Abstract
A general theme that has emerged from studies of DNA tumor viruses is that otherwise unrelated oncoproteins encoded by these viruses often target the same important cellular factors. Major oncogenic determinants for human adenovirus type 9 (Ad9) and high-risk human papillomaviruses (HPV) are the E4-ORF1 and E6 oncoproteins, respectively, and although otherwise unrelated, both of these viral proteins possess a functional PDZ domain-binding motif that is essential for their transforming activity and for binding to the PDZ domain-containing and putative tumor suppressor protein DLG. We report here that the PDZ domain-binding motifs of Ad9 E4-ORF1 and high-risk HPV-18 E6 also mediate binding to the widely expressed cellular factor MUPP1, a large multi-PDZ domain protein predicted to function as an adapter in signal transduction. With regard to the consequences of these interactions in cells, we showed that Ad9 E4-ORF1 aberrantly sequesters MUPP1 within the cytoplasm of cells whereas HPV-18 E6 targets this cellular protein for degradation. These effects were specific because mutant viral proteins unable to bind MUPP1 lack these activities. From these results, we propose that the multi-PDZ domain protein MUPP1 is involved in negatively regulating cellular proliferation and that the transforming activities of two different viral oncoproteins depend, in part, on their ability to inactivate this cellular factor.
Human adenovirus type 9 (Ad9) is a unique oncogenic virus that generates estrogen-dependent mammary tumors in rats (22). Whereas the viral E1A and E1B oncoproteins are responsible for tumorigenesis by most human adenoviruses (44), the primary oncogenic determinant for Ad9 is its E4-ORF1 (9ORF1) transforming protein (21, 23, 52, 59). Mutational analyses of the 125-amino-acid (aa) 9ORF1 protein implicate three separate regions (regions I, II, and III) as being critical for transformation (56). Although the activities associated with regions I and II have not been determined, region III at the extreme carboxyl terminus of 9ORF1 mediates interactions with multiple cellular polypeptides (p220, p180, p160, p155, and p140/p130) (57). This carboxyl-terminal 9ORF1 domain was recently discovered to define a functional PDZ domain-binding motif (28) and, consistent with this finding, 9ORF1-associated protein p140/130 was identified as the cellular PDZ protein DLG (28), a mammalian homolog of the Drosophila discs large tumor suppressor protein dlg-A (29, 33).
In humans, infections with human T-cell leukemia virus type 1 and high-risk human papillomaviruses (HPV) are associated with the development of adult T-cell leukemia and cervical carcinoma, respectively (5, 43). Finding a functional PDZ domain-binding motif at the carboxyl terminus of 9ORF1 subsequently led us to discover that human T-cell leukemia virus type 1 Tax and high-risk but not low-risk HPV E6 oncoproteins possess similar binding motifs at their carboxyl termini and, in addition, bind DLG (28). Although it is well established that transformation by high-risk HPV E6 proteins depends in part on an ability to target the tumor suppressor protein p53 for degradation (42), other E6 functions are also known to be important (27, 38, 47). In this regard, high-risk HPV-16 E6 mutant proteins having a disrupted PDZ domain-binding motif lose the capacity to oncogenically transform rat 3Y1 fibroblasts (26). Moreover, we recently showed that high-risk HPV E6 proteins target the PDZ protein DLG for degradation in cells (11). Therefore, a common ability of several different human virus oncoproteins to complex with cellular PDZ domain proteins probably contributes to their transforming potentials.
PDZ domains are approximately 80-aa modular units that mediate protein-protein interactions (6, 7). PDZ domain-containing proteins represent a diverse family of polypeptides that contain single or multiple PDZ domains, other types of protein-protein interaction modules including SH3, WW, PTB or pleckstrin homology domains, and protein kinase or phosphatase domains (35, 40). Consistent with such domain structures, many PDZ proteins play a role in signal transduction. In this capacity, these cellular factors serve to localize receptors and cytosolic signaling proteins to specialized membrane sites in cells and, in addition, to act as scaffolding proteins to organize these cellular targets into large supramolecular complexes (6, 8, 37). The PDZ domains of these cellular factors typically recognize specific peptide sequence motifs located at the extreme carboxyl termini of their target proteins (48), although PDZ domains can also mediate other types of protein interactions (3, 30, 63). To date, three different types of carboxyl-terminal PDZ domain-binding motifs have been identified (31, 48, 50), and at their extreme carboxyl-termini, the Ad 9ORF1, HTLV-1 Tax, and high-risk HPV E6 oncoproteins possess a type I binding motif with the consensus sequence -(S/T)-X-(V/I/L)-COOH (where X is any residue) (28).
Although our findings (28, 56, 57) and those of others (26) suggest that DLG is an important cellular target for transformation by both human Ad E4-ORF1 and high-risk HPV E6 oncoproteins, our previous results with the 9ORF1 protein also argue for the existence of additional important cellular PDZ protein targets. Specifically, disruption of the 9ORF1 PDZ domain-binding motif abolishes the interaction of 9ORF1 with DLG, as well as with several other unidentified cellular proteins (p220, p180, p160, and p155) (57). Consistent with this observation, we now report that the 9ORF1-associated protein p220 is the multi-PDZ domain protein MUPP1 (55) and that 9ORF1 abnormally sequesters this cellular factor within the cytoplasm of cells. We further show that the high-risk HPV-18 E6 (18E6) oncoprotein likewise complexes with MUPP1 but instead targets this cellular factor for degradation in cells. These findings suggest that the transforming potentials of the human Ad E4-ORF1 and 18E6 proteins depend on their ability to block the function of MUPP1, a large multi-PDZ domain protein predicted to function as a scaffolding factor in cell signaling.
MATERIALS AND METHODS
Cells.
NIH 3T3 (20), CREF (9), COS7 (13), TE85 (32), and 293 (14) cell lines, as well as the Ad9-induced rat mammary tumor cell line 20-8, were maintained in culture medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum) under a 5% CO2 atmosphere in a humidified incubator at 37°C. CREF cell pools (group 16) stably expressing wild-type or mutant 9ORF1 protein (56) and a CREF cell pool stably expressing an influenza virus hemagglutinin (HA) epitope-tagged 9ORF1 protein (58) were maintained in culture medium supplemented with G418 (Gibco BRL).
Plasmids.
The partial murine 9BP-1 cDNA (28) encoding the carboxyl-terminal 526 aa of 9BP-1 was inserted between the BamHI and HindIII sites of plasmid pQE9 (Qiagen) to make plasmid pQE9-9BP1-CT526. The full-length rat MUPP1 cDNA from pBSK-MUPP1 (55) was introduced between either the SacII and EcoRV sites of plasmid pSL301 (Invitrogen) or the HindIII and EcoRI sites of cytomegalovirus expression plasmid GW1 (British Biotechnology) to create plasmid pSL301-MUPP1 or GW1-MUPP1, respectively. Plasmid GW1-HAMUPP1 was derived from GW1-MUPP1 by introducing an HA epitope tag at the amino terminus of MUPP1 by PCR methods. Plasmids GW1-HAMUPP1ΔPDZ7, GW1-HAMUPP1ΔPDZ10, and GW1-HAMUPP1ΔPDZ7/10 were derived from GW1-HAMUPP1 by deletion of MUPP1 sequences coding for either PDZ7 (aa 1166 to 1232) or PDZ10 (aa 1616 to 1656) or both of these PDZ domains, respectively, by PCR methods.
Plasmids GW1-9ORF1wt, GW1-9ORF1IIIA, GW1-9ORF1IIIC, GW1-9ORF1IIID, and GW1-18E6 contain the respective wild-type or mutant 9ORF1 (56) or wild-type 18E6 gene inserted between the HindIII and EcoRI sites of plasmid GW1. The wild-type 18E6 cDNA was also introduced between the HindIII and EcoRI sites of plasmid pSP64 (Promega) to make plasmid pSP64-18E6. Substitution of 18E6 valine residue 158 with alanine (18E6-V158A) or of threonine and valine residues 156 and 158 with aspartic acid and alanine (18E6-T156D/V158A), as well as introduction of an HA epitope tag at the amino termini of 18E6, HPV-11 E6 (11E6), rat DLG (33), and human ZO-1 (61) proteins, was accomplished by PCR methods. Altered E6 and DLG cDNAs were introduced between the HindIII and EcoRI sites of plasmid GW1 to make plasmids GW1-HA18E6, GW1-HA18E6-V158A, GW1-HA18E6-T156D/V158A, GW1-HA11E6, and GW1-HADLG, whereas the HAZO-1 cDNA was inserted between the KpnI and BglII sites of plasmid GW1 to make plasmid GW1-HAZO-1. Plasmids pcDNA3-DLG and pSP64-p53 were described previously (11, 36).
For the construction of glutathione S-transferase (GST) fusion protein expression plasmids, cDNA sequences coding for 9BP1-US9/10 (aa 179 to 251), MUPP1-NT (aa 1 to 123), MUPP1-PDZ1–3 (aa 118 to 504), MUPP1-PDZ4-5 (aa 489 to 785), MUPP1-US5/6 (aa 780 to 990), MUPP1-PDZ6 (aa 985 to 1110), MUPP1-PDZ7 (aa 1105 to 1312), MUPP1-PDZ8–9 (aa 1307 to 1611), MUPP1-PDZ10 (aa 1606 to 1706), MUPP1-PDZ11 (aa 1701 to 1830), MUPP1-PDZ12–13 (aa 1825 to 2054), 18E6-V158A, and 18E6-T156D/V158A were PCR amplified and introduced in frame with the GST gene of plasmid pGEX-2T or pGEX-4T-1 (Pharmacia). pGEX-2T plasmids containing wild-type or mutant E4-ORF1 genes have been described previously (57). pGEX-2T plasmids containing wild-type 18E6 and 11E6 cDNAs were kindly provided by P. Howley.
