Abstract
Objective
To survey the epidemiology of respiratory pathogens during the coronavirus disease 2019 (COVID-19) pandemic using multiplex polymerase chain reaction (PCR).
Methods
Specimens were assayed using multiplex nested PCR.
Materials
Specimens were obtained from outpatients who presented with symptoms of upper respiratory tract infection and asymptomatic outpatients who had contact with patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection at Tohoku Medical and Pharmaceutical University Hospital in Sendai, Japan, from November 1, 2020, to May 31, 2023. The analysis included multiple specimens collected from the same patients at different time-points. Data were collected from the electronic records after testing.
Results
This study included 8,335 patients (4,311 men) with a median age of 59 years old, and 11,741 total specimens were collected. At least 1 positive SARS-CoV-2 result was obtained for 1,710 (14.6%) specimens. Furthermore, 15 pathogens were identified in the positive specimens, and rhinovirus/enterovirus was detected more frequently than other viruses. We identified a larger number of SARS-CoV-2-positive specimens in patients ≥10 years old. In contrast, in patients 0-9 years old, we identified a larger number of specimens positive for rhinovirus/enterovirus than for other viruses.
Conclusion
In this study, we examined the epidemiology of circulating respiratory pathogens during the COVID-19 pandemic era.
Keywords: epidemiology, multiplex polymerase chain reaction assay, COVID-19, respiratory pathogens, SARS-CoV-2
Introduction
Coronavirus disease 2019 (COVID-19) was first reported as an outbreak of pneumonia in Wuhan, China, in December 2019 (1), and the number of infected individuals continues to increase worldwide (2). The symptoms of COVID-19 are similar to those of other respiratory diseases. Therefore, it is difficult to distinguish between COVID-19 and other upper respiratory infections, such as influenza (3), making the epidemiology of upper respiratory syndrome in the COVID-19 era unclear.
The early diagnosis of patients with respiratory symptoms is necessary to distinguish between COVID-19 and other serious illnesses in the COVID-19 era. The gold standard for the diagnosis of COVID-19 is the direct detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA using reverse-transcription polymerase chain reaction (RT-PCR) (4). A multiplex nested PCR assay (BioFireⓇ Respiratory Panel (RP) 2.1; BioFire Diagnostics/bioMérieux, Salt Lake City, USA) targeting various viruses, including SARS-CoV-2, and three bacterial species was approved for use in Japan in October 2020. This multiplex PCR assay can detect adenoviruses, human coronaviruses (HCoV) (229E, HKU1, NL63, and OC43), human metapneumoviruses, human rhinoviruses/enteroviruses, influenza virus A (H1, H1-2009, and H3), B, parainfluenza viruses (types 1-4), respiratory syncytial virus (RSV), SARS-CoV-2, Mycoplasma pneumoniae, Chlamydia pneumoniae, and Bordetella pertussis (5).
We surveyed the epidemiology of respiratory pathogens during the COVID-19 pandemic using a multiplex nested PCR assay.
Materials and Methods
This study was approved by the Institutional Review Board of Tohoku Medical and Pharmaceutical University Hospital (Approval No. 2022-2-083-1).
Patients and specimens
This study included the following patients: i) outpatients with upper respiratory tract infection symptoms who visited Tohoku Medical and Pharmaceutical University Hospital in Sendai, Japan, and ii) asymptomatic outpatients who had come in contact with patients infected with SARS-CoV-2 from November 1, 2020, to May 31, 2023.
The pre-alpha, alpha, delta, and omicron variants were predominant in Japan during the study period (6). We reviewed one or more of the following symptoms suggestive of acute upper respiratory infection: a fever, headache, fatigue, nasal congestion, nasal discharge, sneezing, sore throat, and cough. Data were collected from electronic records after the examination. In some cases, more than one sample was collected from the same patient at different time points; thus, 11,741 nasopharyngeal swab specimens were collected. Previous studies have shown an increasing number of children (0-9 years old) infected with rhinoviruses since the COVID-19 pandemic (7,8). Therefore, we investigated whether or not there were differences in the pathogens detected between children <10 years old and those ≥10 years old.
Multiplex nested PCR assay
To survey the epidemiology of respiratory pathogens, we used a multiplex nested PCR assay (BioFireⓇ RP 2.1; BioFire Diagnostics/bioMérieux) in accordance with the manufacturer's instructions. Nasopharyngeal swab specimens (300 μL) were injected into the Viral Transport Medium in the FilmArray pouch, and the reaction proceeded automatically.
This system performs automated nucleic acid extraction, reverse transcription, nucleic acid amplification, and automated result analysis in approximately 45 minutes per specimen. If any of the internal controls fail, the software program automatically returns results of “invalid" for all panel analytes. Analytes are qualitatively reported as “detected,” “not detected,” or “equivocal" (for influenza A only). The BioFireⓇ RP 2.1 contains two independent assays for the identification of SARS-CoV-2 (one targeting the M gene and another targeting the S gene). A positive result in either assay indicates a positive result for SARS-CoV-2.
