Abstract
Rheumatoid arthritis (RA) is an inflammatory autoimmune disease characterized by synovitis, bone and cartilage destruction, and increased fracture risk with bone loss. Although disease–modifying antirheumatic drugs have dramatically improved clinical outcomes, these therapies are not universally effective in all patients because of the heterogeneity of RA pathogenesis. Therefore, it is necessary to elucidate the molecular mechanisms underlying RA pathogenesis, including associated bone loss, in order to identify novel therapeutic targets. In this study, we found that Budding uninhibited by benzimidazoles 1 (BUB1) was highly expressed in RA patients’ synovium and murine ankle tissue with arthritis. As CD45+CD11b+ myeloid cells are a Bub1 highly expressing population among synovial cells in mice, myeloid cell–specific Bub1 conditional knockout (Bub1ΔLysM) mice were generated. Bub1ΔLysM mice exhibited reduced femoral bone mineral density when compared with control (Ctrl) mice under K/BxN serum–transfer arthritis, with no significant differences in joint inflammation or bone erosion based on a semi–quantitative erosion score and histological analysis. Bone histomorphometry revealed that femoral bone mass of Bub1ΔLysM under arthritis was reduced by increased osteoclastic bone resorption. RNA-seq and subsequent Gene Set Enrichment Analysis demonstrated a significantly enriched nuclear factor-kappa B pathway among upregulated genes in receptor activator of nuclear factor kappa B ligand (RANKL)–stimulated bone marrow–derived macrophages (BMMs) obtained from Bub1ΔLysM mice. Indeed, osteoclastogenesis using BMMs derived from Bub1ΔLysM was enhanced by RANKL and tumor necrosis factor-α or RANKL and IL-1β treatment compared with Ctrl. Finally, osteoclastogenesis was increased by Bub1 inhibitor BAY1816032 treatment in BMMs derived from wildtype mice. These data suggest that Bub1 expressed in macrophages plays a protective role against inflammatory arthritis–associated bone loss through inhibition of inflammation–mediated osteoclastogenesis.
Keywords: rheumatoid arthritis, bone metabolism, osteoclasts, Bub1, NF-κB
Lay Summary
Rheumatoid arthritis (RA) is a disease caused by an abnormal immune system, resulting in inflammation, swelling, and bone destruction in the joints, along with systemic bone loss. While new medications have dramatically improved treatment efficacy, these therapies are not universally effective for all patients. Therefore, we need to understand the regulatory mechanisms behind RA, including associated bone loss, to develop better therapies. In this study, we found that Budding uninhibited by benzimidazoles 1 (Bub1) was highly expressed in inflamed joints, especially in myeloid cells, which are a type of immune cells. To explore its role, we created myeloid cell–specific Bub1 conditional knockout (cKO) mice and induced arthritis to analyze its role during arthritis. The cKO mice exhibited lower bone mineral density when compared with control mice under inflammatory arthritis because of increased osteoclastic bone resorption, without significant differences in joint inflammation or bone erosion. Further investigation showed that Bub1 prevents excessive osteoclast differentiation induced by inflammation in bone marrow macrophages. These data suggest that Bub1 in macrophages protects against bone loss caused by inflammatory arthritis, offering potential insights for developing treatments that focus on bone health.
Graphical Abstract
Graphical Abstract.

Introduction
Rheumatoid arthritis (RA) is an autoimmune disease with chronic inflammation, progressive bone erosion in the joint and systemic bone loss.1–4 Despite significant advances in the development of biologic disease–modifying antirheumatic drugs that inhibit proinflammatory cytokines such as tumor necrosis factor-α (TNFα) and interleukin-6 (IL-6), recent studies have suggested that a certain population of RA patients show resistance to any therapeutics5,6 and lose responsiveness over time.7 In addition, systemic comorbidities such as osteoporosis further exacerbate the symptoms of RA.1–3 It has been shown that reduced bone mineral density (BMD) in patients with RA results in increased fracture risk compared with those without RA.3,8,9 In the steady state, healthy bone homeostasis is tightly regulated by the balances between bone resorption by osteoclasts and bone formation by osteoblasts. However, the disruption of this balance in pathological conditions such as RA causes excessive bone loss. These bone characteristics are mediated by activated osteoclasts, which are differentiated from myeloid lineage cells by stimulation of macrophage–colony stimulating factor (M-CSF) and receptor activator of nuclear factor kappa B ligand (RANKL).10–12 RANKL binds to its receptor RANK that is expressed on pre–osteoclasts and activates downstream signaling pathways such as nuclear factor-kappa B (NF-κB), mitogen–activated protein kinases and activator protein-1 followed by activation of nuclear factor of activated T cells 1 (NFATc1), which is known as a master regulator of osteoclast differentiation.13–16 Furthermore, it has been reported that inflammatory cytokines, such as TNF, IL-6, and IL-1β, also drive osteoclastogenesis and enhance RANKL–mediated osteoclastogenesis in vitro, suggesting their contribution to osteoclast formation under inflammatory pathological conditions.17–19 Meanwhile, endogenous negative regulators for osteoclastogenesis such as IRF8,20 BCL6, and BLIMP121 have been identified and are important to further understand the inhibitory mechanisms of osteoclastogenesis especially during inflammatory conditions for the treatment of RA–mediated bone erosion and RA–associated osteoporosis. The downstream signaling of RANK inside pre-osteoclasts may be a possible therapeutic target for bone erosion in the joint and bone loss in RA patients.
In this study, to identify the novel molecules that regulate RA pathogenesis, we reanalyzed multiple high–throughput sequencing data sets of synovium derived from human RA patients and from murine arthritis models. Our results indicated that Budding uninhibited by benzimidazoles 1 (BUB1), which is known as a serine/threonine protein kinase,22,23 was upregulated in arthritis tissues in both groups. We also identified that Bub1 was highly expressed in myeloid cells. It was previously reported that BUB1 acts as a mitotic checkpoint to regulate cell cycle progression and thus is essential for embryonic development.24–26 However, its molecular functions in myeloid cells are largely unknown. Here, we demonstrate that Bub1 negatively controls NF-κB signaling during inflammatory arthritis in bone marrow–derived macrophages (BMMs) and suggest the possible regulatory mechanisms to inhibit osteoclast activation to protect from systemic bone loss caused by RA.
Materials and methods
Gene expression data analyses
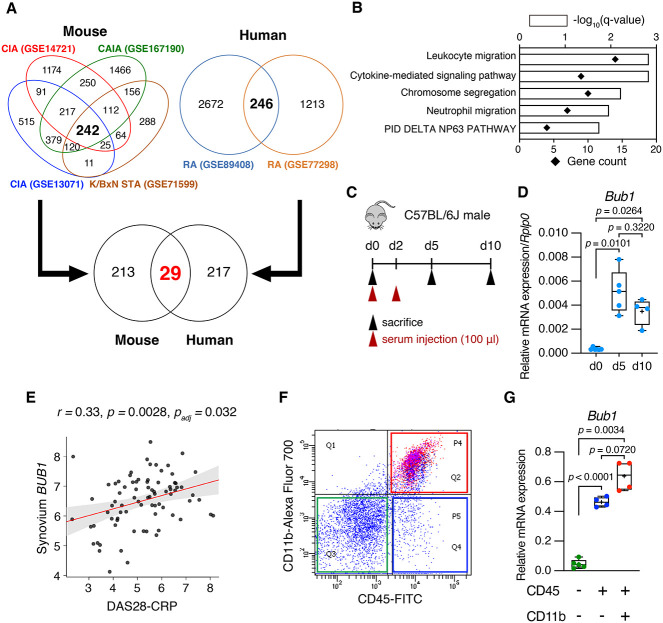
Expression profiling by high throughput sequencing data sets was obtained from Gene Expression Omnibus (GEO). GSE14721: Articular tissue from collagen–induced arthritis (CIA) (vs. adjuvant ctrl)27; GSE13071: Knee joint synovium from CIA (vs. naïve ctrl)28; GSE167190: Whole ankle tissue from collagen antibody–induced arthritis (CAIA) (vs. LPS ctrl)29; GSE71599: K/BxN serum–transfer arthritis (STA) synovium (vs. PBS ctrl)30; GSE89408: RA synovial biopsies (vs. Osteoarthritis)31; GSE77298: RA synovial biopsies (vs. healthy ctrl)32 (Figure 1A). Enrichment analysis was performed using Metascape33 (https://metascape.org/) (Figure 1B).
