Abstract
Human herpesvirus 8 (HHV-8), or Kaposi's sarcoma-associated herpesvirus, is a gammaherpesvirus implicated in all forms of Kaposi's sarcoma and certain lymphomas. HHV-8 has been extensively characterized, both biochemically and immunologically, since its first description in 1994. However, its three-dimensional (3D) structure remained heretofore undetermined largely due to difficulties in viral purification. We have used log-phase cultures of body cavity-based lymphoma 1 cells induced with 12-O-tetradecanoylphorbol-13-acetate to obtain HHV-8 capsids for electron cryomicroscopy and computer reconstruction. The 3D structure of the HHV-8 capsids revealed a capsid shell composed of 12 pentons, 150 hexons, and 320 triplexes arranged on a T=16 icosahedral lattice. This structure is similar to those of herpes simplex virus type 1 (HSV-1) and human cytomegalovirus (HCMV), which are prototypical members of alpha- and betaherpesviruses, respectively. The inner radius of the HHV-8 capsid is identical to that of the HSV-1 capsid but is smaller than that of the HCMV capsid, which is consistent with the relative sizes of the genomes they enclose. While the HHV-8 capsid exhibits many structural similarities to the HSV-1 capsid, our reconstruction shows two major differences: its hexons lack the “horn-shaped” VP26 densities bound to the HSV-1 hexon subunits, and the HHV-8 triplexes appear smaller and less elongated than those of HSV-1. These differences are in excellent agreement with our sequence comparisons of HHV-8 and HSV-1 capsid proteins. This gammaherpesvirus capsid structure complements previous structural studies on alpha- and betaherpesviruses in providing an account of structural similarities and differences among capsids representing all human herpesvirus subfamilies.
Human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma (KS)-associated herpesvirus, has been implicated in all forms of KS, in primary effusion lymphoma (PEL), and in multicentric Castleman's disease, based on serological analyses and epidemiological studies (8, 9, 24, 37, 40). HHV-8 was initially identified from two novel herpesvirus-like DNA fragments cloned from AIDS-associated KS (AIDS-KS), an angiogenic neoplasm composed of endothelial and spindle cells (9). Subsequent sequencing of an AIDS-KS genome library fragment hybridizing to fragment KS330Bam demonstrated that HHV-8 belongs to the Gammaherpesvirinae subfamily, which also includes two other DNA tumor viruses: Epstein-Barr virus and Herpesvirus saimiri (25). More recently, two new viruses with more homology to HHV-8 than any other previously known herpesviruses have been isolated from monkeys (15, 38). To date, the virus cannot be successfully propagated in vitro, although a susceptible cell line with limited viral culturing capability has been reported (26). Instead, several tumor cell lines that appear to harbor forms of HHV-8 DNA, such as body cavity-based lymphoma 1 (BCBL-1), BC-1, and BC-3, have been established. Cell-free virus can be generated by induction of lytic replication with tumor-promoting agents such as 12-O-tetradecanoylphorbol-13-acetate (TPA) (31).
Pulsed-field gel electrophoresis of DNA extracted from virions purified from TPA-induced BCBL-1 cells revealed that the HHV-8 genome consists of 165 to 170 kb (30, 50). This genome size is slightly larger than the 153-kb genome of herpes simplex virus type 1 (HSV-1), the prototypical herpesvirus and a member of the Alphaherpesvirinae subfamily (32), but significantly smaller than the 230-kb genome of human cytomegalovirus (HCMV), a member of the Betaherpesvirinae subfamily (23). The HHV-8 genome contains at least 85 open reading frames, of which 66 are homologous to those of other herpesviruses; the others are unique to HHV-8 and are designated with the prefix K (27, 33).
While extensive biochemical and immunological efforts have been undertaken to characterize HHV-8 infection and pathogenesis since its first description (9), structural studies of HHV-8 particles have been limited. The morphology and structure of this human pathogen are difficult to study because it remains refractory to cultivation in either tissue culture systems or animals. In contrast to the detailed descriptions of the three-dimensional (3D) structures of HSV-1 (5, 36, 53) and HCMV capsids (7, 10, 47), the only available structural description of HHV-8 is a comparison of electron microscopy images of HHV-8 and HCMV in virus-infected endothelial cells (35). Based on negative-stain images of thin sections, it was suggested that the morphological features of the HHV-8 and HCMV nucleocapsids are similar (35). However, the HCMV virion is generally larger (150 to 200 nm) than that of HHV-8 (120 to 150 nm), and its tegument layer is denser than that of HHV-8 (35). Here we report the 3D structure of the HHV-8 capsid, reconstructed from electron cryomicroscopy (cryoEM) images of HHV-8 capsids purified from TPA-induced BCBL-1 cells, and show its structural similarities and differences with the HSV-1 capsid.
MATERIALS AND METHODS
Cell lines.
An HHV-8-positive BCBL-1 PEL cell line (31) was obtained from the National Institutes of Health AIDS Research and Reagent Program (Rockville, Md.). BCBL-1 cells were propagated in T150 flasks containing RPMI 1640 medium (Sigma) with 10% fetal bovine serum (HyClone), 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 50 μM 2-mercaptoethanol (Sigma) in 5% CO2 at 37°C.
