ABSTRACT
Commensal bacteria are crucial in maintaining host physiological homeostasis, immune system development, and protection against pathogens. Despite their significance, the factors influencing persistent bacterial colonization and their impact on the host still need to be fully understood. Animal models have served as valuable tools to investigate these interactions, but most have limitations. The bacterial genus Neisseria, which includes both commensal and pathogenic species, has been studied from a pathogenicity to humans perspective but lacks models that study immune responses in the context of long-term persistence. Neisseria musculi, a recently described natural commensal of mice, offers a unique opportunity to study long-term host-commensal interactions. In this study, for the first time, we have used this model to study the transcriptional, phenotypic, and functional dynamics of immune cell signatures in the mucosal and systemic tissue of mice in response to N. musculi colonization. We found key genes and pathways vital for immune homeostasis in palate tissue, validated by flow cytometry of immune cells from the lung, blood, and spleen. This study offers a novel avenue for advancing our understanding of host-bacteria dynamics and may provide a platform for developing efficacious interventions against mucosal persistence by pathogenic Neisseria.
KEYWORDS: Neisseria, immune dynamics, vaccine, mucosal immunity, immune homeostasis, host-microbes interactions, NK cells, monocytes
INTRODUCTION
Commensal bacteria are an important component of a host’s physiological environment and play a critical role in its functional homeostasis, immune system development, and protection against pathogens (1, 2). Despite their importance, the factors influencing persistent bacterial colonization and its impact on the host’s immune system remain understudied. To delve deeper into commensal-host interactions and their consequences, animal models have traditionally served as valuable tools.
Neisseria has been investigated for its commensal nature within mammalian hosts. It encompasses many species that are genetically related (3). These can colonize a broad range of hosts ranging from rodents to non-human primates to humans (4). The upper respiratory tract is considered the commonest anatomical location from which Neisseria species have been cultured (5); other niches where commensals thrive include the tongue dorsum and gingival plaque (6). The two primary human pathogens, Neisseria gonorrhoeae and Neisseria meningitidis, are associated with high levels of asymptomatic colonization of the upper respiratory mucosal tissue and can straddle the border between commensalism and pathogenicity (7). Both pathogens are frequently found at anorectal sites (8, 9). Given the absence of an effective vaccine for gonococci, various infection models have been created to enhance our comprehension of pathogen-host dynamics and uncover insights into the immune response and potential vaccine-induced protection mechanisms. Indeed, several mouse models have been proposed to study N. gonorrhoeae genital tract infections, but due to its tropism for humans, these models require several external manipulations to maintain transitory infections and, therefore, may not mimic natural conditions in which host-pathogen interactions can be fully assessed (10). Pharyngeal infection models are limited, yet this site facilitates persistent colonization by both pathogenic Neisseria species for several months (11, 12). We have recently developed a mouse model to explore Neisseria colonization, using the newly characterized mouse commensal Neisseria musculi (13). Retaining crucial host interaction genes in N. musculi, shared with N. gonorrhoeae, makes it an excellent surrogate species for modeling gonococcal pharyngeal colonization (13–15). Furthermore, given that N. musculi is a commensal of wild mice, its oral inoculation into laboratory-bred mice (i.e., non-natural host) likely represents the first exposure of this bacterial species to a naïve host in a manner that can mimic N. gonorrhoeae colonization of previously uninfected human oropharyngeal tissue.
In this study, we found multiple changes in the transcriptional profile of palate tissue colonized with N. musculi and the enrichment of the gene pathways associated with diverse biological processes. For the first time, we have unraveled the dynamic alterations in both innate and adaptive immune signatures within the systemic tissue of colonized mice, fostering the establishment and maintenance of immune homeostasis. Our findings may advance the knowledge toward developing potential vaccine candidates efficacious against mucosal carriage of pathogenic Neisseria species.
RESULTS
Oropharyngeal colonization by N. musculi was established following oral inoculation of the bacterium
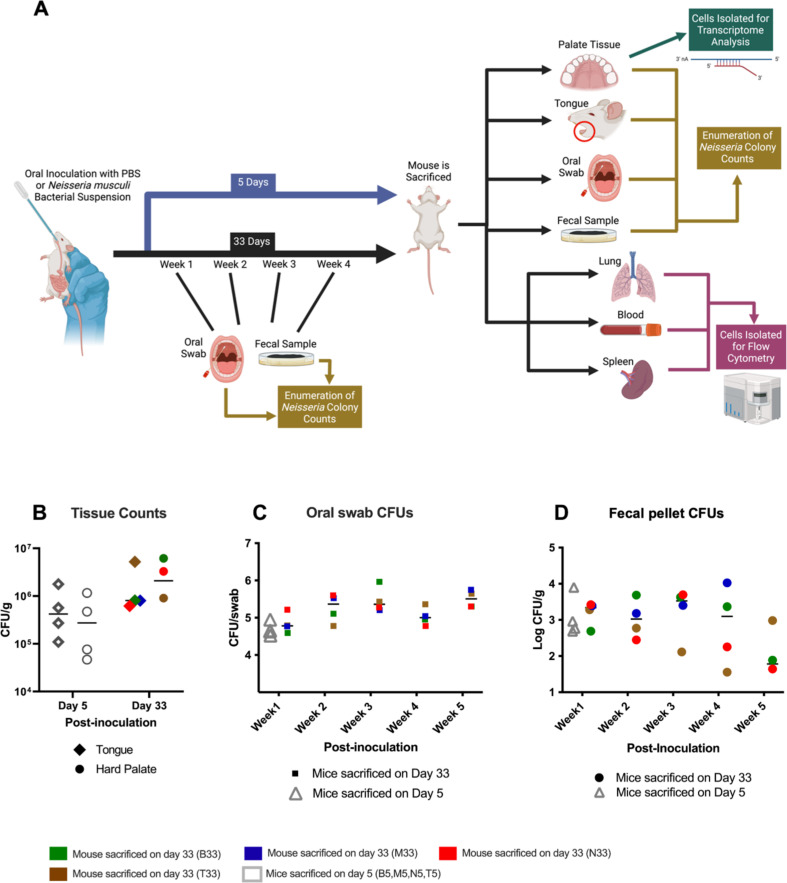
A total of 16 A/J mice were divided into two groups of eight mice each consisting of an inoculated group and a control group. A slow oral inoculation of 50 µL of N. musculi bacterial suspension was administered to each mouse in the inoculated group, while control mice were mock inoculated with 50 µL of phosphate-buffered saline (PBS) into the oral cavity (Fig. 1A). N. musculi colonies were enumerated for the following sample types: oral swabs, fecal pellets, tongue, and hard palate (lung, spleen, and blood samples were used exclusively for immunophenotyping studies; see Materials and methods). Here, we found that following inoculation, the bioburden of N. musculi in colonized tissues of the tongue and hard palate was higher at day 33 (late) compared with day 5 (early; Fig. 1B). However, our serial monitoring of colony counts obtained through a weekly sampling of oral swabs (Fig. 1C) and fecal pellets (Fig. 1D) suggests that bacterial colonies continue to increase and peak around the third week after inoculation with no further increases, suggesting a degree of homeostasis and tolerance between host tissue and commensal bacteria has been reached.
Fig 1.
Experimental design and establishment of colonization following oral inoculation with N. musculi. (A) Mice from the experimental group (n = 8) were orally inoculated with N. musculi followed by a subset (n = 4) being euthanized after 5 days. Inoculated mice left for 33 days (n = 4) before euthanasia were sampled weekly for enumeration of oral and fecal colony counts. Blood, lung, and spleen were harvested at sacrifice for flow cytometric analysis of immune signatures between inoculated and control mice. Enumeration of oral colonization was done using colony counts of the tongue and palate. Bulk RNA sequencing was performed on palate tissue to observe differences in tissue transcriptome following colonization. Control mice were administered PBS orally. (B) Dot plot depicting the differential abundance of N. musculi colonies in colonized tongue and hard palate tissue at days 5 and 33 after inoculation. Dot plots represent weekly changes in N. musculi colony counts obtained from the culture of (C) oral swabs and (D) fecal pellets of inoculated mice. CFU/g, colony-forming units per gram of tissue. Animal M33 did not have palate colony counts and fecal counts for week 5.