Figure 1.
Bub1 expression was upregulated in human RA and the murine arthritis model. (A) Venn diagram showing the number of upregulated genes in the murine arthritis models and in human RA. GSE14721: Articular tissue from CIA (vs. adjuvant ctrl); GSE13071: Knee joint synovium from CIA (vs. naïve ctrl); GSE167190: Whole ankle tissue from CAIA (vs. LPS ctrl); GSE71599: K/BxN STA synovium (vs. PBS ctrl); GSE89408: RA synovial biopsies (vs. Osteoarthritis); GSE77298: RA synovial biopsies (vs. healthy ctrl). (B) Gene ontology and pathway enrichment analysis of 29 upregulated genes in (A) by Metascape. (C) Schematic of the time schedule to induce K/BxN STA. In all, 100 μl K/BxN serum was injected twice (i.p.). (D) The expression level of Bub1 gene categorized as “chromosome segregation” was verified using the K/BxN STA model (d0: n = 6, d5: n = 5, d10: n = 4). Whole ankle tissue was collected and analyzed by RT-qPCR. (E) The PEAC study showed a significant correlation between DAS (DAS28-CRP) and BUB1 expression in synovium. (F, G) Synovial tissue was obtained from K/BxN mice and digested by type IV collagenase. Each cell population was isolated by flow cytometry. Bub1 expression in isolated cells was analyzed by RT-qPCR (CD45-CD11b-: n = 5, CD45+CD11b-: n = 4, CD45+CD11b+: n = 4). Statistical significance was determined by Brown–Forsythe and Welch ANOVA tests followed by post hoc Dunnett’s T3 multiple comparisons tests. Symbols represent individual mice.
Generation of Bub1 flox mice
To generate Bub1 flox mice, we designed two guide RNAs (gRNAs) targeting introns 2 and 3, as well as two single–stranded oligodeoxynucleotide (ssODN) containing loxP sequence with homologous sequence to each side of each gRNA (Supplementary Table S1). The gRNAs made up of a target complementary CRISPR RNA (crRNA) and an auxiliary trans–activating crRNA were synthesized using GeneArt Precision gRNA Synthesis Kit (A29377, Life Technologies, Carlsbad, CA, United States). Using electroporation (Pulse Generator CUY21EDIT II, BEX CO., LTD., Tokyo, Japan), synthesized gRNA and Cas9 protein (A36499, Life Technologies) and HPLC purified ssODN (Eurofins Genomics, Bayern, Germany) were introduced to zygotes derived from C57BL/6J mice (CLEA Japan, Tokyo, Japan). The zygotes were incubated overnight, and embryos developed to the two–cell stage were transferred to the oviduct of pseudopregnant ICR females (CLEA Japan).
Animals
To generate Bub1 conditional knockout (cKO) mice, Bub1–floxed mice were crossed with LysM-Cre mice (B6.129P2-Lyz2tm1(cre)Ifo/J; strain 004781, The Jackson Laboratory), Prrx1-Cre mice (B6.Cg-Tg (Prrx1-cre)1Cjt/J, The Jackson Laboratory), and CMV-Cre mice, kindly provided by Prof. Pierre Chambon (IGBMC). To generate K/BxN mice, KRN transgenic mice, kindly provided by Profs. C. Benoist and D. Mathis (Harvard Medical School, Boston, MA),34 were crossed with NOD/ShiJcl mice (CLEA Japan). Serum obtained from K/BxN mice was collected at 9-14 weeks of age and stored at -80°C. C57BL/6J male mice were purchased from CLEA Japan to induce K/BxN STA. All mice were housed in a specific pathogen–free facility under climate–controlled conditions with a 12–h light/12–h dark cycle and were provided with water and standard diet (MF, Oriental Yeast Co. Ltd., Tokyo, Japan) ad libitum. Animal experiments were conducted with permission of the Animal Experiment Committee of Ehime University (approval no. 37-A1-1,16 and 37A11-16) and were performed in accordance with Ehime University Guidelines for Animal Experiments.
Induction and evaluation of K/BxN STA
K/BxN STA was induced in mice (7-9 weeks old) by injecting 100 μl of K/BxN serum intraperitoneally twice to induce severe inflammation (Figure 1C and D) or once to avoid saturation of joint swelling (Figures 2D-I and 3). The development of arthritis was evaluated for 10 days by measuring hind paw thickness and a clinical score with a semiquantitative 5–point scale scoring system (0 = no evidence of erythema and swelling, 1 = erythema and mild swelling confined to the tarsals or ankle joint, 2 = erythema and mild swelling extending from the ankle to the tarsals, 3 = erythema and moderate swelling extending from the ankle to metatarsal joints, 4 = erythema and severe swelling encompass the ankle, foot and digits, or ankylosis of the paw). The data represent the mean of either paw that showed more severe phenotype as some mice showed joint swelling only one side of paw. Male mice were mainly used for the in vivo analysis because female mice did not show significant difference in BMD (Figure 3B).
Figure 2.
Bub1ΔLysM showed no difference in arthritis severity under K/BxN STA. (A) Schematic illustration showing the breeding strategy to generate myeloid cell–specific Bub1 cKO mice. (B) BMMs were collected from tibiae and KO efficiency was analyzed in BMMs by RT-qPCR (Ctrl: n = 6, Bub1ΔLysM: n = 7). (C) Cell proliferation of Ctrl (n = 13) and Bub1ΔLysM (n = 8) derived BMMs was determined by MTT assay. (D) Schematic of the time schedule to induce K/BxN STA. Overall, 100 μl K/BxN serum was injected at day 0 (i.p.). (E) Evaluation of clinical score and hind paw thickening of Ctrl (n = 11) and Bub1ΔLysM (n = 9) male mice after K/BxN STA induction. (F) Representative 3D reconstructions of the ankle joints of mice induced with K/BxN STA (d10). Scale bars: 100 μm. (G) Semiquantitative analysis of bone erosion score (Ctrl: n = 7, Bub1ΔLysM: n = 8). The scoring method is shown in Materials and Methods. (H) Representative images of histological sections of the ankle obtained from Ctrl and Bub1ΔLysM male mice induced with K/BxN STA (d10). The sections were stained to show TRAP activity and bone tissue was counterstained by fast green. Scale bars: 500 μm. (I) Quantitative bone histomorphometric analysis of the tibiae and calcaneus bone derived from Ctrl (n = 8) and Bub1ΔLysM (n = 9) mice induced with K/BxN STA. Oc.S/Cortical BS and N.Oc/Cortical B.Pm were scored and statistically compared. Statistical significance was determined by two–tailed Welch’s t-tests. Data shown as mean ± SD or box plots for each group. Symbols represent individual mice.
Figure 3.
Trabecular bone mass was decreased in Bub1ΔLysM under K/BxN STA. (A) The body weight of male (Left: Ctrl: n = 11, Bub1ΔLysM: n = 9) and female (Right: Ctrl: n = 9, Bub1ΔLysM: n = 8) after K/BxN STA (d10). (B) Femoral BMD of Ctrl and Bub1ΔLysM mice induced with K/BxN STA (d10) was analyzed by DXA (Left: male, Ctrl: n = 11, Bub1ΔLysM: n = 9, Right: female, Ctrl: n = 9, Bub1ΔLysM: n = 8). (C) Femoral BMD distribution in male (Ctrl: n = 11, Bub1ΔLysM: n = 9). (D) Representative 3D reconstructions of trabecular bone of male mice induced with K/BxN STA (d10). Scale bars: 100 μm. (E) Quantification of trabecular and cortical bone in male mice was performed using μCT (Ctrl: n = 11, Bub1ΔLysM: n = 9). (F) Representative images of histological sections of the distal femurs obtained from Ctrl and Bub1ΔLysM male mice induced with K/BxN STA (d10). The sections were stained to show TRAP activity. Scale bars: 50 μm. (G) Quantitative bone histomorphometric analysis of the trabecular bone in the distal femurs derived from Ctrl (n = 8) and Bub1ΔLysM (n = 8) male mice induced with K/BxN STA. Oc.S/BS and N.Oc/B.Pm were scored and statistically compared. (H) Representative images of toluidine blue staining. Scale bars: 50 μm. (I) Quantitative bone histomorphometric analysis. Ob.S/BS and N.Ob/B.Pm were scored and statistically compared (Ctrl: n = 8, Bub1ΔLysM: n = 8). Statistical significance was determined by two–tailed Welch’s t-tests. Data shown as mean ± SD or box plots for each group. Symbols represent individual mice.