Purification of HHV-8 capsids.
A log-phase culture of BCBL-1 cells (1,000 ml) at a density of ∼2 × 106 cells/ml was induced with 20 ng of TPA (Sigma)/ml for 6 days to obtain the maximum production of extracellular virus. The cells were pelleted by centrifugation at 4,000 × g for 30 min, and the supernatant was collected. Polyethylene glycol 6000 (Sigma) was dissolved in the supernatant to a final concentration of 6% (wt/vol). After incubation overnight at 4°C, the precipitate was sedimented by centrifugation at 4,000 × g for 30 min and the pellet was resuspended in 10 ml of Tris-buffered saline (0.05 M Tris–0.15 M NaCl, pH 7.4) containing 0.25% Nonidet P-40 (NP-40). The remaining solution represented approximately a 100-fold concentration of the original culture fluid.
Density gradient purification of HHV-8 capsids and virions was performed as described previously for cytomegalovirus (17). Aliquots of the concentrated capsids (2 ml each) were loaded onto 10-ml gradients of 20 to 60% sucrose with Tris-buffered saline and centrifuged at 110,000 × g for 20 h at 4°C in a Beckman SW 41Ti rotor. Two visible bands, corresponding to 35 and 50% sucrose, respectively, were collected and separated into two pools. Each pool was diluted with the original buffer and centrifuged at 75,000 × g for 60 min. The final pellet was resuspended in 0.2 ml of Tris-HCl buffer (0.05 M, pH 7.4). Initial negative-stain electron microscopy indicated that the upper band contained virions and a significant amount of cellular debris while the lower band contained predominantly capsids at approximately 5 × 107 particles/ml with less cellular debris. The capsid-containing fraction was further concentrated using a Microcon YM-100 centrifugal filter device (Millipore) and subsequently used for cryoEM observation.
cryoEM.
cryoEM images of freeze-hydrated HHV-8 capsids were recorded using standard procedures as described previously (42, 57). Briefly, 3 μl of a purified HHV-8 capsid sample was applied to carbon-coated holey grids and quickly frozen in liquid ethane so that the capsids were suspended in a thin layer of vitreous ice across the holes of the carbon support film. Most micrographs were taken as focal pairs at ×30,000 magnification with a dosage of ∼9 electrons/Å2 for each micrograph in a JEOL JEM1200 electron cryomicroscope operated at 100 kV with a cold stage at −167°C. Selected micrographs were digitized with a SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.) using a step size equivalent to 4.67 Å/pixel and subsequently averaged by combining adjacent pixels to yield a step size of 9.34 Å/pixel on the specimen scale.
3D reconstruction and visualization.
Data processing and visualization were carried out on SGI Octane dual-processor workstations (Silicon Graphics, Inc.) using parallel programs for refinement (52) and reconstruction (20), which are based on Fourier common lines (12, 18) and Fourier-Bessel synthesis (13). Individual HHV-8 capsid particles were boxed out from digitized micrographs into separate image files. A list of 28 initial orientation estimates was generated for each particle image using a program based on self common lines. A preliminary 3D model was then computed at 45 Å from 10 particles that showed the best self-common-line phase residuals and that had been refined by cross-common-line phase residual minimization among all 10 particles (52). The incorrect orientations were eliminated from the list of initial orientation estimates of each particle by evaluating the cross-common-line phase residual between the particle and projection images of the preliminary model. The selected orientations were then refined, first by a global refinement that minimized the cross-common-line phase residuals across all selected particles (52) and then by a projection-based refinement that optimized the match between the image and the projections computed from the preliminary 3D model. The new model reconstructed from these refinements was used as a template for the next round of particle selection and refinement, resulting in a further improved model. This process was iterated for several cycles utilizing the entire data set until no more particles with correct orientation parameters could be obtained and no improvement was evident in the cross-common-line phase residual between particle images and the computed projections. The icosahedral (5-3-2-2) symmetry was imposed in the final reconstruction.
The final reconstruction was calculated by merging data up to 22-Å resolution from 431 particle images with defocus values ranging from 0.8 to 3.5 μm, which were determined from the incoherently averaged Fourier transforms of particle images in each micrograph (54). The contrast transfer function was corrected as described previously (51) with an estimated temperature factor (or B factor) of 500 Å2 and an amplitude contrast of 7% (39). The effective resolution of the final map was estimated to be 24 Å, where the cross-correlation coefficient between two independent reconstructions reaches 0.5. For comparison, an HSV-1 B-capsid map was reconstructed similarly at 24-Å resolution from 431 particle images; the particles were a subset of those selected from images used in the determination of a previously published HSV-1 capsid structure (53). The magnification difference between the two electron cryomicroscopes used for imaging HSV-1 and HHV-8 capsids was estimated to be 2.0%, using the HCMV particle as a calibration standard, by matching the diameters of HCMV reconstructions independently determined from images taken in both microscopes (not shown) (10).
The 3D visualization was carried out using Iris Explorer (NAG, Inc., Downers Grove, Ill.) with custom-designed modules (16). The maps were displayed using a contour level of 1 standard deviation above the mean density of the map unless otherwise indicated. The radial density distribution profiles were obtained by spherically averaging the 3D density maps, and their relative densities were scaled by matching the height of the floor density peaks.
