Summary
For healthcare datasets, it is often impossible to combine data samples from multiple sites due to ethical, privacy, or logistical concerns. Federated learning allows for the utilization of powerful machine learning algorithms without requiring the pooling of data. Healthcare data have many simultaneous challenges, such as highly siloed data, class imbalance, missing data, distribution shifts, and non-standardized variables, that require new methodologies to address. Federated learning adds significant methodological complexity to conventional centralized machine learning, requiring distributed optimization, communication between nodes, aggregation of models, and redistribution of models. In this systematic review, we consider all papers on Scopus published between January 2015 and February 2023 that describe new federated learning methodologies for addressing challenges with healthcare data. We reviewed 89 papers meeting these criteria. Significant systemic issues were identified throughout the literature, compromising many methodologies reviewed. We give detailed recommendations to help improve methodology development for federated learning in healthcare.
Keywords: federated learning, machine learning, healthcare, applications, systematic review, privacy, security, methodological advances, deployment, best practices
The bigger picture
Federated learning (FL) promises to solve the challenges of applying machine learning methods within healthcare, such as isolated datasets, ethical, privacy, and logistical concerns with data sharing, and the lack of diversity in single-center datasets. By connecting multiple sites and keeping data at their source, FL can help address these issues. An increasing number of novel FL methodologies have been developed, and there is an urgent need for the consolidation of this knowledge to address the unique challenges of healthcare data. This review focuses on literature published between 2015 and 2023 that methodologically advances FL for healthcare applications.
Federated learning (FL) is a technique for privacy-preserving machine learning. FL promises to enable large-scale deployment of diagnostic and prognostic tools in healthcare settings. This systematic review of new methodological contributions within FL research highlights innovations in the literature, identifies pitfalls and issues in recent studies, and presents recommendations for future FL practitioners in the development of FL applications in healthcare.
Introduction
Healthcare data are abundant, representing approximately 30% of the entire global data volume,1 and are becoming increasingly available to researchers to allow for such interrogations as trend analysis, pattern recognition, and predictive modeling. This is helped primarily by the increased adoption of electronic health record (EHR) systems in hospitals, with most UK NHS Trusts currently using one and all expected to have one by 2025.2 In parallel, there has been a revolution in the capabilities of machine learning (ML) methods, allowing for the efficient analysis of high-dimensional clinical and imaging data.
There are different types and formats of healthcare data, including text from medical notes, imaging data, medical device outputs, wearable signals data, and genomic data. These are usually stored in distinct silos, with EHR data often in a database structure, imaging in a picture archiving and communication system, and medical device/wearable data stored locally. Although there are some well-documented challenges to reproducibility,3,4 ML methods have shown great utility for performing both single and multiple modality modeling of healthcare data.5,6
To create high-quality models that generalize across different data sources, it is most common to pool datasets from different locations and train using the combined dataset (centralized learning). However, this is a serious challenge in the healthcare setting, as there are ethical, privacy, logistical, legal, and security concerns regarding transferring clinical data outside the hospital environment. Additionally, there are logistical issues to ensure data security is maintained in the transfer of such large-scale healthcare data. Finally, each hospital might have its own data transfer and sharing rules, making research across multiple hospitals problematic.
While confidential computing methods, e.g., homomorphic encryption (HE) and secure multi-party computation, are utilized for protecting data privacy during computations, HE alone leads to significant additional computational demands,7 and secure multi-party computation faces scalability challenges when handling large datasets.8 However, federated learning (FL) incorporates the benefits of secure compute methods while also effectively reducing the computational load and improving scalability. FL also offers a solution to the problems encountered through centralized learning by permitting data to remain locally at each hospital site, with only the ML model being transferred between them. An FL network can be either decentralized or have a central aggregator communicating with all nodes. In the decentralized setting, a model is trained at individual sites. The updated model is passed around to other sites in the network in a defined order, who then initialize from the model, train, and pass it on again. In the centralized aggregator scenario, an ML model is trained at each site, and information about the final model is transferred from all nodes to the aggregator. The aggregator summarizes the model updates from each site and generates a new global model, which is then redistributed to all sites for the local training process to start again. This continues until a pre-defined convergence criterion is met for the global model.
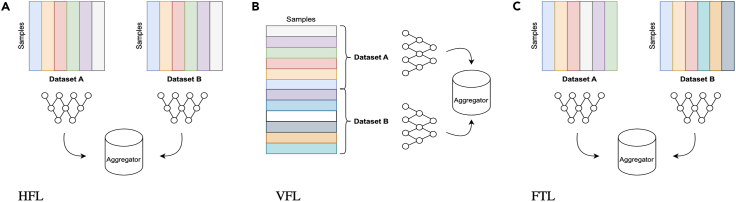
FL methods can be broadly categorized into three groups: horizontal, vertical, and transfer9 (see Figure 1). For horizontal FL (HFL) methods, each site holds the same features for different data samples, whereas for vertical FL (VFL), the sites hold different features related to the same samples. For federated transfer learning (FTL), each site has different feature sets that are related to different samples.9 Each of these has high relevance for healthcare data, where HFL is akin to learning across different hospitals for common variables and VFL allows for linking of different data silos for, e.g., imaging, EHR, and genomic data. FTL applies most to the real-world clinical environment where different hospitals collect different variables on different people, often dependent on their local clinical protocols.
Figure 1.
Types of FL
Schematic illustrating the feature and sample distributions for horizontal federated learning (HFL), vertical FL (VFL), and federated transfer learning (FTL).
There are several practical use cases for FL in healthcare including building models for low-prevalence diseases where individual hospitals only have few samples, which may be highly identifiable, while preserving patient privacy. FL also enables cross-institutional collaboration of hospitals in a secured manner, where each member benefits from the access to models trained using data of the other members. Outside of the hospital environment, FL methods can be trained using Internet of Things devices at patients’ homes to collect data and train a model to predict health outcomes.10,11,12,13 Finally, FL can be applied in real time for population monitoring, allowing for early disease outbreak detection.14 Such a large-scale system can only be achieved through FL, as data from hospitals are often legally prohibited from being shared with other international sites.
With so many diverse application areas, there is now significant literature describing FL methods applied to healthcare challenges such as breast cancer diagnosis,15,16,17,18,19,20,21 COVID-19 detection,22,23,24,25,26,27,28,29,30,31,32,33 length of hospital stay prediction,34 and depression diagnosis.35,36 Indeed, in this systematic review, we identified a corpus of 220 such papers between 2015 and 2023.
However, it is also imperative to recognize that healthcare data inherently contain many issues that require special consideration and adjustments to FL methodologies to address. Crucially, in a network of hospitals, each hospital can potentially serve a fundamentally different patient population. This bias can lead to downstream modeling issues if there are, for example, different disease prevalences at different sites or significant differences in the patient numbers at each location.37 Each hospital may also follow different clinical practices, leading to differences in data collection and, consequently, issues such as missing data38 and non-standardized variable name mappings.39 Differences between sites can also manifest in variations in data quality, posing a unique challenge in the best possible aggregation of locally learned parameters.40 In addition to the data challenges, privacy is a top concern for all users of healthcare data, and it is critical that FL methods for healthcare are considerate of this when transferring model parameters between the sites or to the aggregator.41 Finally, we highlight that hospital environments are typically not equipped with high-performance computational environments, so FL methods must factor in the computational cost of training, transferring, and evaluating models, especially as changes in patient populations or treatment protocols may require retraining of the global FL model and thereby incur additional computational costs.
Therefore, it is surprising that within the corpus of 220 papers, 131 apply existing off-the-shelf FL methods to healthcare datasets, with only 89 papers describing modifications of the FL methodology to address the challenges unique to healthcare data. It is the latter group that this review focuses on, as we are keen to identify which areas of methodology advancement are receiving the focus to address healthcare challenges and whether there are any systematic pitfalls in the way that these new models are being developed.
Earlier systematic reviews have considered FL applications in healthcare, including Antunes et al.,42 Prayitno et al.,43 and Crowson et al.44 Our review builds on previous works in several respects. Firstly, this review is much larger than prior reviews, covering 89 papers compared to 44 of the next-largest review.42 Secondly, this review has a much larger scope than prior reviews, which focused particularly on EHR data,42 data management,43 or reproducibility and risk of bias.44 Thirdly, we systematically dissect FL into five distinct components (see Figure 2) and highlight the methodological approaches and recent advances in each. Finally, based on our findings, we give direct and practical recommendations (Tables 1 and 2) for each identified challenge such that they might be of immediate use to healthcare practitioners and researchers seeking to use FL in healthcare contexts.
Figure 2.
Typical FL workflow
The five key components are identified that form the basis of our analysis.
Table 1.
Component-wise recommendations
| Component | Recommendations |
|---|---|
| Local data processing | The types of missingness found in datasets should be detailed and any approaches to imputation stated. Where imaging data are used, extreme care should be taken to ensure that biases in the data are understood and the best practice is followed in the local optimization by using checklists, e.g., CLAIM.139 For real-world deployment, it would be preferable for, e.g., feature value normalization to be based on globally exchanged parameters or transformations.140 We would encourage authors to consider obscuring their datasets at source before use in the training network, e.g., by a shared hashing function,115 noise addition,141 or slicing,142 to mitigate against data reconstruction attacks. Class imbalances across nodes and within datasets at nodes should be factored into the methodologies, as they are a primary source of bias for FL in healthcare, and it is essential that authors disclose and discuss their strategies to address imbalances, considering potential privacy concerns of data owners. Clinical data can inherently contain large biases and issues from various sources, such as impossible values, missing values, and corruption. Without direct access to the data at each node, automated pipelines should be integrated that can perform a quality and integrity check and remove known sources of bias.143 Securely confirming, across nodes, that the demographics are similar and within a tolerance range is also encouraged.144 For applying inclusion and exclusion criteria to EHRs or tabular datasets at nodes, authors should disclose the filters used to curate the cohort used in the experiments with, e.g., a structured query language (SQL) query.145 We encourage more widespread adoption of internal validation and holdout cohorts to ensure local performance is not overstated due to overfitting of the training data. |
| Local optimization | It is important for authors to consider whether their solutions are of practical use in the hospital environment, where computing capabilities are more limited than in simulated experiments. The required hardware, in particular whether a GPU is required, should be disclosed in manuscripts and stated as a limitation if not widely available. Outside of FL settings, local optimization is best terminated when the loss converges for a validation cohort of the data. FL, in particular, would benefit significantly from early stopping, as the local node compute times can be highly variable, depending on the sample number and model architecture at each site. |
| Communication | The cost of sharing data between nodes is a key concern for FL methods in development and deployment. Practitioners must pragmatically balance their concerns around privacy, with the increased computational cost. We therefore give some general advice that may be factored in to the decision-making process. The number of samples at each node may be sensitive and can pose privacy risks if exploited by hostile actors. Sharing this information should be tailored, carefully weighing privacy against utility. Practical strategies include secure sharing through encrypted channels or utilizing aggregation methods that do not require the sample size. In the context of healthcare data, FL networks should use an established cryptography technique146 to encrypt as much of the data and communication as possible. It is incredibly important for the aggregator to authenticate and authorize all nodes from which data are accepted and to which data are redistributed. Without this, the network is vulnerable to many different attacks.147,148 |
| Aggregation | Authors should consider whether synchronous or asynchronous updates are preferable for their use case, especially if the local optimizations have very different training times or not all nodes are always available. When the nodes supply updates to the global model, they should be assessed for consistency with other nodes. It should then be investigated if a node shares very different model updates than the others. Of the papers we reviewed, none performed such comparisons. |
| Redistribution | Communication rounds should be terminated in a principled way. This could be based on the performance of the global model at each node, on a validation or holdout cohort, or the performance of the global model on evaluation data held at the central aggregator. |
Table 2.