Fluorescence–activated cell sorting
To identify Bub1 expressing inflamed synovial cells from murine ankle tissue, synovial cells were isolated as previously reported.35 Briefly, K/BxN mice (8-10 weeks old) were anesthetized and then rapidly euthanized with reflux flow of PBS. Inflamed ankles were harvested and muscle tissues were removed. Then, ankles were treated with 1–mg/mL collagenase type IV (C5138, Merck KGaA, Darmstadt, Germany) in Dulbecco’s minimum essential medium GlutaMax (DMEM GlutaMax, 10569-010, Life Technologies) supplemented with 10% fetal bovine serum (FBS, 173012, Life Technologies) and 1% antibiotic–antimycotic solution (15240-062, Life Technologies) for 3 h before filtration with a Falcon 40–μm cell strainer (352 340, Corning, NY, United States). The collected cells were treated with anti-CD45-FITC (2.5 μg/mL) (103107, BioLegend, San Diego, CA, United States) and anti-CD11b-Alexa Fluor 700 (5 μg/mL) (101222, BioLegend) for 30 min on ice. After washing the cells, each cell population (CD45+CD11b+, CD45+CD11b-, CD45-CD11b-) was defined by the fluorescent minus one (FMO) controls, and sorted using flow cytometer fluorescence–activated cell sorting (FACS) Aria (BD Biosciences, Franklin Lakes, NJ, United States).
To analyze the expression level of Bub1 in blood myeloid cells, peripheral blood was collected from control (Ctrl) and Bub1ΔLysM mice in MiniCollect with EDTA-2K (450532, Greiner Bio-One) and treated with anti-CD45-FITC and anti-CD11b-Alexa Fluor 700 for 30 min. Then, red blood cells were lysed using RBC Lysis Buffer (TNB-4300, TONBO biosciences) and sorted using FACS Aria. The CD45+CD11b+ cell population was defined by the FMO controls. Data were analyzed by BD FACS Diva software.
Radiological examination
Ankle tissues and femurs were collected from mice induced with K/BxN STA or from normal mice at 7-9 weeks old after the reflux flow of PBS and fixed with 4% paraformaldehyde (PFA) in PBS (163-20145, FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for 1 day. The BMD of femurs was measured by dual–energy X–ray absorptiometry (DXA) using a bone mineral analyzer (DCS-600EX, ALOKA, Tokyo, Japan). Hind paws and femurs were scanned with micro–computed tomography (μCT) using a Scanco Medical μCT35 System (SCANCO Medical AG, Brüttisellen, Switzerland) as previously described.36 Briefly, scans were conducted in 70% ethanol using an X–ray tube potential of 70 kVp, X–ray intensity of 114 μA, and an integration time of 400 ms with an isotropic voxel size of 6 μm. The image slices were reconstructed using Scanco μCT Version 6.1 software (Scanco Medical AG). Four bones in the ankle joint, such as the talus, calcaneus, navicular, and medial cuneiform, were scored by three blinded observers (S.Y., A.I., and Y.Y.). Erosions were scored based on a 0-3 semiquantitative scale as previously reported18 with slight modifications: 0—normal cortical bone; 1—a large cortical erosion without perforation; 2—a large cortical erosion with a small perforation; 3—a large cortical erosion with a large perforation or multiple small perforations.
Two hundred slices (1.2 mm) of the distal part of the femurs were analyzed starting 0.6 mm from the end of the growth plate according to the manufacturer’s instructions and the standardized guidelines of the American Society for Bone and Mineral Research (ASBMR).37
Histological analysis
For histological analysis, arthritic ankle tissues were fixed with 4% PFA for 1 day and then decalcified with 0.5 M EDTA (345-01865, FUJIFILM Wako Pure Chemical Corporation) for 2 weeks. The samples were embedded in paraffin after dehydration and the paraffinized samples were cut into 5–μm–thick sections with a microtome (LeicaRM2255, Leica BIO SYSTEMS, Tokyo, Japan). Tartrate-Resistant Acid Phosphatase (TRAP) staining was performed using a TRAP Stain kit (294-67 001, FUJIFILM Wako Pure Chemical Corporation). Histomorphometry was completed with the OsteoMeasure analysis system (OsteoMetrics, Inc., Decatur, GA, United States). Tibiae for six optical fields (500 × 800 μm) and whole–calcaneus bone were subjected to analyze osteoclast surface per cortical bone surface (Oc.S/Cortical BS) and osteoclast number per cortical bone perimeter (N.Oc/Cortical B.Pm).
For femoral bone histomorphometry, undecalcified femurs were embedded in methyl methacrylate (139-02726, FUJIFILM Wako Pure Chemical Corporation). Longitudinal 5–μm–thick sections were cut using a microtome. TRAP staining was performed to measure osteoclast surface per bone surface (Oc.S/BS) and osteoclast number per bone perimeter (N.Oc/B.Pm). Toluidine blue staining (T3260, Merck KGaA) was performed and osteoblast surface per bone surface (Ob.S/BS) and osteoblast number per bone perimeter (N.Ob/B.Pm) were measured. Histomorphometry of the secondary spongiosa was completed in the same region as μCT analysis for nine optical fields (750 × 1200 μm) with the OsteoMeasure analysis system according to ASBMR guidelines.38
in vitro osteoclast differentiation
Murine bone marrow (BM) cells were isolated from tibiae and femurs of 7-9–week–old mice and cultured in Minimum Essential Medium α (MEM-α, 12571-063, Life Technologies) supplemented with 10% CELLect FBS (2917354, MP Biomedicals, Santa Ana, CA, United States), and 1% antibiotic–antimycotic solution, 20–ng/mL M-CSF (139-14394, FUJIFILM Wako Pure Chemical Corporation) at 37°C in 5% CO2 humidified air for 14 h. Non–adherent cells were cultured for another 3 days with M-CSF to induce the differentiation into BMMs. Fifty– or 150–ng/mL sRANKL (47197900, Oriental Yeast Co. Ltd.) with or without 25–ng/mL mTNFα (315-01A, Life Technologies), 25–ng/mL mIL-1β (211-11B, Life Technologies), and 25–ng/mL mIL-6 (406-ML, R&D Systems, Minneapolis, MN, United States) were used to induce osteoclast differentiation. BAY1816032 (HY-103020, MedChem Express, NJ, United States), a Bub1 inhibitor, was utilized (100 nM or 1 μM, vehicle, DMSO) to test the ability to promote osteoclastogenesis. Both male and female mice were used for the in vitro analysis.
MTT assay
MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide) assay was performed using the MTT cell count kit (23506-80, Nacalai Tesque, Inc., Kyoto, Japan) according to the manufacturer’s instructions. Briefly, 1.5 × 105 of BM cells in MEM-α containing 10% CELLect FBS and 1% antibiotic–antimycotic, 20–ng/mL M-CSF solution were seeded to a 96–well plate. After 24–, 48–, or 72–h incubation, cells were treated with 0.5–mg/mL MTT solution for 2 h and lysed with 0.04 M HCl in isopropyl alcohol. Absorbance at 570 nm was measured (reference wavelength: 650 nm) using a Multiskan SkyHigh Microplate Reader (Life Technologies).