Transcriptome analysis reveals dynamic changes in gene expression and immune cell composition during N. musculi colonization in palate tissue
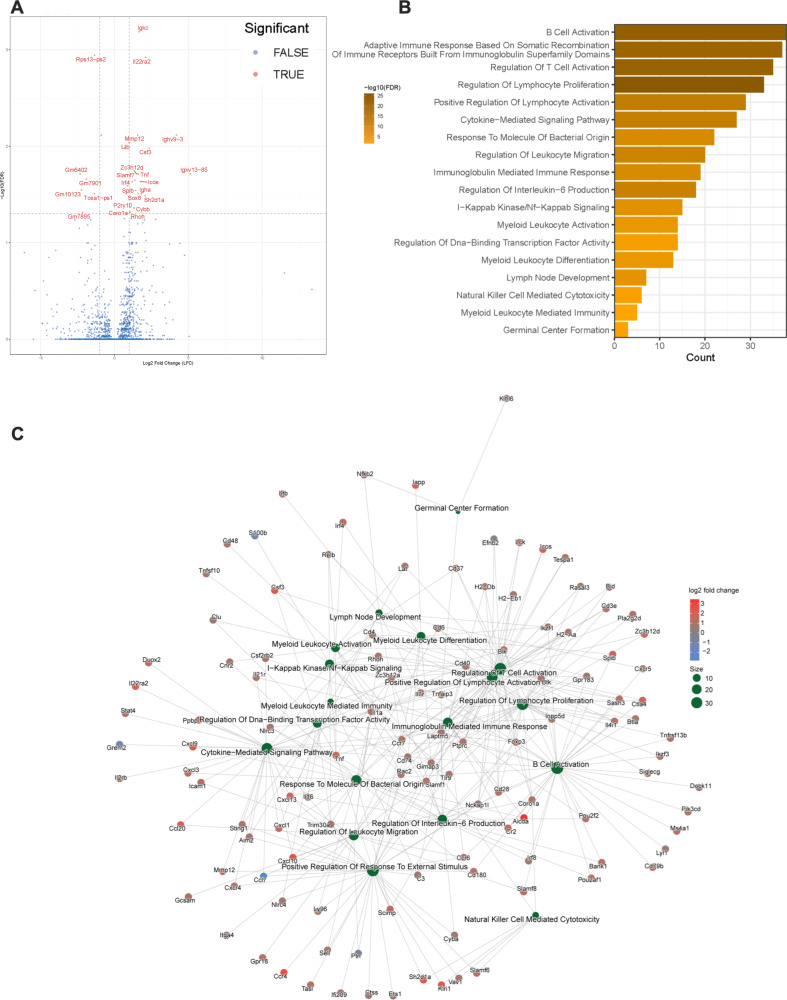
We performed bulk RNA sequencing (RNA-seq) on the palate tissue of mice inoculated with N. musculi and then sacrificed on day 5 and day 33 post-inoculation, as well as their control mice. Based on the expression of selected biomarkers from our transcriptome analysis, we observed that host gene expression patterns distinguish mice colonized with N. musculi from their controls (Fig. S1). While no major differences in gene expression were noted by day 5 after inoculation, at day 33, a total of 26 differentially expressed genes (DEGs), including 20 upregulated DEGs and 6 downregulated DEGs (Table S1), were identified in the tissues of the palate of mice colonized with N. musculi compared with those of control mice (Fig. 2A). Here, we observed several DEGs linked to inflammation (Ltb, Csf3, Tnf, and Slamf7) and mucosal immunity (IL22ra and Igha), likely attributed to colonization with N. musculi.
Fig 2.
DEGs and enriched transcriptional pathways of cellular biological processes in palates of mice 33 days after being orally inoculated with N. musculi vs mock-inoculated controls. (A) Volcano plot of DEGs between inoculated mice (n = 4) and controls (n = 4; screening thresholds: adjusted P value < 0.05). (B) Gene Ontology (GO) analysis was performed and revealed significantly enriched pathways in palate tissue from inoculated mice vs controls related to biological processes (screening threshold: adjusted P value < 0.05). (C) Cnet plot of GO biological process enrichment of DEGs of palate tissue of inoculated vs control mice shows distinct connections between biological processes resulting from colonization with N. musculi.
To analyze the biological importance of these DEGs, Gene Ontology (GO) enrichment analysis was performed (Table S2). Based on the enriched GO terms, we observed significantly enriched pathways involving the biological activity of several immune cells including T cells, B cells, cells of the myeloid lineage, and natural killer (NK) cells (Fig. 2B). Of particular interest were enriched pathways involving myeloid leukocyte activation and differentiation, NK cell-mediated cytotoxicity, T- and B-cell activation, lymphocyte proliferation, immunoglobulin-mediated immune response, IL-6 production, germinal center formation, and response to bacteria. We also created network visualizations of selected enriched GO pathways using Cnet plots (Fig. 2C; Fig. S2). Connections between pathways in the network imply shared genes, suggesting possible crosstalk or cooperation in enriched biological processes particularly related to leukocyte immune activation due to colonization with N. musculi.
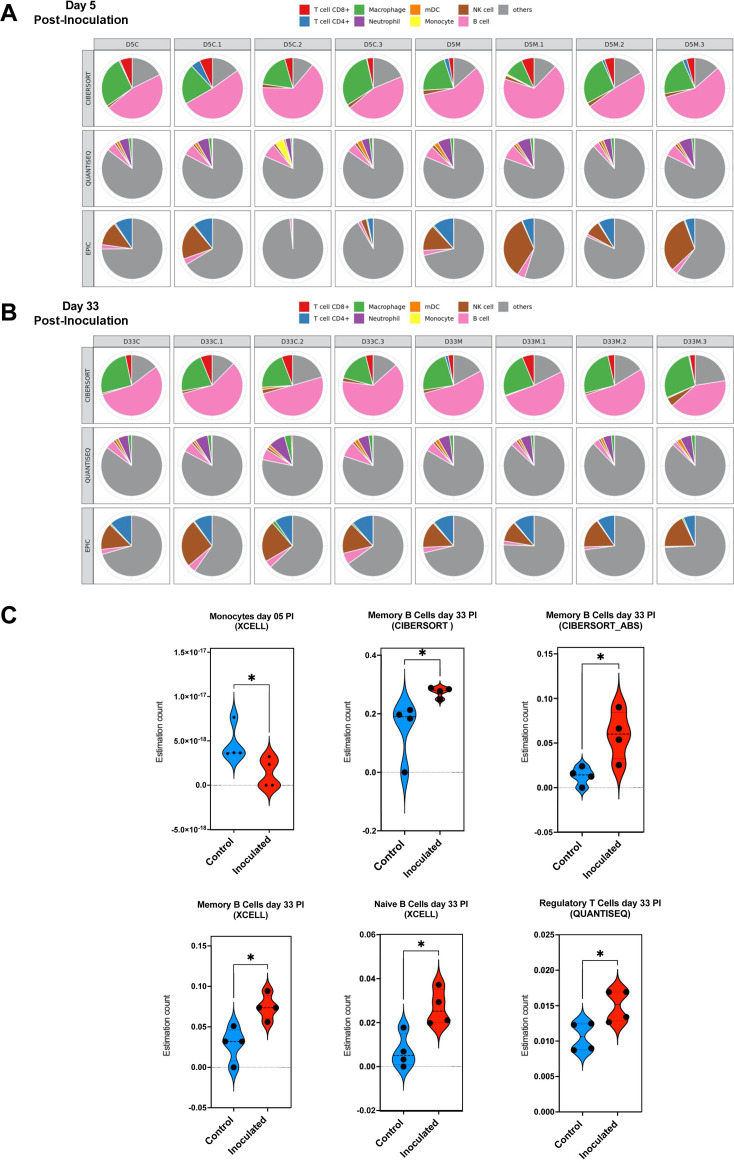
We also performed deconvolution of our bulk transcriptome data of palate tissue of mice inoculated with N. musculi and control mice using TIMER2.0 (16). Using six algorithms, we generated differential estimations of the abundance of immune cell subsets in each sample of the two groups (Fig. 3A and B; Fig. S3). Here, we found significant differences in the expression of immune cells at both time points (day 5 and day 33) following bacterial colonization of palate tissue (Fig. 3C). At day 5 after inoculation, our data indicated significantly reduced expression of monocytes, the main producers of IL-6, which mediates partial resistance to bacterial colonization, in the palates of mice colonized with N. musculi. Conversely by day 33 after inoculation, data from deconvolution of the palate transcriptome indicated an overall enhancement of the adaptive immune response reflected by an altered tissue transcriptome suggesting increased expression of memory B cells, naïve B cells, and regulatory T cells in the palates of mice colonized with N. musculi compared with palate tissue of controls.
Fig 3.
Deconvolution of transcriptome data from palate tissue of mice 5 and 33 days after being inoculated with N. musculi vs mock-inoculated control mice. The TIMER2.0 database was used to generate estimations of the abundance of immune cell subsets in the palate tissue of each mouse belonging to both time points using six algorithms. Multi-panel pie plots for three algorithms that generate comparable values show the proportions of major immune cell types in each sample at (A) day 5 and (B) day 33 after inoculation (including those of controls). D5C/D33C, mice colonized with N. musculi sacrificed on day 5/day 33. D5M/D33M, mock-inoculated mice sacrificed at day 5/day 33. (C) Violin plots based on algorithm-generated abundance scores depict the differential expression of selected immune cell subsets at day 5 and day 33 after inoculation compared with control mice in corresponding time points. The P value was calculated using the Wilcoxon rank-sum test *P < 0.05. PI, post-inoculation.
