Abstract

Interactions among biomacromolecules, predominantly noncovalent, underpin biological processes. However, recent advancements in biospecific chemistry have enabled the creation of specific covalent bonds between biomolecules, both in vitro and in vivo. This Review traces the evolution of biospecific chemistry in proteins, emphasizing the role of genetically encoded latent bioreactive amino acids. These amino acids react selectively with adjacent natural groups through proximity-enabled bioreactivity, enabling targeted covalent linkages. We explore various latent bioreactive amino acids designed to target different protein residues, ribonucleic acids, and carbohydrates. We then discuss how these novel covalent linkages can drive challenging protein properties and capture transient protein–protein and protein–RNA interactions in vivo. Additionally, we examine the application of covalent peptides as potential therapeutic agents and site-specific conjugates for native antibodies, highlighting their capacity to form stable linkages with target molecules. A significant focus is placed on proximity-enabled reactive therapeutics (PERx), a pioneering technology in covalent protein therapeutics. We detail its wide-ranging applications in immunotherapy, viral neutralization, and targeted radionuclide therapy. Finally, we present a perspective on the existing challenges within biospecific chemistry and discuss the potential avenues for future exploration and advancement in this rapidly evolving field.
1. Introduction
There exist four major classes of biomacromolecules in life forms on earth: proteins, nucleic acids, carbohydrates, and lipids. These biomacromolecules engage in intricate interactions, collaborating to execute diverse biological functions essential for life. Their interactions, which encompass electrostatic forces, van der Waals forces, π-effects, and hydrophobic effects, are primarily noncovalent in nature. Covalent connections between biomacromolecules usually require enzymatic catalysis, such as the attachment of ubiquitin and ubiquitin-like proteins to other proteins,1 as well as the attachment of glycans to proteins, lipids, and RNAs.2 Interaction through spontaneous covalent bonding is rare, with the formation of disulfide bonds in proteins between cysteine residues being a notable exception.3 Although other covalent bonds, such as the isopeptide bond and N–O–S bridge, have been identified in certain proteins,4,5 their formation demands a specialized protein microenvironment, limiting their general applicability across diverse proteins. Noncovalent interactions are typically characterized by their relative weakness, transience, and reversibility. In contrast, covalent bonding offers a more robust, selective, and stable form of connection. Although evolutionary pressure has not favored the proliferation of additional covalent linkages in biomolecules, exploring new covalent bonds holds the potential to unlock novel structures, properties, and functions.6 This exploration is particularly valuable for advancing the research, control, and utilization of biological activities.
For covalent reactions with biomacromolecules, bio-orthogonal click chemistry has gained prominence.7,8 These reactions avoid interference with biological processes and efficiently occur under physiological conditions between two abiotic bio-orthogonal functional groups. One bio-orthogonal functional group is introduced into target biomolecules through metabolic or genetic engineering, while the complementary group is integrated into the probe molecule.9 This setup allows for the selective formation of covalent bonds between the probe and the target biomolecule in the presence of other biomolecules. While bio-orthogonal click chemistry has transformed chemical biology and significantly advanced biotechnology, its application in living organisms may be hindered by the impracticality of introducing exogenous components in certain situations, particularly for in vivo applications involving disease-related biomacromolecules in humans.
Over the past decade, biospecific chemistry has flourished, enabling the selective covalent targeting of endogenous biomacromolecules in cells and organisms without altering the target biomacromolecules themselves. This approach requires only one latent bioreactive functional group in the reactant biomacromolecule, which selectively reacts with the target biomacromolecule upon binding.6,10,11 Unlike bio-orthogonal chemistry, which is designed not to react with endogenous biomacromolecules, biospecific chemistry is tailored to react with native biomacromolecules under cellular or physiological conditions with high specificity. While bio-orthogonal chemistry has primarily been used for labeling biomacromolecules with small-molecule probes, biospecific chemistry aims to create covalent linkages within or between biomacromolecules.
Here, we provide an account of the development of biospecific chemistry, tracing its initiation in proteins. We discuss the design and introduction of different latent bioreactive functional groups into proteins to covalently target various amino acid residues of proteins, ribonucleic acids, and carbohydrates. Additionally, we showcase how these new covalent linkages among biomacromolecules facilitate the engineering of challenging protein properties, enable the capture of elusive biomolecular interactions in situ for subsequent identification, and support the development of peptide and protein therapeutics that operate in a covalent mode. We conclude with a discussion of the future development of biospecific chemistry and directions to be explored.
2. Biospecific Chemistry to Target Different Classes of Biomacromolecules
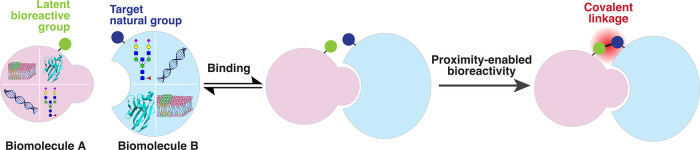
The foundational methodology of biospecific chemistry involves the integration of a latent bioreactive functional group into biomolecule A that is designed to selectively target a specific natural functional group in biomolecule B (Figure 1). The interaction between biomolecules A and B brings the latent bioreactive group into close proximity with its target natural group. Such proximity leads to an increased effective concentration and a reduction in entropy loss, thereby activating the latent bioreactive group’s reaction toward its target group and resulting in the formation of a precise covalent bond. We refer to this process as “biospecific chemistry”, which enables the specific covalent targeting of biomolecules without requiring modifications to the target molecules themselves. Furthermore, biomolecules A and B may belong to different classes; the reaction is also viable within a single biomacromolecule and is applicable across diverse environments, including in vitro conditions, live cells, and whole organisms.
Figure 1.
Biospecific chemistry for introducing covalent linkages between biomacromolecules.
2.1. Biospecific Chemistry through Proximity-Enabled Bioreactivity
Selective introduction of new covalent linkages via biospecific chemistry was initially enabled in proteins.10 The approach involves the genetic incorporation of an unnatural amino acid (Uaa), whose side chain bears a functional group capable of reacting with the side chain of the target natural residue, into proteins,12 facilitating the spontaneous formation of a covalent linkage.11 A suitable chemical reaction for this process must be efficient enough to react with the relatively unreactive natural amino acid side chains and must also proceed under mild conditions to ensure biocompatibility. Given the prevalence of natural amino acids and residues within proteins and cells, achieving selectivity in targeting a specific residue within the vast proteome presented a significant challenge. Additionally, off-target reactions involving bioreactive Uaas could lead to cytotoxic effects, and side reactions with the translational machinery might hinder the ribosomal incorporation of these Uaas.
The paradox of balancing bioreactivity, selectivity, and genetic encoding was addressed through the concept of proximity-enabled bioreactivity (Figure 2).10 This approach fine-tunes the reactivity of the Uaa to prevent undesirable reactions with free amino acids, protein residues, and other biomolecules inside cells under physiological conditions, thus permitting genetic incorporation. Once the Uaa is incorporated into a protein and positioned close to its target natural residue, the proximity effect reduces entropy loss and increases the effective concentration of reactants. This significantly accelerates the reaction rate, thereby facilitating the specific formation of the covalent bond between the Uaa and its target residue. The term “latent bioreactive Uaa” was coined to describe this specific type of Uaa.6 This designation highlights its potential for reactivity toward biomolecules in contrast to the majority of genetically encoded Uaas, which are bio-orthogonal or chemically inert,13−15 and underscores its hidden reactivity, which is triggered by proximity.
Figure 2.

Building new covalent bonds in proteins through proximity-enabled bioreactivity. The process involves the proximity-enabled activation of a latent bioreactive Uaa toward its target natural amino acid residue. The proximity can be achieved either through intramolecular protein folding or conformational changes or via intermolecular protein interactions. The proximity effect accelerates the reaction between the Uaa and the target residue, leading to the specific formation of a covalent bond. Adapted with permission from ref (6). Copyright 2021 John Wiley and Sons.
The breakthrough occurred in 2013, with the development of latent bioreactive Uaa p-2′-fluoroacetyl-phenylalanine (Ffact) to target Cys selectively in proteins (Figure 3).10 This success was driven by the strategic expectation that the sulfhydryl group of Cys, possessing the highest nucleophilicity among natural amino acid side chains, would selectively react with a weak electrophilic group when in close proximity. The fluoromethyl ketone in Ffact was chosen as the weak electrophile because the C–F bond is strong and F is a poor leaving group but the α-carbonyl group increases its reactivity. Indeed, Ffact demonstrated reactivity only at an elevated Cys concentration (10 mM) in vitro, which is substantially higher than intracellular Cys concentrations (<1 mM). Ffact was incorporated into proteins in Escherichia coli cells through genetic code expansion. Mass spectrometric (MS) analysis of these expressed proteins confirmed the absence of adducts involving Cys, glutathione, or imidazole or any modifications to Ffact, indicating that Ffact remained stable and unaltered during protein biosynthesis and purification. Ffact and Cys were subsequently introduced into an affibody and the Z protein, respectively, at proximal sites of their binding interface. Incubation of these two proteins at biocompatible mild conditions led to covalent complex formation with a 63% yield. Noteworthy is the observation that substituting Ffact with the isosteric Fact, lacking only fluorine and thus being unreactive, or altering the position of Cys yielded no detectable covalent complexes. This underscores the inherently proximity-dependent and chemoselective nature of the reaction.
Figure 3.
Genetic encoding of the bioreactive Uaa Ffact and its reaction with Cys through proximity-enabled bioreactivity. (A) Structure of Ffact alongside its isosteric control, Fact. (B) LC-MS analysis shows that 1 mM Ffact reacts completely with 10 mM Cys, but less than 5% reacts at 1 mM Cys in PBS (pH 7.4) at 37 °C for 24 h. (C) Site-specific incorporation of Ffact into an affibody in E. coli, analyzed via SDS-PAGE. (D) ESI-MS analysis of the Ffact mutant affibody from (C), confirming the exclusive incorporation of Ffact at site 36 without modification. (E) Structure of the affibody–Z complex (PDB 1LP1), highlighting the proximal Asp36 and Asn6 sites for introducing Ffact and Cys, respectively. (F) Incubation of affibody(Ffact36) with Z(Cys6) leads to the formation of a covalent affibody–Z complex (indicated by a red arrow) as evidenced by SDS-PAGE analysis. C3, Cys at site 3; N6, Asn at site 6. Incubation time: 1 h. Reproduced with permission from ref (11). Copyright 2017 Elsevier.
Besides covalently linking the affibody–Z protein pair intermolecularly, Ffact was further shown to react with Cys intramolecularly. Ffact was incorporated in the chromophore and Cys was introduced to a proximal site in the β-barrel in various fluorescent proteins. After expressing these proteins in E. coli cells, the chromophore was found to covalently attach to the β-barrel via the intramolecular Ffact–Cys reaction almost quantitatively. In addition to working inside E. coli cells, the Ffact–Cys reaction is also compatible for use on mammalian cell surface to allow a GPCR to capture its peptide ligand.10,16 In short, unlocking the latent reactivity of Ffact toward Cys through the proximity effect paves the way for endowing proteins with new covalent bonding abilities, a strategy that has been generally embraced for developing new biospecific chemistry in ensuing years.
This methodology, which involves the genetic encoding of a latent bioreactive Uaa into a protein to covalently target a natural amino acid residue through proximity-enabled bioreactivity, was later referred to as genetically encoded chemical cross-linking (GECX).17 Extensions of this method, GECX-RNA and GECX-sugar,18,19 have been developed to target RNA nucleotides and carbohydrates, respectively. GECX can be applied intramolecularly to create covalent linkages within proteins, a technique that has been utilized to engineer protein properties, as detailed in section 3. Additionally, GECX can also be used intermolecularly between two proteins, or between a protein and another type of biomacromolecule, to study biomolecular interactions in situ, as described in section 4.
2.2. Covalently Targeting Various Amino Acid Residues in Proteins
In addition to the fluoromethyl ketone contained in Ffact, diverse functional groups installed on different Uaa scaffolds have subsequently been proven suitable for genetic encoding and reactions with various natural amino acid residues through proximity-enabled bioreactivity.
2.2.1. Nucleophilic Substitution of Alkyl or Aryl Halide
To target Cys, a range of haloalkane Uaas were designed and synthesized, with halogen atoms (I, Br, and Cl) connected with aliphatic chains of varying lengths (Figure 4).20 These Phe-scaffold Uaas were incorporated into proteins in both E. coli and mammalian cells using an evolved Methanosarcina mazei tRNAPyl/MmXYRS pair. MS analysis of the mutant proteins confirmed that the Uaas were specifically incorporated without modifications or side reactions. Upon incorporation into the affibody, these haloalkane Uaas reacted with a proximal Cys on the Z protein at the binding interface, resulting in irreversible cross-linking of the two proteins. When installed on the same alkyl chain, the cross-linking efficiency follows the order of I > Br > Cl, consistent with the halide leaving ability in substitution reactions. Meanwhile, benzyl chloride was installed on an azobenzene to develop Uaa Cl-PSCaa, which was demonstrated to react with a proximal Cys intramolecularly to form a photoswitchable bridge on proteins, as detailed in section 3.4.21
Figure 4.
Latent bioreactive Uaas with alkyl or aryl halides that primarily target Cys, as illustrated at the bottom. Target residues are listed in orange.
Substitution of fluorine is generally difficult, which is mainly attributed to its high electronegativity and limited leaving ability. In the context of the fluoromethyl ketone in Ffact, activation is facilitated by the α-carbonyl group.10 On the other hand, the fluoroacetamide group has traditionally been deemed biologically inert and applied in diagnostic applications. However, when fluoroacetamide was installed on Uaa FAcK and incorporated into proteins, it exhibited notable reactivity with Cys in close proximity.22 This observation underscores the significant impact of proximity on enhancing reactivity. Employing this protein-confined proximity strategy holds promise for precisely assessing the chemical reactivity of small molecules toward biomolecules, thereby mitigating the risk of undesirable side reactions in drug development.
In addition to nucleophilic substitution reactions, nucleophilic aromatic substitution (SNAr) has also proven effective in harnessing proximity-enabled reactivity. The genetically incorporated F-PSCaa, characterized by a perfluoro-benzene moiety, demonstrated reactivity with a proximal Cys residue within the protein, which facilitated the in situ construction of a photoswitchable bridge on the protein, as detailed in section 3.4.23
Cys is not always available at desired positions or within target proteins. To enhance the applicability of proximity-enabled bioreactivity across a broader spectrum of proteins, an alkyl bromide was introduced onto a lengthy linear alkyl side chain to augment its orientation flexibility.24 The resulting Uaa BrC6K, upon incorporation into proteins, exhibited the capability to react not only with proximal Cys but also with proximal His and Lys residues. By specifically targeting His490 on HER2, the ZHER2 affibody, featuring BrC6K at site 37, formed covalent cross-links with the endogenous HER2 receptor on cancer cells.24 Interestingly, subsequent investigations showed that BrC6K could also cross-link with proximal Glu and Asp residues.25 In another comprehensive study, MS analysis unveiled a broader reactivity spectrum for bromoalkyl-containing BprY.26 In addition to Cys, His, and Lys, BprY was found to react with Tyr, Ser, Thr, Asp, and Glu in proximity, albeit with lower efficiency.
2.2.2. Addition to Michael Acceptors or Isothiocyanate
The addition of a nucleophilic side chain with a Michael acceptor such as N-arylacrylamide and vinyl sulfonamide has been widely used for the site-specific covalent inhibition of proteins.27,28 Uaas containing acrylamide, namely AcrK and AcrF, were genetically incorporated into proteins and demonstrated reactivity with a proximal Lys residue, albeit with modest yields under basic conditions (Figure 5).29 Vinyl sulfonamides exhibit enhanced reactivity when compared to their acrylamide counterparts. Notably, the incorporation of the Uaa VSF, containing a vinyl sulfonamide, into proteins exhibited remarkable efficiency (86%) in the reaction with a proximal Lys in vitro. When VSF was incorporated into the Herceptin Fab at site 92, the mutant Fab demonstrated the ability to cross-link the native HER2 receptor on mammalian cells, achieving complete labeling within 2 h. It is worth noting that 10 μM N-arylacrylamide has been shown to react with 100 μM Cys in PBS under mild conditions,30 implying that these Michael acceptors may not remain entirely latent within cells.
Figure 5.
Latent bioreactive Uaas with Michael acceptors or isothiocyanate that primarily target Lys, as illustrated at the bottom. Target residues are listed in orange.
Phenyl isothiocyanates are used to modify N-terminal amino group of proteins during Edman degradation.31 Uaa p-isothiocyanate phenylalalnine (pNCSF) was thus developed and genetically incorporated into proteins, allowing for selective conjugation of proteins to amine-containing probes under basic conditions.32 In addition, pNCSF enables the formation of intramolecular and intermolecular thiourea cross-links through addition with a proximal Lys side chain in proteins.
2.2.3. Acyl Transfer via Aryl Carbamate or Aryl Triazole
Aryl carbamates function as moderate electrophiles. An aryl carbamate-containing Uaa, FPheK, has been incorporated into proteins in both E. coli and mammalian cells (Figure 6).33 Incorporation of FPheK into thioredoxin facilitated high-efficiency intramolecular cross-linking with proximal Lys, Cys, or Tyr, with low efficiency observed for His and Ser. In addition, cross-links are formed between FPheK on the heavy chain and a Lys residue on the light chain of the Herceptin Fab. Notably, FPheK-mediated cross-linking in these experiments required a weakly basic pH of 8.5.
Figure 6.
Latent bioreactive Uaas reacting via acyl transfer. The reaction with Lys is shown as an example. Target residues are listed in orange.
The development of aryl-1,2,3-triazole Uaa CATK1 further advances acyl transfer reactions under neutral conditions owing to the triazole’s acidity (pKa 9.4) as an excellent leaving group.34 CATK1, when incorporated into proteins, demonstrated high-efficiency cross-linking with proximal Lys and Tyr and lower efficiency with His and Cys at neutral pH and within E. coli cells. CATK1 is nontoxic to HEK293T cells at concentrations below 0.5 mM.
2.2.4. Sulfur Fluoride Exchange (SuFEx)
SuFEx represents a cutting-edge click chemistry approach, which is distinguished by the exchange of the fluoride connected to the sulfur(VI) center with incoming nucleophiles.35 The robust sulfur–fluoride bonds in high oxidation states exhibit remarkable resistance to oxidation, reduction, hydrolysis, and thermolysis.35 Activation of the S(VI)–F bond for nucleophilic exchange occurs reliably only when the appropriate catalyst–reagent combination is applied.35,36 Aryl fluorosulfates are unreactive toward free amino acids and biomolecules under physiological conditions.30 When serving as warheads in chemical probes that bind proteins specifically, aryl fluorosulfates form covalent linkages with Tyr, Lys, and Ser in the protein binding pocket through SuFEx.37−39
Due to its exceptional biocompatibility and click SuFEx nature, aryl fluorosulfate was implemented as the side chain in the Uaa fluorosulfate-l-tyrosine (FSY), which was genetically incorporated into proteins in both E. coli and mammalian cells (Figure 7).40 FSY (1 mM) was nontoxic to E. coli or mammalian cells. The incorporated FSY efficiently reacted with proximal Lys, His, and Tyr in proteins via SuFEx, both intra- and intermolecularly, and in vitro and in live cells, generating stable linkages resistant to hydrolysis. Furthermore, FSY exhibited reactivity with proximal Cys, Ser, and Thr, yet yielded unstable linkages.41 Ser and Thr are converted to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively; Dha and Dhb can be subsequently used for protein labeling or conjugation. The introduction of an electron-withdrawing fluorine substitution on FSY led to Uaa FFY, accelerating the SuFEx reaction rate by 2.4-fold.42 Additionally, fluorosulfonyloxybenzoyl-l-lysine (FSK), another aryl fluorosulfate-containing Uaa, was genetically encoded in E. coli and mammalian cells, demonstrating a reactive specificity similar to FSY.43 FSK’s longer and more flexible side chain facilitated reactions with residues inaccessible to FSY. Covalent nanobodies were generated that irreversibly bound to epidermal growth factor receptor (EGFR) on mammalian cells, with FSK and FSY targeting distinct positions on EGFR.43 Moreover, meta-fluorosulfate-l-tyrosine (mFSY), genetically encoded in E. coli and mammalian cells, presented an alternate approach with fluorosulfate at the meta position.44 This allowed mFSY to target residues elusive to FSY through a different side chain orientation. Nanobodies incorporating mFSY at specific sites demonstrated the ability to covalently cross-link either EGFR or HER2, a capability not observed with FSY incorporation at the same site, and vice versa. Together, FSY, FSK, and mFSY complement each other in terms of side chain length and orientation.
Figure 7.
Latent bioreactive Uaas capable of the proximity-enabled SuFEx reaction. They all target Tyr, Lys, and His. The reaction of FSY with Tyr is shown as an example.
Another SuFEx-capable latent bioreactive Uaa, SFY, has also been genetically encoded in E. coli and mammalian cells.18,19 SFY is designed to contain an aryl sulfonyl fluoride group, which is more reactive than the aryl fluorosulfate found in other SuFEx-capable Uaas. This enhanced reactivity has been utilized to covalently target carbohydrates, as detailed in section 2.4. To mitigate potential cytotoxicity, a methoxy group has been added to the phenyl ring, moderating the reactivity of the sulfonyl fluoride. SFY has also been shown to react with Lys, His, and Tyr residues when they are in proximity within proteins in both E. coli and mammalian cells. Given that these target residues, namely Lys, His, and Tyr, are often abundant at or near protein binding interfaces, these SuFEx-based latent bioreactive Uaas significantly broaden the scope of proximity-enabled bioreactivity in diverse applications.
