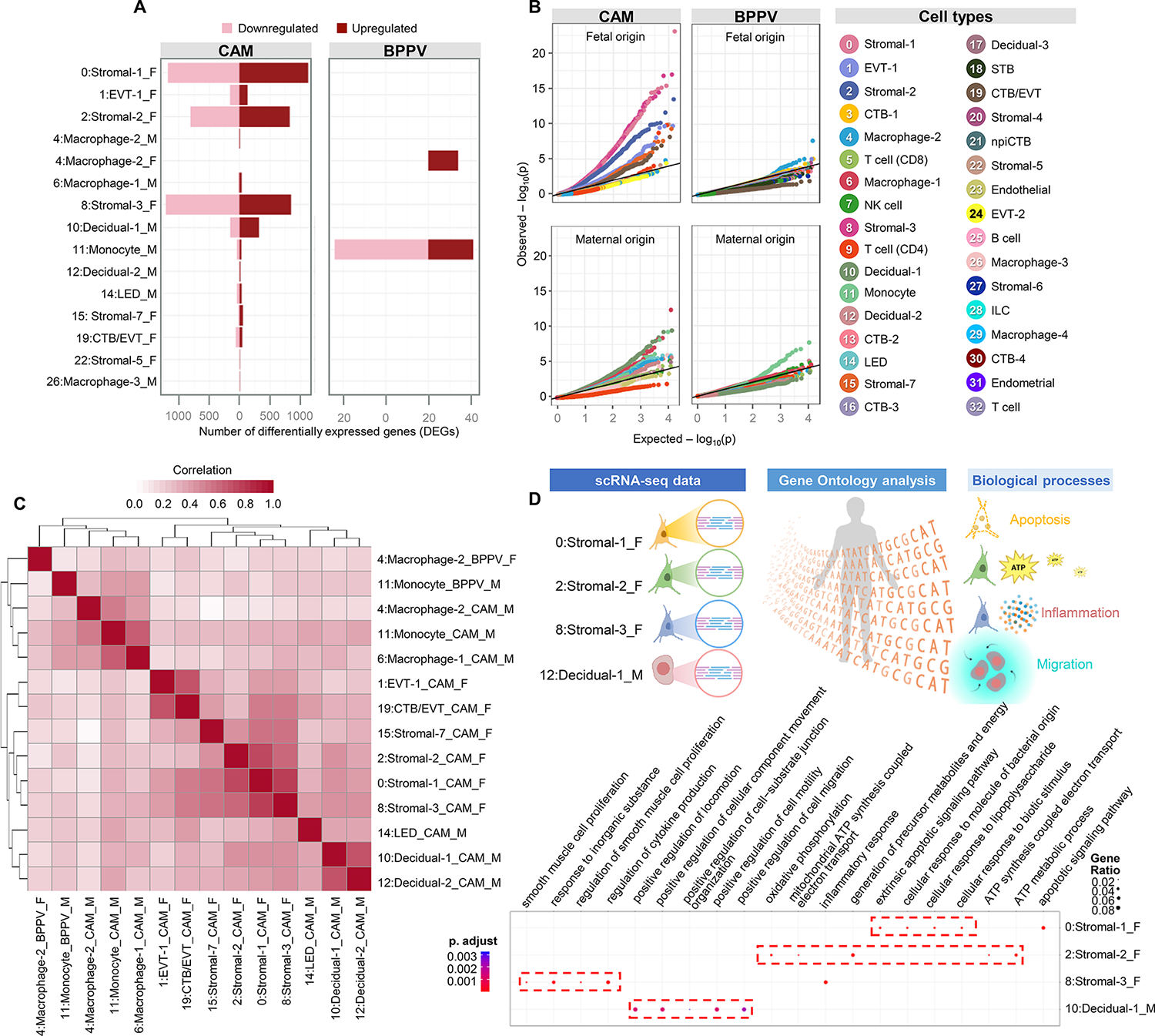
(A) Bar plots show the numbers of differentially expressed genes (DEGs) associated with labor for each cell type in the CAM and BPPV, where red and pink indicate upregulated or downregulated genes, respectively. (B) Quantile-quantile (Q-Q) plots showing the DEGs for selected enriched cell types of maternal or fetal origin from the CAM and BPPV. Deviation above 1:1 (solid black line) indicates enrichment. (C) Heatmap showing the log2(fold change) correlation among cell types of maternal (M) or fetal (F) origin from the CAM and BPPV, where red represents increasing correlation. (D) ClusterProfiler dot plots showing the Gene Ontology (GO) pathways that are enriched with labor in the fetal Stromal-1, Stromal-2, Stromal-3, and maternal Decidual-1 cell types of the CAM and/or BPPV based on over-representation analysis (ORA). The size and color of dots represent enrichment score and significance level, respectively. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; ILC, innate lymphoid cell; npiCTB, non-proliferative interstitial cytotrophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell; STB, syncytiotrophoblast.
We then evaluated the Gene Ontology (GO) biological processes enriched across all cell types in the CAM and BPPV (fig. S1, A and B). In the CAM, enriched processes during labor included those related to cell motility and migration, angiogenesis, and responsiveness (fig. S1A). By contrast, in the BPPV, the biological processes enhanced during labor were related to neutrophil and myeloid cell functions such as degranulation (fig. S1B), suggesting tissue-specific processes during parturition.
We also explored the over-representation analysis in GO biological processes within cell types that displayed the highest numbers of DEGs with labor (Fig. 2A) to infer functionality, a previously utilized approach (66). The fetal Stromal-1 showed enrichment of apoptotic signaling, fetal Stromal-2 was enriched for ATP metabolism, and Stromal-3 for inflammatory response including cytokine production (Fig. 2D), suggesting distinct putative functions of these fetal stromal subsets to the process of labor. Furthermore, maternal Decidual-1 showed enrichment of processes related to cell motility and migration (Fig. 2D), suggesting that decidual cells participate in the leukocyte infiltration that accompanies labor (10, 15, 34, 74–76).
These data provide further insight into the labor-specific transcriptomic activity by maternal and fetal cell types, particularly those undergoing substantial changes in gene expression such as structural cells in the CAM and immune cells in the BPPV. Moreover, we demonstrated cell type-specific enrichment of pathways pertaining to cell migration and proliferation, leukocyte recruitment, and cytokine/chemokine signaling during the process of parturition. The most highly represented cell types, such as trophoblast cells, did not undergo drastic changes in gene expression with labor, pointing to more transcriptionally active subsets, rather than more abundant subsets, as major contributors to labor-associated signaling.
Labor comprises inflammatory pathways orchestrated by maternal-fetal cell-cell crosstalk
Having established the maternal and fetal cell types that are most affected by labor, we then sought to infer the resulting changes in maternal-fetal crosstalk in the CAM (Fig. 3) and BPPV (Fig. 4) using CellChat (77). We first visualized incoming and outgoing interactions for CAM cell types, where the direction and length of each arrow indicate labor-driven predominance of incoming or outgoing interactions and their strength, respectively, as shown in the corresponding legend (Fig. 3A). Fetal stromal cell types showed the strongest increases (represented by the length of the arrow) in both incoming and outgoing interactions, implicating these cells as key participants in labor-associated signaling in the CAM (Fig. 3A). By contrast, maternal T cells, NK cells, and decidual cells showed diminished incoming interactions (Fig. 3A). This interaction strength analysis provided information as to which cells are undergoing changes in outgoing and/or incoming signaling with labor but did not inform us as to the cellular targets or origins of such signaling. Therefore, we next considered each cell type as a signaling sender or receiver and evaluated the interaction strength between cell type pairs, as displayed using the heatmap in Fig. 3B and the network plot in Fig. 3C. The fetal stromal cell types exhibited strong labor-associated interactions with each other as well as non-immune and immune cells as senders or receivers (Fig. 3, B and C). Moreover, multiple stromal cell types displayed self-signaling, suggesting positive feedback loops implicated in labor (Fig. 3C). Consistent with the arrow plot, incoming signaling to maternal CD4 T cells was diminished with labor (Fig. 3B). Given the substantial changes in labor-specific cell-cell signaling in the CAM, we then selected specific signaling pathways affected by labor to infer the top contributing cell types (Fig. 3, D to F and fig. S2, A and B). The collagen signaling pathway showed substantial involvement of fetal stromal and maternal decidual cell types (Fig. 3D), potentially indicating their participation in structural changes in the CAM associated with labor. The C-X-C motif ligand (CXCL) signaling pathway was largely driven by maternal Decidual-3, Macrophage-2, and Macrophage-1 cell types, with many interactions being directed toward the fetal EVT-1 subset as well as other fetal and maternal cell types (Fig. 3E), indicating maternal-fetal crosstalk between maternal decidual cells and fetal trophoblasts. Maternal Monocyte and Macrophage-1 cell types drove TNF signaling that likely contributed to labor-associated inflammation (Fig. 3F). The IL-1 signaling pathway was primarily driven by maternal Monocyte and Macrophage-1 cell types, which displayed outgoing signaling to fetal stromal cell types and decidual/CTB/EVT, respectively (fig. S2A). Both maternal (Monocyte) and fetal (Stromal-1, Stromal-2) cell types contributed to the IL-6 signaling pathway in the CAM, with maternal macrophages being the top recipients of these interactions (fig. S2B).
Figure 3. Maternal-fetal crosstalk in the CAM during parturition.
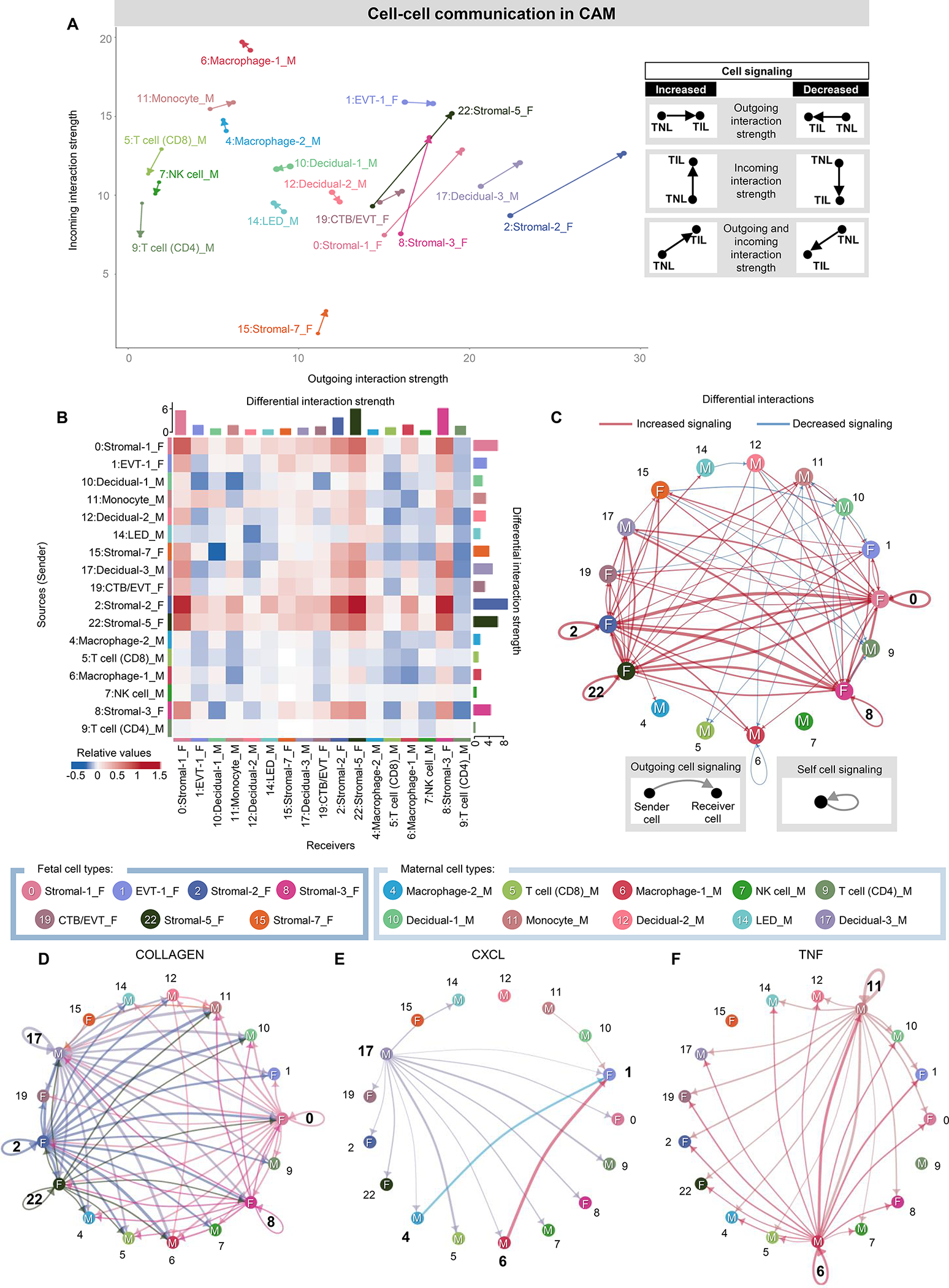
(A) Arrow plot showing changes in outgoing and incoming interaction strength between the labor (point of the arrow) and no labor (base of the arrow) conditions for specific maternal or fetal cell types in the CAM. F, fetal origin. M, maternal origin. (B) Heatmap showing the differential interaction strength among cell types (as senders or receivers) in the CAM with labor. Red and blue shading indicate increased or decreased signaling, respectively, in labor compared to no labor. (C) Circle plot showing the top 25% increased (red) or decreased (blue) signaling interactions in the CAM with labor compared to no labor among maternal and fetal cells. (D to F) Circle plots representing the top 25% aggregated interactions between maternal and fetal cell types in the CAM for the (D) Collagen, (E) CXCL, and (F) TNF signaling pathways in labor. Cell cluster numbers in bold indicate top participants in labor or in each signaling pathway. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell.
