Highlights
-
•
The study identified strains of the A/H5N1 virus in analyzed samples of subsistence poultry, wild birds and mammals, belonging to clade 2.3.4.4b, with genetic similarity to strains from Chile, Uruguay and Argentina
-
•
Genetic mutations, particularly in the PB2 gene, were observed in Brazilian strains, potentially indicating adaptation to mammals and raising zoonotic concerns.
-
•
No mutations related to resistance to neuraminidase inhibitors (NAIs) were detected.
Keywords: Highly pathogenic avian influenza virus; Outbreaks; Complete genetic characterization, Phylodynamics, Point mutations
Abstract
Our study identified strains of the A/H5N1 virus in analyzed samples of subsistence poultry, wild birds, and mammals, belonging to clade 2.3.4.4b, genotype B3.2, with very high genetic similarity to strains from Chile, Uruguay, and Argentina. This suggests a migratory route for wild birds across the Pacific, explaining the phylogenetic relatedness. The Brazilian samples displayed similarity to strains that had already been previously detected in South America. Phylogeographic analysis suggests transmission of US viruses from Europe and Asia, co-circulating with other lineages in the American continent. As mutations can influence virulence and host specificity, genomic surveillance is essential to detect those changes, especially in critical regions, such as hot spots in the HA, NA, and PB2 sequences. Mutations in the PB2 gene (D701N and Q591K) associated with adaptation and transmission in mammals were detected suggesting a potential zoonotic risk. Nonetheless, resistance to neuraminidase inhibitors (NAIs) was not identified, however, continued surveillance is crucial to detect potential resistance. Our study also mapped the spread of the virus in the Southern hemisphere, identifying possible entry routes and highlighting the importance of surveillance to prevent outbreaks and protect both human and animal populations.
1. Introduction
Highly pathogenic avian influenza viruses (HPAIVs) have attracted attention due to concerns over both animal and public health. Viruses bearing the hemagglutinin (HA) gene of the H5 subtype and H7 subtype have caused thousands of human cases around the world, including deaths. Those viruses have caused numerous disease outbreaks in wild birds, marine mammals, and domestic poultry and are responsible for the loss of millions of domestic birds (Shi et al., 2023).
The first known strain of HPAI A(H5N1) (called A/chicken/Scotland/59) killed two flocks of chickens in Scotland in 1959, but that strain was very different from the highly pathogenic H5N1 strain. Since the emergence of the HPAIV H5N1 gs/Gd lineage in China in 1996, the virus has shown rapid genomic evolution, with lineages classified into at least ten clades and multiple subclades. Currently, the dominant H5 viruses circulating worldwide are the clade 2.3.4.4 viruses, which have formed eight subclades (clade 2.3.4.4a–h) and multiple antigenic groups (Lee et al., 2017) One of those genetic clades, 2.3.4.4b, represents the main type of avian influenza virus that is causing current outbreaks around the world. In South America, detections of HPAIV H5N1 have been made in various countries such as Argentina, Brazil, Chile, Peru, and Uruguay (Rimondi, et al., 2024; Reischak et al., 2023; Jimenez-Bluhm et al., 2023; Fernández-Díaz et al., 2023; Marandino et al, 2023).
HPAI H5N1 is a viral disease that affects birds, and in rare cases, spillover events occur for mammals, including transmission to humans. The increased detection in diverse mammalian species may indicate viral adaptation, increasing the risk for human infection (Sutton et al., 2018; WHO, 2023).
In South America, Leguia et al., 2023 reported the genomic characterization of HPAI/H5N1 in five species of marine mammals and seabirds in Peru (dolphins, sea lions, sanderlings, pelicans, and cormorants. Starting in January 2023, an increase in mass marine mammal strandings and mortality caused by HPAI H5N1 spread along the coast of Chile and Peru (Leguia et al., 2023), reaching small odontocetes, mustelids and especially impacting South American sea lions (Otaria flavescens) (Ulloa et al., 2023). In Argentina, the first case in South American sea lions was reported in August 2023, and the virus then moved north, along the Atlantic coast, affecting pinnipeds (includes seals, sea lions, etc) in Argentina and Uruguay. (WHOA, 2024). At least 24,000 sea lions died in Peru, Chile, Argentina, Uruguay, and Brazil between January–October 2023 (Plaza et al., 2024).
In Brazil, the first case of infection in a wild bird with the highly pathogenic avian influenza virus (HPAI A/H5N1) was reported in May 2023. Since then, cases in backyard poultry, wild bird species, and marine mammals have been detecting and monitoring with the presence of hundreds of outbreaks (https://mapa-indicadores.agricultura.gov.br/publico/extensions/SRN/SRN.html]). Nevertheless, until date, no cases in commercial poultry or human cases have been reported in Brazilian territory. HPAI represents a risk to the poultry industry, food security, and public health; and biosafety measures, including quarantine, culling of infected birds, disinfection, and vaccination have been appling to mitigate the disease. Furthermore, the use of vaccination against avian influenza may be recommended under specific conditions. Vaccination can be used as an effective complementary control tool when a stamping-out policy alone is not sufficient. Whether to vaccinate or not should be decided by the Veterinary Authority on the basis of the avian influenza situation, as well as the ability of the Veterinary Services to implement the vaccination strategy (WOAH, 2022). Vaccination has successfully eliminated H7N9 viruses in China (Zeng et al., 2018), and it has been discussing in Europe and the United States to control H5 viruses.
By identifying genetic recombination events, especially those involving lineages adapted to birds and humans, the likelihood of zoonotic transmission and potential pandemic threats can be assessed (Blagodatski et al., 2021). Genetic recombination plays a significant role in the evolution of influenza viruses since it can result in the emergence of lineages with altered antigenic properties, leading to the evasion of pre-existing immunity in both birds and humans. Identifying recombination events helps to both monitor viral evolution and understand the potential risks associated with the emergence of new strains HPAI, possible spillover events with adaptation to mammalian hosts, and pandemic potential (Petrova & Russell, 2018). Although genetic recombination plays an important role in the evolution of influenza viruses and can even result in the emergence of lineages with altered antigenic properties, in most cases for both H5 and H7 HPAIVs, the antigenic evolution of HPAIVs is mainly caused by point mutations (Ohkawara et al., 2017).
The hemagglutinin (HA) gene codes for one of the two surface glycoproteins and is central to species specificity because it is responsible for virus attachment and fusion with host cells. In your turn, the neuraminidase (NA) gene encodes the other surface protein of the virus. The major role of the NA is to release new progeny virions from an infected cell by enzymatically cleaving sialic acid receptors, which aids virus spread to uninfected cells within an infected host. In addition to the HA and NA, the RNA transcription and replication complex (PB2, PB1, PA, NP) also have species-specific determinants that impact efficient replication in humans and other mammals, particularly polymerase basic protein 2 (PB2). PB1, PA, and NP also have important markers of mammalian adaptation (CDC, 2024).
Regarding public health, resistance to antivirals in the avian influenza H5N1 virus have been the major concern due to its high pathogenicity in birds and its occasional abilityto infect humans. Antivirals, such as neuraminidase inhibitors, are often used to treat influenza, but resistance can arise due to genetic mutations, making the drugs less effective (Scheibner et al., 2023).
Therefore, the propagation of the H5N1 virus within avian populations and the potential for zoonotic transmission to humans is a significant public health concern. Implementing efficient antiviral strategies, enhancing biosecurity measures in avian habitats, and conducting comprehensive genomic monitoring are critical interventions to reduce the risk of antiviral resistance and effectively manage epidemic events.