Additional recommendations
| Component | Recommendations |
|---|---|
| Deployment | The deployment strategy should consider the entire process, from node distribution to aggregator interaction, ensuring seamless communication and efficient training rounds. Furthermore, the authors should consider integrating machine learning operations (MLOps) practices, as these enhance automation, monitoring, and security; ensure seamless integration and deployment; encourage collaboration; and increase the efficiency and reliability of the FL platform. None of the papers we reviewed discussed version control of the global model. Implementing version control in FL enhances traceability, supports asynchronous communication, enables A/B testing, and provides a rollback mechanism. Storing copies of model artifacts across different versions strengthens auditability and facilitates benchmarking and tracking model performance over time. |
| Reproducibility | As FL is a rapidly evolving field of innovative research, we recommend that the community work together to develop an FL methodology checklist to improve the documentation of future studies. In the absence of such a checklist, we recommend that authors and reviewers use existing checklists, such as CLAIM,139 for assessing the completeness of the data and model descriptions in a medical imaging context. Additionally, tools such as PROBAST149 are recommended for assessing the biases in the data and models. Practitioners should only develop a new FL codebase when the existing frameworks fundamentally do not accomplish their aim, otherwise there is a risk of coding errors due to the complexity of the FL system. Codebases, and trained models, should be released publicly if possible to allow the community to easily apply the model and validate the performance. |
Findings in the literature
This section will first give a general overview of the methodological advances in the reviewed studies, followed by a separate analysis of each of the five distinct components of FL depicted in Figure 2. These findings are also summarized in Table S1.
Study selection
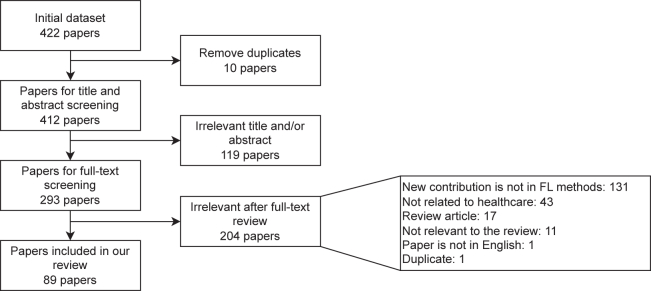
The initial search identified 422 papers that met the search criteria (see Figure 3 and the methods section). After eliminating 10 duplicate papers and filtering to only abstracts and titles focusing on a new approach of FL in healthcare, we retained 293 papers for full-text screening. In this systematic review, we focus on the 89 papers that were relevant to our review question, namely those that introduce a new methodology for applying FL in the context of healthcare.
Figure 3.
PRISMA flowchart
Study selection for our systematic review highlighting the reasons for exclusion of manuscripts at different stages.
Methodology advances
We consider the methodological contributions made to the different FL components identified in Figure 2. The majority of papers (68/89) contribute to a single component, with 18/89 contributing to two and 1/89 contributing to three. Most papers focus on improving the aggregation component (37/89), followed by the communication (35/89) and local optimization (26/89) components. The local data processing component is improved in 5/89 papers and model redistribution in 4/89.
Types of FL
HFL was the most popular among the three approaches, with 80/89 studies exclusively considering it.10,11,12,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,34,35,36,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100 In contrast, VFL101,102,103,104,105 is considered in only five papers and FTL33,106 in four, while two studies13,107 considered both HFL and VFL together.
Data sources are often fragmented across sites. VFL was used when the joining of features across locations was discouraged due to privacy or logistical concerns. For example, when imaging in different modalities was pooled,102 different sensors recorded data about the same subject,104 or genotype and phenotype data in different sites were linked. FTL was primarily applied to clinical data collected under different protocols where the set of available variables differed between nodes.
Applications considered
The majority of papers (70/89) applied FL to classification problems; segmentation was addressed in 7/89 reviewed studies, and the remainder focused on problems such as anomaly detection,78,93 tensor factorization,83,94 feature selection,103 and regression.20,25,33,56,65,69,90 See Table S1 for full details.
Use of existing frameworks
We find that most papers considered in this review (83/89) develop their own FL framework for FL rather than building on existing platforms such as Flower,108 TensorFlow Federated,109,110 PySyft,111 FATE,112 and NVIDIA FLARE.113 Six papers used existing frameworks, namely FATE,101 Flower,32 NVIDIA FLARE,99,102 and PySyft.16,81,82
Public codebases
Only 10/89 studies publicly released their code,10,17,20,25,27,46,50,54,64,98 and no papers shared their trained model.
FL component analysis
We now review each component of the FL pipeline (outlined in Figure 2) as it was applied to healthcare in the 89 considered studies.
Component 1: Local data processing
Methodological advances
In 5/89 papers, the authors focus on improving the data pre-processing. Of these, 4/5 were motivated by problems of class imbalance, with three12,56,79 using a generative model to create new data samples and one26 using augmentation to reduce sample size imbalances across nodes. The remaining paper91 used anonymization techniques (such as the quantization of some continuous features) to enhance the privacy of the raw data samples.
Datasets
A wide range of data types and sources were considered in the papers reviewed. The most popular sources were imaging data (31/89), sensor data (17/89), and EHR or tabular clinical data (21/89). Other studies considered more niche data sources such as medical devices (7/89), insurance claims data (4/89), and genomic data (2/89). For the imaging studies, a variety of modalities were considered. Chest X-ray (9/31),22,23,26,27,28,29,30,31,32 retinal (6/31),31,56,62,63,66,70 microscopy (5/31),16,19,45,55,72 dermoscopy (4/31),31,73,79,96 and magnetic resonance (3/31)71,77,102 imaging constituted the majority. Sensor data were collected from wearable technologies (14/17)10,11,12,17,30,53,60,64,74,82,86,95,98,100 and ambient sensors (4/17).13,59,82,104 For papers using EHR or tabular clinical datasets, MIMIC-III114 was used in 7/21,34,46,83,90,91,94,107 synthetic EHR data in 4/21,65,83,94,105 and proprietary data in 2/21.87,91 Offline medical device data were sourced from electrocardiograms (5/7)51,57,97,101,106 and electromyograms (2/7),59,86 while insurance claims data were sourced from the Centers for Medicare & Medicaid Services,83,94 UnitedHealth Group Clinical Research,25 and MarketScan Research.25,49 Twelve papers also used popular non-medical datasets, including MNIST (10/12),23,35,45,63,64,76,79,80,88,95 CIFAR10 (6/12),23,63,76,79,89,95 Fashion-MNIST (2/12),27,45 and STL10 (1/12)23 in order to benchmark their proposed method’s performance.
Outcomes of interest
The scope of the applications considered in the papers was very diverse. A large number (51/89) focused on the diagnosis of diseases such as COVID-19 (12/51),22,23,24,25,26,27,28,29,30,31,32,33 lung cancer (3/51),15,45,52 breast cancer (7/51),15,16,17,18,19,20,21 skin disease (6/51),31,66,72,73,79,96 eye disease (4/51),31,62,63,66 heart disease (5/51),85,97,101,105,106 brain tumor (4/51),63,71,77,102 diabetes (2/51),56,70 neurodegenerative disorders (2/51),86,104 Alzheimer’s disease (2/51),58,80 colorectal carcinoma (2/51),52,55 and sepsis.54 There are also applications varying from using bone imaging to predict age56 and detecting emotion in speech67 to identifying depression through social networking interactions and posts.35
Data pre-processing
Before training models, data are typically transformed through normalization, transformation, and cleaning. This process was only mentioned in 41/89 papers, performed centrally (before data were allocated to the nodes for synthetic FL experiments) or at a node-by-node level. Commonly used techniques include feature value normalization,23,31,36,56,75,77,84,85 dimensionality reduction,11,23,34,46,85 feature engineering through a data-specific transformation,11,12,13,23,53,89,91 and data filtering11,30,49,54,61,75,77,84,85,107 for tabular data. For video data, resampling was used,69 and for imaging data, resizing,55,56,62,66,96 intensity windowing,60 sliding window,12,30,60,86 and cropping19,48,56,77 were employed.
Most papers do not mention performing quality or integrity checks on the data before or after pre-processing, the exceptions being Gad et al.,10 where samples with impossible values are excluded (e.g., negative heart rates) or where inconsistencies in feature values are identified, and Shaik et al.,11 who performed principal-component analysis to filter out noise. One paper105 examined genotype data for discrepancies and anomalies to ensure reliability and accuracy. Within the 89 papers reviewed, 131 datasets were considered, with the smallest dataset having 116 samples21 and the largest having 3.7 million.91 None of the papers applied hashing or encryption to the raw data before training.115
Imbalances in data
There are two potential sources of imbalanced data for FL. Firstly, different nodes in the network can be associated with highly varying sample sizes, and secondly, the prevalence of the outcome variable (class imbalance) can also vary between nodes. In the studies reviewed, imbalances in sample sizes were addressed in only three papers, which employed downsampling84 and augmentation.26,73
Imbalance in outcome prevalence was addressed in only four studies. SMOTE116 oversampling was used in two papers12,22 to generate a balanced dataset at each node, and deep reinforcement learning was used by Zhang et al.82 to encourage devices with balanced class labels to participate more frequently in the local updates. A weighted random forest, assigning higher weights to the less prevalent classes, was used by Gencturk et al.70
Consistency across nodes
Almost all papers performed their FL experiments in controlled environments without determining whether datasets or their feature values were comparable across the different nodes. The exceptions were Tong et al.,25 who computed a surrogate pairwise likelihood function to account for bias between the model parameters from each site and used this to adjust the final model predictions, and Chen et al.,86 who used the aggregator to compare each node’s data distribution with a benchmark dataset to estimate the label quality, reflecting the reliability and accuracy of each node’s local data labels. In addition, no papers discussed how the dataset used at each node is curated for use in the experiments, i.e., the systematic collection, organization, and verification of data before the pre-processing stage. Accurate data curation is fundamental in ensuring the reliability and consistency of data cohorts across different nodes in FL systems.
Missing data
Most studies did not highlight whether datasets contained any missing values, with only two considering an imputation method while also deleting features with high missingness rates.46,104
Component 2: Local optimization
Methodological advances
In 26/89 papers, the authors focus on improving local optimization. In 6/26 papers, the authors focused on improving the training procedure by using gradient clipping,102 modifying the activation function to better tolerate data heterogeneity,53 regularizing the model by penalizing deviations from earlier time-series data points,17 using contrastive learning for federated model pre-training,46 using parameter sharing to reduce model size,100 and reordering the data samples to process the most difficult samples at the end of training.90 Six papers focus on improving the model architecture to allow for multi-modal data75 by tuning the architecture to local datasets,18 using smaller binarized neural networks for resource constrained settings,98 using extremely randomized trees for privacy preservation,23 using an extreme learning machine to directly find the model parameters in one iteration,104 and introducing a new method for survival analysis to factor in time-varying covariates.32 In 9/26 papers,12,20,27,57,66,72,89,96,107 the authors decompose the model into parts that are trained on the local nodes and parts trained on the central server. This gives some aspects of the model, which is fine-tuned to local data. In 4/26 papers, the authors design approaches to perform federated semi-supervised learning to utilize unlabeled data samples. In one paper, the authors design a highly stratified cross-validation strategy based on confounding factors to overcome distribution differences between local datasets.
