Abstract
Minus-strand DNA transfer, an essential step in retroviral reverse transcription, is mediated by the two repeat (R) regions in the viral genome. It is unclear whether R simply serves as a homologous sequence to mediate the strand transfer or contains specific sequences to promote strand transfer. To test the hypothesis that the molecular mechanism by which R mediates strand transfer is based on homology rather than specific sequences, we examined whether nonviral sequences can be used to facilitate minus-strand DNA transfer. The green fluorescent protein (GFP) gene was divided into GF and FP fragments, containing the 5′ and 3′ portions of GFP, respectively, with an overlapping F fragment (85 bp). FP and GF were inserted into the 5′ and 3′ long terminal repeats, respectively, of a murine leukemia virus-based vector. Utilization of the F fragment to mediate minus-strand DNA transfer should reconstitute GFP during reverse transcription. Flow cytometry analyses demonstrated that GFP was expressed in 73 to 92% of the infected cells, depending on the structure of the viral construct. This indicated that GFP was reconstituted at a high frequency; molecular characterization further confirmed the accurate reconstitution of GFP. These data indicated that nonviral sequences could be used to efficiently mediate minus-strand DNA transfer. Therefore, placement and homology, not specific sequence context, are the important elements in R for minus-strand DNA transfer. In addition, these experiments demonstrate that minus-strand DNA transfer can be used to efficiently reconstitute genes for gene therapy applications.
All retroviruses replicate their genome using an RNA form to generate a DNA form in a process called reverse transcription (42). The viral RNA is characterized by short repeat (R) regions at the 5′ and 3′ ends (7, 14). The R region at the 5′ end is immediately followed by a unique 5′ sequence named U5. Because the viral RNA is the mRNA or the plus strand, the first strand of DNA synthesized is complementary to the viral RNA and is referred to as the minus-strand DNA. Viral DNA synthesis initiates near the 5′ end of the viral RNA, using a tRNA primer that binds to the primer-binding site (PBS) in the viral RNA (7). Reverse transcriptase (RT) copies R and U5 and quickly reaches the 5′ end of the RNA template. This short stretch of DNA that contains R and U5 is referred to as minus-strand strong-stop DNA. It is thought that the RNase H activity of RT degrades the RNA template in the RNA-DNA hybrid and exposes the strong-stop DNA. The newly synthesized R in the viral DNA is complementary to the R near the 3′ end of the viral RNA. Presumably, the complementarity facilitates alignment and hybridization of the two nucleic acids and allows RT to continue DNA synthesis, using sequences near the 3′ end of the viral RNA as a template. This switching of the RT complex from the 5′ end to near the 3′ end of the viral RNA, known as minus-strand DNA transfer, is an essential step in reverse transcription (7, 14). Minus-strand DNA transfer is primarily mediated by the strong-stop DNA (7, 14); however, it has been observed that DNA containing U5 and only a portion of R, referred to as minus-strand weak-stop DNA, can also mediate minus-strand DNA transfer, although at a lower frequency (27, 28, 36, 45).
Minus-strand DNA transfer is accomplished through complex interactions between the viral proteins and nucleic acids. At least two viral proteins, RT and nucleocapsid (NC), play important roles in this transfer. The RNase H activity of RT is essential for minus-strand DNA transfer (5, 29, 34). Through various in vitro assays, NC was also clearly demonstrated to facilitate the efficiency of minus-strand DNA transfer (2, 8, 12, 16, 20, 25, 33, 35, 38, 39, 43, 46).
Less is known about the requirement of cis-acting sequences for minus-strand DNA transfer. The R regions vary significantly in length and sequence among different viruses. The length of R can vary up to 16-fold (from the shortest R, 15 nucleotides [nt], in mouse mammary tumor virus to the longest R, 247 nt, in human T-cell leukemia virus type 2) (4, 7). Furthermore, it was shown that in some viruses, such as murine leukemia virus (MLV), spleen necrosis virus, and human immunodeficiency virus type 1 (HIV-1), minus-strand DNA synthesis was not required to reach the end of R before strand transfer occurred; therefore, only a portion of R is needed to mediate minus-strand DNA transfer (26–28, 36, 45). In a recent study using a viral vector-cell culture system, we defined the relationship between the length of homology and the efficiency of minus-strand DNA transfer (Q. Dang and W.-S. Hu, submitted for publication). We found that 12 nt of homology is sufficient to mediate efficient minus-strand DNA transfer.
Although homology length can play a role in the efficiency of minus-strand transfer, it was not clear whether there is a requirement for specific sequences in the R region for this transfer. The R regions of various retroviruses do not have apparent conserved motifs. However, in an in vitro assay using purified proteins and RNA, it was demonstrated that R sequences from MLV, Rous sarcoma virus, and HIV-1 could mediate minus-strand transfer, whereas a nonviral sequence failed to mediate strand transfer (3). This finding led to the hypothesis that the sequence context in the viral R region promotes strand transfer and viral sequences are required for efficient minus-strand DNA transfer.
Recent data from our laboratory indicated that efficient minus-strand DNA transfer can be mediated by short stretches of homology (45) (Dang and Hu, submitted). Therefore, we hypothesized that homology in the R region, rather than sequence context, is key to promoting this transfer. To test this hypothesis, we investigated whether nonviral sequences can mediate efficient minus-strand DNA transfer in vivo.
MATERIALS AND METHODS
Construction of vectors.
Plasmids pSR2-2GFP, pSR5-FP-GF, pSR6-2wtLTR, and pMS2-FP-GF-no3R were derived from pAR2 (45), an MLV-based vector that contains the hygromycin phosphotransferase B gene (hygro) (15). For the nomenclature used here, plasmid names begin with “p,” whereas the names of viruses derived from the plasmids do not (e.g., pSR5-FP-GF refers to the plasmid construct, whereas SR5-FP-GF refers to the virus derived from this plasmid).
pAR2 was digested to completion with AatII and self-ligated to generate pTR1, a plasmid that contained a portion of hygro and the downstream long terminal repeat (LTR). The green fluorescent protein (GFP) gene (6) from pGreen-Lantern-1 (Gibco) was amplified by PCR using primers GFPp1-AscI and PFG2p-csA. Sequences of the primers are shown in Table 1. The resulting DNA was digested with AscI and inserted into the AscI site between U3 and R in the upstream LTR of pAR2 to generate pCM1. GFP was amplified by PCR using primers GFPp3-EheI and PFG4p-ehEI; the amplified product was digested with EheI and inserted into the EheI site between U3 and R in the pTR1 LTR to generate pCM2. pCM1 and pCM2 were digested with ScaI, and the pCM1 DNA fragment containing the upstream LTR with GFP was ligated to the pCM2 DNA fragment containing the downstream LTR. The resulting plasmid, pSR1, contained hygro, and both LTRs had one copy of GFP. pSR1 was digested with BstEII and ClaI to excise hygro, which was replaced with the simian virus 40 (SV40) promoter-hygro fragment from pMSM2 to generate pSR2-2GFP.