2. Materials and methods
2.1. Sample collection
Samples were obtained from the central nervous system, oropharyngeal swabs (sample 12) and lung and trachea pool (sample 26) from animals as part of the passive surveillance for avian influenza. During January 2023 until May 2024 7.860 samples were collected in 850 investigations with sample collection, totalization 164 outbreaks positive for H5N1 HPAI. These samples were collected by the Official Veterinary Service of the Brazilian states. Following notification of a suspected case of HPAI, an Official Veterinarian went to the property within 12 hours to perform an investigation of the probable case in domestic and wild birds and aquatic mammals. The samples collected were then sent to the Federal Agricultural Defense Laboratory of São Paulo (LFDA-SP), which is the national reference laboratory for the diagnosis of avian influenza in Brazil.
2.2. Confirmation for HPAIV H5N1 - PCRs and Sanger sequencing
The samples were initially confirmed as positive for A/H5 by three RT-qPCR techniques (VetMAX Gold AIV, Thermo Fisher; Lee et al., 2015; NVSL/APHIS/USDA, 2022) followed by Sanger sequencing (Sanger et al., 1977). The partial sequencing of the HA gene (segment 4) was used to confirm the presence of a polybasic motif at the cleavage site, (PLREKRKKR/GLF or PLREKRRKR/GLF) to assess the pathogenicity profile of the identified viruses (Slomka et al., 2007). Additionally, positive samples for influenza A/H5 underwent RT-qPCR for neuraminidase subtyping (Naguib et al., 2017).
H5N1 positive samples were inactivated and sent to the Federal Laboratory of Agropecuary Defense in Goiás (LFDA-GO) for sample enrichment by PCR amplification of influenza A genomic segments accordingly to the Thermofisher's protocol (https://assets.thermofisher.com/TFS-Assets/CSD/Application-Notes/pcr-amplification-influenza-a-whole-genome-sequencing-app-note.pdf) previously to the NGS. The primer sequences were as follows: 5´-CTGGATACGCCAGCRAAAGCAGG-3´ (sense) and 5´-GACCTGATGCGGAGTAGAAACAAGG-3´ (antisense).
All tests were performed in accordance with the general requirements for the competence of testing and calibration laboratories of ISO/IEC 17,025/2017 (International Organization for Standardization / International Electrotechnical Commission) and Chapter 3.3.4. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals of WOAH (World organisation for Animal Health).
2.3. NGS sequencing
Sample selection to perform NGS were considered when their Ct values are <28, the cold chain was maintained during transport, and RNA samples stored at least ≤ -20 °C, with different species of birds and all mammal samples possible to be sequencing. The amplicons were sent to the Federal Laboratory of Agropecuary Defense in Minas Gerais (LFDA-MG) for next-generation complete genome sequencing on a solid phase using the Nextera XT DNA Library Prep Kit (Illumina Inc.). The library preparation process involved several steps and was adapted from the protocol described by Gauthier et al. (2021). The resulting cDNA was then amplified, prepared for sequencing, and used to construct the libraries. Library construction involved tagmentation of cDNA using a transposome to fragment it and then marking the DNA with adapter sequences. Subsequent PCR cycles used the adapters to amplify the DNA. The PCR step also added index adapter sequences to both ends of the DNA, enabling dual indexing for sequencing multiple libraries in different samples. Amplicons were pooled and purified using AMPure XP (Beckman Coulter, Indianapolis, IN, USA). The resulting libraries were quality controlled using D1000 ScreenTape (Agilent 5067–5582) on a TapeStation System instrument and subsequently sequenced by using the MiSeq Reagent Kit v2 (300-cycle paired-end) on a MiSeqTM platform by Illumina Inc.
2.4. Bioinformatics analyses
The raw data generated from the amplicon sequencing were analyzed in Varsmetagen (https://varsomics.com/varsmetagen/), an online platform for bioinformatic analysis and interpretation of NGS data for infectious diseases. Varsmetagen implements a viromic pipeline with the following steps: (1) quality control of raw sequences with metrics and removal of low-quality sequences; (2) mapping of the obtained sequences to the host genome to remove contaminants (Gallus gallus, assembly: GCF_016699485.2 in the case of avian-derived samples and Homo sapiens, assembly: GRCh38.p14 in case of mammal samples); (3) taxonomic identification using Kraken2 (Wood et al., 2019) with a custom database; (4) assembly with Spades 3.13 to recover the viral genome (Antipov et al., 2020); (5) search for distant homologous sequences using Hidden Markov Models (HMM) of viral proteins implemented in the eegNOG database (Huerta-Cepas et al., 2019); and (6) complementary analyses involving confirmation and genome coverage metrics. Specifically for Influenza samples, the last step involves taking a suitable reference genome to perform target mapping (Usually A/California/07/2009(H1N1), or any other closer strain identified by BLASTN tophits to NCBI Refseq database). Metrics as sequencing depth and coverage breadth were analyzed, and consensus sequences for each segment were assembled. Finally, consensus sequences and de novo contigs were inspected to generate final segments and the complete genome. Once consensus genomes were obtained, they were annotated by the NCBI Influenza Virus Sequence Annotation Tool https://www.ncbi.nlm.nih.gov/genomes/FLU/annotation/ (Bao et al., 2007).
The obtained sequences for the hemagglutinin (HA) (segment 4) were used to infer the phylogenetic relationships. H5Nx sequences were obtained from the GenBank and GISAID (Global Initiative on Sharing All Influenza Data) databases. Additionally, we included on the phylogenetic analysis the top 250 closely related sequences to the sequences obtained in Brazil that were determined by sequence similarity searches at nucleotide level using BLAST (Zhang et al., 2000 and Morgulis et al., 2008). Sequences with coverage breadth <90% of the total length were excluded from the analysis. The Flusurver genotyping tool (https://flusurver.bii.a-star.edu.sg/), enabled by GISAID data (Elbe & Buckland-Merrett, 2017) was used to confirm the classification clade.
Multiple sequence alignment was performed using the MAFFT v7.407 software (Katoh and Standley, 2013), built-in on the software Geneious Prime® 2024.0.3. All analyses were performed online using the Los Alamos Science Gateway (Miller et al. 2011). Maximum likelihood (ML) analysis was performed in IQ-TREE multicore v.1.6.12 (Ngyuen et al. 2015), with the best-fitted substitution model (previously obtained) and generating 1000 bootstrap replicates. The outgroup for rooting the tree was determined as the reference sequence for HA from the first outbreak in China (AF148678.1/A/goose/Guangdong/1/96/H5N1).
Genotypes were classified using the eight segments of each assigned sample according to the scheme described in Youk et al. (2023) and using the GenoFLU tool that uses BLAST to identify segments from a curated database. Pre-defined genotypes are cross-referenced with the top segment identifications, and a genotype is assigned. A cutoff of 2% difference from the closest curated sequence identifies new reassortment. New reassortment is reviewed using segment-based phylogenetic trees. (https://github.com/USDA-VS/GenoFLU).
2.5. Phylogeography
Beast 2.0 (Bouckae et al., 2014) was used for phylogeographic analysis. The sequences for the hemagglutinin (HA) (segment 4), with respective metadata of the samples (latitude, longitude, and the date of registration of the outbreak), were inserted in Beauti in which the file was generated for the following analyses: (a) strict molecular clock with coalescent constant population, (b) strict molecular clock with coalescent Bayesian skyline, (c) strict with coalescent exponential population, (d) relaxed clock log normal with coalescent constant population, (e) relaxed clock log-normal coalescent Bayesian skyline, and (f) relaxed clock log normal with coalescent exponential population. The string size was 1000,000 with a minimum ESS of 200. The results were compared in Tracer to find the best model. The most suitable model was used for the construction of trees and visualization of phylogeographic data in SPREAD with a chain of 1000,000,000 (Bielejec et al, 2011).