Results
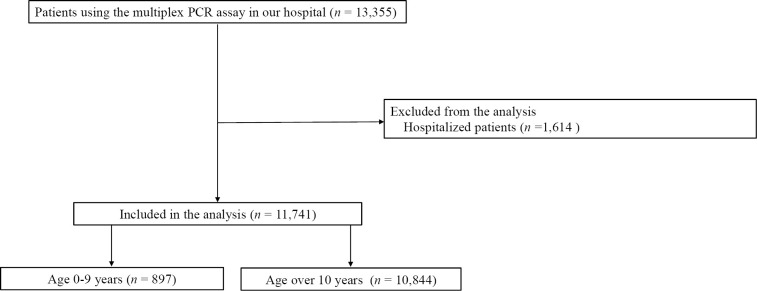
A total of 11,741 nasopharyngeal swab specimens were collected from 8,335 patients from November 1, 2020, to May 31, 2023. This included 897 specimens from patients 0-9 years old and 10,844 specimens from patients ≥10 years old (Fig. 1,Table).
Figure 1.
Patient flow chart.
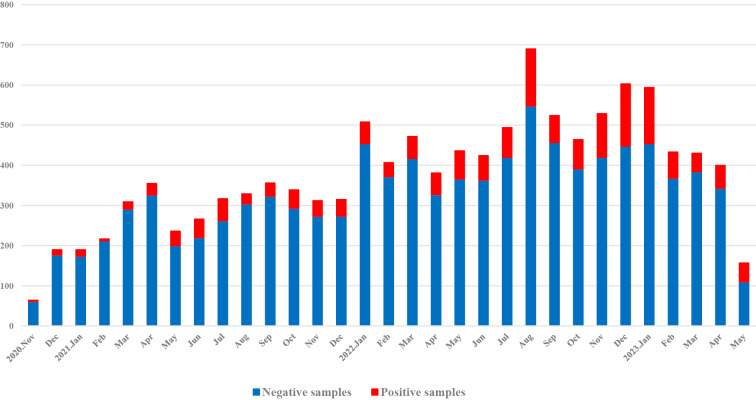
At least one positive result was obtained for 1,710 (14.6%) specimens during the study period (November 1, 2020 to May 31, 2023) (Fig. 2, Table). Compared to patients ≥10 years old, the percentage of positive specimens was higher (73.2%) in the 0-9-year-old group (Table).
Figure 2.
Trends in the number of samples that tested positive for at least one pathogen. The numbers of specimens obtained are presented by month. The number of positive results obtained using multiplex PCR is shown in red, and the number of negative results is shown in blue.
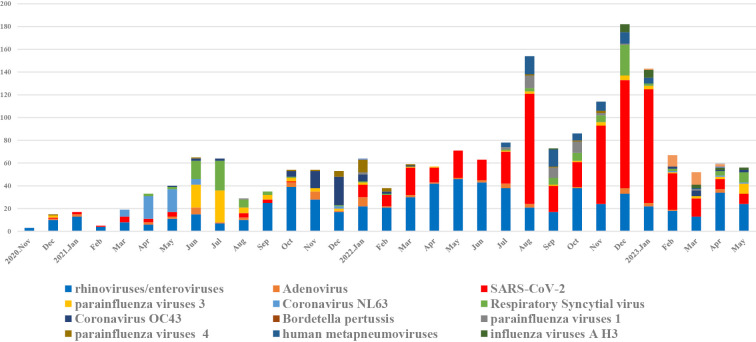
Fig. 3 shows the trends in the circulation of SARS-CoV-2 and other respiratory pathogens during the COVID-19 pandemic. Fifteen pathogens were detected, as follows: rhinovirus (n=682), SARS-CoV-2 (n=624), RSV (n=127), parainfluenza virus 3 (n=100), HCoV-OC43 (n=69), human metapneumovirus (n=65), adenovirus (n=63), HCoV-NL63 (n=56), parainfluenza virus 1 (n=39), parainfluenza virus 4 (n=29), HCoV-HKU1 (n=24), influenza A H3 (n=22), HCoV-229E (n=2), influenza virus A H1-2009 (n=1), and influenza virus A unclassified (n=1). We also examined trends in the detection rates of each pathogen; SARS-CoV-2 and rhinovirus/enterovirus were detected more frequently than other pathogens (Supplementary material 1-3). The highest detection rates of SARS-CoV-2 were observed in August 2022 (62.9%) and January 2023 (69.9%). In contrast, the highest detection rates for rhinovirus/enterovirus were observed in February 2021 (80%), September 2021 (71.4%), October 2021 (72.2%), and April 2022 (73.6%). Other viruses with high detection rates were HCoV-NL63 in April 2021 (60.6%), parainfluenza virus 3 in July 2021 (43.7%), RSV in July 2021 (40.6%), and HCoV-OC43 in December 2021 (47%). Influenza viruses were not detected for approximately two years during the study period, with H3 beginning in September 2022, peaking during the study period in March 2023, and accounting for 5.7% of all specimens in March 2023 (Supplementary material 1-13).