Model architectures
In the 82/89 studies that described their model, there was a wide range of complexity, ranging from highly intricate and parameterized models to simpler, traditional ML methods. The most popular choice for model architecture was a convolutional neural network (CNN), with 46/82 studies considering them. Most papers (see Table S1) discuss their own custom CNN architectures (27/46), while many of these papers (19/46) compare several different established architectures such as ResNet (9/46),16,27,29,48,56,62,63,72,96 DenseNet (3/46),22,45,48 MobileNet (2/46),27,101 U-Net (46),32,66,71,102 AlexNet (4/46),16,30,59,89 and LeNet (2/46).30,59 Recurrent neural networks (RNNs) were also popular, with long short-term memory (LSTM) and Bi-LSTM backbones found in 11/82 studies10,11,13,34,46,51,53,57,80,93,107 and five papers using both CNNs and RNNs.10,11,51,57,80 Multi-layer perceptrons (MLPs) were used in 11 studies, with custom architectures for a vanilla MLP employed in 1018,20,21,24,55,67,74,75,84,87 and an attention layer incorporated in one.98 Some studies also considered FL with more traditional ML algorithms such as gradient-boosted trees,21,90,103 support vector machines,21,85,91,103 fuzzy clustering,61 logistic regression,21,25,49,58,65,91,105 and random forests.17,21,70,92,103 The remaining studies either used custom algorithms or focused on other aspects of the FL pipeline.
Optimizers
Of the 59 papers that mentioned their optimization algorithm, stochastic gradient descent is the most widely used method, found in 35/59 papers, while Adam was used in 20/59 papers (see Table S1). Other methods such as RMSprop,10,13,106 SAGA,63 Adadelta,26 maximum likelihood estimation,65 or Newton’s method105 were also considered. One paper developed their own optimizer.70
Training initialization
In the 68/89 studies that mentioned the use of initialization strategies for their model parameters, initialization with random weights was the most common (45/67), while 16 papers utilized pre-determined parameters (identified by the authors), 7 used pre-trained weights,28,30,31,32,35,48,97 and another used the parameters obtained by the node whose dataset had the highest number of features.61 See Table S1 for additional details.
Dataset partitioning
Of the papers, 47/89 stated that they partitioned their dataset into internal validation or holdout cohorts. Only two papers45,46 used both an internal and an external holdout cohort. Thirteen papers used an internal validation dataset to avoid model overfitting by using between 5% and 25% of the local dataset.19,22,46,48,52,55,56,67,71,73,99,101,102 15 studies performed 3-,17,49 5-,17,20,34,53,84,90,91,100,106 or 10-fold51,57,58,70 cross-validation, while two papers12,75 mentioned the use of cross-validation without specifying the number of folds. In 23 papers,10,11,15,26,28,29,30,31,32,33,35,36,59,62,63,64,74,77,88,96,97,106,107 there is a holdout cohort used for model evaluation after a fixed number of rounds of local optimizations. Three papers mentioned the use of test datasets85,104 or validation datasets86 but not their sizes.
Hardware
Only 40/89 papers mentioned the hardware used in their study. The computational requirements for model optimization varied significantly, and consequently, the hardware used was highly diverse. Most FL methods required either hardware with GPU(s) attached (26/40) or simple CPU machines (15/40). Some studies use edge devices such as a Raspberry Pi26,46,69,93,106 or a smartphone.59
Privacy-preserving optimization
Six papers23,55,58,78,84,89 applied differential privacy117 during the optimization. This aimed to preserve the privacy of individual samples in the training data. Five papers accomplished this by adding Gaussian noise to the exchanged data, with the sole exception of Hilberger et al.,55 who used TensorFlow Privacy118 instead.
Training termination
Criteria for ending the local optimization are mentioned in 75/89 papers, most of which (67/75) used a fixed number of local training epochs for each node (see Table S1), while four papers47,53,65,100 terminated on convergence but did not provide details on how this was determined. Two papers71,104 only required one epoch for training by design.
Decentralized training
In seven papers,18,31,36,53,60,74,93 the training of the model itself was not entirely performed on the nodes. Three studies trained using both the nodes and edge devices,18,31,93 and four studies trained small clusters of nodes, which then communicate to the aggregator.36,53,60,74
Novel developments
One paper removed the need to perform iterations of an algorithm by use of a one-layer MLP, whose weights can be directly solved for,104 and another paper partitioned the model architecture and then trained the computationally intensive part on the central aggregator.89 Knowledge distillation was used in two studies10,48 where more powerful “teacher” nodes train less powerful “student” nodes. One paper introduced a method where all but the batch normalization layers are exchanged between each node.19
Component 3: Communication
Methodological advances
In 35/89 papers, there are contributions to improve the communication component, with five papers even describing contributions to several aspects. Primarily, authors focused on improving encryption methods (11/35), aimed to reduce either the amount of data communicated (12/35) or the number of communication rounds (7/35), and also introduced methods for fully decentralized communication (5/35).31,71,77,85,87 Encryption improvements tended to focus on methods for sharing secret keys among nodes,54,88 encryption mechanisms for the data exchanged,58,68,73,89,92,100,101,103 and also a technique for perturbing model outputs at each node using a secret key.28 A reduction in the amount of data communicated is achieved by transferring a subset of the model parameters,19,30,32,51,60,74,83,106 or by compressing, masking and quantizing of gradients or model outputs before exchange.76,84,94,101 The number of rounds of communication with the server can be reduced by the inherent design of the model,25,34,56,65 by aggregating based on the elapsed time (rather than the epochs),76,94 and by assessing whether a proposed update is beneficial to the network before communicating it.88 The remaining papers focus on techniques for detecting attacks during communication78 and developing an authentication system for nodes in the network47,81 and systems for client/node management.64,73
Data exchanged
In the studies we considered, the data most commonly shared with the aggregator were the model weights (35/89) and gradients (13/89) (see also Table S1). Many papers simply state that they are exchanging “model parameters” (20/89)13,19,20,23,30,33,34,46,47,52,53,61,62,64,66,67,78,84,85,99 or “model updates” (2/89)22,86 without specifically stating if or how these relate to weights or gradients. The outputs of the local models were shared in 7/8910,11,21,48,49,101,107 of the papers reviewed. Some papers encrypted the weights before sharing (9/35),12,15,16,35,58,68,76,77,100 some encrypted the gradients (4/13),54,88,89,92 and four papers perturbed the model parameters before sharing.18,23,78,84
Beyond the conventional gradient and weight data shared in FL methods, some papers also shared additional information about the data or the training process or procedure. Metadata such as the model architecture, optimizer and loss function,55 and training time and maximum performance,22,77 along with the initial learning rate and thresholds used for classification and learning strategy,51 were among the extra information exchanged.
Encryption
Data were exchanged unencrypted for most of the studies we considered (70/89). Most papers that did encrypt the exchanged data primarily used HE methods (13/19),12,15,16,17,54,68,73,76,89,92,100,101,103 while Chen et al.88 relied on a symmetric-key algorithm for encrypting the gradients when exchanged. Only three papers50,77,81 discussed the encryption of the communication channel between the node and the aggregator rather than direct data encryption before exchange. Two papers35,58 stated that they encrypted their data without detailing the method used.
Partial model communication
In 12/89 papers, only a subset of the model layers19,27,30,32,51,57,60,74,89,95,106,107 are communicated. This allows for reduced communication overhead along with improved privacy and more personalized models, as only generic features are exchanged. Other papers accomplish this by sharing only some intermediate model outputs104 or randomly shuffling the model outputs.28
Authentication of nodes
Only two papers47,81 ensured that the node is authenticated as part of the approved FL network before the aggregator accepts the exchanged data. The local model parameters were shared, along with a ring signature, to prove that the data originated from the node.
Fully decentralized communication
There were nine papers describing fully decentralized FL methods.31,48,59,61,71,77,85,87,94 These require communication between nodes rather than a central aggregator, using gradient and weight updates of neighbor nodes to update the local model. Knowledge distillation between the local nodes is employed in two studies10,48 where more powerful nodes behave as teachers to the less powerful student nodes. None of the papers considered the significance of the sequence of updates in the fully decentralized scenario.
Optimizing efficiency
Two papers considered approaches to reduce the number of rounds of communication with the aggregator by sharing embeddings of the data with the aggregator (which then trains the model)104 and by modeling the pairwise relationships between the data at each node directly,25 both of which allow for only a single round of communication. Communication time is optimized in one paper,31 where an optimal ring structure between nodes is obtained by solving a version of the traveling salesman problem. Additionally, eight papers focused on reducing the amount of data transferred between the nodes and the aggregator at each iteration. This can be accomplished by sharing only a subset of model parameters27,51,57,60,89 or compressing gradients.83,84,94 One paper focused on training for several tasks simultaneously to avoid training independent models.34
Component 4: Aggregation
Methodological advances
In 37/89 papers, the authors focus on improving the aggregation component. Most commonly, papers consider the weighting of the contributions of the nodes in the network (11/37) by considering the local training loss,29,99 local classification performance,70 signal-to-noise ratio of the data,69 node similarity,30 data quality,73 fairness,45 Shapley values,63 model performance on aggregator test data,86 and how each local model performs at all other nodes.21
Aggregation methods are often improved by measuring and correcting for the distribution differences between nodes,25,33,65,105 allowing for multi-modal data aggregation across different nodes.13,101 In 5/37 papers, the authors cluster nodes together based on their similarity before aggregation, and 5/37 papers focus on improving the aggregation strategy. This is accomplished using hierarchical aggregation,93 performing some training rounds on the aggregator,62 dynamically selecting participating nodes and scheduling of aggregation,22 and the ensembling of the local models to obtain a final model.49,102 The remaining papers focus on methods for asynchronous aggregation (3/37),35,59,97 using knowledge distillation (2/37),10,48 secure aggregation (2/37),68,92 feature selection in a federated manner,103 attack detection during aggregation,80 and aggregation of heterogeneous model architectures.11
Model aggregation techniques
In 78/89 papers, the methods for aggregating the contributions from individual nodes were mentioned. Most papers focused on improving the aggregation strategy, with 10/78 developing custom simple averaging strategies and 31/78 developing customized weighted averaging methods. The well-known FedAvg119 aggregation method was used in 14/78 papers, with a modified formulation of it employed in a further 10/78. The remainder of the studies used non-averaging-based methods such as knowledge distillation,10,48 stacking,11,49 split learning,102 feature fusion,101 federated goal programming,21 and training the model one node at a time.71 See Table S1 for additional details.
Synchronous vs. asynchronous updates
Only four studies35,59,77,97 developed FL methods that allowed for asynchronous updates to the global model, while all others required the local nodes to finish their optimization before the updates were applied. Asynchronous updates were made after comparing the performance of the model with the new and previous parameters and only updating the model if the update performed better.
Node weighting
A weighting factor was discussed in 50/89 papers to balance the node contributions during aggregation. The majority of papers (38/50) weighted the contribution based on the sample size at each node. Two papers10,70 assigned weights based on the classification accuracy performance of each node, one weighted by considering the credibility of each node86 (giving higher weight to nodes whose local model loss on a benchmark dataset lies closer to the global model loss), and another dynamically adjusted the weights based on the variation of loss values from the previous round.99 Some papers did not aggregate the local contribution from a node if it was not beneficial to the overall network35,88 or did not satisfy defined performance criteria.70 Model training time was considered in one paper22 to limit the number of devices transmitting their updates to the aggregator. A reward mechanism was used in one paper82 to select optimal nodes, considering the quality of their data and associated energy costs. One paper49 compared multiple means from which to determine the weighting during aggregation.
Distribution comparisons
In order to address data heterogeneity between sites, only three papers30,36,95 mentioned that the model distributions between nodes were compared for clustering purposes before aggregation.