3. Results
The sequences with the complete eight segments of A/H5N1 were deposited in GenBank under the accession codes indicated in Table 1. Analysis using segments 4 - HA and 6 - NA confirmed that all the viral strains reconstituted from the analyzed samples belong to the same clade 2.3.4.4b. The eight segments of all Brazilian samples were classified within the Genotype B3.2 (PB2:am2.1, PB1:am1.2, PA:ea1, HA:ea1, NP:am1.4.1, NA:ea1, MP:ea1, NS:am1.1).
Table 1.
Overview of the samples of wild birds, backyard poultry, captive wild birds and marine mammals analyzed in this study distributed by GenBank number and description, date of outbreak.
| Sample | Lab sample number | Nº Genbank | Genbank Description | Date outbreak detection | Latitude/ Longitude |
|---|---|---|---|---|---|
| 1 | LDDV-2023–0389–0002 | OR269884-OR269891 | A/Thalasseus cuflavidus/EspiritoSanto/1339_N2/2023 | 15May2023 | 20.33124 S 40.35964 W |
| 2 | LDDV-2023–0787–0001 | OR853701-OR853708 | A/Megascops choliba/Serra/1409-N/2023 | 26May2023 | 20.15440 S 40.26940 W |
| 3 | LDDV-2023–0510–0002 | OR420089-OR420096 | A/Fregata magnificens/RJ/1532-N/2023 | 03Jun2023 | 22.95947 S 43.06116 W |
| 4 | LDDV-2023–0787–0002 | OR853748-OR853755 | A/Thalasseus maximus/Ubatuba/1546-N/2023 | 05Jun2023 | 23.431739 S 45.074222 W |
| 5 | LDDV-2023–0787–0004 | OR860358-OR860365 | A/Thalasseus maximus/Itapoa/1941-N/2023 | 25Jul2023 | 26.184561 S 48.608717 W |
| 6 | LDDV-2023–0787–0005 | OR881250-OR881257 | A/Numida meleagris/Maracaja/1843-N3/2023 | 15Jul2023 | 28.8211 S 49.4211 W |
| 7 | LDDV-2023–0787–0006 | OR878085-OR878092 | A/Thalasseus maximus/Antonina/1775-N/2023 | 23Jun2023 | 25.460489 S 48.692789 W |
| 8 | LDDV-2023–0787–0007 | OR889488-OR889495 | A/Buteogallus urubitinga/Itapemirim/1630-SC/2023 | 07Jun2023 | 20.999540 S 40.941583 W |
| 9 | LDDV-2023–0807–0001 | OR839997-OR840004 | A/Gallus gallus/Bonito/2108-SN52/2023 | 22Sep2023 | 21.183888 S 56.526019 W |
| 10 | LDDV-2023–0807–0003 | OR857851-OR857858 | A/Sula leucogaster/Sao Francisco do Sul/2122-N/2023 | 27Sep2023 | 26.216361 S 48.588686 W |
| 11 | LDDV-2023–0807–0004 | OR844537-OR844544 | A/Otaria flavescens/RioGrande/2148-N/2023 | 03Oct2023 | 32.174091 S 52.137250 W |
| 12 | LDDV-2023–0807–0005 | OR852412-OR852419 | A/Otaria flavescens/Torres/2165-SO/2023 | 11Oct2023 | 29.428016 S 49.793198 W |
| 13 | LDDV-2024–0001–0001 | PP386937-PP386944 | A/Sterna hirundo/EspiritoSanto/1455-N/2023 | 27May2023 | 20.839900 S 40.722100 W |
| 14 | LDDV-2024–0001–0005 | PP390163-PP390170 | A/Thalasseus maximus/RioGrandeSul/2177-N/2023 | 13Oct2023 | 32.011622 S 51.949222 W |
| 15 | LDDV-2024–0418–0001 | PP768657-PP768664 | A/Pluvialis dominica/BertiogaBR/2252-N/20,239 | 08Nov2023 | 23.791761 S 45.991416 W |
| 16 | LDDV-2024–0418–0002 | PP768247-PP768254 | A/Procellaria aequinoctialis/SaoSebastiaoBR/2259-N/2023 | 09Nov2023 | 23.7896666667 S 45.5671666667 W |
| 17 | LDDV-2024–0418–0003 | PP769611-PP769618 | A/S. hirundo/PenhaBR/2261-N/2023 | 09Nov2023 | 26.792738 S 48.61598 W |
| 18 | LDDV-2024–0418–0004 | PP769736-PP769743 | A/Procellaria aequinoctialis/UbatubaBR/2271-N/2023 | 10Nov2023 | 23,519,727 S 45,194,599 W |
| 19 | LDDV-2024–0418–0005 | PP776749-PP776756 | A/Thalasseus acuflavidus/MatinhosBR/2277-N/2023 | 14Nov2023 | 25.809500 S 48.538417 W |
| 20 | LDDV-2024–0418–0006 | PP777455-PP777462 | A/Thalasseus acuflavidus/IlhaCompridaBR/2280-N/2023 | 16Nov2023 | 24.697327 S 47.4514 W |
| 21 | LDDV-2024–0418–0007 | PP778502-PP778509 | A/Thalasseus maximus/PraiaGrandeBR/2339-N/2023 | 13Dec2023 | 24.0071444444 S -46.4202305556 W |
| 22 | LDDV-2024–0418–0008 | PP779057-PP779064 | A/S. hirundo/ItapemirimBR/0155-N/2024 | 02Feb2024 | 20.906211 S 40.780175 W |
| 23 | LDDV-2024–0418–0009 | PP779930-PP779937 | A/S. hirundo/MacaeBR/0177-N/2024 | 09Feb2024 | 22.341605 S 41.744273 W |
| 24 | LDDV-2024–0418–0010 | PP780171-PP780178 | A/T.acuflavidus/BertiogaBR/0291-N/2024 | 22Feb2024 | 23.75938611 S 45.906852777 W |
| 25 | LDDV-2024–0418–0011 | PP780443-PP780450 | A/S. hirundo/PiumaBR/0448-N/2024 | 02Mar2024 | 20.838072 S 40.708908 W |
| 26 | LDDV-2024–0418–0013 | PP782202-PP782209 | A/S. hirundo/SaoFranciscodeItabapoanaBR/0481-R/2024 | 02Mar2024 | 21.321255 S 40.961377 W |
| 27 | LDDV-2024–0418–0014 | PP782336-PP782343 | A/S. hirundo/Cabo FrioBR/0613-N/2024 | 09Mar2024 | 22.5978 S 41.994 W |
| 28 | LDDV-2024–0418–0015 | PP785018-PP785025 | A/S. hirundo/RiodasOstrasBR/0721-N/2024 | 15Mar2024 | 22.5219611111 S -41.9440111111 W |
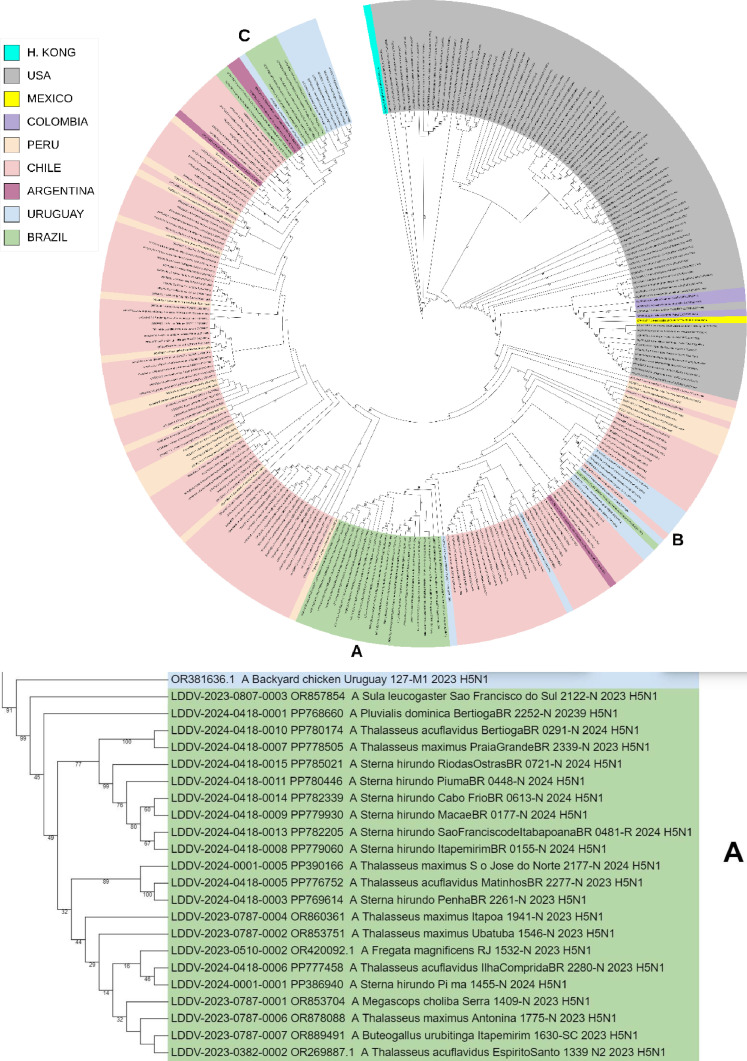
Brazilian A/H5N1 lineages jointly grouped with Chilean, Uruguayan and Argentinian 2022–2024 viral lineages forming a clade with 82–92% bootstrap support value, which confirms its closest phylogenetic relatedness to those sequences from South American countries facing the Pacific Ocean (Chile and Argentina) (Fig. 1).