Dynamic and temporal changes are observed in the immune response of mucosal tissue that is anatomically adjacent to tissue colonized with N. musculi
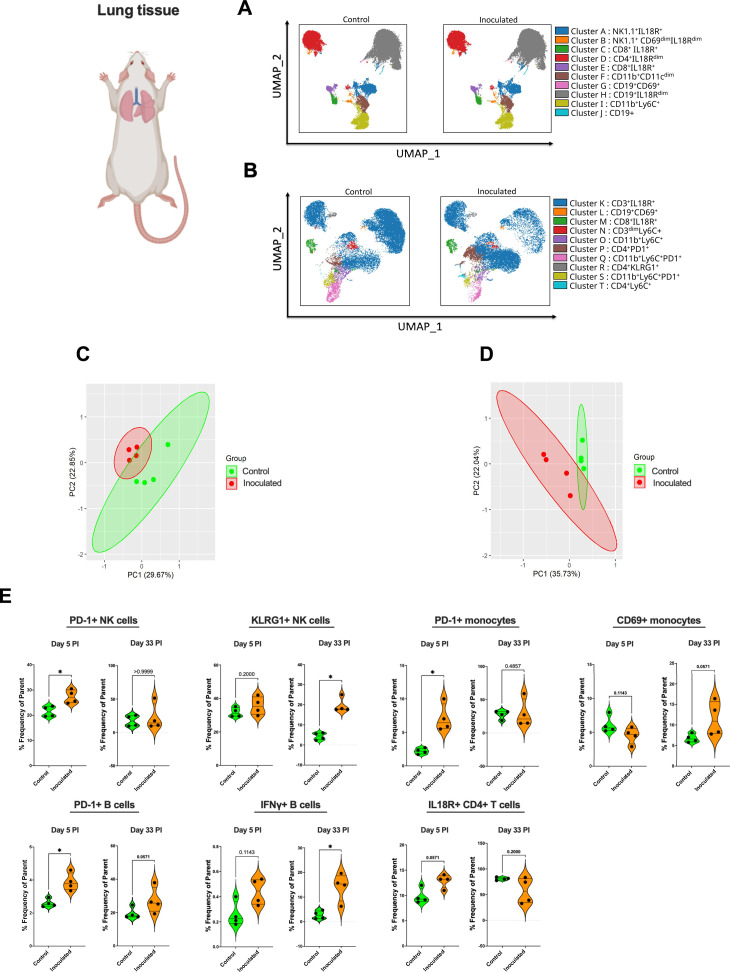
Of the tissues analyzed by flow cytometry, we considered tissue of the respiratory system (i.e., lung) as mucosal tissue anatomically most adjacent to the inoculated (and therefore colonized) oral cavity (17). We initially performed unbiased clustering of lung immune cells at both day 5 (Fig. 4A) and day 33 (Fig. 4B) after inoculation by utilizing FlowSOM-based automatic clustering algorithms and Uniform Manifold Approximation and Projection (UMAP) for dimensional reduction visualization. This analysis was conducted on CD45+ cells to discern changes in immune cell clusters between mice inoculated with N. musculi and controls. While no major changes in clustering were observed between these two groups on day 5 after inoculation (Fig. 4A), we observed changes in monocyte and T cell clusters at day 33 (Fig. 4B) after inoculation (late colonization) suggesting differences in the phenotypic expression of these immune cells in the lung mucosal tissue influenced by Neisseria colonization. Principal component analysis (PCA) performed using flow cytometric data revealed distinct clustering between inoculated mice and control mice at both day 5 (Fig. 4C) and day 33 (Fig. 4D) after bacterial inoculation, suggesting that colonization with N. musculi drastically alters the immunophenotype and functionality of mucosal immune cells in adjacent tissue. To understand changes induced by bacterial colonization in specific immune cell subsets, we performed manual gating (Fig. S4) of our flow cytometry data (Fig. 4E). Many of the immune signatures significantly altered by bacterial colonization were dynamic and temporal, in that these alterations were limited to occur either early on (day 5) or later (day 33) during colonization. Interestingly, we observed upregulation of PD-1 on B cells, which was consistently elevated in the lung tissue of inoculated mice throughout colonization. Given that these are unswitched, IgM-producing memory B cells, colonization therefore results in a persistently enhanced mucosal humoral immune response capable of generating antibodies on exposure to related antigens (18). We also observed increased PD-1 expression in monocytes and NK cells of inoculated mice compared with controls, but only early on in colonization and reaching baseline later on day 33, suggesting tolerance and immune homeostasis developing following early persistent antigenic stimulation due to bacterial colonization (19). Nevertheless, our results indicated that the activity of tissue-resident monocytes may still be enhanced with a more mature phenotype seen among NK cells even late into colonization as evidenced by increased CD69 (also a marker of tissue residency) expression in monocytes (20, 21) and increased KLRG1 expression in NK cells (22), respectively. The enhanced activity of monocytes in lung tissue may be at least partially explained by significantly higher proportions of IL-18 receptor (IL18R) expressing CD4+ T cells (found to express more IFNγ) (23) and IFNγ producing innate B cells in this tissue of colonized mice (24) which also suggests a more pro-inflammatory environment occurring in lung mucosal tissue as a result of N. musculi colonization.
Fig 4.
Alterations in the immune landscape of adjacent mucosal tissue in mice following oral colonization with N. musculi. FlowSOM-based clustering of CD45+ cells in mucosal lung tissue of mice (A) 5 days and (B) 33 days after inoculation with N. musculi, depicted on UMAPs, reveal changes in phenotypic clusters compared with that of controls. PCA was performed using selected immune markers and revealed distinct clustering of mice inoculated with N. musculi at (C) day 5 and (D) day 33, compared with controls. (E) Violin plots depicting differential expression of immune signatures between inoculated and control mice at day 5 and day 33 after inoculation. The P value was calculated using the Wilcoxon rank-sum test *P < 0.05. PI, post-inoculation.
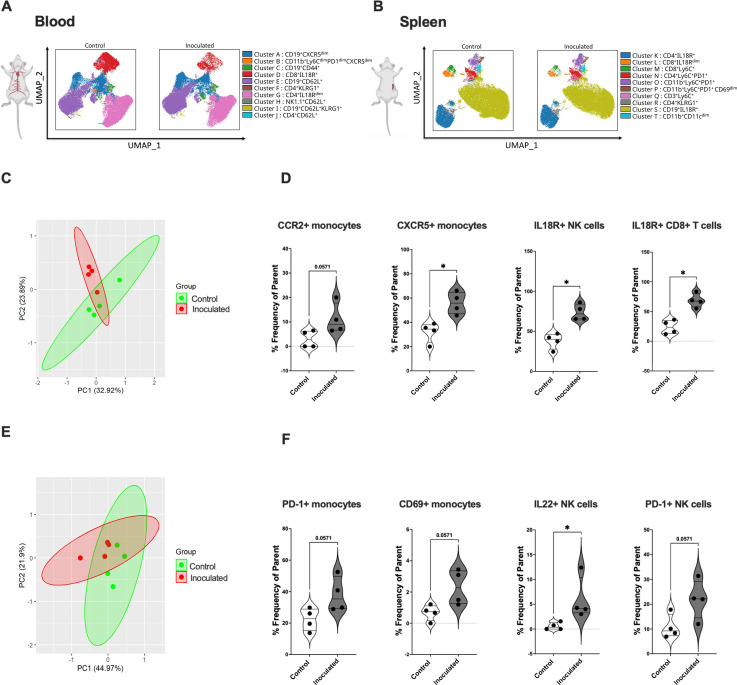
Oral inoculation with N. musculi alters the systemic immune response in mice toward a pro-inflammatory phenotype early on after exposure to the bacterium
Alterations in immune signatures in the blood and spleen were representative of the systemic immune response in mice following colonization with N. musculi. FlowSOM-based unbiased clustering visualized on UMAP revealed changes in the expression of CD45+ cell clusters in both the blood and spleen in mice 5 days after bacterial inoculation vs controls. In terms of immune responses in the blood, we specifically observed differential expression of monocyte clusters and T cell clusters between the two groups (Fig. 5A), whereas in the spleen, changes were most apparent in monocyte clusters (Fig. 5B). To further delineate more specific differences in immune cell subsets brought on by N. musculi oral inoculation, we performed manual gating of our flow cytometry data from immune cells of blood and spleen. Based on PCA, overall changes in immune cell markers of the blood revealed distinct clustering of mice inoculated with N. musculi compared to controls (Fig. 5C). Detailed analysis of peripheral blood immune signatures revealed that colonization with N. musculi early on (day 5) increased the migratory capacity of blood monocytes to the inflamed mucosal tissue and secondary lymphoid organs as evidenced by increased surface expression of CCR2 (25) and CXCR5 (26), respectively (Fig. 5D). Furthermore, we found that oral inoculation resulted in peripheral cytotoxic immune cells including NK cells and CD8+ T cells displaying a more pro-inflammatory phenotype, as evidenced by their increased expression of IL-18R compared with controls, suggesting more IFNγ production by these cells (27, 28). In the spleen, we observed significant differences in immune phenotypes between mice inoculated with N. musculi compared to control mice (Fig. 5E). Here, we found increased PD-1 expression among NK cells and monocytes following inoculation, suggesting their response to persistent antigenic stimulation (29), coupled with enhanced activation of tissue-resident monocytes suggested by increased CD69 expression. Interestingly, inoculation with N. musculi also increased IL-22 production by NK cells of the spleen, suggesting their role in maintaining mucosal immunity during the early phase of colonization (Fig. 5F) (30).
Fig 5.