Component 5: Redistribution
Methodological advances
In only 4/89 papers, the authors focus on improving the way models are redistributed to local nodes. Two papers personalize the model that is redistributed to the local nodes based on their similarity to other nodes in the network30 and by comparing the loss of the global model on a benchmark dataset to the local node losses.36 The other two papers focus on improving access to the global model by allowing local nodes or registered third-party researchers to run encrypted inference directly on the global model.16,54
Training termination
The criteria for ending model redistribution were specified in 79/89 papers. The majority (69/79) used a pre-determined number of update rounds before termination. Three papers11,25,104 required only a single round of communication by design. Two papers97,105 used a minimum loss threshold to claim convergence of the global model and terminate redistribution.
FL evaluation criteria
Model efficacy
Common ML performance metrics were reported in most papers, such as the model accuracy (59/89), area under the receiver operator characteristic curve (17/89), precision (15/89), F1-score (20/89), sensitivity/recall (18/89), specificity (6/89), Dice score (8/89), and loss value (12/89).
There were 34 papers that benchmarked their proposed methods against other FL methods, while 27 studies compared their simulated FL approaches with classic centralized ML. Given that the focus of this review was on papers introducing novel contributions to the FL methodology, ablation studies were commonly performed to assess the performance impact of including and excluding their proposed modifications.11,13,22,28,30,32,46,48,55,71,81,84,97,103 Seven papers compared the performance of different local model architectures while keeping the FL framework the same.11,31,52,67,81,101,103 Furthermore, 34 studies showcase the performance for multiple hyperparameter configurations for their proposed adaptations.
Communication efficiency
Key metrics for measuring the communication overhead were considered in 18/89 papers, such as communication costs (13/18),12,25,31,45,47,60,73,83,85,88,94,101,103 number of communication rounds (4/18),60,61,85,87 and the latency (4/18).17,22,59,101
Resource consumption
Time was an important consideration in several papers, including training time (5/89),18,59,76,103,104 model parameter encryption time (2/89),16,89 and authentication signature compute time.47 Energy consumption was measured in four papers.18,59,76,82 One paper18 focused on CPU processing time, total training time, memory usage, and energy consumption for different numbers of offloaded layers in mobile devices.
Issues identified, recommendations, and remaining challenges
Our systematic review has identified a keen appetite in the research community for developing FL methods with application to diverse healthcare problems. Most studies explore HFL, while VFL is under-explored in the literature. This is surprising, as VFL holds immense potential for addressing healthcare problems, especially considering that healthcare data often have the inherent challenges of being siloed due to logistical, legal, ethical, and privacy concerns related to extensive data linking. In the following, we highlight the many systemic issues in the FL literature that we have identified and give recommendations for corrective action to address them in Tables 1 and 2.
Datasets: Missing values, modalities, encryption, class imbalances, and partitioning
Careful curation and preparation of data are crucial in ensuring data consistency and quality across nodes; however, we identified many data-related issues throughout the literature. Firstly, given the nature of healthcare data, where different variables will be recorded/available at different sites, it is surprising that no studies discussed structural or informative missingness,120,121 with only two considering the imputation of missing data. One paper36 even mentions missing values within their data without describing how the missingness was addressed. It has been shown in other studies that poor quality imputation, and imputation for non-random missingness, can bias a model trained using it.122,123 Only one paper discussed how the authors derived the particular cohort at each node for their analysis. For example, if each node has EHR data, then for each use case, the EHR at each node must be filtered with defined inclusion and exclusion criteria informed through discussions with healthcare professionals. Secondly, we found 32/89 studies applying FL using clinical imaging to resolve the problem that these studies are often limited by low patient numbers due to low disease prevalence at individual sites.124 However, given the known challenges of applying ML to imaging data,125,126 FL also has the potential to amplify these issues, as the inherent data biases cannot be explored. Thirdly, where pre-processing of data is used, it is always performed at local nodes or centrally before distribution to the nodes for artificial FL setups. This local approach, however, propagates any biases in feature values through to the pre-processed data. Additionally, while many authors enhanced the network’s privacy by encrypting the exchanged data, all models were trained using pre-processed raw data. No paper considered encrypting data before performing training115 or a one-way hashing. This is alarming given that model weights can be highly informative about the raw patient data, whereas hashing of the data at the source allows for breakage of this link.127 Fourth, imbalances and distribution shifts are a reality for healthcare data and represent a critical challenge for ML methods. FL methods compound these, as disease prevalence may also vary between nodes.128 However, this issue was only considered and addressed in six of the papers we reviewed. We also identified issues in node consistency, a challenge unique to FL. Only one study checked for consistency in the data distributions between sites. In a real-world deployment scenario, biases between different sites are a reality that must be considered.129 For example, in an FL network of hospitals, it would be important to understand whether data for pediatric or maternity hospitals were hosted at particular nodes to fully appreciate the age and sex biases inherent to the data. Finally, only two papers employed both an internal validation and a holdout cohort. Most papers considered exclusively either a validation cohort or a holdout cohort. In the wider ML literature, using validation data to mitigate the risks of model overfit and also a holdout cohort for evaluation is standard practice. It is surprising that the FL literature does not echo this.
Local optimization: Limitations in training and termination
Systemic issues were identified with local optimization methodologies with respect to hardware requirements and training termination. Firstly, in most papers reviewed, the local optimization of models required GPU compute capability attached to the data source, which is not currently found in most hospital environments. There is some progress toward this with the increasing adoption of cloud computing capabilities in clinical environments along with Trusted Research Environments.130 However, the lack of widespread computational capability remains a barrier to the mass adoption of FL in real-world healthcare settings. Secondly, most papers train their models for only a fixed number of iterations, with only one mentioning early stopping and two others specifying “until convergence.” This is highly irregular, with principled stopping criteria widespread in the non-FL ML literature, such as early stopping.131
Communication: Metadata, encryption, and authentication
Securely exchanging information among distributed nodes is essential for effective collaborative learning. Nevertheless, this component is fraught with some challenges. Firstly, it becomes apparent from many studies that metadata, beyond the model parameters, were often communicated in parallel to the aggregator. For instance, nearly all papers that relied on FedAvg as an aggregation method required sharing the sample number at each node. This can severely compromise the network’s security if an attacker intercepts the communication or compromises the aggregator by highlighting those nodes that contain a large amount of data to a hostile actor. Secondly, given our focus on healthcare applications, where privacy is a primary concern, it was surprising that most papers developed FL networks that do not encrypt the model parameters when being exchanged with the aggregator. It has been shown that private information contained within the training data can leak into the learned parameters of a model.132,133,134 Finally, FL networks are susceptible to attacks by a node exchanging inauthentic or false weights or by allowing the network to train on poisoned data.135 Only two papers authenticated nodes before accepting their update parameters.
Aggregation: Central vs. decentralized, synchronous vs. asynchronous, and weighting individual contributions
Effectively combining the knowledge from individual nodes to give an optimal model for a particular use case is the core challenge in FL development. We identified several areas of concern with model aggregation approaches that were systemic to the literature. Firstly, most papers required the use of a central aggregator, with only eight studies exploring a fully decentralized architecture. Decentralization allows for a network that is more robust to failure and attack,136 with updates performed mutually between the nodes themselves. Secondly, only three papers discussed a method that allows for asynchronous updates to the global model. Almost all aggregation techniques are simple or a weighted averaging of the contributions from each node. Requiring synchronous updates is a limitation for real-world deployment, as differing local optimization speeds will result in nodes lying idle, waiting for the aggregator to return the global model only once all nodes have finished training. Finally, with real-world data, it is to be expected that nodes may suggest very different updates to the global model. Most papers that disclosed their method simply weighted the contributions by sample size alone. However, it is useful to understand whether those updates are consistent with one another or whether one or more nodes are suggesting updates that are vastly different from the others, as this may be symptomatic of issues with the data or training.137,138 Two papers introduced methods that allow for ignoring or minimizing the contributions of those nodes that are not beneficial to the overall network.
Redistribution: Termination criteria
Surprisingly, redistribution of the global model was always performed for a fixed number of epochs rather than terminating with a principled stopping criterion. Compared to the wider ML literature, this is aberrant, where early stopping and validation data are used to terminate training.
Deployment: No real-world setup
It is crucial that real-world deployment is carefully considered and planned for. None of the papers provided evidence about the deployment of their FL platform in a healthcare environment. It needs to be clarified how each node is set up in individual hospitals, how the local model is delivered to these nodes, and how these nodes connect to existing databases. The communication protocols, enabling interactions between nodes and aggregators, were largely unspecified. Moreover, the mechanism or event that triggers new training rounds was not described.
Reproducibility: Poor documentation and code
Conducting and documenting reproducible research is a cornerstone of the scientific method. However, we identified that the available manuscript documentation and software codebase were mostly not sufficient to allow for the reproduction of the study findings. In particular, we found that key details such as the data pre-processing techniques, data imputation methods, model initialization strategies, and optimizers employed were missing in a large proportion of the papers reviewed. These basic details are crucial to allow for the reproduction of the results that the papers describe. There is also poor documentation of the data exchanged from nodes to the aggregator, with the terms “model parameters” and “model updates” used interchangeably without specifying if these are the gradients, model weights, or some other parameter. It is also very surprising that most papers develop their own implementation of the FL codebase rather than leveraging and building upon existing FL frameworks. The extreme complexity of FL systems leads to concerns that individual implementations are likely to suffer from issues of correctness unless carefully developed.4 Also, no papers released trained models, and therefore it is not possible to assess the performance of models independently.
Limitations of this review
The scope of this review leads it to focus only on those papers whose contribution is to the methodology of FL applied to healthcare. We do not consider manuscripts that apply off-the-shelf FL methods directly to the data. This is a limitation to the review, as many manuscripts have not been considered that claim to have been successfully applied to healthcare data. Additionally, we have not assessed manuscript quality using checklists or performed a review of bias (ROB) assessment. Currently, we have been unable to identify an approach appropriate for FL papers, and it is unclear how to fairly generalize checklists such as CLAIM139 or ROB frameworks such as PROBAST149 to an FL setting.
Challenges and future directions
There are numerous challenges to the widespread adoption of FL within the healthcare setting, many of which we have described. We focus on two significant future challenges. Firstly, convergence rates for different FL aggregation methods will continue to be derived, adding to those identified for FedAvg.150,151 Li et al.150 have shown that convergence slows significantly if the ratio of the sample numbers between the most prominent and most minor nodes is large. They also demonstrate that the convergence rate of the global model is dependent on the number of local optimization steps. In real-world scenarios, there will be large differences in sample numbers and disease prevalence between the nodes. Deriving aggregation methods that give strong convergence rates of FL, despite these issues, will be a necessary future direction of research. Secondly, the deployment of FL algorithms in healthcare relies on satisfying legal restrictions such as General Data Protection Regulation (GDPR)152 and Health Insurance Portability and Accountability Act (HIPAA),153 along with regulatory approval for “software as a medical device.” This leads to a potential conflict, as device regulation is expensive and best suited to commercial entities. However, building these FL networks requires strong collaborations and incentivization of hospitals, which is often achieved through academic collaborations. Finally, approaches for dataset curation, pre-processing, and imputation in the federated setting will be further developed along with the widespread adoption of standards and checklists for the development of high-quality FL methods.
Conclusions
This review focused on the literature describing FL methods for healthcare applications where there were methodological advances. We considered the different areas in which methodological advancements are being made while systematically exploring the application areas and how the FL components were developed. We identified systemic pitfalls in each component and gave recommendations to support practitioners in developing FL methods in healthcare. Specifically, significant improvements are required in areas such as documentation quality, addressing imbalanced and missing data, and sharing non-encrypted updates. The community must also work together to design appropriate checklists for FL methods in healthcare and review bias frameworks for this setting. FL will become a more common and significant tool for healthcare analytics in the future, and by following these best-practice recommendations, we increase the likelihood of adopting these tools in clinical practice.
Methods
Review strategy and selection criteria
We performed a search of published works using Scopus for phrases shown in the search terms section covering the period from January 2015 to February 2023. The review was performed using the Covidence154 systematic review platform.