Figure 3.
Trends in the circulation of SARS-CoV-2 and other respiratory pathogens. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2, RSV: respiratory syncytial virus
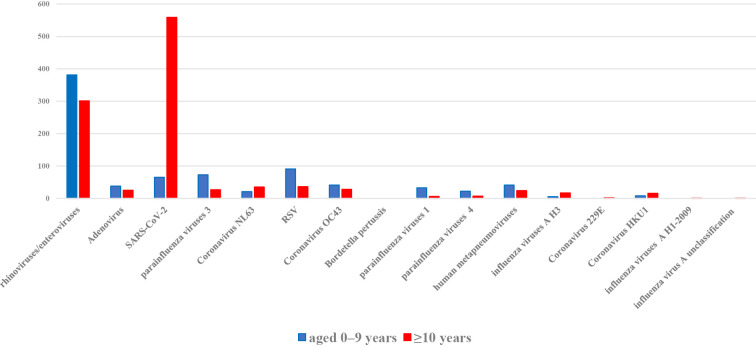
Fig. 4 shows that rhinovirus/enterovirus, RSV, adenovirus, human metapneumovirus, parainfluenza virus 1, parainfluenza virus 3, parainfluenza virus 4, and HCoV-OC43 were detected more frequently in the 0-9-year-old population than in the ≥10-year-old population. In contrast, SARS-CoV-2 was detected more frequently in the ≥10-year-old population than in the 0-9-year-old population.
Figure 4.
Number of respiratory pathogens detected in patients <10 and ≥10 years old. The numbers of respiratory pathogens detected in samples from patients <10 and ≥10 years old are shown. The number of respiratory pathogens detected in patients 0-9 years old is shown in blue, and those detected in patients ≥10 years old is shown in red. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2, RSV: respiratory syncytial virus
Discussion
In this study, we assessed the epidemiology of respiratory pathogens, including SARS-CoV-2, from 11,741 nasopharyngeal specimens obtained during approximately 2.5 years of the COVID-19 pandemic. More specimens tested positive for SARS-CoV-2 in patients ≥10 years old, whereas in patients 0-9 years old, more specimens tested positive for rhinovirus/enterovirus than other viruses.
The COVID-19 pandemic may influence the seasonality of respiratory pathogens. For example, seasonal coronavirus outbreaks in the United States were slower at the beginning of the COVID-19 pandemic than in previous season (9). Although the trends of seasonal coronaviruses in Japan prior to the COVID-19 pandemic are unknown, it is possible that they followed a course similar to that in the United States. This might be attributed to changes in human behavior associated with the COVID-19 pandemic, such as decreased domestic and international travel, mask use, school and office closures, and physical distancing (10). As the pandemic dragged on, human behavior gradually reverted to pre-pandemic conditions, which may have resulted in the detection of influenza viruses that remained undetected for approximately two years.
In this study, before the outbreak of the omicron strain, the number of patients <10 years old who were infected with SARS-CoV-2 was lower than that of patients ≥10 years old. Since the emergence of the omicron strain, the number of children <10 years old infected with SARS-CoV-2 has increased (11). Why there were fewer pediatric infections in the period leading up to the emergence of the delta strain is unclear, although factors such as exposure patterns (12) and differences in gene expression (13) are thought to be responsible.
Whether or not the number of patients infected with rhinoviruses/enteroviruses has increased compared to those infected with other viruses is controversial. Preschool children are reservoirs of rhinoviruses (14). Our finding that rhinovirus/enterovirus is more common in children 0-9 years old than in those ≥10 years old is consistent with previous reports (7,8). This may be because it is difficult to control rhinovirus infection in homes with preschool children.
Strengths and limitations
Our study has several strengths. First, the results of our study reflect a real-world clinical setting for COVID-19 in Japan. Second, our study included the largest number of patients with upper respiratory symptoms (n=11,741 specimens) for investigating the epidemiology of circulating respiratory pathogens using a multiplex PCR assay among such studies.