2.2.5. Phosphorus Fluoride Exchange (PFEx)
Recent advancements in click chemistry have prominently featured PFEx reactions alongside SuFEx. PFEx reactions enable the substitution of P(V)–F bonds with incoming nucleophiles, leading to the formation of stable tetrahedral P(V)–O and P(V)–N bonds. In small-molecule chemistry, these reactions require a Lewis base catalyst and a silicon-based additive.45 In protein applications, two latent bioreactive amino acids characterized by phosphoramidofluoridate groups, namely, PFY and PFK, have been effectively incorporated into proteins in both E. coli and mammalian cells through genetic code expansion (Figure 8).46 Once integrated, these amino acids can covalently target proximal His, Tyr, Lys, and Cys residues via a proximity-driven PFEx reaction, both in vitro and in vivo, without the need for external reagents. This demonstrates that the proximity of reactants alone is sufficient to activate PFEx in proteins. Interestingly, a water-soluble silicon reagent, Na2SiO3, enhances the PFEx reaction between PFY and Cys/Tyr but diminishes it between PFY and His in proteins. Additionally, the P–N linkage formed between PFY and His in proteins is reversible at temperatures above 50 °C. PFY also shows greater durability in proteins compared with FSY, its SuFEx counterpart, which is advantageous for in vivo applications. The genetic facilitation of PFEx click chemistry in proteins not only enables covalent protein engineering using nature’s preferred phosphate connectors but also lays the groundwork for PFEx’s broader application in biological and biomedical research.
Figure 8.
Latent bioreactive Uaas capable of the proximity-enabled PFEx reaction. They both target Tyr, Lys, His, and Cys. The reaction of PFY with Tyr is shown as an example.
2.2.6. Inducible Proximity-Enabled Bioreactivity
The proximity-enabled bioreactivity of Uaas can be initially constrained and subsequently unleashed through light or chemical conversion, adding an extra layer of control over covalent bond formation in terms of spatial and temporal aspects.
Traditional photo-cross-linking Uaas, utilizing azide, benzophenone, or diazirine, typically progress through radical intermediates upon light activation, lacking specificity for amino acid residues.47,48 This nonspecific reactivity leads to the formation of complex cross-linked products and unpredictable sites of cross-linking, rendering them unsuitable for the precise introduction of covalent bonds into proteins. Recently developed and genetically encoded photoactivatable bioreactive Uaas release chemical groups that selectively react with specific amino acid residues upon being triggered by light.49
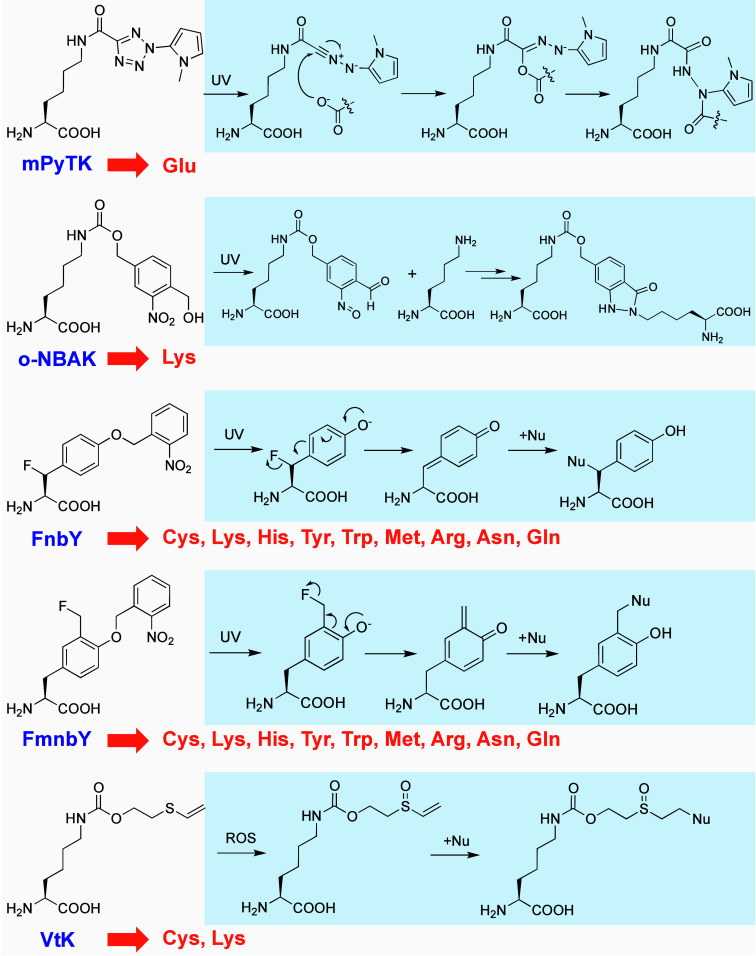
The integration of 2-aryl-5-carboxytetrazole into small-molecule drugs has demonstrated the ability to cross-link target proteins through a photoreleased carboxynitrile imine reacting with a Glu residue near the active site.50 Uaa mPyTK was thus designed to bear 2-aryl-5-carboxytetrazole and incorporated into GST in E. coli, demonstrating more efficient photo-cross-linking of GST into a homodimer compared to a diazirine-based Uaa (Figure 9).51 Mutagenesis studies suggest that mPyTK cross-links with Glu92 at the GST dimer interface. In addition, incorporation of mPyTK in Grb2 also enables cross-linking with EGFR in mammalian cells.
Figure 9.
Uaas with proximity-enabled bioreactivities inducible by UV light or ROS. Target residues are listed in orange.
A photoactivatable Uaa, o-nitrobenzyl alcohol-containing o-NBAK, has been developed for cross-linking with proximal lysine residues.52 o-NBAK generates aryl-nitroso intermediates upon photoactivation, which have half-lives of ∼30 min and react with Lys within GST dimers both in vitro and in E. coli cells. When incorporated into the active site of the lysine acetyltransferase SlPatA, o-NBAK enables the cross-linking of this enzyme with its substrate protein SeAcs in vitro, as confirmed by MS.
Amino acid identities at protein–protein interfaces vary due to diverse molecular interactions, making it valuable to develop photoactivatable Uaas capable of specifically targeting a broader range of natural amino acid residues. The multitargeting reactivity is particularly useful when identifying unknown protein–protein interactions without information on the target natural residue. FnbY, a photoactivatable para-quinone methide (QM) generating Uaa, was developed and encoded in both E. coli and mammalian cells.53 In the GST model protein, FnbY, upon UV light exposure, can cross-link multiple nucleophilic residues, including Cys, His, Lys, Tyr, Trp, Asn, Gln, Glu, and Met. The QM half-life extends to a duration measured in seconds, significantly surpassing the brief lifetimes of radical intermediates typical in conventional photo-cross-linking Uaas. Correspondingly, FnbY demonstrates enhanced cross-linking efficiency when compared to p-azido-phenylalanine. Another Uaa, FmnbY, that releases an ortho-QM exhibits similar selectivity but higher reactivity than the para-QM of FnbY.54 Beyond protein cross-linking, both FnbY and FmnbY can be employed for photocontrolled protein conjugation with widely available amine or thiol reagents and feature rapid kinetics, with reactions completing in tens of seconds.54
A vinyl thioether-containing Uaa, VtK, has been designed to drive oxidation-induced proximity-enabled bioreactivity in proteins.55,56 Upon oxidation, vinyl thioether is chemically converted into vinyl sulfoxide, acting as a Michael acceptor that selectively reacts with proximal Cys or Lys residues. In vitro experiments with sfGFP incorporating VtK reveal nearly complete oxidation by 5 mM H2O2 and 0.5 mM Na2WO4, followed by successful labeling using the amine-containing fluorescent probe N-dansylethylenediamine.56 Moreover, VtK incorporation into GST facilitates GST cross-linking into a dimer, triggered by H2O2/Na2WO4 oxidation either in vitro or within E. coli cell lysates.56 Additionally, human Trx expressed with VtK at positions 60, 72, or 73 in HEK293T cells demonstrates spontaneous covalent dimerization (15%) and trimerization (3%) of Trx without the addition of external oxidants, suggesting intrinsic cellular reactive oxygen species are adequate to activate VtK for protein cross-linking.55 VtK thus can be a promising tool for probing reactive-oxygen-species-associated cell signaling proteins and processes.
2.2.7. General Considerations for Choosing a Latent Bioreactive Uaa
The reactivity of a functional group is typically inversely correlated with its specificity. In the design of latent bioreactive Uaas, a delicate balance between reactivity and specificity is maintained. Despite this, each latent bioreactive Uaa exhibits a distinct profile of properties. In vivo applications demand a higher level of specificity compared to in vitro applications. Assessing the toxicity of Uaas to cells or organisms serves as an indicator of the nonspecific reactivity of the functional group. The assessment of off-target reactions of Uaa-incorporated proteins has been conducted in cells and serum for a limited number of Uaas to date, specifically the SuFEx-based Uaas.57 Uaas featuring bulky leaving groups may encounter challenges fitting into the tight binding interfaces of biomolecules. To address this, Uaas with small leaving groups (e.g., F– in SuFEx Uaas) or those undergoing addition reactions are preferable to avoid potential interference with target interactions. Additionally, for various applications, factors such as reaction kinetics and cross-linking yield play important roles. The study of the SuFEx reaction in a protein context reveals that kinetics and yields are influenced by the binding affinities of interacting proteins, the chemical reactivity of the Uaa’s functional group, and the identity of the target residue, among other factors.57 The side chain length and orientation of Uaas also impact the reaction rate, as exemplified by FSY, FSK, and mFSY,40,43,44 which share the same fluorosulfate functional group. Consequently, data on kinetics and cross-linking yields reported in separate papers for different latent bioreactive Uaas are not directly comparable given variations in protein pairs or sites employed during determination.
2.3. Covalently Targeting Ribonucleic Acids
Beyond protein–protein interactions, protein–nucleic acid interactions are also vital for life. Protein–RNA interactions regulate almost all facets of RNA molecules, encompassing pre-mRNA splicing, RNA modification, translation, and degradation.58 Dysfunctional RNA-binding proteins (RBPs) may cause disorders like neurodegeneration and cancer.59,60 Many RBPs bind RNA with disordered regions.61 To comprehend these complex regulatory mechanisms, identifying protein–RNA interactions in vivo with single nucleotide and amino acid resolution is critical. Various methods are available for detecting protein–RNA interactions, which can be categorized into noncovalent and covalent approaches based on the chemical nature of these interactions.62 Among these, nucleoside-based UV cross-linking is a prevalent methodology.63 This technique utilizes UV-induced nucleoside radicals to cross-link with nearby amino acids, enabling the determination of RNA targets via immunoprecipitation and high-throughput sequencing in a variety of CLIP methods.64 Despite its efficacy, UV cross-linking exhibits nucleotide bias and has difficulty achieving amino acid resolution,65,66 making precise engineering of covalent linkages infeasible. In response, GECX-RNA has been developed (Figure 10A).19 This approach introduces a latent bioreactive Uaa at the RBP binding interface. Upon RNA binding, the Uaa reacts specifically with RNA nucleophilic groups, forming a covalent linkage between protein and bound RNA. Targeting the ribose 2′-hydroxyl group allows for unbiased application to all RNA nucleotides. This proximity-enabled reactivity, not dependent on an external trigger, enables in vivo use and promises single amino acid and nucleotide resolution.
Figure 10.
Development of GECX-RNA to covalently target RNA. (A) The principle of GECX-RNA. (B) Structure of the BzoCas13b–crRNA binary complex showing positively charged amino acids (yellow sticks) involved in precursor CRISPR RNA (pre-crRNA) cleavage. crRNA is shown as the salmon-colored stick (PDB 6AAY). FSY was incorporated to target the 2′-OH group of the cleavage nucleotide. (C) Scheme showing Cas13b processing pre-crRNA into mature crRNA, with the cleavage site indicated by the red arrow. EMSA on denaturing urea–PAGE demonstrates that the Cas13b(FSY) mutant cross-linked with all four RNA nucleotides. Panels (B) and (C) adapted with permission from ref (19). Copyright 2022 Springer Nature.
To develop the method, FSY was incorporated into Cas13b, a class 2 type VI RNA-guided RNA-targeting CRISPR-Cas effector.19 When incorporated at the catalytic His133 site of Cas13b, the resultant mutant protein, after incubation with guide and target RNAs, demonstrated efficient cross-linking with the RNAs. In addition, R380 was identified as being involved in precursor guide RNA cleavage in Cas13b. Incorporation of FSY at this site would position the FSY side chain toward the 2′-hydroxyl group on the ribose of the cleavage nucleotide (Figure 10B). Subsequently, incubation with this FSY mutant of Cas13b showcased efficient cross-linking of the precursor guide RNA, regardless of the base identity of the cleavage nucleotides, demonstrating RNA cross-linking without exhibiting nucleotide bias (Figure 10C).
GECX-RNA was further assessed for capturing in vivo interactions between RBPs and their endogenous RNA targets.19 Hfq is a conserved RNA chaperone interacting with numerous sRNAs and mRNAs in Gram-negative bacteria.67 FSY was strategically incorporated into Hfq at specific sites, and the resulting mutant proteins efficiently cross-linked with RNAs in E. coli, as confirmed by Western blot and RNase treatment.19 Notably, Hfq-25FSY and Hfq-30FSY exhibited enhanced enrichment of the known target RNA, rpoS, compared to Hfq-WT, validating that FSY cross-linked to enrich the target RNA specifically in E. coli cells. The methodology was further advanced through GRIP (GECX-RNA with immunoprecipitation), enabling the identification of amino acid-specific binding sites on RNA.19 Using Hfq-25FSY, the study provided direct in vivo evidence of site 25 of Hfq interacting with the (AAN)4 element on rpoS RNA and the (ARN)4 element on the ptsG RNA (where R represents A or G, and N represents any nucleotide), offering a powerful tool for probing protein–RNA interactions with extraordinary amino acid specificity.
GECX-RNA was also able to capture protein–RNA interactions in mammalian cells with single amino acid and nucleotide resolution, a detailed account of which is outlined in section 4.2 below. In summary, GECX-RNA surmounts the constraints associated with nucleoside-based UV cross-linking, offering a robust methodology for studying protein–RNA interactions both in vitro and in vivo with exceptional precision and broad applicability.
2.4. Covalently Targeting Carbohydrates
Carbohydrate–protein interactions are indispensable in biological systems, impacting processes such as cell–cell communication, organism development, cancer metastasis, the invasion of bacteria and virus, and immune responses.68,69 Despite their centrality, studying these interactions remains intricate due to their dynamic and transient nature, along with challenges in achieving monosaccharide specificity. Carbohydrate structures, lacking genetic encoding, often exhibit low-affinity interactions,70 impeding the development of high-affinity protein binders.71 Covalently cross-linking proteins with carbohydrates would offer an innovative solution. However, the predominant weak nucleophilic hydroxyl groups of carbohydrates pose challenges for selective targeting with biospecific chemistry.
The success achieved in targeting amino acid side chains and RNA through proximity-enabled bioreactivity opens up a promising avenue for extending this approach to carbohydrate targeting. To identify functional groups capable of reacting with carbohydrates via proximity-enabled bioreactivity, plant-and-cast cross-linkers were employed (Figure 11A).72 These cross-linkers feature a succinimide ester that reacts with Lys side chains, first planting on proteins by rapidly reacting with Lys and then casting the other end to react with the bound carbohydrate. Five cross-linkers, incorporating sulfonyl fluoride, benzyl bromide, fluorosulfate, photocaged quinone methide (QM), and homo-QM, were designed and synthesized.18 In protein–carbohydrate cross-linking studies, Siglec-7, a transmembrane receptor involved in immune function, was selected. The extracellular V-set domain of Siglec-7 (Siglec-7v) was incubated with the tumor-associated carbohydrate antigen GD3. Notably, the NHSF cross-linker successfully formed cross-links between Siglec-7v and GD3, highlighting the capability of sulfonyl fluoride to react with carbohydrates under mild conditions.18 Exploring variations in the planting site on Siglec-7v and the length of the NHSF cross-linker revealed that the reactivity is indeed proximity-enabled.
Figure 11.
Development of GECX–sugar to covalently target carbohydrates. (A) Identification of sulfonyl fluoride as the functional group to react with carbohydrate via proximity-enabled bioreactivity through the plant-and-cast cross-linking strategy. NHSF, a plant-and-cast cross-linker, facilitated the cross-linking of Siglec-7v with GD3. An azido group on GD3 enabled subsequent biotin attachment, allowing for the detection of the cross-linked GD3. (B) Covalent targeting of the carbohydrate via GECX. Latent bioreactive Uaa SFY was incorporated into Siglec-7v to cross-link sialoglycan. (C) Cross-linking of azido-GD3 with Siglec-7v, with SFY incorporated at the indicated Lys sites. (D) Flow cytometric quantification of the Siglec-7v protein bound on the SK-MEL-28 cell surface. Adapted with permission from ref (18). Copyright 2022 Springer Nature.
To introduce the identified sulfonyl fluoride group into proteins, a latent bioreactive Uaa SFY was designed, featuring sulfonyl fluoride and a stabilizing methoxy group (Figure 11B). A new Methanomethylophilus alvus tRNAPyl/SFYRS pair was evolved, enabling the genetic encoding of SFY with high specificity in both E. coli(18) and mammalian cells.19 Successful cross-linking with the bound GD3 ligand in vitro was achieved by incorporating SFY at specific Lys sites of Siglec-7v (Figure 11C).18 Notably, Siglec-7v(127SFY) exhibited enhanced binding to sialylated SK-MEL-28 melanoma cells compared to WT Siglec-7v (Figure 11D). Additionally, sialidase pretreatment of these cells reduced the amount of bound Siglec-7v, confirming that the enhanced cell binding of Siglec-7v(127SFY) primarily resulted from its covalent cross-linking with cell-surface sialoglycans.
Siglec-7v(127SFY) has been further utilized to cross-link with sialoglycan on the cancer cell surface to enhance cancer cell killing by natural killer cells, which is described in section 5.3.6. In short, this GECX-sugar technology enables the precise incorporation of covalent linkages between proteins and carbohydrates, overcoming the persistent challenges of low affinity and weak interactions. This breakthrough holds the potential to advance the exploration of glycobiology and inspire innovative approaches for protein diagnostics and therapeutics through efficient glycan targeting.
3. Engineering Protein Properties via Intramolecular GECX
Genetically incorporated during translation, the latent bioreactive Uaa can selectively form covalent linkages with target natural residues within the same protein through intramolecular GECX, similar to disulfide bonds. These novel linkages provide unique properties, such as irreversibility and increased length, which distinguish them from traditional disulfide bonds. Customizing the Uaa allows for the integration of additional features into the covalent linkage, thereby enhancing the protein’s functionality. By exploiting these new covalent linkages, it is possible to enhance existing protein properties and facilitate the engineering of novel functionalities.
3.1. Recombinantly Bridging or Stapling Proteins
Chemical stapling of α-helical peptides improves target affinity, proteolytic resistance, serum half-life, and membrane permeability.73,74 Utilizing latent bioreactive Uaas, α-helix stapling in both peptides and proteins is now achievable through recombinant expression within cells.24 The incorporation of BrC6K into an affibody with a Cys mutation at the i + 4 site leads to effective stapling, reaching nearly quantitative efficiency without requiring additional treatment following the expression of the mutant affibody in live E. coli cells (Figure 12A).24 This approach not only staples α-helices but also permits the installation of bridges spanning different secondary structures in proteins. Incorporating BrC6K at position 30 in helix 2 of an affibody results in a quantitative reaction with Cys47 in helix 3, forming a bridge that spans the two helices upon expression of the mutant affibody in E. coli.24 In another study, the incorporation of the Uaa CATK-1 into the monobody Nsa1 also leads to the quantitative formation of a bridge with Tyr92 upon expression in E. coli cells (Figure 12B).34 This bridge has been demonstrated to enhance the uptake of the positively supercharged monobody into HeLa cells by 40% and extend the monobody’s proteolytic stability against cathepsin B by threefold. Recombinant expression of staples and bridges avoids the need for chemical catalysts, which should facilitate the generation of staple libraries for selection and large-scale production.
Figure 12.
(A) Recombinantly building staples or bridges on a protein affibody through BrC6K reacting with Cys. (B) Bridge on a monobody Nsa1 formed through CATK-1 reacting with Tyr.
3.2. Enhancing Photostability
In single-molecule imaging, spatial and temporal resolution hinge on the fluorophore’s photo output, which is intrinsically tied to its photostability.75,76 Despite their vital role in conventional and super-resolution biological imaging, enhancing the photostability of fluorescent proteins remains a challenge.77 The fluorophore of these proteins is tethered to the central α-helix, leaving one end free.78 It is hypothesized that enhancing the rigidity of the fluorophore by covalently attaching its free end to the β-barrel could improve its photostability.