Fig. 1.
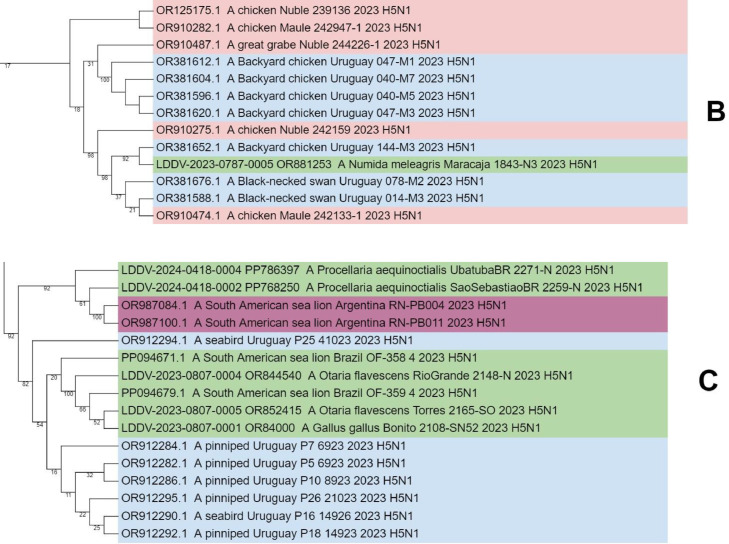
Phylogenetic analysis of highly pathogenic avian influenza A(H5N1) from wild birds, poultry, and mammals, Brazil. Maximum-likelihood method was used for phylogeny of hemagglutinin H5 sequences from avian influenza viruses in IQ-TREE multicore v.1.6.12. Phylogenetic tree was generated with Geneious Prime® 2024.0.3 software and edited with iTOL tool (https://itol.embl.de/). Sequences were aligned by using the MAFFT program in the Geneious Prime® 2024.0.3 software and editor for the same software. We used the best-fitted evolutionary model based on BIC = Bayesian Information Content models; robustness of tree topology was assessed with 1000 bootstrap replicates Green color indicate clustering of strains from Brazil and sequences from this study; “LDDV” indicates the sequences from this study. Non–goose/Guangdong lineage virus strains from Eurasia were outgroups. A, B and C shares a most recent common ancestor with viral lineages of Chile, Uruguay, and Argentina (2022–2024). With 82–92% bootstrap support values, confirming its closest phylogenetic relatedness with the sequences from South American.
The estimates of genetic divergence among Brazilian viral sequences of HA-characterized genomes indicate that the 22 samples (samples no. 1–5, 7–8, 10, 13–15, 17, 19–28) are highly phylogenetically related with the Chilean sample OR910409–1 A/domestic duck/OHiggins/245,741–2/ 2023. Moreover, two Brazilian samples (samples no. 16 and 18) are highly phylogenetically related with the Argentinian samples OR987100.1 A/South_American_sea_lion/Argentina/RN-PB011/2023 and OR987084.1 A/South_American_sea_lion/Argentina/RN-PB004/2023. Besides, other two Brazilian samples (no. 9 and 12) are highly phylogenetically related with the Uruguayan sample A/pinniped/Uruguay/P7_6923/2023. Additionally, the Brazilian sample 6 was very similar (with the same number of base differences per site) with the Chilean A/chicken/Maule/242,133–1/2023 and Uruguayans (A/Black-necked_swan/Uruguay/078-M2/2023 and A/Black-necked_swan/Uruguay/014-M3/2023) samples. Likewise, the sample 1 was very similar (the same number of base differences per site) with the Chilean A/black_skimmer/Chile/C61962/2022 and Uruguayans A/pinniped/Uruguay/P7_6923/2023 sequences (Table 2).
Table 2.
Mutational analysis of Brazilian HPAI A/H5N1 viruses (1 to 14). Additional reference sequences used are available through GenBank and GISAID.Sequences A/domestic duck/OHiggins/245741-2/2023 and A/Black-necked swan/Uruguay/ 014-M3/2023(H5N1) are the most similar with Brazilian sequences was using a Matriz Distance with Geneious Prime® 2024.0.3 software. The Sequence A/Mink/Spain/3691-8_22VIR10586-10/2022 was illustration a mutation T271A no observed in Brazilian sequences. Mutations PB2 Q591K and D701N has been previously linked to mammalian host adaptation and enhanced transmission was observed in Brazilian sequences. Mutations PA M86I and D26K, reported in recently human case in Chile was observed in Brazilian sequences. n.a: not available.