Alterations in the systemic immune landscape of mice early on following oral colonization with N. musculi. FlowSOM-based clustering of CD45+ cells in (A) blood and (B) spleen of mice 5 days after inoculation with N. musculi, depicted on UMAPs, reveals changes in their phenotypic clusters compared with that of controls. PCA was performed using selected immune markers in (C) blood and (E) spleen and revealed distinct clustering of mice inoculated with N. musculi at day 5 compared with controls. Violin plots depicting differential expression of immune signatures in (D) blood and (F) spleen between inoculated and control mice at day 5 after inoculation. The P value was calculated using the Wilcoxon rank-sum test *P < 0.05.
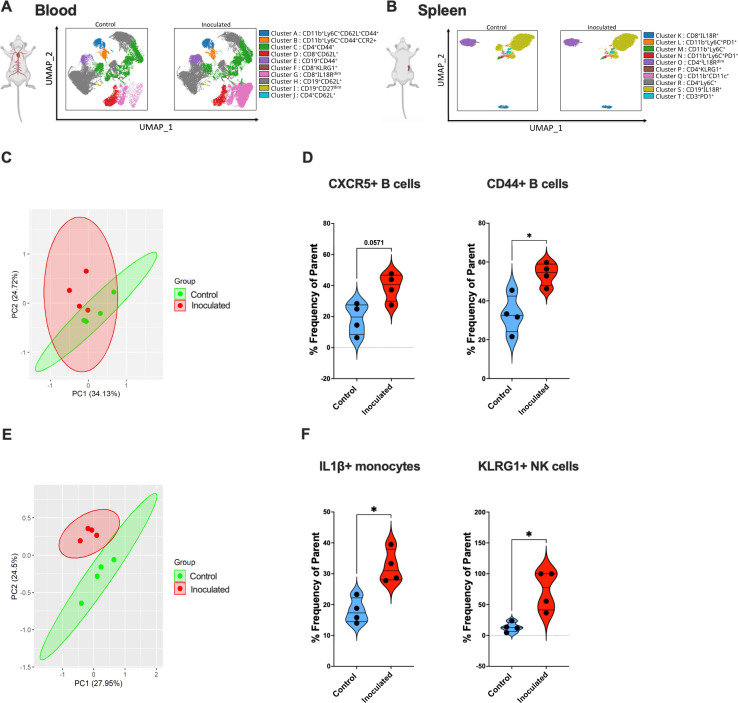
Colonization with N. musculi maintains a pro-inflammatory immune environment within systemic tissue even toward the later stages following inoculation
Our immune data obtained from the blood at day 33 after inoculation with N. musculi indicated notable changes in the expression of B and T cell clusters (Fig. 6A), whereas in the spleen, it was evident that monocytes and B cell clusters were the most prominently affected (Fig. 6B). Based on PCA, we also observed distinct immune responses in the blood (Fig. 6C) and spleen (Fig. 6E) of mice, at day 33 after oropharyngeal colonization with N. musculi. However, it is noteworthy that the frequency of prominent changes in immune signatures within both these tissues at day 33 post-inoculation compared to their corresponding controls (Fig. 6D and F) was comparatively lower than the frequency of changes in immune signatures (between inoculated and control groups) observed at day 5 post-inoculation. These findings suggest that over time, a certain degree of immune homeostasis and tolerance is established within these systemic tissues, supported by the patterns identified through PCA. However, we still observed increased migratory capabilities of B cells in the blood toward secondary lymphoid tissue and inflamed tissue as indicated by their increased expression of CXCR5 and CD44 (31) possibly due to the effects of continuous bacterial antigen exposure. Additionally, immune data from spleen tissue at day 33 after inoculation with N. musculi revealed more activated pro-inflammatory immune cells indicated by increased expression of IL1β from monocytes compared with controls (32). At the same stage of colonization, splenic NK cells were found to be of a more mature phenotype indicated by their increased expression of KLRG1, further confirming the impact of N. musculi colonization on the innate immune compartment.
Fig 6.
Alterations in the systemic immune landscape of mice in later stages following oral colonization with N. musculi. FlowSOM-based clustering of CD45+ cells in (A) blood and (B) spleen of mice 33 days after inoculation with N. musculi, depicted on UMAPs, reveals changes in their phenotypic clusters compared with that of controls. PCA was performed using selected immune markers in (C) blood and (E) spleen and revealed distinct clustering of mice inoculated with N. musculi at day 33 compared with controls. Violin plots depicting differential expression of immune signatures in (D) blood and (F) spleen between inoculated and control mice at day 33 after inoculation. The P value was calculated using the Wilcoxon rank-sum test *P < 0.05.
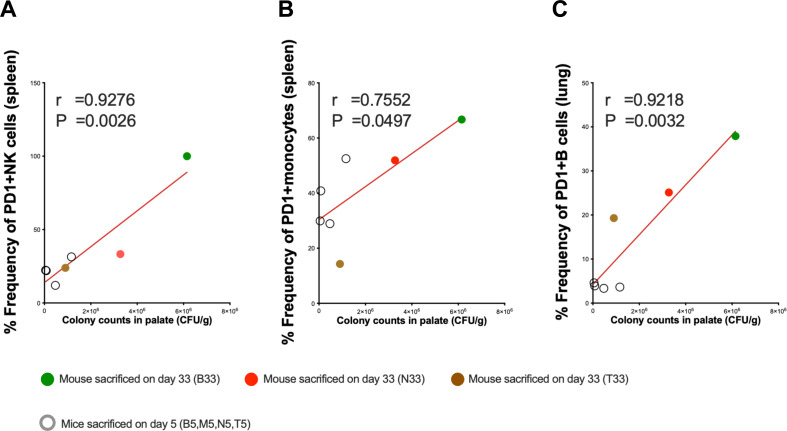
N. musculi colonization is associated with PD-1-expressing immune cells
We performed Pearson correlation coefficient analyses between colony counts of N. musculi in palate tissue at both days 5 and 33 and corresponding immune cell subsets in tissues. This revealed that the magnitude of colonization of palate tissue correlated with several immune cell subsets expressing the immune checkpoint marker PD-1, including splenic NK cells (Fig. 7A), splenic monocytes (Fig. 7B), and B cells of the lung (Fig. 7C). Given that PD-1 expression is associated with persistent antigenic stimulation and that the above immune subsets were found to be increasingly expressed at one or both time points following inoculation, this indicates that N. musculi directly influences the systemic and mucosal tissue immune landscape of the host.
Fig 7.
Correlational analysis between N. musculi colonization and immune signatures. Pearson correlation coefficient analysis reveals correlations between the magnitude of colonization of N. musculi in the palate and expression of PD-1 in (A) splenic NK cells, (B) splenic monocytes, and (C) B cells in lung. r, Pearson correlation coefficient. P, P value.
We did not assess the presence of live bacteria or bacterial antigens in tissues of the lung, spleen, or blood following oropharyngeal colonization with N. musculi. Nevertheless, our findings collectively indicate systemic alterations in the host immune system in response to oropharyngeal colonization of our laboratory mice with this Neisseria species to which they have been previously unexposed. Much like how intestinal colonization regulates the immune response both locally and systemically by microbial metabolites and other methods of immune modulation (20, 33), we speculate that our model of oropharyngeal and gut colonization would influence the immune response in a similar way. Nevertheless, understanding these determinants requires further investigation and is beyond the scope of this study.
DISCUSSION
The primary focus of existing Neisseria colonization studies has been on N. gonorrhoeae and N. meningitidis which cause human infections, with a goal to identify targets for vaccine antigens that can induce protective immune responses against these Neisseria species (34, 35). Many homologs of pathogenic Neisseria-host interaction factors and candidate vaccine antigens have been identified in the mouse commensal N. musculi. Our study unveiled notable changes in the expression of both systemic and mucosal innate and adaptive immune signatures following oral inoculation with N. musculi. As demonstrated by Powell et al., cytokines like IL-6 are pivotal in conferring partial resistance against N. musculi colonization (36). While several cell types, including stromal cells, epithelial cells, and fibroblasts, produce IL-6, monocytes stand out as one of the major immune cells responsible for IL-6 secretion (37). Interestingly, in this study, we observed reduced expression of monocyte signatures in the locally colonized palate tissue revealed by our transcriptomic deconvolution data in early colonization. Furthermore, at this early-stage colonization, we also noted increased expression of monocytes with upregulated chemokine receptors (i.e., CCR2 and CXCR5) (25, 26), suggesting their enhanced capacity to subsequently migrate from the circulation and enter tissue, and markers of activation and tissue residency (i.e., CD69) (20), in both the blood and the spleen. Therefore, we suggest that the innate immune response promotes local tissue colonization while, at the same time, likely led by tissue homing and tissue resident monocytes/macrophages, walling off the colonized tissue and limits systemic spread.