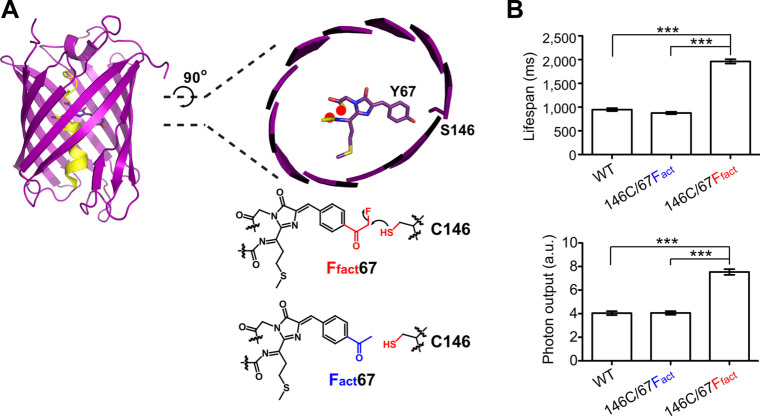
To examine this, a mutant of red fluorescent protein mPlum,79 denoted as mPlum(S146C/Y67Ffact), was expressed in E. coli (Figure 13A).10 The mutant featured the replacement of Tyr67 in the fluorophore with Ffact and the replacement of Ser146 on the β-barrel with Cys. Mass spectrometry confirmed the near-quantitative formation of the Ffact67–Cys146 bond postexpression. Single-molecule analysis revealed that mPlum(S146C/Y67Ffact) exhibited a 2.24-fold longer lifespan and a 1.86-fold increase in photon output compared to the control mPlum(S146C/Y67Fact) lacking this bond (Figure 13B). Moreover, it displayed a 1.92-fold increase in lifespan and a 1.86-fold increase in photon output compared to WT mPlum. Similarly, introducing the Ffact-Cys bond into another red fluorescent protein, mKate2(S143C/Y64Ffact), improved its single-molecule lifespan by 1.49-fold and its photon output by 2.31-fold compared to mKate2(S143C/Y64Fact). In comparison to WT mKate2, mKate2(S143C/Y64Ffact) exhibited a 2.04-fold increase in lifespan and a 2.46-fold increase in photo output. These findings underscore the substantial improvement in the photostability and photon output of fluorescent proteins through the spontaneous formation of a single Ffact–Cys covalent bond.
Figure 13.
A single new covalent bond linking the fluorophore to the β-barrel increases the photostability of red fluorescent proteins. (A) Structure of red fluorescent protein mPlum and the cross section of the fluorophore (PDB 2QLG), showing Tyr67 for mutation to Ffact or the isosteric control Fact and Ser146 for mutation to Cys. (B) Single molecular measurements indicate that the covalent bond formation between Ffact67 and C146 in mPlum increased the lifespan and photon output compared to those of the isosteric control Fact67/C146 mutant and the WT mPlum.
3.3. Enhancing Thermostability
The disulfide bond plays a crucial role in protein stability, yet its fixed length poses challenges in accommodating diverse positions. The locations within proteins that can undergo cross-linking through a cystine disulfide are generally limited to a separation of approximately 5.5 Å between the two β-carbons and an almost perpendicular 90° dihedral angle for the disulfide bond.80,81 Latent bioreactive Uaas have been explored to create covalent linkages to overcome such geometrical constrains, thus increasing protein thermostability.
Haloalkane Uaas with varying side chain lengths were incorporated into an affibody protein, allowing reactions with an introduced Cys residue in a distant helix (Figure 14A).20 After expression in E. coli and subsequent purification, mass spectrometry confirmed the successful intramolecular cross-linking of mutant proteins Afb(30BetY-47C) and Afb(30BprY-47C) at positions beyond the reach of a disulfide bond, with efficiencies exceeding 98%. Circular dichroism (CD) analysis revealed that while WT Afb and mutant Afb(30BprY-47C) displayed similar melting temperatures (Tm) at 46.7 and 46.5 °C, respectively, mutant Afb(30BetY-47C) showed a significantly elevated Tm of 60.4 °C.20 Notably, the sole difference between BetY and BprY is a single methylene group, indicating that an intramolecular covalent bridge of an appropriate length substantially enhances Afb’s thermal stability. This study suggests that the strategy of exploring various sites for bridge formation and concurrently engineering multiple bonds holds promise for further enhancing protein thermostability.
Figure 14.
Increasing protein thermostability through proximity-enabled cross-linking of protein residues. (A) Intramolecular cross-linking of BetY/BprY with Cys in the affibody. CD meting curves are shown in the bottom panel. (B) Structures of the alkyl thiol Uaas. (C) CD melting curves of WT MetA (green) and F264pNCSF mutant (blue and red) (left) and structure of WT MetA (PDB 6MTG), showing the two monomers in blue and beige (right). F264, the site for pNCSF incorporation, is colored magenta; Pro2, cross-linked by pNCSF, is colored red. Panel (C) reproduced from ref (84). Copyright 2019 American Chemical Society.
By substituting halides in haloalkane Uaas with thiol, alkyl thiol Uaas (SetY, SprY, and SbuY) featuring varying side chain lengths were also genetically encoded (Figure 14B).82 These were designed to react with Cys to construct disulfide bridges with extended lengths. Specifically, one Cys was mutated at 12 different sites within the high-mobility regions of TEM-1 β-lactamase, and alkyl thiol Uaas were incorporated at 144 random sites. This mutant library was expressed in E. coli and selected for growth under a nonpermissive temperature of 40 °C. A single mutant, R65C/A184SbuY, was identified. The formation of an extended disulfide bridge between 184SbuY and 65Cys was confirmed by mass spectrometry, which increased the Tm of TEM-1 β-lactamase by 9 °C from 48.6 (WT) to 57.4 °C.82
Mutation and selection were subsequently conducted with E. coli homoserine O-succinyltransferase (MetA), leveraging its vital role in methionine biosynthesis to rescue E. coli growth at 44 °C.83 Twelve Uaas, including three latent bioreactive Uaas, were incorporated at 261 random sites out of 308 in MetA. Growth selection revealed that substituting Phe21 with (p-benzoylphenyl)alanine (pBzF) drastically increased MetA’s Tm by 21 °C from 53 (WT) to 74 °C for the F21pBzF mutant.83 Mutagenesis and 13C NMR studies suggested that pBzF formed a reversible hemithioketal covalent cross-link with a proximal Cys90 from the other monomer, stabilizing the dimeric form of MetA. Following that, a comparable selection process was carried out by incorporating the amine-reactive latent bioreactive Uaa pNCSF into MetA.84 The F264pNCSF mutant was identified, demonstrating a striking 24 °C increase in Tm (Figure 14C). Mass spectrometry analysis indicated that pNCSF formed a thiourea cross-link with the N-terminal proline of the other monomer, even though these two positions were >30 Å apart in the X-ray crystal structure of the WT MetA. The authors hypothesize that pNCSF is able to irreversibly cross-link two distal sites that are likely conformationally flexible, resulting in a covalently trapped dimer that exhibits a large increase in Tm.
Covalently linking two interacting proteins has also been demonstrated to enhance the stability of transient, low-affinity protein complexes, facilitating their crystallization and structure determination. The bromoalkyl Uaa BrCnK was incorporated into the small G-protein Rab1b to cross-link a Cys residue mutated in the guanine nucleotide exchange factor (GEF) domain of DrrA.25 This cross-linking of Rab1b with DrrA-GEF stabilized the transient ternary complex of Rab1b:GDP:DrrA-GEF, leading to the visibility of GDP electron density.25
Collectively, these findings highlight the potential of tailored covalent bridges as a versatile tool for enhancing protein stability.
3.4. Optical Nanoswitch for Photomodulation
The ability to manipulate protein function using light provides precise temporal and spatial resolution, revolutionizing neuroscience and extending to general biology.85−89 Existing optogenetic methods encounter challenges, particularly their limited applicability to diverse proteins and low flexibility in selecting modulation sites, thereby constraining study precision. To overcome these hurdles, photoswitchable click amino acids (PSCaas) incorporate an azobenzene photoswitch and a latent bioreactive group (Figure 15).21,23 This group selectively reacts with a strategically positioned natural amino acid residue to form an in situ covalent optical nanoswitch that can reversibly modulate protein secondary structures or domains, thus controlling protein function in response to light.
Figure 15.
Photoswitchable click amino acids (PSCaas) react with Cys to build optical nanoswitch onto proteins in situ, which allows reversible optical modulation of protein structure and function with high resolution up to the amino acid residue level. Cl-PSCaa and F-PSCaa react with proximal Cys through nucleophilic substitution and nucleophilic aromatic substitution (SNAr), respectively.
The Cl-PSCaa isomerizes with 365/405 nm light,21 and the F-PSCaa isomerizes with visible light at 405/540 nm.23 Incorporating F-PSCaa into calmodulin (CaM) at site 76 and placing a Cys at site 83 leads to the precise construction of an optical nanoswitch on CaM by covalently connecting the two sites.23 Illumination with green light (540 nm) induces photoisomerization, transitioning the nanoswitch from the trans to cis configuration. Subsequent exposure to blue light (405 nm) reverts the conformation from cis to trans, establishing the photostationary state of the trans configuration. Sequential illumination with green or blue light enables reversible transformation between the two states. The photoisomerization of the nanoswitch induces a significant reversible conformational change in CaM, as detected by circular dichroism. Consequently, the conformational alteration in CaM modulates its binding activity to the CaM-binding domain of the neuronal nitric oxide synthase.23
Compared to light-sensitive protein domains, optical nanoswitches built with latent bioreactive PSCaas offer smaller sizes and enhanced site flexibility for installation. These characteristics minimize interference and enable high-resolution control at the level of individual amino acid residues. Capable of structural modulation, this nanoswitch is even applicable to proteins with unknown functions. As accurate protein structure prediction rapidly advances,90,91 the nanoswitch holds promise as a versatile optical controller for regulating diverse protein positions and secondary features, addressing research and engineering needs with unparalleled spatiotemporal resolution.
4. Studying Biomolecular Interactions In Situ via Intermolecular GECX
GECX employs a latent bioreactive Uaa genetically incorporated into the protein of interest.17 When this protein binds to an unknown biomolecule (e.g., a peptide, protein, or RNA), the Uaa reacts with a functional group on the bound molecule, covalently linking the two molecules together through intermolecular GECX. This robust connection withstands harsh processing and detection conditions. Notably, GECX requires no modification of the target biomolecule, allowing the analysis of native biomolecules in their physiological context. GECX spontaneously cross-links interacting biomolecules, eliminating the need for external triggers like light activation.17 This enables the capture of interactions whenever they occur, making GECX particularly suitable for in vivo applications where external triggers are difficult to deliver or their timing is crucial. The extended reaction window of the latent bioreactive Uaa further enhances the cross-linking efficiency and detection sensitivity. Additionally, the reaction specificity of the Uaa leads to predictable and defined covalent linkages, facilitating MS analysis. Identifying cross-linked fragments through MS also provides evidence of direct interaction, minimizing false positives from indirect binders.
4.1. GECX to Study Protein–Protein Interactions
GECX initially emerged as an innovative method for investigating receptor–peptide ligand interactions directly on live mammalian cell surfaces, offering structural insights into protein complexes in native settings (Figure 16).10,16 This approach circumvents limitations associated with artificial membrane compositions, in vitro reconstitution, and incomplete systems, providing a more accurate representation of biological processes. The biocompatibility and genetic encodability of latent bioreactive Uaas like Ffact pave the way for their use in live-cell studies. Ffact was employed to elucidate the interaction between corticotropin releasing factor receptor type 1 (CRF1R), a class B GPCR, and its peptide ligand urocortin-1 (Ucn1) in mammalian cells.16 To pinpoint Ucn1’s interaction sites on CRF1R, the photo-cross-linking Uaa p-azido-phenylalanine (Azi) was incorporated at various positions within the receptor expressed in HEK293T cells.16,92 Upon UV light activation, Azi cross-linked Ucn1, allowing the detection of the covalent CRF1R-Ucn1 complex in Western blot analyses. Positive cross-linking indicates that Ucn1 interacts with CRF1R at the Azi incorporation site. However, the specific positioning of the peptide ligand relative to the receptor remained elusive due to the nonspecific nature of Azi’s photo-cross-linking reaction. To solve this challenge, Ffact was incorporated into CRF1R, and Cys residues were strategically positioned at different sites on Ucn1. Ffact’s reaction with Cys is both residue- and distance-dependent, providing precise spatial constraints on the receptor–ligand interaction. By combining these spatial constraints with the separate known structures of CRF1R’s transmembrane and extracellular domains, a complete conformational model for the peptide–receptor complex was generated.16 This model revealed the binding path of the peptide agonist within the activation domain of the class B receptor, offering valuable insights into the receptor activation mechanism. Overall, this GECX-based approach provides comprehensive information derived from the full-length receptor in its native live-cell environment. This information complements data obtained from crystallographic characterization of isolated receptors in artificial settings, offering a more holistic understanding of receptor–ligand interactions.
Figure 16.
GECX to probe the ligand–receptor interaction on mammalian cell surface. Photo-cross-linking Uaa Azi was incorporated into CRF1R to reveal the binding pocket (in purple) for its peptide agonist Ucn1; latent bioreactive Uaa Ffact was incorporated into CRF1R together with Cys in Ucn1 to determine reciprocal spatial constraints (circles with the same colors) of the receptor–ligand complex. These data were then integrated with structural information on the receptor’s separate domains to build a conformation model for full-length CRF1R in complex with Ucn1. Reproduced with permission from ref (6). Copyright 2021 John Wiley and Sons.
Taking a bold leap from the cell surface, GECX next ventured into the cellular interior to capture weak and transient protein–protein interactions, including the elusive enzyme–substrate binding, for identification (Figure 17A).17 This strategy was exemplified using the bromoalkyl-containing Uaa BprY and its alkyne derivative EB3 (Figure 17B). These Uaas react with proximal Cys residues, and EB3 further allows biotin labeling via click chemistry on alkyne, enabling the enrichment of cross-linked peptides to enhance the MS detection sensitivity.17 The strategy involves genetically incorporating the Uaa into the target protein at a chosen site in live cells.17 Upon protein interaction, this site is positioned close to a Cys residue on the interacting protein. The subsequent reaction between the Uaa and Cys covalently captures the interacting protein to the target within the living cell. Following affinity purification of the target protein, the copurified cross-linked proteins undergo protease digestion and MS analysis, revealing both the protein identity and the cross-linking site. GECX has proven remarkably effective in capturing weak protein binding.17 For instance, GECX successfully cross-linked the affibody–Z complex directly within E. coli cells, whose Kd is ∼6 μM.93 Notably, GECX exhibited a higher cross-linking efficiency compared to the conventional photo-cross-linker Azi. Additionally, incorporation of EB3 around the ubiquitination site of the substrate protein PCNA allowed for the cross-linking of the ubiquitin-conjugating enzyme UBE2D3 in live cells. Moreover, to explore whether GECX could capture and identify unknown protein interactions in cells, BprY was incorporated into the active site of the enzyme thioredoxin 1 (Trx1),17 which is a ubiquitous oxidoreductase catalyzing the reduction of disulfide bonds of proteins.94 Expressing Trx1-containing BprY at position 32 in E. coli resulted in the covalent capture of multiple proteins directly in E. coli cells, shown on Western blot as Trx1-containing bands with higher molecular weights (Figure 17C).17 A total of 91 endogenous E. coli proteins were identified by MS, including known and previously unknown substrate proteins of Trx1. For GECX-cross-linked proteins in this study, tandem MS spectra were obtained to verify the cross-linked residue, substantiating direct interaction. These remarkable results highlight the potential of GECX in uncovering both enzyme–substrate relationship and interacting proteins within the dynamic environment of live cells.
Figure 17.

(A) GECX to capture protein–protein interactions in live cells for subsequent identification by MS. (B) Initial development of GECX used Uaas BprY and EB3 to target the Cys residue. (C) Western blot of cell lysate of E. coli cells expressing Trx1(32BprY-33S-36A), showing multiple endogenous proteins cross-linked to Trx1. (D) Use of FSY and FSK in GECX enables His, Lys, or Tyr targeting and Uaa to be placed at the binding periphery, thus expanding the diversity of targetable proteins. Adapted with permission from ref (6). Copyright 2021 John Wiley and Sons.
The combination of photo-cross-linking and GECX has been successfully employed to elucidate structural information on various protein complexes both on the surface and within mammalian cells. Photo-cross-linking serves as a mapping tool for interacting sites, guiding the strategic incorporation of latent bioreactive Uaa in GECX and significantly reducing the combinatorial matrix. In a specific study,95 Azi-based photo-cross-linking was initially employed to map the footprints of peptide agonists and antagonists on CRF1R. Subsequently, GECX was conducted using α-chloroacetamide introduced in the peptides and Cys mutations in CRF1R. The spatial constraints derived from these experiments were then utilized to construct 3D models for both agonist- and antagonist-bound CRF1R, revealing distinct folds and stabilizing unique conformations of the transmembrane domain. In another investigation,96 the interactions between GPCRs and β-arrestins were explored in mammalian cells. Photo-cross-linking Uaa Bpa or bromoalkyl-containing Uaa BetY was incorporated into β-arrestin at different sites in HEK293T cells, followed by the study of their cross-linking with coexpressed GPCRs. This study highlights that each GPCR receptor leaves a distinctive footprint on arrestins and defines the orientation of the arrestin relative to the GPCR. In a recent comprehensive scan,97 BetY was incorporated at 24 sites in β-arrestin 2 against the GPCR secretin-like parathyroid hormone 1 receptor (PTH1R), with Cys mutations at 120 sites in HEK293T cells. Remarkably, 136 intermolecular proximity points were identified, enabling the construction of energy-optimized models for the PTH1R-arrestin 2 complex. These models provided intricate structural details, unveiling flexible elements absent in existing structures and offering new insights into the dynamics of the system.
GECX has been crucial in elucidating specific details of diverse protein–protein interactions, enhancing the understanding of their biological functions. For instance, in the case of NleE, a methyltransferase in enteropathogenic Escherichia coli known to interfere with autophagy, the incorporation of Azi in NleE followed by photo-cross-linking in HEK239T cells identified PSMD10 as an interaction partner.98 Knockout of PSMD10 revealed its essential role in NleE-mediated suppression of host autophagy. To delve into the mechanism of NleE’s impact on PSMD10 function, BetY was incorporated into NleE, and cross-linking with PSMD10 was performed through GECX targeting Cys in mammalian cells. This analysis revealed that NleE binds with the N-terminus of PSMD10, impeding PSMD10 homodimerization and, consequently, attenuating host autophagosome formation. Using a similar GECX strategy, the same team incorporated BetY into NleE in mammalian cells and discovered that NleE interacts with the host zinc finger protein ZPR1.99 The NleE–ZPR1 interaction facilitates the bacterial pathogen’s ability to attenuate the host’s unfolded protein response. In another investigation,100 full-length PD-L1 was observed to form homodimers and tetramers in cells through photo-cross-linking. BetY and BrC7K, featuring bromoalkyl groups of different lengths, were incorporated into different domains of PD-L1, and GECX was performed with Cys introduced at different sites. The results of pairwise chemical cross-linking unveiled that PD-L1 homodimerizes asymmetrically through the transmembrane domain, intracellular domain, and extracellular domain. In addition, it was discovered that homodimerization at the intracellular domain regulates PD-L1 glycosylation, subsequently influencing PD-1 binding and T cell toxicity.
For protein identification, the GECX strategy has been applied to identify protein tyrosine phosphatase (PTP) in mammalian cells.101 Various latent bioreactive Uaas were incorporated into substrate proteins at their phospho-tyrosine (pY) site to examine cross-linking with the conserved active-site Cys of PTPs. Upon incorporation into substrate protein ABL1-SH3 (SH3 domain of Abelson murine leukemia viral oncogene homologue 1) or HER2, the fluoromethyl ketone-containing FpAcF (previously known as Ffact10) and the bromoalkyl-containing BetY20 displayed efficient cross-linking with SHP2-PTP in vitro and in HEK293 cells, respectively.101 Furthermore, when BetY was incorporated at pY site 1221 of HER2 in HEK293 cells, silver-stained SDS PAGE revealed distinct cross-linking bands. MS analysis of these bands unveiled 116 unique proteins, among which only one PTP enzyme (PTP1B) was identified. Coexpression of PTP1B and HER2(Y1221BetY) in HEK293 cells confirmed their cross-linking, while BetY incorporation at other pY sites did not yield robust cross-linking. These results suggest that PTP1B dephosphorylates HER2 on Y1221.
GECX has proven its versatility in cell lysates as well, exemplified by its application in identifying proteins interacting with small ubiquitin-like modifiers (SUMOs). These small proteins are reversibly conjugated to target proteins regulating genome stability and transcription. To identify SUMO2-interacting proteins, BprY was incorporated into SUMO2 at its binding groove.26 The SUMO2 mutant was expressed in E. coli cells, then affinity-purified onto resin and incubated with cell lysates from 293T cells. This process enabled the GECX-mediated covalent capture of SUMO2 binders. Subsequent on-bead trypsin digestion and MS analysis successfully identified 264 SUMO2-interacting proteins. The compatibility of GECX with pre-prepared samples can be valuable for studying cells or tissues isolated from animals or patients. In a different study, BprY was incorporated into ubiquitin and ubiquitin-like proteins, which function as activity-based protein probes and cross-link with deconjugating enzymes both in vitro and in vivo.102 Additionally, upon expressing BprY-modified ubiquitin in HEK293T cells, quantitative MS analysis identified 57 deubiquitinating enzymes and a substantial array of ubiquitin-interacting proteins.