| PB2 |
PB1 | PB1-F2 | PA |
NA |
NS1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
T271A |
Q591K |
E627K |
D701N |
K702R |
L378M |
N66S | M86I | 224P | 383D |
H275Y |
N295S | T438I | D26K | |||
| 1 | LDDV-2023–0389–0002 | A/Thalasseus cuflavidus/EspiritoSanto/1339-N2/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 2 | LDDV-2023–0787–0001 | A/Megascops choliba/Serra/1409-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 3 | LDDV-2023–0510–0002 | A/Fregata magnificens/RJ/1532-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 4 | LDDV-2023–0787–0002 | A/Thalasseus maximus/Ubatuba/1546-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 5 | LDDV-2023–0787–0004 | A/Thalasseus maximus/Itapoa/1941-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 6 | LDDV-2023–0787–0005 | A/Numida meleagris/Maracaja/1843-N3/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 7 | LDDV-2023–0787–0006 | A/Thalasseus maximus/Antonina/1775-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 8 | LDDV-2023–0787–0007 | A/Buteogallus urubitinga/Itapemirim/1630-SC/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 9 | LDDV-2023–0807–0001 | A/Gallus gallus/Bonito/2108-SN52/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 10 | LDDV-2023–0807–0003 | A/Sula leucogaster/Sao Francisco do Sul/2122-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 11 | LDDV-2023–0807–0004 | A/Otaria flavescens/RioGrande/2148-N/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 12 | LDDV-2023–0807–0005 | A/Otaria flavescens/Torres/2165-SO/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 13 | LDDV-2024–0001–0001 | A/S. hirundo/EspiritoSanto/1455-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 14 | LDDV-2024–0001–0005 | A/Thalasseus maximus/RioGrandeSul/2177-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 15 | LDDV-2024–0418–0001 | A/P. dominica/BertiogaBR/2252-N/20,239 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 16 | LDDV-2024–0418–0002 | A/Procellaria aequinoctialis/SaoSebastiaoBR/2259-N/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 17 | LDDV-2024–0418–0003 | A/S. hirundo/PenhaBR/2261-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 18 | LDDV-2024–0418–0004 | A/Procellaria aequinoctialis/UbatubaBR/2271-N/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 19 | LDDV-2024–0418–0005 | A/Thalasseus acuflavidus/MatinhosBR/2277-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 20 | LDDV-2024–0418–0006 | A/Thalasseus acuflavidus/IlhaCompridaBR/2280-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 21 | LDDV-2024–0418–0007 | A/Thalasseus maximus/PraiaGrandeBR/2339-N/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 22 | LDDV-2024–0418–0008 | A/S. hirundo/ItapemirimBR/0155-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 23 | LDDV-2024–0418–0009 | A/S. hirundo/MacaeBR/0177-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 24 | LDDV-2024–0418–0010 | A/Thalasseus acuflavidus/BertiogaBR/0291-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 25 | LDDV-2024–0418–0011 | A/S. hirundo/PiumaBR/0448-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 26 | LDDV-2024–0418–0013 | A/S. hirundo/SaoFranciscoItabapoanaBR/0481-R/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 27 | LDDV-2024–0418–0014 | A/S. hirundo/Cabo FrioBR/0613-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 28 | LDDV-2024–0418–0015 | A/S. hirundo/RiodasOstrasBR/0721-N/2024 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 29 | Argentina | A/South American sea lion/Argentina/RN-PB011/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 30 | Argentina | A/South American sea lion/Argentina/RN-PB004/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 31 | Chile | A/domestic duck/OHiggins/245,741–2/2023 | T | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 32 | Chile | A/black_skimmer/Chile/C61962/2022 | T | K | E | N | K | M | S | M | S | D | H | N | T | E |
| 33 | Chile (Human) | A/Chile/25,945/2023 | T | K | E | N | K | M | S | I | S | D | H | N | T | K |
| 34 | Uruguay | A/Black-necked swan/Uruguay/ 014-M3/2023 | I | Q | E | D | K | M | S | M | S | D | H | N | T | E |
| 35 | Uruguay | A/pinniped/Uruguay/P7_6923/2023 | T | K | E | N | K | n.a. | n.a. | I | S | D | H | N | T | K |
| 36 | Spain | A/Mink/Spain/3691–8_22VIR10586–10/2022 | A | Q | E | D | K | L | S | T | S | D | H | N | T | D |
| 37 | Hong Kong | A/Goose/Guangdong/1/1996 | T | Q | E | D | K | L | N | M | S | D | H | N | T | D |
In hemagglutinin, the observed variations represented differences in five amino acids (HA S16G 8 <0,01%, D104 0,02%, M242I 0,04%, S304N 0,02%, D503Y 0,03%) among the sequences of greatest similarity A/domestic duck/OHiggins/245,741–2/2023, and only 3 amino acids (HA D104 0,02%, L225M <0,01% e M242I 0,04%) among the sequences of greatest similarity A/Black-necked swan/Uruguay/014-M3/2023. Despite the changes observed in HA, the Residue Conservation Score among the evaluated 568 amino acids remains with values between 0.728 and 0.919 using BLOSUM62 background distribution (https://shin-kinos.github.io/cons-capra07-wasm/).
Hotspots of variation in the sequence encoding hemagglutinin may be in various genomic regions. The variations observed in the Brazilian H5N1 samples are related to the HA1 and HA2 regions. There were 12 Single Nucleotide Polymorphisms (SNP) with significant P-value (see the specific value and which test, P < 0.001) among the Brazilian samples.
Point mutations were also evaluated to infer the possibility of a probable higher adaptation to the mammalian host and improved transmission (Table 2). In order to confirm the presence of markers already related to H5N1 strains associated with higher virulence in human infections, such as the recent human case of H5N1 in Chile.
4. Discussion
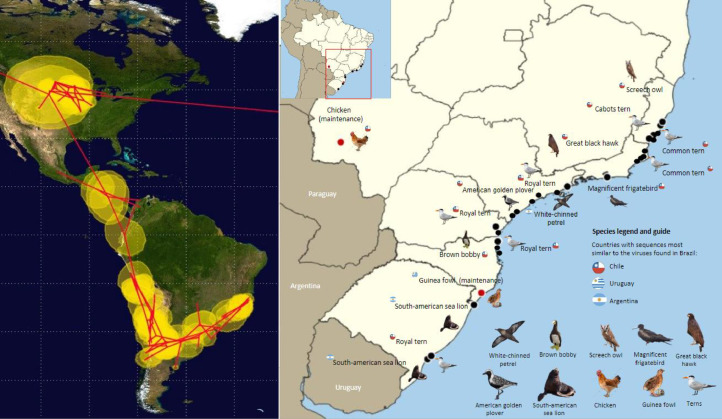
The phylogeographic analysis strongly suggests that the virus spread from the Northern Hemisphere to the Southern Hemisphere, following Central America to the Andes, spreading to southern Brazil and moving northeast (to the state of Espírito Santo) and northwest (to the state of Mato Grosso do Sul) (Table 2 - left side). The Pacific and Atlantic routes are well known and followed by several species of migratory birds (Shi et al., 2023). Although birds from different routes have difficulty crossing the Andes, there is a geographic connectivity between different populations due to significant intra-regional movements. These populations, composed of non-migratory or vagrant species, may act as steppingstones, and transmit the virus between these two routes. So far and with the available sequences, it has been possible to determine that the Pacific route seems to be more closely related to the H5N1 cases observed in Brazil (Fig. 2).
Fig. 2.
Left: Spatial representation of the possible dissemination routes of H5N1 using the 250 closely related sequences to the Brazilian variant using the Beast 2.0 and SPREAD software. Right: Distribution among Brazilian states of bird and mammal species with NGS analysis.
Non-synonymous substitutions, also known as missense mutations can alter the amino acid sequence of a protein. In the context of avian influenza virus, which has a segmented RNA genome, such mutations can affect different viral genes, including the HA gene (Smith et al., 2009). In the sample 6, Guinea fowl (Numida meleagris) from Southern Brazil (Santa Catarina state), the hemagglutinin showed high similarity to the that encoded by the genome recovered from Uruguay A/Black-necked swan/Uruguay/ 014-M3/2023(H5N1). It is worth noting that some genetic alterations have been detected in comparison with this isolate, with 12 altered aminoacids, reinforcing the importance of continuous surveillance in the coming months to closely monitor viral evolution in our region. Nonetheless, the changes observed in Brazilian samples are not in major antigenic epitopes on hemagglutinin in our study.
Point mutations may have significant implications for the virulence, transmissibility, and host specificity of the avian influenza virus. For example, mutations in hemagglutinin can lead to changes in the viral affinity for host cells, affecting its ability to infect birds or potentially adapt to mammals, including humans (Shaw and Palese, 2013).
A significant obstacle to interspecies transmission is the inability of avian-origin viral polymerases to replicate efficiently in mammalian cells. Studies have shown that about half of the mammalian viruses from the current panzootic contain at least one of three mutations that enhance the viral polymerase ability to replicate in mammalian cells (Adlhoch et al., 2023). The PB2 subunit is involved in the initial mechanism of viral mRNA synthesis during multiplication (Miaomiao et al., 2021), and the most frequently observed mutation is PB2-E627K. Moreover, when PB2-E627K is absent, the mutations PB2-T271A and PB2-D701N are believed to provide similar functionality. Mutation of the polymerase complex is crucial for adaptation and transmission to a new host (Lagan et al., 2023).
