ABSTRACT
Monocytes play a crucial role in the immune response against pathogens. Here, we sought to determine COVID-19 and the vaccine Gam-COVID-Vac induce long-term changes in the phenotype and cytokine production of circulating monocytes. Monocytes were purified from peripheral blood mononuclear cells of healthy donors who had not had COVID-19 or vaccination, who had received two doses of Gam-COVID-Vac, and who had mild/moderate COVID-19 in the last 6 months and evaluated by flow cytometry. To investigate the effect of SARS-CoV-2 proteins, monocytes were cultured for 2 days with or without stimulation with recombinant SARS-CoV-2 S1 and N peptides. Monocytes obtained from vaccinated and recovered individuals showed increased basal expression of HLA-DR, CD63, CXCR2, and TLR7. We also observed an increased frequency of CD63+ classical monocytes in both groups, as well as an increased frequency of HLA-DR+ non-classical monocytes in the COVID-19-recovered group compared to the control group. Monocytes from vaccinated and recovered donors produced higher basal levels of IL-6, IL-1β, and TNF-α cytokines. Ex vivo stimulation with SARS-CoV-2 antigens induced increased expression of HLA-DR and TLR7 on monocytes obtained from the control group. The challenge with SARS-CoV-2 antigens had no effect on the production of IL-6, IL-1β, and TNF-α cytokines by monocytes. The acquired data offer compelling evidence of enduring alterations in both the phenotype and functional status of circulating monocytes subsequent to vaccination with Gam-COVID-Vac and mild/moderate COVID-19 infection. At least some of these changes appear to be a consequence of exposure to SARS-CoV-2 S1 and N antigens.
KEYWORDS: monocytes, COVID-19, Gam-COVID-Vac vaccine, SARS-CoV-2 antigens, phenotype, cytokines
INTRODUCTION
Coronavirus disease 2019 (COVID-19), an emerging infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV2), is characterized by a broad range of symptoms and is associated with excessive inflammation driven by the innate immune system, including monocytes (1). Since the beginning of the COVID-19 pandemic, more than 770 million cases have been reported worldwide. Efforts to find a vaccine against SARS-CoV-2 infection have been unprecedented with more than 13 million vaccine doses administered (2). However, the long-term effects of COVID-19 infection and vaccination against SARS-CoV-2 on the immune system are unclear.
SARS-CoV-2 infection can lead to activation of immune cells and elevated levels of various proinflammatory cytokines that can persist for weeks or even months after recovery (3, 4). While monocytes are known to play a pivotal role in inflammation during acute SARS-CoV-2 infection, their fate after recovery from COVID-19 is not yet well described. Recent studies have shown that the number of circulating monocytes increases 1–10 months post-infection (5). One study identified an increase in circulating intermediate and non-classical monocytes, but not classical monocytes, up to 3 months after recovery from COVID-19 (6). In contrast, another study showed a decreased frequency of total monocytes due to a reduction in intermediate and non-classical monocytes in recovered patients (7). Monocytes from patients recovered from COVID-19 have been shown to express increased levels of inflammatory genes (8), have an increased type I IFN signature (9), and are more responsive to toll-like receptor 1/2 (TLR1/2) ligation (10). An altered expression of antigen presentation and activation molecules, as well as chemokine receptors, was also found in monocytes obtained from recovered patients (5, 7, 11). Similar findings were observed in long COVID-19 (5, 11, 12), a heterogeneous syndrome characterized by long-lasting symptoms such as extreme fatigue, shortness of breath, myalgias, brain fog, depression, fibrotic lung disease, and pulmonary vascular disease (13).
Information on the long-term effects of vaccination against SARS-CoV-2 on monocytes is sparser. Recently, it was shown that the frequency of total monocytes, monocytes co-expressing CD14 and CD16, and monocytes expressing HLA-DR is elevated in peripheral blood up to 3 months after vaccination with an adenovirus vector-based anti-SARS-CoV-2 vaccine (14). In the group of individuals who had received the inactivated CoronaVac vaccine, an increase in the relative frequency of classical monocytes was observed up to 150 days after vaccination (15).
To further expand our understanding of the long-lasting changes in monocytes mediated by SARS-CoV-2, we examined the phenotype and cytokine production of monocytes derived from vaccinated and recovered individuals and analyzed whether SARS-CoV-2 antigens are able to induce similar alterations in monocytes derived from a control group.
MATERIALS AND METHODS
Ethical aspects and donors
EDTA-anticoagulated peripheral blood samples were obtained from individuals who agreed to participate in the study and signed an informed consent form in accordance with the Declaration of Helsinki. Individuals were recruited between January 2021 and December 2022 and divided into three groups: a control group of unvaccinated healthy donors without a history of confirmed COVID-19 (Control, n = 13); a group of vaccinated individuals who had received two doses of the non-replicating adenovirus-vectored vaccine Gam-COVID-Vac (Gamaleya Research Institute of Epidemiology and Microbiology, Russia) at least 1 month and up to 180 days ago and had no prior COVID-19 in the anamnesis (Gam-COVID-Vac, n = 11); and the group of donors who had PCR-confirmed COVID-19 of mild or moderate severity (clinical manifestations including fever, cough, malaise, headache, loss of taste and smell, nausea with respiratory symptoms with an oxygen saturation of ≥94% on room air) and had recovered completely with no residual symptoms at least 1 month and up to 180 days ago (COVID-19, n = 11). Individuals who had received another vaccine in the last 6 months, or individuals who had any infection other than COVID-19, or who had been diagnosed with COVID-19 more than 180 days ago, individuals who had been vaccinated against COVID-19 more than 180 days ago, and those who had not consented to participate in the study were not enrolled in this study. Peripheral blood samples were collected from the donors and analyzed once. The demographic characteristics (gender, age) of the donors are presented in Table 1.