TABLE 1.
Primers used for vector construction, PCR amplification, and DNA sequencing
| Primer name | Sequence |
|---|---|
| GFPp1-AscI | 5′-TCTCCGAATGGCGCGCCGCCACCATGAGCAAGGGC-3′ |
| PFG2p-csA | 5′-CCTATCTCGAGGCGCGCCTCACTTGTACAGCTCGTCCATG-3′ |
| GFPp3-EheI | 5′-ATCCCGAATCGGCGCCGCCACCATGAGCAAGGGC-3′ |
| PFG4p-ehEI | 5′-GTCATGTCAAGGCGCCTCACTTGTACAGCTCGTCCATG-3′ |
| FPAscp1 | 5′-ATATAGGCCTGGCGCGCCATGCCCGAGGGCTATG-3′ |
| FG-ehE-4p | 5′-CGGTAGCAATGGCGCCAACTTGACTTCAGCGCGGGTC-3′ |
| MLVU3 | 5′-ATGTTTCCAGGGTGCCCCAAGGACC-3′ |
| ispVLM5′ | 5′-TCAAACATAGACACTAGACAATCGG-3′ |
| GF101 | 5′-CAGCGGAGAGGGTGAAGGTG-3′ |
| 5UVLM | 5′-TCGTGGGTAGTCAATCACTCAGAGG-3′ |
| R/3UM-629 | 5′-CGACGCAGTCTATCGGAAGACT-3′ |
| PFG-695 | 5′-CATGCCATGTGTGATCCCAGC-3′ |
| PFG-464 | 5′-TTGTGCGCCATGATGTACAC-3′ |
The portion of GFP containing the 3′ 462-bp fragment, termed FP, was amplified by PCR using primers FPAscp1 and PFG2-pcsA (Table 1). The PCR product was digested with AscI and inserted into the AscI site in the 5′ LTR of pAR2 to generate pTR4. The portion of GFP containing the 5′ 350-bp fragment, termed GF, was amplified using primers FG-ehE-4p and GFPp3-EheI (Table 1). The PCR product was digested with EheI and inserted into the EheI site of pTR1 to generate pTR3. The pTR4 DNA fragment containing the upstream LTR with FP was isolated and the DNA fragment containing the downstream LTR with GF was isolated from pTR3. These two DNA fragments were ligated to form pTR5. pTR5 was digested with BstEII and ClaI to excise hygro, which was replaced with the 1.7-kb DNA fragment containing SV40 promoter-hygro to generate pSR3. DNA sequencing of pSR3 revealed the presence of an inactivating mutation in GF (data not shown). To remove this mutation from the plasmid, pCM2 was digested with ClaI and MscI to excise the 3′ hygro, U3, and GF fragments, which were then ligated into the eluted 6.1-kb backbone of pSR3 that was generated by digestion with ClaI and partial digestion with MscI. In the resulting plasmid, pSR5-FP-GF, the GF region contained the 5′ 350-bp of GFP and the FP region contained the 3′ 462-bp of GFP with a 85-bp F region shared by both GF and FP fragments.
pSR6-2wtLTR was generated by replacing the 1.4-kb ClaI-BstEII fragment of pAR2 between the Ψ packaging signal and the 3′ end of hygro with the 1.7-kb BstEII-ClaI fragment containing SV40-hygro from pMSM2.
pJD220SVhy (13) was digested with ClaI and BstZ17I to isolate a 409-bp DNA fragment containing the SV40 termination signal. pCM2 was digested with BstBI and BstZ17I to delete the 3′ 351-bp fragment containing a portion of GFP, R, and U5 and then was treated with the Klenow fragment of Escherichia coli DNA polymerase I and ligated with the 409-bp ClaI-BstZ17I DNA fragment from JD220SVhy. The resulting plasmid, pMS1, was digested with ScaI, and the fragment containing the 3′ LTR was isolated and ligated to ScaI-digested pSR5-FP-GF to generate pMS2-FP-GF-no3R. pMP1 was derived from pWH390 (9) by the insertion of GFP upstream of the internal ribosomal entry site (IRES) from encephalomyocarditis virus (21, 22).
Standard cloning techniques were used to construct all of the vectors (30). Plasmid structures were analyzed by restriction enzyme mapping. All PCR-amplified DNA fragments that were cloned into plasmids were further analyzed by DNA sequencing to detect inadvertent mutations generated during the PCR procedures.
Cell culture, DNA transfection, and virus infection.
PG13 cells (American Type Culture Collection) were derived from NIH 3T3 cells expressing MLV gag-pol and gibbon ape leukemia virus (GaLV) env (31). D17 (American Type Culture Collection) is a dog osteosarcoma cell line permissive for MLV infection (37). PG13 and D17 cells were maintained at 37°C in Dulbecco modified Eagle medium supplemented with penicillin (50 U/ml; Gibco), streptomycin (50 μg/ml; Gibco), and bovine calf serum (10% for PG13 and 6% for D17). G418, a neomycin analog, was used for selection at a final concentration of 600 μg/ml for PG13 cells and 400 μg/ml for D17 cells. Hygromycin was used at final concentrations of 300 μg/ml in PG13 cells and 120 μg/ml in D17 cells.
Transfections were performed using the calcium phosphate precipitation method as previously described (30) or Transfast transfection reagents from Promega as recommended by the manufacturer. PG13 cells were plated at a density of 105 cells per 60-mm-diameter dish; 5 or 10 μg of vector DNA was used per dish for the Transfast or calcium phosphate transfection, respectively. Transfected cells were placed on the appropriate drug selection; drug-resistant colonies were pooled, expanded, and plated at a density of 5 × 106 cells per 100-mm-diameter dish. Virus was harvested from each transfected cell pool 48 h later and centrifuged at 3,000 × g for 10 min to remove cellular debris. Serial dilutions of the supernatants were used to infect D17 cells that were plated at 2 × 105 cells per 60-mm dish. Infected D17 cells were then placed on appropriate drug selection. Viral titers were calculated based on the number of drug-resistant colonies and standardized to RT activities.
RT assay.