PCR amplifications were performed with pfu polymerase (Stratagene), and plasmids were verified by restriction enzyme and limited sequence analyses.
Lambda phage cDNA library screening.
A DNA fragment from the 9BP-1 cDNA (nucleotides 134 to 544) (28) was radiolabeled by the random-priming method (51) and used to screen a mouse pancreatic cell λgt11 cDNA library, kindly provided by S. Tsai, by standard methods (49).
Antisera and antibodies.
The His6-tagged 9BP1-CT526, GST-9BP1-US9/10, and GST-MUPP1-US5/6 fusion proteins were expressed in bacteria and purified with either Ni-nitrilotriacetic acid agarose (Qiagen) or glutathione beads (Pharmacia) (46), as recommended by the manufacturers. Rabbits were immunized with purified 9BP1-CT526 or MUPP1-US5/6 fusion proteins to generate polyclonal antisera by standard methods (17). The purified GST-9BP1-US9/10 fusion protein was covalently linked to Affi-Gel 10 beads (Bio-Rad) and used to affinity purify 9BP-1 antibodies by standard methods (17). Commercially available HA (12CA5) monoclonal antibodies (BABCO), horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG antibodies (Southern Biotechnology Associates), fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Gibco BRL), and Texas red-conjugated goat anti-mouse IgG antibodies (Molecular Probes, Eugene, Oreg.) were utilized. p53, DLG, and 9ORF1 antisera were described previously (1, 11, 23).
Transfections and cell extracts.
COS7 cells were transfected with Lipofectin or Lipofectamine (Gibco BRL) as recommended by the manufacturer and harvested 48 h posttransfection. For preparation of cell extracts, cells were washed with ice-cold phosphate-buffered saline (4.3 mM Na2HPO4, 1.4 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) and either lysed in sample buffer (0.065 M Tris-HCl [pH 6.8], 2% [wt/vol] sodium dodecyl sulfate (SDS), 10% [vol/vol] β-mercaptoethanol, 0.005% bromophenol blue) and boiled immediately or lysed for 10 min on ice in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% [vol/vol] Nonidet P-40, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS) containing protease inhibitors (300 μg of phenylmethylsulfonyl fluoride per ml, 6 μg each of aprotinin and leupeptin per ml) and phosphatase inhibitors (50 mM NaF, 0.1 mM Na3VO4) and cleared by centrifugation (14,000 × g for 20 min at 4°C). Protein concentrations of cell extracts were determined by the Bradford method (45). For crude cell fractionation assays, the pellet recovered after centrifugation of RIPA buffer-lysed cells was solubilized in sample buffer using an equivalent volume to that originally used to lyse the cells in RIPA buffer.
GST pulldown, immunoprecipitation, and immunoblot assays.
For both GST pulldown and immunoprecipitation assays, glutathione- or protein A-Sepharose beads (Pharmacia) bound to GST fusion proteins or antibodies, respectively, were incubated with cell extracts in RIPA buffer (3 h at 4°C), washed extensively with RIPA buffer, and boiled in sample buffer. Recovered proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). For each GST pulldown reaction, 5 μg of each GST fusion protein was used and verified in each experiment by staining relevant portions of the protein gel with Coomassie brilliant blue dye. Each immunoprecipitation reaction was carried out with 1 or 12 μg of p53 or HA antibodies, respectively, 1 μl of DLG antiserum, or 5 μl of either 9ORF1, 9BP-1, or MUPP1 antiserum or the corresponding matched preimmune serum.
For immunoblot assays, proteins separated by SDS-PAGE were electrotransferred to a polyvinylidene difluoride membrane, which was incubated for 1 h with blocking buffer, consisting of TBST (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 0.2% [vol/vol] Tween 20) containing 5% nonfat dry milk, for 2 h with the appropriate primary antiserum or antibody (1:5,000 dilution of either 9ORF1, 9BP-1, or MUPP1 antiserum or 1.2 μg of HA antibodies per ml), and then for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG secondary antibodies (1:5,000). Antibodies were diluted in TBST containing 0.5% nonfat dry milk, and incubations were performed at room temperature. After extensive washes in TBST buffer, the membranes were developed by enhanced chemiluminescence methods (Pierce).
Protein-blotting assays.
Protein-blotting assays and preparation of 32P-radiolabeled 9ORF1 fusion protein probes were performed as described previously (57). Briefly, MUPP1 fusion proteins were separated by SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane. The membranes were incubated with blocking buffer and then for 12 h at 4°C with the 32P-labeled GST-9ORF1 protein probe (5 × 105 cpm/ml) in TBST, washed extensively with RIPA buffer, and developed by autoradiography.
In vitro translation and degradation assays.
pcDNA3-DLG, pSP64-p53, pSL301-MUPP1, or pSP64-18E6 was transcribed and translated in vitro using the TNT-coupled rabbit reticulocyte system (Promega) and 10 μCi of [35S]cysteine (1,000 Ci/mmol) (Amersham), as specified by Promega. In vitro degradation assays were performed as described previously (11). Briefly, the specified in vitro translation reaction mixtures were mixed and incubated at 30°C for the indicated times and proteins were immunoprecipitated, separated by SDS-PAGE, and detected by autoradiography.
Pulse-chase labeling of cell proteins.
Transfected COS7 cells were preincubated for 30 min in culture medium lacking methionine and cysteine, metabolically labeled for 10 min with 0.4 mCi of EXPRE35S35S [35S]protein-labeling mix (New England Nuclear) in 1.5 ml of culture medium lacking methionine and cysteine, and chased with culture medium containing excess unlabeled methionine (15 mg/liter) (2). At various times postchase, cells were harvested and lysed in RIPA buffer and cell proteins were immunoprecipitated with HA antibodies, separated by SDS-PAGE, and developed by autoradiography. The amount of radioactivity present within each protein band of interest was quantified using a Storm Molecular Dynamics PhosphorImager.
IF microscopy.
Indirect-immunofluorescence (IF) microscopy assays were performed by standard methods (17). Cells were grown on glass coverslips, fixed in methanol for 20 min at −20°C, blocked with IF buffer (TBS [50 mM Tris-HCl, pH 7.5, 200 mM NaCl] containing 10% goat serum), and reacted first with either preimmune serum or MUPP1 antiserum (1:500) and then with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG secondary antibodies (1:250) (Gibco BRL). For double-labeling IF experiments, cells on coverslips were incubated with both MUPP1 antiserum (1:500) and HA monoclonal antibodies (66 μg/ml) and then with both fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (1:250) (Gibco BRL) and Texas red-conjugated goat anti-mouse IgG secondary antibodies (1:300) (Molecular Probes). All antibodies were diluted in IF buffer, and incubations were performed at 37°C. Coverslips with attached cells were rinsed briefly in a 0.5 mg of 4′,6-diamidino-2-phenylindole (DAPI) solution per ml to stain nuclei and affixed to slides with mounting medium (VectorShield). Images were collected with a Zeiss Axiophot fluorescence microscope and digitally processed using Adobe PhotoShop software.
Sequence alignment.
Pairwise sequence alignments were performed using the Align algorithm of the BCM Search Launcher web browser (http://dot.imgen.bcm.tmc.edu:9331/seq-search/alignment.html). Mouse, rat, and human MUPP1 sequences (accession numbers AJ131869, AJ001320, and AJ001319, respectively) were obtained from GenBank.
RESULTS
9BP-1 and 9ORF1-associated protein p220 have similar characteristics.
By screening λgt11 cDNA expression libraries with a 9ORF1 protein probe, we previously isolated a partial cDNA coding for the carboxyl-terminal 526 aa of the novel mouse multi-PDZ domain protein 9BP-1 (28). Subsequent rescreening of the same λgt11 library with a 9BP-1 DNA probe led to the isolation of a cDNA coding for a larger partial carboxyl-terminal 688-aa 9BP-1 polypeptide that contains five PDZ homology domains (Fig. 1A).
FIG. 1.
The partial mouse protein 9BP-1 represents the carboxyl terminus of the mouse multi-PDZ protein MUPP1. (A) PDZ domain organizations of the partial mouse protein 9BP-1 (688 aa) and the mouse multi-PDZ domain protein MUPP1 (2,055 aa). Note that the domain organization of 9BP-1 is identical to that of the carboxyl-terminal region of MUPP1. The unique protein region used to generate MUPP1 antisera is indicated. (B) The partial mouse protein 9BP-1 exhibits 99% amino acid sequence identity to the carboxyl-terminal region of the mouse MUPP1 protein (aa 1368 to 2055). Highlighted sequences denote PDZ homology domains. Sequence alignment was performed using the Align Global Sequence Alignment algorithm from the Baylor College of Medicine Search Launcher Web site.