TABLE 1.
Donors’ characteristics
| Parameters | Indicators | ||
|---|---|---|---|
| Control group | Gam-COVID-Vac group | COVID-19 group | |
| Average age ± SD (range) | 62.3 ± 27.0 (21–86) |
35.9 ± 22.6 (19–83) |
54.9 ± 29.7 (21–87) |
| Sex | |||
| Men (average age ± SD) | 2 (49.5 ± 26.0) | 5 (40.6 ± 22.8) | 4 (72.0 ± 24.7) |
| Women (average age ± SD) | 11 (64.6 ± 27.1) | 6 (32.0 ± 22.6) | 7 (45.1 ± 28.6) |
Cell separation
Monocytes were isolated by plastic adhesion as described previously (16). In brief, peripheral blood was diluted 1 × 1 with RPMI-1640 (Sigma-Aldrich), and peripheral blood mononuclear cells (PBMCs) were obtained by density gradient centrifugation on Hystopaque-1077 (Sigma-Aldrich). The PBMCs were resuspended in complete medium based on RPMI-1640 supplemented with 10% fetal bovine serum (FBS) (Serva), 2 mМ glutamine, and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) (Sigma-Aldrich) 2 × 106 cells/mL and cultured in Nuclon Delta surface Petri dishes (ThermoFisher Scientific) in a humidified incubator containing 5% CO2 at 37°C for 18 h. Non-adherent cells were removed by thorough washing with RPMI-1640.
Cell culture
Adherent cells were incubated at 2 × 106 cells/mL in complete medium in Petri dishes in a humidified incubator with 5% CO2 at 37°C for additional 48 h alone or in the presence of SARS-CoV-2 proteins (100 ng/mL). After incubation, the monocytes were detached using TrypLEX (Invitrogen). The SARS-CoV-2 proteins, hFc-His Taq recombinant SARS-CoV-2 Spike protein S1 (S antigen) (RP-87689), and His Taq recombinant SARS-CoV-2 nucleocapsid protein (N antigen) (RP-87669) were purchased from Thermo Fisher Scientific. The proteins were resuspended in phosphate-buffered saline (PBS), pH 7.4, and stored at −20°C prior to use.
Analysis of interleukin production
The concentrations of pro-inflammatory cytokines in the cell supernatants after 48 h of incubation of monocytes with or without SARS-CoV-2 proteins were determined using an IL-8 Human ELISA kit (ThermoFisher Scientific), IL-6-IFA-BEST, IL-1β-IFA-BEST, and TNFα-IFA-BEST human ELISA kits (VektorBest) according to the manufacturer’s instructions.
Flow cytometry
For surface and intracellular staining, monocytes obtained by plastic adhesion were detached from the plastic and stained with fluorochrome-conjugated mAbs specific for surface markers according to the manufacturer’s protocols. For intracellular staining, the detached monocytes were fixed and permeabilized with Fixation/Permeabilization solution (BD Biosciences). Cells were then washed with Perm/Wash buffer (BD Biosciences) and subsequently labeled with mAbs specific for intracellular molecules. Afterward, cells were washed with PBS and immediately analyzed by flow cytometry on a FACSCalibur using CellQuest Pro software (BD Biosciences) or AttuneNxT using AttuneNxT software v3.2.1 (Thermo Fisher Scientific).
The following anti-human mAbs were used for flow cytometry analysis: CD14-PerCP, CD16-FITC (Miltenyi Biotec); CD14-APC, CD16-PerCP, HLA-DR-PE, CD11b-FITC, CD15-FITC, CD54-PE, CD63-PE, CD181(CXCR1)-PE, CD182(CXCR2)-PE, CD282(TLR2)-PE, CD287(TLR7)-PE, CD288(TLR8)-PE, HLA-DR/DP/DQ-PE, CD49d-PE, E4Bp4-PE (Thermo Fisher Scientific).
Forward and side scatter gating was used to analyze the phenotype of the total population of monocytes. Expression of markers was also analyzed in monocytes gated on СD14+, CD16+, as well as on CD14+CD16− classical, CD14+CD16+ intermediate, and CD14lowCD16+ non-classical subsets of monocytes, as previously described in detail (17) (Fig. 1A).
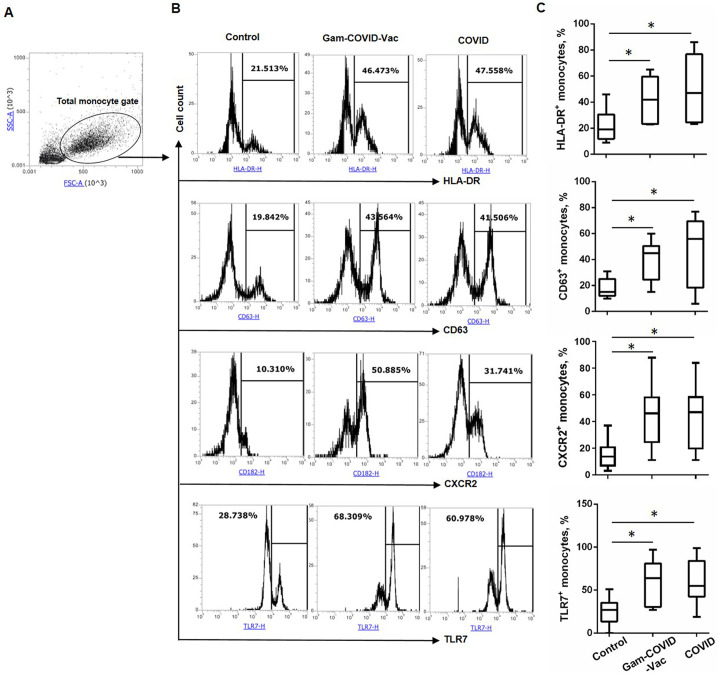
Fig 1.