Western blotting
Cultured BMMs were washed with PBS and lysed with RIPA buffer (182-02451, FUJIFILM Wako Pure Chemical Corporation) containing a cocktail of protease inhibitor (25955-11, Nacalai Tesque, Inc.) and phosphatase inhibitor (07574-61, Nacalai Tesque, Inc.). Following lysis, samples were vortexed and incubated on ice for 30 min, then centrifuged at 20 000 g for 20 min at 4°C, and supernatants were collected. Protein concentration was determined using the Pierce BCA protein assay kit (23225, Life Technologies). Protein extract was mixed with sample buffer solution with 3-mercapto-1,2-propanediol (196-16142, FUJIFILM Wako Pure Chemical Corporation) and boiled at 95°C for 5 min. Samples were applied to SuperSep Ace polyacrylamide gels (195-14951, 192-14961, FUJIFILM Wako Pure Chemical Corporation) and subjected to SDS–polyacrylamide gel electrophoresis. Proteins were transferred to PVDF membranes (1620177, Bio-Rad Laboratories, Hercules, CA, United States) using a mini transblot cell (Bio-Rad Laboratories). The membranes were blocked with 5% bovine serum albumin (BSA, 013-27054, FUJIFILM Wako Pure Chemical Corporation) in TBS containing 0.05% Tween20 (TBS-T) for 1 h at room temperature. The membranes were reacted with primary antibodies at 4°C overnight. The primary antibodies were: anti-IκBα (4814, Cell Signaling Technology (CST), MA, United States, 1:1000), anti-Phospho-IκBα (Ser32) (2859, CST, 1:500), and anti-β-actin (M177-3, MEDICAL & BIOLOGICAL LABORATORIES CO., LTD., Tokyo, Japan, 1:10 000). After washing with TBS-T, membranes were incubated with secondary antibodies, including horseradish peroxidase (HRP)–conjugated anti–rabbit IgG (7074, CST, 1:2000) and HRP–conjugated anti–mouse IgG (7076, CST, 1:2000), for 1 h at room temperature. ECL Prime Western Blotting Detection Reagent (RPN2232, Cytiva, Tokyo, Japan) was used to detect signals, and membranes were imaged using Amersham ImageQuant800 (Cytiva). Quantification of the intensity was determined using Image J wand tool.
Immunocytochemistry
Cells were fixed with 4% PFA in PBS for 10 min, followed by permeabilization with PBS containing 0.5% Triton X-100 for 5 min. Cells were blocked using 1% BSA, 0.02% Triton X-100 containing PBS. The following primary antibodies were used: anti-Ki67 (14-5698-82, Life technologies, 1:200) and anti-NF-κB p65 (8242, CST, 1:200). The signals were visualized using the following fluorochrome–coupled secondary antibodies: Alexa fluor 488 goat anti–rabbit IgG (A11008, Life Technologies, 1:400) and Alexa fluor 568 goat anti–rat IgG (A11077, Life Technologies, 1:400). Nuclei were counterstained with DAPI. Stained cells were photographed with Axio Observer (Carl Zeiss Co., Ltd., Oberkochen, Germany) and analyzed using Fiji (v2.14.0) (https://imagej.net/Fiji). The numbers of Ki67+ or NF-κB p65+ cells were determined using the Fiji Analyze Particles function.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using ISOGEN (319-90211, Nippon Gene CO., LTD., Tokyo, Japan) and the RNeasy spin column kit (74106, Qiagen, Venlo, The Netherlands). Isolated total RNA was reverse-transcribed into first–strand cDNA using Prime Script RT Master Mix (R045A, Takara Bio Inc., Kusatsu, Japan). RT-qPCR was performed using TB Green Premix Ex Taq II (RR820L, Takara Bio Inc.) with the Thermal Cycler Dice Real–Time System (Takara Bio Inc.). Gene expression was normalized to that of Rplp0 as a housekeeping gene. Primer sequences are listed in Supplementary Table S2.
RNA-seq analysis
BM cells were isolated from tibiae and treated with M-CSF and RANKL (Figure 4A). High–quality total RNA was obtained using RNeasy spin column kits and verified using an Agilent 2100 Bioanalyzer. RNA-seq was performed by Kazusa DNA Research Institute using an Illumina NextSeq 500 with a read configuration of 75 bp for single reads; 1 million reads were generated per sample. Obtained FASTQ files were analyzed by RaNA-seq with the DESeq2 package.39 The P value cutoff was set to 0.05. Data were registered in GEO with the accession number GSE241409. Gene Set Enrichment Analysis (GSEA) was conducted by RaNA-seq. For the Ingenuity Pathway Analysis (IPA), upregulated and downregulated genes (log2FC < -1, 1 < log2FC) in Bub1ΔLysM–derived BMMs obtained from RNA-seq analysis were analyzed.
Figure 4.
NF-κB pathway was enhanced in Bub1ΔLysM–derived BMMs. (A) Schematic illustrating the induction of osteoclast differentiation. (B) Representative images of TRAP staining of BMMs. In all, 150–ng/mL RANKL was treated. Scale bars: 200 μm. (C) The number of multinucleated TRAP+ cells (Nuclei ≥ 5) (Ctrl: n = 3, Bub1ΔLysM: n = 3). (D) Expression of osteoclast differentiation marker genes were analyzed by RT-qPCR. In all, 50–ng/mL RANKL was treated (Ctrl: n = 3, Bub1ΔLysM: n = 3). (E) PCA of gene expression profiles obtained by RNA-seq analysis (Ctrl: n = 3, Bub1ΔLysM: n = 3). (F) Heatmap of expression values normalized as transcripts per million (TPM) in DEGs of Ctrl and Bub1ΔLysM–derived BMMs on day 2 after RANKL (50 ng/mL) treatment. (G) Volcano plot showing log2 fold change (log2 FC) and statistical significance (Padj value) of differences between Ctrl and Bub1ΔLysM BMMs. Padj value cutoff was set to 0.05. (H) Significantly enriched pathways among upregulated genes by GSEA. Statistical significance was determined by two–tailed Welch’s t-tests (C) or two–way ANOVA tests followed by post hoc Šídák’s multiple comparison tests (D). Data represent means ± SD for each group. Symbols represent individual mice.
Statistical analysis
Statistical analysis was performed using Graphpad Prism version 10.0. Two–tailed unpaired Welch’s t-tests were conducted to analyze differences between two groups. Brown–Forsythe and Welch ANOVA tests followed by post hoc Dunnett’s T3 tests or two–way ANOVA tests followed by post hoc Šídák’s tests were applied for multiple comparisons. The in vivo data are presented as box plots where the horizontal line represents the median, the box outlines the interquartile range, and the bars represent maximum and minimum (means are shown by the plus mark). The in vitro data are presented as scatter dot plots with mean ± SD. Each dot corresponds to an individual mouse. Statistical significance was accepted when P values were <0.05.
Results
Bub1 was upregulated in RA synovium and highly expressed in myeloid cells among inflamed synovial cells
To explore upregulated genes in RA pathogenesis, we reanalyzed multiple high–throughput sequencing data sets of synovium derived from human RA patients and murine arthritis models that were deposited in GEO. In inflamed murine synovium and RA patients’ synovium, 242 genes and 246 genes were upregulated, respectively, compared with the Ctrl group of each data set. We identified 29 genes that were commonly upregulated in mouse and human synovial tissue (Figure 1A). Enrichment analysis for these 29 genes showed “leukocyte migration” and “cytokine-mediated signaling pathway,” which are well–established RA regulatory pathways, followed by “chromosome segregation” (Figure 1B). Because the relationship between genes involved in chromosome segregation and RA pathogenesis was unclear, we decided to focus on genes involved in “chromosome segregation,” which contained 10 genes (BUB1, BUB1B, MKI67, TOP2A, NCAPG, NUF2, RRM2, CCNB2, HELLS, S100A9) as candidate genes to be analyzed. To verify expression levels of these genes in inflamed synovial tissue, we generated the K/BxN STA mouse model by injecting serum derived from K/BxN mice, which spontaneously develop autoimmune arthritis, to wildtype C57BL/6J male mice at days 0 and 2 (Figure 1C). We collected whole–inflamed ankle tissue at days 5 and 10. RT-qPCR data showed that Bub1, which is a serine/threonine protein kinase,22,23 was the most highly expressed at day 5 and sustained to express until day 10 compared with day 0 (Figure 1D) among the candidate genes. Furthermore, the Pathobiology of Early Arthritis Cohort (PEAC) study (https://peac.hpc.qmul.ac.uk) showed that BUB1 expression in RA patients’ synovium was positively correlated with disease activity score (DAS) 28-CRP, which is a common RA disease activity scoring scale (r = 0.33, P = .0028, Padj = .032) (Figure 1E). Because BUB1 was significantly correlated with disease and was the most upregulated expression among genes involved in chromosome segregation, we analyzed Bub1’s functions in arthritis pathogenesis. To identify synovial cells that highly express Bub1 in arthritis, we collected inflamed synovial cells from K/BxN mice, isolated CD45+CD11b+ myeloid cells, CD45+CD11b- lymphocytes and CD45-CD11b- cells by FACS (Figure 1F), and conducted RT-qPCR using sorted cells. This showed that Bub1 expression was the highest in CD45+CD11b+ myeloid cells among the sorted cell populations (Figure 1G).