Protein sequence analyses.
Pairs of HHV-8 and HSV-1 capsid proteins were assigned based on homologies among their amino acid sequences and/or the relative positioning of their genes among HHV-8 (33), HSV-1 (22), and other gammaherpesviruses (1). The amino acid sequences for these capsid proteins were downloaded from GenBank. Homologies were calculated using a variety of programs, including the CLUSTALW (45) and LALIGN (19) modules in Biology Workbench, version 3.0 (http://biology.ncsa.uiuc.edu) (11), the BESTFIT program of the Genetics Computer Group package (Wisconsin package; Genetics Computer Group, Inc., Madison, Wis., 1998) in BioNavigator (http://www.bionavigator.com), and the BLASTP program of the BLAST 2 tool (44) (http://www.ncbi.nlm.nih.gov/gorf/b12.html). A preliminary comparison was made using the default parameters given by Biology Workbench for both CLUSTALW and LALIGN. In order to improve the alignments for the ORF62 and VP19C and ORF26 and VP23 pairs, LALIGN was rerun using different gap penalties to get the optimum length of aligned sequence. The open and extended gap penalties were 12 and 2, respectively, for ORF25 and VP5 and ORF17 and VP40 and 5 and 3, respectively, for ORF26 and VP23 and ORF62 and VP19c.
RESULTS
Purification and cryoEM of HHV-8 capsids.
Our initial attempt to purify HHV-8 capsids from BC3 cells (2) induced either by TPA or by a combination of TPA and butyric acid yielded barely detectable viral particles using a density gradient procedure, though a higher percentage of cells was induced into lytic replication in BC3 cells (40 to 60%) than in BCBL-1 cells (30 to 40%), as confirmed by an immunofluorescence assay using a mouse monoclonal antibody against the late antigen glycoprotein K8.1. It appeared possible that, prior to achieving maximum yield of virions in the medium, BC3 cells lysed into debris, which could have interfered with the subsequent purification of viral particles. Our best yield was obtained by inducing lytic replication of HHV-8 in BCBL-1 cells with TPA. It is noted, however, that the HHV-8 capsid preparation, including capsids from BCBL-1 cells, was much less homogeneous and had a significantly lower yield than preparations routinely obtained for HSV-1 and HCMV capsids, although various techniques, including those used for HSV-1 and HCMV in previous studies, were tried to optimize the isolation procedure.
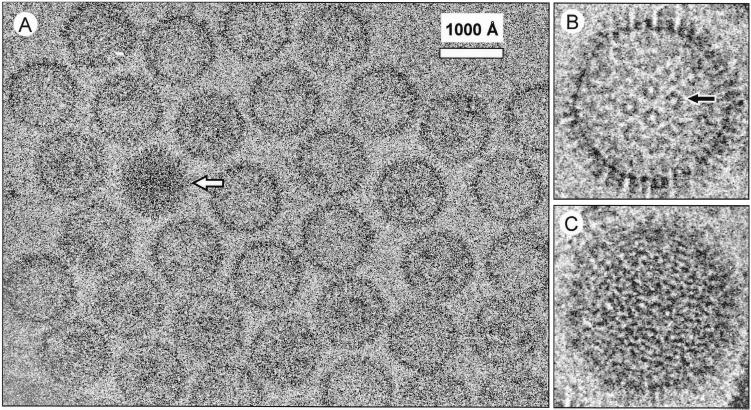
Our subsequent cryoEM effort was focused on imaging the lower band of the HHV-8 preparation from BCBL-1 cells, which contained HHV-8 capsids with the least cellular debris. The cryoEM micrographs of the ice-embedded HHV-8 capsids (Fig. 1) show that the capsids have a polyhedral shape with characteristic capsomer protrusions, similar to those seen in HSV-1 (5, 36), HCMV (7, 10, 47), and bovine herpesvirus (3) capsids. The HHV-8 capsids are approximately 1,250 Å in diameter, similar to HSV-1 capsids. Two types of HHV-8 capsids can be seen, designated intermediate and full capsids based on their resemblance to cryoEM images of the intermediate and full HSV-1 capsids (5, 36). Most capsids are of the intermediate type, which are less electron opaque (Fig. 1A and B). It is possible to identify the hexagonally arranged capsomers and capsomer channels (Fig. 1B) from the images of the intermediate capsids but not the full capsids (Fig. 1C). The full HHV-8 capsids (Fig. 1A) have a striated, fingerprint-like appearance (Fig. 1C) similar to that caused by the presence of viral DNA in HSV-1 (5, 51) and HCMV (4) C-capsids. Since the HSV-1 full and intermediate capsids were shown to have similar structures (5, 36, 57), we subsequently focused on the more prevalent intermediate capsids for in-depth data analyses.
FIG. 1.
cryoEM of HHV-8 capsids. (A) An area of a micrograph of the ice-embedded HHV-8 capsids showing predominantly intermediate capsids and one full capsid (arrow). The underfocus value of this image was estimated to be 1.6 μm based on incoherently averaged Fourier transforms (54). The enlarged view of an intermediate capsid (B) shows the capsomers (e.g., arrow) that form a characteristic hexagonal pattern with adjacent capsomers. The full-capsid (C) image reveals the characteristic “fingerprint” pattern.