A portion of the virus harvested from transfected PG13 cells was subjected to RT assays as previously described (17). Briefly, harvested virus was centrifuged in a SW28 rotor (Beckman) or a Surespin 630 rotor (Sorvall) at 25,000 rpm for 90 min. Viral pellets were resuspended in serum-free medium and stored at −80°C. Exogenous RT activities were determined by incubating 10 μl of virus with 50 μg of 20-mer Oligo T (Integrated DNA Technologies) per ml, 100 μg of poly(A) (Pharmacia) per ml, 60 mM NaCl, 50 mM Tris (pH 8.0), 1 U of RNase inhibitor per μl, 10 mM dithiothreitol, 0.6 mM MnCl2, 80 μM dTTP, 0.5% IGE Pal (Sigma), and 10 μCi of [3H]dTTP (72 Ci/mmol; ICN). The samples were incubated at 37°C for 90 min. The reaction mixtures were precipitated with 10% trichloroacetic acid (Sigma) and filtered through 0.45-μm-pore-diameter Metricel membranes (Gelman Sciences, Inc.); the amount of 3H incorporated was determined using a scintillation counter.
Detection of GFP expression by flow cytometry and fluorescence microscopy.
The number of cells in the transfected pools that expressed GFP was measured using flow cytometry (FACScan; Becton Dickinson); results were analyzed using CellQuest software (Becton Dickinson). GFP expression in infected, drug-resistant D17 cells was analyzed by two methods. Drug-resistant cell colonies from plates containing 100, 10−1, and 10−2 viral dilutions were separately pooled for flow cytometry analyses. Drug-resistant colonies from 10−3, 10−4, and 10−5 viral dilution plates were analyzed by fluorescence microscopy (Axiovert inverted fluorescence microscope; Zeiss). GFP expression in individual cell clones was also analyzed by flow cytometry and microscopy.
Analyses of proviral structure by PCR and DNA sequencing.
Hygromycin-resistant cell clones were isolated and lysed for use as PCR substrates. The upstream LTR of the proviruses was amplified using primers in the U3 (MLVU3) and 5′ Ψ regions (ispVLM5′) (Table 1). PCR products were analyzed by DNA sequencing with an automated sequencer (PE Biosystem) using one or more of the following primers: GF101, 5UVLM, R/3UM-629, PFG695, and PFG464 (Table 1).
RESULTS
Vectors used to determine the primary sequence requirement for minus-strand DNA transfer.
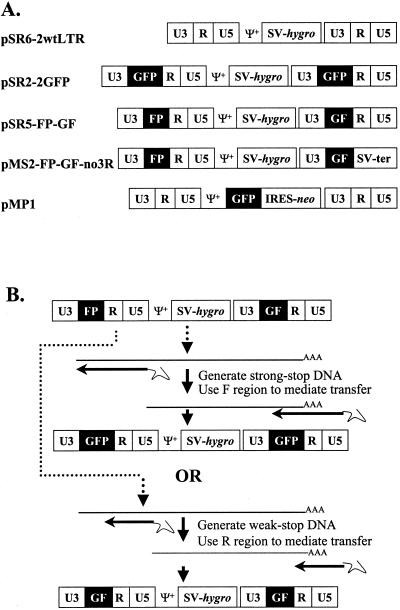
A series of MLV-based retroviral vectors was used to test the hypothesis that nonviral sequences can mediate minus-strand DNA transfer and that this process can reconstitute genes; the structures of these vectors are shown in Fig. 1A. All of these vectors contained the cis-acting sequences necessary for retroviral replication, such as the PBS, packaging signal (Ψ+), polypurine tract, and attachment sites. In addition, most of the gag- and the entire pol- and env-coding regions were deleted from these vectors. pSR2-2GFP, pSR5-FP-GF, pSR6-2wtLTR, and pMS2-FP-GF-no3R each contained an SV40 promoter upstream of hygro between the two LTRs. pSR6-2wtLTR contained two unmodified LTRs, whereas pSR2-2GFP had two modified LTRs, each containing the full-length GFP between U3 and R. pSR5-FP-GF also had two modified LTRs; the upstream LTR contained the 3′ 462-bp FP fragment of GFP between U3 and R, whereas the downstream LTR contained the 5′ 350-bp GF fragment of GFP between U3 and R. FP and GF share a homology stretch of 85 bp (the F region). pMS2-FP-GF-no3R also had two modified LTRs similar to those in pSR5-FP-GF, except that the R and U5 regions in the downstream LTR were replaced by a DNA fragment containing the SV40 termination signal. pMP1 contained two unmodified LTRs with GFP, IRES, and the neomycin phosphotransferase gene (neo) (23) between the LTRs.
FIG. 1.
MLV-based vectors and strategy used to study the requirement of sequence context in mediating minus-strand DNA transfer. (A) Structures of the MLV-based vectors. SV, SV40 promoter; hygro, hygromycin phosphotransferase B gene; GFP, green fluorescent protein gene; GF, the 5′ 350-bp fragment of GFP; FP, the 3′ 462-bp fragment of GFP; SV-ter, SV40 termination signal; Ψ+, the extended MLV packaging signal; IRES, internal ribosomal entry site; neo, neomycin phosphotransferase gene. (B) Strategy used to test the ability of nonviral sequences to mediate minus-strand DNA transfer. The structure of pSR5-FP-GF is illustrated at the top. Either minus-strand DNA transfer would be mediated by strong-stop DNA and the GFP would therefore be reconstituted, or minus-strand DNA transfer would be mediated by weak-stop DNA and the GFP would not be reconstituted.
pSR6-2wtLTR and pMP1 contained unmodified LTRs and were expected to produce RNAs with R at the two ends of the viral sequences that could mediate minus-strand DNA transfer. The LTRs in pSR5-FP-GF were modified; the viral RNA should contain FP-R-U5 at the 5′ end and U3-GF-R at the 3′ end of the viral sequences (Fig. 1B). The minus-strand strong-stop DNA from this vector should contain FP-R-U5 (Fig. 1B). If minus-strand DNA transfer can be mediated by nonviral sequences such as the F region, then GFP should be reconstituted during this process. At a lower frequency (∼1 to 10%), minus-strand DNA synthesis terminates early to form weak-stop DNA (27, 28, 36, 45), which may contain only U5 and R; when the weak-stop DNA transfers to the 3′ end of viral RNA, the resulting DNA will contain U3-GF-R and cannot reconstitute GFP.
The LTRs in pMS2-FP-GF-no3R were modified in a manner similar to those in pSR5-FP-GF except that R and U5 were deleted from the 3′ LTR. The MS2-FP-GF-no3R viral RNA was also expected to have FP-R at the 5′ end and U3-GF, but not the R region, at the 3′ end of the viral sequences. Weak-stop DNA containing only R-U5 would not have sequence complementary to the 3′ viral RNA and should not be able to transfer efficiently. Therefore, if the F region could not mediate efficient minus-strand DNA transfer, MS2-FP-GF-no3R would not be able to replicate well and would have a severely reduced viral titer.