Transcriptomic and immunophenotyping readouts for animal models of infectious disease frequently focus on short periods of infection. An acute murine model of uterine infection identified hundreds of differentially expressed genes in response to 6 hours of exposure to the gonococcus (38). Great efforts have been made to monitor murine immune responses within 2 weeks of bacterial colonization in the hopes of identifying immunomodulatory processes (39). Some bacteria stimulate immune responses that result in their clearance. Citrobacter rodentium is eliminated from the murine gut within 30 days by the production of IL-22, an IL-10 family cytokine that can stimulate the production of proinflammatory cytokines (40). Murine spleen cells and vaginal tissue release IL-22 in response to N. gonorrhoeae (41). We observed increased systemic cytokine responses in early colonization, including upregulation of IL-22 production by splenic NK cells, a crucial cytokine involved in maintaining epithelial integrity and promoting bacterial colonization in lymphoid tissue, contributing to more robust systemic immune responses (32). We were intrigued that 33 days after N. musculi inoculation the gene (IL22RA2), which encodes the IL-22 binding protein (IL22BP), is upregulated in palate tissue. IL22BP is a soluble IL-22 receptor that can dampen IL-22 cell signaling and suggests a systemic response geared to regulate the colonization of N. musculi and promote a lymphoid tissue response to the bacteria. This type of data makes us curious to know if the pathogenic Neisseria species influence IL22BP expression at extragenital sites in a manner that influences persistence dynamics. We hope our N. musculi data sets will be useful in comparing how different modes of infection or colonization contribute to immune homeostasis and systemic immunity whether those modes are acute, chronic, transient, or commensal in nature.
Our data also suggest an overall pro-inflammatory response taking place in the early phase of colonization indicated by increased expression of peripheral blood T cells expressing IL18R which in turn are known to produce high levels of IFNγ (23). Similar to the discoveries by Baldridge et al., (42) wherein they revealed that IFNγ initiates the activation of dormant hematopoietic stem cells within the backdrop of chronic infection, we propose a similar phenomenon occurring during early colonization by N. musculi. Here, increased systemic IFNγ response could promote hematopoietic stem cell activation and enhance the immune response. Thus, we propose a local and systemic immune response occurring in the early phase of colonization geared toward promoting the establishment of N. musculi colonization in local inoculated tissue. Here, the systemic immune response, possibly influenced by circulating bacterial antigens and/or a systemically altered cytokine milieu, also builds up its defenses to ensure the host is protected from the potential systemic effects of the bacteria.
The immune response we observed during the late colonization differs significantly from the early phase of colonization. Specifically, we noted that by day 33 of colonization, many of the initially altered systemic immune responses return to levels similar to control mice with only a few immune signatures being altered during this late stage of colonization. During the later stages of colonization (i.e., day 33 after inoculation), our specific emphasis was directed toward examining the immune response within the mouse lung. Given its anatomical adjacency to the colonized palate, we identified the lung as an instrumental system profoundly influenced by N. musculi colonization. Hence, we deem the lung as a surrogate marker, providing insights into the immune response observed not only in mucosal tissues but also in anatomically neighboring sites affected by colonization. Given that pathogens like N. gonorrhoeae also colonize mucosal tissues of the upper respiratory tract, the data we gather by analyzing these tissue types will likely be invaluable during future vaccine development. The proposed link between colonized palate tissue and lung tissue was partially confirmed by deconvoluting transcriptome data from the palate, which showed similar signature changes to those observed in the late stages of lung tissue colonization. Here, we found the immune response to be protective and geared up to defend against further bacterial invasion. In the monocytes of lung tissue, we found increased expression of markers of activation and tissue residency such as CD69, suggesting increased production of IL-6 by monocytes in the lung micro-environment to limit the spread of bacterial colonization (as described earlier). Enrichment of GO terms corresponding to myeloid cell activation and differentiation further confirmed our results. A higher population of unswitched, IgM-producing memory B cells, signified by their increased expression of PD-1 (18), and confirmed by enrichment of GO terms corresponding to the humoral response means that colonization likely stimulates local neutralizing antibody production against the offending antigen. In late colonization, an increase in IFNγ-producing B cells suggests the sustained presence of a pro-inflammatory environment within this tissue. We also observed increased expression of KLRG1 on the NK cells of lung tissue suggesting that even innate immune cells in this tissue adjacent to colonized regions display a more mature phenotype and also contribute to immune homeostasis (43). This was likely primed by signals generated from adjacent colonized tissue, the transcriptome data of which showed enrichment of related pathways like leukocyte differentiation. We also observed upregulated immune markers associated with increased maturity (KLRG1+ splenic NK cells), pro-inflammatory activity (IL1β-secreting monocytes in blood), and lymphoid tissue homing capability (CXCR5 expressing B cells in blood) in the systemic immune compartment even late into colonization. A β-estradiol mouse model which was used to investigate gonococcal infection found that the infection, through unknown mechanisms, induces the expression of IL-10 which leads to the expansion of regulatory T cells (44). The fact that we also saw a significant expansion of regulatory T cells in the colonized palate through deconvolution of the transcriptome at day 33 further adds to the relevance of our model to N. gonorrhoeae. N. musculi appears to promote an immunosuppressive environment in the palate during late colonization by diminishing Th1 and Th2 responses, perhaps through regulatory T cell-mediated induction of IL-10 expression (45). This may be at least partly compensated by robust IFNγ and B cell responses we observed, both in colonized tissue and those adjacent to colonization at this similar stage (day 33), since these responses have been associated with N. gonorrhoeae clearance and protection from reinfection for at least 6 months (46).
The significant correlations we observed between the magnitude of colonization with N. musculi in palate tissue and the expression of the immune marker PD-1 in many systemic immune cells further confirm the influence of local bacterial colonization on the systemic immunophenotype. While the phenotypic and functional attributes of PD-1 in conventional T cells are well established (47), less is known about the significance of PD-1 expression in other immune cell types. Indeed, from a T cell perspective, PD-1 is found to be expressed during activation and shows high and sustained expression during persistent encounters with antigen, much like during persistent bacterial colonization. The PD-1 pathway limits overactivation during initial priming and fine-tunes effector cell differentiation when exposed to antigen. Furthermore, in tissues such as the lung during acute infection, the PD-1 pathway protects tissues from immunopathology, regulates memory cell formation, and mediates a return to immune homeostasis (19). Limited studies on PD-1 expression on monocytes confirm that elevated levels of microbial products and inflammatory cytokines in the blood cause the upregulation of PD-1 on monocytes. Furthermore, it has been shown that PD-1 expression on monocytes correlated with IL-10 concentrations in the blood (48). IL-10 has potent anti-inflammatory properties and plays a key role in limiting host immune response to microbes, thereby preventing damage to the host and maintaining normal tissue (particularly mucosal) homeostasis. While we have already described the importance of maintaining immune homeostasis during bacterial colonization, further research is needed to delineate the more specific roles of the PD-1 pathways in the other immune cell subsets.
Through our study, we have attempted to describe the local and systemic immunological determinants that are likely to be involved in the maintenance of N. musculi tissue colonization while effectively limiting its pathogenic potential within the host. However, the host immune system is just one of many factors that are important in the maintenance of this host-bacterial homeostasis. Just as important are the bacterial factors that play a role in determining the balance between commensalism and pathogenicity. For example, N. musculi lacks orthologs for transferrin and lactoferrin receptors found in human pathogenic Neisseria species. This may indicate that N. musculi uses alternative host iron sources and limits its ability to cause invasive disease (49). Though we did not investigate bacterial factors that influence nutritional immunity in our present study, it is, without doubt, an important consideration when determining the relevance of this model for informing vaccine development against pathogenic Neisseria in humans.
Despite such limitations, this murine model can still be regarded as an excellent platform for investigating the immunological determinants of N. musculi colonization and persistence. Its similarities with human pathogenic Neisseria species make N. musculi an ideal candidate species to investigate the transcriptional and immunological events surrounding long-term persistent neisserial colonization in humans. Although N. musculi does not cause disease, it does encode many pathogenic Neisseria-host interaction factors important for colonization that can be considered candidate vaccine antigens, such as components of type IV pili, or outer membrane proteins like LctP, a lactate permease (50, 51). In vitro studies have found that type IV pili of N. gonorrhoeae are involved in reprogramming the host transcriptional profile and activation of immune signaling pathways (52). Furthermore, studies on natural colonization of humans with N. gonorrhoeae have identified robust IL-6, IL-10, and IFNγ responses, which is comparable with our immune data (53, 54).
Although, in this study, we only measured responses up to 33 days (to observe the late adaptive immune responses), we and others have shown persistent colonization from 10 weeks up to 1 year. (13) Thus our immunophenotyping and transcriptomic results can be considered to have been obtained in the context of an upper respiratory tract long-term persistence model, influenced by similar antigenic factors. The fact that human pathogenic species like N. gonorrhoeae also colonize related tissue like the nasopharynx makes our data on the consequences of oral colonization by N. musculi very relatable and significant. In our experience with this model, A/J mice oral colonization levels stabilize 3 weeks after oral inoculation to above 105 colony-forming unit (CFU) per oral swab, but fecal pellet counts remain highly variable (55). One of this model’s strengths is that it allows the study of host and bacterial factors that influence long-term persistence in the upper respiratory tract (36, 55, 56). While the N. musculi mouse model is not equivalent to studies of the pathogenic Neisseria species in mice or humans, it acts as a surrogate for investigating conserved mechanisms that contribute to stable pharyngeal persistence in humans. Additional pathogenic neisserial antigens could be engineered into N. musculi and tested in our model for their effect on interactions with the host and ensuing immune responses. Identification of safe and effective immune profiles and transcriptome changes induced by infection have been used as surrogates for “ideal” vaccine-elicited responses (57). Similarly, our new model and associated data will be a useful tool for characterizing the in vivo transcriptional and immunological endpoints of Neisseria-host mucosal colonization and would likely be useful for the development of vaccine candidates against neisserial pathogens. This model system could be used to test the efficacy of N. musculi or gonococcal antigen vaccination on mucosal persistence by N. musculi or N. musculi strains serving as expression vectors for antigens from Neisseria species pathogenic to humans. The model could also study how enhancement or suppression of host immune responses influences N. musculi-induced innate or adaptive immune responses that impact vaccine efficacy against persistent asymptomatic carriage.