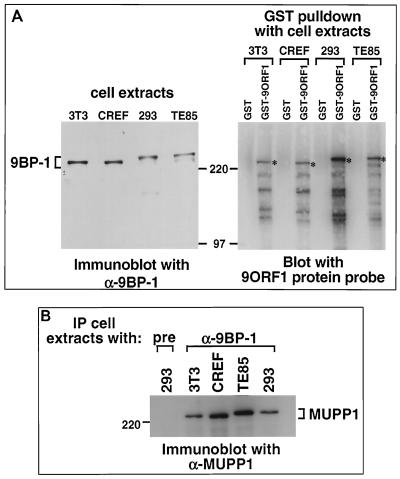
To assess initially whether 9BP-1 may represent one of the unidentified 9ORF1-associated cellular proteins (p220, p180, p160, or p155), we raised polyclonal antisera to the carboxyl-terminal 526 aa of this partial polypeptide (28). By immunoblot analysis, two independent 9BP-1 antisera specifically recognized an approximately 250-kDa protein which, in cell lines derived from various species, exhibited slightly different gel mobilities (Fig. 2A and data not shown). The latter observation presumably reflects species-specific differences for this polypeptide. More important, 9BP-1 was found to be complexed with 9ORF1 in lysates of 9ORF1-expressing cells (see below; data not shown). Additionally, comparison of the gel mobility of 9BP-1 with that of each 9ORF1-associated protein showed that 9BP-1 and 9ORF1-associated protein p220 comigrate and exhibit identical species-specific gel mobilities (Fig. 2A), suggesting that these proteins are the same.
FIG. 2.
9ORF1-associated protein p220 displays similar properties to both 9BP-1 and MUPP1. (A) 9BP-1 and 9ORF1-associated protein p220 comigrate and exhibit identical species-specific gel mobilities. Proteins from RIPA buffer-lysed mouse NIH 3T3, rat CREF, human 293, and human TE85 cell lines were either immunoblotted with 9BP-1 antiserum (left) or first subjected to a GST pulldown assay with the indicated fusion protein and then blotted with a radiolabeled 9ORF1 protein probe (right). For the experiment shown in the left or right panel, 100 or 2.5 mg of cell proteins was used, respectively, and the protein gels were run in parallel. Asterisks indicate 9ORF1-associated protein p220. (B) MUPP1 antiserum cross-reacts with 9BP-1 protein derived from several different species. Cell proteins (2.5 mg) in RIPA buffer from the indicated cell lines were first immunoprecipitated (IP) with either 9BP-1 antiserum (α-9BP-1) or the matched preimmune serum (pre) and then immunoblotted with MUPP1 antiserum. Also note that the 9BP-1 protein detected in panel A (left) and the MUPP1 protein detected here exhibited identical species-specific gel mobilities.
9BP-1 is the multi-PDZ domain protein MUPP1.
From BLAST searches of protein sequence databases, we subsequently found that the partial mouse 9BP-1 polypeptide exhibits 99% amino acid sequence identity to the carboxyl-terminal region of the 2,055-aa mouse multi-PDZ domain protein MUPP1 (Fig. 1), as well as 94 or 82% amino acid sequence identity to the carboxyl-terminal region of rat MUPP1 (2,054 aa) or human MUPP1 (2,042 aa), respectively (data not shown). MUPP1 is a widely expressed polypeptide, containing 13 PDZ domains and no other recognizable protein motifs, and was isolated by virtue of its ability to bind the cytoplasmic domain of the 5-HT2C serotonin receptor in yeast two-hybrid screens (55).
To confirm that 9BP-1 and MUPP1 are indeed the same polypeptide, we generated polyclonal antisera to a unique 210-aa rat MUPP1 region that lies between PDZ5 and PDZ6 and that lacks sequence similarity to other known proteins (Fig. 1A). As with the 9BP-1 antisera, these MUPP1 antisera specifically recognized an approximately 250-kDa cellular protein in cell lysates (data not shown) and, in addition, cross-reacted with 9BP-1 protein immunoprecipitated from human, rat, and mouse cell lines (Fig. 2B).
9ORF1 binds MUPP1.
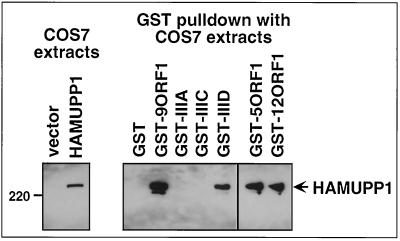
We utilized GST pulldown assays to show that the wild-type 9ORF1 protein can bind to MUPP1. In these assays, we found that the GST-9ORF1 fusion protein bound both to HA epitope-tagged rat MUPP1 (HAMUPP1) transiently expressed in COS7 cells (Fig. 3) and to endogenous MUPP1 from CREF rat embryo fibroblasts (data not shown), as did GST fusion proteins of the related wild-type Ad5 and Ad12 E4-ORF1 transforming proteins (GST-5ORF1 and GST-12ORF1, respectively) (58). To assess whether these binding results with 9ORF1 were specific, we also examined in these same assays three different transformation-defective 9ORF1 mutant proteins having disrupted (mutant IIIA) or altered (mutants IIIC and IIID) carboxyl-terminal PDZ domain-binding motifs (Table 1) (28, 56). With respect to the residues mutated in 9ORF1 mutants IIIC and IIID, such sequences surrounding the conserved residues of type I PDZ domain-binding motifs are known to influence the binding to some PDZ domains (48). In GST pulldown assays with the three 9ORF1 mutant proteins, we found that only mutant IIID was able to bind to MUPP1, albeit at substantially reduced levels compared to that of wild-type 9ORF1 (Fig. 3). This binding profile of MUPP1 to these 9ORF1 mutants is distinct from that of DLG (28) and, as expected, is identical to that previously observed for 9ORF1-associated protein p220 (57).
FIG. 3.
9ORF1 binds MUPP1 in vitro. GST-9ORF1 binds HA epitope-tagged rat MUPP1 protein (HAMUPP1) expressed in COS7 cells. Cells were lipofected with 4 μg of either empty GW1 plasmid (vector) or GW1-HAMUPP1 plasmid, and cell proteins in RIPA buffer were either immunoblotted with HA antibodies (left) or first subjected to a GST pulldown assay with the indicated wild-type or mutant E4-ORF1 fusion protein (Table 1) and then immunoblotted with HA antibodies (right). COS7 cell proteins at 100 μg or 1 mg were used in the experiment shown in the left and right panels, respectively.
TABLE 1.
Carboxyl-terminal amino acid sequences of human Ad E4-ORF1 and HPV E6 proteinsa
| Proteinb | Carboxyl-terminal amino acid sequence with respect to the consensus type I PDZ domain-binding motifc:
|
|||
|---|---|---|---|---|
| X | (S/T) | X | (V/I/L)-COOH | |
| wt 9ORF1 | A | T | L | V |
| IIIA 9ORF1 | A | P | ||
| IIIC 9ORF1 | D | T | L | V |
| IIID 9ORF1 | A | T | P | V |
| wt 5ORF1 | A | S | N | V |
| wt 12ORF1 | A | S | L | I |
| wt 18E6 | E | T | Q | V |
| 18E6-V158A | E | T | Q | A |
| 18E6-T156D/V158A | E | D | Q | A |
| wt 11E6 | D | L | L | P |
The sequence of the last 4 aa at the carboxyl terminus of E4-ORF1 proteins from Ad9 (9ORF1), Ad5 (5ORF1), and Ad12 (12ORF1) and the HPV-18 E6 (18E6) protein define a consensus type I PDZ domain-binding motif, whereas the HPV-11 E6 (11E6) protein lacks such a motif. Also shown are 9ORF1 and 18E6 mutant proteins having altered or disrupted PDZ domain-binding motifs.
wt, wild type.
Substitution mutations are depicted as bold amino acid residues.
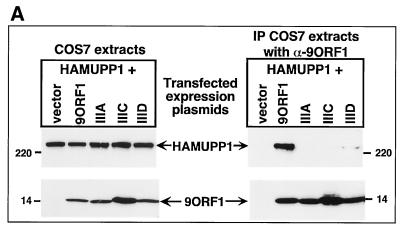
The physical association between 9ORF1 and MUPP1 in cells was examined by performing coimmunoprecipitation assays. From lysates of COS7 cells transiently expressing HAMUPP1 and either wild-type or mutant 9ORF1 protein, we found that MUPP1 coimmunoprecipitated with wild-type 9ORF1 and with mutant IIID 9ORF1 at reduced levels but failed to coimmunoprecipitate with either mutant IIIA or IIIC 9ORF1 (Fig. 4A). Identical results were obtained for endogenous MUPP1 from CREF cell lines stably expressing comparable levels of wild-type or mutant 9ORF1 protein (Fig. 4B). These findings with COS7 and CREF cells were fully concordant with the GST pulldown assay results (Fig. 3). It was also noteworthy that MUPP1 similarly coimmunoprecipitated with wild-type 9ORF1 from lysates of an Ad9-induced rat mammary tumor cell line (Fig. 4C). Taken together, the results of GST pulldown and coimmunoprecipitation assays demonstrated that 9ORF1 utilizes its PDZ domain-binding motif to mediate a specific interaction with the multi-PDZ domain protein MUPP1 in cells.
FIG. 4.