Accurately identifying peptides cross-linked by the latent bioreactive Uaa, rather than non-cross-linked peptides, from MS data provides compelling evidence of direct protein–protein interaction. To address this critical need, OpenUaa, a new database search engine specifically designed for analyzing Uaa-mediated cross-linking at the proteomic scale, has been developed.103 Unlike traditional search engines that treat the Uaa-incorporated peptide as a large modification, overlooking fragment information, OpenUaa preserves all fragment details. First, OpenUaa employs in silico digestion to identify Uaa-incorporated peptides, reducing redundant candidate generation. Second, these identified peptides are searched against a global protein database to pinpoint the interacting protein. This inclusive and open search strategy dramatically enhances sensitivity and coverage. By employing OpenUaa to analyze the GECX data of BprY-incorporated Trx cross-linking in E. coli cells, a remarkable 289 cross-linked peptides were identified, corresponding to 205 proteins directly binding to Trx.103 The integration of GECX with OpenUaa will greatly facilitate the identification of the direct interactome of various proteins and enzymes in live cells.
The SuFEx-based FSY and FSK exhibit reactivity with proximal Lys, His, and Tyr residues. The incorporation of FSY/FSK in GECX should broaden the spectrum for capturing interacting proteins, especially those lacking Cys but featuring accessible Lys, His, or Tyr residues for targeting. Moreover, FSY and FSK can be strategically incorporated at the periphery, steering clear of the active site or binding interface to minimize interference with protein interactions (Figure 17D). To explore this versatility, FSY and FSK were introduced into Trx at site 62, positioned outside Trx’s active site, within E. coli cells to capture substrate proteins through GECX.43 Western blot analysis of cell lysates revealed more cross-linking bands compared to GECX with Cys-targeting BprY in the active site. MS analysis of the cross-linked proteins further unveiled a greater number of Trx binders with identified cross-linked peptides. Intriguingly, while some substrates exhibited overlap between FSK and FSY, they cross-linked different residues, underscoring their distinct reaction radii. These findings highlight the utility of both FSK and FSY in GECX within live cells, breaking free from Cys limitations and enabling cross-linking at the periphery of protein binding.
4.2. GECX-RNA to Map RNA Modifications
GECX-RNA showcases the ability to selectively cross-link proteins with interacting RNAs in cells, displaying both amino acid and nucleotide specificity. This unique attribute enables the high-resolution identification of protein–RNA interactions, a feature leveraged to map RNA modifications in live cells. The focus of this application is on N6-methyladenosine (m6A), a prevalent RNA modification crucial for mRNA regulation.104 Precise identification of m6A sites becomes imperative for a comprehensive understanding of its functional impact. Traditional methods for detecting m6A face limitations either in achieving single-nucleotide resolution or by relying on m6A-specific antibodies for in vitro recognition.105−107 Additionally, the chemical conversion of m6A for detection lacks protein specificity and might not be universally applicable to other RNA modifications.108
The GECX-RNA-based GRIP-seq approach has emerged as a solution that captures m6A sites on RNA in vivo while providing protein specificity.19 The strategy involves the expression of an m6A reader protein recognizing m6A in mammalian cells, along with a strategically incorporated bioreactive Uaa in proximity to the recognition site (Figure 18). Through the GECX-RNA mechanism, this Uaa cross-links the RNA nucleotide adjacent to the m6A. The ensuing immunoprecipitation of the cross-linked reader–RNA complex, followed by digestion with proteinase K, releases the captured RNAs. Subsequent reverse transcription terminates at the cross-linking site due to the presence of the cross-linked Uaa. The resulting reverse transcripts undergo high-throughput sequencing, revealing all cross-linking sites, with m6A sites expected to be immediately adjacent. Specifically, the YTH domain of the human m6A reader YTHDF1,109 capable of recognizing m6A, was utilized to probe m6A in HEK293T cells. Uaa SFY was strategically incorporated at site 397, where it was positioned proximal to the m6A binding site.
Figure 18.

Principle of GRIP-seq to detect RNA modifications in vivo using m6A as an example. Adapted with permission from ref (19). Copyright 2022 Springer Nature.
Analysis of GRIP-seq data unveiled sequencing peaks enriched with the m6A consensus motif.19 Notably, 80.4% and 9.3% of cross-linked nucleotides were identified at the −3 position and the −4 position, respectively, relative to m6A, demonstrating single-nucleotide resolution. A total of 13968 m6A sites were predicted from the GRIP-seq data. In comparison with known human m6A sites,110 6072 sites aligned with annotations from other methods, validating the approach. Intriguingly, the remaining 7896 sites had not been reported previously, showcasing GRIP-seq’s capacity to unveil novel m6A sites. Significantly, GRIP-seq demonstrated effectiveness in capturing m6A modifications on low-abundance RNAs, highlighting its potential to reveal previously overlooked facets of m6A-mediated RNA regulation.
The innovative GRIP-seq strategy, utilizing a reader protein and a bioreactive Uaa, distinguishes itself as an antibody-free in vivo method for m6A identification.19 Its capability to attain single-nucleotide resolution, exhibit reader protein specificity, and precisely capture m6A sites within the intricate transcriptome landscape underscores its significance in advancing our understanding of RNA modifications. The versatility of GRIP-seq extends beyond m6A, suggesting potential applications for mapping other RNA modifications in vivo, contingent upon the availability of specific readers or binders.
5. Peptide and Protein Therapeutics Utilizing Biospecific Chemistry
In recent years, the field of drug discovery has been revolutionized by the development of covalent small-molecule drugs, now representing about 30% of all marketed drugs across various therapeutic areas.111−115 Unlike traditional drugs, these covalent drugs work in two steps: they first bind to their target in a reversible way and then form a strong, permanent bond. This unique mechanism leads to prolonged action and increased effectiveness.116 As a result, they are generally more potent and selective than noncovalent drugs, effectively overcoming certain resistance mechanisms and fully inactivating their targets.117,118 Traditionally, drug development has focused on small molecules due to their diverse chemical makeup and ability to precisely target specific areas within proteins.119 However, they fall short in binding to larger, flatter protein surfaces or in blocking protein–protein interactions (PPIs), which are key players in most cellular processes.119,120 To address this limitation, there is a growing shift toward developing peptide and protein drugs. These larger molecules are better suited for interacting with extensive surface areas,121 as seen in the clinical success of antibody drugs and the increasing interest in peptides and peptidomimetics.122−124 Despite these advances, peptide and protein drugs made of canonical amino acids still lack the covalent binding ability that small molecules possess. This gap is being bridged by incorporating Uaas with biospecific chemical reactivity. This innovative approach has led to the creation of covalent peptides and proteins for therapeutic use, extending the unique advantages of covalent binding, once exclusive to small-molecule drugs, to a broader range of drug types.125
5.1. Covalent Peptides
Peptide binders are typically derived from segments of a PPI interface or innovatively designed from scratch.126 They can be engineered to traverse cell membranes, thereby targeting both intracellular and membrane-bound proteins.127 Despite their potential, peptides grapple with issues like diminished binding affinity and short lifespans, often due to rapid degradation and clearance. Introducing a covalent binding mechanism holds promise in markedly boosting the efficiency of peptide drugs by enhancing their stability and affinity. Photoreactive moieties were initially attempted to develop covalent peptides,128 yet they are incompatible with in vivo applications.
A more straightforward approach involves incorporating warheads used in covalent small-molecule drugs into peptides, enabling them to form covalent bonds with natural amino acid residues such as cysteine. One study by Stebbins et al. identified a 13-mer peptide (BI-107D1) targeting the E3 ubiquitin ligase Siah, a key player in cancer development and progression.129 They converted BI-107D1 into a covalent peptide by adding a Lys-acrylamide warhead to one of its residues. This modification allowed the peptide to form a covalent bond with an endogenous Cys residue located near its binding pocket. This covalent peptide, after appending a cell penetrating peptide, more potently inhibited Siah activity in cells compared to its noncovalent counterpart. Another research group incorporated multiple electrophilic warheads into the native peptide BIM to target a specific Cys residue of the oncogenic protein Bcl2A1.130 When acrylamide was used as the warhead, the modified BIM peptide formed a permanent covalent bond with a single Cys residue within Bcl2A1’s helix-binding groove, highlighting its selectivity. However, using more reactive warheads like chloroacetamide or propiolamide resulted in undesirable reactions with other exposed Cys residues on Bcl2A1’s surface, emphasizing the importance of achieving reaction specificity.
Unpaired Cys residues, though critical in active sites and binding pockets, are infrequently found at protein–protein interaction interfaces. Targeting other residues like Lys, Tyr, and His, commonly found on protein surfaces and interaction interfaces, is highly sought after, along with a mechanism ensuring reaction specificity. The use of latent bioreactive Uaas in proteins to target various natural residues has significantly progressed this field.11 In 2016, Hoppmann and colleagues introduced the concept of proximity-enabled bioreactivity to develop covalent peptide binders (Figure 19A).131 This method involves incorporating a latent bioreactive Uaa into a peptide. This Uaa becomes reactive only when the peptide binds to a protein, placing the Uaa in close proximity to a target residue of the protein. The Uaa’s chemical inertness toward other molecules minimizes nonspecific reactions and potential cytotoxicity, thereby ensuring a high degree of target specificity.
Figure 19.

(A) Covalent cross-linking of a peptide to its target protein via proximity-enabled bioreactivity. An electrophilic warhead is installed onto the peptide, which reacts with a specific amino acid residue on the target protein upon peptide binding with the protein. (B) List of warheads and their target residues.
To validate this concept, the team designed a peptide with an aryl sulfonyl fluoride-containing Uaa targeting nucleophilic Lys or His residues.131 This Uaa was introduced into the SAHp53–8 peptide,132 which inhibits p53-Mdm2/4 interactions. Blocking these interactions can restore p53 activity to suppress cancer growth. The resulting mSF-SAH peptide efficiently cross-linked with MDM2 and MDM4 and enhanced the inhibition of the p53-MDM4 interaction over 10-fold in mammalian cell lysate. Additionally, the mSF-SAH peptide was nontoxic to the p53-null Saos-2 cells, in contrast to the noncovalent SAHp53–8 peptide. Notably, covalent binding occurred only when sulfonyl fluoride was in the meta-position, not the para-position, of the Uaa, highlighting the importance of side chain orientation for specificity. Furthermore, unlike the noncovalent SAHp53–8, the mSF-SAH peptide showed a sixfold greater selectivity for MDM4 over MDM2, a challenging goal in MDM2/4 inhibition. This research underscores the potential of latent bioreactive Uaas in targeting noncatalytic noncysteine residues through proximity-enabled bioreactivity and demonstrates how covalence can enhance peptide drug efficacy and specificity.
Building on proximity-enabled bioreactivity, Spring and colleagues adopted a unique method for creating covalent stapled peptides.133 In their technique, rather than attaching the latent bioreactive group directly to a peptide residue, they positioned it within a staple core. This core acts as a bridge, connecting two residues in the peptide to form the staple. This “two-component” strategy simplifies the production of stapled peptides with electrophiles, offering a versatile “one-size fits all” solution. They demonstrated this approach by attaching an activated sulfotetrafluorophenyl ester to the staple of peptide P1, targeting a Lys residue on MDM2. The resulting peptide formed a covalent bond with MDM2, exhibiting a progressively improved dissociation constant over time, in contrast to its noncovalent counterpart.
Selective inhibition of X-linked inhibitor of apoptosis protein (XIAP) can trigger programmed death of cancer cells. A SMAC-derived tetrapeptide binder of XIAP was modified at position 2 with various substituents, and adding a sulfonyl fluoride enabled it to covalently target a proximal Lys311 residue on the XIAP protein.134 The resultant covalent peptide not only selectively engaged with XIAP but also exhibited increased effectiveness in destroying various cancer cell lines, including those resistant to the leading noncovalent peptides. Advancements in SuFEx technology have highlighted aryl sulfonyl fluoride and aryl fluorosulfate as superior warheads for engaging other nucleophilic residues such as Lys, Tyr, His, Thr, and Ser within proteins.38−40 These warheads were introduced in the SMAC-derived peptidic inhibitor and tested against the XIAP protein with different nucleophilic residues mutated at the target Lys311 site, confirming that both benzyl sulfonyl fluoride and benzyl fluorosulfate effectively target Lys, Tyr, and His residues.135,136 The aryl fluorosulfate derivative of the inhibitor was identified to be the most suitable for its cell permeability, stability in aqueous buffer and plasma, and cellular efficacy in inducing caspase-3 activation. Subsequently, the design principles were applied to target the related protein melanoma-IAP (ML-IAP).137 The Lys135 site in ML-IAP is analogous to the Lys311 site in XIAP. By introducing a modified aryl sulfonyl fluoride group at the second position of the peptide inhibitor, a potent covalent inhibitor was created, effectively targeting ML-IAP with cellular activity.
The oncogenic Ras G12C mutant, successfully targeted by covalent small-molecule drugs,115 has also been targeted by covalent peptides specifically.138 Different electrophilic groups, including acrylamides, vinyl sulfonamides, and vinyl sulfones, were attached via the side chain of a lysine residue to SOS-mimic peptides to generate covalent peptide inhibitors. Among these modifications, acrylamides were too weak, while vinyl sulfones were too reactive. The peptide with a vinyl sulfonamide group exhibited the highest effectiveness. In comparison to control peptides, this vinyl sulfonamide-containing peptide significantly reduced cancer cell viability and inhibited ERK downstream signaling in a Ras G12C-dependent manner.
In a strategy to inhibit FtsQ, a protein crucial in bacterial cell division, a stapled peptide was crafted from its binding partner, FtsB.139 To this peptide, 11 different electrophiles were installed at the Thr83 position, aiming to react with Lys239 on FtsQ. Of these variants, the peptide modified with bromoacetamide exhibited the strongest inhibition of the E. coli lptD4213 strain, which has a permeable outer membrane. Furthermore, this peptide significantly enhanced survival and slowed the progression of infection in a zebrafish larvae model infected with the multidrug resistant E. coli 87 strain.
An innovative approach to cysteine targeting involves proximity-enabled reactivity via a sulfonium group on a peptide tether, integrating peptide stapling and bioreactive group installation in a single step.140 This was demonstrated in inhibiting PDZ-RGS3, a key player in ephrin-B signaling, which is known to influence chemotaxis induced by stromal derived factor 1. A peptide, mimicking the C-terminal sequence of ephrin-B, was macrocyclized by bis-alkylating cysteine and methionine, creating a sulfonium at the methionine site. When the resultant peptide binds to the PDZ domain of GRS3, it positions the sulfonium near PDZ’s target cysteine residue, triggering a rapid nucleophilic substitution that covalently links the peptide to the protein. This Cys-targeting approach has shown high specificity both in vitro and in cell lysate. Subsequently, this method was adapted to covalently target BFL-1,141 a member of the BCL-2 protein family. By binding at the BH3-binding pockets of these antiapoptotic proteins, it is possible to reinitiate apoptosis in BCL-2-dependent cancers. Here, a BIM BH3 helical peptide was tethered at its N-terminus to form a sulfonium, designed to target Cys55 in BFL-1. Among the inhibitors tested, B4-MC showed selective covalent binding to BFL-1 in both in vitro and in cells, effectively inhibiting the growth of BFL-1-expressing cancer cells.
Peptide inhibitors that form reversible covalent bonds with target proteins have been developed, offering potential benefits like reducing immunogenicity by avoiding permanent protein modification. 2-Acetylphenylboronic acid (APBA) forms a reversible iminoboronate with lysine side chains. This warhead was chemically attached onto peptides displayed on bacteriophage surfaces, creating linear and cyclic peptide libraries142,143 that were then screened against S. aureus sortase A (SrtA) and the coronavirus spike protein. These screenings identified peptide binders for both targets, demonstrating single-digit micromolar affinity and high specificity in cellular environments (for SrtA) and in saliva and serum (for the spike protein).143 To extend residence time of the binder, a new warhead named RMR1 was developed.144 Unlike APBA, which equilibrates with lysine instantaneously, RMR1 reacts with lysine to form a diazaborine bond that dissociates slowly, having a rate constant (k–1) of 2.6 × 10–5 s–1. Integrating RMR1 into a cyclic peptide that binds to SrtA increased its efficacy approximately threefold in S. aureus cells by targeting Lys173 of SrtA. Additionally, the RMR1-modified peptide continued to inhibit SrtA’s transpeptidase activity in S. aureus cells even after washout, unlike its APBA-modified or wild-type counterparts.
In summary, the development of covalent peptide-based inhibitors targeting complex PPI interfaces, a challenge for small molecules, is gaining traction. The design process often involves structure-based methods or variant screenings to pinpoint sites for covalent modifications and target residues on proteins. Balancing the reactivity and selectivity of the covalent inhibitor in cellular contexts is vital. Despite hurdles such as predicting target residues, the limited scope of amino acids that can be targeted, and the low uptake of peptides by cells, advancements in this emerging field are notable. The need for more in vivo studies is clear to assess the inhibitors’ efficacy and to identify optimal warheads.
5.2. Site-Specific Conjugation of Native Antibodies
Antibodies conjugated with a range of chemical or biological molecules, such as biophysical probes, cytotoxins, enzymes, and cytokines, have found extensive use in in vitro assays, diagnostics, and targeted therapies.145 Traditional methods of labeling antibodies often employ highly reactive N-hydroxysuccinimide (NHS) esters to acylate lysine residues or maleimides to alkylate cysteine residues. These methods, however, lack control over the modification site and the number of payloads, leading to heterogeneous antibody conjugates.146 Site-specific antibody conjugation has been achieved by engineering antibodies with Uaas containing bio-orthogonal groups, peptide tags for enzymatic modification, or through glycan remodeling.146−148 Nonetheless, these approaches necessitate genetic, chemical, or enzymatic modification of the antibody. To conjugate unmodified antibodies, researchers have introduced photo-cross-linkers into protein domains or peptides that bind to the antibody.149,150 Upon UV irradiation, the protein domain or peptide cross-links to the antibody. This method, however, suffers from a lack of chemical specificity, and prolonged UV exposure can damage proteins.
In recent years, proximity-enabled reactivity has facilitated the site-specific conjugation of native antibodies. This strategy involves using a peptide that binds specifically to an antibody’s Fc region (Figure 20). This peptide is modified with a reactive functional group together with the desired payload or a bio-orthogonal functionality allowing for subsequent modification. When incubated with a native antibody, the peptide’s reactive functional group is positioned close to a natural residue of the antibody, enabling a specific cross-link and, consequently, the conjugation of the peptide to the antibody. This method shifts the engineering effort from the antibody to the peptide. Payload installation on the peptide is precisely controlled through chemical synthesis, and the proximity-enabled reactivity between the peptide and the antibody ensures site-specific conjugation and homogeneity in the final product.
Figure 20.

Site-specific conjugation of native antibodies. (A) Conjugation of an antibody with the FPheK-modified FB protein or ssFB peptide. (B) Conjugation of an antibody with NHS-modified IgG-binding peptide. (C) Conjugation of an antibody with NHS-modified Fc-binding peptides, followed with TCEP cleavage of the disulfide linkage. (D) An Fc-binding peptide was used to attach dirhodium complexes, catalyzing the labeling of a proximal Asn residue with the diazo reagent. (E) Fc-III peptide-guided proximity-enabled lysine acetylation to transfer an azide group from the affinity peptide to a lysine residue on the antibody.
High-efficiency direct peptide conjugation to antibodies has been successfully achieved.151 A latent bioreactive Uaa, FPheK, capable of reacting with proximal lysine residues was incorporated at the Glu25 site in the B domain of Staphylococcus aureus protein A (referred to as FB protein). The conjugation occurs when the FB protein binds to an antibody, allowing FPheK to react with a nearby lysine residue on the antibody (Figure 20A). The cross-linking process has demonstrated efficiencies ranging from 91% to 99%. This method is applicable to both human and mouse IgGs. For instance, a fluorescein-labeled FB mutant was attached to Trastuzumab, a HER2-specific antibody, enabling fluorescence imaging of HER2 on cancer cells. Subsequently, the 66 amino acid FB protein was further refined to a shorter 33 amino acid FB peptide (ssFB).152 The ssFB peptide includes FPheK and a C-terminal azide moiety. After achieving conjugation yields greater than 95%, the azide moiety facilitated the attachment of the microtubule toxin MMAE. This process led to the creation of a trastuzumab–MMAE conjugate, which exhibited an EC50 in the nanomolar range in eradicating HER2-positive cancer cells. Additionally, a PSMA-specific small-molecule inhibitor, DUPA, was conjugated to muromonab, an anti-CD3 antibody.152 This resulted in a bispecific conjugate that selectively directed T cells toward PSMA-expressing cancer cells, inducing cytotoxicity at picomolar concentrations in vitro. Furthermore, this conjugate significantly inhibited prostate tumor growth in mouse xenograft models. In another study, this conjugation technology was used to generate bone-targeting antibodies through coupling a bone-targeting bisphosphonate moiety to therapeutic antibodies.153 Trastuzumab was conjugated with the azide-bearing ssFB peptide, followed by a reaction with bicyclo[6.1.0]nonyne-functionalized Alendronate (ALN). ALN is bisphosphonate drug used for bone-targeting and the treatment of osteoporosis and bone metastasis. The resulting trastuzumab–ALN conjugate specifically targets the bone metastatic niche to eliminate bone micrometastases and also prevents the seeding of multiorgan metastases from bone lesions in mouse models. These results together suggest that the conjugated ssFB peptide moiety minimally perturbs the binding of the antibody–ssFB conjugate with its antigen or to FcγRIII, the receptor responsible for activating antibody-dependent cell-mediated cytotoxicity.