MATERIALS AND METHODS
Bacterial strains and growth conditions
A naturally occurring RifR smooth morphotype, strain NW742, of N. musculi, was used for oral inoculations (55). Our experience to date with the N. musculi colonization model has observed that mice inoculated with the smooth morphotype upon culture of oral and fecal longitudinal samples yield only the inoculated morphotype. Future studies are needed to determine if the N. musculi genome undergoes within-host variation during colonization. N. musculi was cultivated on Gonococcal Base (GCB; Difco) agar plates containing rifampin (40 mg/L) and Kellogg’s supplements routinely incubated at 37°C with 5% CO2 for 48 hours. Forty-eighty-hour-old colonies were spread onto GCB plates, grown for 18 hours, then suspended in PBS, and adjusted to an optical density at 600 nm of 1.0 for oral inoculations of mice.
Mouse inoculations
Sixteen female A/J mice, 5 weeks old, procured from the Jackson Laboratory (Bar harbor, ME) were acclimatized for 1 week within the animal facility at Ohio University. Two days prior to N. musculi inoculation, oral swabs and fecal pellets were collected to screen for pre-existing Neisseria flora. No Neisseria spp. were detected after 48 hours of incubation. Mice were evenly divided into two groups of eight mice each, consisting of an inoculated group and a control group. Mice in the inoculated group were manually restrained, and a slow oral inoculation of 50 µL bacterial suspension was administered, while the control group was mock inoculated with 50 µL of PBS into the oral cavity.
Confirmation and sample collection
Three days post-inoculation, oral swabs and fecal pellets were collected and subsequently plated on media containing rifampin as described earlier to confirm the colonization of N. musculi in the inoculated group, with no detection of bacteria in the control group (pre-inoculation swab samples and fecal pellets were found to be negative for growth on plates with rifampicin). Enumerated N. musculi colonies longitudinally retrieved from mice uniformly had a smooth colony morphology. Following confirmation, on day 5, four mice each from both the inoculated and control groups were humanely euthanized using CO2 asphyxiation. Lung, spleen, and blood samples were used exclusively for immunophenotyping studies. N. musculi counts in the lung and spleen have been low or undetected in past studies (unpublished observation Thapa, Zia, and Weyand). Counts in blood have not been monitored. Blood collection was conducted through cardiac puncture using a 20-gauge hypodermic needle (BD), resulting in the collection of approximately 2 mL of blood, which was placed into a vacutainer tube. Lungs and spleens were harvested by making dorsal incisions, and the respective organs were carefully extracted using sterile forceps. The collected organs were each placed in a tube with an organ transport medium (RPMI with 10% fetal bovine serum) containing penicillin and streptomycin and transported to the Ohio State University on wet ice. The tongues of mice were harvested as previously described (55) by making incisions made on both sides of the mouth of euthanized mice. The oral cavity was opened, and the tongue was held with sterile forceps from the tip and cut at the base with sterile scissors. Each tongue was cut into small pieces, homogenized with a Mini Beadbeater-16 using 2.3 mm Zirconia beads (BioSpec Products, Bartlesville, OK), diluted in Hanks’ Balanced Salt Solution with 0.01 mM HEPES and 0.3% (wt/vol) bovine serum albumin, and plated to enumerate CFUs per gram of tissue. The hard palate was collected and one-third of each hard palate was homogenized as above and plated for CFU enumeration. The remaining two-thirds were placed in RNA protect (0.5 mL/palate) and snap-frozen in liquid nitrogen.
Final sampling
The remaining eight mice were monitored for 33 days. Weekly fecal and oral swabs were collected to quantify the colonization burden in the inoculated group and confirm the absence of Neisseria in the control group. Oral swabs were collected as described previously and suspended in GCB + 20% glycerol, vortexed for 1 minute, and serially diluted in GC broth, for plating on GCB agar plates containing rifampin. Fecal pellets were collected in sterile microfuge tubes and weighed, and each pellet was mixed with 1 mL of GCB + 20% glycerol using a sterile plain wooden applicator. The pellet suspensions were vortexed for 1 minute and then plated on GCB agar plates containing rifampin to enumerate CFUs per gram. After 33 days, the remaining mice were euthanized using CO2 asphyxiation. Blood, lungs, spleens, tongues, and hard palates were collected as described above; blood, lungs, and spleens were used exclusively for immunophenotyping.
RNA extraction and sequencing
Mouse palates were removed from RNA Protect, rinsed with cold PBS, and then subjected to bead beating using two to three 3 mm glass beads on a TIssueLyser II (4 minutes each side, 20 Hz) in 350 µL of buffer RLT supplemented with 1% betamercaptoethanol and 20 ng of cRNA as a carrier. Debris were pelleted and discarded, and total RNAs were extracted from the supernatant with the Qiagen Micro RNeasy kit. RNAs were quantitated using a Bioanalyzer, and then ribosomal RNAs were depleted using the NEBNext rRNA Depletion Kit with bacterial and human/mouse/rat probes mixed at a ratio of 30%:70%, respectively. Strand-specific, dual unique indexed libraries were made using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA). Manufacturer protocol was modified by diluting adapter 1:30 and using 3 µL of this dilution. Library size selection was performed with AMPure SPRI-select beads (Beckman Coulter Genomics, Danvers, MA). Glycosylase digestion of the adapter and the second strand was done in the same reaction as the final amplification. Libraries were QC’ed using the DNA High Sensitivity Assay on the LabChip GX Touch (Perkin Elmer, Waltham, MA). Library concentrations were also assessed by qPCR using the KAPA Library Quantification Kit (Complete, Universal; Kapa Biosystems, Woburn, MA). Pooled libraries were sequenced on an Illumina NovaSeq 6000 using 150 bp PE reads (Illumina, San Diego, CA). Reads were mapped to the Mus musculus GRCm39 genome using HISAT (58).
RNA-seq data analysis
Gene expression counts were estimated using HTseq (59). Normalized count data and variance-stabilized transformation (VST) counts were generated using the R DESeq2 package (60). For DEG estimation, inoculated samples were compared to their respective control samples using DESeq2 with an FDR ≤ 0.05 and an absolute Log2 Fold Change ≥ 1 (“Significant” DEGs, Table S1). A heatmap (Fig. S1) and a volcano plot of immunological biomarkers were generated based on Z-scores of VST counts (Fig. 2A). GO analysis was performed using DEGs (with a reduced significance cutoff of P-value ≤ 0.01) with the R package ClusterProfiler v4.0, and Cnet plots were generated using the cnetplot function (61). All FASTQ files and associated metadata are uploaded to the Gene Expression Omnibus repository (see Data availability).
Deconvolution of transcriptome data
The TIMER2.0 database (16) was utilized to perform deconvolution of bulk transcriptome data to explore the differential expression of immune cell subsets within tissue colonized by N. musculi. Using six algorithms (TIMER, CIBERSORT-ABS, QUANTISEQ, XCELL, EPIC, and MMCPCOUNTER), we generated differential estimations of the abundance of immune subsets from colonized palate tissue of inoculated mice at day 5 and day 33 of sacrifice and that of control mice. Statistical comparisons using abundance scores of immune cells were performed using the Wilcoxon rank-sum test (P < 0.05).
Tissue processing for flow cytometry staining
Tissue processing and flow cytometry staining were performed as previously described (62). Briefly, mice were euthanized, and their lungs, spleens, and blood were processed. Tissues were filtered, suspended, and centrifuged. Following lysis and washing steps, cells were resuspended in R-10 media. One hundred microliter of whole blood was stained for flow cytometry using the below protocol. The cells were then fixed with 2% paraformaldehyde and filtered into polystyrene FACS tubes using filter caps.
Flow staining and data acquisition
Flow cytometry staining and data acquisition were conducted using previously described methods (63). In brief, splenic and lung tissue samples were divided into two equal portions (1 million cells/mL each). One portion was stimulated with a cocktail of phorbol 12-myristate 13-acetate and ionomycin, while the other remained unstimulated. Stimulation included Golgi-Plug and Golgi-Stop, followed by a 20-hour incubation. After fixation and permeabilization, cells were stained and analyzed on a Cytek Aurora flow cytometer. Data were processed using FlowJo v.10.6.2, with “fluorescence minus 1” controls for each marker.
Graphing and statistics
Graphs were prepared using Graph Pad Prism (version 9.3.1). FlowSOM-based automatic clustering algorithms and UMAP for dimensional reduction visualization were performed using OMIQ. Data were statistically analyzed using their bundled software. Comparisons between the two groups were performed using the Wilcoxon rank-sum test. Pearson correlation coefficient analysis was used for correlations between immune signatures and tissue colony counts.