Bub1 flox mice were generated to create Bub1 cKO mice
To generate Bub1 cKO mice, we took advantage of the CRISPR Cas9 system and created Bub1 flox mice (Supplementary Figure S1A). Generated Bub1 flox mice were crossed with CMV-Cre mice to confirm the recombination capacity. As expected, the exon3 region between two loxP sites was nicely excised from CMV-Cre;Bub1flox/+ mice (Supplementary Figure S1B). CMV-Cre;Bub1flox/+ mice were further crossbred with WT mice to generate Bub1+/Δ mice and they were intercrossed with each other to confirm the embryonic lethality that is a common feature of systemic Bub1 KO mice.24–26 The births of Bub1+/+ and Bub1+/Δ occurred at an approximate ratio of 1:2 according to Mendel's laws, whereas the birth of Bub1Δ/Δ was not observed (Supplementary Figure S1C). We also crossed Bub1 flox mice with Prrx1-Cre mice, which express Cre specifically in the limb mesenchyme, to further confirm specific deletion of Bub1 in mesenchymal lineage cells. We could not confirm any living Bub1ΔPrx-1 neonate mice, indicating that Bub1ΔPrx-1 mice are embryonic or neonatally lethal. Therefore, we observed Bub1ΔPrx-1 at E18.5, which showed almost undeveloped forelimbs and tiny hindlimbs (Supplementary Figure S1D). These data suggested that the establishment of Bub1 flox mice was successful and we coincidentally found that Bub1 is indispensable for normal limb development.
Bub1ΔLysM showed no difference in arthritis severity under K/BxN STA
To analyze physiological functions of Bub1 in myeloid cells during inflammatory arthritis, we crossed Bub1 flox mice with LysM-Cre mice to generate Bub1 cKO mice (Bub1ΔLysM) (Figure 2A). As macrophages among myeloid cells regulate inflammation, and osteoclasts derived from macrophage lineage cells control bone homeostasis and bone erosion in arthritis, we first evaluated the KO efficiency of BMM–like cells and analyzed their proliferation to confirm the Bub1 function as a mitotic checkpoint. BM cells were treated with M-CSF to induce the differentiation to BMMs. RT-qPCR analysis demonstrated that Bub1 expression was significantly lower in BMMs obtained from Bub1ΔLysM compared with Ctrl (Bub1flox/+, Bub1flox/flox, LysM-Cre;Bub+/+) (Figure 2B). On the other hand, Bub1 expression in peripheral blood myeloid cells (Supplementary Figure S2A) were low and observed no significant difference in normal condition and even in the K/BxN STA day 10 (Supplementary Figure S2B). In addition, there was no significant difference in the ratio of CD45+CD11b+ cells between Ctrl and Bub1ΔLysM mice (Supplementary Figure S2C). MTT assay of BMMs was performed to confirm Bub1’s contribution to cell proliferation, but there was no significant difference between cells obtained from Ctrl and Bub1ΔLysM (Figure 2C). In addition, there was no significant difference in the Ki67+ cell ratio after 3 days of culture between the cells obtained from Ctrl and Bub1ΔLysM (Supplementary Figure S3A and B). These results suggested that Bub1’s function in cell division as a mitotic checkpoint did not affect BMM–like cells. Next, K/BxN STA was induced to Ctrl and Bub1ΔLysM mice to analyze the severity of arthritis. However, there was no significant difference in the hind paw clinical score or thickening between Ctrl and Bub1ΔLysM mice (Figure 2D and E). We further analyzed bone erosion of the hind paw by μCT. Semiquantitative analysis of 3D reconstructed images showed no significant difference between Ctrl and Bub1ΔLysM mice (Figure 2F and G). We also performed TRAP staining in inflamed ankle tissue followed by histomorphometry on cortical bone surface. However, no significant difference was observed in Oc.S/Cortical BS and the N.Oc/Cortical B.Pm (Figure 2H and I). These data suggest that the effect of Bub1 functions in myeloid cells is limited in the joint swelling and hind paw bone erosion of cortical bone surface under K/BxN STA.
Trabecular bone mass was decreased in Bub1ΔLysM under K/BxN STA
To analyze the contribution to bone homeostasis of Bub1 expressed in pre-osteoclasts and osteoclasts, we analyzed femoral BMD at K/BxN STA day 10. There was no significant difference in body weight (Figure 3A) but DXA analysis showed that femoral BMD was significantly lower in Bub1ΔLysM compared with Ctrl in male, whereas there was no significant difference in female mice (Figure 3B). Furthermore, BMD was reduced in the distal femurs in male Bub1ΔLysM (Figure 3C). Therefore, the bone microstructure of the distal femoral region was analyzed using μCT. Trabecular BMD (Tb. BMD), bone volume/tissue volume, and trabecular thickness were significantly lower in Bub1ΔLysM compared with Ctrl, whereas there were no significant changes in cortical bone parameters including BMD, cortical area fraction (Ct.Ar/Tt.Ar), and cortical thickness (Figure 3D and E). Furthermore, the structure model index was higher in Bub1ΔLysM, suggesting trabecular bone became structurally more rod–like rather than plate–like compared with Ctrl mice during arthritis pathogenesis. To determine whether this difference was caused by osteoclasts in vivo, TRAP staining of the distal femurs was performed. Bone histomorphometry analysis revealed higher Oc.S/BS and the N.Oc/B.Pm in Bub1ΔLysM compared with Ctrl (Figure 3F and G). We also performed toluidine blue staining to analyze osteoblastic bone formation, but there was no significant difference in Ob.S/BS and the N.Ob/B.Pm between Ctrl and Bub1ΔLysM mice (Figure 3H and I). Although body weight was significantly higher in male Bub1ΔLysM mice than Ctrl, bone loss was not observed in Bub1ΔLysM mice without arthritis in both male and female (Supplementary Figure S3C-E), indicating osteoclast activation in Bub1ΔLysM was distinctive to arthritis pathology.
NF-κB pathway was enhanced in Bub1ΔLysM–derived BMMs
To examine the changes in expression level of osteoclastic marker genes, we isolated BM cells from Ctrl and Bub1ΔLysM, and induced osteoclast differentiation by RANKL stimulation after pre-cultivation with M-CSF (Figure 4A). M-CSF and RANKL treatment induced osteoclast differentiation in Ctrl and Bub1ΔLysM and there was no significant difference in the number of TRAP positive multinucleated cells (Figure 4B and C). Although expression of Bub1 was significantly lower, with the exception of Acp5 at day 4, there were no differences in osteoclast differentiation marker genes between BMMs from Ctrl and Bub1ΔLysM (Figure 4D). Next, to analyze the comprehensive gene expression profiles of Ctrl and Bub1ΔLysM mice–derived BMMs, BMMs were collected at day 2 after RANKL stimulation and RNA-seq was performed (Figure 4A) because expression of Bub1 in Ctrl BMMs tended to decrease at day 4 (Figure 4D). Principal components analysis (PCA) and hierarchical clustering analysis showed differential gene expression profiles between BMMs isolated from Ctrl and Bub1ΔLysM mice (Figure 4E and F). Differentially expressed genes (DEGs) were visualized with a volcano plot (18 genes upregulated and 11 genes downregulated) (Figure 4G). Subsequent GSEA indicated significantly enriched NF-κB pathways among upregulated genes in RANKL–stimulated BMMs obtained from Bub1ΔLysM (Figure 4H), whereas limited pathways were enriched among downregulated genes (Supplementary Figure S4A). Furthermore, IPA predicted indirect activation of several inflammatory pathways linked to IL-1B (Supplementary Figure S4B).