Each LTR in pSR2-2GFP contained a copy of GFP between U3 and R, the viral RNA should contain GFP-R-U5 at the 5′ end and U3-GFP-R at the 3′ end of the viral sequences. Regardless of the regions used for minus-strand DNA transfers, the resulting viral DNA would have LTRs with GFP between the U3 and R sequences.
Experimental protocol.
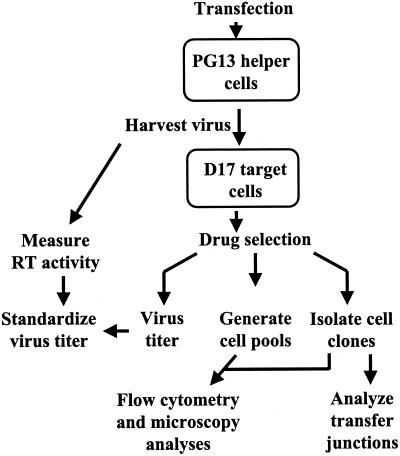
The outline of the experimental protocol is shown in Fig. 2. These vectors were separately transfected into PG13 helper cells that expressed MLV gag-pol and GaLV env. Transfected cells were subjected to appropriate drug selection, and resistant cell colonies were pooled. All of the pools contained at least 250 colonies. Viruses were harvested from these pools; for each sample, a portion of the virus was used to measure the RT activity and another portion was serially diluted and used to infect D17 cells. RT activity was measured to monitor the level of virion production from each vector-transfected cell pool. Within each experiment, the RT activities from cell pools transfected with different vectors were generally very similar (within 1.5-fold), indicating that similar amounts of virions were produced from these cells pools. After the infected D17 cells were subjected to appropriate drug selection, the numbers of drug-resistant colonies were determined and were used to calculate the viral titers generated by these vectors. Infected, drug-resistant cells were either pooled or used to isolate individual cell clones; GFP expression from these drug-resistant cell pools and cell clones was analyzed by flow cytometry and/or fluorescence microscopy. To examine the molecular nature of the minus-strand DNA transfer, a portion of the proviruses containing minus-strand DNA transfer junctions was amplified from the infected cell clones by PCR and characterized by DNA sequencing.
FIG. 2.
Experimental protocol. Virus was harvested from transfected PG13 helper cells and used to infect D17 target cells. Viral titers were determined by the number of drug-resistant colonies and standardized to the RT activities. GFP expression in infected target cells was analyzed by flow cytometry and fluorescence microscopy. Minus-strand DNA transfer junctions were analyzed in infected cell clones.
In this system, viral titers and minus-strand DNA transfers were examined in a single viral replication cycle, from the viruses produced in the PG13 cells to the proviruses generated in the D17 cells. Viruses produced from the murine-derived PG13 cells contained GaLV Env, which could not efficiently infect murine cells (31). Therefore, reinfection had little opportunity to occur in PG13 cells. D17 target cells did not express gag-pol and env needed to produce virions containing these vectors to go through another round of viral replication. Therefore, only one round of retroviral replication was allowed in this system.
Viral titers after one round of retroviral replication.
The titers for vectors MP1, SR2-2GFP, SR5-FP-GF, SR6-2wtLTR, and MS2-FP-GF-no3R are listed in Table 2. The pSR6-2wtLTR titer varied between 8.3 × 104 and 130 × 104 CFU/ml. The SR5-FP-GF titer varied between 1.6 × 104 and 24 × 104 CFU/ml. The average difference between the SR6-2wtLTR titers and SR5-FP-GF titers was 5.4-fold. The reductions in viral titers could have been caused by inserting sequences in the R region, similar to the six- to eightfold decreases in titers previously observed by another laboratory (1). Alternatively, it was possible that the F region could not mediate the minus-strand DNA transfer and caused the decrease in the viral titer. This would suggest that most of the strong-stop DNA could not perform strand transfer and did not produce viral DNA capable of integrating into the host genome. Most of the viral titer would be generated from viral DNA produced by the transfer of minus-strand weak-stop DNA. Because the weak-stop DNA was expected to be generated at a frequency of ∼1 to 10% (27, 28, 36, 45), we could not rule out the possibility that the 5.6-fold decrease in viral titers was caused by the inability of the strong-stop DNA to perform minus-strand DNA transfer.
TABLE 2.
Viral titers generated by PG13 cells transfected with vector plasmidsa
| Expt | Viral titers (104 CFU/ml)
|
||||
|---|---|---|---|---|---|
| MP1 | SR6-2wtLTR | SR2-2GFP | SR5-FP-GF | MS2-FP-GF-no3R | |
| A | 74 | 12 | ND | 2.4 | 1.1 |
| B | 44 | 19 | 6.1 | 1.6 | 4.8 |
| C | 16 | 8.3 | 4.1 | 13 | 11 |
| D | 43 | 17 | 5.9 | 4.0 | 3.2 |
| E | 130 | 130 | 14 | 24 | 21 |
In general, within each set of experiments, RT activities were within 1.5-fold among cell pools transfected with different vectors. ND, not determined.
The SR2-2GFP and MS2-FP-GF-no3R titers varied from 4.1 × 104 to 14 × 104 and 1.1 × 104 to 21 × 104 CFU/ml, respectively. There was no significant difference among the SR2-2GFP, SR5-FP-GF, and MS2-FP-GF-no3R titers (two-sample t test; P > 0.9 for SR5-FP-GF and MS2-FP-GF-no3R, P > 0.8 for SR5-FP-GF and SR2-2GFP). MS2-FP-GF-no3R RNA only contained the 5′ R and thus could not use R to mediate minus-strand DNA transfer. Therefore, in order for MS2-FP-GF-no3R to generate viral titers similar to those of SR2-2GFP and SR5-FP-GF, the F region had to be able to mediate minus-strand DNA transfer. The proviruses generated by F region-mediated transfer would have reconstituted GFP, which could be confirmed by GFP expression or structural analyses.
GF and FP fragments cannot confer positive GFP expression.
GFP expression in all of the transfected PG13 helper cell pools was examined by flow cytometry analyses. As expected, pMP1- or pSR2-2GFP-transfected, drug-resistant cell pools contained significant numbers of GFP-expressing cells, generally between 45 and 70%. As expected, cell pools transfected with pSR6-2wtLTR did not contain a significant percentage of fluorescent cells (<2%), since these plasmids lacked GFP. Cell pools transfected with pSR5-FP-GF or pMS2-FP-GF-no3R also did not contain a significant percentage of fluorescent cells (<2%), indicating that neither FP nor GF could express functional fluorescent proteins.
GFP expression of infected cells.