In lieu of the latent bioreactive FPheK, a highly reactive N-hydroxysuccinimide (NHS) ester was incorporated into peptides and utilized at an acidic pHs, aiming to reduce NHS ester hydrolysis and enhance selectivity toward the proximal lysine residue.154,155 In one study, a 17 amino acid IgG-binding peptide (IgGBP) was modified by introducing an NHS ester at the Arg8 site (Figure 20B).154 When IgGBP binds to human IgG, the NHS ester is positioned near the Lys248 of the antibody, as inferred from the crystal structure. This strategic positioning allows the peptide and the antibody to conjugate rapidly, typically within 15 min. Mass spectrometry analysis confirmed the specificity of this conjugation at Lys248. A trastuzumab–IgGBP–DM2 conjugate was successfully prepared using this method. This conjugate demonstrated an EC50 in the nanomolar range, effectively inhibiting HER2-positive cancer cells. Additionally, conjugating trastuzumab with NHS–IgGBP–azide and subsequently reacting it with a dibenzocyclooctyne (DBCO)-labeled nanobody resulted in the formation of a bispecific antibody. Notably, this NHS–IgGBP method has shown effectiveness in conjugating various IgGs from human, mouse, and rabbit origins.
In another study, three distinct peptides, each binding to different Fc regions of antibodies, were modified with NHS ester (Figure 20C).155 When conjugated at pH 5.5, they demonstrated targeted conjugation to specific lysine residues on the antibodies. Furthermore, a disulfide linkage between the NHS ester and the peptide was introduced. This linkage allows for the peptide to be cleaved from the antibody postconjugation using tris(2-carboxyethyl)phosphine (TCEP), leaving only a small molecular thiol group on the antibody. This residual thiol group facilitates subsequent modifications through thiol–maleimide reactions. For instance, a toxin (DM-1) conjugated to trastuzumab using this method selectively eliminated HER2-positive cells in vitro and showed promising results in xenograft mouse models.
A different strategy to facilitate site-specific conjugation of antibodies, without covalently attaching an affinity peptide, involves catalysis.156 Specifically, an Fc-binding peptide, derived from a minimized Z domain of S. aureus protein A, was modified with three dirhodium complexes through solid-phase peptide synthesis (Figure 20D). Upon the peptide binding to an antibody, these dirhodium complexes act as reactive catalytic centers. They effectively promote the labeling of an adjacent Asn residue on the antibody with an added diazo reagent, achieving efficiencies greater than 90%. The diazo reagent is equipped with an alkyne functional group, allowing for the subsequent attachment of various molecules, such as fluorophores and toxins, to the antibody through copper-catalyzed azide–alkyne cycloaddition (CuAAC) click chemistry. This method conjugates a small-molecule linker, which possesses a bio-orthogonal handle, directly onto the antibody. In contrast, the binding peptide itself is not conjugated. This approach is likely to minimize any potential interference that might occur when binding with secondary antibodies.
Another advancement involves peptide-guided, proximity-enabled lysine acetylation (Figure 20E).157 This technique transfers an azide group from the affinity peptide to a lysine residue on the antibody, also without conjugating the peptide. Specifically, an azide-bearing phenolic ester is attached to the Fc-interacting Fc-III peptide. Upon the peptide binding to the Fc domain of an antibody, the phenolic ester is positioned close to the Lys248 residue. This proximity facilitates a nucleophilic reaction, transferring the azide-bearing acetyl group to Lys248. The modified Lys248 then becomes a site for bio-orthogonal labeling with various payloads. For example, fluorophores have been successfully attached to atezolizumab using this method, allowing for the imaging of HER2 on cancer cells. Similarly, lipids have been conjugated onto trastuzumab to create immunoliposomes targeting HER2-positive cells. Additionally, the functionalization of azide-labeled antibodies with bifunctional linkers has enabled the construction of bispecific antibody complexes. These complexes consist of two complete antibodies and have demonstrated T-cell-mediated cytotoxicity at nanomolar concentrations in vitro. This approach is noteworthy for introducing only a minimal-sized azide group onto Lys248 of the native antibody, potentially minimizing any interference with the antibody’s function. While the final modification yield currently stands at about 80%, there is scope for further enhancement and optimization.
5.3. Proximity-Enabled Reactive Therapeutics (PERx) to Develop Covalent Protein Therapeutics
Covalent small-molecule drugs have the unique ability to establish a covalent bond with their targets upon binding. This feature offers significant advantages over noncovalent drugs, including increased potency, extended duration of action, enhanced therapeutic index, and the capacity to target molecules previously deemed “undruggable”.118,158,159 Since 1947, roughly 7% of small molecule drugs approved by the FDA function via this covalent mechanism.114 In the past decade, these drugs have shown particular efficacy as targeted covalent inhibitors for kinases in cancer therapy. Notable examples include drugs targeting EGFR, BTK, HER2, and KRAS, which are used in treating lung cancer, lymphoma, and breast cancer.114,115 In contrast, the realm of covalent protein drugs remains largely unexplored. This underutilization stems from the natural limitations of proteins in forming covalent bonds with other biomolecules, with the exception of disulfide bonds. Nevertheless, proteins are ideally suited for covalent action due to their expansive interactive interfaces. They can effectively bind to targets with flat and smooth surfaces or engage in complex protein–protein interactions, which pose a challenge for small-molecule drugs. Additionally, the high target specificity of proteins could greatly reduce off-target reactions, a prominent issue with covalent small molecule drugs.
The advent of latent bioreactive Uaas and the concept of proximity-enabled bioreactivity have given rise to proximity-enabled reactive therapeutics (PERx) technology.160 This innovative platform is designed for the development of covalent protein therapeutics. The fundamental principle of PERx involves incorporating a latent bioreactive Uaa into a protein drug near its binding interface (Figure 21). The interaction between the protein drug and its target brings the Uaa proximal to an adjacent natural amino acid residue in the target protein. This proximity activates the Uaa, enabling it to selectively react with the target natural residue. The reaction facilitates the formation of a covalent bond, effectively transforming the protein drug into a covalent protein therapeutic. Since its initial development in 2020,160 PERx technology has been successfully utilized to generate a variety of covalent protein drug candidates, each designed to treat different diseases through distinct mechanisms of action.
Figure 21.
Principle of proximity-enabled reactive therapeutics (PERx) for developing covalent protein drugs.
5.3.1. The Advent of PERx in Immunocheckpoint Inhibition
The development of PERx technology commenced with the human immune checkpoint proteins: programmed cell death protein-1 (PD-1) and its ligand, PD-L1, serving as a foundational model system.160 PD-1, a transmembrane receptor, plays a critical role in regulating T cell activity. PD-L1, one of PD-1’s ligands, is frequently overexpressed in various tumors. The interaction between PD-1 and PD-L1 leads to reduced proliferation and T-lymphocyte activity, resulting in the exhaustion and eventual apoptosis of tumor-specific T cells.161 Blocking this PD-1/PD-L1 interaction can rejuvenate the suppressed immune response against tumors.162 Consequently, several antibodies targeting either PD-1 or PD-L1 have been developed for cancer therapy. However, these antibodies, typically large (around 150 kDa), often exhibit limited tissue and tumor penetration, and patient responses to these treatments vary significantly.
In an effort to inhibit the PD-1/PD-L1 interaction using proteins of smaller molecular weights, the researchers proposed transforming the ectodomain of human PD-1 (approximately 15 kDa) into an efficient covalent binder targeting PD-L1 (Figure 22A).160 This was achieved by genetically incorporating a latent bioreactive Uaa, FSY,40 into PD-1 at its binding interface. Based on the structural analysis of the PD-1/PD-L1 complex,163 site Ala129 was chosen for FSY incorporation, aiming to target His69 of PD-L1. The modified PD-1(FSY) demonstrated high efficiency and specificity in covalently binding to human PD-L1, both in vitro and on cancer cell surfaces, as well as in tumor xenograft models in mice. Mass spectrometry analysis of the cross-linked protein samples validated that FSY reacted specifically with His69, as intended.
Figure 22.
(A) The covalent PD-1(FSY) potently inhibits tumor growth in mice via the PERx mechanism. Using its FSY to react with His69 of PD-L1, PD-1(FSY) irreversibly binds with PD-L1 on tumor cells, efficiently blocking the interaction of PD-L1 (on tumor cells) with PD-1 (on T cells). The blockage revives the proliferation of T cells and their activity to kill tumor cells. (B) Weight comparison of human H460 tumors dissected from Hu-mice that were treated with the indicated protein drugs. PD-1(FSY) shows a more potent antitumor effect than PD-1(WT) and atezolizumab. Panel (A) adapted with permission from ref (6). Copyright 2021 John Wiley and Sons.
To evaluate whether this covalent binding capability of PD-1(FSY) could boost T cell activation by disrupting the PD-1/PD-L1 interaction, mixed lymphocyte reactions were conducted using dendritic cells and T cells, as well as between chimeric antigen receptor T (CAR-T) cells and target cancer cells.160 The studies found that PD-1(FSY) effectively enhanced the functional activities of both human T cells and CAR-T cells in vitro, matching the efficiency of atezolizumab, an FDA-approved anti-PD-L1 antibody for cancer treatment. In contrast, the noncovalent wildtype PD-1(WT) showed no such effect.
Further investigations involved injecting PD-1(FSY) into xenograft mouse models humanized either with human peripheral blood mononuclear cells or human CAR-T cells or via systematic bone marrow-liver-thymus humanization (Hu-mice).160 In all three models, PD-1(FSY) significantly inhibited tumor growth compared to PD-1(WT), displaying a therapeutic effect that was comparable to, or even surpassed, that of atezolizumab (Figure 22B).
Beyond addressing natural protein interactions such as PD-1/PD-L1, PERx technology has also shown effectiveness with engineered protein binders, offering a versatile pathway to covalently target native proteins. For example, introducing FSY into a HER2-specific affibody transforms it into a covalent binder for HER2, effective both in vitro and on breast cancer cell surfaces.160 Similarly, the integration of FSY or FSK into EGFR-specific nanobodies enables these nanobodies to become efficient covalent binders for EGFR, demonstrating efficacy in vitro and on various cancer cells.43
PERx technology is elegantly simple, requiring only the introduction of a single mutation—a latent bioreactive Uaa—into the protein drug. This streamlined approach not only simplifies the conversion process for covalent protein drugs but also minimizes potential immunogenicity risks. PERx-equipped protein drugs establish irreversible bonds with their targets, effectively decoupling the drug’s pharmacodynamic effects from its pharmacokinetics. While small proteins are typically cleared rapidly in vivo, they are advantageous for certain conditions like extravasation and tissue penetration. Traditional methods to prolong protein half-lives often involve increasing their size, which can diminish their effectiveness. PERx, however, allows for the direct utilization of small proteins in vivo, eliminating the need for additional modifications to extend their half-life. A major concern with covalent drugs is off-target reactions. However, PERx offers exceptional target specificity, as its covalent reactivity relies on both the binding of the drug to its target and a unique pairing between unnatural and natural amino acids.57,160 In summary, PERx represents a versatile platform technology that transforms various interactive proteins into covalent binders. Its straightforward process and wide applicability to different proteins hold great promise for accelerating the development of covalent protein drugs to treat a diverse range of diseases.
5.3.2. Neutralization of SARS-CoV-2
Tumor suppression typically unfolds over days to weeks. Recognizing this, the efficacy of PERx technology has been explored in scenarios demanding rapid and immediate action. For instance, covalent nanobodies have been developed to counteract SARS-CoV-2 infections in human cells, showing significantly enhanced potency compared to conventional noncovalent nanobodies (Figure 23A).42 The infection mechanism of SARS-CoV-2 hinges on the interaction of its spike protein with the human cellular angiotensin-converting enzyme 2 (ACE2) receptor.164 Noncovalent drugs that bind to the spike protein can dissociate, allowing the virus to reaccess and infect cells, thereby making complete inhibition a challenge. A covalent nanobody that irreversibly binds to the spike protein is anticipated to achieve more effective inhibition and reduce the likelihood of viral escape.
Figure 23.
(A) Whereas the conventional nanobody can dissociate from the spike RBD, the covalent nanobody binds with the spike RBD irreversibly, permanently preventing viral binding with ACE2 receptor and thus blocking infection more effectively. (B) Compared to the mNb6(WT), the covalent mNb6(108FFY) showed a 36-fold increase in potency in inhibiting SARS-CoV-2 pseudovirus infection and a 40-fold increase in potency in inhibiting the authentic SARS-CoV-2. Adapted with permission from ref (42). Copyright 2022 Elsevier.
To develop a covalent nanobody, 30 sites within the three complementarity-determining regions (CDRs) of the spike-specific nanobody mNb6165 were assessed for FSY incorporation.42 Several of these sites enabled effective covalent cross-linking with the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein in vitro. To enhance the PERx reaction rate, the electron-withdrawing fluorine was introduced into the phenyl ring of FSY, creating a new latent bioreactive Uaa, FFY, which boosted the reaction rate by 2.4 times.42 The most effective variant mNb6(108FFY) achieved rapid cross-linking with the spike RBD within 10 min.
In neutralization assays using lentivirus pseudotyped with the viral spike protein, the WT mNb6 nanobody had an IC50 of 36 nM.42 In contrast, the covalent mNb6(FFY) variant showed a dramatically improved IC50 of 1 nM, marking a 36-fold increase in potency (Figure 23B). Furthermore, against authentic SARS-CoV-2, the covalent nanobody exhibited a 40-fold improvement in IC50 compared to the WT control. Besides the WT virus, mNb6 also bound to SARS-CoV-2 variants alpha, delta, epsilon, and lambda. The mNb6(FFY) mutant covalently cross-linked with these variant spike proteins, significantly enhancing its neutralization potency and showing 23-, 39-, 38-, and 24-fold increases for each variant, respectively.
However, mNb6 was ineffective against later-emerging omicron variants. A different nanobody, Nb70, that binds omicron and other variants with high affinity was modified into a covalent form using the same approach.42 The covalent Nb70,166 with FFY incorporation, demonstrated an 8–10-fold increase in neutralization potency against the omicron variants BA.1 and BA.2 compared to its WT counterpart. In summary, these covalent nanobodies exhibited markedly enhance neutralization potency against both WT SARS-CoV-2 and its various variants compared to their WT equivalents.
Beyond nanobodies, researchers have also transformed the native receptor, human ACE2, into a covalent binder for the spike protein.42 This was achieved by incorporating FSY into the soluble extracellular domain of ACE2. Given that SARS-CoV-2 relies on ACE2 for cell entry, this soluble ACE2 therapy is anticipated to offer broad coverage, as the virus cannot easily evade neutralization through ACE2.
The faster reaction kinetics of FFY compared to FSY have proven crucial in effectively inhibiting SARS-CoV-2 infection. In a study, FSY was incorporated into spike-specific minibinders.167,168 The resulting minibinders required a 2 h incubation to cross-link with the spike RBD in vitro and 2 h incubation with the virus, yielding a modest sixfold potency increase against a single SARS-CoV-2 variant.167 However, their effectiveness against the WT virus and other variants remains unverified, and longer incubation times may allow the virus more opportunity to infect human cells. In contrast, the FFY-augmented covalent nanobody mNb6(FFY) quickly cross-linked with the spike RBD in just 10 min.42 When incubated with the virus for only 1 h, mNb6(108FFY) showed a remarkable 41-fold increase in neutralization potency against SARS-CoV-2 and 23–39-fold increase in potency against SARS-CoV-2 variants, indicating its robust and broad-spectrum efficacy.
Covalent nanobodies can be manufactured in large-scale through bacterial expression. Their high stability simplifies storage and distribution, and their suitability for aerosolization allows for self-administered inhalation treatments targeting nasal and lung epithelia. PERx-enabled ACE2 drugs could become a vital reserve for combating future outbreaks of SARS-CoV, SARS-CoV-2, and any novel coronavirus that utilizes the ACE2 receptor for entry. The irreversible binding characteristic of covalent protein drugs not only promises complete viral inhibition but also helps to reduce the risk of viral resistance. Furthermore, the PERx methodology for creating covalent binders or soluble receptor inhibitors has the potential in the development of effective covalent protein drugs for a variety of infectious diseases, including influenza, hepatitis, AIDS, and anthrax.
5.3.3. Targeted Protein Degradation
Over the past two decades, targeted protein degradation has evolved as a promising therapeutic strategy, offering a significant alternative to conventional inhibition methods.169−173 This approach, applicable to intracellular, membrane, and extracellular proteins, utilizes various technologies including PROTACs, molecular glues, LYTACs, and KineTACs. Research indicates that the efficiency of protein degradation is largely contingent on the binding affinity between the degrader molecule and the target protein. A notable advancement in this field is the application of the PERx principle, where a covalent linkage is established between the degrader and its target, demonstrating efficient degradation of membrane protein PD-1 in cancer cells (Figure 24A).174
Figure 24.
(A) Nb(FSY)–CPP–LSS covalently binds to PD-L1 on the cancer cell surface via PERx, leading to internalization and lysosomal degradation of the cross-linked complex. (B) A mixture of A375 cells expressing PD-L1-EGFP and fresh pBMCs at a 1:1 ratio was injected into NOG mice. The mice were then treated with the specified proteins every 2 days. Post-treatment, the weights of the dissected tumors were compared. Adapted from ref (174). Copyright 2021 American Chemical Society.
The PD-L1-specific nanobody was modified by incorporating the latent bioreactive Uaa FSY,40 resulting in a covalent nanobody capable of cross-linking PD-L1 both in vitro and on the surface of cancer cells.174 This covalent nanobody was further conjugated with a cell-penetrating peptide and a lysosome-sorting sequence (CPP-LSS), which are known to enhance cell internalization and lysosome-mediated degradation, respectively.175,176 The resulting conjugate, Nb(FSY)–CPP–LSS, demonstrated greater efficiency in degrading PD-L1 on cancer cells compared to its noncovalent counterpart, Nb–CPP–LSS.174 This led to a more pronounced activation of T-cells. Additionally, in xenograft mouse models injected with human peripheral blood mononuclear cells, Nb(FSY)–CPP–LSS showed a slight, yet statistically significant, improvement in inhibiting tumor growth compared to Nb(FSY) alone (Figure 24B). This result implies that the CPP–LSS component enhances the overall effectiveness of tumor inhibition by facilitating the degradation of PD-L1, building upon the already highly efficient tumor suppression effect achieved through the covalent blockage of Nb(FSY).
A notable advantage of the Nb(FSY)-CPP-LSS design lies in its cell-type-independent degradation mechanism. This approach does not rely on a specific lysosome-targeting receptor or membrane E3 ligase, making it potentially effective in addressing heterogeneous tumor cells. To enhance the understanding of this strategy’s effectiveness, exploring its application to a target where covalent nanobody binding does not dominantly influence the measured biological function in vivo would be highly advantageous. Anticipated to provide a more definitive demonstration, this exploration could elucidate the in vivo therapeutic efficacy mediated by CPP–LSS degradation more clearly.
5.3.4. Selective Targeting of Interleukin-2 Receptor Subunit
Cytokines, crucial in regulating cell-mediated pro-inflammatory and anti-inflammatory responses, are potential therapeutic targets. Their function can be modulated by altering the trafficking properties of cytokine–receptor complexes, which are internalized after cell surface interaction and either degraded or recycled. Interleukin-2 (IL-2), a key cytokine, orchestrates immune responses by differentially expanding regulatory T cells (Tregs) and effector T cells (Teffs) to maintain immune homeostasis.177 IL-2 signals through a trimeric IL-2 receptor (IL-2R) with high affinity (Kd ∼ 10 pM), consisting of IL-2Rα (CD25), IL-2Rβ, and the common γc receptor subunits, or through a dimeric IL-2R with intermediate affinity (Kd ∼ 1 nM), made up of IL-2Rβ and γc.177 While low-dose IL-2 preferentially activates Tregs, which express IL-2Rα, to treat inflammatory and autoimmune diseases (ADs), higher doses can inadvertently activate Teffs, exacerbating ADs.178,179 For the purpose of targeted cell population activation and to improve the in vivo properties, IL-2 has been engineered using various strategies. These include the utilization of antibodies and PEGylation for selective modulation of IL-2 effects,180,181 as well as the development of high-affinity IL-2Rα-binding IL-2 mutants and variants with impaired internalization to enhance its potency.182−184 PERx has now enabled the engineering of a covalent IL-2 that irreversibly binds to IL-2Rα, leading to preferential and sustained activation of Tregs (Figure 25A).185
Figure 25.
(A) When IL-2 binds to its high-affinity trimeric receptor on Tregs, the resultant IL-2–receptor complex is internalized and later dissociates in the early endosome. While IL-2Rα is recycled to the cell surface, the other components with IL-2 are directed to lysosomal degradation. The addition of FSY to IL-2 enables it to form a covalent bond with IL-2α, infinitely enhancing its affinity. This modification significantly boosts IL-2 recycling and prolongs IL-2 signaling, particularly favoring Tregs. (B) The effects of IL-2 mutants on a xeno-GvHD mouse model. A lethal dose of activated hPBMCs was injected into NSG mice, followed by subcutaneous injection of IL-2 mutants. The weight curves and Kaplan–Meier survival curves of the grafted mice are shown. Adapted with permission from ref (185). Copyright 2023 Springer Nature.