9ORF1 complexes with MUPP1 in cells. (A) 9ORF1 complexes with HA epitope-tagged rat MUPP1 protein (HAMUPP1) expressed in COS7 cells. Cells were lipofected with 6 μg of GW1-HAMUPP1 plasmid and 2 μg of either empty GW1 plasmid (vector) or a GW1 plasmid expressing wild-type or the indicated mutant 9ORF1 protein. Cell proteins in RIPA buffer were either immunoblotted with HA antibodies or 9ORF1 antiserum (left) or first immunoprecipitated (IP) with 9ORF1 antiserum (α-9ORF1) and then immunoblotted with HA antibodies or 9ORF1 antiserum (right). Cell proteins at 100 or 800 μg were used in the experiment shown in the left and right panels, respectively. (B) 9ORF1 complexes with endogenous MUPP1 of CREF cells. Cell proteins in RIPA buffer were either immunoblotted with MUPP1 or 9ORF1 antiserum (top) or first immunoprecipitated (IP) with 9ORF1 antiserum or the matched preimmune serum (pre) (bottom left) or, alternatively, with either MUPP1 antiserum (α-MUPP1) or the matched preimmune serum (pre) (bottom right) and then immunoblotted with either MUPP1 or 9ORF1 antiserum. CREF cell proteins at 100 μg, 3 mg, and 3 mg were used in the experiments in the top, bottom left, and bottom right panels, respectively. (C) 9ORF1 complexes with MUPP1 in the Ad9-induced rat mammary tumor cell line 20-8. This tumor cell line contains a single integrated copy of the entire Ad9 genome (unpublished results). Cell proteins (3 mg) in RIPA buffer were first immunoprecipitated (IP) with either 9BP-1 antiserum (α-9BP-1) or the matched preimmune serum and then immunoblotted with either 9ORF1 or 9BP-1 antiserum.
9ORF1 binds selectively to MUPP1 PDZ7 and PDZ10.
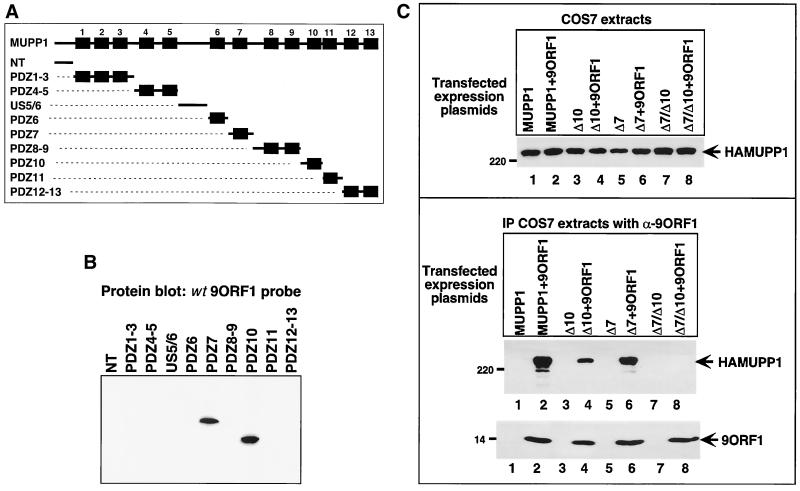
To reveal which of the 13 MUPP1 PDZ domains interacted with 9ORF1, we constructed a panel of fusion proteins containing 10 different, nonoverlapping MUPP1 protein fragments (Fig. 5A), which collectively represented the entire full-length MUPP1 polypeptide. A similar quantity of each MUPP1 fusion protein was immobilized on a membrane (data not shown) and blotted with a radiolabeled 9ORF1 protein probe. In these experiments, 9ORF1 bound to MUPP1 PDZ7 and PDZ10 but not to any other region of this cellular protein (Fig. 5B). A functional 9ORF1 PDZ domain-binding motif was required for these interactions, since a mutant GST-IIIA 9ORF1 protein probe failed to react with any of the MUPP1 fusion proteins in similar assays (data not shown).
FIG. 5.
MUPP1 PDZ7 and PDZ10 mediate binding to 9ORF1. (A) Illustration of the full-length MUPP1 polypeptide and 10 different MUPP1 GST fusion protein constructs used in protein blotting assays. (B) 9ORF1 binds MUPP1 PDZ7 and PDZ10 in vitro. Approximately 1 μg of each indicated MUPP1 GST fusion protein was immobilized on a membrane and protein blotted with a radiolabeled 9ORF1 protein probe. As a control, the membrane was stained with Coomassie brilliant blue dye to verify that an equivalent amount of each fusion protein was used in the experiment (data not shown). (C) A MUPP1 deletion mutant lacking both PDZ7 and PDZ10 fails to complex with 9ORF1 in COS7 cells. Cells were lipofected with 6 μg of a GW1 plasmid expressing wild-type or the indicated deletion mutant MUPP1 protein together with 2 μg of either empty GW1 plasmid (vector) or the GW1-9ORF1wt plasmid. Cell proteins in RIPA buffer were either immunoblotted with HA antibodies (top) or first immunoprecipitated (IP) with 9ORF1 antiserum (α-9ORF1) and then immunoblotted with HA antibodies or 9ORF1 antiserum (bottom). Cell proteins at 50 and 750 μg were used in the experiments in the top and bottom panels, respectively.
To relate the in vitro binding results to the formation of 9ORF1-MUPP1 protein complexes in cells, we constructed MUPP1 deletion mutants lacking PDZ7 (HAMUPP1ΔPDZ7), PDZ10 (HAMUPP1ΔPDZ10), or both domains (HAMUPP1-ΔPDZ7/10) and tested these MUPP1 mutants for their ability to coimmunoprecipitate with 9ORF1 from COS7 cell lysates. The results showed that HAMUPP1ΔPDZ7 coimmunoprecipitated with 9ORF1 at wild-type levels and that HAMUPP1- ΔPDZ10 coimmunoprecipitated with 9ORF1 at slightly reduced levels but that HAMUPP1ΔPDZ7/10 failed to coimmunoprecipitate with 9ORF1 in these assays (Fig. 5C). These findings corroborated our in vitro binding results in showing that, among the 13 MUPP1 PDZ domains, only PDZ7 and PDZ10 are capable of mediating the binding of MUPP1 to 9ORF1 in vivo.
9ORF1 aberrantly sequesters MUPP1 within punctate bodies in the cytoplasm of cells.
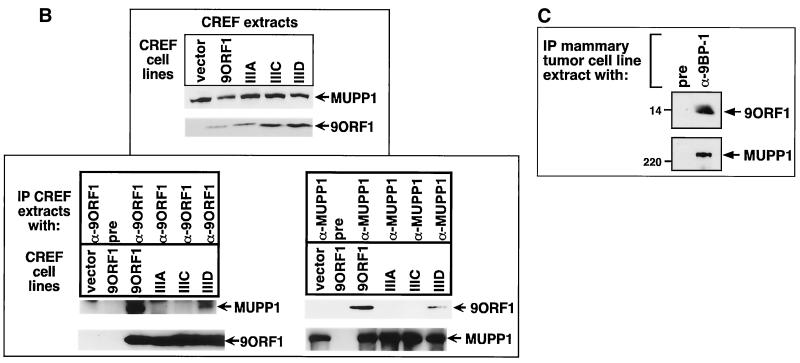
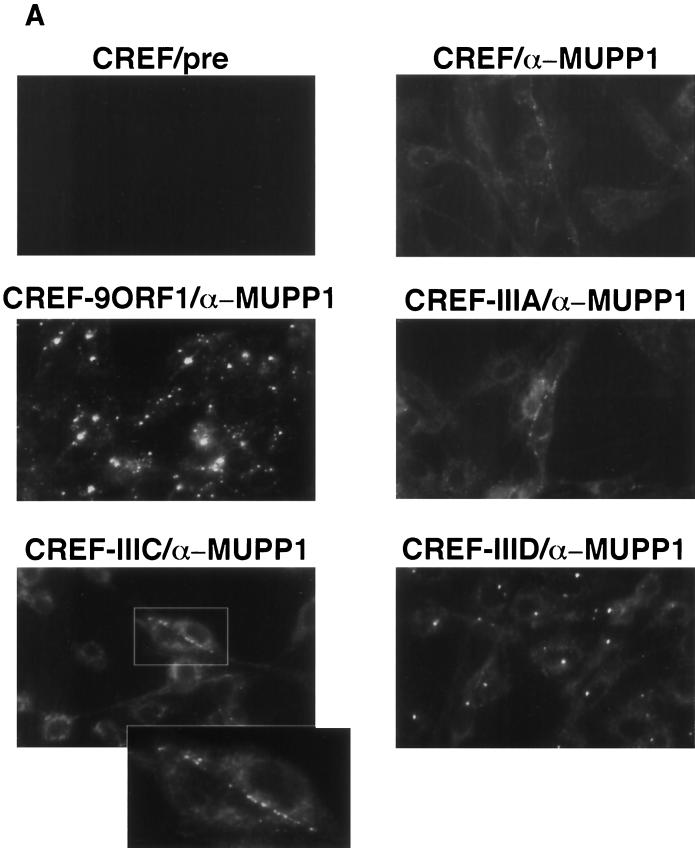
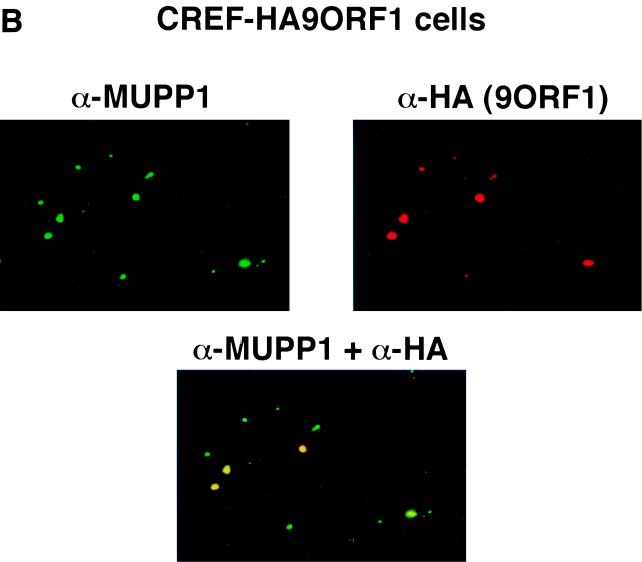
Using IF microscopy assays, we sought to ascertain the subcellular distribution of MUPP1 in normal CREF cells, as well as in CREF cell lines stably expressing wild-type or mutant 9ORF1 protein. In normal CREF cells, MUPP1 displayed mostly diffuse and somewhat perinuclear staining in the cytoplasm, although some MUPP1 protein was also detected at discrete points of cell-cell contact (Fig. 6A). The latter finding is consistent with the observation that PDZ proteins frequently localize to membranes at specialized regions of cell-cell contact in epithelial cells (8). Because 9ORF1 exists primarily within punctate bodies in the cytoplasm of cells (59), we reasoned that the subcellular localization of MUPP1 may be perturbed in 9ORF1-expressing CREF cells. Significantly, in contrast to results obtained with normal CREF cells, MUPP1 was found to be sequestered within punctate bodies in the cytoplasm of more than 95% of CREF cells expressing wild-type 9ORF1 (Fig. 6A), similar to the staining pattern observed for 9ORF1 (59). Using a CREF cell line stably expressing an HA epitope-tagged 9ORF1 protein, we were able to demonstrate that 9ORF1 and MUPP1 colocalize within these cytoplasmic bodies (Fig. 6B).