Vaccination with Gam-COVID-Vac and mild/moderate COVID-19 induce long-term changes in the expression of HLA-DR, CD63, CXCR2, and TLR7 by the total subset of monocytes. Monocytes isolated from the peripheral blood of the Control, Gam-COVID-Vac, and COVID-19 groups were analyzed by flow cytometry. Cells were gated on the monocyte gate based on cell size and complexity using FSC and SSC parameters and the frequency of HLA-DR+, CD63+, CXCR2+, and TLR7+ cells were evaluated. (A) Gating strategy of monocytes, (B) representative, and (C) cumulative data are shown. Student’s t-test showed that there was a significant difference between the control and experimental groups, as indicated: * Р ˂ 0.05.
Statistical analysis
Statistical analysis was carried out using GraphPad Prizm 7.03 software. Data are presented as bars and columns with mean values ± standard deviation. To explore the existence of statistically significant differences between the groups, a two-tailed Student’s t-test was used. Differences were considered statistically significant if P values ≤ 0.05.
RESULTS
Expression of HLA-DR, CD63, CXCR2 and TLR7 is increased on monocytes in the Gam-COVID-Vac and COVID-19 groups
To determine whether the vaccine Gam-COVID-Vac and SARS-CoV-2 infection induce long-lasting changes in monocytes, we evaluated the relative frequency of peripheral blood monocytes and the expression of activation markers, chemokine receptors and toll-like receptors (TLRs) in the Control, Gam-COVID-Vac and COVID-19 groups. The monocyte gating strategy is shown in Fig. 1A. Monocyte subpopulations were assessed by gating on CD14+CD16– classical monocytes, CD14+CD16+ intermediate monocytes, and CD14lowCD16+ non-classical monocytes (Fig. 2A).
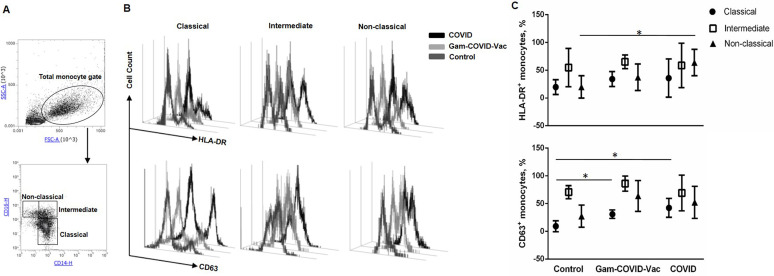
Fig 2.
Vaccination with Gam-COVID-Vac and mild/moderate COVID-19 induce long-term changes in the expression of CD63 by classical monocytes and of HLA-DR by non-classical monocytes, respectively. Monocytes isolated from the peripheral blood of the Control, Gam-COVID-Vac, and COVID-19 groups were analyzed by flow cytometry. (A) Gating strategy of monocyte subsets, (B) representative and (C) cumulative data on the frequency of HLA-DR- and CD63-expressing CD14+CD16− classical, CD14+CD16+ intermediate and CD14lowCD16+ non-classical monocytes are shown. Student’s t-test showed that there was a significant difference between the control and experimental groups, as indicated: * Р ˂ 0.05.
The proportion of CD14+, CD14+CD16–, CD14+CD16+, and CD14lowCD16+ monocytes did not differ significantly between the groups (data not shown). Immunophenotyping revealed that the frequency of HLA-DR- and CD63-expressing monocytes was significantly increased in the Gam-COVID-Vac and COVID-19 groups compared to the Control group (Fig. 1B and C). Additionally, we observed an increase in the percentage of CD63-expressing classical monocytes in the Gam-COVID-Vac and COVID-19 groups and HLA-DR-expressing non-classical monocytes in the COVID-19 group as compared to Control group (Fig. 2), while no statistical differences were observed between the Gam-COVID-Vac and COVID-19 groups.
Flow cytometric analysis also revealed that monocytes in the Gam-COVID-Vac and COVID-19 groups exhibited increased expression of surface CXCR2 and intracellular TLR7 compared to the Control group (Fig. 1B and C). No differences in the frequencies of CXCR2- and TLR7-expressing monocytes were observed between the Gam-COVID-Vac and COVID-19 groups (Fig. 1B and C).
For other markers analyzed, including CD11b, CD15, CD54, CXCR1, TLR2, TLR8, HLA-DR/DP/DQ, CD49d, E4Bp4, we found no differences between the Control, Gam-COVID-Vac, and COVID-19 groups (Fig. S1).
These observations indicate that the Gam-COVID-Vac vaccine and COVID-19 infection can induce long-term changes in the phenotype of circulating monocytes.