Structural organization of the HHV-8 capsid.
We analyzed two sets of cryoEM images recorded for two separate HHV-8 capsid preparations, which were obtained through independent purifications. The 3D reconstructions obtained independently from these two sets of images show almost identical structural features except for slightly different resolutions, which were most likely due to differences in the number of capsid particles used for the two reconstructions. The final reconstruction was computed from 431 particle images of the second preparation and had an effective resolution of 24 Å based on a cross-correlation coefficient evaluation.
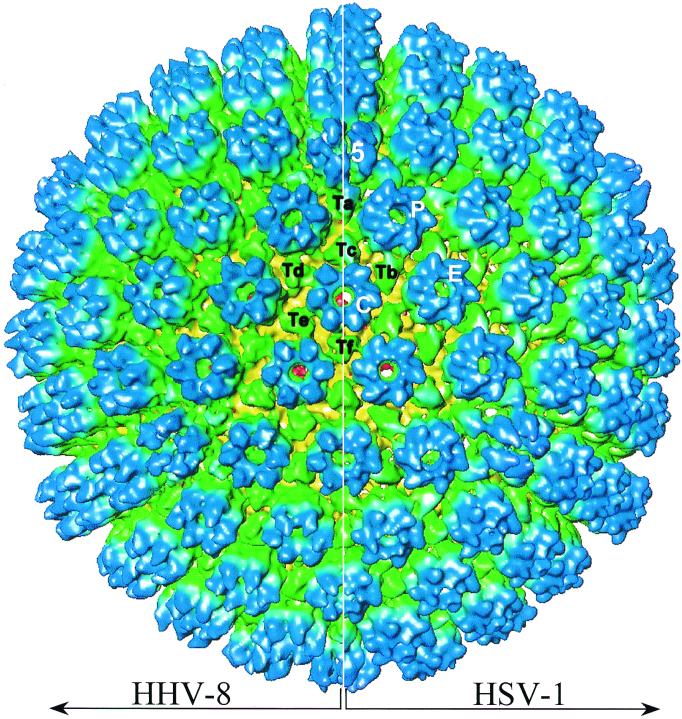
Each capsid, as shown in the shaded surface view along an icosahedral twofold axis (Fig. 2A), contains 12 pentons (blue, denoted by 5) and 150 hexons (blue, denoted by P, E, and C), which are interconnected by 320 triplexes (green). These capsomers or structural components are arranged in a T=16 icosahedral lattice with 20 triangular faces, one of which is outlined in Fig. 2A. Each asymmetric unit (one-third of a triangular face) of the capsid contains one-fifth of a penton at the vertex; one each of P, C, and one-half E hexons; and one each of Ta, Tb, Tc, Td, Te, and one-third Tf triplexes. These structural components were designated following the nomenclature established for HSV-1 (41, 57). Each penton and hexon contain an axial channel that connects the inside of the capsid to the outside (Fig. 2B). The penton and E hexon channels coincide with the fivefold and twofold axes, respectively, as indicated by Fig. 2B. Triplex Tf lies on the icosahedral threefold axis, while the other triplexes are located at the local threefold positions (Fig. 2B). The triplexes are triangularly pyramidal in shape and are positioned around small holes (∼10 Å diameter) penetrating the capsid floor (Fig. 2B). They appear to be connected to the capsid floor by small legs (Fig. 2B) and interact with adjacent pentons and hexons with “head” and “tail” domains (Fig. 2A), in a manner similar to triplexes of HSV-1 (34, 53, 57) and HCMV (7, 10, 47) capsids. The inside of the capsid contains some discontinuous densities (Fig. 2B, left, red), but these are not icosahedrally ordered as judged by their lack of reproducibility among independent reconstructions. This lack of icosahedral symmetry inside the capsid is similar to what was observed in HSV-1 capsids (56).
FIG. 2.
3D structure of the HHV-8 capsid at 24-Å resolution as viewed along the icosahedral twofold axis from the outside (A) and inside (B) of the capsid. The map was color coded according to the particle radius (see color bar at the bottom right), such that the upper domains of the pentons and hexons are in blue (between radii of 570 and 650 Å), the connecting triplexes are in green (between radii of 510 and 560 Å), the shell is in yellow (between radii of 460 and 510 Å), and the densities inside the capsid shell are in red (<460-Å radius). The capsid has a T=16 icosahedral symmetry (3 of the 12 fivefold axes are labeled 5, and 1 of the 20 triangular faces is outlined by a red dashed line in panel A), with the unique structural components in one asymmetric unit labeled, following the nomenclature established for HSV-1 (41, 57). These components include one-fifth of a penton (labeled 5), two and one-half hexons (one P, one C, and one-half of an E), and five and one-third triplexes (one each of the Ta, Tb, Tc, Td, and Te triplexes and one-third of the Tf triplex). The inside view in panel B is the same as that in panel A except that the upper half of the capsid was computationally removed to show the cutaway side views of some of the triplexes (dashed red arrows) and the inner floor of the HHV-8 capsids. Dot-dashed lines indicate icosahedral five-, three-, and twofold axes, which pass through a penton channel, a Tf triplex, and an E hexon channel, respectively. The densities inside the capsid shells (red) lack structural information because they are not icosahedrally disposed and thus have been removed computationally in the right half of panel B to show the internal surface of the capsid shell.