Figure 4. Maternal-fetal crosstalk in the BPPV during parturition.
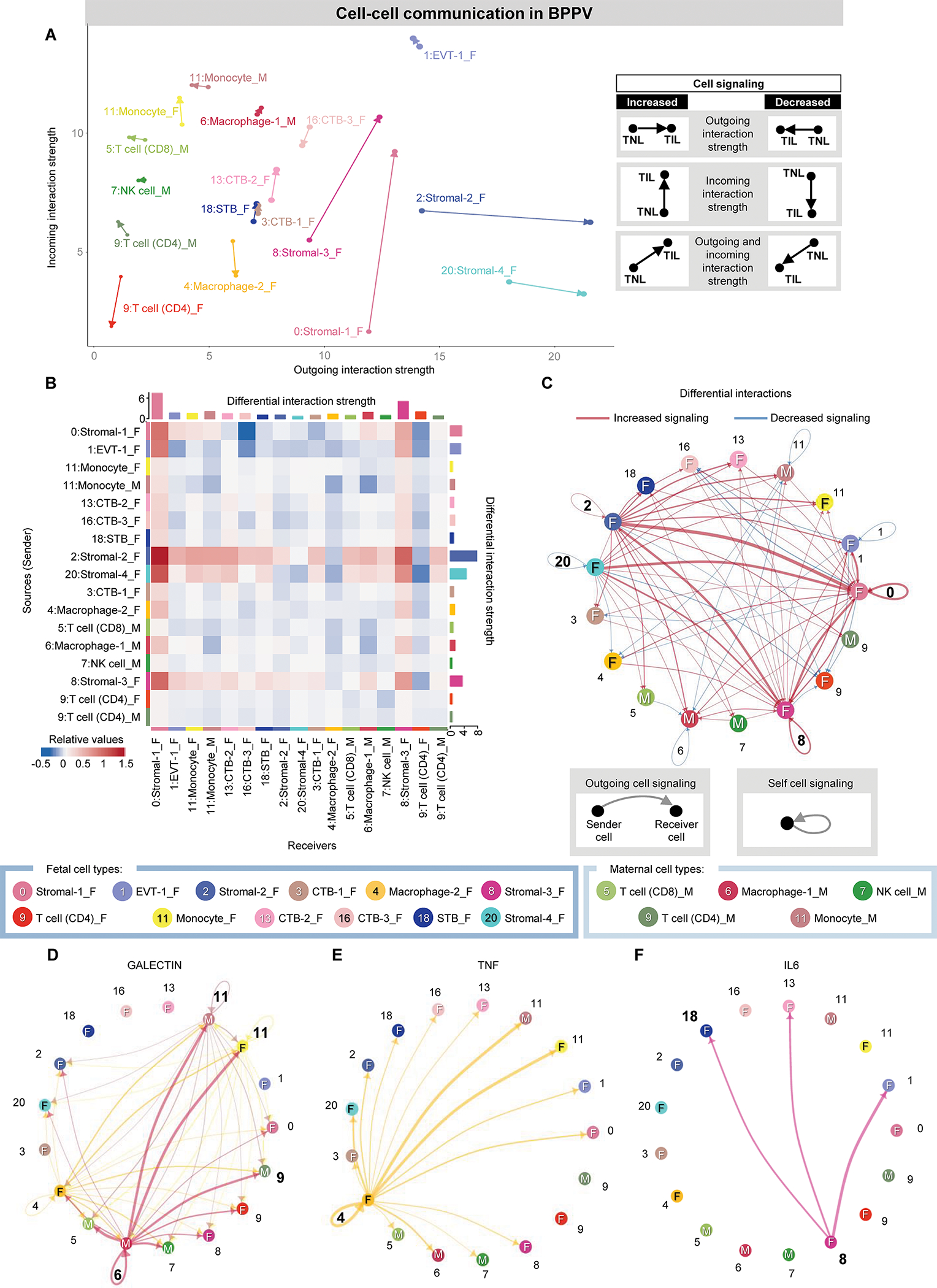
(A) Arrow plot showing changes in outgoing and incoming interaction strength between the labor (point of the arrow) and no labor (base of the arrow) conditions for specific maternal or fetal cell types in the BPPV. F, fetal origin. M, maternal origin. (B) Heatmap showing the differential interaction strength among cell types (as senders or receivers) in the BPPV with labor. Red and blue shading indicate increased or decreased signaling, respectively, in labor compared to no labor. (C) Circle plot showing the top 25% increased (red) or decreased (blue) signaling interactions in the BPPV with labor compared to no labor among maternal and fetal cells. (D to F) Circle plots representing the top 25% aggregated interactions between maternal and fetal cell types in the BPPV for the (D) Galectin, (E) TNF, and (F) IL6 signaling pathways in labor. Cell cluster numbers in bold indicate top participants in labor or in each signaling pathway. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; NK cell, natural killer cell.
In the BPPV, the strongest labor-associated changes in cell-cell interactions were again observed in fetal stromal cell types, with Stromal-1/Stromal-3 showing predominantly increased incoming interaction strength and Stromal-2/Stromal-4 showing predominantly increased outgoing interaction strength (Fig. 4A). By contrast, the fetal CD4 T cell, Macrophage-2, and CTB-3 subsets showed decreased incoming interaction strength with labor (Fig. 4A); these results were reflected in the heat map by the fetal Stromal-2 subset showing increased outgoing interaction strength with immune cells as receivers (Fig. 4B and were in line with the prominent differential gene expression in fetal Macrophages and maternal Monocytes in the BPPV (Fig. 2A) and the enriched biological processes observed in the BPPV (fig. S1B), which were primarily associated with inflammatory responses.
Consistent with the arrow plot, the network plot indicated that fetal stromal cell types displayed stronger incoming or outgoing interactions (Fig. 4C). Top cell-cell signaling pathways affected by labor included Galectin, Tumor Necrosis Factor (TNF), Interleukin (IL)-6, C-C motif ligand (CCL), CD45, Type II Interferon (IFN-II), and Complement, as represented by network plots (Fig. 4, D to F and fig. S2, C to F). A key participant in the Galectin pathway was maternal Macrophage-1, which showed interaction with maternal and fetal monocytes as well as maternal T cells (Fig. 4D). By contrast, the top contributors to TNF and IL-6 signaling in the BPPV were fetal Macrophage-2 and Stromal-3, respectively (Fig. 4, E and F), further indicating a strong fetal contribution to cell-cell interactions associated with labor. The CCL and CD45 pathways showed similar involvement of maternal and fetal myeloid cell types as well as the EVT-1 subset (fig. S2, C and D). The IFN-II pathway was dominated by maternal CD8 T cell-derived signaling directed at maternal Monocyte and fetal EVT-1 (fig. S2E). Last, the Complement pathway involved maternal Macrophage-1 and NK cell together with fetal Stromal-3, with signaling directed at fetal and maternal myeloid cells (fig. S2F). Taken together, these cellular interaction analyses indicated that labor is characterized by maternal-fetal signaling involving inflammatory pathways that are orchestrated by cell-cell crosstalk between immune and non-immune cell types.
Single-cell maternal and fetal signatures derived from the placental tissues can be monitored in the maternal circulation throughout gestation and during labor
We next evaluated whether cell type-specific transcriptomic activity could be tracked in the maternal circulation throughout gestation. We first utilized a previously generated bulk transcriptomic dataset from the peripheral blood (leukocytes) of pregnant women (n = 49) collected throughout gestation (78) and evaluated the trajectories of cell type-specific transcripts (Fig. 5A). Multiple cell type signatures underwent modulation with gestational age; namely, Macrophage-1, Macrophage-2, Macrophage-3, Macrophage-4, CD4 T cell, CD8 T cell, B cell, Stromal-2, Stromal-3, Stromal-4, Stromal-5, Stromal-7, Decidual-1, Endothelial, LED, npiCTB, EVT-2, and STB (Fig. 5B). The observed patterns of change were inconsistent across individual clusters of specific cell types; for example, whereas Macrophage-1 steadily increased with gestational age, Macrophage-2 and Macrophage-3 decreased and Macrophage-4 followed a parabolic trajectory (Fig. 5B), hinting at the differing roles of each cell type cluster. To further refine this analysis in the context of labor, we considered the top 20 upregulated labor-associated DEGs from each placental cell type and tracked these labor-specific signatures in the maternal peripheral blood using the same bulk transcriptomic dataset (fig. S3). We found that labor-specific signatures corresponding to Macrophage-1, Stromal-3, CD4 T cell, Decidual-1, Monocyte, Decidual-2, and Stromal-7 were modulated with gestational age, with the sharpest increase of labor-associated genes occurring in CD4 T cell (fig. S3).
Figure 5. Placental single-cell signatures can be monitored in the maternal circulation during pregnancy.