Bub1ΔLysM mice–derived BMMs were sensitive to TNFα stimulation
As TNFα greatly contributes to RA pathogenesis40 and is the predominant factor together with RANKL that stimulates the NF-κB pathway to induce osteoclast differentiation, we tried to verify whether Bub1 KO macrophages are sensitive to TNFα stimulation. We collected BMMs from Ctrl and Bub1ΔLysM mice and treated them with RANKL and TNFα (Figure 5A). TRAP staining revealed more multinucleated TRAP positive cells in Bub1ΔLysM mice than in Ctrl mice (Figure 5B and C). The number of osteoclasts, which contain 5-9 nuclei, was significantly higher in Bub1ΔLysM, whereas there was no significant difference in the number of osteoclasts which have 10 nuclei or more (Figure 4D). Furthermore, BMMs from Bub1ΔLysM mice showed significantly higher expression of osteoclastic markers such as Nfatc1, Dcstamp, Acp5, and Ctsk at day 4 (Figure 5E). There was no significant change in the expression of Tnfrsf11a even after RANKL and TNFα stimulation (Figure 5E). In addition, Bub1 expression level was significantly elevated in Ctrl but not in Bub1ΔLysM–derived BMMs when TNFα was treated (Figure 5F), suggesting Bub1 expression was also increased in the BMMs under inflammatory conditions. To analyze the activation of NF-κB, we evaluated phosphorylation of IκBα (Ser32), which induces its proteasomal degradation followed by release and nuclear translocation of active NF-κB.41 RANKL and TNFα treatment along with M-CSF significantly promoted the phosphorylation of IκBα in Bub1ΔLysM than Ctrl–derived BMMs (Figure 5G and H). Furthermore, the localization of NF-κB component p65 to the nucleus was significantly greater in Bub1ΔLysM–derived BMMs than in Ctrl BMMs (Supplementary Figure 5I and J). We also treated BMMs with IL-1β or IL-6, which are important proinflammatory cytokines for RA pathogenesis,40,42 along with RANKL to determine the effect of these cytokines for Bub1–related osteoclast differentiation (Supplementary Figure S5). RANKL and IL-1β treatment also promoted osteoclast differentiation with greater number of nuclei per cells in Bub1ΔLysM–derived BMMs as indicated in IPA (Supplementary Figure S5B-D). Furthermore, the expression of Ctsk was significantly higher and the other markers also tended to be high in Bub1ΔLysM than Ctrl–derived BMMs with no significant difference in Tnfrsf11a expression (Supplementary Figure S5E). On the other hand, RANKL and IL-6 treatment did not induce significant difference between Ctrl and Bub1ΔLysM in the number of TRAP positive multinucleated cells (Supplementary Figure S5G and H) and the expression levels of osteoclast differentiation marker genes (Supplementary Figure S5I). These data suggested that proinflammatory signaling pathway such as NF-κB but not JAK–STAT pathway was enhanced in Bub1ΔLysM BMMs.
Figure 5.
BMMs derived from Bub1ΔLysM mice are sensitive to TNFα stimulation. (A) Schematic illustrating the induction of osteoclast differentiation. (B) Representative images of TRAP staining of BMMs. Scale bars: 200 μm. (C) The number of multinucleated TRAP+ cells (Nuclei ≥ 5) (Ctrl: n = 6, Bub1ΔLysM: n = 6). (D) The number of multinucleated TRAP+ cells in each nuclei number group (Ctrl: n = 6, Bub1ΔLysM: n = 6). (E) Expression levels of osteoclast differentiation marker genes were analyzed by RT-qPCR (Ctrl: n = 3, Bub1ΔLysM: n = 3). (F) Expression level of Bub1 in Ctrl and Bub1ΔLysM BMMs with or without TNFα was analyzed by RT-qPCR (d4, Ctrl: n = 3, Bub1ΔLysM: n = 3). (G, H) BMMs were treated with RANKL and TNFα for 30 min and phosphorylation of IκBα (Ser32) was analyzed by western blot (Ctrl: n = 8, Bub1ΔLysM: n = 8). (I) Immunostaining of BMMs after 3 h of RANKL and TNFα treatment. Scale bars: 50 μm. (J) The ratio of nuclear localizing p65+ cells (Ctrl: n = 6, Bub1ΔLysM: n = 6). Statistical significance was determined by two–tailed Welch’s t-tests (C, E, H, J) or by two–way ANOVA tests followed by post hoc Šídák’s multiple comparison tests (D, F). Data represent means ± SD for each group. Symbols represent individual mice.
Bub1–inhibited BMMs were sensitive to TNFα stimulation
Aneuploidy is one of the common features among many cancer cells and an excessive degree of aneuploidy caused by chromosome mis-segregation leads to cell cycle arrest and cell death. In this context, the cell cycle checkpoint regulation may be a novel approach to cancer treatment and BAY1816032 was identified as a catalytic Bub1 inhibitor that directly binds to its ATP–binding pocket.43 Treatment of BMMs with BAY1816032 significantly increased osteoclastogenesis at a concentration of 1 μM but not at the lower concentration of 100 nM compared with the DMSO ctrl (Figure 6A-C). Taken together, these data showed that Bub1 negatively regulates NF-κB signaling in response to proinflammatory cytokines such as TNFα and IL-1β stimulation. Thus, osteoclast differentiation is increased in Bub1 KO macrophages and macrophages treated with Bub1 inhibitor under inflammatory condition.
Figure 6.
Bub1 inhibitor BAY1816032 promotes osteoclastogenesis in vitro. (A) Schematic illustrating the induction of osteoclast differentiation with the treatment of Bub1 inhibitor BAY1816032. (B) Representative images of TRAP staining of BMMs derived from wild–type mice at different concentrations of BAY1816032 (BAY). Scale bars: 200 μm. (C) Number of multinucleated TRAP+ cells (Nuclei ≥ 5) at different concentrations of BAY (n = 3). Statistical significance was determined by Brown–Forsythe and Welch ANOVA tests followed by post hoc Dunnett’s T3 multiple comparisons tests. Data represent means ± SD for each group. Symbols represent individual mice.
Discussion
We have explored upregulated genes in RA pathogenesis to further understand the mechanisms underlying arthritis pathology and identified Bub1 as an inhibitory factor of TNFα or IL-1β–mediated NF-κB signaling in BMMs that suppresses their differentiation into osteoclasts to attenuate bone loss during inflammatory arthritis. Bub1 is a protein kinase for the spindle assembly checkpoint (SAC) that is a surveillance mechanism to ensure proper chromosome alignment and segregation to prevent aneuploidy.22,23 In mammalian cells such as HeLa, Bub1 directly phosphorylates Cdc20, an Anaphase promoting complex/cyclosome coactivator,44 and Histone H2A45 to promote SAC inhibition of metaphase to anaphase progression until all chromosomes are correctly attached to microtubules. However, the functional analysis of SAC in vivo is limited because of the early embryonic lethality of SAC components including Bub1, BubR1, Bub3, Mad1, and Mad2.24–26,46–48 Therefore, the detailed functions of Bub1 in physiological or pathological conditions, and cell type–dependent mechanisms especially in adults are poorly understood. In this study, we identified that the expression of Bub1 mRNA was significantly elevated in inflamed synovium but failed to detect increased Bub1 protein level because of poor anti–murine Bub1 antibody efficacy. We generated Bub1 flox mice to determine the cell type specific molecular functions of Bub1. Viable CMV-Cre–mediated systemic KO mice were never observed as previously reported24–26 and shown in this study (Figure S1C). On the other hand, Bub1ΔLysM mice were viable, fertile, and showed no significant difference in cell proliferation of cultured BMMs (Figures 2C and S3A and B), indicating that the distinctive role of Bub1 is dependent on the cell type. LysM positive myeloid cells include the macrophage and granulocyte lineages. However, because of the difficulty in culturing granulocytes because of the short lifetime, we mainly focused on macrophages and osteoclasts in this study. The involvement of Bub1 in granulocytes such as Ly6G+ neutrophils in pathological conditions needs to be elucidated. While Bub1ΔLysM mice exhibited a normal arthritis phenotype, Bub1ΔLysM mice showed significantly lower femoral Tb. BMD only in K/BxN STA, but not under physiological conditions (Figures 3 and S3C-E). These data suggest that the trabecular bone loss was caused by activated osteoclastic bone resorption (Figure 3F and G). We also identified that the expression of Bub1 was higher in Ctrl mice–derived BMMs when TNFα was treated (Figure 5F). These data suggest Bub1 expression was elevated in osteoclasts or osteoclast progenitor cells as well as synovium to attenuate the systemic bone loss during arthritis. The bone phenotype was only observed in male mice that may be because of the sexual difference in the K/BxN STA–mediated severity of arthritis.