The montage of the HHV-8 capsid reconstruction with that of an HSV-1 B capsid at the same resolution (Fig. 3) clearly shows that they have identical sizes and capsomer organizations. However, some notable differences are seen on closer inspection. The HHV-8 capsid appears slightly more spherical than the HSV-1 capsid, which exhibits a somewhat angular, polyhedral shape (Fig. 3). When viewed from the top (e.g., the C hexons surrounding Tf at the center of Fig. 3), the hexons in the HHV-8 capsid appear flower shaped, whereas those of HSV-1 have slightly tilted subunits and as a result appear more gear shaped (see below). Also, the HHV-8 triplexes are slightly smaller and deviate less from threefold symmetry than the much-elongated triplexes in the HSV-1 capsid. The color differences in the upper domains of HSV-1 and HHV-8 triplexes indicate that the HSV-1 triplexes are slightly taller (Fig. 3).
FIG. 3.
Structural comparison of HHV-8 capsid and HSV-1 B capsid. The two capsid maps are radially colored as in Fig. 2 and are shown in a montage as viewed along the icosahedral threefold axis. The HSV-1 B capsid was reconstructed similarly to the same resolution (24 Å) from a subset of images selected from those used in a structure published previously (53). One penton (5), three types of hexon (P, E, and C), and six types of triplexes (Ta to Tf) are labeled.
Radial distribution of capsid components.
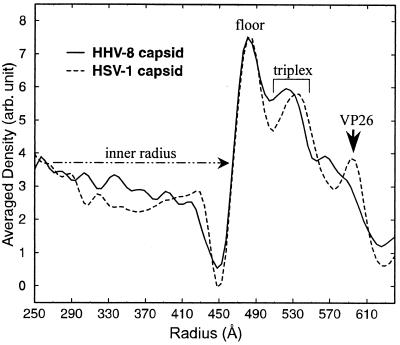
The 3D maps of the HHV-8 and HSV-1 capsids were spherically averaged to generate their radial density distribution profiles as a function of particle radius (Fig. 4). These profiles show that the HHV-8 and HSV-1 capsids have identical inner radii of 460 Å. Because both viruses also have similar genome sizes (30), their identical inner radii suggest that their DNA packing densities inside the capsids are similar. In contrast, betaherpesvirus capsids, such as those of HCMV, have a somewhat larger internal volume than HSV-1 or HHV-8 capsids (7, 10, 47). However, the increase in volume is disproportionate to the large increase in the size of the HCMV genome over the HSV-1 and HHV-8 genomes. This implies that the viral DNA is more densely packed into HCMV virions (4) than into HSV-1 or HHV-8 virions.
FIG. 4.
Radial density distributions of the HHV-8 capsid and HSV-1 B-capsid reconstructions. The density profiles were generated by spherically averaging the HHV-8 and HSV-1 capsid maps and were plotted as a function of particle radius. The HSV-1 and HHV-8 capsids have identical inner radii of about 460 Å. For the HSV-1 capsid, the locations of the four capsid proteins have been established (29, 34, 48, 55) and the density peaks corresponding to radial locations of the capsid floor, the triplexes, and the smallest capsid protein, VP26, are indicated accordingly. The HHV-8 capsid profile lacks the prominent peak corresponding to that attributed to VP26 in the HSV-1 capsid. The triplex peak is narrower and shifted to a lower radius in HHV-8 than in HSV-1. arb, arbitrary.
The profiles also show peaks corresponding to the radial positions and heights of the three major components of the capsid: the floor (460- to 510-Å radius), the triplexes (510- to 540-Å radius), and the upper domains of the pentons and hexons (beyond 540-Å radius) (Fig. 4). The floor peaks of HHV-8 and HSV-1 capsids have similar shapes and radii, but the triplex peaks are quite different. The HHV-8 triplex peak is narrower and at a smaller radius than that of HSV-1, which coincides with the observation from Fig. 3 that the HHV-8 triplexes are shorter than the HSV-1 triplexes. Finally, the peak attributed to the VP26 protein, which makes the HSV-1 hexon gear shaped by attaching to the upper domain of the HSV-1 hexon subunit (see below), is not present in the HHV-8 profile (Fig. 4).
Comparison of the pentons and hexons of HHV-8 and HSV-1.
A penton and an E hexon were computationally extracted from the capsid structures for direct structural comparison. In HHV-8, both the penton (Fig. 5A) and hexon (Fig. 5B) have a cylindrical shape (140-Å diameter, 160-Å height) with a central, axial channel approximately 25 Å in diameter. The penton and hexon subunits both have an elongated shape with multiple domains, including upper, middle, lower, and floor domains (subunit views in Fig. 5A and B). The middle domains of the subunits interact with the triplexes, while the lower domains connect the subunits to each other and form the axial channels. While the upper domains of adjacent hexon subunits interact with one another, adjacent penton subunits are disconnected at their upper domains, resulting in the V-shaped side view of the penton (Fig. 5A). Another major difference between the penton and hexon concerns their floor domains. These domains play an essential role in maintaining capsid stability, as suggested by the higher-resolution structural studies of the HSV-1 capsid (53), where a long α helix inserts into the floor domain of the adjacent subunit (penton side views in Fig. 5A and C). The relative angle between the floor and lower domains is about 110° in the penton subunit and becomes less than 90° in the hexon subunit (Fig. 5A and B, subunit views). In addition, unlike what is found for the penton channel, a ring-like constriction exists in the middle of the hexon channel (Fig. 2B; Fig. 5B, top view); the constriction is formed by the association of a density (Fig. 5B, subunit) protruding from the lower domain of each subunit toward the center of the channel.