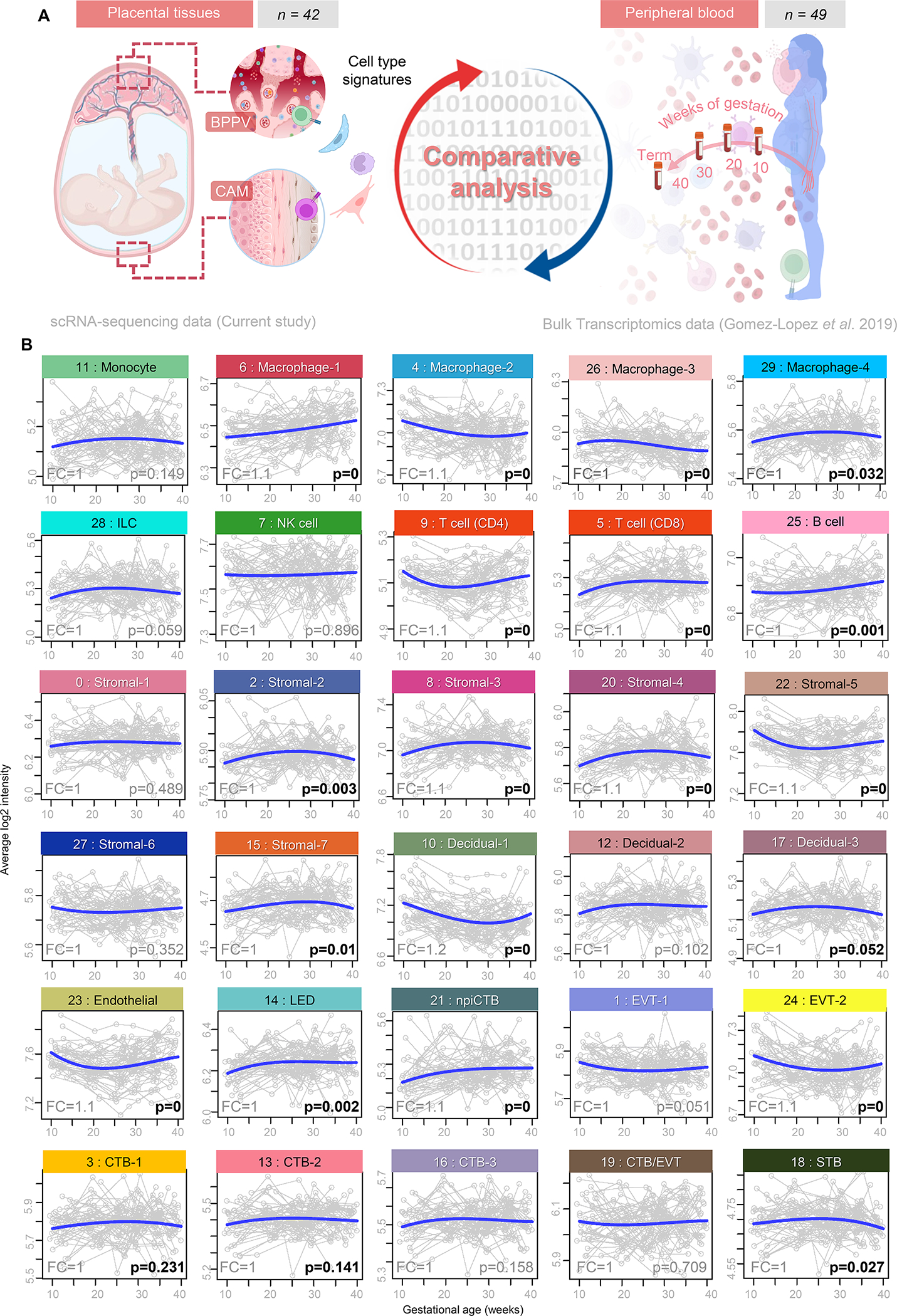
(A) Experimental design for the comparative analysis between cell type-associated signatures from the CAM and BPPV obtained using scRNA-seq (current study) and bulk transcriptomic data from maternal peripheral blood collected throughout normal pregnancy from women who delivered at term (n = 49) (78). (B) Signature analysis plots based on the top 20 placental cell type markers showing changes in the average expression (blue line) in the maternal peripheral blood throughout gestation. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; ILC, innate lymphoid cell; npiCTB, non-proliferative interstitial cytotrophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell; STB, syncytiotrophoblast. Significant P values are shown in bold.
Given that cell type signatures could be tracked in the maternal peripheral blood, we then assessed differences between samples collected at term from women who underwent term labor (n = 21) or who delivered at term without labor (n = 28) in the same set of patients for whom the longitudinal analysis was performed (79) (Fig. 6A). To compare these datasets, CAM scRNA-seq-derived DEGs were used to create a pseudobulk dataset that could be directly contrasted with the bulk transcriptomic dataset while still considering the maternal or fetal origin of each cell type (Fig. 6A). Correlation of cell type-specific transcripts between the CAM and the maternal peripheral blood indicated positive correlations for fetal Stromal-2 as well as maternal Macrophage-1, Macrophage-2, Decidual-1, and Monocyte (Fig. 6B). By contrast, several cell type signatures showed negative correlation between the CAM and maternal peripheral blood: fetal EVT-2, maternal CD8 T cell, and maternal Endothelial (Fig. 6B). When using cell type signatures derived from the BPPV, we noted additional correlations between this tissue and the maternal peripheral blood (Fig. 6C). Indeed, fetal EVT-1, CTB-1, Macrophage-2, CD4 T cell, CTB-3, STB, and npiCTB all showed positive correlation between compartments, as did maternal Macrophage-1, Macrophage-2, Monocyte, and Decidual-2 (Fig. 6C). The signatures of fetal Stromal-1 as well as fetal and maternal NK cell were negatively correlated between the BPPV and maternal peripheral blood (Fig. 6C).
Figure 6. Single-cell signatures derived from the placental tissues can be monitored in the maternal circulation during labor.
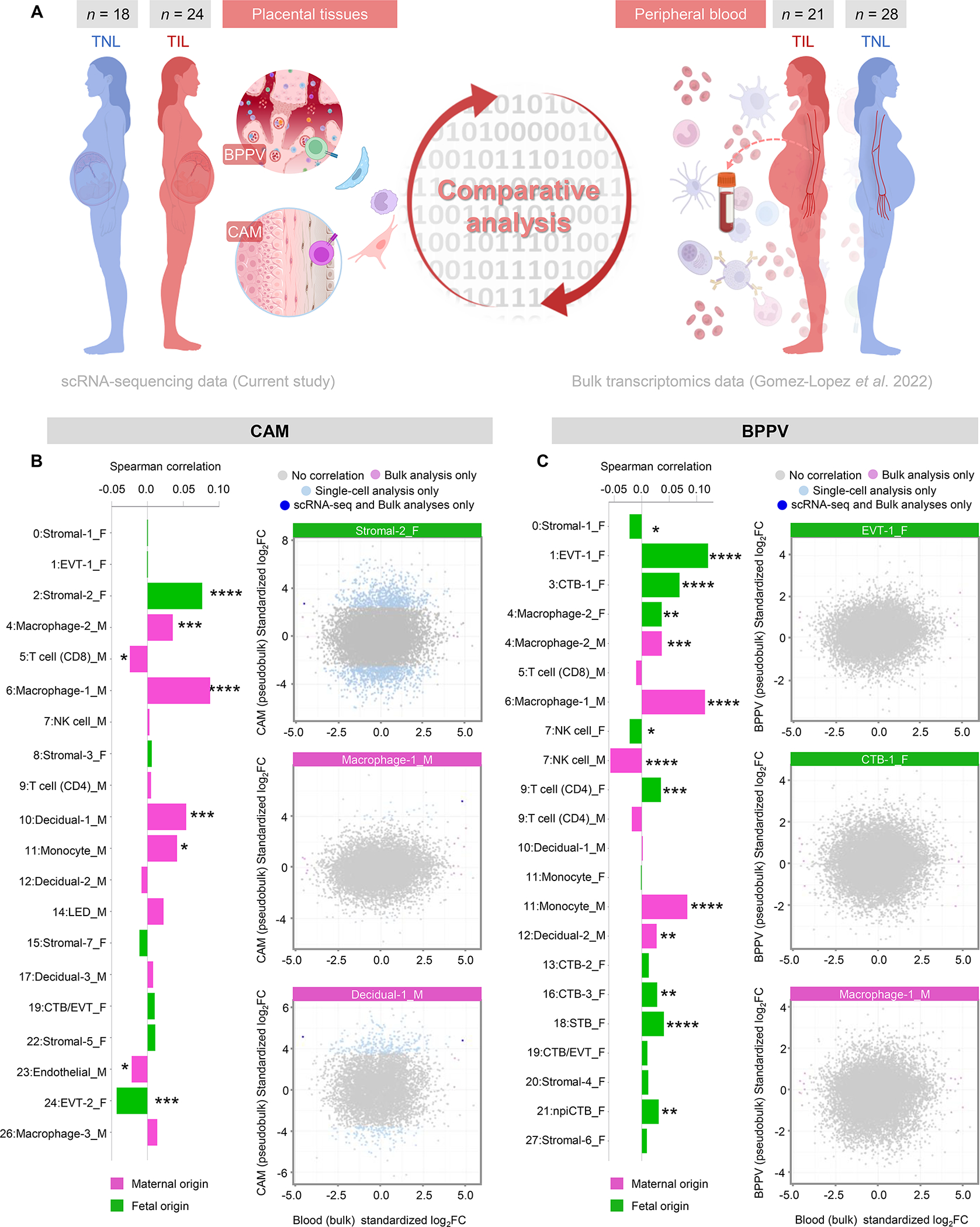
(A) Experimental design for the comparative analysis of labor-associated gene expression changes in maternal (M) and fetal (F) cell types between the placental tissues of women that delivered at term obtained with scRNA-seq (current study) and bulk transcriptomic data of the maternal peripheral blood from women with (n = 21) and without (n = 28) term labor (79). (B) Bar plots showing the correlation between the log2(Fold change) gene expression from CAM single-cell and maternal peripheral blood bulk transcriptomic data (significant with q < 0.1) using the Spearman correlation test. Pink indicates maternal origin, and green indicates fetal origin. Scatter plots showing the log2(Fold change) gene expression associated with labor in bulk microarray (x-axis) and single-cell (y-axis) analyses in the Stromal-2, Macrophage-1 and Decidual-1 cell types from the CAM. Differentially expressed genes (DEGs) shown were obtained using only single-cell analysis (light blue), only bulk analyses (lavender), were shared by both single-cell and bulk analyses (dark blue) or were not differentially expressed between the two contrasts (gray). (C) Bar plots showing the correlation between the log2(Fold change) gene expression from BPPV single-cell and maternal peripheral blood bulk transcriptomic data (significant with q < 0.1) using the Spearman correlation test. Pink indicates maternal origin, and green indicates fetal origin. Scatter plots showing the log2(Fold change) gene expression associated with labor in bulk microarray (x-axis) and single-cell (y-axis) analyses in the EVT-1, CTB-1 and Macrophage-1 cell types from the BPPV. DEGs shown were obtained using only single-cell analysis (light blue), only bulk analyses (lavender), were shared by both single-cell and bulk analyses (dark blue) or were not differentially expressed between the two contrasts (gray). Correlations between data sets were determined using two-sided Spearman’s correlation tests. P values are considered significant when P < 0.05. NS = not significant, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; npiCTB, non-proliferative interstitial cytotrophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell; STB, syncytiotrophoblast.
This comparative analysis supports the concept that placental cells are shed into the maternal circulation (80–82) and/or that labor-associated signaling drives transcriptomic changes in peripheral leukocytes that are at least partially shared with those in the placental tissues. Our data further suggest that maternal and fetal signaling can be non-invasively tracked for monitoring normal gestation and parturition and may potentially serve to generate biomarkers for obstetrical disease.
Term labor-associated maternal-fetal single-cell signatures in the CAM partially overlap with those from women with spontaneous preterm labor
Our results indicated that the CAM seem to undergo more drastic cell type-specific changes in transcriptomic activity with labor (Fig. 2A), which is consistent with the involvement of these tissues in the common pathway of labor (1, 3–5, 83). Although considered a pathological process distinct from term labor, spontaneous preterm labor also includes activation of the common pathway of parturition (3, 4, 83). Therefore, we next evaluated whether the term labor-associated gene expression changes in the CAM were also observed in the tissues of women who underwent spontaneous preterm labor and birth (term labor vs. PTL). For this purpose, we utilized a previous bulk transcriptomic dataset of the CAM (84) from women who delivered preterm and underwent spontaneous preterm labor with differing inflammatory and microbiological status of the amniotic cavity as diagnosed by amniocentesis: without intra-amniotic inflammation [low concentrations of IL-6 (<2.6 ng/mL) without detectable microbes via cultivation and molecular microbiological methods] (PTL, n = 12); with sterile intra-amniotic inflammation [elevated concentrations of IL-6 (≥2.6 ng/mL) without detectable microbes] (PTL+SIAI, n = 17); or with intra-amniotic infection (elevated concentrations of IL-6 with detectable microbes) (PTL+IAI, n = 11) (85–92) (Fig. 7A). To obtain the cell type-specific PTL-associated changes, the bulk transcriptomic profiles of the CAM from each group were compared to the control group [women who experienced an episode of spontaneous preterm labor but delivered at term (n = 15)] (Fig. 7A). Term labor-associated signatures were positively correlated with PTL signatures from fetal Stromal-1, Stromal-3, Stromal-5, Stromal-7, EVT-1, and CTB/EVT in the CAM (Fig. 7B). Modest positive correlations of signatures corresponding to maternal decidual cell types and macrophages were also observed between term labor and PTL (Fig. 7B). Only specific signatures were positively correlated between term labor and PTL+SIAI; namely, EVT-1, CTB/EVT, and stromal cell types, whereas the rest showed only mild or minimal positive correlations (Fig. 7B). Moreover, the term labor vs. PTL+IAI comparison showed substantial correlation of specific signatures, particularly fetal EVT-1 and CTB/EVT, as well as mild correlation of myeloid signatures such as maternal macrophages and fetal stromal cells (Fig. 7B). Thus, our results indicated that maternal-fetal signaling in the CAM during the physiologic process of term labor overlaps with that observed during spontaneous preterm labor; yet the inflammatory and microbiological status of the amniotic cavity drives specific transcriptomic activity in maternal-fetal cell types.