FIG. 6.
9ORF1 aberrantly sequesters MUPP1 within punctate bodies in the cytoplasm of cells. (A) Determination of the subcellular localization of MUPP1 in normal CREF cells (CREF) or CREF cell lines stably expressing wild-type (CREF-9ORF1) or the indicated mutant 9ORF1 protein (CREF-IIIA, CREF-IIIC, and CREF-IIID). IF microscopy assays were performed with either MUPP1 antiserum (α-MUPP1) or the matched preimmune serum (pre). Although all of the CREF cell lines expressed similar amounts of MUPP1 protein (see Fig. 7A), the MUPP1 staining for CREF-9ORF1 cells appeared brighter than that for the other CREF lines. This effect probably resulted from the large amounts of MUPP1 protein concentrated within the cytoplasmic punctate bodies. Discontinuous cell-cell contact staining for MUPP1 was most evident in normal CREF cells, and CREF-IIIA and CREF-IIIC lines, all of which exhibited similar MUPP1 staining patterns. As an example of this cell-cell contact staining, two adjacent CREF-IIIC cells within the delimited rectangular region are shown offset at higher magnification. (B) 9ORF1 and MUPP1 colocalize within punctate bodies in the cytoplasm of CREF cells. Double-labeling IF microscopy assays using both MUPP1 antiserum and HA antibodies (α-HA) were performed with CREF cells stably expressing HA epitope-tagged 9ORF1 protein (CREF-HA9ORF1). Each of the three panels shows the identical field containing the same three cells. The top left and top right panels show the MUPP1 and 9ORF1 staining patterns, respectively, whereas the bottom panel shows the merged images.
Additional IF assay results indicated that the cytoplasmic sequestration of MUPP1 by 9ORF1 depended on an ability of 9ORF1 to complex with this cellular PDZ protein. Specifically, CREF cell lines expressing 9ORF1 mutants IIIA and IIIC, which fail to bind MUPP1, showed a MUPP1 staining pattern similar to that seen in normal CREF cells. Moreover, the CREF cell line expressing 9ORF1 mutant IIID, which exhibits weak binding to MUPP1, showed some cytoplasmic punctate staining for MUPP1, although substantially less than that observed in the CREF cell line expressing wild-type 9ORF1 (Fig. 6A). It is worth mentioning that, similar to the wild-type 9ORF1 protein, these three 9ORF1 mutant proteins also display punctate staining in the cytoplasm of CREF cells (56).
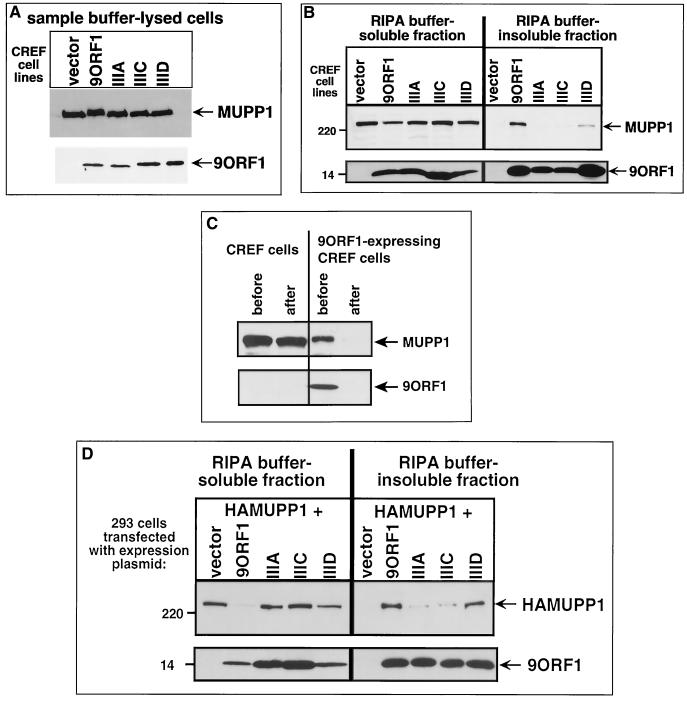
To corroborate the IF assay results, we performed crude cell fractionation experiments with the same CREF cell lines. Following direct lysis in 2% SDS, each cell line was found to express comparable levels of both MUPP1 and 9ORF1 proteins (Fig. 7A). CREF cell lysates were also prepared in RIPA buffer and separated by centrifugation into a RIPA buffer-soluble supernatant fraction and a RIPA buffer-insoluble pellet fraction. Immunoblot analyses of these two fractions with MUPP1 antiserum revealed that wild-type-9ORF1-expressing CREF cells contain substantially less RIPA buffer-soluble MUPP1 protein and concomitantly more RIPA buffer-insoluble MUPP1 protein than do normal CREF cells (Fig. 7B). The fact that the portion of MUPP1 protein retained in the RIPA buffer-soluble fraction of the wild-type-9ORF1-expressing cells could be depleted by quantitative immunoprecipitation of 9ORF1 (Fig. 7C) indicated that the vast majority of MUPP1 protein in these cells is complexed with 9ORF1.
FIG. 7.
9ORF1 aberrantly redistributes MUPP1 into the RIPA buffer-insoluble fraction of cells. (A) Similar amounts of MUPP1 and 9ORF1 proteins within CREF cell lines stably expressing wild-type and mutant 9ORF1 proteins. Cell proteins (100 μg) extracted with sample buffer were immunoblotted with MUPP1 antiserum or 9ORF1 antiserum. (B) Wild-type 9ORF1 specifically redistributes MUPP1 into the RIPA buffer-insoluble fraction of CREF cells. Cells from the indicated CREF lines were lysed in RIPA buffer and centrifuged to yield a RIPA buffer-soluble supernatant fraction and a RIPA buffer-insoluble pellet fraction (see Materials and Methods). Cell proteins from an equivalent volume of either the soluble or insoluble fraction were immunoblotted with MUPP1 or 9ORF1 antiserum. (C) Most MUPP1 protein is complexed with 9ORF1 in 9ORF1-expressing CREF cells. Cell proteins (3 mg) in the RIPA buffer-soluble fraction of normal CREF cells or wild-type 9ORF1-expressing CREF cells were subjected to five serial immunoprecipitations with 9ORF1 antiserum. Relative amounts of MUPP1 and 9ORF1 protein remaining in this fraction (100 μg of protein) “before” and “after” performing the serial immunoprecipitations were determined by immunoblot analysis. (D) Wild-type 9ORF1 also specifically redistributes HA epitope-tagged rat MUPP1 (HAMUPP1) into the RIPA buffer-insoluble fraction of 293 cells. Cells were lipofected with 1 μg of GW1-HAMUPP1 plasmid and 3 μg of either empty GW1 plasmid (vector) or a GW1 plasmid expressing wild-type or the indicated mutant 9ORF1 protein. Cell fractionation assays were performed as described for panel B, except that cell proteins were immunoblotted with HA antibodies or 9ORF1 antiserum.