Search terms
An initial search was performed to extract papers containing one of “federated learning” or “distributed learning” along with one of “classify,” “predict,” “prediction,” “identify,” “predictive,” “prognosticate,” “diagnosis,” “diagnostic,” “diagnose,” “outlier,” “anomaly,” “detect,” or “detecting” in the title or abstract. We also required that “healthcare,” “health,” or “health care” appeared in the abstract. We excluded articles focusing on blockchain development, intrusion detection, remote teaching, and systematic reviews by excluding those papers that included “blockchain,” “block chain,” “classroom,” “class room,” or “attack” in the title or abstract or had “intrusion,” “intrude,” “review,” or “survey” in the title.
Title and abstract screening
A team of eight reviewers screened the titles and abstracts for each paper. Each paper was independently assessed by two reviewers, and conflicts were resolved by the consensus of all reviewers.
Full-text screening
Nine reviewers performed the full-text screening, with each paper independently assessed by two reviewers. Any conflicts were resolved by the consensus of all reviewers.
Data extraction
A team of four extracted the data from each paper used to write the manuscript and assemble Table S1.
Constoria
The members of the BloodCounts! consortium are Martijn Schut, Folkert Asselbergs, Sujoy Kar, Suthesh Sivapalaratnam, Sophie Williams, Mickey Koh, Yvonne Henskens, Bart de Wit, Umberto D’Alessandro, Bubacarr Bah, Ousman Secka, Parashkev Nachev, Rajeev Gupta, Sara Trompeter, Nancy Boeckx, Christine van Laer, Gordon A. Awandare, Kwabena Sarpong, Lucas Amenga-Etego, Mathie Leers, Mirelle Huijskens, Samuel McDermott, Willem H. Ouwehand, James Rudd, Carola-Bibiane Schӧnlieb, Nicholas Gleadall, and Michael Roberts.
Acknowledgments
The authors are grateful for the following funding: The Trinity Challenge (F.Z., D.K., S.T., BloodCounts! consortium, C.-B.S., N.G., M.R.); the EU/EFPIA Innovative Medicines Initiative project DRAGON (101005122) (T.S., C.-B.S., M.R.); AstraZeneca (Y.C.); the EPSRC Cambridge Mathematics of Information in Healthcare Hub EP/T017961/1 (J.H.F.R., J.A.D.A., C.-B.S., M.R.); the Cantab Capital Institute for the Mathematics of Information (C.-B.S.); the European Research Council under the European Union’s Horizon 2020 research and innovation programme grant agreement no. 777826 (C.-B.S.); the Alan Turing Institute (C.-B.S.); the Wellcome Trust (J.H.F.R.); the Cancer Research UK Cambridge Centre (C9685/A25177) (C.-B.S.); the British Heart Foundation (J.H.F.R.); the NIHR Cambridge Biomedical Research Centre (J.H.F.R.); and HEFCE (J.H.F.R.). In addition, C.-B.S. acknowledges support from the Leverhulme Trust project on “Breaking the non-convexity barrier,” the Philip Leverhulme Prize, EPSRC grants EP/S026045/1 and EP/T003553/1, and the Wellcome Innovator Award RG98755. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or the writing of the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Author contributions
Paper search and collection, F.Z.; paper screening, review, and data extraction, F.Z., D.K., Y.C., S.D., S.T., and M.R.; validation, F.Z. and D.K.; writing and editing, F.Z., D.K., and M.R.; visualization, M.R.; supervision, M.R.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2024.101006.
Contributor Information
Michael Roberts, Email: mr808@cam.ac.uk.
BloodCounts! consortium:
Martijn Schut, Folkert Asselbergs, Sujoy Kar, Suthesh Sivapalaratnam, Sophie Williams, Mickey Koh, Yvonne Henskens, Bart de Wit, Umberto D’Alessandro, Bubacarr Bah, Ousman Secka, Parashkev Nachev, Rajeev Gupta, Sara Trompeter, Nancy Boeckx, Christine van Laer, Gordon A. Awandare, Kwabena Sarpong, Lucas Amenga-Etego, Mathie Leers, Mirelle Huijskens, Samuel McDermott, Willem H. Ouwehand, James Rudd, Carola-Bibiane Schӧnlieb, Nicholas Gleadall, and Michael Roberts
Supplemental information
References
- 1.Thomason J. Big tech, big data and the new world of digital health. J. Glob. Health. 2021;5:165–168. [Google Scholar]
- 2.A plan for digital health and social care. 2022. https://www.gov.uk/government/publications/a-plan-for-digital-health-and-social-care/a-plan-for-digital-health-and-social-care
- 3.Kapoor S., Narayanan A. Leakage and the reproducibility crisis in machine-learning-based science. Patterns. 2023;4 doi: 10.1016/j.patter.2023.100804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmer S., Roberts M., Gilbey J., Biguri A., Selby I., Breger A., Thorpe M., Weir-McCall J.R., Gkrania-Klotsas E., Korhonen A., et al. Navigating the development challenges in creating complex data systems. Nat. Mach. Intell. 2023;5:681–686. doi: 10.1038/s42256-023-00665-x. [DOI] [Google Scholar]
- 5.De Fauw J., Ledsam J.R., Romera-Paredes B., Nikolov S., Tomasev N., Blackwell S., Askham H., Glorot X., O'Donoghue B., Visentin D., et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 6.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 7.Pulido-Gaytan B., Tchernykh A., Cortés-Mendoza J.M., Babenko M., Radchenko G., Avetisyan A., Drozdov A.Y. Privacy-preserving neural networks with Homomorphic encryption: Challenges and opportunities. Peer Peer Netw. Appl. 2021;14:1666–1691. doi: 10.1007/s12083-021-01076-8. [DOI] [Google Scholar]
- 8.Volgushev N., Schwarzkopf M., Getchell B., Varia M., Lapets A., Bestavros A. Proceedings of the Fourteenth EuroSys Conference 2019, EuroSys ’19. Association for Computing Machinery; 2019. Conclave: secure multi-party computation on big data. [DOI] [Google Scholar]
- 9.Yang Q., Liu Y., Chen T., Tong Y. Federated machine learning: Concept and applications. ACM Trans. Intell. Syst. Technol. 2019;10:1–19. doi: 10.1145/3298981. [DOI] [Google Scholar]
- 10.Gad G., Fadlullah Z. Federated Learning via Augmented Knowledge Distillation for Heterogenous Deep Human Activity Recognition Systems. Sensors. 2022;23 doi: 10.3390/s23010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaik T., Tao X., Higgins N., Gururajan R., Li Y., Zhou X., Acharya U.R. FedStack: Personalized activity monitoring using stacked federated learning. Knowl. Base Syst. 2022;257 doi: 10.1016/j.knosys.2022.109929. [DOI] [Google Scholar]
- 12.Wu Q., Chen X., Zhou Z., Zhang J. FedHome: Cloud-Edge Based Personalized Federated Learning for In-Home Health Monitoring. IEEE Trans. Mob. Comput. 2022;21:2818–2832. doi: 10.1109/TMC.2020.3045266. [DOI] [Google Scholar]
- 13.Che S., Kong Z., Peng H., Sun L., Leow A., Chen Y., He L. Federated Multi-view Learning for Private Medical Data Integration and Analysis. ACM Trans. Intell. Syst. Technol. 2022;13:1–23. doi: 10.1145/3501816. [DOI] [Google Scholar]
- 14.Stevens J. BloodCounts! Consortium wins Trinity Challenge Prize for breakthrough in infectious disease detection. https://www.cmih.maths.cam.ac.uk/news/bloodcounts-consortium-wins-trinity-challenge-prize-breakthrough-infectious-disease-detection
- 15.Wang Q., Zhou Y. FedSPL: federated self-paced learning for privacy-preserving disease diagnosis. Brief. Bioinform. 2022;23 doi: 10.1093/bib/bbab498. [DOI] [PubMed] [Google Scholar]
- 16.Kalapaaking A.P., Stephanie V., Khalil I., Atiquzzaman M., Yi X., Almashor M. SMPC-Based Federated Learning for 6G-Enabled Internet of Medical Things. IEEE Network. 2022;36:182–189. doi: 10.1109/MNET.007.2100717. [DOI] [Google Scholar]
- 17.Aminifar A., Shokri M., Rabbi F., Pun V.K.I., Lamo Y. Extremely Randomized Trees with Privacy Preservation for Distributed Structured Health Data. IEEE Access. 2022;10:6010–6027. doi: 10.1109/ACCESS.2022.3141709. [DOI] [Google Scholar]
- 18.Guo, Y., Liu, F., Cai, Z., Chen, L., and Xiao, N. (2020). FEEL: A federated edge learning system for efficient and privacy-preserving mobile healthcare. In Proceedings of the 49th International Conference on Parallel Processing (1–11).