Figure 7. Term labor-associated single-cell signatures in the CAM partially overlap with those from women with spontaneous preterm labor and birth.
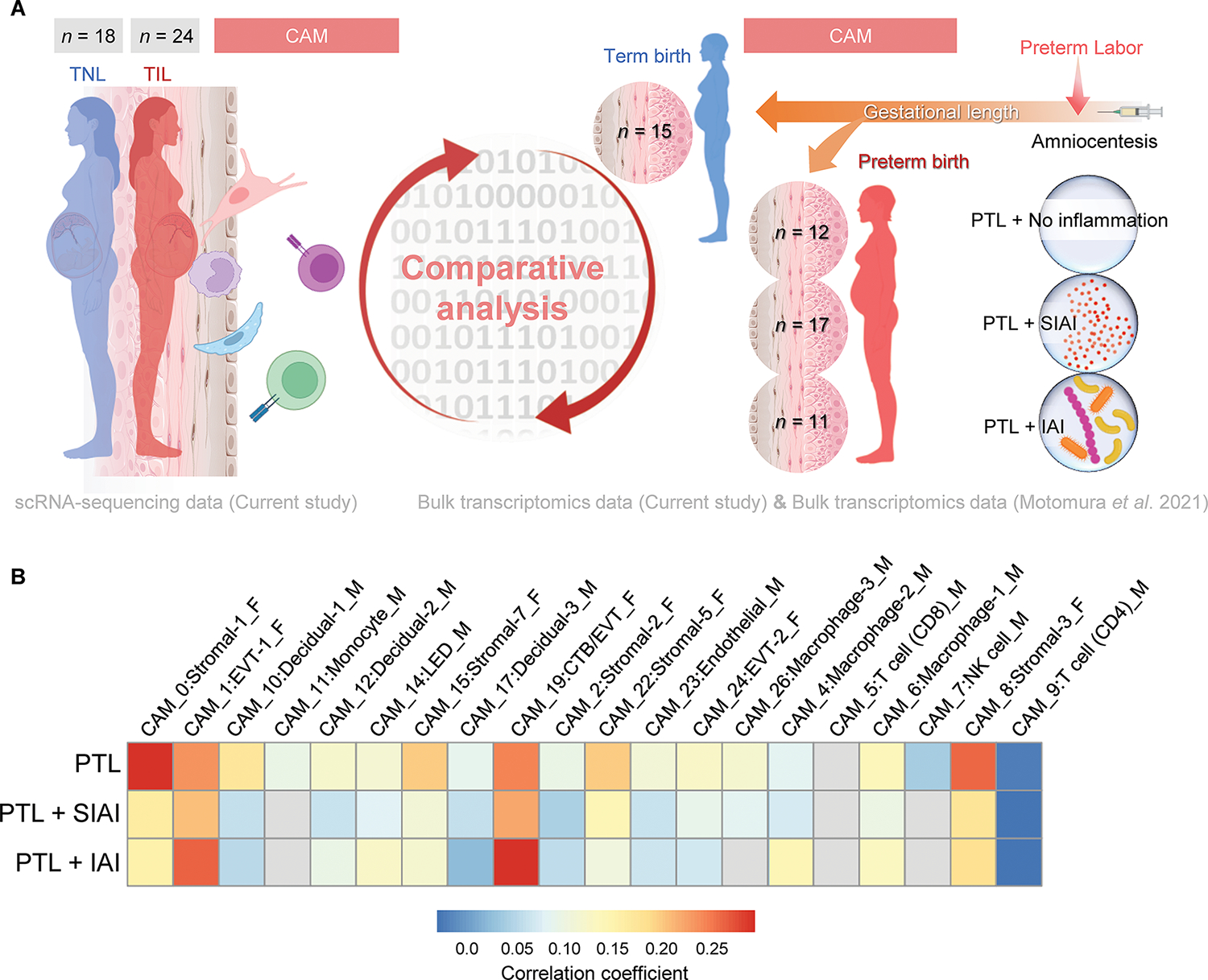
(A) Experimental design for the comparative analysis of term labor-associated gene expression changes in maternal (M) and fetal (F) cell types from the CAM (scRNA-seq, current study) and bulk transcriptomic changes in preterm labor (PTL) of the CAM from women with PTL that delivered at term (n = 15); PTL that delivered preterm without intra-amniotic inflammation (PTL+No Inflammation) (n = 12); PTL with sterile intra-amniotic inflammation (PTL + SIAI) that delivered preterm (n = 17); and PTL with intra-amniotic infection (PTL + IAI) that delivered preterm (n = 11) (84). (B) Heatmap showing the correlations of gene expression between term labor (current study) and PTL, PTL + SIAI, or PTL + IAI, where red and blue indicate increased and decreased correlation, respectively. Gray indicates correlations that are not significantly different from 0. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell.
Labor-associated single-cell CAM-derived signatures can serve as non-invasive biomarkers to identify women who are destined to undergo spontaneous preterm birth
We last sought to determine whether labor-specific CAM-derived scRNA-seq signatures could be tracked during the second and third trimester in the peripheral blood of women who ultimately underwent preterm labor, which could potentially allow for the generation of predictive biomarkers for impending preterm birth. First, we used a previous bulk transcriptomic dataset generated from a cross-sectional study including samples from the peripheral blood of: i) women who experienced an episode of spontaneous preterm labor and delivered preterm (cases); and ii) gestational age-matched controls who delivered at term (93) (fig. S4A). Principal component (PC) analysis of the bulk transcriptome demonstrated modest separation of cases (red dots) from controls (blue dots), which seemed to be driven by PC2 for at least a subset of the cases (fig. S4B). We then overlaid the labor-specific CAM-derived scRNA-seq signatures with the maternal blood transcriptome and evaluated the contribution and correlation of each cell type to PC1 and PC2. All cell types contributed to PC1; Monocytes, LEDs, and stromal cell types showed positive alignment with PC2, whereas NK cell, CD8 T cell, and Macrophage-3 showed a negative alignment with PC2 (fig. S4C). By establishing a receiver operating characteristic (ROC) curve for individual single-cell signatures, we found that some of these (such as maternal Decidual-2) showed value [area-under-the-curve (AUC) = 0.757] for identifying women with preterm labor who ultimately underwent preterm birth (fig. S4D). Thus, labor-specific CAM-derived scRNA-seq signatures were detectable in the maternal circulation, some of which were positively modulated in women with spontaneous preterm labor and birth.
Next, we sought to utilize our labor-specific CAM-derived scRNA-seq signatures in maternal blood samples collected during the second and third trimester (before the clinical presentation of an episode of preterm labor) to predict two distinct subsets of spontaneous preterm birth [spontaneous preterm labor with intact membranes (sPTL) and preterm prelabor rupture of membranes (PPROM)]. We utilized previously generated datasets from a prospective bulk transcriptomic study including two external validation cohorts of the maternal peripheral blood collected from 17 to 33 weeks of gestation from women who ultimately underwent sPTL or PPROM (94) (Fig. 8A). sPTL- and PPROM-associated transcriptomic activity was obtained by comparing the gene expression profiles between blood samples collected from women who ultimately underwent preterm birth and those from gestational-age matched controls who delivered at term. Such sPTL- and PPROM-associated transcriptomic activity was then correlated to the labor-specific CAM-derived scRNA-seq signatures generated in the study herein (Fig. 8A). Several labor-specific CAM-derived scRNA-seq signatures were strongly detected in the circulation of women who ultimately underwent sPTL or PPROM (Fig. 8B). Multiple labor-specific CAM-derived scRNA-seq signatures of maternal and fetal origin — most prominently maternal Decidual-1 and fetal Stromal cell types — showed strong positive correlations (>0.10 correlation coefficient) with PPROM-associated transcriptomic activity, with the correlation strength varying according to the sampling time point (Fig. 8B). Moderate positive correlations (>0.05) were also observed for maternal macrophage cell types and other fetal non-immune cell types (Fig. 8B). The intensity of the correlations between term labor- and sPTL-associated transcriptomic activities was less than that observed for PPROM; yet the main contributors were also maternal Decidual-1 and fetal Stromal cell types (Fig. 8B). Labor-specific CAM-derived scRNA-seq signatures from maternal Macrophage-2 and fetal EVT-1 were negatively correlated with PPROM- and sPTL-associated transcriptomic activity (Fig. 8B).
Figure 8. Labor-associated single-cell signatures as non-invasive biomarkers of spontaneous preterm birth in two external validation cohorts.
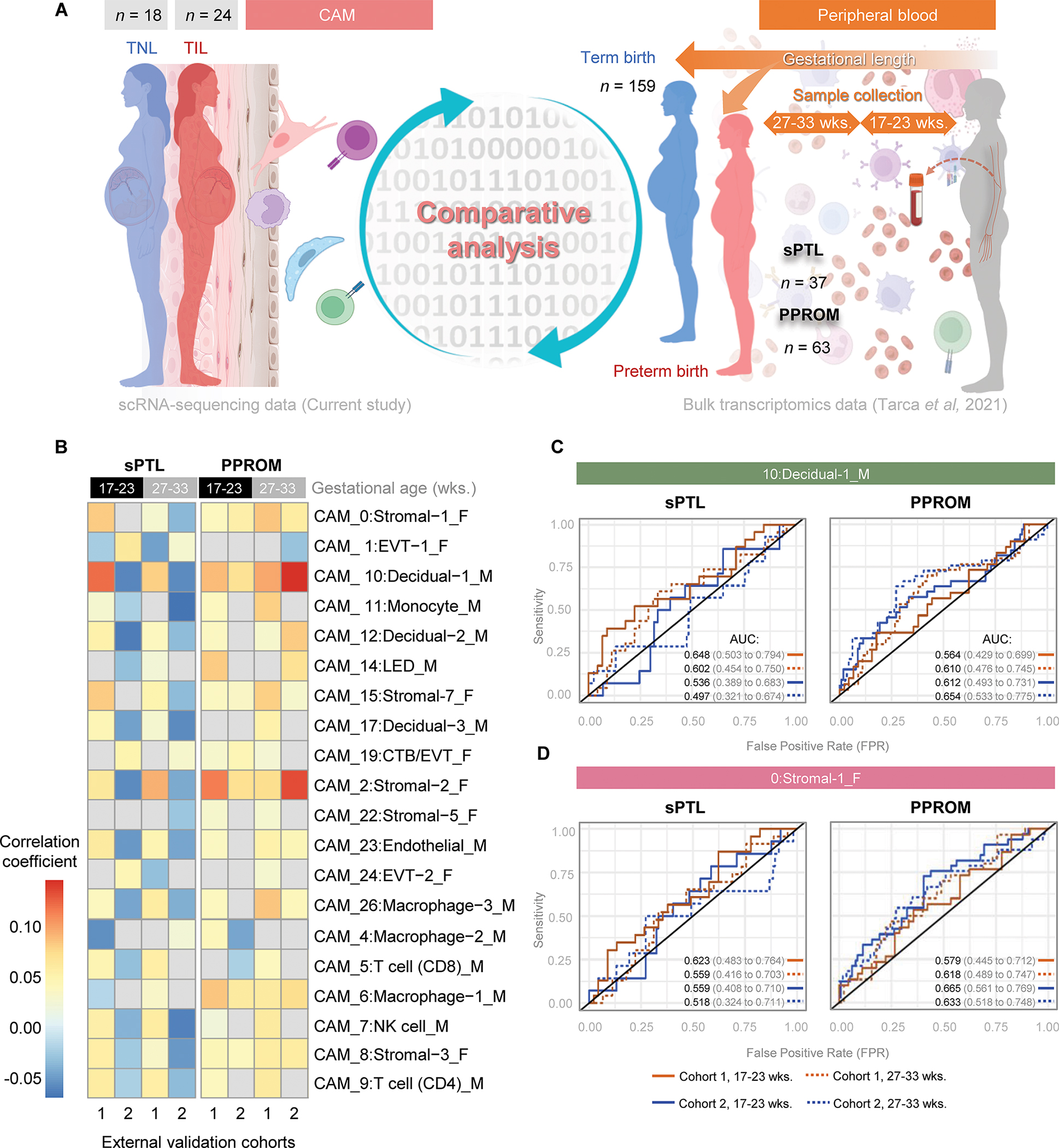
(A) Experimental design for the comparative analysis of term labor-associated gene expression changes in maternal and fetal cell types from the CAM (scRNA-seq, current study) and bulk transcriptomic changes in the peripheral blood of women who ultimately underwent premature prelabor rupture of membranes (PPROM) (n = 63) or spontaneous preterm labor (sPTL) (n = 37), and controls that delivered at term (n = 159). Maternal peripheral blood was collected at two points before delivery: at 17 – 23 weeks of gestation and at 23 – 33 weeks of gestation (94). (B) Heatmap showing the correlation of gene expression according to cell types of fetal (F) or maternal (M) origin between the term CAM and the preterm maternal peripheral blood sampled at 17 – 33 or 27 – 33 weeks of gestation. Red and blue indicate increased and decreased correlation, respectively. Gray indicates correlations that are not significantly different from 0. (C and D) Receiver operating characteristic (ROC) curves showing the value of the (C) maternal Decidual-1 and (D) fetal Stromal-1 single-cell signatures for predicting sPTL or PPROM using the bulk transcriptomes of maternal peripheral blood in two external validation cohorts. Orange ROC lines indicate Cohort 1 and blue ROC lines indicate Cohort 2. Solid and dotted ROC lines represent maternal blood samples collected at 17 – 23 or 27 – 33 weeks of gestation, respectively. Area Under the Curve (AUC) values of the ROC are reported next to its 95% confidence interval in parentheses. A lower bound of the AUC statistics >0.5 represents a significant prediction. Abbreviations used: CTB, cytotrophoblast; EVT, extravillous trophoblast; LED, lymphoid endothelial decidual cell; NK cell, natural killer cell.