A covalent IL-2 variant, L72-FSY, was developed by incorporating the latent bioreactive Uaa FSY at site L72. This mutant selectively cross-links with IL-2Rα, both in vitro and on YT cell surfaces expressing IL-2Rα.185 Treating Tregs with L72-FSY enhances the recycling and cell surface retention of covalent IL-2. Notably, L72-FSY preferentially activates Tregs over Teffs, as observed in vitro and in vivo. In human peripheral blood mononuclear cells (hPBMCs), it increases the proportion of Treg without impacting CD8+ T or natural killer cells and upregulates Treg activation markers. In a mouse model injected with hPBMCs and an IL-2Rα-humanized B6 mouse model, L72-FSY significantly boosted Treg numbers compared to WT IL-2, promoting a central memory phenotype with reduced exhaustion markers like LAG-3 and increased PD-1 expression. Unlike WT IL-2, it induces Tim-3 expression while maintaining CTLA-4 levels, showing enhanced stability and suppressive function in Tregs. These results demonstrate L72-FSY’s effectiveness in expanding functionally superior Tregs without causing terminal differentiation.
To increase its half-life, researchers PEGylated L72-FSY at the N-terminus. Its therapeutic potential was evaluated in a pristane-induced lupus model in B-hIL2RA mice and a xenogeneic graft-versus-host disease (xeno-GvHD) mouse model.185 Both L72-FSY and its PEGylated form significantly reduced lupus severity, showing improvements in kidney damage and reduced autoantibody levels compared to WT IL-2. Moreover, they diminished Teff activity while increasing Treg ratios and activation markers. In the xeno-GvHD model, L72-FSY variants provided more protection than WT IL-2, indicated by reduced body weight loss and increased survival rates (Figure 25B). Overall, the researchers conclude that L72-FSY and PEG-L72FSY are more effective than WT IL-2 in suppressing inflammatory diseases like systemic lupus erythematosus (SLE) and GvHD. Therefore, PERx-enabled covalent cytokines may lead to a distinct class of immunomodulatory therapies.
5.3.5. Targeted Radionucleotide Therapy
Molecularly targeted radionuclide therapies (TRTs) are isotopically labeled drugs delivering ionizing radiation to tumors, exploiting cancer’s vulnerability to genetic damage.186 Recently, the focus has shifted to low-molecular-weight TRTs targeting overexpressed cancer proteins to minimize toxicity. These include FDA-approved therapies like Pluvicto for prostate cancer and Lutathera for neuroendocrine tumors. The transition from high-molecular-weight radiopharmaceuticals such as antibodies, which caused toxicity due to prolonged blood residence, to low-molecular-weight ones was driven by the need for safer yet effective treatments.187 However, low-molecular-weight radioligand therapies (RLTs) face challenges: they are rarely curative and must target highly overexpressed proteins. Additionally, their instability and rapid clearance from tumors limit their effectiveness. Strategies to enhance tumor retention include incorporating hydrophobic binding groups and antibody pretargeting,188,189 but these can increase toxicity or treatment complexity.
To address the safety vs efficacy dilemma in the TRT field, the ideal radiopharmaceutical should possess high specificity, a short blood residence time, and prolonged tumor retention. In pursuit of this, PERx has thus been utilized to develop small covalent protein-based radiopharmaceuticals (Figure 26A).190 These proteins are designed to bind irreversibly to their targets upon interaction, contrasting with their WT counterparts that clear rapidly from the blood but lack persistent binding. This innovative approach enables the covalent small protein radiopharmaceuticals to maintain rapid blood clearance while achieving sustained tumor retention through irreversible target binding.
Figure 26.

Covalent small protein radiopharmaceuticals to enhance efficacy and safety for TRT. (A) When the covalent Nb(FSY) binds to HER2, FSY undergoes a proximity-enabled SuFEx reaction with a target Lys residue on HER2, cross-linking Nb(FSY) with HER2. This irreversible binding increases radioisotope levels in the tumor and extends its residence time while still ensuring rapid systemic clearance. (B) The covalent 124I–Nb(FSY) enabled clear imaging of the tumor distinct from the background. 3D PET image reconstruction of mice 24–72 h after the injection of 124I–Nb(WT) or 124I–Nb(FSY) are shown. (C) Growth curves of engrafted NCI-N87 tumors indicate that the covalent 225Ac–Nb(FSY) inhibited tumor growth, while 225Ac–Nb(WT) did not (top). Body weights of the mice remained stable over the course of the therapy study (bottom). Reproduced from ref (190). Copyright 2023 American Chemical Society.
To implement this strategy, researchers developed a covalent nanobody targeting the HER2 receptor by incorporating the latent bioreactive Uaa FSY into the nanobody at its binding interface (Figure 26A).190 The resulting Nb(FSY) demonstrated rapid cross-linking to the HER2 receptor, achieving this within 10 min in vitro. It effectively targeted endogenous HER2 on the surface of cancer cells and HER2 in tumors within mice.
Both Nb(WT) and Nb(FSY) were labeled with 124I and injected into mice xenografted with HER2-expressing tumors, then visualized using PET/CT imaging.190 The clearance from blood circulation and half-lives of 124I–Nb(WT) and 124I–Nb(FSY) were comparably rapid. Additionally, the uptakes of the radiotracers in the liver, kidney, thyroid, and skeletal muscle were qualitatively similar for both, indicating a similar biodistribution in normal organs devoid of HER2. However, a significant difference emerged in tumor targeting. Between 3 and 10 h postinjection, the on-tumor activity levels of both nanobodies were similar. The striking divergence was observed from 24 to 72 h postinjection: 124I–Nb(FSY) remained detectable in the tumor, whereas 124I–Nb(WT) did not. The total radiation dose accumulated in the tumor by 124I–Nb(FSY) was 81% higher compared to that by 124I–Nb(WT). Three-dimensional maximum intensity projection images further illustrated this distinction (Figure 26B). In mice injected with 124I–Nb(FSY), the tumor was distinctly visible against a background of virtually no signal in normal tissues, except the thyroid, a natural site for iodine metabolism.
To assess whether the enhanced tumor retention of the covalent Nb(FSY) translated into a significant antitumor effect, both Nb(WT) and Nb(FSY) were labeled with actinium-225 (225Ac), a potent α-emitter.190 Mice bearing HER2-expressing NCI-N87 tumors were treated twice (day 0 and day 7), with either form of the Ac-labeled nanobody. Compared to the saline control group, treatment with 225Ac–Nb(WT) showed no significant tumor inhibition. In contrast, 225Ac–Nb(FSY) administration resulted in a considerable slowdown in tumor growth (Figure 26C). End point analysis further revealed that the tumors in mice treated with 225Ac–Nb(FSY) were significantly lighter compared to those in mice treated with 225Ac–Nb(WT). Importantly, there was no significant change in body weight across all three groups, indicating a lack of systemic toxicity. Histopathological analysis of liver, kidney, heart, and bone marrow tissues revealed no abnormalities, suggesting the absence of toxicity in these organs post-treatment.
In summary, by facilitating a highly specific and prolonged retention of radionuclides within tumors while minimizing exposure to normal tissues, these small covalent protein-based radiopharmaceuticals substantially improve both the efficacy and safety of TRT. The transition from noncovalent to covalent binding in protein-based TRT opens up new possibilities for treating a wider range of targets and diseases with greater precision.
5.3.6. Inhibition of the Glycol Immunocheckpoint
The PERx strategies outlined above hinge on the capacity to covalently target proteins. With the emergence of GECX-RNA and GECX-sugar technologies,18,19 this capability of covalent targeting has broadened to encompass other families of biomacromolecules beyond proteins. This expansion opens up exciting opportunities for the development of novel classes of covalent therapeutics focusing on these diverse biomolecules.
Glycol immunocheckpoints, particularly the interaction between sialoglycans on tumor cells and Siglecs (sialic acid-binding immunoglobulin-like lectins) on immune cells, are crucial in the dynamics of cancer immunity.191,192 Sialoglycans are often overexpressed on the surfaces of tumor cells. This overexpression is a strategic evasion tactic, enabling these cells to engage with, for instance, Siglec-7 found on NK cells. The binding of tumor cell sialoglycans to Siglec-7 effectively dampens the NK cells’ cytotoxic response, a key mechanism by which tumors avoid immune detection and destruction.193 Interrupting this specific interaction is a promising strategy in cancer immunotherapy, as it could potentially unleash the innate cytotoxic capability of NK cells against the tumor.194 Antibodies and sialic acid mimetics to target Siglecs have been developed for such purposes,195 and antibody–sialidase conjugates have been used to remove cancer cell surface sialic acids to improve NK cell activity.196
The recent advancements in GECX-sugar technology have facilitated the ability to covalently cross-link glycans using glycan binding proteins through proximity-enabled bioreactivity. A Siglec protein, specific for sialoglycan binding, has been engineered to irreversibly cloak sialoglycans on cancer cell surface so as to block the inhibitory effect of sialoglycans on NK cells (Figure 27A).18 Specifically, the latent bioreactive Uaa SFY was incorporated at position 127 of the extracellular V-set domain of Siglec-7, resulting in the creation of a covalent variant, Siglec-7v(127SFY). This modified Siglec-7v(127SFY) demonstrates enhanced binding to sialoglycans on cancer cell surfaces, as evidenced by flow cytometric analysis of the bound Siglec-7v protein. To investigate whether this enhanced binding could effectively block the interaction between tumor cell surface sialoglycans and Siglec-7 on NK cells, experiments were conducted using Siglec-7v(127SFY) and the WT Siglec-7v (Siglec-7v(WT)). These were incubated with three hypersialylated cancer cell lines (SK-MEL-28, BT-20, MCF-7), followed by exposure to NK-92 cells. The results showed that Siglec-7v(127SFY) significantly increased the killing efficacy of NK-92 cells against all three cancer cell lines in a concentration-dependent manner compared to Siglec-7v(WT) (Figure 27B).18 While these promising results have been obtained in cell-based studies and have yet to be validated in animal models in vivo, they suggest that covalently targeting glycans could be a viable and innovative approach for cancer immunotherapy.
Figure 27.

(A) The application of covalent Siglec-7v(127SFY) aims to block the interaction between sialoglycans on the surface of tumor cells and Siglec-7 on NK cells. This blockage is anticipated to reduce the inhibitory signal of Siglec-7, potentially enhancing the ability of NK cells to kill tumor cells. (B) Cytotoxicity assay of three hypersialylated cancer cell lines demonstrated that Siglec-7v(127SFY) enhanced NK-92 cell killing compared to WT Siglec-7v. The results for breast carcinoma BT-20 cells are shown. Reproduced with permission from ref (18). Copyright 2022 Springer Nature.
6. Conclusions and Outlook
Biospecific chemistry has emerged and advanced significantly, enabling the creation of specific covalent linkages between proteins and various biomacromolecules both in vitro and in vivo. This innovation is rooted in the genetic incorporation of latent bioreactive Uaas into proteins.10,11 Such incorporation facilitates the formation of specific covalent bonds through proximity-enabled bioreactivity when proteins interact with biomacromolecules. Diverse latent bioreactive Uaas have been designed for this purpose, targeting various natural amino acid residues in proteins.6 This methodology has further been extended to the covalent targeting of RNA and carbohydrates.18,19 These novel covalent linkages, created by intramolecular GECX, have introduced unique protein properties previously unattainable with conventional amino acids. The application of GECX intermolecularly17 allows for the exploration of transient protein–protein interactions and high-resolution mapping of protein–RNA interactions within cells. Furthermore, biospecific chemistries have facilitated the development of covalent peptides for therapeutic applications and the conjugation of payloads to native antibodies. A notable advancement in this field is proximity-enabled reactive therapeutics (PERx) technology,160 which produces covalent protein therapeutics with enhanced efficacy compared to their noncovalent counterparts in various applications, including immune-checkpoint inhibition, viral neutralization, targeted protein degradation, receptor subunit targeting, and targeted radionucleotide therapy.
Biospecific chemistry is distinct from bio-orthogonal chemistry. Unlike bio-orthogonal chemistry,7,9 which requires two orthogonal (nonbioreactive) functional groups integrated separately into the probe molecule and target biomolecule, biospecific chemistry only necessitates one bioreactive functional group within a single biomolecule. This approach permits interactions with target biomolecules in their native state, unmodified. Moreover, biospecific chemistry employs latent bioreactive functional groups that remain inert until activated by the proximity effect, making it suitable for live cell applications and in vivo studies. These unique characteristics render biospecific chemistry invaluable for investigating native biomolecules in vivo and for targeted therapeutic applications.
Biospecific chemistry transcends the traditional understanding that biomacromolecular interactions are largely noncovalent, facilitating specific, stable, and irreversible covalent connections between these molecules. Its high specificity stems from a dual reaction requirement: correct binding of biomacromolecules and appropriate pairing of latent bioreactive groups with their targets. This specificity allows for high-resolution analysis at the level of individual amino acids, nucleotides, or carbohydrates, elucidating protein–protein and protein–RNA interactions in living cells with exceptional detail. Such precision is vital for minimizing off-target reactions, a crucial consideration in covalent drug development. Covalent protein therapeutics that utilize the PERx mechanism can achieve heightened target specificity compared to covalent small-molecule drugs, thereby reducing potential off-target effects. Additionally, the stable and irreversible nature of these covalent bonds enhances various protein properties, such as thermostability and photostability, and generates novel abilities such as photomodulation of protein functions. These bonds enable experimentation under more stringent conditions, improving detection sensitivity and reducing false positives. In therapeutics, irreversible linkages between protein drugs and their targets extend the duration of action, achieve receptor subtype selectivity, and enhance drug potency, offering promising applications across diverse diseases.
As new biospecific chemistries emerge and are rediscovered, there is growing anticipation for their integration into other classes of biomolecules beyond proteins, enabling the covalent targeting of a broader range of biomolecules through proximity-enabled bioreactivity. The rapid kinetics of covalent linkage formation are critical for the effectiveness of many applications, and thus enhancing the reaction rates while maintaining biocompatibility is desirable. Advances in protein structure prediction and interaction analysis based on amino acid sequences are poised to facilitate the identification of optimal sites for incorporating latent bioreactive Uaas.90,91 Further, computational methods such as molecular dynamics simulations and machine learning may refine Uaa site selection, accelerate reaction rates, and minimize undesired cross-linking. The expanding diversity of covalent linkages in biomolecules may lead to the emergence of novel properties and functions through directed evolution. Moreover, inducible proximity-enabled bioreactivities could make covalently linked biomolecules more responsive to environmental and external stimuli, enhancing the flexibility of design and achieving higher target specificity. Beyond the genetic encoding of latent bioreative Uaas, alternative approaches are also being explored to enable biospecific chemistry in biomacromolecules. For example, small-molecule cross-linkers equipped with latent bioreactive functional groups have been developed, demonstrating improved precision in cross-linking proteins for structural modeling.49,72
The field of covalent protein therapeutics, developed through latent bioreactive Uaas, is nascent but holds significant promise. This potential is underscored by the successes observed with covalent small-molecule drugs. Given that these protein therapeutics form covalent bonds, specialized evaluation methods distinct from those traditionally used for protein drugs may be necessary to accurately assess their efficacy and side effects. Quantitative measurements and comparative analyses are crucial to elucidate the advantages these therapeutics may offer. Proteins could potentially offer higher specificity than small molecules due to their larger interaction interfaces. However, the possibility of undesired off-target reactions and the long-term effects of such permanent modifications to targets necessitate thorough in vivo investigations. The effectiveness and the subsequent impact on target biology of these drugs are likely to vary with each target, requiring tailored exploration. The research community is keenly awaiting further studies that could establish general principles and provide guidance on the design and application of these covalent protein therapeutics. Moreover, several critical issues concerning covalent protein therapeutics remain insufficiently explored, including potential immunogenicity, idiosyncratic characteristics specific to individual therapeutics, in vivo stability, and biodistribution. These aspects must be rigorously assessed through comprehensive animal studies to ensure safety and functionality. Current applications of PREx have primarily focused on enhancing potency and selectivity. Expanding research into novel applications of covalent linkage197 could significantly broaden the scope and enhance the potential of PERx-based covalent protein drugs. This exploration is vital for pioneering new therapeutic strategies that leverage the unique properties of covalent interactions for clinical benefit.
The rapid advancements and expansion in the field suggest that biospecific chemistry, which shifts biomolecule interactions from noncovalent to covalent binding modes, is poised to significantly advance areas such as basic biological research, biotherapeutics, and synthetic biology. This paradigm shift is enhancing our understanding of and ability to harness biomolecular interactions fundamental to biology.
Acknowledgments
L.W. acknowledges support from NIH (R01GM118384 and R01CA258300).
Biographies
Li Cao received her Ph.D. degree from Nankai University in 2020. She is currently a postdoctoral researcher in the group of Prof. Lei Wang at the University of California San Francisco. Her research focuses on designing and genetically encoding bioreactive unnatural amino acids and utilizing PERx technology for the development of covalent protein therapeutics.
Lei Wang received his B.S. and M.S. degrees from Peking University and a Ph.D. from UC Berkeley (mentored by Peter G. Schultz). His graduate research resulted in the first expansion of the genetic code to incorporate unnatural amino acids (Uaas) in 2001, for which he was awarded the Young Scientist Award by the Science magazine. After postdoctoral training with Roger Y. Tsien, Wang established his research group at the Salk Institute for Biological Studies in 2005 and later transitioned to UCSF in 2014. His group has developed new methods for the expansion of the genetic code in a variety of cells and animals, discovered that release factor one is nonessential in E. coli, and engineered autonomous bacteria capable of incorporating Uaas at multiple sites. In recent years, Wang’s group conceptualized and implemented proximity-enabled bioreactivity. This methodology has opened new avenues for creating covalent linkages among biomolecules in vivo, advancing research and engineering in biological functions. Wang is a Top Young Innovator (by MIT Technology Review), a Beckman Young Investigator, a Searle Scholar, an NIH Director’s New Innovator Awardee, and a recipient of the Emil Thomas Kaiser Award.
Author Contributions
CRediT: Li Cao writing-original draft, writing-review & editing; Lei Wang conceptualization, funding acquisition, supervision, writing-original draft, writing-review & editing.
The authors declare the following competing financial interest(s): Lei Wang is a Scientific Advisor for Enlaza Therapeutics.
Special Issue
Published as part of Chemical Reviewsvirtual special issue “Noncanonical Amino Acids”.