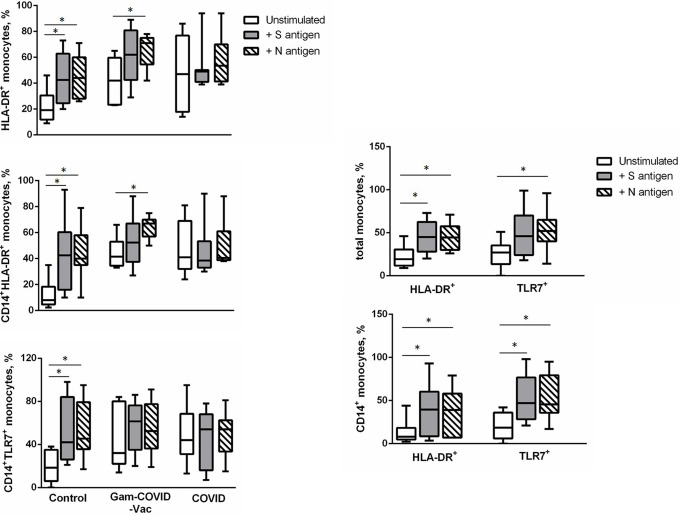
SARS-CoV-2 N and S1 antigens induce the expression of HLA-DR and TLR7 by monocytes
To determine whether the alterations observed in the monocytes obtained from the COVID-19 and Gam-COVID-Vac groups were induced by SARS-CoV-2 antigens, we evaluated the expression of activation markers, chemokine receptors, and TLRs on monocytes obtained from the Control group after in vitro challenge with SARS-CoV-2 antigens. For this purpose, the monocytes were incubated in the presence of SARS-CoV-2 S and N proteins. As a result, both the SARS-CoV-2 S and N antigens induced an increase in HLA-DR expression on the entire subset of monocytes and on CD14+ monocytes as well as an increase in TLR7 expression on CD14+ monocytes. The SARS-CoV-2 N antigen also induced TLR7 expression on the entire population of monocytes (Fig. 3). We did not detect any differences in other analyzed markers after activation of monocytes with SARS-CoV-2 S and N proteins (data not shown).
Fig 3.
SARS-CoV-2 S and N antigens induce the expression of HLA-DR and TLR7 by monocytes. Monocytes were isolated from the peripheral blood of a Control group and incubated for 48 h in the presence of SARS-CoV-2 S and N proteins. The cells were then gated on the monocyte gate and plotted on HLA-DR and TLR7 or CD14 versus HLA-DR and TRL7. Cumulative data on the frequency of HLA-DR+ and TLR7+ cells are shown. Student’s t-test showed that there was a significant difference, as indicated: * Р ˂ 0.05.
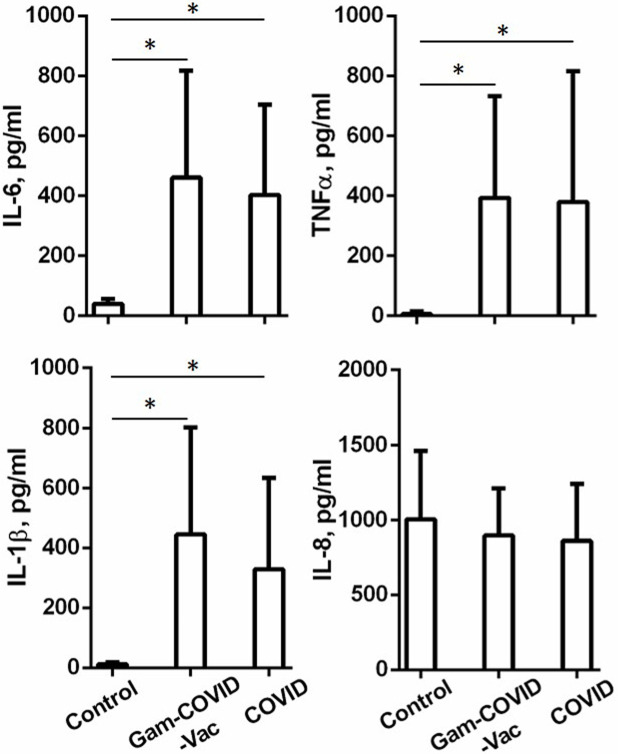
The Gam-COVID-Vac vaccine and mild/moderate COVID-19 infection induce long-term modulation of cytokines produced by monocytes
To understand whether Gam-COVID-Vac and the earlier COVID-19 modulate the cytokine-producing capacity of monocytes, we evaluated the production of IL-6, IL-1b, tumor necrosis factor alpha (TNF-α), and IL-8. The monocytes from the Gam-COVID-Vac and COVID-19 groups showed increased basal production of IL-6, IL-1b, and TNF-α when compared to the Control group. No differences were observed in the production of IL-8 (Fig. 4). There were also no differences in the production of IL-6, IL-1b, TNF-α, and IL-8 between the Gam-COVID-Vac and COVID-19 groups (Fig. 4).
Fig 4.
Vaccination with Gam-COVID-Vac and mild/moderate COVID-19 lead to long-term changes in the production of IL-6, IL-1β, and TNF-α cytokines by monocytes. Monocytes isolated from the peripheral blood of the Control, Gam-COVID-Vac, and COVID-19 groups were incubated for 48 h in a complete medium. The concentrations of pro-inflammatory cytokines in the cell supernatants were determined by ELISA. Cumulative data on the levels of IL-6, IL-1β, TNF-α, and IL-8 are shown. Student’s t-test showed that there was a significant difference between the groups as indicated: * P ˂ 0.05.
However, stimulation with S and N antigens did not induce the production of IL-6, IL-1b, TNF-α, or IL-8 cytokines by monocytes derived from the Control group (data not shown).