- 19.Andreux M., du Terrail J.O., Beguier C., Tramel E.W. Vol. 12444. LNCS; 2020. Siloed Federated Learning for Multi-centric Histopathology Datasets; pp. 129–139. (Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics)). [DOI] [Google Scholar]
- 20.Zhang D.K., Toni F., Williams M. A Federated Cox Model with Non-proportional Hazards. Int. J. Comput. Intell. Stud. 2023;1060:171–185. doi: 10.1007/978-3-031-14771-5_12. [DOI] [Google Scholar]
- 21.Repetto M., La Torre D. 2022 5th International Conference on Signal Processing and Information Security. ICSPIS; 2022. Federated Learning through Goal Programming: a Computational Study in Cancer Detection; pp. 80–85. [DOI] [Google Scholar]
- 22.Malik H., Naeem A., Naqvi R.A., Loh W.-K. DMFL_net: A Federated Learning-Based Framework for the Classification of COVID-19 from Multiple Chest Diseases Using X-rays. Sensors. 2023;23 doi: 10.3390/s23020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akter M., Moustafa N., Lynar T., Razzak I. Edge Intelligence: Federated Learning-Based Privacy Protection Framework for Smart Healthcare Systems. IEEE J. Biomed. Health Inform. 2022;26:5805–5816. doi: 10.1109/JBHI.2022.3192648. [DOI] [PubMed] [Google Scholar]
- 24.Kandati D.R., Gadekallu T.R. Genetic Clustered Federated Learning for COVID-19 Detection. Electronics (Switzerland) 2022;11:2714. doi: 10.3390/electronics11172714. [DOI] [Google Scholar]
- 25.Tong J., Luo C., Islam M.N., Sheils N.E., Buresh J., Edmondson M., Merkel P.A., Lautenbach E., Duan R., Chen Y. Distributed learning for heterogeneous clinical data with application to integrating COVID-19 data across 230 sites. npj Digital Medicine. 2022;5 doi: 10.1038/s41746-022-00615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cetinkaya A.E., Akin M., Sagiroglu S. Improving Performance of Federated Learning based Medical Image Analysis in Non-IID Settings using Image Augmentation. In 2021 International Conference on Information Security and Cryptology (ISCTURKEY) (69–74). 10.1109/ISCTURKEY53027.2021.9654356. [DOI]
- 27.Yang Q., Zhang J., Hao W., Spell G.P., Carin L. Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining. 2021. Flop: Federated Learning on Medical Datasets using Partial Networks; pp. 3845–3853. [DOI] [Google Scholar]
- 28.Park S., Ye J.C. Multi-Task Distributed Learning using Vision Transformer with Random Patch Permutation. IEEE Trans. Med. Imaging. 2023;42:2091–2105. doi: 10.1109/TMI.2022.3218783. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Xu X., Cao X., Liu W., Zhang Y., Chen D., Dai H. Integrated CNN and Federated Learning for COVID-19 Detection on Chest X-Ray Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022 doi: 10.1109/TCBB.2022.3184319. [DOI] [PubMed] [Google Scholar]
- 30.Lu W., Wang J., Chen Y., Qin X., Xu R., Dimitriadis D., Qin T. Personalized Federated Learning with Adaptive Batchnorm for Healthcare. IEEE Trans. Big Data. 2024:1. doi: 10.1109/TBDATA.2022.3177197. [DOI] [Google Scholar]
- 31.Lian Z., Yang Q., Wang W., Zeng Q., Alazab M., Zhao H., Su C. DEEP-FEL: Decentralized, Efficient and Privacy-Enhanced Federated Edge Learning for Healthcare Cyber Physical Systems. IEEE Trans. Netw. Sci. Eng. 2022;9:3558–3569. doi: 10.1109/TNSE.2022.3175945. [DOI] [Google Scholar]
- 32.Park. S., Kim. G., Kim. J., Kim B., Ye J.C. Federated Split Vision Transformer for COVID-19 CXR Diagnosis using Task-Agnostic Training. Adv. Neural Inf. Process. Syst. 2021;29:24617–24630. [Google Scholar]
- 33.Wang X., Zhang H.G., Xiong X., Hong C., Weber G.M., Brat G.A., Bonzel C.-L., Luo Y., Duan R., Palmer N.P., et al. SurvMaximin: Robust federated approach to transporting survival risk prediction models. J. Biomed. Inform. 2022;134 doi: 10.1016/j.jbi.2022.104176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakur A., Sharma P., Clifton D.A. Dynamic Neural Graphs Based Federated Reptile for Semi-Supervised Multi-Tasking in Healthcare Applications. IEEE J. Biomed. Health Inform. 2022;26:1761–1772. doi: 10.1109/JBHI.2021.3134835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Jiang M., Qin Y., Zhang R., Ling S.H. Intelligent depression detection with asynchronous federated optimization. Complex Intell. Systems. 2023;9:115–131. doi: 10.1007/s40747-022-00729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo J.H., Son H.M., Jeong H., Jang E.-H., Kim A.Y., Yu H.Y., Jeon H.J., Chung T.-M. International Conference on ICT Convergence. 2021. Personalized Federated Learning with Clustering: Non-IID Heart Rate Variability Data Application; pp. 1046–1051. [DOI] [Google Scholar]
- 37.Roland T., Böck C., Tschoellitsch T., Maletzky A., Hochreiter S., Meier J., Klambauer G. Domain Shifts in Machine Learning Based Covid-19 Diagnosis From Blood Tests. J. Med. Syst. 2022;46 doi: 10.1007/s10916-022-01807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haneuse S., Arterburn D., Daniels M.J. Assessing Missing Data Assumptions in EHR-Based Studies: A Complex and Underappreciated Task. JAMA Network Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.0184. [DOI] [PubMed] [Google Scholar]
- 39.Overhage J.M., Ryan P.B., Reich C.G., Hartzema A.G., Stang P.E. Validation of a common data model for active safety surveillance research. J. Am. Med. Inform. Assoc. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verma A.A., Pasricha S.V., Jung H.Y., Kushnir V., Mak D.Y.F., Koppula R., Guo Y., Kwan J.L., Lapointe-Shaw L., Rawal S., et al. Assessing the quality of clinical and administrative data extracted from hospitals: The General Medicine Inpatient Initiative (GEMINI) experience. J. Am. Med. Inform. Assoc. 2021;28:578–587. doi: 10.1093/jamia/ocaa225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nass S.J., Levit L.A., Gostin L.O. Beyond the HIPAA Privacy Rule: Enhancing Privacy, Improving Health Through Research. National Academies Press (US); 2009. The Value and Importance of Health Information Privacy. [DOI] [PubMed] [Google Scholar]
- 42.Antunes R.S., André da Costa C., Küderle A., Yari I.A., Eskofier B. Federated learning for healthcare: Systematic review and architecture proposal. ACM Trans. Intell. Syst. Technol. 2022;13:1–23. doi: 10.1145/3501813. [DOI] [Google Scholar]
- 43.Prayitno. Shyu C.R., Putra K.T., Chen H.C., Tsai Y.Y., Hossain K.S.M.T., Jiang W., Shae Z.Y. A systematic review of federated learning in the healthcare area: From the perspective of data properties and applications. Appl. Sci. 2021;11 doi: 10.3390/app112311191. [DOI] [Google Scholar]
- 44.Crowson M.G., Moukheiber D., Arévalo A.R., Lam B.D., Mantena S., Rana A., Goss D., Bates D.W., Celi L.A. A systematic review of federated learning applications for biomedical data. PLOS Digital Health. 2022;1 doi: 10.1371/journal.pdig.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosseini S.M., Sikaroudi M., Babaie M., Tizhoosh H.R. Proportionally Fair Hospital Collaborations in Federated Learning of Histopathology Images. IEEE Trans. Med. Imaging. 2023;42:1982–1995. doi: 10.1109/TMI.2023.3234450. [DOI] [PubMed] [Google Scholar]
- 46.Alam M.U., FedSepsis R.R. A Federated Multi-Modal Deep Learning-Based Internet of Medical Things Application for Early Detection of Sepsis from Electronic Health Records Using Raspberry Pi and Jetson Nano Devices. Sensors. 2023;23 doi: 10.3390/s23020970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., Li X., Qiu X., Zhang X., Brusic V., Zhao J. A privacy preserving framework for federated learning in smart healthcare systems. Information Processing and Management. 2023;60 doi: 10.1016/j.ipm.2022.103167. [DOI] [Google Scholar]
- 48.Nguyen T.V., Dakka M.A., Diakiw S.M., VerMilyea M.D., Perugini M., Hall J.M.M., Perugini D. A novel decentralized federated learning approach to train on globally distributed, poor quality, and protected private medical data. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-12833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reps J.M., Williams R.D., Schuemie M.J., Ryan P.B., Rijnbeek P.R. Learning patient-level prediction models across multiple healthcare databases: evaluation of ensembles for increasing model transportability. BMC Medical Informatics and Decision Making22. 2022;22 doi: 10.1186/s12911-022-01879-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foley P., Sheller M.J., Edwards B., Pati S., Riviera W., Sharma M., Narayana Moorthy P., Wang S.-H., Martin J., Mirhaji P., et al. OpenFL: the open federated learning library. Physics in Medicine and Biology. 2022;67 doi: 10.1088/1361-6560/ac97d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paragliola G. Evaluation of the trade-off between performance and communication costs in federated learning scenario. Future Gener. Comput. Syst. 2022;136:282–293. doi: 10.1016/j.future.2022.06.006. [DOI] [Google Scholar]
- 52.Liu X., Zhao J., Li J., Cao B., Lv Z. Federated Neural Architecture Search for Medical Data Security. IEEE Trans. Ind. Inform. 2022;18:5628–5636. doi: 10.1109/TII.2022.3144016. [DOI] [Google Scholar]
- 53.Gong Q., Ruan H., Chen Y., Su X. EMDL 2022 - Proceedings of the 6th International Workshop on Embedded and Mobile Deep Learning, Part of MobiSys 2022. 2022. CloudyFL: a cloudlet-based federated learning framework for sensing user behavior using wearable devices; pp. 13–18. [DOI] [Google Scholar]
- 54.Sav S., Bossuat J.-P., Troncoso-Pastoriza J.R., Claassen M., Hubaux J.-P. Privacy-preserving federated neural network learning for disease-associated cell classification. Patterns. 2022;3 doi: 10.1016/j.patter.2022.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilberger H., Hanke S., Bodenler M. Federated Learning with Dynamic Model Exchange. Electronics. 2022;11:1530. doi: 10.3390/electronics11101530. [DOI] [Google Scholar]
- 56.Qu L., Balachandar N., Zhang M., Rubin D. Handling data heterogeneity with generative replay in collaborative learning for medical imaging. Medical Image Analysis. 2022;78 doi: 10.1016/j.media.2022.102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paragliola G., Coronato A. Definition of a novel federated learning approach to reduce communication costs. Expert Systems with Applications. 2022;189 doi: 10.1016/j.eswa.2021.116109. [DOI] [Google Scholar]
- 58.Li J., Meng Y., Ma L., Du S., Zhu H., Pei Q., Shen X. A Federated Learning Based Privacy-Preserving Smart Healthcare System. IEEE Trans. Ind. Inform. 2022;18:2021–2031. doi: 10.1109/TII.2021.3098010. [DOI] [Google Scholar]
- 59.Zhang Y., Zhang M., Xie Z., Ding Y., Huang J., Yao J., Lv Y., Zuo J. Research Progress and Direction of Novel Organelle-Migrasomes. IEEE/ACM Transactions on Networking. 2022;15:134–147. doi: 10.1109/TNET.2021.3103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu D., Jaafar W., Yanikomeroglu H. GLOBECOM 2022-2022 IEEE. Global Communications Conference; 2022. On the Design of Communication-Efficient Federated Learning for Health Monitoring. [DOI] [Google Scholar]
- 61.Balkus S.V., Fang H., Wang H. Proceedings - 2022 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies. Vol. 2022. CHASE; 2022. Federated Fuzzy Clustering for Longitudinal Health Data; pp. 128–132. [Google Scholar]
- 62.Shen Z., Ding F., Yao Y., Bhardwaj A., Guo Z., Yu K. A Privacy-Preserving Social Computing Framework for Health Management Using Federated Learning. IEEE Trans. Comput. Soc. Syst. 2023;10:1666–1678. doi: 10.1109/TCSS.2022.3222682. [DOI] [Google Scholar]
- 63.Kumar S., Lakshminarayanan A., Chang K., Guretno F., Mien I.H., Kalpathy-Cramer J., Krishnaswamy P., Singh P. International Workshop on Distributed, Collaborative, and Federated Learning. Springer; 2022. Towards more efficient data valuation in healthcare federated learning using ensembling; pp. 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L., Yu X., Cai X., He X., Liu Y. Contract Theory Based Incentive Mechanism for Federated Learning in Health CrowdSensing. IEEE Internet Things J. 2023;10:4475–4489. doi: 10.1109/JIOT.2022.3218008. [DOI] [Google Scholar]