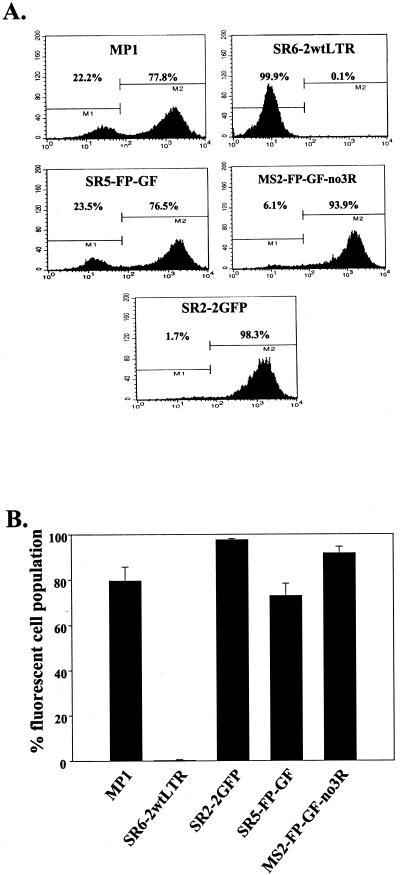
Flow cytometry analyses were also performed on D17 cells infected with virus produced by different transfected cell pools. Drug-resistant D17 cells were pooled and analyzed by flow cytometry; a representative set of flow cytometry analyses are shown in Fig. 3A, and the data from five independent experiments are summarized in Fig. 3B. MP1 and SR2-2GFP both contained intact GFP; most of the D17 cells infected with these viruses were positive for GFP expression, with an average of 79.6 ± 2.7% (standard error [SE]) and 97.7 ± 0.2% (SE), respectively. In contrast, very few SR6-2wtLTR-infected D17 cells (0.3 ± 0.1% [SE]) were positive for GFP expression since this virus did not contain GFP. In all experiments, a high proportion of cells infected with SR5-FP-GF were positive in GFP expression, ranging from 65 to 78.1% with an average of 72.9 ± 2.4% (SE) (Fig. 3). Most of the MS2-FP-GF-no3R-infected cells were positive for GFP expression (88.4 to 95.2%, with an average of 91.7 ± 1.2% [SE]). These experiments demonstrated that, most of the time, GFP was reconstituted during reverse transcription of SR5-FP-GF and MS2-FP-GF-no3R RNA. Therefore, the F region was used to mediate minus-strand DNA transfer during reverse transcription of these proviruses.
FIG. 3.
Flow cytometry analyses of infected cells. (A) Representative set of flow cytometry analyses using cells infected with MP1, SR6-2wtLTR, SR2-2GFP, SR5-FP-GF, and MS2-FP-GF-no3R. In each plot, the y axis is the number of events scored, which is interpreted as the number of cells, and the x axis is the intensity of the fluorescence. (B) Proportion of fluorescent cells infected with various vectors from five independent sets of infections. The error bars represent the SE of the average.
In addition to the flow cytometry analyses, GFP expression of the cell colonies was also examined using fluorescent microscopy. As expected, GFP expression was found in 0 of 369 (<0.1%) SR6-2wtLTR-infected colonies. GFP expression was found in 667 of 828 (80.6%) MP1-infected colonies, 305 of 341 (89.4%) SR2-2GFP-infected colonies, 629 of 856 (73.5%) SR5-FP-GF-infected colonies, and 1,002 of 1,092 (91.8%) MS2-FP-GF-no3R-infected colonies. The frequencies of GFP expression detected by microscopy were similar to those observed by flow cytometry analyses.
Molecular characterization of minus-strand DNA transfer in SR5-FP-GF and MS2-FP-GF-no3R.
To directly analyze the molecular nature of minus-strand DNA transfer, we isolated drug-resistant cell clones. From these clones, a portion of the proviral genome containing the upstream LTR was amplified and sequenced to characterize the molecular nature of the transfer events (Fig. 4). Both GFP-positive and GFP-negative cell clones were characterized; proviruses in GFP-positive cell clones were analyzed to ensure that they contained correctly reconstituted GFP. Multiple mechanisms could have caused the proviruses to fail to express GFP. For example, GFP might not have been reconstituted during minus-strand DNA transfer, GFP might have contained an inactivating mutation introduced either during the process of minus-strand DNA transfer or during elongation steps of reverse transcription, or the U3 promoter might have contained mutations to silence GFP expression. Therefore, the proviral structures in GFP-negative cells were also examined to characterize the mechanisms of GFP inactivation.
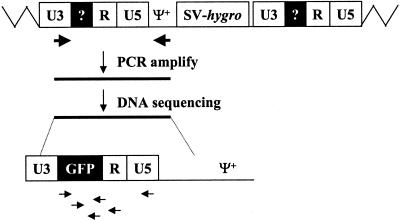
FIG. 4.
Strategy for PCR amplification of proviral LTRs and DNA sequencing. All abbreviations are the same as in Fig. 1. Zigzag lines, host DNA sequences; large arrows, PCR primers; small horizontal arrows, sequencing primers. Because the primers were located in U3 and in Ψ+, only sequences from the upstream LTR and a small portion of the Ψ+ were amplified.
Partial proviral structures from 13 SR5-FP-GF-infected cell clones were characterized; of the 13 clones, 4 were positive and 9 were negative for GFP expression as measured by fluorescence microscopy and flow cytometry. All of the four proviruses that expressed GFP contained the expected structures and had U3-reconstituted GFP-R-U5 in the LTRs. None of the nine proviruses that were negative for GFP expression had the reconstituted GFP in their LTRs. Six of the nine proviruses had U3-GF-R-U5 in the LTRs, and the other three contained U3-FP-R-U5 in their LTRs.