DISCUSSION
In this study, we observed significantly increased expression of surface HLA-DR, CD63, CXCR2, and intracellular TLR7 by the total subset of monocytes in the group of COVID-19 recovered and Gam-COVID-Vac vaccinated individuals, which persisted for up to 6 months. Interestingly, similar alterations in the expression of HLA-DR and TLR-7 were also observed in monocytes derived from the peripheral blood of individuals in the Control group after in vivo stimulation with SARS-CoV-2 antigens. These data may indicate that at least some of the long-lasting changes in monocytes observed in COVID-19-recovered and Gam-COVID-Vac-immunized individuals were induced by exposure to SARS-CoV-2. Similar results were found in other studies. Increased expression of HLA-DR by human monocytes was observed after vaccination with another vector vaccine against COVID-19 (ChAdOx1 nCoV-19) (14) and during acute mild COVID-19 (18, 19). Moreover, elevated TLR7 mRNA expression was previously observed in COVID-19 patients (20). However, we cannot confirm the specificity of the observed phenotypic changes induced by SARS-CoV-2 antigens as we did not evaluate heterologous challenges. For instance, studies have demonstrated that various stimuli can upregulate the expression of HLA-DR on monocytes. These stimuli include immunization with Candida albicans (21) and β-glucan (22). An increased frequency of HLA-DR+ monocytes was observed in patients with primary HIV infection (23) and in patients with acute bacterial intestinal infections (24). Increased expression of TLR7 by monocytes was shown in chronic active Epstein-Barr virus infection (25).
In addition to the phenotypic changes, we also observed functional changes manifested by increased spontaneous production of IL-6, IL-1b, and TNF-α cytokines by peripheral blood monocytes in Gam-COVID-Vac immunized and COVID-19 recovered individuals. However, we failed to detect any changes in the production of these cytokines by monocytes in the Control group after stimulation with SARS-CoV-2 S and N antigens. These results are in contrast to a previously published study by Karwaciak et al. who demonstrated increased IL-6 production and IL-6 mRNA expression by peripheral blood monocytes from healthy donors after stimulation with SARS-CoV-2 S and N antigens (26). This discrepancy can be explained by the different stimulation duration. In our study, we cultured monocytes with the SARS-CoV-2 antigens for 2 days, whereas Karwaciak et al. activated monocytes for 3 days. However, the authors also found no differences in the production of IL-1b and TNF-α in response to the SARS-CoV-2 S and N antigens (26). Possibly other SARS-CoV-2 antigens or immunological triggers provoke elevated expression of IL-1b and TNF-α in individuals vaccinated with Gam-COVID-Vac- and those who had recovered from COVID-19.
The observed long-term phenotypic and functional changes in monocytes in the Gam-COVID-Vac or COVID-19 groups can be explained by “trained immunity.” It has been previously shown that COVID-19 can trigger central-trained immunity characterized by enhanced granulopoiesis. It was shown that CD34+ hematopoietic stem and progenitor cells enriched from the peripheral blood of COVID-19-recovered patients showed higher expression of S100A8/A9 and CSF3R and increased chromatin accessibility of these loci four to 12 months after COVID-19. Moreover, mature monocytes from recovered patients retained the altered chromatin accessibility found in progenitor cells and exhibited an increased type I IFN signature (9). Single-cell sequencing revealed the trained and activated epigenomic state of a population of T-bet-enriched CD16+ and IRF1-enriched CD14+ monocytes circulating in the peripheral blood of individuals convalescing from COVID-19 (27). The inactivated vaccines CoronaVac (Sinovac/Butantan) (15) and Vero Cell (China Biotechnology Group Corporation) (28) as well as the non-replicating viral vector vaccine ChAdOx1 nCoV-19 (Oxford/AstraZeneca/FioCruz) (14, 15) have also been shown to induce trained immunity by activating chromatin remodeling, resulting in a different phenotype and metabolic changes in human monocytes. The persistent alterations in monocytes from COVID-19-recovered and immunized individuals impact subsequent immune responses to pathogens and vaccines and could contribute to long-term clinical symptoms, such as long COVID-19, post-acute sequelae of SARS-CoV-2 infection, and COVID-19-associated multisystem inflammatory syndrome in adults and children (29–31).
An increase in the frequency of HLA-DR-expressing monocytes in both experimental groups in our study may indicate an improved antigen presentation capacity. Similarly, increased HLA-DR expression on trained monocytes triggered by other microbial stimuli was suggested to enhance the T cell response via enhanced antigen presentation by monocytes (32). Moreover, the tetraspanin CD63 serves as an activation-induced reinforcing element that promotes sustained and effective T cell activation (33). The increase in CD63-expressing classical monocytes seen in individuals immunized with the Gam-COVID-Vac and those who had recovered from COVID-19 suggests that SARS-CoV-2 stimuli augment the antigen-presenting functions of classical monocytes. The significantly increased expression of surface CXCR2 and intracellular TLR7 by the total subset of CD14+ cells in the Gam-COVID-Vac and COVID-19 groups may indicate an increased capacity of monocytes to migrate to the injured or inflamed tissues (34, 35) and to recognize single-stranded RNA (36), respectively. A high capacity of cytokine production observed in individuals from the Gam-COVID-Vac and COVID-19 groups could be beneficial for future immune defense and lead to a rapid local antimicrobial response and subsequent elimination of the pathogen. However, the clinical consequences of such long-term changes in monocytes are not clear. Inappropriately strong induction of pro-inflammatory cytokines by monocytes could contribute to the long-term inflammatory complications of COVID-19. Indeed, patients with long COVID-19 display transcriptional dysregulation in their innate immune cells (37) as well as immunological dysfunction characterized by highly activated innate immune cells with high production of IL-6, IL-1β, and TNF-α that persists for more than 6 months after COVID-19 (38, 39). Further research is warranted to elucidate whether the enduring phenotypic and functional alterations in monocytes observed in individuals vaccinated with Gam-COVID-Vac and those who had recovered from COVID-19 benefit immune defense or potentially contribute to COVID-19-related complications.