We also performed RNA-seq and GSEA to analyze DEGs and identified significantly enriched NF-κB signaling and increased osteoclast formation in response to RANKL and TNFα or RANKL and IL-1β, but not IL-6, in Bub1 KO BMMs (Figures 5 and S5). Recent studies have shown that Bub1 kinase activity promotes TGF-β signaling in murine tumor tissues, leading to the activation of SMADs followed by an epithelial mesenchymal transition or proliferation of liver cancer cells.49,50 Thus, these data suggest the novel roles of Bub1 independent from cell cycle regulation and also propose a possible mechanism to regulate several types of signal transduction. In this context, Bub1 emerges as a potential negative regulator of TNFα or IL-1β–mediated NF-κB signaling and osteoclastogenesis in BMMs during inflammatory arthritis (Figure 5). However, the exact substrate of Bub1 in NF-κB signaling has not been reported and we could not identify the substrate in this study. This is one limitation of this study.
While there was no significant difference in periarticular bone erosion in our experiment (Figure 2F-I), a question remains whether Bub1 does not affect periarticular bone erosion during arthritis because inflammation converges within a relatively short period of time in the K/BxN STA model. To address this issue, CIA is much more suitable. However, the CIA model shows lower incidence and severity in C57BL/6 mouse lines than those of susceptible strains such as DBA1.51 Thus, this is another limitation of this study. On the other hand, it has been shown that osteoclasts in pannus exclusively originate from circulating BM–derived cells in arthritis (arthritis–associated osteoclastogenic macrophages: AtoMs) and the differentiation of AtoMs into osteoclasts is regulated by Foxm1.52 Moreover, we show that blood myeloid lineage cells including monocytes scarcely express Bub1 even in pathological conditions (Supplementary Figure S2). Our data and this report suggest that the characteristics of macrophages in blood, BM, and synovium are diversely different. These differences may explain why Bub1ΔLysM mice did not exhibit less bone erosion, whereas femoral bone mass was decreased. Also, Hasegawa et al.52 revealed that Foxm1 KO mice did not have an abnormal bone phenotype under physiological conditions, similar to Bub1ΔLysM mice. This evidence suggests that it is important to target the molecule only under pathological conditions to avoid off–target adverse effects of the treatment. Targeting the molecule to suppress inflammation for RA patients is essential, but at the same time it is important to reduce the fracture risks to RA patients.
In summary, we identified Bub1 as one of the upregulated genes in inflammatory arthritis that may have potential to inhibit osteoporosis in RA. Moreover, the expression level of BUB1 in synovium was positively correlated with DAS (Figure 1E), suggesting BUB1 was upregulated to attenuate the bone loss in RA. Further studies are required to confirm the possibility whether Bub1 could be a novel candidate for the treatment of RA–associated bone loss and bone erosion.
Supplementary Material
Acknowledgments
We gratefully thank the members of the Advanced Research Support Center (ADRES) and the members of the Division of Integrative Pathophysiology, Proteo-Science Center (PROS) of Ehime University for their technical assistance and helpful support.
Contributor Information
Shuhei Yoshida, Department of Pathophysiology, Ehime University Graduate School of Medicine, Toon, Ehime, 791-0295, Japan.
Aoi Ikedo, Division of Integrative Pathophysiology, Proteo-Science Center (PROS), Ehime University, Toon, Ehime, 791-0295, Japan.
Yuta Yanagihara, Division of Integrative Pathophysiology, Proteo-Science Center (PROS), Ehime University, Toon, Ehime, 791-0295, Japan.
Tomohisa Sakaue, Department of Cardiovascular and Thoracic Surgery, Ehime University Graduate School of Medicine, Toon, Ehime, 791-0295, Japan; Division of Cell Growth and Tumor Regulation, Proteo-Science Center (PROS), Ehime University, Toon, Ehime, 791-0295, Japan.
Noritaka Saeki, Division of Integrative Pathophysiology, Proteo-Science Center (PROS), Ehime University, Toon, Ehime, 791-0295, Japan; Division of Medical Research Support, Advanced Research Support Center, Ehime University, Toon, Ehime, 791-0295, Japan.
Yuuki Imai, Department of Pathophysiology, Ehime University Graduate School of Medicine, Toon, Ehime, 791-0295, Japan; Division of Integrative Pathophysiology, Proteo-Science Center (PROS), Ehime University, Toon, Ehime, 791-0295, Japan.
Author contributions
Shuhei Yoshida (Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing—original draft, visualization), Aoi Ikedo (Methodology, Formal analysis, Investigation, Writing—review and editing), Yuta Yanagihara (Methodology, Formal analysis, Investigation, Writing—review and editing), Tomohisa Sakaue (Resources), Noritaka Saeki (Conceptualization, Supervision, Writing—review and editing), and Yuuki Imai (Conceptualization, Methodology, Resources, Funding acquisition, Project administration, Supervision, Writing—review and editing)
Funding
This study was supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI grants JP23689066, JP17K19728, JP19H03786, and JP22H03203 and Chugai Foundation for Innovative Drug Discovery Science: C-FINDs (to Y.I.).
Conflicts of interest
None declared.