Agüero et al., 2023 observed that mink viruses in Spain present an alanine (A) at position 271 of PB2 (T271A), which increases the polymerase activity of influenza A viruses in mammalian and mouse host cells. The PB2 mutations E627K and K702R reported by Bordes et al., 2023 are specifically linked to mammalian host adaptation and enhanced transmission. The PB2 D701N mutation (observed in samples 9, 11, 12, 16 and 18) is associated with increased transmission among mammals (Czudai-Matwich et al., 2014), while the Lysine at position 591 of PB2 (PB2 Q591K), also observed in these same Brazilian samples, confers efficient replication to an H5N1 influenza virus in mammals (Wang et al., 2016).
The PB2 d701n and Q591K mutations detection Brazilian samples 9, 11, 12, 16 and 18 were observed in highly phylogenetically related Argentinian samples 29 and 30 (Rimondi et al., 2024), Uruguayan sample 35 (Tomáz et al., 2024) and Chilean samples 32 and 33 (Jimenez-Bluhm et al., 2022; Castillo et al., 2023). Besides, other two Chilean (31) and Uruguayan (34) samples exhibited high phylogenetically relatedness with the Brazilian ones.
The involvement of this species of mammal, South American sea lion (Otaria flavescens), observed in Brazilian samples 11 and 12, has already been reported in South American countries causing the death of thousands of individuals, such as in Peru, Chile, Argentina, Uruguay, and Brazil (Ulloa et al., 2023 and Plaza et al., 2024). Specific mutations in PB2 have also been reported to affect mammals alike. The PB2 D701N mutation had already been observed in sea lions from Chile and Peru (Leguia et al., 2023) and in the case reported in humans in Chile (WHO, 2023). Nevertheless, it is important to consider that transmission and adaptation generally require a consecutive number of mutations that allow the virus to become more efficiently infectious for a given species.
The adaptive mutations in the PA protein of the H5N1 avian influenza virus also contribute to mammalian adaptation. There is a synergistic effect of the PA 224P + 383D of H5N1 avian influenza viruses and its ability to enhance the pathogenicity and viral replication in a mammalian mouse model (Song et al., 2015). The mutation of PA 224P + 383D was not identified in the Brazilian samples, but the mutation of PA 383D was clearly detected. This mutation is highly conserved in avian influenza viruses, increases the polymerase activity in both avian and human cells, may have roles in maintaining the avian influenza virus in their avian reservoirs, and may also jump species to infect humans (Song et al., 2015).
All Brazilian samples with available PB1-F2 sequence exhibit the PB1 N66S mutation (Table 2), which is associated with increased virulence in human infections with H5N1 (Conenello et al., 2007).
The confirmed human case of HPAI H5N1 in Chile in 2023 illustrates the risk to humans. In this instance, high environmental exposure significantly increased the threat to humans, as there were many dead wild animals near the residence of the affected individual. This was demonstrated by the fact that the sequenced virus from the patient was identical to the strains found in local wild birds. (WHO, 2023). Indeed, the PB2 D701N and PA M86I mutations in the Brazilian samples 9, 11, 12, 16 and 18 were also reported in the human case in Chile.
Two additional mutations (PA M861I and NS1 D26K) in the same sea lion samples (11 and 12), one domestic chicken (9), and two white-chinned petrels (16 and 18) were also identified. The same mutations in sea lions were reported by Leguia et al. (2023), and in the human case from Chile (WHO, 2023). This may indicate the possibility that the virus may be adapting to the mammalian host. If viruses more adapted to mammals are circulating again in wild birds and possibly infecting domestic birds, this fact demonstrates great concern for industrial poultry farms to maintain adequate biosecurity to prevent the entry of HPAI H5N1 into commercial flocks.
The protein products from the M (M1 and M2) genes lacked markers associated with mammalian adaptation (CDC, 2024). Although it was not detected any alteration in HA of Brazilian samples that would possibly indicate an evolutionary adaptation to mammals, those samples had acquired substitutions in PB2 that probably would increase the replication in mammal cells, illustrating that we need to remain vigilant and continuously characterize zoonotic viruses.
Neuraminidase inhibitors (such as oseltamivir, zanamivir, and peramivir) are an important class of drugs used in the treatment of influenza. They act by inhibiting the activity of neuraminidase, an enzyme that is present on the surface of the influenza virus and is essential for the release of new viral particles from infected cells. By interrupting this process, NAIs help to reduce the spread of the virus in the body, and, thus, reducing the severity and duration of flu symptoms (Chan and Hui, 2023).
Earhart et al., 2009 reported the NA N294S mutation (N295S according to N1 numbering) in A(H5N1) virus from two human patients previously associated with resistance to oseltamivir, and that was indeed one of the first reports associated with this type of resistance. Kobayashi et al., 2017 also reported mutations in neuraminidase in mice infected with the H5N1 virus, containing the NA H275Y mutation. Antiviral activity of peramivir against this variant was lower when compared with H5N1 virus without the mutation. Furthermore, the NA substitution T438I in A(H5N1) clade 2.3.4.4b leads to reduced inhibition by zanamivir and peramivir, but not oseltamivir, which continues to be detected all around the world (WHO, 2024).
In the Brazilian samples, no mutations related to NAIs in the hot spots NA N295S, NA H275Y, and NA T438I were detected. These results demonstrate an indication to continue the use of antivirals such as oseltamivir, peramivir and zanamivir in people infected with strains of HPAI A(H5N1) (Earhart et al., 2009; Kobayashi et al., 2017). Nonetheless, it is extremely important the continuous genomic surveillance to detect and inform potential antiviral resistance risks to H5N1 viruses, given the possible exposure to the human population.
5. Conclusion
Our phylogeographic analysis showed more one evidence that H5N1 was transmitted to the USA from Europe and Asia, and from the USA to South America via Pacific route. It is well known that the circulating H5N1 viruses are reassortants of viruses from different origins, and these events must continue in South America. The Pacific route seems to be more closely related to the H5N1 cases observed in Brazil, and other bird populations, mainly composed of non-migratory or vagrant species, may act as steppingstones and can indeed transmit the virus between different routes and regions inside the continent.
We have identified common mutations in avian and human influenza viruses PA 383D and PB1-F2 N66S, respectively. Other more important mutations were detected in one domestic chicken, two South American sea lion, and two /White-chinned petrel that relation with adaptative mammals, such as Q591K and D701N. Additionally, the same mutations (PA M86I and D26K) was also recently reported for a human case in Chile.
The presence of previously reported mutations may have public health implications because of their associations with increased virulence, viral replication, and mammal host adaptation. Furthermore, HPAI H5N1 has already caused outbreaks in commercial and backyard poultry with spillover resulting in sporadic infections in mammals.
Therefore, continuous genomic surveillance is needed to identify markers associated with mammal adaptation and potential human-to-human transmission, provide data to drive public health measures, avoid mass animal deaths, and protect human populations.