We then generated ROC curves for each validation cohort and according to gestational age at sampling to determine how well these signatures could predict sPTL or PPROM (data file S3). For the prediction of impending sPTL, the maternal Decidual-1 signature showed greater value when using samples from Cohort 1 compared to Cohort 2, independent of whether these patients were sampled at 17 – 23 [Cohort 1: AUC 0.648 (0.503–0.794) vs. Cohort 2: AUC 0.536 (0.389–0.683)] or 27 – 33 [Cohort 1: AUC 0.602 (0.454–0.750) vs. Cohort 2: AUC 0.497 (0.321–0.674)] weeks of gestation (Fig. 8C). By contrast, for PPROM the maternal Decidual-1 signature showed a slightly improved predictive value in Cohort 2 at 17 – 23 [Cohort 2: AUC 0.612 (0.493–0.731) vs. Cohort 1: AUC 0.564 (0.429–0.699)] and 27 – 33 [Cohort 2: AUC 0.654 (0.533–0.775) vs. Cohort 1: AUC 0.610 (0.476–0.745)] weeks of gestation (Fig. 8C). Such differences in performance between cohorts may be due to the differing populations included (Cohort 1 was derived from Detroit, Michigan, USA and Cohort 2 was derived from Calgary, Alberta, Canada). The predictive value of the maternal Decidual-1 signature for PPROM was slightly increased at later gestational ages [Cohort 1, 27 – 33 weeks: AUC 0.610 (0.476–0.745) vs. Cohort 1, 17 – 23 weeks: AUC 0.564 (0.429–0.699); Cohort 2, 27 – 33 weeks: AUC 0.654 (0.533–0.775) vs. Cohort 2, 17 – 23 weeks: AUC 0.612 (0.493–0.731)], indicating that a diminished time between sampling and outcome improved prediction. The fetal Stromal-1 signature displayed a more distinct pattern for the prediction of sPTL than for PPROM; yet its predictive performance was improved at 17 – 23 weeks compared to 27 – 33 weeks for sPTL [Cohort 1, 17 – 23 weeks: AUC 0.623 (0.483–0.764) vs. Cohort 1, 27 – 33 weeks: AUC 0.559 (0.416–0.703); Cohort 2, 17 – 23 weeks: AUC 0.559 (0.408–0.710) vs. Cohort 2, 27 – 33 weeks: AUC 0.518 (0.324–0.711)] (Fig. 8D). For PPROM, the predictive performance of the fetal Stromal-1 signature was improved for the Cohort 2 at both sampling times [Cohort 2, 17 – 23 weeks: AUC 0.665 (0.561–0.769) vs. Cohort 1, 17 – 23 weeks: AUC 0.579 (0.445–0.712); Cohort 2, 27 – 33 weeks: AUC 0.633 (0.518–0.748) vs. Cohort 1, 27 – 33 weeks: AUC 0.618 (0.489–0.747)] (Fig. 8D and data file S3). This proof-of-concept analysis suggests that the tracking of relevant single-cell signatures in the maternal circulation may provide a means of predicting impending sPTL or PPROM.
DISCUSSION
Here, we applied scRNA-seq to show that the basal plate, placental villous tree, and chorioamniotic membranes includes immune and non-immune cell types of maternal and fetal origin yet displays distinct cellular composition. Previous single-cell surveys of the placental tissues provided an overview of their cellular composition in early (95, 96), mid (97), and late gestation (65) and allowed for the identification of previously undescribed cell types (65). Our data further emphasize the differences in cell type composition and relative abundance between the placental (placental villous tree and basal plate) and extraplacental (chorioamniotic membranes) tissues driven by their differing functions. Indeed, comparative analysis of the villous (placental) chorion and smooth (extraplacental) chorion in mid-gestation indicated the dissimilar abundance of specific cell types, such as stromal cells, as well as spatially dependent functional differences between similar cell clusters (97). Single-cell exploration during different periods of pregnancy has made it clear that the cellular composition of these compartments is not static but rather undergoes dynamic changes throughout gestation.
Here, we showed that the chorioamniotic membranes displayed a greater number of cell types highly modulated with labor compared to the placental tissues, which is consistent with the participation of membrane activation as an essential component of the common pathway of parturition (1, 4, 5). The clusters most prominently influenced by labor in the chorioamniotic membranes were fetal stromal cells, maternal decidual cells, and maternal macrophages. The participation of decidual cells (65, 98–103) and maternal macrophages (65, 76, 104–107) in the process of labor has been investigated; however, a role for fetal stromal cells in parturition is not well established. Previous studies have explored the utility of mesenchymal stromal cells (MSC) derived from the chorioamniotic membranes for cellular therapy applications (108, 109). Stromal cells derived from the amnion and chorion layers display distinct transcriptomic profiles with enriched pathways suggestive of differing functionality (110), further highlighting the compartmentalization and heterogeneity of the chorioamniotic membranes. Mesenchymal stromal cells could contribute to the maintenance of maternal-fetal tolerance by multiple mechanisms (111) such as suppression of T-cell responses through cell-cell contact and the release of IL-10, VEGF, and PGE2 (112, 113). Moreover, a comparison of amnion MSC from term and preterm labor cases noted differential expression of genes enriched for inflammatory pathways in preterm labor, suggesting active participation in this pathologic process (114). A recent single-cell investigation of the chorioamniotic membranes in spontaneous term labor identified a large subset of amnion-derived fibroblasts (115), which likely included mesenchymal stromal cells (116, 117), and showed that amnion fibroblast cells collected proximal to the rupture zone from spontaneous term labor cases displayed strong upregulation of genes associated with inflammation compared to non-labor controls (115). Together, these data point to an underappreciated and highly active role for fetal stromal cells in the process of labor that warrants further investigation.
Here, we reported that the cell types most impacted by labor in the placental tissues are fetal macrophages and maternal monocytes. Placental tissues include a substantial fraction of fetus-derived macrophages or Hofbauer cells (118, 119). Current evidence suggests that such cells predominantly display an M2-like phenotype, similar to their maternal counterparts (107), which hints at their homeostatic role in normal gestation (120). The engagement of fetal macrophages in inflammatory signaling pathways during labor could indicate that these cells undergo a shift towards a pro-inflammatory phenotype as part of the normal process of parturition, as demonstrated for maternal macrophages present in the decidua (106, 107). The maternal monocyte population detected in the placental tissues likely represents circulating cells present in the intervillous space (121, 122). Previous reports have noted the enhanced activation of maternal circulating monocytes in the context of labor (93, 123, 124), likely driven by signals derived from the placental tissues (125), as such innate immune cells display upregulated expression of activation markers and can phagocytose placenta-derived particles (126).
Placental tissues shed extracellular vesicles, particles, and other debris into the maternal circulation (80). Due to such shedding, the maternal circulation has proven to be a source of cell-free DNA and RNA with diagnostic potential (127–129), and markers in the maternal peripheral blood can be utilized to monitor pregnancy and labor (40, 42, 43, 45, 65, 72, 79, 130, 131). Here, we leveraged this concept for the monitoring and potential prediction of pathologic labor (such as preterm labor). In the absence of intra-amniotic inflammation, the positive correlations between term labor and preterm labor involved a broad array of cell types. Preterm labor in the absence of intra-amniotic inflammation is typically considered idiopathic, and this study group may therefore include multiple underlying mechanisms. Indeed, our previous work has implicated macrophage activation (106) as a potential immune mechanism for idiopathic preterm labor and birth (132). By contrast, cases of preterm labor with intra-amniotic inflammation seemed to involve similar cell types, regardless of the sterile or microbial nature. Both stromal cells (133, 134) and macrophages (135) are known players in the host response against pathogens. The positive correlations by maternal macrophages may reflect the presence of acute histologic chorioamnionitis, a placental lesion driven by infiltration of innate immune cells into the chorioamniotic membranes in response to microbial invasion of the amniotic cavity (136). These results indicate that transcriptomic changes occurring in the CAM of women during physiologic labor overlap with those of women who underwent spontaneous preterm labor; however, the inflammatory and microbiological status of the amniotic cavity also drives placental transcriptomic activity, as previously hinted at (84).
The two subsets of spontaneous preterm birth included herein, sPTL and PPROM, involve differing disease mechanisms (83, 137) and clinical management (138, 139) and thus their prediction and prevention warrants separate investigation. Indeed, we noted distinct patterns of dysregulation between sPTL and PPROM that were at least partially dependent on the window of sampling. Specifically, women who ultimately underwent PPROM displayed stronger positive correlations with labor-specific cell type signatures than those who underwent sPTL. Nonetheless, we found that both subsets shared stromal and decidual signatures, further confirming a common pathway of parturition that leads to premature birth. In the second trimester, women who ultimately underwent spontaneous preterm birth showed dysregulation of specific signatures, primarily maternal decidual cells, which is in line with the concept of a decidual clock (11). Moreover, our findings highlight two key facts: first, the underlying mechanisms of sPTL and PPROM involve alterations in differing cell type signatures, which can potentially enable their distinction using predictive models; second, it is important to consider the differences between sPTL and PPROM for future investigations, as the combination of these subsets may diminish the power to detect specific biomarkers. Importantly, these results suggest that the best single-cell signatures to be used for predictive modeling may differ between sPTL and PPROM, further supporting the multi-etiological nature of the preterm labor syndrome.
The current study has some limitations. Gene expression analysis alone may not completely capture labor processes; however, establishing a transcriptomic foundation for parturition and identifying the specific cell types involved hold substantial value for future studies. Furthermore, analytical modeling techniques can provide robust inferences regarding the signaling processes modulated with labor as well as the participating cell types. Regardless, we acknowledge that additional validation of such predictions is required to verify their accuracy. In addition, certain highly fragile cell types, such as neutrophils, are underrepresented when using conventional single-cell approaches in the human placenta (73), and therefore any potential contribution of these cell types to the labor-associated processes described herein cannot be ruled out. Last, our findings also suggest that placenta-derived predictive models should be tailored to the target population, given our observation that environment/ethnicity may be a contributing factor. This caveat further underscores the significance of customizing biomarkers to suit different subsets of obstetrical diseases.
In conclusion, the findings generated herein provide new insights into maternal-fetal signaling and crosstalk during term parturition. Our findings highlight the utility of single-cell technologies for shedding light on new cell types and signaling pathways that are implicated in the process of labor in the human placenta, and indicate the potential use of placenta-derived single-cell signatures for developing non-invasive biomarkers to predict obstetrical diseases such as preterm labor and birth, the leading cause of neonatal morbidity and mortality worldwide.