Data availability
All RNA-seq data generated and analyzed during this study are available in the NCBI Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/gds. The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;55(12):2255–2270. [DOI] [PubMed] [Google Scholar]
- 2.Haugeberg G, Uhlig T, Falch JA, Halse JI, Kvien TK. Bone mineral density and frequency of osteoporosis in female patients with rheumatoid arthritis: results from 394 patients in the Oslo County rheumatoid arthritis register. Arthritis Rheum. 2000;43(3):522–530. . [DOI] [PubMed] [Google Scholar]
- 3.Wright NC, Lisse JR, Walitt BT, Eaton CB, Chen Z, Women's Health Initiative Investigators . Arthritis increases the risk for fractures — results from the women’s health initiative. J Rheumatol. 2011;38(8):1680–1688. 10.3899/jrheum.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8(11):656–664. 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamanaka H, Tanaka E, Nakajima A, et al. A large observational cohort study of rheumatoid arthritis, IORRA: providing context for today’s treatment options. Mod Rheumatol. 2020;30(1):1–6. 10.1080/14397595.2019.1660028. [DOI] [PubMed] [Google Scholar]
- 6.Bécède M, Alasti F, Gessl I, et al. Risk profiling for a refractory course of rheumatoid arthritis. Semin Arthritis Rheum. 2019;49(2):211–217. 10.1016/j.semarthrit.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 8.Juhász B, Gulyás K, Horváth Á, et al. Comparison of peripheral quantitative computed tomography forearm bone density versus DXA in rheumatoid arthritis patients and controls. Osteoporos Int. 2017;28(4):1271–1277. 10.1007/s00198-016-3850-x. [DOI] [PubMed] [Google Scholar]
- 9.Kocijan R, Finzel S, Englbrecht M, Engelke K, Rech J, Schett G. Decreased quantity and quality of the periarticular and nonperiarticular bone in patients with rheumatoid arthritis: a cross-sectional HR-pQCT study. J Bone Miner Res. 2014;29(4):1005–1014. 10.1002/jbmr.2109. [DOI] [PubMed] [Google Scholar]
- 10.Arai F, Miyamoto T, Ohneda O, et al. Commitment and differentiation of osteoclast precursor cells by the sequential expression of C-Fms and receptor activator of nuclear factor κb (rank) receptors. J Exp Med. 1999;190(12):1741–1754. 10.1084/jem.190.12.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda H, Shima N, Nakagawa N, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci. 1998;95(7):3597–3602. 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacey DL, Timms E, Tan H-L, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. 10.1016/S0092-8674(00)81569-X. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto K, Nakashima T, Shinohara M, et al. Osteoimmunology: the conceptual framework unifying the immune and skeletal systems. Physiol Rev. 2017;97(4):1295–1349. 10.1152/physrev.00036.2016. [DOI] [PubMed] [Google Scholar]
- 14.Tsukasaki M, Takayanagi H. Osteoimmunology: evolving concepts in bone–immune interactions in health and disease. Nat Rev Immunol. 2019;19(10):626–642. 10.1038/s41577-019-0178-8. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu N, Takayanagi H. Mechanisms of joint destruction in rheumatoid arthritis — immune cell–fibroblast–bone interactions. Nat Rev Rheumatol. 2022;18(7):415–429. 10.1038/s41584-022-00793-5. [DOI] [PubMed] [Google Scholar]
- 16.Takayanagi H, Kim S, Koga T, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 17.Yokota K, Sato K, Miyazaki T, et al. Combination of tumor necrosis factor α and interleukin-6 induces mouse osteoclast-like cells with bone resorption activity both in vitro and in vivo. Arthritis Rheumatol. 2014;66(1):121–129. 10.1002/art.38218. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien W, Fissel BM, Maeda Y, et al. RANK-independent osteoclast formation and bone erosion in inflammatory arthritis. Arthritis Rheumatol. 2016;68(12):2889–2900. 10.1002/art.39837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Z, Getting SJ, Locke IC. Regulation of TNF-induced osteoclast differentiation. Cell. 2021;11(1):132. 10.3390/cells11010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, Takami M, Yamada A, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15(9):1066–1071. 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyauchi Y, Ninomiya K, Miyamoto H, et al. The Blimp1–Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207(4):751–762. 10.1084/jem.20091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams GL, Roberts TM, Gjoerup OV. Bub1: escapades in a cellular world. Cell Cycle. 2007;6(14):1699–1704. 10.4161/cc.6.14.4493. [DOI] [PubMed] [Google Scholar]
- 23.Kim T, Gartner A. Bub1 kinase in the regulation of mitosis. Anim Cells Syst. 2021;25(1):1–10. 10.1080/19768354.2021.1884599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179(2):255–267. 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, Taylor SS. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13(4):566–579. 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Tilston V, Taylor SS, Perera D. Inactivating the spindle checkpoint kinase Bub1 during embryonic development results in a global shutdown of proliferation. BMC Res Notes. 2009;2(1):190. 10.1186/1756-0500-2-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godina-Gonzalez S, Furuzawa-Carballeda J, Utrera-Barillas D, et al. Amebic monocyte locomotion inhibitory factor peptide ameliorates inflammation in CIA mouse model by downregulation of cell adhesion, inflammation/chemotaxis, and matrix metalloproteinases genes. Inflamm Res. 2010;59(12):1041–1051. 10.1007/s00011-010-0224-2. [DOI] [PubMed] [Google Scholar]
- 28.Geurts J, Joosten LA, Takahashi N, et al. Computational design and application of endogenous promoters for transcriptionally targeted gene therapy for rheumatoid arthritis. Mol Ther. 2009;17(11):1877–1887. 10.1038/mt.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeki N, Inoue K, Ideta-Otsuka M, et al. Epigenetic regulator UHRF1 suppressively orchestrates pro-inflammatory gene expression in rheumatoid arthritis. J Clin Invest. 2022;132(11):e150533. 10.1172/JCI150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda Y, Farina NH, Matzelle MM, Fanning PJ, Lian JB, Gravallese EM. Synovium-derived MicroRNAs regulate bone pathways in rheumatoid arthritis. J Bone Miner Res. 2017;32(3):461–472. 10.1002/jbmr.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh AM, Wechalekar MD, Guo Y, et al. Triple DMARD treatment in early rheumatoid arthritis modulates synovial T cell activation and plasmablast/plasma cell differentiation pathways. PLoS One. 2017;12(9):e0183928. 10.1371/journal.pone.0183928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broeren MGA, VM d, Bennink MB, et al. Disease-regulated gene therapy with anti-inflammatory interleukin-10 under the control of the CXCL10 promoter for the treatment of rheumatoid arthritis. Hum Gene Ther. 2016;27(3):244–254. 10.1089/hum.2015.127. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouskoff V, Korganow A-S, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87(5):811–822. 10.1016/S0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 35.Saeki, Imai Y. Reprogramming of synovial macrophage metabolism by synovial fibroblasts under inflammatory conditions. Cell Commun Signal. 2020;18(1):188. 10.1186/s12964-020-00678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto Y, Yoshizawa T, Fukuda T, et al. Vitamin D receptor in osteoblasts is a negative regulator of bone mass control. Endocrinology. 2013;154(3):1008–1020. 10.1210/en.2012-1542. [DOI] [PubMed] [Google Scholar]
- 37.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro–computed tomography. J Bone Miner Res. 2010;25(7):1468–1486. 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 38.Dempster DW, Compston JE, Drezner MK, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR histomorphometry nomenclature committee. J Bone Miner Res. 2013;28(1):2–17. 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prieto C, Barrios D. RaNA-Seq: interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics. 2019;36(6):1955–1956. 10.1093/bioinformatics/btz854. [DOI] [PubMed] [Google Scholar]
- 40.Kondo N, Kuroda T, Kobayashi D. Cytokine networks in the pathogenesis of rheumatoid arthritis. Int J Mol Sci. 2021;22(20):10922. 10.3390/ijms222010922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267(5203):1485–1488. 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 42.Franco P, Laura F, Valentina C, Simona A, Gloria A, Eleonora N. Interleukin-6 in rheumatoid arthritis. Int J Mol Sci. 2020;21(15):5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siemeister G, Mengel A, Fernandez-Montalvan AE, et al. Inhibition of BUB1 kinase by BAY 1816032 sensitizes tumor cells toward Taxanes, ATR, and PARP inhibitors in vitro and in vivo. Clin Cancer Res. 2019;25(4):1404–1414. 10.1158/1078-0432.CCR-18-0628. [DOI] [PubMed] [Google Scholar]
- 44.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16(3):387–397. 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing Shugoshin. Science. 2010;327(5962):172–177. 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Liu T, Fang Y, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103(4):1278–1285. 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 47.Kalitsis P, Earle E, Fowler KJ, Choo KHA. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14(18):2277–2282. 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwanaga Y, Chi Y-H, Miyazato A, et al. Heterozygous deletion of mitotic arrest–deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67(1):160–166. 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 49.Nyati S, Schinske-Sebolt K, Pitchiaya S, et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-β signaling. Sci Signal. 2015;6;8(358):ra1. 10.1126/scisignal.2005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu L-J, Pan Y, Chen X-Y, Hou P-F. BUB1 promotes proliferation of liver cancer cells by activating SMAD2 phosphorylation. Oncol Lett. 2020;19(5):3506–3512. 10.3892/ol.2020.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christensen AD, Haase C, Cook AD, Hamilton JA. K/BxN serum-transfer arthritis as a model for human inflammatory arthritis. Front Immunol. 2016;7:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa T, Kikuta J, Sudo T, et al. Identification of a novel arthritis-associated osteoclast precursor macrophage regulated by FoxM1. Nat Immunol. 2019;20(12):1631–1643. 10.1038/s41590-019-0526-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data generated and analyzed during this study are available in the NCBI Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/gds. The data underlying this article will be shared on reasonable request to the corresponding author.