The redistribution of MUPP1 into the RIPA buffer-insoluble fraction of wild-type-9ORF1-expressing CREF cells was also related to the ability of 9ORF1 to bind this cellular protein, because mutant IIIA and IIIC 9ORF1 largely failed to aberrantly redistribute MUPP1 in CREF cells whereas mutant IIID 9ORF1 retained a reduced capacity to induce this effect (Fig. 7B). These differences are not likely to be due to the smaller amounts of RIPA buffer-insoluble mutant IIIA and IIIC 9ORF1 proteins present in these CREF cells (Fig. 7B), since transiently transfected 293 cells contained equivalent amounts of RIPA buffer-insoluble wild-type and mutant 9ORF1 proteins but still yielded a pattern of MUPP1 redistribution similar to that of the CREF cell lines (Fig. 7D). That 9ORF1 redistributed MUPP1 into the RIPA buffer-insoluble fraction of 293 cells more effectively than it did in CREF cells may be due to the higher protein levels attained for 9ORF1 and MUPP1 in transient transfections of the 293 cells. Together, the results of IF and crude cell fractionation assays argued that 9ORF1 aberrantly sequesters MUPP1 within RIPA buffer-insoluble complexes in the cytoplasm of cells.
The high-risk 18E6 oncoprotein binds MUPP1 and targets this cellular protein for degradation in cells.
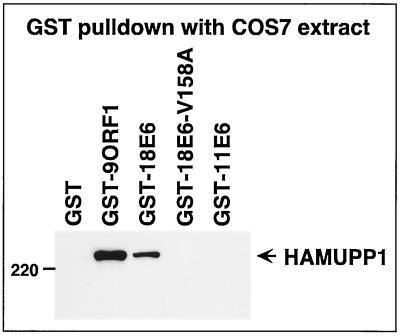
Because, like 9ORF1, high-risk HPV E6 oncoproteins possess a functional PDZ domain-binding motif and complex with DLG (27, 29), we next explored the possibility that such HPV E6 proteins likewise bind to MUPP1. In GST pulldown assays, the wild-type high-risk 18E6 protein associated both with HAMUPP1 protein expressed in COS7 cells (Fig. 8) and with endogenous MUPP1 from CREF cells (data not shown). This binding was specific and dependent on a functional PDZ domain-binding motif because in these assays the 18E6-V158A and 18E6-T156D/V158A mutant proteins, which have disrupted PDZ-domain binding motifs (Table 1), failed to complex with MUPP1 (Fig. 8 and data not shown), as did the wild-type low-risk HPV-11 E6 (11E6) protein, which lacks a PDZ domain-binding motif (Table 1). It is notable that HPV-16 E6 mutants having functionally disrupted PDZ domain-binding motifs, like those of 18E6-V158A and 18E6-T156D/V158A, are no longer able to oncogenically transform rodent fibroblasts (26).
FIG. 8.
18E6 binds MUPP1 in vitro. GST-18E6 binds HA epitope-tagged rat MUPP1 (HAMUPP1) exogenously expressed in COS7 cells. Cells were lipofected with 8 μg of GW1-HAMUPP1 plasmid, and cell proteins (250 μg) in RIPA buffer were first subjected to a GST pulldown assay with the indicated fusion protein and then immunoblotted with HA antibodies.
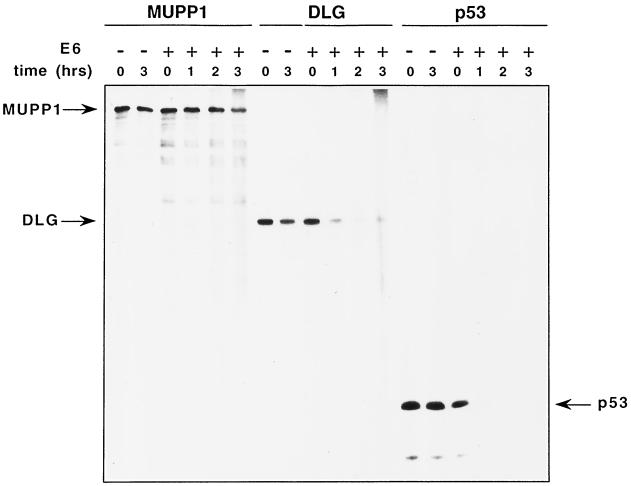
The fact that high-risk HPV E6 oncoproteins promote the degradation of several cellular factors (10, 15, 53), including the tumor suppressor protein p53 (42) and DLG (11), prompted us to test whether 18E6 has similar effects on MUPP1. Incubation of in vitro-translated high-risk HPV E6 proteins with p53 leads to degradation of this cellular factor (42), and so we first examined MUPP1 in similar assays. Although 18E6-induced degradation of both p53 and DLG was more efficient, a modest reduction in MUPP1 protein levels was reproducibly observed following a 3-h incubation with 18E6 (Fig. 9). This effect was also consistently greater than that observed in control water-primed in vitro translation reactions.
FIG. 9.
18E6 promotes the degradation of the MUPP1 protein in vitro. In vitro-translated MUPP1, DLG, or p53 protein was incubated for the indicated times with a 5- to 10-fold molar excess of in vitro-translated 18E6 protein (+) or with an equivalent volume of a water-primed in vitro translation reaction mixture (−). Proteins from each reaction were subjected to immunoprecipitation with MUPP1, DLG, or p53 antibodies, respectively, and detected by autoradiography.
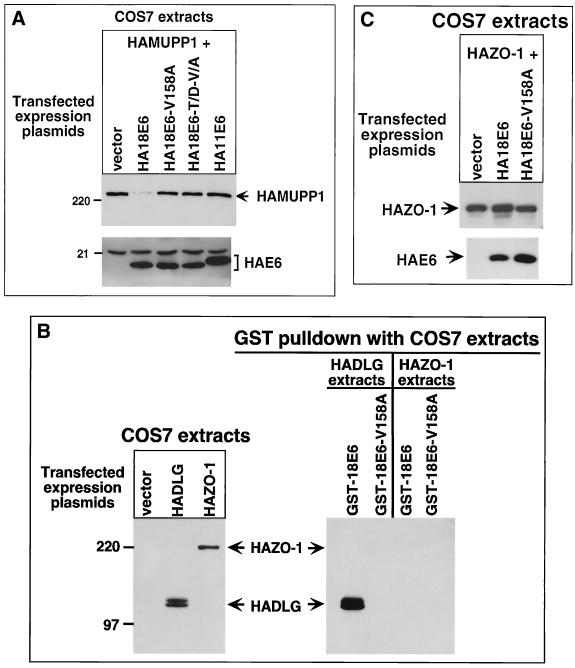
Whether 18E6 may target MUPP1 for degradation in cells was examined by expressing HAMUPP1 alone or together with 18E6 in COS7 cells. We found that in these assays, compared to cells expressing MUPP1 alone, cells coexpressing MUPP1 and 18E6 showed substantially lower steady-state levels of MUPP1 protein (Fig. 10A). It is important to mention that this effect is distinct from that seen in the RIPA buffer-soluble fraction of 9ORF1-expressing cells (Fig. 7B and D), since MUPP1 was not sequestered within the RIPA buffer-insoluble fraction of 18E6-expressing cells (data not shown). To produce this effect, 18E6 required a functional PDZ domain-binding motif because mutants 18E6-V158A and 18E6-T156D/V158A, as well as wild-type 11E6, failed to reduce MUPP1 protein levels in COS7 cells. Additionally, only specific cellular PDZ proteins were affected by 18E6, since 18E6 neither bound the DLG-related PDZ-protein ZO-1 (Fig. 10B) (61) nor reduced its protein levels in these cells (Fig. 10C).
FIG. 10.
18E6 reduces the steady-state levels of MUPP1 protein in cells. (A) 18E6 reduces the steady-state levels of HA epitope-tagged rat MUPP1 (HAMUPP1) protein expressed in COS7 cells. Cells were lipofected with 1 μg of GW1-HAMUPP1 plasmid and 4 μg of either empty GW1 plasmid (vector) or a GW1 plasmid expressing HA epitope-tagged wild-type or the indicated mutant 18E6 protein or expressing HA epitope-tagged wild-type 11E6 protein. Cell proteins (30 μg) in RIPA buffer were immunoblotted with HA antibodies. (B) 18E6 does not bind HA epitope-tagged PDZ-protein ZO-1 (HAZO-1). Cells were lipofected with 3 μg of either empty GW1 plasmid (vector), GW1-HADLG plasmid, or GW1-HAZO-1 plasmid, and cell proteins in RIPA buffer were either immunoblotted with HA antibodies (left) or first subjected to a GST pulldown assay with the indicated fusion protein and then immunoblotted with HA antibodies (right). COS7 cell proteins at 10 and 75 μg were used in the experiments in the left and right panels, respectively. HADLG was included as a positive control in these binding assays (28). (C) 18E6 does not reduce HAZO-1 protein levels in COS7 cells. COS7 cells were lipofected with 0.01 μg of GW1-HAZO-1 plasmid and 4 μg of either empty GW1 plasmid (vector) or a GW1 plasmid expressing HA epitope-tagged wild-type or the indicated mutant 18E6 protein. Cell proteins (30 μg) in RIPA buffer were immunoblotted with HA antibodies.
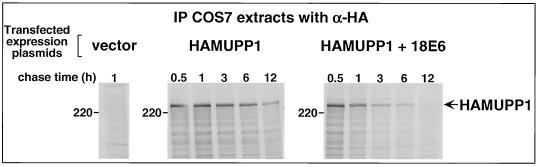
To verify that the 18E6-mediated reduction in MUPP1 steady-state protein levels was due to decreased stability of this cellular protein in cells, we performed pulse-chase experiments with COS7 cells either expressing HAMUPP1 alone or coexpressing HAMUPP1 and 18E6. The results showed that MUPP1 protein levels modestly declined after a 6-h chase period in the absence of 18E6 whereas they were more extensively reduced after only a 3-h chase period in the presence of 18E6 (Fig. 11). By quantifying the amounts of radioactivity present in MUPP1 protein bands at each time point, we estimated that the half-life of the MUPP1 protein was shortened from 5.7 h in control COS7 cells to 1.3 h in 18E6-expressing COS7 cells. This greater than fourfold decrease in the half-life of the MUPP1 protein argues that 18E6 targets this cellular factor for degradation in cells.