MATERIALS AND METHODS
Study design
This prospective cross-sectional study included women who delivered at term (≥ 37 weeks of gestation) and were divided into those who underwent elective cesarean delivery (term not in labor, n = 18) and those with spontaneous term labor (term in labor, n = 24). Women with multiple gestations, or those who had a fetus with chromosomal and/or sonographic abnormalities were excluded. Maternal and neonatal data were obtained by retrospective clinical chart review. The demographic and clinical characteristics of the study groups are shown in tables S1 and S2. Placental tissues were collected from eligible women at term who were enrolled in our research protocols at the Detroit Medical Center, Wayne State University School of Medicine, and the Pregnancy Research Branch, an intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Detroit, MI, USA. The collection and use of human materials for research purposes were approved by the Institutional Review Boards of Wayne State University and the NICHD (WSU IRB 031318MP2F, NICHD 18-CH-N063, and WSU IRB 061217MP2F). Before sample collection, written informed consent was provided by all participating women.
More information regarding the wet and dry laboratory procedures including scRNA-seq data analysis can be found in the Supplementary Materials and Methods.
Statistical analysis
Statistical analyses were performed in GraphPad Prism (v9.0.2; GraphPad), SPSS v19.0 (IBM) or the R package (see Supplementary Materials and Methods for details). Human demographic data were compared using two-tailed Fisher’s exact tests for proportions and Mann-Whitney U-tests for non-normally distributed continuous variables.
Supplementary Material
Acknowledgments:
The figures include artwork created using a licensed version of BioRender.com.
The study was conducted at the Perinatology Research Branch, NICHD/NIH/DHHS, in Detroit, Michigan; the Branch has since been renamed as the Pregnancy Research Branch, NICHD/NIH/DHHS.
Funding:
Pregnancy Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS) Contract No. HHSN275201300006C (R.R.)
Wayne State University Perinatal Research Initiative in Maternal, Perinatal and Child Health (N.G.-L. and A.L.T.)
Footnotes
Competing interests:
The authors declare that they have no competing interests.
Data and materials availability:
All data for this study are included in the paper or the Supplementary Materials and Methods. The scRNA-seq data reported in this study have been submitted to the NIH dbGAP repository (phs001886.v5.p1). All software and R packages used herein are detailed in the Supplementary Materials and Methods. Scripts detailing the single-cell analyses are also available at (140) and at https://github.com/piquelab/Parturition_paper.
REFERENCES
- 1.Norwitz ER, Robinson JN, Challis JR, The control of labor. N Engl J Med 341, 660–666 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Challis JR, Bloomfield FH, Bocking AD, Casciani V, Chisaka H, Connor K, Dong X, Gluckman P, Harding JE, Johnstone J, Li W, Lye S, Okamura K, Premyslova M, Fetal signals and parturition. J Obstet Gynaecol Res 31, 492–499 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Romero R, Mazor M, Munoz H, Gomez R, Galasso M, Sherer DM, The preterm labor syndrome. Ann N Y Acad Sci 734, 414–429 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, Mazor M, The preterm parturition syndrome. BJOG 113 Suppl 3, 17–42 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith R, Parturition. N Engl J Med 356, 271–283 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Liggins CG, in Cervix in Pregnancy and Labour, Elwood DA, Andersson ABM, Eds. (Churchill Livingstone, Edinburgh, 1981), pp. 1 – 9. [Google Scholar]
- 7.Haddad R, Tromp G, Kuivaniemi H, Chaiworapongsa T, Kim YM, Mazor M, Romero R, Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol 195, 394 e391–324 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero R, Gotsch F, Pineles B, Kusanovic JP, Inflammation in pregnancy: its roles in reproductive physiology, obstetrical complications, and fetal injury. Nutr Rev 65, S194–202 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Norman JE, Bollapragada S, Yuan M, Nelson SM, Inflammatory pathways in the mechanism of parturition. BMC Pregnancy Childbirth 7 Suppl 1 S7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Lopez N, Guilbert LJ, Olson DM, Invasion of the leukocytes into the fetal-maternal interface during pregnancy. J Leukoc Biol 88, 625–633 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Norwitz ER, Bonney EA, Snegovskikh VV, Williams MA, Phillippe M, Park JS, Abrahams VM, Molecular Regulation of Parturition: The Role of the Decidual Clock. Cold Spring Harb Perspect Med 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson AJ, Telfer JF, Young A, Campbell S, Stewart CJ, Cameron IT, Greer IA, Norman JE, Leukocytes infiltrate the myometrium during human parturition: further evidence that labour is an inflammatory process. Hum Reprod 14, 229–236 (1999). [PubMed] [Google Scholar]
- 13.Mackler AM, Iezza G, Akin MR, McMillan P, Yellon SM, Macrophage trafficking in the uterus and cervix precedes parturition in the mouse. Biol Reprod 61, 879–883 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Young A, Thomson AJ, Ledingham M, Jordan F, Greer IA, Norman JE, Immunolocalization of proinflammatory cytokines in myometrium, cervix, and fetal membranes during human parturition at term. Biol Reprod 66, 445–449 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Osman I, Young A, Ledingham MA, Thomson AJ, Jordan F, Greer IA, Norman JE, Leukocyte density and pro-inflammatory cytokine expression in human fetal membranes, decidua, cervix and myometrium before and during labour at term. Mol Hum Reprod 9, 41–45 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Yellon SM, Mackler AM, Kirby MA, The role of leukocyte traffic and activation in parturition. J Soc Gynecol Investig 10, 323–338 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Esplin MS, Fausett MB, Peltier MR, Hamblin S, Silver RM, Branch DW, Adashi EY, Whiting D, The use of cDNA microarray to identify differentially expressed labor-associated genes within the human myometrium during labor. Am J Obstet Gynecol 193, 404–413 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Shynlova O, Tsui P, Dorogin A, Lye SJ, Monocyte chemoattractant protein-1 (CCL-2) integrates mechanical and endocrine signals that mediate term and preterm labor. J Immunol 181, 1470–1479 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Mittal P, Romero R, Tarca AL, Gonzalez J, Draghici S, Xu Y, Dong Z, Nhan-Chang CL, Chaiworapongsa T, Lye S, Kusanovic JP, Lipovich L, Mazaki-Tovi S, Hassan SS, Mesiano S, Kim CJ, Characterization of the myometrial transcriptome and biological pathways of spontaneous human labor at term. J Perinat Med 38, 617–643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittal P, Romero R, Tarca AL, Draghici S, Nhan-Chang CL, Chaiworapongsa T, Hotra J, Gomez R, Kusanovic JP, Lee DC, Kim CJ, Hassan SS, A molecular signature of an arrest of descent in human parturition. Am J Obstet Gynecol 204, 177 e115–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shynlova O, Lee YH, Srikhajon K, Lye SJ, Physiologic uterine inflammation and labor onset: integration of endocrine and mechanical signals. Reprod Sci 20, 154–167 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Chaemsaithong P, Madan I, Romero R, Than NG, Tarca AL, Draghici S, Bhatti G, Yeo L, Mazor M, Kim CJ, Hassan SS, Chaiworapongsa T, Characterization of the myometrial transcriptome in women with an arrest of dilatation during labor. J Perinat Med 41, 665–681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuya K, Singh N, Sooranna SR, Johnson MR, Myatt L, Epigenetics of human myometrium: DNA methylation of genes encoding contraction-associated proteins in term and preterm labor. Biol Reprod 90, 98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan YW, van den Berg HA, Moore JD, Quenby S, Blanks AM, Assessment of myometrial transcriptome changes associated with spontaneous human labour by high-throughput RNA-seq. Exp Physiol 99, 510–524 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Sharp GC, Hutchinson JL, Hibbert N, Freeman TC, Saunders PT, Norman JE, Transcription Analysis of the Myometrium of Labouring and Non-Labouring Women. PLoS One 11, e0155413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lui S, Duval C, Farrokhnia F, Girard S, Harris LK, Tower CL, Stevens A, Jones RL, Delineating differential regulatory signatures of the human transcriptome in the choriodecidua and myometrium at term labor. Biol Reprod 98, 422–436 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Stanfield Z, Lai PF, Lei K, Johnson MR, Blanks AM, Romero R, Chance MR, Mesiano S, Koyutürk M, Myometrial Transcriptional Signatures of Human Parturition. Front Genet 10, 185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokstrom H, Brannstrom M, Alexandersson M, Norstrom A, Leukocyte subpopulations in the human uterine cervical stroma at early and term pregnancy. Hum Reprod 12, 586–590 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Hassan SS, Romero R, Haddad R, Hendler I, Khalek N, Tromp G, Diamond MP, Sorokin Y, Malone J Jr., The transcriptome of the uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol 195, 778–786 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Hassan SS, Romero R, Tarca AL, Draghici S, Pineles B, Bugrim A, Khalek N, Camacho N, Mittal P, Yoon BH, Espinoza J, Kim CJ, Sorokin Y, Malone J Jr., Signature pathways identified from gene expression profiles in the human uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol 197, 250 e251–257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmons BC, Fairhurst AM, Mahendroo MS, Temporal changes in myeloid cells in the cervix during pregnancy and parturition. J Immunol 182, 2700–2707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fidel PL Jr., Romero R, Ramirez M, Cutright J, Edwin SS, LaMarche S, Cotton DB, Mitchell MD, Interleukin-1 receptor antagonist (IL-1ra) production by human amnion, chorion, and decidua. Am J Reprod Immunol 32, 1–7 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Keelan JA, Marvin KW, Sato TA, Coleman M, McCowan LM, Mitchell MD, Cytokine abundance in placental tissues: evidence of inflammatory activation in gestational membranes with term and preterm parturition. Am J Obstet Gynecol 181, 1530–1536 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Lopez N, Estrada-Gutierrez G, Jimenez-Zamudio L, Vega-Sanchez R, Vadillo-Ortega F, Fetal membranes exhibit selective leukocyte chemotaxic activity during human labor. J Reprod Immunol 80, 122–131 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Arenas-Hernandez M, Gomez-Lopez N, Garcia-Flores V, Rangel-Escareno C, Alvarez-Salas LM, Martinez-Acuna N, Vazquez-Perez JA, Vega-Sanchez R, Choriodecidual leukocytes display a unique gene expression signature in spontaneous labor at term. Genes Immun 20, 56–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sacks GP, Studena K, Sargent K, Redman CW, Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am J Obstet Gynecol 179, 80–86 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Naccasha N, Gervasi MT, Chaiworapongsa T, Berman S, Yoon BH, Maymon E, Romero R, Phenotypic and metabolic characteristics of monocytes and granulocytes in normal pregnancy and maternal infection. Am J Obstet Gynecol 185, 1118–1123 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY, Quake SR, Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A 111, 7361–7366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsui NB, Jiang P, Wong YF, Leung TY, Chan KC, Chiu RW, Sun H, Lo YM, Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem 60, 954–962 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Aghaeepour N, Ganio EA, McIlwain D, Tsai AS, Tingle M, Van Gassen S, Gaudilliere DK, Baca Q, McNeil L, Okada R, Ghaemi MS, Furman D, Wong RJ, Winn VD, Druzin ML, El-Sayed YY, Quaintance C, Gibbs R, Darmstadt GL, Shaw GM, Stevenson DK, Tibshirani R, Nolan GP, Lewis DB, Angst MS, Gaudilliere B, An immune clock of human pregnancy. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aghaeepour N, Lehallier B, Baca Q, Ganio EA, Wong RJ, Ghaemi MS, Culos A, El-Sayed YY, Blumenfeld YJ, Druzin ML, Winn VD, Gibbs RS, Tibshirani R, Shaw GM, Stevenson DK, Gaudilliere B, Angst MS, A proteomic clock of human pregnancy. Am J Obstet Gynecol 218, 347 e341–347 e314 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Tarca AL, Romero R, Xu Z, Gomez-Lopez N, Erez O, Hsu CD, Hassan SS, Carey VJ, Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci Rep 9, 848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TTM, Lee-McMullen B, Lehallier B, Robaczewska A, McIlwain D, Rosenberg-Hasson Y, Wong RJ, Quaintance C, Culos A, Stanley N, Tanada A, Tsai A, Gaudilliere D, Ganio E, Han X, Ando K, McNeil L, Tingle M, Wise P, Maric I, Sirota M, Wyss-Coray T, Winn VD, Druzin ML, Gibbs R, Darmstadt GL, Lewis DB, Partovi Nia V, Agard B, Tibshirani R, Nolan G, Snyder MP, Relman DA, Quake SR, Shaw GM, Stevenson DK, Angst MS, Gaudilliere B, Aghaeepour N, Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35, 95–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang L, Rasmussen MH, Piening B, Shen X, Chen S, Rost H, Snyder JK, Tibshirani R, Skotte L, Lee NC, Contrepois K, Feenstra B, Zackriah H, Snyder M, Melbye M, Metabolic Dynamics and Prediction of Gestational Age and Time to Delivery in Pregnant Women. Cell 181, 1680–1692 e1615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stelzer IA, Ghaemi MS, Han X, Ando K, Hedou JJ, Feyaerts D, Peterson LS, Rumer KK, Tsai ES, Ganio EA, Gaudilliere DK, Tsai AS, Choisy B, Gaigne LP, Verdonk F, Jacobsen D, Gavasso S, Traber GM, Ellenberger M, Stanley N, Becker M, Culos A, Fallahzadeh R, Wong RJ, Darmstadt GL, Druzin ML, Winn VD, Gibbs RS, Ling XB, Sylvester K, Carvalho B, Snyder MP, Shaw GM, Stevenson DK, Contrepois K, Angst MS, Aghaeepour N, Gaudilliere B, Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci Transl Med 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipovich Y, Klein J, Zhou Y, Hirsch E, Maternal and fetal roles in bacterially induced preterm labor in the mouse. Am J Obstet Gynecol 214, 386.e381–389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendelson CR, Montalbano AP, Gao L, Fetal-to-maternal signaling in the timing of birth. J Steroid Biochem Mol Biol 170, 19–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown AG, Maubert ME, Anton L, Heiser LM, Elovitz MA, The tracking of lipopolysaccharide through the feto-maternal compartment and the involvement of maternal TLR4 in inflammation-induced fetal brain injury. Am J Reprod Immunol 82, e13189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cappelletti M, Doll JR, Stankiewicz TE, Lawson MJ, Sauer V, Wen B, Kalinichenko VV, Sun X, Tilburgs T, Divanovic S, Maternal regulation of inflammatory cues is required for induction of preterm birth. JCI Insight 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motomura K, Romero R, Galaz J, Tao L, Garcia-Flores V, Xu Y, Done B, Arenas-Hernandez M, Miller D, Gutierrez-Contreras P, Farias-Jofre M, Aras S, Grossman LI, Tarca AL, Gomez-Lopez N, Fetal and maternal NLRP3 signaling is required for preterm labor and birth. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galaz J, Motomura K, Romero R, Liu Z, Garcia-Flores V, Tao L, Xu Y, Done B, Arenas-Hernandez M, Kanninen T, Farias-Jofre M, Miller D, Tarca AL, Gomez-Lopez N, A key role for NLRP3 signaling in preterm labor and birth driven by the alarmin S100B. Transl Res 10.1016/j.trsl.2023.04.004, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry SM, Romero R, Gomez R, Puder KS, Ghezzi F, Cotton DB, Bianchi DW, Premature parturition is characterized by in utero activation of the fetal immune system. Am J Obstet Gynecol 173, 1315–1320 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Frascoli M, Coniglio L, Witt R, Jeanty C, Fleck-Derderian S, Myers DE, Lee TH, Keating S, Busch MP, Norris PJ, Tang Q, Cruz G, Barcellos LF, Gomez-Lopez N, Romero R, MacKenzie TC, Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-gamma and TNF-alpha. Sci Transl Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomez-Lopez N, Romero R, Xu Y, Miller D, Arenas-Hernandez M, Garcia-Flores V, Panaitescu B, Galaz J, Hsu CD, Para R, Berry SM, Fetal T Cell Activation in the Amniotic Cavity during Preterm Labor: A Potential Mechanism for a Subset of Idiopathic Preterm Birth. J Immunol 203, 1793–1807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petroff MG, Immune interactions at the maternal-fetal interface. J Reprod Immunol 68, 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Sargent IL, Borzychowski AM, Redman CW, NK cells and human pregnancy--an inflammatory view. Trends Immunol 27, 399–404 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Gellersen B, Brosens IA, Brosens JJ, Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med 25, 445–453 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Erlebacher A, Immunology of the maternal-fetal interface. Annu Rev Immunol 31, 387–411 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Mori M, Bogdan A, Balassa T, Csabai T, Szekeres-Bartho J, The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin Immunopathol 38, 635–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ander SE, Diamond MS, Coyne CB, Immune responses at the maternal-fetal interface. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turco MY, Moffett A, Development of the human placenta. Development 146, (2019). [DOI] [PubMed] [Google Scholar]
- 62.Holder B, Aplin JD, Gomez-Lopez N, Heazell AEP, James JL, Jones CJP, Jones H, Lewis RM, Mor G, Roberts CT, Robertson SA, Zenclussen AC, ‘Fetal side’ of the placenta: anatomical mis-annotation of carbon particle ‘transfer’ across the human placenta. Nat Commun 12, 7049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Megli CJ, Coyne CB, Infections at the maternal-fetal interface: an overview of pathogenesis and defence. Nat Rev Microbiol 20, 67–82 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller D, Garcia-Flores V, Romero R, Galaz J, Pique-Regi R, Gomez-Lopez N, Single-Cell Immunobiology of the Maternal-Fetal Interface. J Immunol 209, 1450–1464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, Gomez-Lopez N, Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pique-Regi R, Romero R, Garcia-Flores V, Peyvandipour A, Tarca AL, Pusod E, Galaz J, Miller D, Bhatti G, Para R, Kanninen T, Hadaya O, Paredes C, Motomura K, Johnson JR, Jung E, Hsu CD, Berry SM, Gomez-Lopez N, A single-cell atlas of the myometrium in human parturition. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gustavii B, Labor: a delayed menstruation? Lancet 2, 1149–1150 (1972). [DOI] [PubMed] [Google Scholar]
- 68.Brunk U, Gustavii B, Lability of human decidual cells. In vitro effects of autolysis and osmotic stress. Am J Obstet Gynecol 115, 811–816 (1973). [DOI] [PubMed] [Google Scholar]
- 69.Gustavii B, Sweeping of the fetal membranes by a physiologic saline solution: effect on decidual cells. Am J Obstet Gynecol 120, 531–536 (1974). [DOI] [PubMed] [Google Scholar]
- 70.Pavlicev M, Norwitz ER, Human Parturition: Nothing More Than a Delayed Menstruation. Reprod Sci 25, 166–173 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF, Cheng YKY, Chiu RWK, Lo YMD, Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci U S A 114, E7786–E7795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarca AL, Romero R, Erez O, Gudicha DW, Than NG, Benshalom-Tirosh N, Pacora P, Hsu CD, Chaiworapongsa T, Hassan SS, Gomez-Lopez N, Maternal whole blood mRNA signatures identify women at risk of early preeclampsia: a longitudinal study. J Matern Fetal Neonatal Med 34, 3463–3474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Flores V, Xu Y, Pusod E, Romero R, Pique-Regi R, Gomez-Lopez N, Preparation of single-cell suspensions from the human placenta. Nat Protoc 10.1038/s41596-022-00772-w, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, Vadillo-Ortega F, Evidence for a role for the adaptive immune response in human term parturition. Am J Reprod Immunol 69, 212–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamilton SA, Tower CL, Jones RL, Identification of chemokines associated with the recruitment of decidual leukocytes in human labour: potential novel targets for preterm labour. PLoS One 8, e56946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shynlova O, Nedd-Roderique T, Li Y, Dorogin A, Nguyen T, Lye SJ, Infiltration of myeloid cells into decidua is a critical early event in the labour cascade and post-partum uterine remodelling. J Cell Mol Med 17, 311–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q, Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gomez-Lopez N, Romero R, Hassan SS, Bhatti G, Berry SM, Kusanovic JP, Pacora P, Tarca AL, The Cellular Transcriptome in the Maternal Circulation During Normal Pregnancy: A Longitudinal Study. Front Immunol 10, 2863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Lopez N, Romero R, Galaz J, Bhatti G, Done B, Miller D, Ghita C, Motomura K, Farias-Jofre M, Jung E, Pique-Regi R, Hassan SS, Chaiworapongsa T, Tarca AL, Transcriptome changes in maternal peripheral blood during term parturition mimic perturbations preceding spontaneous preterm birth. Biol Reprod 106, 185–199 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knight M, Redman CW, Linton EA, Sargent IL, Shedding of syncytiotrophoblast microvilli into the maternal circulation in pre-eclamptic pregnancies. Br J Obstet Gynaecol 105, 632–640 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Ng EK, Tsui NB, Lau TK, Leung TN, Chiu RW, Panesar NS, Lit LC, Chan KW, Lo YM, mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci U S A 100, 4748–4753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taglauer ES, Wilkins-Haug L, Bianchi DW, Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 35 Suppl, S64–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Romero R, Dey SK, Fisher SJ, Preterm labor: one syndrome, many causes. Science 345, 760–765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Motomura K, Romero R, Galaz J, Tarca AL, Done B, Xu Y, Leng Y, Garcia-Flores V, Arenas-Hernandez M, Theis KR, Gershater M, Jung E, Hsu CD, Gomez-Lopez N, RNA Sequencing Reveals Distinct Immune Responses in the Chorioamniotic Membranes of Women with Preterm Labor and Microbial or Sterile Intra-amniotic Inflammation. Infect Immun 89, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol 71, 330–358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romero R, Miranda J, Chaiworapongsa T, Korzeniewski SJ, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Yeo L, Prevalence and clinical significance of sterile intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Reprod Immunol 72, 458–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM, Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med 28, 1394–1409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM, Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J Matern Fetal Neonatal Med 28, 1343–1359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomez-Lopez N, Romero R, Tarca AL, Miller D, Panaitescu B, Schwenkel G, Gudicha DW, Hassan SS, Pacora P, Jung E, Hsu CD, Gasdermin D: Evidence of pyroptosis in spontaneous preterm labor with sterile intra-amniotic inflammation or intra-amniotic infection. Am J Reprod Immunol 82, e13184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhatti G, Romero R, Rice GE, Fitzgerald W, Pacora P, Gomez-Lopez N, Kavdia M, Tarca AL, Margolis L, Compartmentalized profiling of amniotic fluid cytokines in women with preterm labor. PLoS One 15, e0227881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Theis KR, Romero R, Motomura K, Galaz J, Winters AD, Pacora P, Miller D, Slutsky R, Florova V, Levenson D, Para R, Varrey A, Kacerovsky M, Hsu CD, Gomez-Lopez N, Microbial burden and inflammasome activation in amniotic fluid of patients with preterm prelabor rupture of membranes. J Perinat Med 48, 115–131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peiris HN, Romero R, Vaswani K, Reed S, Gomez-Lopez N, Tarca AL, Gudicha DW, Erez O, Maymon E, Mitchell MD, Preterm labor is characterized by a high abundance of amniotic fluid prostaglandins in patients with intra-amniotic infection or sterile intra-amniotic inflammation. J Matern Fetal Neonatal Med 34, 4009–4024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paquette AG, Shynlova O, Kibschull M, Price ND, Lye SJ, Global P Alliance to Prevent, T. Stillbirth Systems Biology of Preterm Birth, Comparative analysis of gene expression in maternal peripheral blood and monocytes during spontaneous preterm labor. Am J Obstet Gynecol 218, 345 e341–345 e330 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Tarca AL, Pataki BA, Romero R, Sirota M, Guan Y, Kutum R, Gomez-Lopez N, Done B, Bhatti G, Yu T, Andreoletti G, Chaiworapongsa T, Consortium DPBPC, Hassan SS, Hsu CD, Aghaeepour N, Stolovitzky G, Csabai I, Costello JC, Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Rep Med 2, 100323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pavlicev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H, Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res 27, 349–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marsh B, Zhou Y, Kapidzic M, Fisher S, Blelloch R, Regionally distinct trophoblast regulate barrier function and invasion in the human placenta. Elife 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Romero R, Mazor M, Wu YK, Avila C, Oyarzun E, Mitchell MD, Bacterial endotoxin and tumor necrosis factor stimulate prostaglandin production by human decidua. Prostaglandins Leukot Essent Fatty Acids 37, 183–186 (1989). [DOI] [PubMed] [Google Scholar]