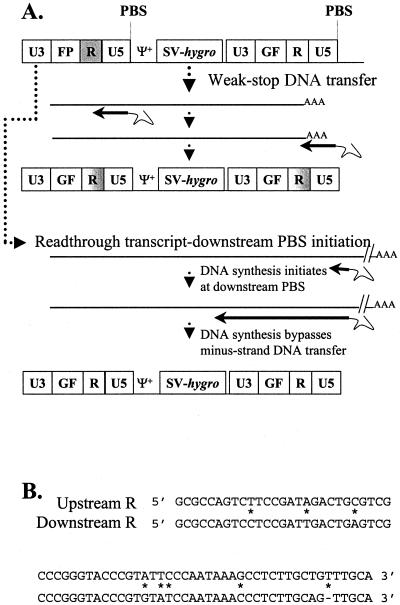
Viruses with U3-GF-R-U5 appeared to be the predicted structures for proviruses generated by weak-stop DNA mediated minus-strand transfers (Fig. 1B). However, upon further analyses, these proviruses were probably generated by a different mechanism. These vectors were derived from the pLN series plasmids (32). With the exception of pMS2-FP-GF-no3R, all of these vectors had an additional PBS directly downstream of the 3′ LTR (Fig. 5A), as well as sequence variation between the 5′ and 3′ R regions (Fig. 5B). In these vectors, there were two possible mechanisms to generate viral DNA with LTRs containing U3-GF-R-U5: weak-stop minus-strand DNA transfer and readthrough RNA transcripts with DNA synthesis initiated from the downstream PBS. Termination of retroviral RNA transcripts is known to be relatively inefficient; 15% of the total transcripts are read through the termination signal and contain sequences downstream of LTR (18). In addition, it has been shown that these readthrough transcripts can be efficiently packaged into viral particles (19, 40, 41). In SR5-FP-GF, these readthrough transcripts would contain the entire downstream LTR and PBS (Fig. 5A). If DNA synthesis initiated from the downstream PBS, RT would copy the entire downstream LTR and proceed to copy the rest of the viral RNA. In this situation, reverse transcription would entirely bypass the step of minus-strand DNA transfer. The resulting viral DNA would have U3-GF-R-U5 in the LTRs (Fig. 5A), similar to the viral DNA generated from the weak-stop DNA transfer. The difference between these two viral DNAs was the origin of R in the LTR. With the weak-stop DNA transfer, a portion of R would be from the upstream R and another portion would be from the downstream R (Fig. 5A). With readthrough RNA transcripts and downstream PBS initiation, however, the entire R sequence would be derived from the downstream LTR. When we compared the R sequences of the six proviruses, all of them were entirely derived from the downstream R, indicating that these proviruses were generated by readthrough RNA transcripts and downstream PBS initiation rather than by weak-stop minus-strand DNA transfer.
FIG. 5.
Two models for the generation of proviruses with U3-GF-R-U5 in the LTRs. (A) Illustration of the two models. All abbreviations are the same as in Fig. 1. Sequences derived from upstream R are shown in gray, whereas sequences from downstream R are shown in white. (B) Sequence comparisons of the upstream and downstream R regions.
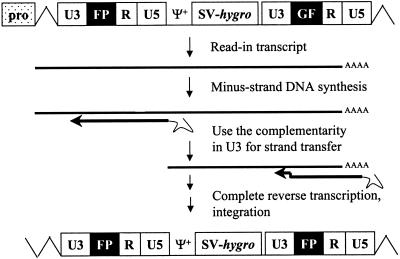
Three of the nine proviruses had U3-FP-R-U5 in the LTRs; these were likely to be generated from RNA transcripts initiating from the promoter upstream of U3, termed read-in RNA transcripts (Fig. 6). We hypothesized that in the virus-producing cells, some transfected pSR5-FP-GF might integrate close to the promoters. Read-in RNA transcripts would therefore contain the upstream U3 sequences along with FP, R, and U5. During reverse transcription, minus-strand DNA synthesis would copy U5, R, FP, and a portion of U3. The complementarity between the newly synthesized U3 DNA and U3 near the 3′ end of RNA could be used to mediate minus-strand DNA transfer. The resulting provirus would have two LTRs containing U3, FP, R, and U5.
FIG. 6.
Model for generating proviruses with U3-FP-R-U5 by using read-in RNA transcripts. All abbreviations are the same as in Fig. 1 and 4. pro, upstream promoter.
The molecular nature of minus-strand DNA transfer was also examined in 18 MS2-FP-GF-no3R-infected cell clones. Of these clones, nine were positive and nine were negative for GFP expression by both fluorescence microscopy and flow cytometry analyses. Similar to the SR5-FP-GF-infected clones, all of the 9 MS2-FP-GF-no3R proviruses that expressed GFP had precisely reconstituted GFP in the LTRs. Of the nine MS2-FP-GF-no3R proviruses that did not express GFP, two had mutations in GFP: one had a reconstituted GFP that contained a G-to-C substitution mutation in the G region that converted a glycine to an arginine and thereby inactivated GFP; the other contained a mutant GFP with a 120-bp deletion in the P region. The structure of the LTR from the latter provirus contained U3, G, F, 52 bp of 5′ P, a 120-bp deletion within P, 205 bp of 3′ P, R, and U5. Because the junction between F and P as well as the junction between P and R remained intact, the deletion was probably independent of minus-strand DNA transfer. The other seven MS2-FP-GF-no3R proviruses had a U3-FP-R-U5 structure in the LTR similar to those in SR5-FP-GF, which were presumably generated from read-in RNA transcripts.
Because the 3′ R, U5, and PBS were deleted in pMS2-FP-GF-no3R, proviruses with U3-GF-R-U5 structure in the LTRs were not observed. This impacted the frequency of the GFP-expressing cells. Approximately 8 and 27% of the cells infected with MS2-FP-GF-no3R and SR5-FP-GF did not express GFP, respectively (Fig. 3). The 19% difference between the cells infected with MS2-FP-GF-no3R and SR5-FP-GF is close to the percentage of SR5-FP-GF proviruses containing the U3-GF-R-U5 structure in LTRs. Thus, the increase in the frequency of gene reconstitution in MS2-FP-GF-no3R was directly correlated to the lack of readthrough transcription and initiation from the downstream PBS which generated proviruses with U3-GF-R-U5 LTRs.
Efficient minus-strand DNA transfers mediated by nonviral sequences.
GFP was reconstituted at 72.9 and 91.7% efficiencies in cells infected with SR5-FP-GF and MS2-FP-GF-no3R, respectively. Molecular characterization of 13 GFP-expressing proviruses demonstrated that GFP was precisely reconstituted, indicating that the F region was used to mediate minus-strand DNA transfer. MS2-FP-GF-no3R generated titers similar to SR5-FP-GF and SR2-2GFP, which could use either the R or F region to mediate minus-strand DNA transfer. Taken together, these data established that the F region could be used to mediate minus-strand DNA transfer in an efficient manner. In addition, these data demonstrated that minus-strand DNA transfer could be used as an effective means to reconstitute genes during virus replication.
DISCUSSION
In this study, we intended to define the requirement of sequence context of the R region in mediating minus-strand DNA transfers. The current model postulates that the role of the R regions is to provide complementarity between the newly synthesized minus-strand DNA and the 3′ R of the viral RNA to align the reverse transcription complex for accurate switching of the RNA template. This view suggests that the two R regions allow the hybridization of the nascent DNA and the 3′ RNA. In this model, it is likely that R is required to exceed a certain length to allow precise and efficient DNA-RNA alignment, but it is unlikely that R must contain specific sequences for the hybridization of nascent DNA and viral RNA. This prediction, however, was contradicted by an in vitro study suggesting that nonviral sequences could not mediate minus-strand DNA transfer (3).