This study has several limitations. One limitation of this study is that no heterologous microbial stimuli were used and the specificity of the SARS-CoV-2 stimuli for the observed phenotypic and functional changes in monocytes remains unclear. Another limitation is that we were unable to find an age-matched Control group of donors; as a result, the mean age of the Control group is considerably older than that of the Gam-COVID-Vac group. The reason for this is the fact that activity and willingness to be vaccinated against COVID-19 was very high among younger individuals in the first year that the vaccines became available in the country. However, the correlation analysis between the age of the donors in all groups and the proportion of HLA-DR+, TLR-7+, CD63+, and CXCR2+ monocytes as well as cytokine production showed no significant associations, suggesting that the observed changes were not age-related. No correlation was also observed between the gender of donors and the parameters mentioned. Additionally, the small cohort of individuals involved in the study and the lack of re-analysis of peripheral blood monocytes at different time points after Gam-COVID-Vac vaccination and COVID-19 may be a limitation of our findings. Moreover, the study did not evaluate the absolute number of monocytes and did not analyze epigenomic changes in monocytes or underlying signaling pathways that could contribute to the observed long-term alterations in monocytes. Furthermore, we did not expose the monocytes obtained from the experimental group to secondary heterologous challenges to confirm whether the observed changes are indicative of trained immunity. Lastly, it would be most interesting to assess the impact of Gam-COVID-Vac and COVID-19 on other effector mechanisms such as phagocytosis and microbial killing capacity of monocytes, but this was beyond the aims of the present study.
Conclusions
Our results reveal long-term changes in the phenotype and functional state of circulating monocytes in individuals vaccinated with Gam-COVID-Vac and those who had recovered from mild/moderate COVID-19, including enhanced expression of HLA-DR, CD63, CXCR2, and TLR7 and increased production of IL-6, IL-1β, and TNF-α. Moreover, our ex vivo experiments demonstrate that SARS-CoV-2 antigens can induce enhanced expression of HLA-DR and TLR7 on monocytes, suggesting that at least some of the observed alterations in vaccinated individuals or those recovered from COVID-19 are provoked by the virus itself. While our study is a pilot investigation and has a descriptive nature, it underscores the need for further studies to understand the potential importance of these changes in subsequent infections.
ACKNOWLEDGMENTS
This study was supported by the Science Committee of Ministry of Higher Education and Science of the Republic of Kazakhstan (grant #AP13067593 “Role of MDSCs in regulation of vaccine-induced immune response in aging and development of pharmacological approaches to improve vaccination efficacy”).
Conceptualization, Y.O.O.; data curation, S.A.K., A.V.L., N.A., Y.O.O., A.K., R.T., A.V.P.; data analysis, Y.O.O.; supervision and writing—original draft preparation, Y.O.O.; writing—review and editing, Y.V.P. All authors have read and agreed to the published version of the paper.
Contributor Information
Yekaterina O. Ostapchuk, Email: katyostapchuk@gmail.com.
Yuliya V. Perfilyeva, Email: perfilyevayulya@gmail.com.
Jeroen P. J. Saeij, University of California Davis, Davis, California, USA
DATA AVAILABILITY
Data supporting the findings of this study are presented in the paper. Additional information will be made available by the corresponding author upon reasonable request.
ETHICS APPROVAL
The study was approved by the Ethics Committee of the M.A. Aitkhozhin’s Institute of Molecular Biology and Biochemistry (Almaty, Kazakhstan).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00216-24.
Expression of CD11b, CD15, CD54, CXCR1, TLR2, TLR8, HLA-DR/DP/DQ, CD49d, and E4Bp4 on monocytes.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Merad M, Martin JC. 2020. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 20:355–362. doi: 10.1038/s41577-020-0331-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO . 2023. World health organization Coronavirus (COVID-19) dashboard (2023). World Health Organization, Geneva. Available from: https://covid19.who.int/. Retrieved 30 Nov 2023. [Google Scholar]
- 3. Bonny TS, Patel EU, Zhu X, Bloch EM, Grabowski MK, Abraham AG, Littlefield K, Shrestha R, Benner SE, Laeyendecker O, Shoham S, Sullivan D, Quinn TC, Casadevall A, Pekosz A, Redd AD, Tobian AAR. 2021. Cytokine and chemokine levels in coronavirus disease 2019 convalescent plasma. Open Forum Infect Dis 8:faa574. doi: 10.1093/ofid/ofaa574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ong SWX, Fong S-W, Young BE, Chan Y-H, Lee B, Amrun SN, Chee R-L, Yeo N-W, Tambyah P, Pada S, Tan SY, Ding Y, Renia L, Leo Y-S, Ng LFP, Lye DC. 2021. Persistent symptoms and association with inflammatory cytokine signatures in recovered coronavirus disease 2019 patients. Open Forum Infect Dis 8:fab156. doi: 10.1093/ofid/ofab156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park J, Dean LS, Jiyarom B, Gangcuangco LM, Shah P, Awamura T, Ching LL, Nerurkar VR, Chow DC, Igno