References
- Welchman R. L.; Gordon C.; Mayer R. J. Ubiquitin and Ubiquitin-like Proteins as Multifunctional Signals. Nat. Rev. Mol. Cell Biol. 2005, 6, 599–609. 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Reily C.; Stewart T. J.; Renfrow M. B.; Novak J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier C. S.; Kaiser C. A. Formation and Transfer of Disulphide Bonds in Living Cells. Nat. Rev. Mol. Cell Biol. 2002, 3, 836–847. 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- Wensien M.; von Pappenheim F. R.; Funk L.-M.; Kloskowski P.; Curth U.; Diederichsen U.; Uranga J.; Ye J.; Fang P.; Pan K.-T.; et al. A Lysine-Cysteine Redox Switch with an NOS Bridge Regulates Enzyme Function. Nature 2021, 593, 460–464. 10.1038/s41586-021-03513-3. [DOI] [PubMed] [Google Scholar]
- Kang H. J.; Baker E. N. Intramolecular Isopeptide Bonds: Protein Crosslinks Built for Stress?. Trends Biochem. Sci. 2011, 36, 229–237. 10.1016/j.tibs.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Cao L.; Wang L. New Covalent Bonding Ability for Proteins. Protein Sci. 2022, 31, 312–322. 10.1002/pro.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed. 2009, 48, 6974–6998. 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem., Int. Ed. Engl. 2001, 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
- Baskin J. M.; Bertozzi C. R. Bioorthogonal Click Chemistry: Covalent Labeling in Living Systems. QSAR Comb. Sci. 2007, 26, 1211–1219. 10.1002/qsar.200740086. [DOI] [Google Scholar]
- Xiang Z.; Ren H.; Hu Y. S.; Coin I.; Wei J.; Cang H.; Wang L. Adding an Unnatural Covalent Bond to Proteins through Proximity-Enhanced Bioreactivity. Nat. Methods 2013, 10, 885–888. 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Genetically Encoding New Bioreactivity. New Biotechnol. 2017, 38, 16–25. 10.1016/j.nbt.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Brock A.; Herberich B.; Schultz P. G. Expanding the Genetic Code of Escherichia Coli. Science 2001, 292, 498–500. 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- Liu C. C.; Schultz P. G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Wang L.; Schultz P. Expanding the Genetic Code. Angew. Chem.-Int. Ed. 2005, 44, 34–66. 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- Wang L.; Xie J.; Schultz P. G. Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Coin I.; Katritch V.; Sun T.; Xiang Z.; Siu F. Y.; Beyermann M.; Stevens R. C.; Wang L. Genetically Encoded Chemical Probes in Cells Reveal the Binding Path of Urocortin-I to CRF Class B GPCR. Cell 2013, 155, 1258–1269. 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.; Tang S.; Ma C.; Li S.-T.; Shao G.-C.; Dang B.; DeGrado W. F.; Dong M.-Q.; Wang P. G.; Ding S.; et al. Spontaneous and Specific Chemical Cross-Linking in Live Cells to Capture and Identify Protein Interactions. Nat. Commun. 2017, 8, 2240. 10.1038/s41467-017-02409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Wang N.; Yu B.; Sun W.; Wang L. Genetically Encoded Chemical Crosslinking of Carbohydrate. Nat. Chem. 2023, 15, 33–42. 10.1038/s41557-022-01059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Wang N.; Liu H.; Yu B.; Jin L.; Ren X.; Shen Y.; Wang L. Genetically Encoded Chemical Crosslinking of RNA in Vivo. Nat. Chem. 2023, 15, 21–32. 10.1038/s41557-022-01038-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z.; Lacey V. K.; Ren H.; Xu J.; Burban D. J.; Jennings P. A.; Wang L. Proximity-Enabled Protein Crosslinking through Genetically Encoding Haloalkane Unnatural Amino Acids. Angew. Chem., Int. Ed. 2014, 53, 2190–2193. 10.1002/anie.201308794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C.; Lacey V. K.; Louie G. V.; Wei J.; Noel J. P.; Wang L. Genetically Encoding Photoswitchable Click Amino Acids in Escherichia Coli and Mammalian Cells. Angew. Chem., Int. Ed. 2014, 53, 3932–3936. 10.1002/anie.201400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T.; Hoppmann C.; Yang B.; Wang L. Using Protein-Confined Proximity To Determine Chemical Reactivity. J. Am. Chem. Soc. 2016, 138, 14832–14835. 10.1021/jacs.6b08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C.; Maslennikov I.; Choe S.; Wang L. In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J. Am. Chem. Soc. 2015, 137, 11218–11221. 10.1021/jacs.5b06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-H.; Xiang Z.; Hu Y. S.; Lacey V. K.; Cang H.; Wang L. Genetically Encoding an Electrophilic Amino Acid for Protein Stapling and Covalent Binding to Native Receptors. ACS Chem. Biol. 2014, 9, 1956–1961. 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigler M.; Müller T. G.; Horn-Ghetko D.; von Wrisberg M.-K.; Fottner M.; Goody R. S.; Itzen A.; Müller M. P.; Lang K. Proximity-Triggered Covalent Stabilization of Low-Affinity Protein Complexes In Vitro and In Vivo. Angew. Chem., Int. Ed. Engl. 2017, 56, 15737–15741. 10.1002/anie.201706927. [DOI] [PubMed] [Google Scholar]
- Shu X.; Asghar S.; Yang F.; Li S.-T.; Wu H.; Yang B. Uncover New Reactivity of Genetically Encoded Alkyl Bromide Non-Canonical Amino Acids. Front. Chem. 2022, 10, 815991. 10.3389/fchem.2022.815991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddick J. J.; Cheng J.; Roush W. R. Relative Rates of Michael Reactions of 2‘-(Phenethyl)Thiol with Vinyl Sulfones, Vinyl Sulfonate Esters, and Vinyl Sulfonamides Relevant to Vinyl Sulfonyl Cysteine Protease Inhibitors. Org. Lett. 2003, 5, 1967–1970. 10.1021/ol034555l. [DOI] [PubMed] [Google Scholar]
- Barf T.; Kaptein A. Irreversible Protein Kinase Inhibitors: Balancing the Benefits and Risks. J. Med. Chem. 2012, 55, 6243–6262. 10.1021/jm3003203. [DOI] [PubMed] [Google Scholar]
- Furman J. L.; Kang M.; Choi S.; Cao Y.; Wold E. D.; Sun S. B.; Smider V. V.; Schultz P. G.; Kim C. H. A Genetically Encoded Aza-Michael Acceptor for Covalent Cross-Linking of Protein-Receptor Complexes. J. Am. Chem. Soc. 2014, 136, 8411–8417. 10.1021/ja502851h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee H.; Debreczeni J.; Breed J.; Tentarelli S.; Aquila B.; Dowling J. E.; Whitty A.; Grimster N. P. A Study of the Reactivity of S(VI)-F Containing Warheads with Nucleophilic Amino-Acid Side Chains under Physiological Conditions. Org. Biomol. Chem. 2017, 15, 9685–9695. 10.1039/C7OB02028G. [DOI] [PubMed] [Google Scholar]
- Edman P.; Högfeldt E.; Sillén L. G.; Kinell P.-O. Method for Determination of the Amino Acid Sequence in Peptides. Acta Chem. Scand. 1950, 4, 283–293. 10.3891/acta.chem.scand.04-0283. [DOI] [Google Scholar]
- Xuan W.; Li J.; Luo X.; Schultz P. G. Genetic Incorporation of a Reactive Isothiocyanate Group into Proteins. Angew. Chem. 2016, 128, 10219–10222. 10.1002/ange.201604891. [DOI] [PubMed] [Google Scholar]
- Xuan W.; Shao S.; Schultz P. G. Protein Crosslinking by Genetically Encoded Noncanonical Amino Acids with Reactive Aryl Carbamate Side Chains. Angew. Chem., Int. Ed. Engl. 2017, 56, 5096–5100. 10.1002/anie.201611841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Rahim A.; Lin Q. Spontaneous Orthogonal Protein Crosslinking via a Genetically Encoded 2-Carboxy-4-Aryl-1,2,3-Triazole. Angew. Chem., Int. Ed. 2022, 61, e202202657 10.1002/anie.202202657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Krasnova L.; Finn M. G.; Sharpless K. B. Sulfur(VI) Fluoride Exchange (SuFEx): Another Good Reaction for Click Chemistry. Angew. Chem., Int. Ed. Engl. 2014, 53, 9430–9448. 10.1002/anie.201309399. [DOI] [PubMed] [Google Scholar]
- Homer J. A.; Xu L.; Kayambu N.; Zheng Q.; Choi E. J.; Kim B. M.; Sharpless K. B.; Zuilhof H.; Dong J.; Moses J. E. Sulfur Fluoride Exchange. Nat. Rev. Methods Primer 2023, 3, 58. 10.1038/s43586-023-00241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeyi O. O.; Hoth L. R.; Choi C.; Feng X.; Gopalsamy A.; Hett E. C.; Kyne R. E.; Robinson R. P.; Jones L. H. Covalent Enzyme Inhibition through Fluorosulfate Modification of a Noncatalytic Serine Residue. ACS Chem. Biol. 2017, 12, 2015–2020. 10.1021/acschembio.7b00403. [DOI] [PubMed] [Google Scholar]
- Chen W.; Dong J.; Plate L.; Mortenson D. E.; Brighty G. J.; Li S.; Liu Y.; Galmozzi A.; Lee P. S.; Hulce J. J.; et al. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc. 2016, 138, 7353–7364. 10.1021/jacs.6b02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortenson D. E.; Brighty G. J.; Plate L.; Bare G.; Chen W.; Li S.; Wang H.; Cravatt B. F.; Forli S.; Powers E. T.; et al. Inverse Drug Discovery” Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates. J. Am. Chem. Soc. 2018, 140, 200–210. 10.1021/jacs.7b08366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Yang B.; Fu C.; Zhu H.; Zheng F.; Kobayashi T.; Liu J.; Li S.; Ma C.; Wang P. G.; et al. Genetically Encoding Fluorosulfate-L-Tyrosine to React with Lysine, Histidine, and Tyrosine via SuFEx in Proteins in Vivo. J. Am. Chem. Soc. 2018, 140, 4995–4999. 10.1021/jacs.8b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.; Wang N.; Schnier P. D.; Zheng F.; Zhu H.; Polizzi N. F.; Ittuveetil A.; Saikam V.; DeGrado W. F.; Wang Q.; et al. Genetically Introducing Biochemically Reactive Amino Acids Dehydroalanine and Dehydrobutyrine in Proteins. J. Am. Chem. Soc. 2019, 141, 7698–7703. 10.1021/jacs.9b02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B.; Li S.; Tabata T.; Wang N.; Cao L.; Kumar G. R.; Sun W.; Liu J.; Ott M.; Wang L. Accelerating PERx Reaction Enables Covalent Nanobodies for Potent Neutralization of SARS-CoV-2 and Variants. Chem. 2022, 8, 2766–2783. 10.1016/j.chempr.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Cao L.; Klauser P. C.; Cheng R.; Berdan V. Y.; Sun W.; Wang N.; Ghelichkhani F.; Yu B.; Rozovsky S.; et al. A Genetically Encoded Fluorosulfonyloxybenzoyl- l -Lysine for Expansive Covalent Bonding of Proteins via SuFEx Chemistry. J. Am. Chem. Soc. 2021, 143, 10341–10351. 10.1021/jacs.1c04259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser P. C.; Berdan V. Y.; Cao L.; Wang L. Encoding Latent SuFEx Reactive Meta-Fluorosulfate Tyrosine to Expand Covalent Bonding of Proteins. Chem. Commun. 2022, 58, 6861–6864. 10.1039/D2CC01902G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Homer J. A.; Smedley C. J.; Cheng Q.-Q.; Sharpless K. B.; Moses J. E. Phosphorus Fluoride Exchange: Multidimensional Catalytic Click Chemistry from Phosphorus Connective Hubs. Chem. 2023, 9, 2128–2143. 10.1016/j.chempr.2023.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Yu B.; Li S.; Zhang P.; Li Q.; Wang L. Genetically Enabling Phosphorus Fluoride Exchange Click Chemistry in Proteins. Chem. 2024, 10, 1868–1884. 10.1016/j.chempr.2024.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M. M.; Olaoye O. O. Recent Advances in Chemical Biology Using Benzophenones and Diazirines as Radical Precursors. Molecules 2020, 25, 2285. 10.3390/molecules25102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N. D.; Parker R. B.; Kohler J. J. Photocrosslinking Approaches to Interactome Mapping. Curr. Opin. Chem. Biol. 2013, 17, 90–101. 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Yang B.; Wang L. Residue Selective Crosslinking of Proteins through Photoactivatable or Proximity-Enabled Reactivity. Curr. Opin. Chem. Biol. 2023, 74, 102285. 10.1016/j.cbpa.2023.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herner A.; Marjanovic J.; Lewandowski T. M.; Marin V.; Patterson M.; Miesbauer L.; Ready D.; Williams J.; Vasudevan A.; Lin Q. 2-Aryl-5-Carboxytetrazole as a New Photoaffinity Label for Drug Target Identification. J. Am. Chem. Soc. 2016, 138, 14609–14615. 10.1021/jacs.6b06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Jacinto M. P.; Zeng Y.; Yu Z.; Qu J.; Liu W. R.; Lin Q. Genetically Encoded 2-Aryl-5-Carboxytetrazoles for Site-Selective Protein Photo-Cross-Linking. J. Am. Chem. Soc. 2017, 139, 6078–6081. 10.1021/jacs.7b02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.; Yuan Y.; Wang C.-H.; Tian H.-T.; Guo A.-D.; Nie H.-J.; Hu H.; Tan M.; Tang Z.; Chen X.-H. Genetically Encoded Residue-Selective Photo-Crosslinker to Capture Protein-Protein Interactions in Living Cells. Chem. 2019, 5, 2955–2968. 10.1016/j.chempr.2019.08.020. [DOI] [Google Scholar]
- Liu J.; Li S.; Aslam N. A.; Zheng F.; Yang B.; Cheng R.; Wang N.; Rozovsky S.; Wang P. G.; Wang Q.; et al. Genetically Encoding Photocaged Quinone Methide to Multitarget Protein Residues Covalently in Vivo. J. Am. Chem. Soc. 2019, 141, 9458–9462. 10.1021/jacs.9b01738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Cheng R.; Van Eps N.; Wang N.; Morizumi T.; Ou W.-L.; Klauser P. C.; Rozovsky S.; Ernst O. P.; Wang L. Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents. J. Am. Chem. Soc. 2020, 142, 17057–17068. 10.1021/jacs.0c06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Lv L.; Tang S.; Zang Y.; Wan T.; Wang D.; Cai L.; Ye H.; Tan R.; Wang N. Oxidation-Induced Protein Cross-Linking in Mammalian Cells. ACS Synth. Biol. 2023, 12, 984–992. 10.1021/acssynbio.3c00052. [DOI] [PubMed] [Google Scholar]
- Shang X.; Chen Y.; Wang N.; Niu W.; Guo J. Oxidation-Induced Generation of a Mild Electrophile for Proximity-Enhanced Protein-Protein Crosslinking. Chem. Commun. 2018, 54, 4172–4175. 10.1039/C8CC01639A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B.; Cao L.; Li S.; Klauser P.; Wang L. Proximity-Enabled Sulfur Fluoride Exchange Reaction in Protein Context. Chem. Sci. 2023, 14, 7913–7921. 10.1039/D3SC01921G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S.; Hafner M.; Tuschl T. A Census of Human RNA-Binding Proteins. Nat. Rev. Genet. 2014, 15, 829–845. 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbacher J. K.; Batra R.; Lagier-Tourenne C.; Yeo G. W. RNA-Binding Proteins in Neurodegeneration: Seq and You Shall Receive. Trends Neurosci. 2015, 38, 226–236. 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A.; Fischer B.; Hentze M. W.; Preiss T. RNA-Binding Proteins in Mendelian Disease. Trends Genet. 2013, 29, 318–327. 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Hentze M. W.; Castello A.; Schwarzl T.; Preiss T. A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Ramanathan M.; Porter D. F.; Khavari P. A. Methods to Study RNA-Protein Interactions. Nat. Methods 2019, 16, 225–234. 10.1038/s41592-019-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenmakers A. J.; Reinders R. J.; van Venrooij W. J. Cross-Linking of mRNA to Proteins by Irradiation of Intact Cells with Ultraviolet Light. Eur. J. Biochem. 1980, 112, 323–330. 10.1111/j.1432-1033.1980.tb07207.x. [DOI] [PubMed] [Google Scholar]
- Lee F. C. Y.; Ule J. Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol. Cell 2018, 69, 354–369. 10.1016/j.molcel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y.; Koenig J.; Hussain S.; Zupan B.; Curk T.; Frye M.; Ule J. Analysis of CLIP and iCLIP Methods for Nucleotide-Resolution Studies of Protein-RNA Interactions. Genome Biol. 2012, 13, R67. 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I.; Matsuura T. Chemical Aspects of UV-Induced Crosslinking of Proteins to Nucleic Acids. Photoreactions with Lysine and Tryptophan. Acc. Chem. Res. 1985, 18, 134–141. 10.1021/ar00113a002. [DOI] [Google Scholar]
- Holmqvist E.; Wright P. R.; Li L.; Bischler T.; Barquist L.; Reinhardt R.; Backofen R.; Vogel J. Global RNA Recognition Patterns of Post-Transcriptional Regulators Hfq and CsrA Revealed by UV Crosslinking in Vivo. EMBO J. 2016, 35, 991–1011. 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S. R.; Ju T.; Cummings R. D. Protein Glycosylation in Cancer. Annu. Rev. Pathol. 2015, 10, 473–510. 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro R. G. Protein Glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Glycopeptide Bonds. Glycobiology 2002, 12, 43R–56R. 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- Sears P.; Wong C. Carbohydrate Mimetics: A New Strategy for Tackling the Problem of Carbohydrate-Mediated Biological Recognition. Angew. Chem., Int. Ed. Engl. 1999, 38, 2300–2324. . [DOI] [PubMed] [Google Scholar]
- Polonskaya Z.; Savage P. B.; Finn M. G.; Teyton L. High-Affinity Anti-Glycan Antibodies: Challenges and Strategies. Curr. Opin. Immunol. 2019, 59, 65–71. 10.1016/j.coi.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.; Wu H.; Schnier P. D.; Liu Y.; Liu J.; Wang N.; DeGrado W. F.; Wang L. Proximity-Enhanced SuFEx Chemical Cross-Linker for Specific and Multitargeting Cross-Linking Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 11162–11167. 10.1073/pnas.1813574115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Chen S.; Zhang W.-D.; Hu H.-G. Stapled Helical Peptides Bearing Different Anchoring Residues. Chem. Rev. 2020, 120, 10079–10144. 10.1021/acs.chemrev.0c00532. [DOI] [PubMed] [Google Scholar]
- Walensky L. D.; Bird G. H. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress: Miniperspective. J. Med. Chem. 2014, 57, 6275–6288. 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.; Davidson M.; Manley S.; Lippincott-Schwartz J. Superresolution Imaging Using Single-Molecule Localization. Annu. Rev. Phys. Chem. 2010, 61, 345–367. 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. E.; Larson D. R.; Webb W. W. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophys. J. 2002, 82, 2775–2783. 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H.; Yang B.; Ma C.; Hu Y. S.; Wang P. G.; Wang L. Cysteine Sulfoxidation Increases the Photostability of Red Fluorescent Proteins. ACS Chem. Biol. 2016, 11, 2679–2684. 10.1021/acschembio.6b00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 1998, 67, 509–544. 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Wang L.; Jackson W.; Steinbach P.; Tsien R. Evolution of New Nonantibody Proteins via Iterative Somatic Hypermutation. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16745–16749. 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wart H. E.; Lewis A.; Scheraga H. A.; Saeva F. D. Disulfide Bond Dihedral Angles from Raman Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 1973, 70, 2619–2623. 10.1073/pnas.70.9.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombkowski A. A.; Sultana K. Z.; Craig D. B. Protein Disulfide Engineering. FEBS Lett. 2014, 588, 206–212. 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Liu T.; Wang Y.; Luo X.; Li J.; Reed S. A.; Xiao H.; Young T. S.; Schultz P. G. Enhancing Protein Stability with Extended Disulfide Bonds. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 5910–5915. 10.1073/pnas.1605363113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. C.; Liu T.; Wang Y.; Mehta A. P.; Schultz P. G. Enhancing Protein Stability with Genetically Encoded Noncanonical Amino Acids. J. Am. Chem. Soc. 2018, 140, 15997–16000. 10.1021/jacs.8b07157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. C.; Nastertorabi F.; Xuan W.; Han G. W.; Stevens R. C.; Schultz P. G. A Single Reactive Noncanonical Amino Acid Is Able to Dramatically Stabilize Protein Structure. ACS Chem. Biol. 2019, 14, 1150–1153. 10.1021/acschembio.9b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.-Y.; Kawaguchi D.; Coin I.; Xiang Z.; O’Leary D. D. M.; Slesinger P. A.; Wang L. In Vivo Expression of a Light-Activatable Potassium Channel Using Unnatural Amino Acids. Neuron 2013, 80, 358–370. 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L.; Yizhar O.; Deisseroth K. The Development and Application of Optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szobota S.; Isacoff E. Y. Optical Control of Neuronal Activity. Annu. Rev. Biophys. 2010, 39, 329–348. 10.1146/annurev.biophys.093008.131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A.; Weiner O. D.; Lim W. A.; Voigt C. A. Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 2009, 461, 997–1001. 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. Z.; Lin M. Z. Optical Control of Biological Processes by Light-Switchable Proteins. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 545–554. 10.1002/wdev.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M.; DiMaio F.; Anishchenko I.; Dauparas J.; Ovchinnikov S.; Lee G. R.; Wang J.; Cong Q.; Kinch L. N.; Schaeffer R. D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin I.; Perrin M. H.; Vale W. W.; Wang L. Photo-Cross-Linkers Incorporated into G-Protein-Coupled Receptors in Mammalian Cells: A Ligand Comparison. Angew. Chem., Int. Ed. 2011, 50, 8077–8081. 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högbom M.; Eklund M.; Nygren P.-Å.; Nordlund P. Structural Basis for Recognition by an in Vitro Evolved Affibody. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 3191–3196. 10.1073/pnas.0436100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Holmgren A. The Thioredoxin Antioxidant System. Free Radic. Biol. Med. 2014, 66, 75–87. 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- Seidel L.; Zarzycka B.; Zaidi S. A.; Katritch V.; Coin I. Structural Insight into the Activation of a Class B G-Protein-Coupled Receptor by Peptide Hormones in Live Human Cells. eLife 2017, 6, e27711 10.7554/eLife.27711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttke T.; Ernicke S.; Serfling R.; Ihling C.; Burda E.; Gurevich V. V.; Sinz A.; Coin I. Exploring GPCR-arrestin Interfaces with Genetically Encoded Crosslinkers. EMBO Rep. 2020, 21, e50437 10.15252/embr.202050437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin Y.; Böttke T.; Lam J. H.; Ernicke S.; Fortmann A.; Tretbar M.; Zarzycka B.; Gurevich V. V.; Katritch V.; Coin I. Structural Details of a Class B GPCR-Arrestin Complex Revealed by Genetically Encoded Crosslinkers in Living Cells. Nat. Commun. 2023, 14, 1151. 10.1038/s41467-023-36797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Guo S.; Chai F.; Sun Q.; Li P.; Gao L.; Dai L.; Ouyang X.; Zhou Z.; Zhou L.; et al. Genetically Incorporated Crosslinkers Reveal NleE Attenuates Host Autophagy Dependent on PSMD10. eLife 2021, 10, e69047 10.7554/eLife.69047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.; Wang X.; Li P.; Huang Q.; Zhou L.; Li J.; Gao L.; Sun Q.; Chai F.; Guo S.; et al. Bacterial Effector Restricts Liquid-Liquid Phase Separation of ZPR1 to Antagonize Host UPRER. Cell Rep. 2023, 42, 112700. 10.1016/j.celrep.2023.112700. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Chai F.; He Y.; Zhou Z.; Guo S.; Li P.; Sun Q.; Zu X.; Liu X.; Huang Q.; et al. Homodimerized Cytoplasmic Domain of PD-L1 Regulates Its Complex Glycosylation in Living Cells. Commun. Biol. 2022, 5, 887. 10.1038/s42003-022-03845-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.; Dai Z.; Qin X.; Cai W.; Hu L.; Huang Y.; Cao W.; Yang F.; Wang C.; Liu T. Proteomic Identification of Protein Tyrosine Phosphatase and Substrate Interactions in Living Mammalian Cells by Genetic Encoding of Irreversible Enzyme Inhibitors. J. Am. Chem. Soc. 2018, 140, 13253–13259. 10.1021/jacs.8b06922. [DOI] [PubMed] [Google Scholar]