FIG. 11.
18E6 decreases the half-life of the MUPP1 protein in cells. A total of 5.5 × 105 COS7 cells were lipofected with 5 μg of empty GW1 plasmid (vector) or 1 μg of GW1-HAMUPP1 plasmid and 4 μg of either empty GW1 plasmid (HAMUPP1) or the GW1-18E6 plasmid (HAMUPP1 + 18E6). At 24 h posttransfection, cells were pulse-labeled and then chased for the indicated times (see Materials and Methods). Cell proteins (200 μg) were immunoprecipitated with HA antibodies (α-HA), and HAMUPP1 protein was detected by autoradiography and quantified with a PhosphorImager.
DISCUSSION
The results presented in this paper demonstrate that the widely expressed multi-PDZ protein MUPP1 is a direct cellular target for the Ad9 E4-ORF1 oncoprotein (9ORF1), as well as for the related E4-ORF1 transforming proteins derived from Ad5 and Ad12 (5ORF1 and 12ORF1, respectively) (Fig. 3). We also showed that interactions between 9ORF1 and MUPP1 are mediated by the carboxyl-terminal PDZ domain-binding motif of 9ORF1 and the PDZ7 and PDZ10 domains of MUPP1 (Fig. 3 to 5). Since 5ORF1 and 12ORF1 also possess carboxyl-terminal PDZ domain-binding motifs, these viral proteins probably complex with MUPP1 in a similar fashion. More important, the fact that transformation-defective 9ORF1 mutants with altered PDZ domain-binding motifs either fail or have reduced capacities to complex with MUPP1 in cells argues that binding of 9ORF1 to MUPP1 is critical for 9ORF1-induced transformation (Fig. 3 and 4). Our finding that 9ORF1 associates with MUPP1 in an Ad9-induced mammary tumor cell line (Fig. 4C) further suggests that this interaction also contributes to Ad9-induced mammary tumorigenesis in rats. With the findings presented in this paper, 9ORF1 has now been shown to complex with two different cellular PDZ proteins, MUPP1 and DLG (28). Since the weakly transforming 9ORF1 mutants IIIC and IIID bind only one of these two PDZ proteins whereas the completely transformation-defective 9ORF1 mutant IIIA fails to bind either PDZ protein, we believe that interaction of 9ORF1 with both MUPP1 and DLG is important for full 9ORF1 transforming activity.
It is also worth noting that we have failed to detect any binding of 9ORF1 to several other cellular PDZ proteins (B. Glaunsinger and R. Javier, unpublished results), suggesting that 9ORF1 interacts with only a select group of these cellular factors. We hypothesize that such selective binding of 9ORF1 is achieved through sequences surrounding its PDZ domain-binding motif. In this model, specific amino acid residues adjacent to the 9ORF1 PDZ domain-binding motif would play differential roles in mediating the binding to each 9ORF1-associated PDZ protein. Consistent with this idea, 9ORF1 mutant IIIC retains wild-type binding to DLG (28) but fails to bind MUPP1 and, conversely, 9ORF1 mutant IIID fails to bind DLG (28) but retains an ability to bind MUPP1 (Table 1 and Fig. 3 and 4).
The domain structure of MUPP1 suggests that this cellular factor functions as an adapter protein in signal transduction (55). Moreover, having the largest number of PDZ domains (i.e., 13) yet reported in a polypeptide, MUPP1 has the capacity to assemble a large array of cellular targets into a multitude of different signaling complexes. As further support for a presumed role in cell signaling, MUPP1 was isolated in yeast two-hybrid screens for its ability to interact with the cytoplasmic carboxyl-terminal domain of the 5-HT2C serotonin receptor (55), which, in the central nervous system, is implicated in a variety sensory, motor, and behavioral processes (18). The putative interaction between the 5-HT2C receptor and MUPP1 in cells is suspected to involve a type I PDZ domain-binding motif at the extreme carboxyl terminus of the 5-HT2C receptor and at least one MUPP1 PDZ domain (55). The fact that overexpression of the 5-HT2C receptor has been shown to confer a transformed state on 3T3 fibroblasts (60) also suggests a possible link between MUPP1 and signaling pathways involved in regulating cellular proliferation.
We found that most of the MUPP1 protein in CREF fibroblasts is present within the cytoplasm, although some MUPP1 protein is also detected at discrete regions of cell-cell contact (Fig. 6). Consistent with the notion that MUPP1 functions in signal transduction, PDZ proteins that play known roles in cell signaling also localize to membranes at regions of cell-cell contact in epithelial cells (8) and, in some cases, within the cytoplasm (62, 64). With respect to how MUPP1 may function in cells, studies with the Drosophila MUPP1-related multi-PDZ protein InaD are likely to provide the most useful paradigm. The polypeptide InaD, consisting of five PDZ domains, functions to regulate the rhodopsin light-activated signaling pathway in retinal neurons. Each of the InaD PDZ domains mediates binding to a different signaling protein, including the calcium channel TRP, phospholipase C-β, and protein kinase C. InaD serves to organize these cellular targets into large signaling complexes and localize them to specialized cell membranes, and, in so doing, it allows rapid and efficient activation and deactivation of the light response (54). Although the PDZ protein-regulated signaling pathways perturbed by 9ORF1 in cells have yet to be identified, we previously showed that two prominent transformed properties of 9ORF1-expressing CREF cells in culture are anchorage-independent growth and an ability to grow to high saturation densities (59). Since proliferation of normal cells is inhibited by either detachment from a substrate matrix or formation of extensive cell-cell contacts (12, 16), one intriguing possibility is that 9ORF1 interferes with the ability of MUPP1 to regulate cell growth-controlling signaling cascades that emanate from these important plasma membrane contact points.
Of the 13 MUPP1 PDZ domains, 9ORF1 specifically targets only 2, namely, PDZ7 and PDZ10 (Fig. 5). This selective interaction may serve to block MUPP1 from associating with cellular targets of these particular domains or, alternatively, to bring 9ORF1 into close proximity with other MUPP1 cellular targets in order to modify their activities. A recently discovered activity for some PDZ proteins is the ability to direct their cellular targets to the proper location within cells (4, 24, 39, 54). One notable example comes from studies of vulval development in Caenorhabditis elegans. In this system, LET-23, an epidermal growth factor receptor-like protein, must be localized to the basolateral membrane of the vulval epithelial cells for this receptor to associate with its growth factor ligand (25). Three different PDZ domain-containing proteins, LIN-7, LIN-2, and LIN-10, which complex with LET-23, are responsible for directing this receptor to its proper site in cells (24). Therefore, considering that 9ORF1 aberrantly sequesters MUPP1 in the cytoplasm of cells (Fig. 6 and 7), it is reasonable to assume that 9ORF1 completely abolishes the function of MUPP1 by preventing this PDZ protein and its cellular targets from reaching their proper destinations in the cell.
We also showed that the high-risk 18E6 oncoprotein utilizes a PDZ domain-binding motif to complex with and promote degradation of the MUPP1 protein in cells (Fig. 8 to 11). Whether 18E6 targets MUPP1 for ubiquitin-mediated, proteasome-dependent proteolysis, as it does for p53 (41), was not determined. The modest 18E6-induced MUPP1 degradation observed after mixing in vitro-translated proteins (Fig. 9) may indicate that 18E6 utilizes a different mechanism to degrade MUPP1 from the one it uses to degrade p53 and DLG. Nevertheless, the degradation of MUPP1 that we observed in 18E6-expressing COS7 cells implies that MUPP1 function is abrogated in cells infected by HPV-18. Because the carboxyl-terminal PDZ domain-binding motif sequence of high-risk HPV-31, HPV-39, HPV-45, and HPV-51 E6 proteins is identical to that of 18E6 (28), MUPP1 is also likely to be targeted for degradation by these and probably other high-risk HPV E6 oncoproteins. Our additional finding that the low-risk 11E6 protein neither binds MUPP1 nor targets this cellular protein for degradation in cells (Fig. 8 and 10) provides additional support for the idea that binding of high-risk E6 proteins to MUPP1 may contribute to HPV-induced carcinogenesis.