F, Shikuma CM, Devendra G. 2023. Elevated circulating monocytes and monocyte activation in COVID-19 convalescent individuals. Front Immunol 14:1151780. doi: 10.3389/fimmu.2023.1151780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rajamanickam A, Kumar NP, Pandiarajan AN, Selvaraj N, Munisankar S, Renji RM, Venkatramani V, Murhekar M, Thangaraj JWV, Kumar MS, Kumar CPG, Bhatnagar T, Ponnaiah M, Sabarinathan R, Saravanakumar V, Babu S. 2021. Dynamic alterations in monocyte numbers, subset frequencies and activation markers in acute and convalescent COVID-19 individuals. Sci Rep 11:20254. doi: 10.1038/s41598-021-99705-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ravkov EV, Williams ESCP, Elgort M, Barker AP, Planelles V, Spivak AM, Delgado JC, Lin L, Hanley TM. 2023. Reduced monocyte proportions and responsiveness in convalescent COVID-19 patients. Front Immunol 14:1329026. doi: 10.3389/fimmu.2023.1329026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Dong L, Cui X, Miao Y, Wang D, Dong J, Xiao C, Chen W, Wang H. 2020. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov 6:31. doi: 10.1038/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheong J-G, Ravishankar A, Sharma S, Parkhurst CN, Grassmann SA, Wingert CK, Laurent P, Ma S, Paddock L, Miranda IC, et al. 2023. Epigenetic memory of coronavirus infection in innate immune cells and their progenitors. Cell 186:3882–3902. doi: 10.1016/j.cell.2023.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brauns E, Azouz A, Grimaldi D, Xiao H, Thomas S, Nguyen M, Olislagers V, Vu Duc I, Orte Cano C, Del Marmol V, Pannus P, Libert F, Saussez S, Dauby N, Das J, Marchant A, Goriely S. 2022. Functional reprogramming of monocytes in patients with acute and convalescent severe COVID-19. JCI Insight 7:e154183. doi: 10.1172/jci.insight.154183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott NA, Pearmain L, Knight SB, Brand O, Morgan DJ, Jagger C, Harbach S, Khan S, Shuwa HA, Franklin M, et al. 2023. Monocyte migration profiles define disease severity in acute COVID-19 and unique features of long COVID. Eur Respir J 61:2202226. doi: 10.1183/13993003.02226-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mohan A, Iyer VA, Kumar D, Batra L, Dahiya P. 2023. Navigating the post-COVID-19 immunological era: understanding long COVID-19 and immune response. Life (Basel) 13:2121. doi: 10.3390/life13112121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. PHOSP-COVID Collaborative Group . 2022. Clinical characteristics with inflammation profiling of long COVID and association with 1-year recovery following hospitalization in the UK: a prospective observational study. Lancet Respir Med 10:761–775. doi: 10.1016/S2213-2600(22)00127-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy DM, Cox DJ, Connolly SA, Breen EP, Brugman AA, Phelan JJ, Keane J, Basdeo SA. 2023. Trained immunity is induced in humans after immunization with an adenoviral vector COVID-19 vaccine. J Clin Invest 133:e162581. doi: 10.1172/JCI162581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Antunes M da S, Sugiyama FHC, Gravina HD, Castro RC, Mercado FJR, de Lima JO, Fontanari C, Frantz FG. 2023. COVID-19 inactivated and non-replicating viral vector vaccines induce regulatory training phenotype in human monocytes under epigenetic control. Front Cell Infect Microbiol 13:1200789. doi: 10.3389/fcimb.2023.1200789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui C, Schoenfelt KQ, Becker KM, Becker L. 2021. Isolation of polymorphonuclear neutrophils and monocytes from a single sample of human peripheral blood. STAR Protoc 2:100845. doi: 10.1016/j.xpro.2021.100845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJM, Liu Y-J, MacPherson G, Randolph GJ, Scherberich J, Schmitz J, Shortman K, Sozzani S, Strobl H, Zembala M, Austyn JM, Lutz MB. 2010. Nomenclature of monocytes and dendritic cells in blood. Blood 116:e74–80. doi: 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]
- 18. Walter LO, Cardoso CC, Santos-Pirath ÍM, Costa HZ, Gartner R, Werle I, Mohr ETB, da Rosa JS, Felisberto M, Kretzer IF, Masukawa II, Vanny P de A, Luiz MC, de Moraes ACR, Dalmarco EM, Santos-Silva MC. 2022. The relationship between peripheral immune response and disease severity in SARS-CoV-2-infected subjects: a cross-sectional study. Immunology 165:481–496. doi: 10.1111/imm.13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Q, Wen Y, Qi F, Gao X, Chen W, Xu G, Wei C, Wang H, Tang X, Lin J, Zhao J, Zhang M, Zhang S, Zhang Z. 2021. Suppressive monocytes impair MAIT cells response via IL-10 in patients with severe COVID-19. J Immunol 207:1848–1856. doi: 10.4049/jimmunol.2100228 [DOI] [PubMed] [Google Scholar]
- 20. El-Hefnawy SM, Eid HA, Mostafa RG, Soliman SS, Omar TA, Azmy RM. 2022. COVID-19 susceptibility, severity, clinical outcome and Toll-like receptor (7) mRNA expression driven by TLR7 gene polymorphism (rs3853839) in middle-aged individuals without previous comorbidities. Gene Rep 27:101612. doi: 10.1016/j.genrep.2022.101612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg B-J, Wijmenga C, Joosten LAB, Xavier RJ, van der Meer JWM, Stunnenberg HG, Netea MG. 2012. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12:223–232. doi: 10.1016/j.chom.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding J, Feng T, Ning Y, Li W, Wu Q, Qian K, Wang Y, Qi C. 2015. β-Glucan enhances cytotoxic T lymphocyte responses by activation of human monocyte-derived dendritic cells via the PI3K/AKT pathway. Hum Immunol 76:146–154. doi: 10.1016/j.humimm.2015.01.009 [DOI] [PubMed] [Google Scholar]