ACKNOWLEDGMENTS
We thank members of the Maryland Genomics Core at the Institute for Genome Sciences (IGS), University of Maryland School of Medicine, for palate sample processing and sequencing.
This study was partially supported by Ohio University’s Infectious and Tropical Disease Institute and Molecular and Cellular Biology Graduate program.
M.A. and M.G. conducted immunological assessments, analyzed data, and led the overall manuscript writing. T.Z. and S.B. conducted mouse colonization studies and tissue harvests. A.D. performed transcriptome analysis and contributed to manuscript writing. W.M., L.T., and T.S. were involved in tissue processing and flow cytometry experiments. S.W., S.S., and A.J.W. conducted deconvolution data analysis and contributed to manuscript writing. D.K. supervised data analysis. H.T. designed the transcriptome analysis and contributed to manuscript writing. N.W. designed the bacterial inoculation study and contributed to manuscript writing. N.P.M.L. designed the immunological assessment, integrated microbiological and transcriptome data, and led the overall manuscript writing.
Contributor Information
Namal P. M. Liyanage, Email: namal.liyanage@osumc.edu.
Kimberly A. Kline, Universite de Geneve, Geneva, Switzerland
ETHICS APPROVAL
All animal protocols were approved by the Ohio University Institutional Animal Care and Use Committee prior to the initiation of the experiments.
DATA AVAILABILITY
All FASTQ files and associated metadata have been uploaded to the NCBI Gene ExpressionOmnibus (GEO) repository under accession number GSE267528.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00048-24.
Heatmap of selected immunological biomarkers.
Expanded Cnet plot of GO biological process enrichment of DEGs of palate tissue of inoculated vs uninoculated mice.
Deconvolution of palate transcriptome.
Gating and FMO controls.
Legends for Tables S1 and S2.
Diagram summarizing the study.
DESeq2 gene lists.
List of Gene Ontology (GO) terms.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Sommer F, Bäckhed F. 2013. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 2. Hancock V, Dahl M, Klemm P. 2010. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J Med Microbiol 59:392–399. doi: 10.1099/jmm.0.008672-0 [DOI] [PubMed] [Google Scholar]
- 3. Diallo K, MacLennan J, Harrison OB, Msefula C, Sow SO, Daugla DM, Johnson E, Trotter C, MacLennan CA, Parkhill J, Borrow R, Greenwood BM, Maiden MCJ. 2019. Genomic characterization of novel Neisseria species. Sci Rep 9:13742. doi: 10.1038/s41598-019-50203-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu G, Tang CM, Exley RM. 2015. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology (Reading) 161:1297–1312. doi: 10.1099/mic.0.000086 [DOI] [PubMed] [Google Scholar]
- 5. Barrett SJ, Sneath PH. 1994. A numerical phenotypic taxonomic study of the genus Neisseria. Microbiology (Reading) 140:2867–2891. doi: 10.1099/00221287-140-10-2867 [DOI] [PubMed] [Google Scholar]
- 6. Donati C, Zolfo M, Albanese D, Tin Truong D, Asnicar F, Iebba V, Cavalieri D, Jousson O, De Filippo C, Huttenhower C, Segata N. 2016. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol 1:16070. doi: 10.1038/nmicrobiol.2016.70 [DOI] [PubMed] [Google Scholar]
- 7. Seifert HS. 2019. Location, location, location-commensalism, damage and evolution of the pathogenic Neisseria. J Mol Biol 431:3010–3014. doi: 10.1016/j.jmb.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 8. Ladhani SN, Lucidarme J, Parikh SR, Campbell H, Borrow R, Ramsay ME. 2020. Meningococcal disease and sexual transmission: urogenital and anorectal infections and invasive disease due to Neisseria meningitidis. Lancet 395:1865–1877. doi: 10.1016/S0140-6736(20)30913-2 [DOI] [PubMed] [Google Scholar]
- 9. Man OM, Ramos WE, Vavala G, Goldbeck C, Ocasio MA, Fournier J, Romero-Espinoza A, Fernandez MI, Swendeman D, Lee S-J, Comulada S, Rotheram-Borus MJ, Klausner JD. 2021. Optimizing screening for anorectal, pharyngeal, and urogenital Chlamydia trachomatis and Neisseria gonorrhoeae infections in at-risk adolescents and young adults in New Orleans, Louisiana and Los Angeles, California, United States. Clin Infect Dis 73:e3201–e3209. doi: 10.1093/cid/ciaa1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jerse AE, Wu H, Packiam M, Vonck RA, Begum AA, Garvin LE. 2011. Estradiol-treated female mice as surrogate hosts for Neisseria gonorrhoeae genital tract infections. Front Microbiol 2:107. doi: 10.3389/fmicb.2011.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weyand NJ. 2017. Neisseria models of infection and persistence in the upper respiratory tract. Pathog Dis 75. doi: 10.1093/femspd/ftx031 [DOI] [PubMed] [Google Scholar]
- 12. Barbee LA, Soge OO, Khosropour CM, Haglund M, Yeung W, Hughes J, Golden MR. 2021. The duration of pharyngeal gonorrhea: a natural history study. Clin Infect Dis 73:575–582. doi: 10.1093/cid/ciab071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma M, Powell DA, Weyand NJ, Rhodes KA, Rendón MA, Frelinger JA, So M. 2018. A natural mouse model for Neisseria colonization. Infect Immun 86:e00839-17. doi: 10.1128/IAI.00839-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weyand NJ, Ma M, Phifer-Rixey M, Taku NA, Rendón MA, Hockenberry AM, Kim WJ, Agellon AB, Biais N, Suzuki TA, Goodyer-Sait L, Harrison OB, Bratcher HB, Nachman MW, Maiden MCJ, So M. 2016. Isolation and characterization of Neisseria musculi sp. nov., from the wild house mouse. Int J Syst Evol Microbiol 66:3585–3593. doi: 10.1099/ijsem.0.001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhodes K, Ma M, So M. 1997. A natural mouse model for Neisseria persistent colonization. Methods Mol Biol:403–412. doi: 10.1007/978-1-4939-9496-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. 2020. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48:W509–W514. doi: 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuburu N, Kweon M-N, Song J-H, Hervouet C, Luci C, Sun J-B, Hofman P, Holmgren J, Anjuère F, Czerkinsky C. 2007. Sublingual immunization induces broad-based systemic and mucosal immune responses in mice. Vaccine 25:8598–8610. doi: 10.1016/j.vaccine.2007.09.073 [DOI] [PubMed] [Google Scholar]
- 18. Thibult M-L, Mamessier E, Gertner-Dardenne J, Pastor S, Just-Landi S, Xerri L, Chetaille B, Olive D. 2013. PD-1 is a novel regulator of human B-cell activation. Int Immunol 25:129–137. doi: 10.1093/intimm/dxs098 [DOI] [PubMed] [Google Scholar]
- 19. Sharpe AH, Pauken KE. 2018. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 18:153–167. doi: 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- 20. Cibrián D, Sánchez-Madrid F. 2017. CD69: from activation marker to metabolic gatekeeper. Eur J Immunol 47:946–953. doi: 10.1002/eji.201646837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sancho D, Gómez M, Sánchez-Madrid F. 2005. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol 26:136–140. doi: 10.1016/j.it.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 22. Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, Smyth MJ, Tarlinton DM, Nutt SL. 2007. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol 178:4764–4770. doi: 10.4049/jimmunol.178.8.4764 [DOI] [PubMed] [Google Scholar]
- 23. Pham OH, O’Donnell H, Al-Shamkhani A, Kerrinnes T, Tsolis RM, McSorley SJ. 2017. T cell expression of IL-18R and DR3 is essential for non-cognate stimulation of Th1 cells and optimal clearance of intracellular bacteria. PLoS Pathog 13:e1006566. doi: 10.1371/journal.ppat.1006566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dallagi A, Girouard J, Hamelin-Morrissette J, Dadzie R, Laurent L, Vaillancourt C, Lafond J, Carrier C, Reyes-Moreno C. 2015. The activating effect of IFN-γ on monocytes/macrophages is regulated by the LIF-trophoblast-IL-10 axis via Stat1 inhibition and Stat3 activation. Cell Mol Immunol 12:326–341. doi: 10.1038/cmi.2014.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serbina NV, Pamer EG. 2006. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 7:311–317. doi: 10.1038/ni1309 [DOI] [PubMed] [Google Scholar]
- 26. Howard OMZ, Dong HF, Su SB, Caspi RR, Chen X, Plotz P, Oppenheim JJ. 2005. Autoantigens signal through chemokine receptors: uveitis antigens induce CXCR3- and CXCR5-expressing lymphocytes and immature dendritic cells to migrate. Blood 105:4207–4214. doi: 10.1182/blood-2004-07-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaix J, Tessmer MS, Hoebe K, Fuséri N, Ryffel B, Dalod M, Alexopoulou L, Beutler B, Brossay L, Vivier E, Walzer T. 2008. Cutting edge: priming of NK cells by IL-18. J Immunol 181:1627–1631. doi: 10.4049/jimmunol.181.3.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomura M, Maruo S, Mu J, Zhou X-Y, Ahn H-J, Hamaoka T, Okamura H, Nakanishi K, Clark S, Kurimoto M, Fujiwara H. 1998. Differential capacities of CD4+, CD8+, and CD4-CD8- T cell subsets to express IL-18 receptor and produce IFN-γ in response to IL-18. J Immunol 160:3759–3765. doi: 10.4049/jimmunol.160.8.3759 [DOI] [PubMed] [Google Scholar]