Using a series of MLV vectors with modified LTRs, we demonstrated that a nonviral sequence, a portion of GFP, could efficiently mediate minus-strand DNA transfer. This finding is in contrast with the in vitro study in which the nonviral sequence was unable to mediate minus-strand DNA transfer. It is possible that this difference reflects the experimental systems employed in the two studies. In the in vitro system, strand transfer depended solely on the interactions between RT, NC, and nucleic acids. In our system, viruses containing the vector RNA were used to infect target cells; most of the DNA synthesis, including the strand transfer steps, was conducted within the reverse transcription complex in the cells. Many factors present in the in vivo but not in the in vitro system, such as the configurations of the RNAs and reverse transcription complexes, were very likely to be important in minus-strand DNA transfer. Therefore, the contrast of the two studies emphasizes the important roles of the elements missing in the in vitro assays.
Readthrough RNA transcripts and downstream PBS initiation.
During the analyses of SR5-FP-GF-generated proviruses, we observed LTRs with U3-GF-R-U5 structures. Despite the resemblance of these LTRs to the structures predicted from weak-stop DNA transfer (Fig. 1B), these proviruses were unlikely to be generated by weak-stop minus-strand DNA transfer. The two R regions in the vectors we used contained sequence variation scattered throughout the length of the two R regions. During minus-strand DNA synthesis, the first difference in the two R sequences that RT would encounter was at nt 6 at the 3′ end of R (T for upstream R and G for downstream R). In order for a provirus to contain R regions with all of the markers from the downstream R, DNA synthesis would have to stop before nt 6 of the upstream R and use less than 6-nt complementarity between nascent DNA and 3′ RNA to mediate minus-strand DNA transfer. However, we have observed in other studies that a 6-nt homology is not sufficient to mediate accurate and efficient minus-strand DNA transfer (Dang and Hu, submitted). Therefore, these proviruses were likely to be generated by other mechanisms.
We have previously observed efficient initiation of DNA synthesis from the downstream PBS in MLV (V. K. Pathak, P. D. Yin, R. J. Teufel II, and W.-S. Hu, unpublished data); downstream PBS initiation was also reported in avian leukosis virus (44). Approximately 27% of the SR5-FP-GF-infected cells did not express GFP (Fig. 3). Of the nine proviruses analyzed, six had the U3-GF-R-U5 structure. Therefore, approximately 18% of the proviruses had the U3-GF-R-U5 structure, close to the observed 15% efficiency of readthrough RNA transcription (18). This result also suggested that DNA synthesis of these readthrough RNA transcripts mostly initiated from the downstream PBS and bypassed the minus-strand DNA transfer event. This was possible because the downstream PBS in these vectors contained a large portion of the sequences proposed to form a secondary structure that was important for efficient initiation of DNA synthesis. In addition, the GF fragment is 350 bp in length, whereas FP is 462 bp. If DNA synthesis initiated from both upstream and downstream PBS simultaneously, there would be a race for the 3′ template usage. By the time DNA synthesis that was initiated from the upstream PBS reached the end of FP, DNA synthesis that was initiated from the downstream PBS would have copied through GF and a portion of U3. Consequently, the RNA templates at the 3′ end of the viral sequences would be degraded, including the F region of the RNA that could be used for minus-strand DNA transfer. Therefore, it was likely that the minus-strand DNA that was initiated from the upstream PBS would lose the “race” during reverse transcription, which may account for the proportion of proviruses that contained sequences derived from the downstream R.
Alternatively, it is possible that in readthrough RNA transcripts, minus-strand DNA synthesis initiated from the upstream PBS, copied a portion of U5, and used the U5 sequences to transfer to near the 3′ end of the viral RNA. RT could then copy the 3′ R, GF, and U3 sequences. In order for this mechanism to account for most of the proviruses containing U3-GF-R-U5 in the LTR, minus-strand DNA transfer had to occur at a high frequency during the copying of the 72-nt U5 region. Because minus-strand DNA transfer mediated by weak-stop DNA is known to occur at a low frequency while copying R, which is 68 nt in length, this mechanism is unlikely to be responsible for generating all of these proviruses (26–28, 36, 45).
Read-in RNA transcripts and minus-strand DNA transfer using the homology region including a portion of U3.
In this study, we also observed proviruses containing U3-FP-R-U5 LTR structures. We hypothesized that these proviruses were generated from RNA transcripts that were initiated from upstream promoters. In another study from our laboratory, modified MLV vectors lacking the downstream U3 were used to examine minus-strand DNA transfer (Dang and Hu, submitted). Some of the proviruses resulting from that study contained portions of U3 sequences, indicating that these proviruses were also the products of read-in RNA transcripts. In addition, RNA species containing the upstream U3 were identified in cellular RNA samples by RNase protection assays, further confirming our hypothesis.
This observation raised the question of whether U3 sequences are occasionally used in minus-strand DNA transfer during the replication of wild-type viruses. If an active promoter is located upstream of a provirus, RNA transcripts containing the upstream U3 could be easily generated. However, most of these transcripts would terminate at the 5′ R; only 15% of the transcripts would read through the 5′ R and terminate at the 3′ R. Similar to the events observed in our experiments, RT could copy a portion of the upstream U3 and use the U3 sequences to align the nascent DNA and 3′ RNA for minus-strand DNA transfer. Because these events depend on both the location of the proviral integration and the RNA transcript extending through 5′ R, if U3 were used to mediate minus-strand DNA transfer in wild-type viruses, it would be likely to occur at a low frequency.
Application of minus-strand DNA transfer-mediated gene reconstitution to gene therapy and inducible gene expression systems.
Previously, direct repeat deletion has been utilized to reconstitute various genes with a high efficiency; these genes included the drug resistance gene neo and a suicide gene encoding the herpes simplex virus thymidine kinase that has been used for cancer therapy (9–11, 24). This strategy uses high-frequency template switching events that occur during reverse transcription to reconstitute genes and delete sequences from the portions of the viral genome that are internal to the LTRs. We describe here gene reconstitution using a different strategy utilizing the obligatory minus-strand DNA transfer step in the reverse transcription process. These experiments demonstrated that this strategy works in principle; GFP was reconstituted at approximately 92% efficiency in the vector pMS2-FG-GF-no3R. This high-frequency reconstitution could be exploited for gene delivery in gene therapy applications or as an inducible gene expression system. In the plasmid vector, a gene of interest or a cassette containing a gene expression unit could be divided into two portions with a small stretch of overlapping sequences in which neither portion could express the gene of interest. During reverse transcription and virus replication, this gene of interest could be reconstituted and expressed in the infected cells. Such an approach could be very useful for the delivery of potentially toxic genes in gene therapy applications in which the toxicity of the gene hampers the production of the viral-vector-containing virions. This approach could also be very useful as an inducible gene expression system in which viral replication activates gene expression. Because the reporter gene is inactive prior to virus replication, there would be little background expression in the system. For example, this strategy could be used to detect the presence of replication-competent retroviruses (RCR). Target cells transfected with pMS2-FP-GF-no3R can be established; these cells would not express GFP and could not produce vector virus because they lack viral proteins. Test samples could be applied to these target cells; if RCR were present in the sample, then the MS2-FP-GF-no3R would be mobilized, GFP would be reconstituted during reverse transcription, and the infected cells would express GFP. This could be performed in a one-step assay to directly detect RCR rather than a lengthy coculture or infection assay.