- 65.Chang C., Bu Z., CEDAR L.Q. communication efficient distributed analysis for regressions. Biometrics. 2022 doi: 10.1111/biom.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wen T., Zhao S., Zhang R. Federated Semi-Supervised Learning Through a Combination of Self and Cross Model Ensembling. Proceedings of the International Joint Conference on Neural Networks. 2022 doi: 10.1109/IJCNN55064.2022.9892834. [DOI] [Google Scholar]
- 67.Feng T., Semi-FedSER N.S. Proceedings of the Annual Conference of the International Speech Communication Association. INTERSPEECH; 2022. Semi-supervised Learning for Speech Emotion Recognition On Federated Learning using Multiview Pseudo-Labeling; pp. 5050–5054. [DOI] [Google Scholar]
- 68.Shin Y.A., Noh G., Jeong I.R., Chun J.Y. Securing a Local Training Dataset Size in Federated Learning. IEEE Access. 2022;10:104135–104143. doi: 10.1109/ACCESS.2022.3210702. [DOI] [Google Scholar]
- 69.Liu X., Zhang M., Jiang Z., Patel S., McDuff D. IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops. 2022. Federated Remote Physiological Measurement with Imperfect Data; pp. 2154–2163. [DOI] [Google Scholar]
- 70.Gencturk M., Sinaci A.A., Cicekli N.K. BOFRF: A Novel Boosting-Based Federated Random Forest Algorithm on Horizontally Partitioned Data. IEEE Access. 2022;10:89835–89851. doi: 10.1109/ACCESS.2022.3202008. [DOI] [Google Scholar]
- 71.Souza R., Tuladhar A., Mouches P., Wilms M., Tyagi L., Forkert N.D. International MICCAI Brainlesion Workshop. 2022. Multi-institutional Travelling Model for Tumor Segmentation in MRI Datasets; pp. 420–432. [Google Scholar]
- 72.Guo K., Chen T., Ren S., Li N., Hu M., Kang J. Federated Learning Empowered Real-Time Medical Data Processing Method for Smart Healthcare. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022 doi: 10.1109/TCBB.2022.3185395. [DOI] [PubMed] [Google Scholar]
- 73.Zhang L., Xu J., Vijayakumar P., Sharma P.K., Ghosh U. Homomorphic Encryption-based Privacy-preserving Federated Learning in IoT-enabled Healthcare System. IEEE Trans. Netw. Sci. Eng. 2023;10:2864–2880. doi: 10.1109/TNSE.2022.3185327. [DOI] [Google Scholar]
- 74.Presotto R., Civitarese G., Bettini C. 2022 IEEE International Conference on Pervasive Computing and Communications, PerCom. Vol. 2022. 2022. FedCLAR: Federated Clustering for Personalized Sensor-Based Human Activity Recognition; pp. 227–236. [DOI] [Google Scholar]
- 75.Oh H., Yu J., Kim N., Kim D., Lee J., Yang J. 4th International Conference on Artificial Intelligence in Information and Communication, ICAIIC 2022 - Proceedings. 2022. A federated binarized neural network model for constrained devices in IoT healthcare services; pp. 241–245. [Google Scholar]
- 76.Wang R., Lai J., Zhang Z., Li X., Vijayakumar P., Karuppiah M. Privacy-Preserving Federated Learning for Internet of Medical Things under Edge Computing. IEEE J. Biomed. Health Inform. 2023;27:854–865. doi: 10.1109/JBHI.2022.3157725. [DOI] [PubMed] [Google Scholar]
- 77.Camajori Tedeschini B., Savazzi S., Stoklasa R., Barbieri L., Stathopoulos I., Nicoli M., Serio L. Decentralized Federated Learning for Healthcare Networks: A Case Study on Tumor Segmentation. IEEE Access. 2022;10:8693–8708. doi: 10.1109/ACCESS.2022.3141913. [DOI] [Google Scholar]
- 78.Cholakoska A., Pfitzner B., Gjoreski H., Rakovic V., Arnrich B., Kalendar M. Adjunct Proceedings of the 2021 ACM International Joint Conference on Pervasive and Ubiquitous Computing and Proceedings of the 2021 ACM International Symposium on Wearable Computers. 2021. Differentially Private Federated Learning for Anomaly Detection in eHealth Networks; pp. 514–518. [DOI] [Google Scholar]
- 79.Rajotte J.-F., Mukherjee S., Robinson C., Ortiz A., West C., Ferres J.M.L., Ng R.T. Proceedings of the 2021 Conference on Information Technology for Social Good. 2021. Reducing bias and increasing utility by federated generative modeling of medical images using a centralized adversary; pp. 79–84. [DOI] [Google Scholar]
- 80.Xi B., Li S., Li J., Liu H., Liu H., Zhu H.B.F.L. 2021 IEEE/ACM 29th International Symposium on Quality of Service, IWQOS 2021. 2021. Backdoor Detection on Federated Learning in e-Health. [DOI] [Google Scholar]
- 81.Papadopoulos P., Abramson W., Hall A.J., Pitropakis N., Buchanan W.J. Privacy and Trust Redefined in Federated Machine Learning. Mach. Learn. Knowl. Ext. 2021;3:333–356. doi: 10.3390/make3020017. [DOI] [Google Scholar]
- 82.Zhang D.Y., Kou Z., Wang D. FedSens: A Federated Learning Approach for Smart Health Sensing with Class Imbalance in Resource Constrained Edge Computing. Proc. - IEEE INFOCOM. 2021 doi: 10.1109/INFOCOM42981.2021.9488776. [DOI] [Google Scholar]
- 83.Ma J., Zhang Q., Lou J., Xiong L., Ho J.C. Communication efficient federated generalized tensor factorization for collaborative health data analytics. Proc. Int. World. Wide. Web Conf. 2021;2021:171–182. doi: 10.1145/3442381.3449832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kerkouche R., Acs G., Castelluccia C., Genevès P. Proceedings of the 2021 ACM Conference on Health, Inference, and Learning. 2021. Privacy-preserving and bandwidth-efficient federated learning: An application to in-hospital mortality prediction; pp. 25–35. [DOI] [Google Scholar]
- 85.Brisimi T.S., Chen R., Mela T., Olshevsky A., Paschalidis I.C., Shi W. Federated learning of predictive models from federated Electronic Health Records. Int. J. Med. Inform. 2018;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Y., Yang X., Qin X., Yu H., Chan P., Shen Z. In: Federated Learning. Yang Q., Fan L., Yu H., editors. Springer; 2020. Dealing with Label Quality Disparity in Federated Learning; pp. 108–121. [Google Scholar]
- 87.Lu S., Zhang Y., Wang Y. Decentralized Federated Learning for Electronic Health Records. 54th Annual Conference on Information Sciences and Systems, CISS 2020. 2020 doi: 10.1109/CISS48834.2020.1570617414. [DOI] [Google Scholar]
- 88.Chen H., Li H., Xu G., Zhang Y., Luo X. Achieving Privacy-preserving Federated Learning with Irrelevant Updates over E-Health Applications. IEEE Int. Conf. Commun. 2020 doi: 10.1109/ICC40277.2020.9149385. [DOI] [Google Scholar]
- 89.Hao M., Li H., Xu G., Liu Z., Chen Z. Privacy-aware and Resource-saving Collaborative Learning for Healthcare in Cloud Computing. IEEE Int. Conf. Commun. 2020 doi: 10.1109/ICC40277.2020.9148979. [DOI] [Google Scholar]
- 90.Bey R., Goussault R., Grolleau F., Benchoufi M., Porcher R. Fold-stratified cross-validation for unbiased and privacy-preserving federated learning. J. Am. Med. Inform. Assoc. 2020;27:1244–1251. doi: 10.1093/jamia/ocaa096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choudhury O., Gkoulalas-Divanis A., Salonidis T., Sylla I., Park Y., Hsu G., Das A. A syntactic approach for privacy-preserving federated learning. Front. Artif. Intell. Appl. 2020;325:1762–1769. doi: 10.3233/FAIA200290. [DOI] [Google Scholar]
- 92.Xie Y., Li P., Zhu X., Wu Q. Federated diabetes mellitus analysis via homomorphic encryption. J. Phys. Conf. Ser. 2020;1684:12033. doi: 10.1088/1742-6596/1684/1/012033. [DOI] [Google Scholar]
- 93.Gupta D., Kayode O., Bhatt S., Gupta M., Tosun A.S. Vol. 2021. CIC; 2021. Hierarchical Federated Learning based Anomaly Detection using Digital Twins for Smart Healthcare; pp. 16–25. (Proceedings - 2021 IEEE 7th International Conference on Collaboration and Internet Computing). [DOI] [Google Scholar]
- 94.Ma J., Zhang Q., Lou J., Xiong L., Bhavani S., Ho J.C. Communication Efficient Tensor Factorization for Decentralized Healthcare Networks. InProc. IEEE Int. Conf. Data Min. 2021;2021:1216–1221. doi: 10.1109/icdm51629.2021.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mocanu I., Smadu R., Dragoi M., Mocanu A., Cramariuc O. 2021 9th E-Health and Bioengineering Conference, EHB 2021. 2021. Testing Federated Learning on Health and Wellbeing Data. [DOI] [Google Scholar]
- 96.Wu Y., Zeng D., Wang Z., Sheng Y., Yang L., James A.J., Shi Y., Hu J. IEEE/ACM International Conference on Computer-Aided Design, Digest of Technical Papers. ICCAD; 2021. Federated Contrastive Learning for Dermatological Disease Diagnosis via On-device Learning. [DOI] [Google Scholar]
- 97.Sakib S., Fouda M.M., Md Fadlullah Z., Abualsaud K., Yaacoub E., Guizani M. 2021 IEEE International Mediterranean Conference on Communications and Networking. MeditCom; 2021. Asynchronous Federated Learning-based ECG Analysis for Arrhythmia Detection; pp. 277–282. [DOI] [Google Scholar]
- 98.Yu H., Chen Z., Zhang X., Chen X., Zhuang F., Xiong H., Cheng X. FedHAR: Semi-Supervised Online Learning for Personalized Federated Human Activity Recognition. IEEE Trans. Mob. Comput. 2023;22:3318–3332. doi: 10.1109/TMC.2021.3136853. [DOI] [Google Scholar]
- 99.Shen C., Wang P., Roth H.R., Yang D., Xu D., Oda M., Wang W., Fuh C.S., Chen P.T., Liu K.L., et al. In: Clinical Image-Based Procedures, Distributed and Collaborative Learning, Artificial Intelligence for Combating COVID-19 and Secure and Privacy-Preserving Machine Learning. Laura C.O., Cardoso M.J., Rosen-Zvi M., Shekhar R., Wesarg S., Erdt M., Drechler K., Chen Y., et al., editors. Springer; 2021. Multi-task Federated Learning for Heterogeneous Pancreas Segmentation; pp. 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng X., Shah S.B.H., Ren X., Li F., Nawaf L., Chakraborty C., Fayaz M. Mobile Edge Computing Enabled Efficient Communication Based on Federated Learning in Internet of Medical Things. Wirel. Commun. Mob. Comput. 2021;2021:1–10. doi: 10.1155/2021/4410894. [DOI] [Google Scholar]
- 101.Ji J., Yan D., Mu Z. Proceedings of the 2022 6th International Conference on Machine Learning and Soft Computing. 2022. Personnel status detection model suitable for vertical federated learning structure; pp. 98–104. [Google Scholar]
- 102.Roth H.R., Hatamizadeh A., Xu Z., Zhao C., Li W., Myronenko A., Xu D. International Workshop on Distributed, Collaborative, and Federated Learning. Springer; 2022. Split-u-net: Preventing data leakage in split learning for collaborative multi-modal brain tumor segmentation; pp. 47–57. [DOI] [Google Scholar]
- 103.Zhang R., Li H., Hao M., Chen H., Zhang Y. IEEE International Conference on Communications; 2022. Secure Feature Selection for Vertical Federated Learning in eHealth Systems; pp. 1257–1262. [DOI] [Google Scholar]
- 104.Sun W., Chen Y., Yang X., Cao J., Song Y. Proceedings - 2021 IEEE International Conference on Bioinformatics and Biomedicine. BIBM; 2021. FedIO: Bridge Inner- and Outer-hospital Information for Perioperative Complications Prognostic Prediction via Federated Learning; pp. 3215–3221. [DOI] [Google Scholar]
- 105.Kim J., Li W., Bath T., Jiang X., Ohno-Machado L. AMIA … Annual Symposium proceedings. AMIA Symposium; 2021. VERTIcal Grid lOgistic regression with Confidence Intervals (VERTIGO-CI) pp. 355–364. [PMC free article] [PubMed] [Google Scholar]
- 106.Raza A., Tran K.P., Koehl L., Li S. Designing ECG monitoring healthcare system with federated transfer learning and explainable AI. Knowl.-Based Syst. 2022;236 doi: 10.1016/j.knosys.2021.107763. [DOI] [Google Scholar]
- 107.Yu C., Shen S., Wang S., Zhang K., Zhao H. Efficient Multi-Layer Stochastic Gradient Descent Algorithm for Federated Learning in E-health. IEEE Int. Conf. Commun. 2022:1263–1268. doi: 10.1109/ICC45855.2022.9839100. [DOI] [Google Scholar]
- 108.Beutel, D. J. Topal, T., Mathur, A., Qiu, X., Fernandez-Marques, J., Gao, Y., Sani, L., Li, K.H., Parcollet, T., de Gusmão, P.P.B. and Lane, N.D. Flower: A Friendly Federated Learning Research Framework. Preprint at arXiv DOI 10.48550/arXiv.2007.14390.