FIG. 5.
Structural comparisons of the penton (A and C), hexon (B and D), and their subunits in HHV-8 capsid (A and B) and HSV-1 B capsid (C and D). The top views of the HHV-8 and HSV-1 pentons (A and C, respectively) and E hexons (B and D, respectively) reveal an axial channel approximately 25 Å in diameter in each penton and E hexon. The side views were generated by rotating the top view about the short edge of the figure by about 80° and show interactions between their adjacent subunits. In the side views of the HHV-8 penton and hexon subunits, the upper (u, blue), middle (m, green), lower (l, green to yellow), and floor (f, yellow) domains of a subunit are labeled, following the same designations used for HSV-1 (52, 57). Arrow in the HHV-8 hexon subunit (B), density protrusion that forms the constriction inside the hexon channel. The red dotted lines on the lower and floor domains of these subunits (A and B) illustrate the angles between the two domains. The two side views of the HHV-8 penton and hexon subunits (A and B, right) are related to each other by a roughly 90° rotation about the long edge of the Figure. (C and D, right) Side views of HSV-1 penton and hexon. The coiled lines drawn on the floor domain of penton side views (A and C) illustrate the location of the long α helix joining the adjacent major capsid proteins together at the floor domains, which was first visualized in the 8.5-Å structure of HSV-1 B capsid (53). (E and F) Superposition of penton (shown in semitransparent red) and hexon subunits (shown as wire frames using the same radial coloring scheme as in panels A to D) extracted from the HHV-8 capsid (E) and from the HSV-1 capsid (F). In the HSV-1 hexon subunit (D and F), a horn-shaped VP26 density attaches to the upper domain of each subunit (arrow) (48, 55). No horn-shaped or other extra density of similar size can be identified at the corresponding location on the HHV-8 hexon subunit (B and E). The pentons and hexons were displayed using a density threshold of 1.2 standard deviations (SD) above the mean, and their extracted subunits were shown at 1.5 SD above the mean density. Except for the penton subunits in panels E and F, the maps are colored according to the capsid radius as in Fig. 2 and 3 (see color bar in Fig. 2).
The HSV-1 penton (Fig. 5C) and hexon (Fig. 5D) subunits have the same basic shape as the HHV-8 subunits. Each consists of upper, middle, lower, and floor domains (Fig. 5C and D, subunit views). However, the upper domains of the HSV-1 penton subunits point inward toward the channel, whereas those of the HHV-8 penton subunits point outward. The upper domain of the HHV-8 subunit has a rectangular shape (subunit view in Fig. 5A), while that of the HSV-1 penton subunit appears as a triangle (subunit views in Fig. 5C). The most striking difference is that the HSV-1 hexon subunits contain an extra horn-shaped density (Fig. 5D), the VP26 protein, which is not found in either the HSV-1 penton (Fig. 5C) (48, 55, 57) or the HHV-8 hexon. This extra density binds to the top of each HSV-1 hexon subunit, forming a ring around the hexon at a radius of approximately 600 Å. This accounts for the tilted or gear-like appearance of the HSV-1 hexon top view (Fig. 5D). The superposition of HHV-8 penton and hexon subunits (Fig. 5E) shows that the domain features of HHV-8 penton and hexon subunits are roughly similar (except for the floor domain, as described above), whereas superposition of the HSV-1 penton and hexon subunits clearly shows an extra density bound to the upper domain of the hexon subunit (Fig. 5F).
Sequence homology between HHV-8 and HSV-1 capsid proteins.
HHV-8 capsid proteins were previously assigned to their HSV-1 counterparts by positional homology. Based on primary sequence analyses using CLUSTALW, the major capsid proteins (HHV-8 ORF25 protein and HSV-1 VP5) are almost identical in size and show significant identity (25%) and similarity (60%) (Table 1). In contrast, the smallest capsid proteins (HHV-8 ORF65 protein and HSV-1 VP26) differ substantially in size and show almost no identity (5%). LALIGN failed to align the ORF65 protein and VP26 pair even after the gap and deletion penalties were relaxed.
TABLE 1.