- 23. Gascon RL, Narváez AB, Zhang R, Kahn JO, Hecht FM, Herndier BG, McGrath MS. 2002. Increased HLA-DR expression on peripheral blood monocytes in subsets of subjects with primary HIV infection is associated with elevated CD4 T-cell apoptosis and CD4 T-cell depletion. J Acquir Immune Defic Syndr 30:146–153. doi: 10.1097/00042560-200206010-00002 [DOI] [PubMed] [Google Scholar]
- 24. Tillinger W, Jilch R, Waldhoer T, Reinisch W, Junger W. 2013. Monocyte human leukocyte antigen-DR expression-a tool to distinguish intestinal bacterial infections from inflammatory bowel disease?. Shock 40:89–94. doi: 10.1097/SHK.0b013e318299ebdd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Wang Y, Wang W, Ying W, Sun B, Wang X, Sun J. 2022. Increased expression of the TLR7/9 signaling pathways in chronic active EBV infection. Front Pediatr 10:1091571. doi: 10.3389/fped.2022.1091571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karwaciak I, Sałkowska A, Karaś K, Dastych J, Ratajewski M. 2021. Nucleocapsid and spike proteins of the coronavirus SARS-CoV-2 induce IL6 in monocytes and macrophages-potential implications for cytokine storm syndrome. Vaccines (Basel) 9:54. doi: 10.3390/vaccines9010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. You M, Chen L, Zhang D, Zhao P, Chen Z, Qin EQ, Gao Y, Davis MM, Yang P. 2021. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat Cell Biol 23:620–630. doi: 10.1038/s41556-021-00690-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J, Wang J, Xu J, Xia H, Wang Y, Zhang C, Chen W, Zhang H, Liu Q, Zhu R, et al. 2021. Comprehensive investigations revealed consistent pathophysiological alterations after vaccination with COVID-19 vaccines. Cell Discov 7:99. doi: 10.1038/s41421-021-00329-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Del Rio C, Collins LF, Malani P. 2020. Long-term health consequences of COVID-19. JAMA 324:1723–1724. doi: 10.1001/jama.2020.19719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris SB, Schwartz NG, Patel P, Abbo L, Beauchamps L, Balan S, Lee EH, Paneth-Pollak R, Geevarughese A, Lash MK, et al. 2020. Case series of multisystem inflammatory syndrome in adults associated with SARS-CoV-2 infection - United Kingdom and United States, March-August 2020. MMWR Morb Mortal Wkly Rep 69:1450–1456. doi: 10.15585/mmwr.mm6940e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gruber CN, Patel RS, Trachtman R, Lepow L, Amanat F, Krammer F, Wilson KM, Onel K, Geanon D, Tuballes K, et al. 2020. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C). Cell 183:982–995. doi: 10.1016/j.cell.2020.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy DM, Mills KHG, Basdeo SA. 2021. The effects of trained innate immunity on T cell responses; clinical implications and knowledge gaps for future research. Front Immunol 12:706583. doi: 10.3389/fimmu.2021.706583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maecker HT, Todd SC, Levy S. 1997. The tetraspanin superfamily: molecular facilitators. FASEB J 11:428–442. doi: 10.1096/fasebj.11.6.9194523 [DOI] [PubMed] [Google Scholar]
- 34. Wong KL, Tai J-Y, Wong W-C, Han H, Sem X, Yeap W-H, Kourilsky P, Wong S-C. 2011. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118:e16–31. doi: 10.1182/blood-2010-12-326355 [DOI] [PubMed] [Google Scholar]
- 35. Weber C, Belge KU, von Hundelshausen P, Draude G, Steppich B, Mack M, Frankenberger M, Weber KS, Ziegler-Heitbrock HW. 2000. Differential chemokine receptor expression and function in human monocyte subpopulations. J Leukoc Biol 67:699–704. doi: 10.1002/jlb.67.5.699 [DOI] [PubMed] [Google Scholar]
- 36. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101:5598–5603. doi: 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryan FJ, Hope CM, Masavuli MG, Lynn MA, Mekonnen ZA, Yeow AEL, Garcia-Valtanen P, Al-Delfi Z, Gummow J, Ferguson C, O’Connor S, Reddi BAJ, Hissaria P, Shaw D, Kok-Lim C, Gleadle JM, Beard MR, Barry SC, Grubor-Bauk B, Lynn DJ. 2022. Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med 20:26. doi: 10.1186/s12916-021-02228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin JX, Agbana YL, Sun ZS, Fei SW, Zhao HQ, Zhou XN, Chen JH, Kassegne K. 2023. Increased interleukin-6 is associated with long COVID-19: a systematic review and meta-analysis. Infect Dis Poverty 12:43. doi: 10.1186/s40249-023-01086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schultheiß C, Willscher E, Paschold L, Gottschick C, Klee B, Henkes SS, Bosurgi L, Dutzmann J, Sedding D, Frese T, Girndt M, Höll JI, Gekle M, Mikolajczyk R, Binder M. 2022. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep Med 3:100663. doi: 10.1016/j.xcrm.2022.100663 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of CD11b, CD15, CD54, CXCR1, TLR2, TLR8, HLA-DR/DP/DQ, CD49d, and E4Bp4 on monocytes.
Data Availability Statement
Data supporting the findings of this study are presented in the paper. Additional information will be made available by the corresponding author upon reasonable request.