- 29. Kulpa DA, Lawani M, Cooper A, Peretz Y, Ahlers J, Sékaly R-P. 2013. PD-1 coinhibitory signals: the link between pathogenesis and protection. Semin Immunol 25:219–227. doi: 10.1016/j.smim.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei HX, Wang B, Li B. 2020. IL-10 and IL-22 in mucosal immunity: driving protection and pathology. Front Immunol 11:1315. doi: 10.3389/fimmu.2020.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Camp RL, Kraus TA, Birkeland ML, Puré E. 1991. High levels of CD44 expression distinguish virgin from antigen-primed B cells. J Exp Med 173:763–766. doi: 10.1084/jem.173.3.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AAB, Cooper MA, Graf T, Hornung V. 2016. Human monocytes engage an alternative inflammasome pathway. Immunity 44:833–846. doi: 10.1016/j.immuni.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 33. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, et al. 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350:1079–1084. doi: 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jerse AE, Bash MC, Russell MW. 2014. Vaccines against gonorrhea: current status and future challenges. Vaccine 32:1579–1587. doi: 10.1016/j.vaccine.2013.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zielke RA, Wierzbicki IH, Baarda BI, Gafken PR, Soge OO, Holmes KK, Jerse AE, Unemo M, Sikora AE. 2016. Proteomics-driven antigen discovery for development of vaccines against gonorrhea. Mol Cell Proteomics 15:2338–2355. doi: 10.1074/mcp.M116.058800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Powell DA, Ma M, So M, Frelinger JA. 2018. The commensal Neisseria musculi modulates host innate immunity to promote oral colonization. Immunohorizons 2:305–313. doi: 10.4049/immunohorizons.1800070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seshadri S, Kannan Y, Mitra S, Parker-Barnes J, Wewers MD. 2009. MAIL regulates human monocyte IL-6 production. J Immunol 183:5358–5368. doi: 10.4049/jimmunol.0802736 [DOI] [PubMed] [Google Scholar]
- 38. Francis IP, Islam EA, Gower AC, Shaik-Dasthagirisaheb YB, Gray-Owen SD, Wetzler LM. 2018. Murine host response to Neisseria gonorrhoeae upper genital tract infection reveals a common transcriptional signature, plus distinct inflammatory responses that vary between reproductive cycle phases. BMC Genomics 19:627. doi: 10.1186/s12864-018-5000-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, Kasper DL. 2017. Mining the human gut microbiota for immunomodulatory organisms. Cell 168:928–943. doi: 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manta C, Heupel E, Radulovic K, Rossini V, Garbi N, Riedel CU, Niess JH. 2013. CX3CR1+ macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium. Mucosal Immunol 6:177–188. doi: 10.1038/mi.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feinen B, Russell MW. 2012. Contrasting roles of IL-22 and IL-17 in murine genital tract infection by Neisseria gonorrhoeae. Front Immunol 3:11. doi: 10.3389/fimmu.2012.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature 465:793–797. doi: 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kamimura Y, Lanier LL. 2015. Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep 10:280–291. doi: 10.1016/j.celrep.2014.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y, Liu W, Russell MW. 2014. Suppression of host adaptive immune responses by Neisseria gonorrhoeae: role of interleukin 10 and type 1 regulatory T cells. Mucosal Immunol 7:165–176. doi: 10.1038/mi.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Belcher T, Rollier CS, Dold C, Ross JDC, MacLennan CA. 2023. Immune responses to Neisseria gonorrhoeae and implications for vaccine development. Front Immunol 14:1248613. doi: 10.3389/fimmu.2023.1248613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y, Perez J, Hammer LA, Gallagher HC, De Jesus M, Egilmez NK, Russell MW. 2018. Intravaginal administration of interleukin 12 during genital gonococcal infection in mice induces immunity to heterologous strains of Neisseria gonorrhoeae. mSphere 3:e00421-17. doi: 10.1128/mSphere.00421-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin HT, Ahmed R, Okazaki T. 2011. Role of PD-1 in regulating T-cell immunity. Curr Top Microbiol Immunol 350:17–37. doi: 10.1007/82_2010_116 [DOI] [PubMed] [Google Scholar]
- 48. Said EA, Dupuy FP, Trautmann L, Zhang Y, Shi Y, El-Far M, Hill BJ, Noto A, Ancuta P, Peretz Y, Fonseca SG, Van Grevenynghe J, Boulassel MR, Bruneau J, Shoukry NH, Routy J-P, Douek DC, Haddad EK, Sekaly R-P. 2010. Programmed death-1-induced interleukin-10 production by monocytes impairs CD4+ T cell activation during HIV infection. Nat Med 16:452–459. doi: 10.1038/nm.2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ewasechko NF, Chaudhuri S, Schryvers AB. 2022. Insights from targeting transferrin receptors to develop vaccines for pathogens of humans and food production animals. Front Cell Infect Microbiol 12:1083090. doi: 10.3389/fcimb.2022.1083090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cehovin A, Kroll JS, Pelicic V. 2011. Testing the vaccine potential of PilV, PilX and ComP, minor subunits of Neisseria meningitidis type IV pili. Vaccine 29:6858–6865. doi: 10.1016/j.vaccine.2011.07.060 [DOI] [PubMed] [Google Scholar]
- 51. Sun Y, Li Y, Exley RM, Winterbotham M, Ison C, Smith H, Tang CM. 2005. Identification of novel antigens that protect against systemic meningococcal infection. Vaccine 23:4136–4141. doi: 10.1016/j.vaccine.2005.03.015 [DOI] [PubMed] [Google Scholar]
- 52. Howie HL, Glogauer M, So MTN. 2005. The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism. PLoS Biol 3:e100. doi: 10.1371/journal.pbio.0030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hedges SR, Sibley DA, Mayo MS, Hook III EW, Russell MW. 1998. Cytokine and antibody responses in women infected with Neisseria gonorrhoeae: effects of concomitant infections. J Infect Dis 178:742–751. doi: 10.1086/515372 [DOI] [PubMed] [Google Scholar]
- 54. Lovett A, Duncan JA. 2018. Human immune responses and the natural history of Neisseria gonorrhoeae infection. Front Immunol 9:3187. doi: 10.3389/fimmu.2018.03187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thapa E, Lauderback L, Simmons C, Holzschu DL, D’Mello A, Ma M, So M, Tettelin H, Weyand NJ. 2022. Phenotypic and transcriptomic variation in Neisseria musculi morphotypes correlate with colonization variability and persistence. bioRxiv. doi: 10.1101/2022.02.03.479073 [DOI]
- 56. Rhodes KA, Ma MC, Rendón MA, So M. 2022. Neisseria genes required for persistence identified via in vivo screening of a transposon mutant library. PLoS Pathog 18:e1010497. doi: 10.1371/journal.ppat.1010497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heinonen S, Velazquez VM, Ye F, Mertz S, Acero-Bedoya S, Smith B, Bunsow E, Garcia-Mauriño C, Oliva S, Cohen DM, Moore-Clingenpeel M, Peeples ME, Ramilo O, Mejias A. 2020. Immune profiles provide insights into respiratory syncytial virus disease severity in young children. Sci Transl Med 12:eaaw0268. doi: 10.1126/scitranslmed.aaw0268 [DOI] [PubMed] [Google Scholar]
- 58. Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anders S, Pyl PT, Huber W. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gunasena M, Shukla RK, Yao N, Rosas Mejia O, Powell MD, Oestreich KJ, Aceves-Sánchez M de J, Flores-Valdez MA, Liyanage NPM, Robinson RT. 2022. Evaluation of early innate and adaptive immune responses to the TB vaccine Mycobacterium bovis BCG and vaccine candidate BCGΔBCG1419c. Sci Rep 12:12377. doi: 10.1038/s41598-022-14935-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shukla RK, Gunasena M, Reinhold-Larsson N, Duncan M, Hatharasinghe A, Cray S, Weragalaarachchi K, Kasturiratna D, Demberg T, Liyanage NPM. 2023. Innate adaptive immune cell dynamics in tonsillar tissues during chronic SIV infection. Front Immunol 14:1201677. doi: 10.3389/fimmu.2023.1201677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heatmap of selected immunological biomarkers.
Expanded Cnet plot of GO biological process enrichment of DEGs of palate tissue of inoculated vs uninoculated mice.
Deconvolution of palate transcriptome.
Gating and FMO controls.
Legends for Tables S1 and S2.
Diagram summarizing the study.
DESeq2 gene lists.
List of Gene Ontology (GO) terms.
Data Availability Statement
All FASTQ files and associated metadata have been uploaded to the NCBI Gene ExpressionOmnibus (GEO) repository under accession number GSE267528.