CRediT authorship contribution statement
Anselmo Vasconcelos Rivetti: Writing – review & editing, Writing – original draft, Visualization, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Dilmara Reischak: Methodology, Formal analysis. Cairo Henrique Sousa de Oliveira: Methodology, Formal analysis. Juliana Nabuco Pereira Otaka: Formal analysis. Christian Steffe Domingues: Formal analysis. Talita de Lima Freitas: Formal analysis. Fernanda Gomes Cardoso: Formal analysis. Lucas Oliveira Montesino: Formal analysis. Ana Luiza Savioli da Silva: Formal analysis. Soraya Cecília Albieri Camillo: Formal analysis. Fernanda Malta: Methodology, Data curation. Deyvid Amgarten: Validation, Software, Methodology, Formal analysis, Data curation, Conceptualization. Aristóteles Goés-Neto: Writing – review & editing, Writing – original draft, Validation, Supervision, Software, Methodology, Formal analysis, Data curation. Eric Roberto Guimarães Rocha Aguiar: Writing – review & editing, Writing – original draft, Validation, Software, Methodology, Data curation. Iassudara Garcia de Almeida: Formal analysis, Data curation. Carla Amaral Pinto: Formal analysis. Antônio Augusto Fonseca: Writing – original draft, Software, Data curation. Marcelo Fernandes Camargos: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Adlhoch C., Fusaro A., Gonzales J.L., Kuiken T., Melidou A., Mirinavičiūtė G., Niqueux É., Ståhl K., Staubach C., Terregino C., Baldinelli F., Broglia A., Kohnle L. Avian influenza overview April - June 2023. EFSA J. 2023;21(7):e08191. doi: 10.2903/j.efsa.2023.8191. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agüero M., Monne I., Sánchez A., Zecchin B., Fusaro A., Ruano M.J., del Valle A., Manuel F.R., Souto A.M., Tordable P., Cañás J., Bonfante F., Giussani E., Terregino C., Orejas J.J. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro. Surveill. 2023;28(3) doi: 10.2807/1560-7917.ES.2023.28.3.2300001. pii=2300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov D., Raiko M., Lapidus A., Pevzner P.A. Metaviralspades: assembly of viruses from metagenomic data. Bioinformatics. 2020;36(14):4126–4129. doi: 10.1093/bioinformatics/btaa490. [DOI] [PubMed] [Google Scholar]
- Blagodatski A., Trutneva K., Glazova O., Mityaeva O., Shevkova L., Kegeles E., Onyanov N., Fede K., Maznina A., Khavina E., Yeo S., Park H., Volchkov P. Avian influenza in wild birds and poultry: dissemination pathways, monitoring methods, and virus ecology. Pathogens. 2021;10(5):630. doi: 10.3390/2Fpathogens10050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Bolotov P., Dernovoy D., Kiryutin B., Tatusova T. FLAN: a web server for influenza virus genome annotation. Nucl. Acid. Res. 2007;35:W280–W284. doi: 10.1093/nar/gkm354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27(20):2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2 a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10(4) doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes L, Vreman S, Heutink R, Roose M, Venema S, Pritz-Verschuren SBE, Rijks JM, Gonzales JL, Germeraad EA, Engelsma M, Beerens N. Highly pathogenic avian influenza H5N1 virus infections in wild red foxes (Vulpes vulpes) show neurotropism and adaptive virus mutations. Microbiol. Spectr. 2023;11:e02867. doi: 10.1128/spectrum.02867-22. –02822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Fasce R, Parra B, Andrade W, Covarrubias P, Hueche A, Campano C, Tambley C, Rojas M, Araya M, Hernández F, Bustos P, Fernández J. The first case of human infection with H5N1 avian Influenza A virus in Chile. J. Travel. Med. 2023;30(5):taad083. doi: 10.1093/2Fjtm/2Ftaad083. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Controls and Prevention - CDC, 2024. Technical Update: Summary Analysis of Genetic Sequences of Highly Pathogenic Avian Influenza A(H5N1) Viruses in Texas. April 2024 https://www.cdc.gov/flu/avianflu/spotlights/2023-2024/h5n1-analysis-texas.htm.
- Chan KKP, Hui DSC. Antiviral therapies for influenza. Curr. Opin. Infect. Dis. 2023;36(2):124–131. doi: 10.1097/QCO.0000000000000910. Apr 1. [DOI] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudai-Matwich V, Otte A, Matrosovich M, Gabriel G, Klenk HD. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J. Virol. 2014;88(16):8735–8742. doi: 10.1128/JVI.00422-14. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earhart KC, Elsayed NM, Saad MD, Gubareva LV, Nayel A, Deyde VM, Abdelsattar A, Abdelghani AS, Boynton BR, Mansour MM, Essmat HM, Klimov A, Shuck-Lee D, Monteville MR, Tjaden JA. Oseltamivir resistance mutation N294S in human influenza A(H5N1) virus in Egypt. J. Infect. Public Health. 2009;2(2):74–80. doi: 10.1016/j.jiph.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Challeng. (Hoboken, NJ) 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Díaz M, Villanueva-Pérez D, Tataje-Lavanda L, Montalvan-Avalos A, Isasi-Rivas G, Lulo-Vargas M, Fernández-Sánchez M. Detection and genomic characterization of an avian influenza virus subtype H5N1 (Clade 2.3.4.4b) strain isolated from a Pelican in Peru. Microbiol. Resour. Announc. 2023;12(6) doi: 10.1128/mra.00199-23. Jun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier NPG, Nelson C, Bonsall MB, Locher K, Charles M, MacDonald C, Krajden M., Chorlton S.D., Manges A.R. Nanopore metagenomic sequencing for detection and characterization of SARS-CoV-2 in clinical samples. PLoS ONE. 2021;16(11) doi: 10.1371/journal.pone.0259712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., von Mering C., Borket P. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucl. Acid. Res. 2019;47 doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Bluhm P., Siegers J.Y., Tan S., Sharp B., Freiden P., Johow M., Orozco K., Ruiz S., Baumberger C., Galdames P., Gonzalez M.A., Rojas C., Karlsson E.A., Hamilton-West C., Schultz-Cherry S. Detection and phylogenetic analysis of highly pathogenic A/H5N1 avian influenza clade 2.3.4.4b virus in Chile, 2022. Emerg. Microb. Infect. 2023 doi: 10.1080/22221751.2023.2220569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecul. Biol. Evolut. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Kodama M, Noshi T, Yoshida R, Kanazu T, Nomura N, Soda K, Isoda N, Okamatsu M, Sakoda Y, Yamano Y, Sato A, Kida H. Therapeutic efficacy of peramivir against H5N1 highly pathogenic avian influenza viruses harboring the neuraminidase H275Y mutation. Antiviral Res. 2017;139:41–48. doi: 10.1016/j.antiviral.2016.12.011. Mar. [DOI] [PubMed] [Google Scholar]
- Lagan P., McKenna R., Baleed S., Hanna B., Barley J., McConnell S., Georgaki A., Sironen T., Kauppinen A., Gadd T., Lindh E., Ikonen N., McMenamy M.J., Lemon K. Highly pathogenic avian influenza A(H5N1) virus infection in foxes with PB2-M535I identified as a novel mammalian adaptation, Northern Ireland. Euro. Surveill. 2023;28(42) doi: 10.2807/1560-7917.ES.2023.28.42.2300526. 2023 Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Bertran K., Kwon J.H., Swayne D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx Clade 2.3.4.4. J. Vet. Sci. 2017;18:269–280. doi: 10.4142/jvs.2017.18.s1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Torchetti M.K., Winker K., Ip H.S., Song C.S., Swayne D.E. Intercontinental spread of Asian-Origin H5N8 to North America through Beringia by migratory birds. J. Virol. 2015;89(12):6521–6524. doi: 10.1128/jvi.00728-15. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguia M., Garcia-Glaessner A., Muñoz-Saavedra B., Juarez D., Barrera P., Calvo-Mac C., Jara J., Silva W., Ploog K., Amaro L., Colchao-Claux P., Johnson C.P., Uhart M.M., Nelson M., Lescano J. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Commun. 2023;14:5489. doi: 10.1038/s41467-023-41182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marandino A., Tomás G., Panzera Y., Leizagoyen C., Pérez R., Bassetti L., Negro R., Rodríguez S., Pérez R. Spreading of the high pathogenicity avian influenza (H5N1) virus of clade 2.3.4.4b into Uruguay. Viruses. 2023;15:1906. doi: 10.3390/v15091906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaomiao Z., Mingbin L., Shimeng B., Chen Z., Zejun L., Jianqing X., Xiaoyan Z. Influenza A virus–host specificity: an ongoing cross-talk between viral and host factors. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.777885. doi:10.3389/fmicb.2021.777885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery. Vol. 41. 2011. The CIPRES science gateway: a community resource for phylogenetic analyses; pp. 1–8. [DOI] [Google Scholar]
- Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schaffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;15:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib M.M., Graaf A., Fortin A., Luttermann C., Wernery U., Amarin N., Hussein H.A., Sultan H., Al Adhadh B., Hassan M.K., Beer M., Monne I., Harder T.C. Novel real-time PCR-based patho- and phylotyping of potentially zoonotic avian influenza A subtype H5 viruses at risk of incursion into Europe in 2017. Euro. Surveill. 2017;22(1):30435. doi: 10.2807/2F1560-7917.ES.2017.22.1.30435. Jan 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawara A., Okamatsu M., Ozawa M., Chu D.H., Nguyen L.T., Hiono T., Matsuno K., Kida H., Sakoda Y. Antigenic diversity of H5 highly pathogenic avian influenza viruses of clade 2.3.4.4 isolated in Asia. Microbiol. Immunol. 2017;61(5):149–158. doi: 10.1111/1348-0421.12478. [DOI] [PubMed] [Google Scholar]
- Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. pages. [DOI] [PubMed] [Google Scholar]
- Plaza P.I., Gamarra-Toledo V., Rodríguez Euguí J., Rosciano N., Lambertucci S.A. Pacific and Atlantic sea lion mortality caused by highly pathogenic Avian Influenza A(H5N1) in South America. Travel. Med. Infect. Dis. 2024;59 doi: 10.1016/j.tmaid.2024.102712. [DOI] [PubMed] [Google Scholar]
- Reischak D., Rivetti, Otaka A.V., Domingues J.N.P., Freitas C.S., Cardoso T.L., Montesino F.G., Silva L.O., Malta A.L.S., Amgarten F., Goés-Neto D., Oliveira A., Camargos A.F., F M. First report and genetic characterization of the highly pathogenic avian influenza A(H5N1) virus in Cabot's tern (Thalasseus acuflavidus), Brazil. Veterin. Anim. Sci. 2023;22 doi: 10.1016/j.vas.2023.100319. ISSN 2451-943X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimondi A, Vanstreels RET, Olivera V, Donini A, Lauriente MM, Uhart MM. Highly pathogenic avian influenza A(H5N1) viruses from multispecies outbreak, Argentina, August 2023. Emerg. Infect. Dis. 2024 doi: 10.3201/eid3004.231725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibner D., Salaheldin A.H., Bagato O., Zaeck L.M., Mostafa A., Blohm U., Müller C., Eweas A.F., Franzke K., Karger A., Schäfer A., Gischke M., Hoffmann D., Lerolle S., Li X., Abd, El-Hamid H.S., Veits J., Breithaupt A., Boons G.J., Matrosovich M., Finke S., Pleschka S., Mettenleiter T.C., de Vries R.P., Abdelwhab E.M. Phenotypic effects of mutations observed in the neuraminidase of human origin H5N1 influenza A viruses. PLoS Pathog. 2023;19(2) doi: 10.1371/journal.ppat.1011135. Feb 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M.L., Palese P. Fields Virology, 5ª Edição DM Knipe e PM Howley, Editores. 2006. Lippencott Williams e Wilkins; Filadélfia: 2013. Orthomyxoviridae: the viruses and their replication; pp. 1691–1740. ISBN-10: 0-7817-6060-7. [Google Scholar]
- Shi J., Zeng X., Cui P., Yan C., Chen H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microb. Infect. 2023;12 doi: 10.1080/22221751.2022.2155072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomka M.J., Coward V.J., Banks J., Löndt B.Z., Brown I.H., Voermans J., Koch G., Handberg K.J., Jørgensen P.H., Cherbonnel-Pansart M., Jestin V., Cattoli G., Capua I, Ejdersund A., Thorén P., Czifra G. Identification of sensitive and specific avian influenza polymerase chain reaction methods through blind ring trials organized in the European Union. Avian Dis. 2007;51(1 Suppl):227–234. doi: 10.1637/7674-063006R1.1. [DOI] [PubMed] [Google Scholar]
- Song J., Xu J., Shi J., Li Y., Chen H. Synergistic effect of S224P and N383D substitutions in the PA of H5N1 avian influenza virus contributes to mammalian adaptation. Sci. Rep. 2015;22(5):10510. doi: 10.1038/2Fsrep10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton T.C. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses. 2018;10(9):461. doi: 10.3390/v10090461. Aug 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.J.D., Vijaykrishna D., Bahl J., Lycett S.J., Worobey M., Pybus O.G., Guan Y. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. ... &. [DOI] [PubMed] [Google Scholar]
- Tomás G., Marandino A., Panzera Y., Rodríguez S., Wallau G.L., Dezordi F.Z., Pérez R., Bassetti L., Negro R., Williman J., Uriarte V., Grazioli F., Leizagoyen C., Riverón S., Coronel J., Bello S., Páez E., Lima M., Méndez V., Pérez R. Highly pathogenic avian influenza H5N1 virus infections in pinnipeds and seabirds in Uruguay: Implications for bird–mammal transmission in South America. Virus Evolut. 2024;10(1):veae031. doi: 10.1093/ve/veae031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa M., Fernández A., Ariyama N., Colom-Rivero A., Rivera C., Nuñez P., Sanhueza P., Johow M., Araya H., Torres J.C., Gomez P., Muñoz G., Agüero B., Alegría R., Medina R., Neira V., Sierra E. Mass mortality event in South American sea lions (Otaria flavescens) correlated to highly pathogenic avian influenza (HPAI) H5N1 outbreak in Chile. Vet. Q. 2023;43(1):1–10. doi: 10.1080/01652176.2023.2265173. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee H,H, Yang Z.F., Mok C.K., Zhang Z. PB2-Q591K mutation determines the pathogenicity of avian H9N2 influenza viruses for mammalian species. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0162163. Sep 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genom. Biol. 2019;20:257. doi: 10.1186/2Fs13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization - WHO, 2023. Human Infection caused by Avian Influenza A (H5N1) - Chile, https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON461.
- World Health Organization - WHO, 2024. Genetic and antigenic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. February 2024. https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-northern-hemisphere-recommendation-2024-2025/202402_zoonotic_vaccinvirusupdate.pdf?sfvrsn=70150120_4.
- World Organisation for Animal Health - WOAH, 2022. Terrestrial Animal Health Code. Chapter 10.4. Article 10.4.1. Infection With High Pathogenicity Avian Influenza Viruses. https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/?id=169&L=1&htmfile=chapitre_avian_influenza_viruses.htm.
- World Organisation for Animal Health - WOAH, 2024. Practical guide for authorised field responders to HPAI outbreaks in marine mammals. February 2024 https://www.woah.org/app/uploads/2024/02/woah-practicalguide-forauthorisedfieldresponders-hpaimarinemammals-feb24.pdf.
- Youk S., Torchetti M.K., Lantz K., Lenoch J.B., Killian M.L., Leyson C., Bevins S.N., Dilione K., Ip H.S., Stallknecht D.E., Poulson R.L., Suarez D.L., Swayne D.E., Pantin-Jackwood M.J. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021-April 2022. Virology. 2023;587 doi: 10.1016/j.virol.2023.109860. 2023. [DOI] [PubMed] [Google Scholar]
- Zeng X., Tian G., Shi J., Deng G., Li C., Chen H. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci. China Life Sci. 2018;61(12):1465–1473. doi: 10.1007/s11427-018-9420-1. 2018 Dec. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.