Sequence comparison of the HHV-8 and HSV-1 capsid shell proteins
| Proteins compareda (HHV-8/HSV-1) | % Identity by CLUSTALW | LALIGN result
|
||
|---|---|---|---|---|
| % Identity | % Similarity | Overlap (no. of aa) | ||
| MCP (1,376, 153, ORF25)/VP5 (1,374, 149, UL19) | 25 | 30.2 | 59.7 | 1,409 |
| TMP (331, 36, ORF62)/VP19c (465, 50, UL38) | 14 | 26.5b | 50.7b | 381b |
| TDP (305, 34, ORF26)/VP23 (318, 34, UL18) | 13 | 27.4 | 51.3 | 318 |
| sVCA (170, 19, ORF65)/VP26 (112, 12, UL35) | 5 | NSc | NS | NS |
| Protease (553, 60, ORF17)/VP40 (635, 66, UL26) | 19 | 25.6 | 51.1 | 575 |
In parentheses are the number of amino acids (aa), the molecular mass (in kilodaltons), and the associated gene, respectively, for each protein. MCP, major capsid protein; TMP, triplex monomer protein; TDP, triplex dimer protein; sVCA, small viral capsid antigen.
Result obtained by deleting aa 322 to 340.
NS, not significant because the gaps in the alignment were too large.
DISCUSSION
Structural proteins of the HHV-8 capsid and their homologs in other herpesviruses.
At least five major proteins are likely to be involved in the assembly of the HHV-8 capsid, including a protease (encoded by ORF17), the major capsid protein (encoded by ORF25), and three other smaller capsid proteins (encoded by ORF62, ORF26, and ORF65) (33) (Table 1). The protease, although not associated with the capsid shell, plays an essential role in capsid assembly and is the most functionally conserved among members of the Herpesviridae (49). Although our structure does not directly indicate which proteins make up the capsomers, this can be inferred from the positional and sequence homology between the HHV-8 and HSV-1 capsid proteins and also by comparing the structures of both capsids. Therefore, our results provide structural evidence that the HHV-8 pentons and hexons are composed of five and six copies, respectively, of the ORF25 protein. The penton and hexon subunits closely resemble those of HSV-1 in shape and size, and the ORF25 protein is homologous to HSV-1 major capsid protein VP5, which makes up the pentons and hexons.
Previous structural studies have shown that the HSV-1 triplex is a monomer of VP19c and a dimer of VP23 (29, 34, 57) and that the HCMV triplex is similarly composed of a monomer and a dimer (10). By analogy, the HHV-8 triplexes are likely also composed of a monomer of the ORF62 protein and a dimer of the ORF26 protein, which are the respective homologs of VP19c and VP23. The molecular masses of the ORF62 and ORF26 proteins are similar, in contrast to the significant molecular mass difference between VP19c and VP23 (Table 1). These differences accord well with our observation that the HHV-8 triplex is smaller and less deviated from threefold symmetry than the HSV-1 triplex.
The ORF65 protein is absent from the HHV-8 capsid reconstruction.
Due to difficulties in purifying homogeneous HHV-8 virions and capsids (see Materials and Methods), it remains technically prohibitive to verify, through biochemical means such as sodium dodecyl sulfate-polyacrylamide gel electrophoresis or Western blotting, whether the ORF65 protein is associated with HHV-8 capsids. Difference imaging of HSV-1 recombinant capsids has unambiguously shown that the extra densities attached to the upper domains of the hexons can be attributed to monomers of the VP26 protein. VP26 binds only to VP5 in hexons and not to VP5 in pentons (48, 55), demonstrating the quasiequivalent structures of identical VP5 proteins in different locations. The HHV-8 hexons do not have this extra density (Fig. 5E), which implies that the VP26 positional homolog, the ORF65 protein (also referred to as small viral capsid antigen or sVCA) (21), is not attached to the hexons in our reconstruction.
At present, the exact role of VP26 in HSV-1 capsid assembly is still unclear. It has been shown that VP26 is not essential for capsid assembly in insect cells infected with recombinant baculoviruses expressing HSV-1 capsid proteins, although B-capsid assembly is more efficient when VP26 is present (43, 46). Moreover, VP26 is not required for HSV-1 infection (14). VP26 might serve to connect the capsid proteins with the tegument layer, given that VP26 is located on the outermost surface of the hexons (51).
We are not able to offer a definitive explanation for the absence of the ORF65 protein from our structure. It may have been lost during the purification procedure if it is not as tightly bound to the hexons as VP26 in HSV-1. In this regard, 2 M guanidine chloride treatment resulted in the dissociation of VP26 from HSV-1 capsids (28). Thus, NP-40 or Triton X-100 may have dissociated the ORF65 protein from the HHV-8 capsids in our preparation. If this is the case, the interaction between the ORF65 protein and HHV-8 major capsid protein must be weaker than that between its counterpart in HSV-1 or HCMV and the major capsid protein, since its homologs in HCMV and simian cytomegalovirus remain bound when treated with 0.5% NP-40 (7, 47) and since VP26 remains bound when HSV-1 is treated with 0.25% NP-40 or 1% Triton X-100 (5, 28). It is also possible that the mature HHV-8 capsid lacks a binding site for the ORF65 protein and, consequently, that the ORF65 protein is not involved in capsid assembly. This situation is similar to that for the channel catfish virus, a related virus that lacks an ORF65 protein or VP26 positional homolog and showed no horn-shaped densities on its hexon subunits (6).
In any event, this first step in overcoming the technical problem of virus isolation has permitted the first view of this gammaherpesvirus capsid and the structural comparisons with other herpesvirus capsids. Our observations should encourage further investigation of the HHV-8 structures at a higher resolution in order to reveal in greater detail structural differences that are important to the understanding of their pathogenesis in different diseases.