- 109.The TensorFlow Federated Authors . 2018. TensorFlow Federated.https://github.com/tensorflow/federated [Google Scholar]
- 110.Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G.S., Davis A., Dean J., Devin M., Ghemawat S. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. 2015. http://download.tensorflow.org/paper/whitepaper2015.pdf
- 111.Ziller A., Trask A., Lopardo A., Szymkow B., Wagner B., Bluemke E., Nounahon J.M., Passerat-Palmbach J., Prakash K., Rose N., et al. Pysyft: A library for easy federated learning. Federated Learning Systems: Towards Next-Generation. 2021;AI:111–139. doi: 10.1007/978-3-030-70604-3_5. [DOI] [Google Scholar]
- 112.Liu Y., Fan T., Chen T., Xu Q., Yang Q. Fate: An industrial grade platform for collaborative learning with data protection. J. Mach. Learn. Res. 2021;22:1–6. [Google Scholar]
- 113.Roth H.R., Cheng Y., Wen Y., Yang I., Xu Z., Hsieh Y.T., Kersten K., Harouni A., Zhao C., Lu K., Zhang Z. Workshop on Federated Learning: Recent Advances and New Challenges (in Conjunction with NeurIPS 2022) 2022. NVIDIA FLARE: Federated learning from simulation to real-world. [Google Scholar]
- 114.Johnson A.E.W., Pollard T.J., Shen L., Lehman L.W.H., Feng M., Ghassemi M., Moody B., Szolovits P., Celi L.A., Mark R.G. Mimic-iii, a freely accessible critical care database. Sci. Data. 2016;3:160035–160039. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee J., Sun J., Wang F., Wang S., Jun C.H., Jiang X. Privacy-preserving patient similarity learning in a federated environment: development and analysis. JMIR Med. Inf. 2018;6:e20. doi: 10.2196/medinform.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. Smote: synthetic minority over-sampling technique. J. Art. Intell. Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 117.Dwork C., McSherry F., Nissim K., Smith A. Theory of Cryptography: Third Theory of Cryptography Conference, TCC 2006, New York, NY, USA, March 4-7, 2006. Proceedings 3. Springer; 2006. Calibrating noise to sensitivity in private data analysis; pp. 265–284. [DOI] [Google Scholar]
- 118.Andrew G., Chien S., Papernot N. Google; 2023. TensorFlow Privacy.https://www.tensorflow.org/responsible_ai/privacy/guide [Google Scholar]
- 119.McMahan B., Moore E., Ramage D., Hampson S., y Arcas B.A. Artificial intelligence and statistics. PMLR; 2017. Communication-efficient learning of deep networks from decentralized data; pp. 1273–1282. [Google Scholar]
- 120.Mitra R., McGough S.F., Chakraborti T., Holmes C., Copping R., Hagenbuch N., Biedermann S., Noonan J., Lehmann B., Shenvi A., et al. Learning from data with structured missingness. Nat. Mach. Intel. 2023;5:13–23. doi: 10.1038/s42256-022-00596-z. [DOI] [Google Scholar]
- 121.Van Buuren S. CRC press; 2018. Flexible Imputation of Missing Data. [Google Scholar]
- 122.Groenwold R.H.H. Informative missingness in electronic health record systems: The curse of knowing. Diagn. Prog. Res. 2020;4:8. doi: 10.1186/s41512-020-00077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shadbahr T., Roberts M., Stanczuk J., Gilbey J., Teare P., Dittmer S., Thorpe M., Torné R.V., Sala E., Lió P., et al. The impact of imputation quality on machine learning classifiers for datasets with missing values. Comm. Med. 2023;3 doi: 10.1038/s43856-023-00356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rieke N., Hancox J., Li W., Milletarì F., Roth H.R., Albarqouni S., Bakas S., Galtier M.N., Landman B.A., Maier-Hein K., et al. The future of digital health with federated learning. NPJ Digital Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., Aviles-Rivero A.I., Etmann C., McCague C., Beer L., et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for covid-19 using chest radiographs and ct scans. Nat. Mach. Intell. 2021;3:199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 126.Simkó A., Garpebring A., Jonsson J., Nyholm T., Löfstedt T. Medical Imaging with Deep Learning. 2023. Reproducibility of the methods in medical imaging with deep learning. [Google Scholar]
- 127.Qiu P., Zhang X., Ji S., Fu C., Yang X., Wang T. Hashvfl: Defending against data reconstruction attacks in vertical federated learning. IEEE Trans. Inform. Forensic. Secur. 2024;19:3435–3450. doi: 10.1109/TIFS.2024.3356164. [DOI] [Google Scholar]
- 128.Wang L., Xu S., Wang X., Zhu Q. Addressing class imbalance in federated learning. Proc. AAAI Conf. Artif. Intell. 2021;35:10165–10173. doi: 10.1609/aaai.v35i11.17219. [DOI] [Google Scholar]
- 129.Fang X., Ye M. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022. Robust federated learning with noisy and heterogeneous clients; pp. 10072–10081. [DOI] [Google Scholar]
- 130.Kavianpour S., Sutherland J., Mansouri-Benssassi E., Coull N., Jefferson E. Next-generation capabilities in trusted research environments: interview study. J. Med. Internet Res. 2022;24 doi: 10.2196/33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prechelt L. Neural Networks: Tricks of the trade. Springer; 2002. Early stopping-but when? pp. 55–69. [DOI] [Google Scholar]
- 132.Geiping J., Bauermeister H., Dröge H., Moeller M. In: Larochelle H., Ranzato M., Hadsell R., Balcan M., Lin H., editors. Vol. 33. Curran Associates, Inc.; 2020. Inverting gradients - how easy is it to break privacy in federated learning? pp. 16937–16947. (Advances in Neural Information Processing Systems). [Google Scholar]
- 133.Zhang Q., Ma J., Xiao Y., Lou J., Xiong L. 2020 IEEE International Conference on Big Data (Big Data) 2020. Broadening Differential Privacy for Deep Learning Against Model Inversion Attacks; pp. 1061–1070. [DOI] [Google Scholar]
- 134.Fredrikson M., Jha S., Ristenpart T. Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security, CCS ’15. Association for Computing Machinery; 2015. Model Inversion Attacks that Exploit Confidence Information and Basic Countermeasures; pp. 1322–1333. [DOI] [Google Scholar]
- 135.Bagdasaryan E., Veit A., Hua Y., Estrin D., Shmatikov V. In: Proceedings of the Twenty Third International Conference on Artificial Intelligence and Statistics, vol. 108 of Proceedings of Machine Learning Research. Chiappa S., Calandra R., editors. PMLR; 2020. How to backdoor federated learning; pp. 2938–2948. [Google Scholar]
- 136.Kairouz P., McMahan H.B., Avent B., Bellet A., Bennis M., Bhagoji A.N., Bonawitz K., Charles Z., Cormode G., Cummings R., et al. Advances and open problems in federated learning. Foundations and Trends in Machine Learning. 2021;14:1–210. [Google Scholar]
- 137.Cho Y.J., Wang J., Joshi G. International Conference on Artificial Intelligence and Statistics. PMLR; 2022. Towards understanding biased client selection in federated learning; pp. 10351–10375. [Google Scholar]
- 138.Abay A., Zhou Y., Baracaldo N., Rajamoni S., Chuba E., Ludwig H. Mitigating Bias in Federated Learning. arXiv. 2020 doi: 10.48550/arXiv.2012.02447. Preprint at. [DOI] [Google Scholar]
- 139.Mongan J., Moy L., Kahn J.,C.E. 2020. Checklist for Artificial Intelligence in Medical Imaging (Claim): A Guide for Authors and Reviewers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Marchand T., Muzellec B., Beguier C., Ogier du Terrail J., Andreux M.S. a safe feature gaussianization protocol for federated learning. Adv. Neural Inf. Process. Syst. 2022;35:36585–36598. [Google Scholar]
- 141.Islam M.Z. University of Newcastle; Brisbane: 2007. Privacy Preservation in Data Mining through Noise Addition. [Google Scholar]
- 142.Li T., Li N., Zhang J., Molloy I. Slicing: A New Approach for Privacy Preserving Data Publishing. IEEE Trans. Knowl. Data Eng. 2012;24:561–574. doi: 10.1109/TKDE.2010.236. [DOI] [Google Scholar]
- 143.Breger A., Selby I., Roberts M., Babar J., Gkrania-Klotsas E., Preller J., Escudero Sánchez L., AIX-COVNET Collaboration. Rudd J.H.F., Aston J.A.D., et al. A pipeline to further enhance quality, integrity and reusability of the nccid clinical data. Scientific Data. 2023;10 doi: 10.1038/s41597-023-02340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou H.H., Singh V., Johnson S.C., Wahba G., Alzheimer’s Disease Neuroimaging Initiative Statistical tests and identifiability conditions for pooling and analyzing multisite datasets. Proc. Natl. Acad. Sci. USA. 2018;115:1481–1486. doi: 10.1073/pnas.1719747115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kotecha D., Asselbergs F.W., Achenbach S., Anker S.D., Atar D., Baigent C., Banerjee A., Beger B., Brobert G., Casadei B., et al. Code-ehr best practice framework for the use of structured electronic healthcare records in clinical research. BMJ. 2022;378 doi: 10.1136/bmj-2021-069048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ma C., Li J., Ding M., Yang H.H., Shu F., Quek T.Q.S., Poor H.V. On safeguarding privacy and security in the framework of federated learning. IEEE network. 2020;34:242–248. doi: 10.1109/MNET.001.1900506. [DOI] [Google Scholar]
- 147.Foley P., Sheller M.J., Edwards B., Pati S., Riviera W., Sharma M., Narayana Moorthy P., Wang S.H., Martin J., Mirhaji P., et al. Openfl: the open federated learning library. Physics in Medicine & Biology. 2022;67 doi: 10.1088/1361-6560/ac97d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li G., Zhao Y., Li Y.C. 2023 IEEE Wireless Communications and Networking Conference (WCNC) IEEE; 2023. Certificateless authentication-based trustworthy federated learning for 6g semantic communications; pp. 1–6. [DOI] [Google Scholar]
- 149.Wolff R.F., Moons K.G.M., Riley R.D., Whiting P.F., Westwood M., Collins G.S., Reitsma J.B., Kleijnen J., Mallett S., PROBAST Group† Probast: a tool to assess the risk of bias and applicability of prediction model studies. Ann. Int. Med. 2019;170:51–58. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- 150.Li X., Huang K., Yang W., Wang S., Zhang Z. International Conference on Learning Representations. 2020. On the convergence of fedavg on non-iid data. [Google Scholar]
- 151.Zhou F., Cong G. Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, IJCAI-18. International Joint Conferences on Artificial Intelligence Organization; 2018. On the convergence properties of a k-step averaging stochastic gradient descent algorithm for nonconvex optimization; pp. 3219–3227. [DOI] [Google Scholar]
- 152.European Commission Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC (General Data Protection Regulation) (Text with EEA relevance) 2016. https://eur-lex.europa.eu/eli/reg/2016/679/oj
- 153.Centers for Medicare & Medicaid Services The Health Insurance Portability and Accountability Act of 1996 (HIPAA) 1996. http://www.hhs.gov/hipaa/
- 154.Veritas Health Innovation Ltd. Covidence Systematic Review Software. https://www.covidence.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.