- 99.Romero R, Mazor M, Manogue K, Oyarzun E, Cerami A, Human decidua: a source of cachectin-tumor necrosis factor. Eur J Obstet Gynecol Reprod Biol 41, 123–127 (1991). [DOI] [PubMed] [Google Scholar]
- 100.Khan H, Ishihara O, Sullivan MH, Elder MG, Changes in decidual stromal cell function associated with labour. Br J Obstet Gynaecol 99, 10–12 (1992). [DOI] [PubMed] [Google Scholar]
- 101.Cha JM, Aronoff DM, A role for cellular senescence in birth timing. Cell Cycle 16, 2023–2031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anders AP, Gaddy JA, Doster RS, Aronoff DM, Current concepts in maternal-fetal immunology: Recognition and response to microbial pathogens by decidual stromal cells. Am J Reprod Immunol 77, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang J, Xie Y, Peng Q, Wang W, Pei C, Zhao Y, Liu R, Huang L, Li T, Nie J, Liu L, Zhang X, Luo X, Luo J, Zhang W, Single-cell transcriptomics analysis showing functional heterogeneity in decidual stromal cells during labor. J Investig Med 10.1136/jim-2020-001616, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gonzalez JM, Dong Z, Romero R, Girardi G, Cervical remodeling/ripening at term and preterm delivery: the same mechanism initiated by different mediators and different effector cells. PLoS One 6, e26877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hamilton S, Oomomian Y, Stephen G, Shynlova O, Tower CL, Garrod A, Lye SJ, Jones RL, Macrophages infiltrate the human and rat decidua during term and preterm labor: evidence that decidual inflammation precedes labor. Biol Reprod 86, 39 (2012). [DOI] [PubMed] [Google Scholar]
- 106.Xu Y, Romero R, Miller D, Kadam L, Mial TN, Plazyo O, Garcia-Flores V, Hassan SS, Xu Z, Tarca AL, Drewlo S, Gomez-Lopez N, An M1-like Macrophage Polarization in Decidual Tissue during Spontaneous Preterm Labor That Is Attenuated by Rosiglitazone Treatment. J Immunol 196, 2476–2491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gomez-Lopez N, Garcia-Flores V, Chin PY, Groome HM, Bijland MT, Diener KR, Romero R, Robertson SA, Macrophages exert homeostatic actions in pregnancy to protect against preterm birth and fetal inflammatory injury. JCI Insight 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Umezawa A, Hasegawa A, Inoue M, Tanuma-Takahashi A, Kajiwara K, Makino H, Chikazawa E, Okamoto A, Amnion-derived cells as a reliable resource for next-generation regenerative medicine. Placenta 84, 50–56 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Torre P, Flores AI, Current Status and Future Prospects of Perinatal Stem Cells. Genes (Basel) 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones B, Li C, Park MS, Lerch A, Jacob V, Johnson N, Kuang JQ, Dhall S, Sathyamoorthy M, Comprehensive Comparison of Amnion Stromal Cells and Chorion Stromal Cells by RNA-Seq. Int J Mol Sci 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Magatti M, Stefani FR, Papait A, Cargnoni A, Masserdotti A, Silini AR, Parolini O, Perinatal Mesenchymal Stromal Cells and Their Possible Contribution to Fetal-Maternal Tolerance. Cells 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roelen DL, van der Mast BJ, in’t Anker PS, Kleijburg C, Eikmans M, van Beelen E, de Groot-Swings GM, Fibbe WE, Kanhai HH, Scherjon SA, Claas FH, Differential immunomodulatory effects of fetal versus maternal multipotent stromal cells. Hum Immunol 70, 16–23 (2009). [DOI] [PubMed] [Google Scholar]
- 113.Yamahara K, Harada K, Ohshima M, Ishikane S, Ohnishi S, Tsuda H, Otani K, Taguchi A, Soma T, Ogawa H, Katsuragi S, Yoshimatsu J, Harada-Shiba M, Kangawa K, Ikeda T, Comparison of angiogenic, cytoprotective, and immunosuppressive properties of human amnion- and chorion-derived mesenchymal stem cells. PLoS One 9, e88319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shin SY, Kim MS, Kim YG, Kang D, Choi HY, Pae JY, Wie JH, Park IY, Characteristics of amniotic mesenchymal stromal cells derived from term and preterm labor. Taiwan J Obstet Gynecol 61, 51–56 (2022). [DOI] [PubMed] [Google Scholar]
- 115.Wang WS, Lin YK, Zhang F, Lei WJ, Pan F, Zhu YN, Lu JW, Zhang CY, Zhou Q, Ying H, Sun K, Single cell transcriptomic analysis of human amnion identifies cell-specific signatures associated with membrane rupture and parturition. Cell Biosci 12, 64 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Soundararajan M, Kannan S, Fibroblasts and mesenchymal stem cells: Two sides of the same coin? J Cell Physiol 233, 9099–9109 (2018). [DOI] [PubMed] [Google Scholar]
- 117.Ugurlu B, Karaoz E, Comparison of similar cells: Mesenchymal stromal cells and fibroblasts. Acta Histochem 122, 151634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sutton L, Mason DY, Redman CW, HLA-DR positive cells in the human placenta. Immunology 49, 103–112 (1983). [PMC free article] [PubMed] [Google Scholar]
- 119.Thomas JR, Appios A, Zhao X, Dutkiewicz R, Donde M, Lee CYC, Naidu P, Lee C, Cerveira J, Liu B, Ginhoux F, Burton G, Hamilton RS, Moffett A, Sharkey A, McGovern N, Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells. J Exp Med 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zulu MZ, Martinez FO, Gordon S, Gray CM, The Elusive Role of Placental Macrophages: The Hofbauer Cell. J Innate Immun 11, 447–456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vega-Sanchez R, Gomez-Lopez N, Flores-Pliego A, Clemente-Galvan S, Estrada-Gutierrez G, Zentella-Dehesa A, Maida-Claros R, Beltran-Montoya J, Vadillo-Ortega F, Placental blood leukocytes are functional and phenotypically different than peripheral leukocytes during human labor. J Reprod Immunol 84, 100–110 (2010). [DOI] [PubMed] [Google Scholar]
- 122.Solders M, Gorchs L, Tiblad E, Gidlöf S, Leeansyah E, Dias J, Sandberg JK, Magalhaes I, Lundell AC, Kaipe H, Recruitment of MAIT Cells to the Intervillous Space of the Placenta by Placenta-Derived Chemokines. Front Immunol 10, 1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paquette AG, Shynlova O, Wu X, Kibschull M, Wang K, Price ND, Lye SJ, MicroRNA-transcriptome networks in whole blood and monocytes of women undergoing preterm labour. J Cell Mol Med 23, 6835–6845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J, Shynlova O, Sabra S, Bang A, Briollais L, Lye SJ, Immunophenotyping and activation status of maternal peripheral blood leukocytes during pregnancy and labour, both term and preterm. J Cell Mol Med 21, 2386–2402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Messerli M, May K, Hansson SR, Schneider H, Holzgreve W, Hahn S, Rusterholz C, Feto-maternal interactions in pregnancies: placental microparticles activate peripheral blood monocytes. Placenta 31, 106–112 (2010). [DOI] [PubMed] [Google Scholar]
- 126.Arenas-Hernandez M, Romero R, Gershater M, Tao L, Xu Y, Garcia-Flores V, Pusod E, Miller D, Galaz J, Motomura K, Schwenkel G, Para R, Gomez-Lopez N, Specific innate immune cells uptake fetal antigen and display homeostatic phenotypes in the maternal circulation. J Leukoc Biol 111, 519–538 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS, Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997). [DOI] [PubMed] [Google Scholar]
- 128.Lee T, LeShane ES, Messerlian GM, Canick JA, Farina A, Heber WW, Bianchi DW, Down syndrome and cell-free fetal DNA in archived maternal serum. Am J Obstet Gynecol 187, 1217–1221 (2002). [DOI] [PubMed] [Google Scholar]
- 129.Farina A, LeShane ES, Romero R, Gomez R, Chaiworapongsa T, Rizzo N, Bianchi DW, High levels of fetal cell-free DNA in maternal serum: a risk factor for spontaneous preterm delivery. Am J Obstet Gynecol 193, 421–425 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Fragiadakis GK, Baca QJ, Gherardini PF, Ganio EA, Gaudilliere DK, Tingle M, Lancero HL, McNeil LS, Spitzer MH, Wong RJ, Shaw GM, Darmstadt GL, Sylvester KG, Winn VD, Carvalho B, Lewis DB, Stevenson DK, Nolan GP, Aghaeepour N, Angst MS, Gaudilliere BL, Mapping the Fetomaternal Peripheral Immune System at Term Pregnancy. J Immunol 197, 4482–4492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tarca AL, Romero R, Bhatti G, Gotsch F, Done B, Gudicha DW, Gallo DM, Jung E, Pique-Regi R, Berry SM, Chaiworapongsa T, Gomez-Lopez N, Human Plasma Proteome During Normal Pregnancy. J Proteome Res 10.1021/acs.jproteome.2c00391, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gomez-Lopez N, Galaz J, Miller D, Farias-Jofre M, Liu Z, Arenas-Hernandez M, Garcia-Flores V, Shaffer Z, Greenberg JM, Theis KR, Romero R, The immunobiology of preterm labor and birth: intra-amniotic inflammation or breakdown of maternal-fetal homeostasis. Reproduction 164, R11–R45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bernardo ME, Fibbe WE, Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell 13, 392–402 (2013). [DOI] [PubMed] [Google Scholar]
- 134.Shaw TD, Krasnodembskaya AD, Schroeder GN, Zumla A, Maeurer M, O’Kane CM, Mesenchymal Stromal Cells: an Antimicrobial and Host-Directed Therapy for Complex Infectious Diseases. Clin Microbiol Rev 34, e0006421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weiss G, Schaible UE, Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 264, 182–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM, Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol 213, S29–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goldenberg RL, Culhane JF, Iams JD, Romero R, Epidemiology and causes of preterm birth. Lancet 371, 75–84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.American College of Obstetricians and Gynecologists, Practice Bulletin No. 171: Management of Preterm Labor. Obstet Gynecol 128, e155–164 (2016). [DOI] [PubMed] [Google Scholar]
- 139.American College of Obstetricians and Gynecologists, Prelabor Rupture of Membranes: ACOG Practice Bulletin, Number 217. Obstet Gynecol 135, e80–e97 (2020). [DOI] [PubMed] [Google Scholar]
- 140.Pique-Regi R, Peyvandipour A. (Zenodo, 2023), 10.5281/zenodo.10279146. [DOI] [Google Scholar]
- 141.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM, Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 143.Melsted P, Booeshaghi AS, Gao F, Beltrame E, Lu L, Hjorleifsson KE, Gehring J, Pachter L, Modular and efficient pre-processing of single-cell RNA-seq. bioRxiv 10.1101/673285, 673285 (2019). [DOI] [Google Scholar]
- 144.Alvarez M, Rahmani E, Jew B, Garske KM, Miao Z, Benhammou JN, Ye CJ, Pisegna JR, Pietilainen KH, Halperin E, Pajukanta P, Enhancing droplet-based single-nucleus RNA-seq resolution using the semi-supervised machine learning classifier DIEM. Sci Rep 10, 11019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, Hemberg M, Lawniczak MKN, Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods 17, 615–620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ, Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McInnes L, Healy J, Melville J, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2018. [Google Scholar]
- 152.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW, Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 10.1038/nbt.4314, (2018). [DOI] [PubMed] [Google Scholar]
- 153.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M, Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, Quake SR, Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 26, 1644–1653 (2020). [DOI] [PubMed] [Google Scholar]
- 155.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barshan E, Ghodsi A, Azimifar Z, Zolghadri Jahromi M, Supervised principal component analysis: Visualization, classification and regression on subspaces and submanifolds. Pattern Recognition 44, 1357–1371 (2011). [Google Scholar]
- 157.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57, 289–300 (1995). [Google Scholar]
- 159.Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, Hall TA, Blyn LB, Eshoo MW, Ranken R, Hofstadler SA, Tang YW, New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn 10, 399–415 (2010). [DOI] [PubMed] [Google Scholar]
- 161.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK, Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol 185, 1130–1136 (2001). [DOI] [PubMed] [Google Scholar]
- 162.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this study are included in the paper or the Supplementary Materials and Methods. The scRNA-seq data reported in this study have been submitted to the NIH dbGAP repository (phs001886.v5.p1). All software and R packages used herein are detailed in the Supplementary Materials and Methods. Scripts detailing the single-cell analyses are also available at (140) and at https://github.com/piquelab/Parturition_paper.