ACKNOWLEDGMENTS
This research was supported in part by Public Health Service grants (AI 46420 to Z.H.Z. and HL42886 to J.K.S.) and the March of Dimes Birth Defect Foundation (5-FY99-852 to Z.H.Z.). Z.H.Z. is a Pew Scholar in the Biomedical Sciences.
We thank L. Oshiro and H. Zhang for initial electron microscopy assessment of sample conditions, John Stewart for preliminary data processing, W. Chiu and F. Rixon for the use of the HSV-1 capsid structure, and T. Dunnebacke-Dixon for comments.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 3.Baker T S, Newcomb W W, Booy F P, Brown J C, Steven A C. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhella D, Rixon F J, Dargan D J. Cryomicroscopy of human cytomegalovirus virions reveals more densely packed genomic DNA than in herpes simplex virus type 1. J Mol Biol. 2000;295:155–161. doi: 10.1006/jmbi.1999.3344. [DOI] [PubMed] [Google Scholar]
- 5.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booy F P, Trus B L, Davison A J, Steven A C. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- 7.Butcher S J, Aitken J, Mitchell J, Gowen B, Dargan D J. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J Struct Biol. 1998;124:70–76. doi: 10.1006/jsbi.1998.4055. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Chen D H, Jiang H, Lee M, Liu F, Zhou Z H. Three-dimensional visualization of tegument/capsid interactions in intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 11.Computational Biology Group, National Center for Supercomputing Applications. Biology workbench 3.0. Urbana: University of Illinois at Urbana-Champaign; 1997. [Google Scholar]
- 12.Crowther R A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc Lond B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 13.Crowther R A, DeRosier D J, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc R Soc London. 1970;317:319–340. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- 14.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 15.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dougherty M T, Chiu W. Using animation to enhance 3D visualization: a strategy for a production environment. In: Bailey G W, Alexander K B, Jerome W G, Bond M G, McCarthy J J, editors. Microscopy and Microanalysis. Atlanta, Ga: Springer; 1998. pp. 452–453. [Google Scholar]
- 17.Forghani B, Schmidt N J. Humoral immune response to virions and dense bodies of human cytomegalovirus determined by enzyme immunofluorescence assay. J Med Virol. 1980;6:119–127. doi: 10.1002/jmv.1890060204. [DOI] [PubMed] [Google Scholar]
- 18.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 19.Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 20.Johnson O, Govindan V, Park Y, Zhou Z H. Proceedings of the 4th International Conference on High Performance Computing. Los Alamitos, Calif: IEEE Computer Society Press; 1997. Custom virtual memory policy for an image reconstruction application; pp. 517–521. [Google Scholar]
- 21.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 23.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 24.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 25.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neipel F, Albrecht J C, Ensser A, Huang Y Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 30.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 32.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 33.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saad A, Zhou Z H, Jakana J, Chiu W, Rixon F J. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J Virol. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Said J W, Chien K, Tasaka T, Koeffler H P. Ultrastructural characterization of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) in Kaposi's sarcoma lesions: electron microscopy permits distinction from cytomegalovirus (CMV) J Pathol. 1997;182:273–281. doi: 10.1002/(SICI)1096-9896(199707)182:3<273::AID-PATH835>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 36.Schrag J D, Prasad B V V, Rixon F J, Chiu W. Three-dimensional structure of the HSV-1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 37.Schulz T F, Chang Y, Moore P S. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) In: McCance D J, editor. Human tumor viruses. Washington, D.C.: ASM Press; 1998. pp. 87–134. [Google Scholar]
- 38.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith M F, Langmore J P. Quantitation of molecular densities by cryo-electron microscopy. J Mol Biol. 1992;226:763–774. doi: 10.1016/0022-2836(92)90631-s. [DOI] [PubMed] [Google Scholar]
- 40.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 41.Steven A C, Roberts C R, Hay J, Bisher M E, Pun T, Trus B L. Hexavalent capsomers of herpes simplex virus type II: symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoops J K, Baker T S, Schroeter J P, Kolodziej S J, Niu X D, Reed L J. Three-dimensional structure of the truncated core of the Saccharomyces cerevisiae pyruvate dehydrogenase complex determined from negative stain and cryoelectron microscopy images. J Biol Chem. 1992;267:24769–24775. [PMC free article] [PubMed] [Google Scholar]
- 43.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 44.Tatusova T A, Madden T L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trus B L, Gibson W, Cheng N, Steven A C. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unal A, Pray T R, Lagunoff M, Pennington M W, Ganem D, Craik C S. The protease and the assembly protein of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:7030–7038. doi: 10.1128/jvi.71.9.7030-7038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z H, Chiu W, Haskell K, Spears H J, Jakana J, Rixon F J, Scott L R. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys J. 1998;74:576–588. doi: 10.1016/S0006-3495(98)77816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z H, Dougherty M, Jakana J, He J, Rixon F J, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Z H, Hardt S, Wang B, Sherman M B, Jakana J, Chiu W. CTF determination of images of ice-embedded single particles using a graphics interface. J Struct Biol. 1996;116:216–222. doi: 10.1006/jsbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Z H, Prasad B V V, Jakana J, Rixon F, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]