However, several limitations associated with the present study also warrant mention. First, our study was a single-center retrospective study in Miyagi Prefecture, and our results were reflected in the local prevalence of upper respiratory tract pathogens during the COVID-19 epidemic. Second, which is the causative organism if multiple pathogens are detected is unclear, and this may have biased our results. Third, the study included patients who were in close contact with confirmed COVID-19 patients, and it was difficult to distinguish between patients with or without close contact with confirmed COVID-19 due to the nature of the database. Thus, the results may have been biased. Fourth, this study was based only on PCR methods. However, the BioFireⓇ RP2.1 PCR assay detected SARS-CoV-2 in nasopharyngeal specimens with a positive agreement rate of 98.4% and a negative agreement rate of 98.9% (15); therefore, these results are reliable for further epidemiological studies. Fifth, we did not use bacterial sputum culture, which may have biased our results; instead, nasopharyngeal specimens were examined using the BioFireⓇ RP 2.1 PCR assay. Finally, information regarding each patient's SARS-CoV-2 vaccination history and SARS-CoV-2 subtype was unknown in this study, which may have influenced our results (16).
Conclusion
In the present study, using the multiplex PCR assay, we acquired epidemiological data for a variety of respiratory viruses circulating during the COVID-19 pandemic.
The authors state that they have no Conflict of Interest (COI).
Supplementary Material
Detection rate of SARS-CoV-2
Detection rate of Rhinoviruses/enteroviruses
Detection rate of RSV
Detection rate of Parainfluenza viruses3
Detection rate of Adenovirus
Detection rate of Coronavirus OC43
Detection rate of Human metapneumoviruses (table 7)
Detection rate of Coronavirus NL63
Detection rate of Parainfluenza viruses 1
Detection rate of Parainfluenza viruses 4
Detection rate of Coronavirus HKU1
Detection rate of Influenza viruses
Detection rate of Coronavirus 229E
References
- 1.Zhu N, Zhang D, Wang W, et al. ;the China Novel Coronavirus Investingating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727-733, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Weekly epidemiological update on COVID-19 - 11 2023 May [Internet]. 2023 May 11 [cited 2023 Mar 15]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---11-may-2023/
- 3.Bai Y, Tao X. Comparison of COVID-19 and influenza characteristics. J Zhejiang Univ Sci B 22: 87-98, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann Intern Med 172: 726-734, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry GJ, Zhen W, Smith E, et al. Multicenter evaluation of the BioFire Respiratory Panel 2.1 (RP2.1) for detection of SARS-CoV-2 in nasopharyngeal swab samples. J Clin Microbiol 60: e0006622, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institute of Infectious Disease. Coronavirus disease (COVID-19) [Internet]. [cited 2023 Mar 15]. Available from: https://www.niid.go.jp/niid/en/
- 7.Takashita E, Kawakami C, Momoki T, et al. Increased risk of rhinovirus infection in children during the coronavirus disease-19 pandemic. Influenza Other Respir Viruses 15: 488-494, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diesner-Treiber SC, Voitl P, Voitl JJM, et al. Respiratory infections in children during a Covid-19 pandemic winter. Front Pediatr 9: 740785, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah MM, Winn A, Dahl RM, Kniss KL, Silk BJ, Killerby ME. Seasonality of common human coronaviruses, United States, 2014-2021. Emerg Infect Dis 28: 1970-1976, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddadin Z, Schuster JE, Spieker AJ, et al. Acute respiratory illnesses in children in the SARS-CoV-2 pandemic: prospective multicenter study. Pediatrics 148: e2021051462, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ministry of Health, Labour and Welfare. Information regarding SARS-CoV-2 vaccination in Japan [Internet]. [cited 2023 Jul 8]. Available from: https://www.cov19-vaccine.mhlw.go.jp/qa/ (in Japanese)
- 12.CDC COVID-19 Response Team. Coronavirus disease 2019 in children - United States, February 12-April 2, 2020. MMWR Morb Mortal Wkly Rep 69: 422-426, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA 323: 2427-2429, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis 197: 382-389, 2008. [DOI] [PubMed] [Google Scholar]
- 15.Creager HM, Cabrera B, Schnaubelt A, et al. Clinical evaluation of the BioFireⓇ Respiratory Panel 2.1 and detection of SARS-CoV-2. J Clin Virol 129: 104538, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Information regarding SARS-CoV-2 vaccination in Sendai City [Internet]. [cited 2023 May 12]. Available from: https://www.city.sendai.jp/covidvaccine/sessyuzyoukyou.html (in Japanese)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection rate of SARS-CoV-2
Detection rate of Rhinoviruses/enteroviruses
Detection rate of RSV
Detection rate of Parainfluenza viruses3
Detection rate of Adenovirus
Detection rate of Coronavirus OC43
Detection rate of Human metapneumoviruses (table 7)
Detection rate of Coronavirus NL63
Detection rate of Parainfluenza viruses 1
Detection rate of Parainfluenza viruses 4
Detection rate of Coronavirus HKU1
Detection rate of Influenza viruses
Detection rate of Coronavirus 229E