In summary, we have established that homology, rather than sequence context, is important in the mediation of efficient minus-strand DNA synthesis. During this study, we also demonstrated that genes could be reconstituted during minus-strand DNA transfer efficiently with a short stretch of homology, a strategy that could also be exploited for the design of gene therapy vectors.
ACKNOWLEDGMENTS
We thank Carrie McBee, Terence Rhodes, Michelle Paulson, and Melanie Sal for the construction of some of the plasmids. We also thank Que Dang and Krista Delviks for critical reading of the manuscript; Anne Arthur for expert editorial revisions of the manuscript; John Coffin, Alan Rein, and Vineet KewalRamani for discussions, intellectual input, and suggestions regarding the manuscript; and V. KewalRamani for help with the flow cytometry.
This work was supported in part by research grants from the NIH and the ACS and also by the HIV Drug Resistance Program, National Cancer Institute. J.A.A. was fully supported and S.R.C. and C.K.H. were partially supported by the Medical Scientist Training Program at West Virginia University.
REFERENCES
- 1.Adam M A, Osborne W R, Miller A D. R-region cDNA inserts in retroviral vectors are compatible with virus replication and high-level protein synthesis from the insert. Hum Gene Ther. 1995;6:1169–1176. doi: 10.1089/hum.1995.6.9-1169. [DOI] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allain B, Rascle J B, de Rocquigny H, Roques B, Darlix J L. CIS elements and trans-acting factors required for minus strand DNA transfer during reverse transcription of the genomic RNA of murine leukemia virus. J Mol Biol. 1998;277:225–235. doi: 10.1006/jmbi.1997.1596. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J Mol Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 5.Blain S W, Goff S P. Effects on DNA synthesis and translocation caused by mutations in the RNase H domain of Moloney murine leukemia virus reverse transcriptase. J Virol. 1995;69:4440–4452. doi: 10.1128/jvi.69.7.4440-4452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M. Retroviridae: the viruses and their replication. Vol. 3. New York, N.Y: Raven Press; 1996. [Google Scholar]
- 8.Darlix J L, Vincent A, Gabus C, de Rocquigny H, Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C R Acad Sci III. 1993;316:763–771. [PubMed] [Google Scholar]
- 9.Delviks K A, Hu W S, Pathak V K. Psi- vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J Virol. 1997;71:6218–6224. doi: 10.1128/jvi.71.8.6218-6224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delviks K A, Pathak V K. Development of murine leukemia virus-based self-activating vectors that efficiently delete the selectable drug resistance gene during reverse transcription. J Virol. 1999;73:8837–8842. doi: 10.1128/jvi.73.10.8837-8842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delviks K A, Pathak V K. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J Virol. 1999;73:7923–7932. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeStefano J J. Interaction of human immunodeficiency virus nucleocapsid protein with a structure mimicking a replication intermediate. Effects on stability, reverse transcriptase binding, and strand transfer. J Biol Chem. 1996;271:16350–16356. [PubMed] [Google Scholar]
- 13.Dougherty J P, Temin H M. A promoterless retroviral vector indicates that there are sequences in U3 required for 3′ RNA processing. Proc Natl Acad Sci USA. 1987;84:1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 15.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halvas E K, Svarovskaia E S, Pathak V K. Development of an in vivo assay to identify structural determinants in murine leukemia virus reverse transcriptase important for fidelity. J Virol. 2000;74:312–319. [PMC free article] [PubMed] [Google Scholar]
- 18.Herman S A, Coffin J M. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986;60:497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman S A, Coffin J M. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science. 1987;236:845–848. doi: 10.1126/science.3033828. [DOI] [PubMed] [Google Scholar]
- 20.Hsu M, Rong L, Rocquigny H, Roques B P, Wainberg M A. The effect of mutations in the HIV-1 nucleocapsid protein on strand transfer in cell-free reverse transcription reactions. Nucleic Acids Res. 2000;28:1724–1729. doi: 10.1093/nar/28.8.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen R A, Rothstein S J, Reznikoff W S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177:65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- 24.Julias J G, Hash D, Pathak V K. E-vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J Virol. 1995;69:6839–46. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J K, Palaniappan C, Wu W, Fay P J, Bambara R A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 26.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulpa D, Topping R, Telesnitsky A. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 1997;16:856–865. doi: 10.1093/emboj/16.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobel L I, Goff S P. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J Virol. 1985;53:447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo G X, Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990;64:4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 33.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 34.Peliska J A, Benkovic S J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 35.Raja A, DeStefano J J. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry. 1999;38:5178–5184. doi: 10.1021/bi9828019. [DOI] [PubMed] [Google Scholar]
- 36.Ramsey C A, Panganiban A T. Replication of the retroviral terminal repeat sequence during in vivo reverse transcription. J Virol. 1993;67:4114–4121. doi: 10.1128/jvi.67.7.4114-4121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggs J L, McAllister R M, Lennette E H. Immunofluorescent studies of RD-114 virus replication in cell culture. J Gen Virol. 1974;25:21–29. doi: 10.1099/0022-1317-25-1-21. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 39.Rong L, Liang C, Hsu M, Kleiman L, Petitjean P, de Rocquigny H, Roques B P, Wainberg M A. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J Virol. 1998;72:9353–9358. doi: 10.1128/jvi.72.11.9353-9358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swain A, Coffin J M. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 41.Swain A, Coffin J M. Polyadenylation at correct sites in genome RNA is not required for retrovirus replication or genome encapsidation. J Virol. 1989;63:3301–3306. doi: 10.1128/jvi.63.8.3301-3306.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Temin H M. The DNA provirus hypothesis. Science. 1976;192:1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- 43.Weiss S, Konig B, Morikawa Y, Jones I. Recombinant HIV-1 nucleocapsid protein p15 produced as a fusion protein with glutathione S-transferase in Escherichia coli mediates dimerization and enhances reverse transcription of retroviral RNA. Gene. 1992;121:203–212. doi: 10.1016/0378-1119(92)90123-7. [DOI] [PubMed] [Google Scholar]
- 44.Whitcomb J M, Ortiz-Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin P D, Pathak V K, Rowan A E, Teufel II R J, Hu W S. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J Virol. 1997;71:2487–2494. doi: 10.1128/jvi.71.3.2487-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You J C, McHenry C S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]