Our results argue that an ability to bind cellular PDZ proteins contributes to the transforming activities of both the Ad E4-ORF1 and high-risk HPV E6 oncoproteins. With respect to possible roles in the life cycles of Ad and HPV, these interactions are expected to help create an optimal environment for viral replication within infected host cells by overcoming normal defense mechanisms that block abnormal progression into S phase. Such an activity is common to the oncoproteins of DNA tumor viruses and is invariably mediated by their interactions with host cell factors intimately involved in regulating cellular proliferation and differentiation (34). In this regard, the fact that two unrelated viral oncoproteins, Ad E4-ORF1 and high-risk HPV E6, have both evolved to target the MUPP1 protein in cells strengthens the hypothesis that interactions with this cellular factor are pertinent to transformation. It is well known that the Ad5 E1B oncoprotein sequesters the tumor suppressor protein p53 in cells (44) whereas the unrelated high-risk HPV E6 oncoproteins promote the degradation of this cellular factor (19). In these examples, functional inactivation of p53 is the ultimate outcome of these interactions, but the mechanisms by which these viral oncoproteins accomplish this effect are distinct. Our findings suggest that the Ad E4-ORF1 and HPV E6 oncoproteins likewise inactivate MUPP1 by different mechanisms. This interesting parallel with the tumor suppressor protein p53, together with the fact that the PDZ protein DLG is a putative tumor suppressor protein, hints that MUPP1 may function to negatively regulate cellular proliferation and thus may represent a novel tumor suppressor protein. Consequently, revealing the cellular functions for this multi-PDZ domain adapter protein may provide new insights into mechanisms that contribute to the development of human malignancies.
ACKNOWLEDGMENTS
We are grateful to Christoph Ullmer and Alan Fanning for generously providing the pBSK-MUPP1 and pSK-ZO-1 plasmids, respectively. We thank Richard Sutton for helpful discussions and critical reading of the manuscript. We also thank Hank Adams and Frank Herbert (Microscopy Core, Department of Cell Biology, Baylor College of Medicine) for assistance.
S.S.L. was the recipient of a U.S. Army Breast Cancer Training Grant (DAMD17-94-J4204), and B.G. was the recipient of a Molecular Virology Training Grant (T32 AI07471). This work was supported by National Institutes of Health (ROI CA58541), American Cancer Society (RPG-97-668-01-VM), and U.S. Army (DAMD17-97-1-7082) grants to R.T.J. and an Associazione Italiana per la Ricerca sul Cancro grant to L.B.
REFERENCES
- 1.Banks L, Matlashewski G, Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986;159:529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino J S. Biosynthetic labeling of proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1991. pp. 10.18.1–10.18.9. [Google Scholar]
- 3.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 4.Butz S, Okamoto M, Sudhof T C. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 5.Cann A J, Chen I S Y. Human T-cell leukemia virus types I and II. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1849–1880. [Google Scholar]
- 6.Craven S E, Bredt D S. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 7.Fanning A S, Anderson J M. PDZ domains and the formation of protein networks at the plasma membrane. Curr Top Microbiol Immunol. 1998;228:209–233. doi: 10.1007/978-3-642-80481-6_9. [DOI] [PubMed] [Google Scholar]
- 8.Fanning A S, Anderson J M. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Investig. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher P B, Babiss L E, Weinstein I B, Ginsberg H S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF) Proc Natl Acad Sci USA. 1982;79:3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiol D, Kuhne C, Glaunsinger B, Lee S S, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 12.Giancotti F G, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 13.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 14.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias K E, Schwartz A L, Kahana C, Ciechanover A. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumbiner B M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies. A laboratory manual. 1988. pp. 55–137. , 283–318, and 359–420. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- 18.Heisler L K, Chu H M, Tecott L H. Epilepsy and obesity in serotonin 5-HT2C receptor mutant mice. Ann N Y Acad Sci. 1998;861:74–78. doi: 10.1111/j.1749-6632.1998.tb10175.x. [DOI] [PubMed] [Google Scholar]
- 19.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2045–2076. [Google Scholar]
- 20.Jainchill J L, Aaronson S A, Todaro G J. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javier R, Raska K, Jr, Shenk T. Requirement for the adenovirus type 9 E4 region in production of mammary tumors. Science. 1992;257:1267–1271. doi: 10.1126/science.1519063. [DOI] [PubMed] [Google Scholar]
- 22.Javier R, Shenk T. Mammary tumors induced by human adenovirus type 9: a role for the viral early region 4 gene. Breast Cancer Res Treat. 1996;39:57–67. doi: 10.1007/BF01806078. [DOI] [PubMed] [Google Scholar]
- 23.Javier R T. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaech S M, Whitfield C W, Kim S K. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S K. Polarized signaling: basolateral receptor localization in epithelial cells by PDZ-containing proteins. Curr Opin Cell Biol. 1997;9:853–859. doi: 10.1016/s0955-0674(97)80088-9. [DOI] [PubMed] [Google Scholar]
- 26.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubbutat M H, Vousden K H. New HPV E6 binding proteins: dangerous liaisons? Trends Microbiol. 1998;6:173–175. doi: 10.1016/s0966-842x(98)01267-0. [DOI] [PubMed] [Google Scholar]
- 28.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lue R A, Marfatia S M, Branton D, Chishti A H. Cloning and characterization of hdlg: the human homologue of the Drosophila discs large tumor suppressor binds to protein 4.1. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maekawa K, Imagawa N, Naito A, Harada S, Yoshie O, Takagi S. Association of protein-tyrosine phosphatase PTP-BAS with the transcription-factor-inhibitory protein IkappaBalpha through interaction between the PDZ1 domain and ankyrin repeats. Biochem J. 1999;337:179–184. [PMC free article] [PubMed] [Google Scholar]
- 31.Maximov A, Sudhof T C, Bezprozvanny I. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- 32.McAllister R M, Gardner M B, Greene A E, Bradt C, Nichols W W, Landing B H. Cultivation in vitro of cells derived from a human osteosarcoma. Cancer. 1971;27:397–402. doi: 10.1002/1097-0142(197102)27:2<397::aid-cncr2820270224>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 33.Muller B M, Kistner U, Veh R W, Cases-Langhoff C, Becker B, Gundelfinger E D, Garner C C. Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs-large tumor suppressor protein. J Neurosci. 1995;15:2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nevins J R, Vogt P K. Cell transformation by viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 301–343. [Google Scholar]
- 35.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 36.Pim D, Storey A, Thomas M, Massimi P, Banks L. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene. 1994;9:1869–1876. [PubMed] [Google Scholar]
- 37.Ranganathan R, Ross E M. PDZ domain proteins: scaffolds for signaling complexes. Curr Biol. 1997;7:R770–R773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 38.Rapp L, Chen J J. The papillomavirus E6 proteins. Biochim Biophys Acta. 1998;1378:F1–F19. doi: 10.1016/s0304-419x(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 39.Rongo C, Whitfield C W, Rodal A, Kim S K, Kaplan J M. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 40.Saras J, Heldin C H. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 42.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 43.Shah K V, Howley P M. Papillomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2077–2109. [Google Scholar]
- 44.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 45.Simonian M H, Smith J A. Quantitation of proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1996. pp. 10.1.1–10.1.10. [Google Scholar]
- 46.Smith D B, Corcoran L M. Expression and purification of glutathione-S-transferase fusion proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1994. pp. 16.7.1–16.7.7. [DOI] [PubMed] [Google Scholar]
- 47.Song S, Pitot H C, Lambert P F. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 49.Strauss W M. Hybridization with radioactive probes. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1993. pp. 6.3.1–6.3.6. [Google Scholar]
- 50.Stricker N L, Christopherson K S, Yi B A, Schatz P J, Raab R W, Dawes G, Bassett D E, Jr, Bredt D S, Li M. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- 51.Tabor S, Struhl K, Scharf S J, Gelfand D H. DNA-dependent DNA polymerases. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1990. pp. 3.5.1–3.5.15. [Google Scholar]
- 52.Thomas D L, Shin S, Jiang B H, Vogel H, Ross M A, Kaplitt M, Shenk T E, Javier R T. Early region 1 transforming functions are dispensable for mammary tumorigenesis by human adenovirus type 9. J Virol. 1999;73:3071–3079. doi: 10.1128/jvi.73.4.3071-3079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 54.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker C S. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 55.Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- 56.Weiss R S, Gold M O, Vogel H, Javier R T. Mutant adenovirus type 9 E4 ORF1 genes define three protein regions required for transformation of CREF cells. J Virol. 1997;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss R S, Javier R T. A carboxy-terminal region required by the adenovirus type 9 E4 ORF1 oncoprotein for transformation mediates direct binding to cellular polypeptides. J Virol. 1997;71:7873–7880. doi: 10.1128/jvi.71.10.7873-7880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss R S, Lee S S, Prasad B V, Javier R T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss R S, McArthur M J, Javier R T. Human adenovirus type 9 E4 open reading frame 1 encodes a cytoplasmic transforming protein capable of increasing the oncogenicity of CREF cells. J Virol. 1996;70:862–872. doi: 10.1128/jvi.70.2.862-872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westphal R S, Sanders-Bush E. Differences in agonist-independent and -dependent 5-hydroxytryptamine 2C receptor-mediated cell division. Mol Pharmacol. 1996;49:474–480. [PubMed] [Google Scholar]
- 61.Willott E, Balda M S, Fanning A S, Jameson B, Van Itallie C, Anderson J M. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu H, Reuver S M, Kuhlendahl S, Chung W J, Garner C C. Subcellular targeting and cytoskeletal attachment of SAP97 to the epithelial lateral membrane. J Cell Sci. 1998;111:2365–2376. doi: 10.1242/jcs.111.16.2365. [DOI] [PubMed] [Google Scholar]
- 63.Xia H, Winokur S T, Kuo W L, Altherr M R, Bredt D S. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang N, Higuchi O, Mizuno K. Cytoplasmic localization of LIM-kinase 1 is directed by a short sequence within the PDZ domain. Exp Cell Res. 1998;241:242–252. doi: 10.1006/excr.1998.4053. [DOI] [PubMed] [Google Scholar]