- Shu X.; Liao Q.-Q.; Li S.-T.; Liu L.; Zhang X.; Zhou L.; Zhang L.; Coin I.; Wang L.; Wu H.; et al. Detecting Active Deconjugating Enzymes with Genetically Encoded Activity-Based Ubiquitin and Ubiquitin-like Protein Probes. Anal. Chem. 2023, 95, 846–853. 10.1021/acs.analchem.2c03270. [DOI] [PubMed] [Google Scholar]
- Liu C.; Wu T.; Shu X.; Li S.; Wang D. R.; Wang N.; Zhou R.; Yang H.; Jiang H.; Hendriks I. A.; et al. Identification of Protein Direct Interactome with Genetic Code Expansion and Search Engine OpenUaa. Adv. Biol. 2021, 5, 2000308. 10.1002/adbi.202000308. [DOI] [PubMed] [Google Scholar]
- Nachtergaele S.; He C. Chemical Modifications in the Life of an mRNA Transcript. Annu. Rev. Genet. 2018, 52, 349–372. 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B.; Grozhik A. V.; Olarerin-George A. O.; Meydan C.; Mason C. E.; Jaffrey S. R. Single-Nucleotide-Resolution Mapping of m6A and m6Am throughout the Transcriptome. Nat. Methods 2015, 12, 767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D.; Saletore Y.; Zumbo P.; Elemento O.; Mason C. E.; Jaffrey S. R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; et al. Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 2012, 485, 201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Hu L.; Liu S.; Peng Y.; Ge R.; Su R.; Senevirathne C.; Harada B. T.; Dai Q.; Wei J.; Zhang L.; et al. m6A RNA Modifications Are Measured at Single-Base Resolution across the Mammalian Transcriptome. Nat. Biotechnol. 2022, 40, 1210–1219. 10.1038/s41587-022-01243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.; Liu K.; Ahmed H.; Loppnau P.; Schapira M.; Min J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J. Biol. Chem. 2015, 290, 24902–24913. 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Chen K.; Song B.; Ma J.; Wu X.; Xu Q.; Wei Z.; Su J.; Liu G.; Rong R.; et al. m6A-Atlas: A Comprehensive Knowledgebase for Unraveling the N 6-Methyladenosine (m6A) Epitranscriptome. Nucleic Acids Res. 2021, 49, D134–D143. 10.1093/nar/gkaa692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F.; Li B. T.; Dy G. K.; Price T. J.; Falchook G. S.; Wolf J.; Italiano A.; Schuler M.; Borghaei H.; Barlesi F.; et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R. A. Covalent Inhibitors in Drug Discovery: From Accidental Discoveries to Avoided Liabilities and Designed Therapies. Drug Discov Today 2015, 20, 1061–1073. 10.1016/j.drudis.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Baillie T. A. Targeted Covalent Inhibitors for Drug Design. Angew. Chem., Int. Ed. 2016, 55, 13408–13421. 10.1002/anie.201601091. [DOI] [PubMed] [Google Scholar]
- Singh J. The Ascension of Targeted Covalent Inhibitors. J. Med. Chem. 2022, 65, 5886–5901. 10.1021/acs.jmedchem.1c02134. [DOI] [PubMed] [Google Scholar]
- Ostrem J. M.; Peters U.; Sos M. L.; Wells J. A.; Shokat K. M. K-Ras(G12C) Inhibitors Allosterically Control GTP Affinity and Effector Interactions. Nature 2013, 503, 548–551. 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A. The Drug-Target Residence Time Model: A 10-Year Retrospective. Nat. Rev. Drug Discovery 2016, 15, 87–95. 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- Smith A. J. T.; Zhang X.; Leach A. G.; Houk K. N. Beyond Picomolar Affinities: Quantitative Aspects of Noncovalent and Covalent Binding of Drugs to Proteins. J. Med. Chem. 2009, 52, 225–233. 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J.; Petter R. C.; Baillie T. A.; Whitty A. The Resurgence of Covalent Drugs. Nat. Rev. Drug Discovery 2011, 10, 307–317. 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Gehringer M.; Laufer S. A. Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology. J. Med. Chem. 2019, 62, 5673–5724. 10.1021/acs.jmedchem.8b01153. [DOI] [PubMed] [Google Scholar]
- Jones S.; Thornton J. M. Principles of Protein-Protein Interactions. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13–20. 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milroy L.-G.; Grossmann T. N.; Hennig S.; Brunsveld L.; Ottmann C. Modulators of Protein-Protein Interactions. Chem. Rev. 2014, 114, 4695–4748. 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- Vinogradov A. A.; Yin Y.; Suga H. Macrocyclic Peptides as Drug Candidates: Recent Progress and Remaining Challenges. J. Am. Chem. Soc. 2019, 141, 4167–4181. 10.1021/jacs.8b13178. [DOI] [PubMed] [Google Scholar]
- Craik D. J.; Fairlie D. P.; Liras S.; Price D. The Future of Peptide-based Drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- Leader B.; Baca Q. J.; Golan D. E. Protein Therapeutics: A Summary and Pharmacological Classification. Nat. Rev. Drug Discovery 2008, 7, 21–39. 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- Berdan V. Y.; Klauser P. C.; Wang L. Covalent Peptides and Proteins for Therapeutics. Bioorg. Med. Chem. 2021, 29, 115896. 10.1016/j.bmc.2020.115896. [DOI] [PubMed] [Google Scholar]
- Bhardwaj G.; Mulligan V. K.; Bahl C. D.; Gilmore J. M.; Harvey P. J.; Cheneval O.; Buchko G. W.; Pulavarti S. V. S. R. K.; Kaas Q.; Eletsky A.; et al. Accurate de Novo Design of Hyperstable Constrained Peptides. Nature 2016, 538, 329–335. 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm P. M.; Spiegel J.; Grossmann T. N. Hydrocarbon Stapled Peptides as Modulators of Biological Function. ACS Chem. Biol. 2015, 10, 1362–1375. 10.1021/cb501020r. [DOI] [PubMed] [Google Scholar]
- Braun C. R.; Mintseris J.; Gavathiotis E.; Bird G. H.; Gygi S. P.; Walensky L. D. Photoreactive Stapled BH3 Peptides to Dissect the BCL-2 Family Interactome. Chem. Biol. 2010, 17, 1325–1333. 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins J. L.; Santelli E.; Feng Y.; De S. K.; Purves A.; Motamedchaboki K.; Wu B.; Ronai Z. A.; Liddington R. C.; Pellecchia M. Structure-Based Design of Covalent Siah Inhibitors. Chem. Biol. 2013, 20, 973–982. 10.1016/j.chembiol.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo A. D.; Lim J.; Good A. C.; Skerlj R. T.; Fairlie D. P. Electrophilic Helical Peptides That Bond Covalently, Irreversibly, and Selectively in a Protein-Protein Interaction Site. ACS Med. Chem. Lett. 2017, 8, 22–26. 10.1021/acsmedchemlett.6b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C.; Wang L. Proximity-Enabled Bioreactivity to Generate Covalent Peptide Inhibitors of P53-Mdm4. Chem. Commun. 2016, 52, 5140–5143. 10.1039/C6CC01226D. [DOI] [PubMed] [Google Scholar]
- Bernal F.; Tyler A. F.; Korsmeyer S. J.; Walensky L. D.; Verdine G. L. Reactivation of the P53 Tumor Suppressor Pathway by a Stapled P53 Peptide. J. Am. Chem. Soc. 2007, 129, 2456–2457. 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenpattarapreeda J.; Tan Y. S.; Iegre J.; Walsh S. J.; Fowler E.; Eapen R. S.; Wu Y.; Sore H. F.; Verma C. S.; Itzhaki L.; et al. Targeted Covalent Inhibitors of MDM2 Using Electrophile-Bearing Stapled Peptides. Chem. Commun. Camb 2019, 55, 7914–7917. 10.1039/C9CC04022F. [DOI] [PubMed] [Google Scholar]
- Baggio C.; Gambini L.; Udompholkul P.; Salem A. F.; Aronson A.; Dona A.; Troadec E.; Pichiorri F.; Pellecchia M. Design of Potent Pan-IAP and Lys-Covalent XIAP Selective Inhibitors Using a Thermodynamics Driven Approach. J. Med. Chem. 2018, 61, 6350–6363. 10.1021/acs.jmedchem.8b00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambini L.; Udompholkul P.; Salem A. F.; Baggio C.; Pellecchia M. Stability and Cell Permeability of Sulfonyl Fluorides in the Design of Lys-Covalent Antagonists of Protein-Protein Interactions. ChemMedChem. 2020, 15, 2176–2184. 10.1002/cmdc.202000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambini L.; Baggio C.; Udompholkul P.; Jossart J.; Salem A. F.; Perry J. J. P.; Pellecchia M. Covalent Inhibitors of Protein-Protein Interactions Targeting Lysine, Tyrosine, or Histidine Residues. J. Med. Chem. 2019, 62, 5616–5627. 10.1021/acs.jmedchem.9b00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udompholkul P.; Baggio C.; Gambini L.; Alboreggia G.; Pellecchia M. Lysine Covalent Antagonists of Melanoma Inhibitors of Apoptosis Protein. J. Med. Chem. 2021, 64, 16147–16158. 10.1021/acs.jmedchem.1c01459. [DOI] [PubMed] [Google Scholar]
- Yoo D. Y.; Hauser A. D.; Joy S. T.; Bar-Sagi D.; Arora P. S. Covalent Targeting of Ras G12C by Rationally Designed Peptidomimetics. ACS Chem. Biol. 2020, 15, 1604–1612. 10.1021/acschembio.0c00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen F. M.; Schouten G. K.; Moertl C.; Verheul J.; Hoekstra I.; Koningstein G. M.; Hutchins G. H.; Alkir A.; Luirink R. A.; Geerke D. P.; et al. Covalent Proteomimetic Inhibitor of the Bacterial FtsQB Divisome Complex. J. Am. Chem. Soc. 2022, 144, 15303–15313. 10.1021/jacs.2c06304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Yu M.; Liu N.; Lian C.; Hou Z.; Wang R.; Zhao R.; Li W.; Jiang Y.; Shi X.; et al. A Sulfonium Tethered Peptide Ligand Rapidly and Selectively Modifies Protein Cysteine in Vicinity. Chem. Sci. 2019, 10, 4966–4972. 10.1039/C9SC00034H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.; Wang D.; Lian C.; Zhao R.; Tu L.; Zhang Y.; Liu J.; Zhu H.; Yu M.; Wan C.; et al. Selective Covalent Targeting of Anti-apoptotic BFL-1 by a Sulfonium-Tethered Peptide. ChemBioChem. 2021, 22, 340–344. 10.1002/cbic.202000473. [DOI] [PubMed] [Google Scholar]
- Chen S.; Lovell S.; Lee S.; Fellner M.; Mace P. D.; Bogyo M. Identification of Highly Selective Covalent Inhibitors by Phage Display. Nat. Biotechnol. 2021, 39, 490–498. 10.1038/s41587-020-0733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Chen F.-J.; Li K.; Reja R. M.; Haeffner F.; Gao J. Lysine-Targeted Reversible Covalent Ligand Discovery for Proteins via Phage Display. J. Am. Chem. Soc. 2022, 144, 15885–15893. 10.1021/jacs.2c07375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reja R. M.; Wang W.; Lyu Y.; Haeffner F.; Gao J. Lysine-Targeting Reversible Covalent Inhibitors with Long Residence Time. J. Am. Chem. Soc. 2022, 144, 1152–1157. 10.1021/jacs.1c12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontet C.; Reichert J. M.; Senter P. D.; Lambert J. M.; Beck A. Antibody-Drug Conjugates Come of Age in Oncology. Nat. Rev. Drug Discovery 2023, 22, 641–661. 10.1038/s41573-023-00709-2. [DOI] [PubMed] [Google Scholar]
- Agarwal P.; Bertozzi C. R. Site-Specific Antibody-Drug Conjugates: The Nexus of Bioorthogonal Chemistry, Protein Engineering, and Drug Development. Bioconjugate Chem. 2015, 26, 176–192. 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline T.; Steiner A. R.; Penta K.; Sato A. K.; Hallam T. J.; Yin G. Methods to Make Homogenous Antibody Drug Conjugates. Pharm. Res. 2015, 32, 3480–3493. 10.1007/s11095-014-1596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H.; Axup J. Y.; Schultz P. G. Protein Conjugation with Genetically Encoded Unnatural Amino Acids. Curr. Opin. Chem. Biol. 2013, 17, 412–419. 10.1016/j.cbpa.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Lee Y.; Ko B. J.; Yoo T. H. Peptide-Directed Photo-Cross-Linking for Site-Specific Conjugation of IgG. Bioconjugate Chem. 2018, 29, 3240–3244. 10.1021/acs.bioconjchem.8b00515. [DOI] [PubMed] [Google Scholar]
- Konrad A.; Eriksson Karlström A.; Hober S. Covalent Immunoglobulin Labeling through a Photoactivable Synthetic Z Domain. Bioconjugate Chem. 2011, 22, 2395–2403. 10.1021/bc200052h. [DOI] [PubMed] [Google Scholar]
- Yu C.; Tang J.; Loredo A.; Chen Y.; Jung S. Y.; Jain A.; Gordon A.; Xiao H. Proximity-Induced Site-Specific Antibody Conjugation. Bioconjugate Chem. 2018, 29, 3522–3526. 10.1021/acs.bioconjchem.8b00680. [DOI] [PubMed] [Google Scholar]
- Cao Y. J.; Yu C.; Wu K.-L.; Wang X.; Liu D.; Tian Z.; Zhao L.; Qi X.; Loredo A.; Chung A.; et al. Synthesis of Precision Antibody Conjugates Using Proximity-Induced Chemistry. Theranostics 2021, 11, 9107–9117. 10.7150/thno.62444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z.; Wu L.; Yu C.; Chen Y.; Xu Z.; Bado I.; Loredo A.; Wang L.; Wang H.; Wu K.-L.; et al. Harnessing the Power of Antibodies to Fight Bone Metastasis. Sci. Adv. 2021, 7, eabf2051 10.1126/sciadv.abf2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto S.; Nakashimada Y.; Yokota R.; Hatanaka T.; Adachi M.; Ito Y. Site-Specific Chemical Conjugation of Antibodies by Using Affinity Peptide for the Development of Therapeutic Antibody Format. Bioconjugate Chem. 2019, 30, 698–702. 10.1021/acs.bioconjchem.8b00865. [DOI] [PubMed] [Google Scholar]
- Yamada K.; Shikida N.; Shimbo K.; Ito Y.; Khedri Z.; Matsuda Y.; Mendelsohn B. A. AJICAP: Affinity Peptide Mediated Regiodivergent Functionalization of Native Antibodies. Angew. Chem., Int. Ed. 2019, 58, 5592–5597. 10.1002/anie.201814215. [DOI] [PubMed] [Google Scholar]
- Ohata J.; Ball Z. T. A Hexa-Rhodium Metallopeptide Catalyst for Site-Specific Functionalization of Natural Antibodies. J. Am. Chem. Soc. 2017, 139, 12617–12622. 10.1021/jacs.7b06428. [DOI] [PubMed] [Google Scholar]
- Yuan D.; Zhang Y.; Lim K. H.; Leung S. K. P.; Yang X.; Liang Y.; Lau W. C. Y.; Chow K. T.; Xia J. Site-Selective Lysine Acetylation of Human Immunoglobulin G for Immunoliposomes and Bispecific Antibody Complexes. J. Am. Chem. Soc. 2022, 144, 18494–18503. 10.1021/jacs.2c07594. [DOI] [PubMed] [Google Scholar]
- Lonsdale R.; Ward R. A. Structure-Based Design of Targeted Covalent Inhibitors. Chem. Soc. Rev. 2018, 47, 3816–3830. 10.1039/C7CS00220C. [DOI] [PubMed] [Google Scholar]
- Johnson D. S.; Weerapana E.; Cravatt B. F. Strategies for Discovering and Derisking Covalent, Irreversible Enzyme Inhibitors. Future Med. Chem. 2010, 2, 949–964. 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Chen Q.; Klauser P. C.; Li M.; Zheng F.; Wang N.; Li X.; Zhang Q.; Fu X.; Wang Q.; et al. Developing Covalent Protein Drugs via Proximity-Enabled Reactive Therapeutics. Cell 2020, 182, 85–97. 10.1016/j.cell.2020.05.028. [DOI] [PubMed] [Google Scholar]
- Sharpe A. H.; Pauken K. E. The Diverse Functions of the PD1 Inhibitory Pathway. Nat. Rev. Immunol. 2018, 18, 153–167. 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- Sanmamed M. F.; Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 2018, 175, 313–326. 10.1016/j.cell.2018.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak K. M.; Kitel R.; Przetocka S.; Golik P.; Guzik K.; Musielak B.; Dömling A.; Dubin G.; Holak T. A. Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure 2015, 23, 2341–2348. 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. B.; Farzan M.; Chen B.; Choe H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M.; Faust B.; Saunders R. A.; Sangwan S.; Rezelj V.; Hoppe N.; Boone M.; Billesbølle C. B.; Puchades C.; Azumaya C. M.; et al. An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike. Science 2020, 370, 1473–1479. 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Ren Y.; Aw Z. Q.; Chen B.; Yang Z.; Lei Y.; Cheng L.; Liang Q.; Hong J.; Yang Y.; et al. Broadly Neutralizing and Protective Nanobodies against SARS-CoV-2 Omicron Subvariants BA.1, BA.2, and BA.4/5 and Diverse Sarbecoviruses. Nat. Commun. 2022, 13, 7957. 10.1038/s41467-022-35642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Yang Z.; Hu H.; Zhang H.; Chen L.; Li K.; Kong L.; Wang Q.; Liu B.; Wang M.; et al. Covalently Engineered Protein Minibinders with Enhanced Neutralization Efficacy against Escaping SARS-CoV-2 Variants. J. Am. Chem. Soc. 2022, 144, 5702–5707. 10.1021/jacs.1c11554. [DOI] [PubMed] [Google Scholar]
- Cao L.; Goreshnik I.; Coventry B.; Case J. B.; Miller L.; Kozodoy L.; Chen R. E.; Carter L.; Walls A. C.; Park Y.-J.; et al. De Novo Design of Picomolar SARS-CoV-2 miniprotein Inhibitors. Science 2020, 370, 426–431. 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pance K.; Gramespacher J. A.; Byrnes J. R.; Salangsang F.; Serrano J.-A. C.; Cotton A. D.; Steri V.; Wells J. A. Modular Cytokine Receptor-Targeting Chimeras for Targeted Degradation of Cell Surface and Extracellular Proteins. Nat. Biotechnol. 2023, 41, 273–281. 10.1038/s41587-022-01456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik S. M.; Pedram K.; Wisnovsky S.; Ahn G.; Riley N. M.; Bertozzi C. R. Lysosome-Targeting Chimaeras for Degradation of Extracellular Proteins. Nature 2020, 584, 291–297. 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Teng P.; Montgomery N. T.; Li X.; Tang W. Development of Triantennary N-Acetylgalactosamine Conjugates as Degraders for Extracellular Proteins. ACS Cent. Sci. 2021, 7, 499–506. 10.1021/acscentsci.1c00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton A. D.; Nguyen D. P.; Gramespacher J. A.; Seiple I. B.; Wells J. A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Crews C. M. PROTACs: Past, Present and Future. Chem. Soc. Rev. 2022, 51, 5214–5236. 10.1039/D2CS00193D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Han Y.; Yang Y.; Lin F.; Li K.; Kong L.; Liu H.; Dang Y.; Lin J.; Chen P. R. Covalently Engineered Nanobody Chimeras for Targeted Membrane Protein Degradation. J. Am. Chem. Soc. 2021, 143, 16377–16382. 10.1021/jacs.1c08521. [DOI] [PubMed] [Google Scholar]
- Verdurmen W. P. R.; Mazlami M.; Plückthun A. A Quantitative Comparison of Cytosolic Delivery via Different Protein Uptake Systems. Sci. Rep. 2017, 7, 13194. 10.1038/s41598-017-13469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Da Y.; Yu M.; Cheng Y.; Wang X.; Xiong J.; Guo G.; Li Y.; Jiang X.; Cai X. Protein Labeling Approach to Improve Lysosomal Targeting and Efficacy of Antibody-Drug Conjugates. Org. Biomol. Chem. 2020, 18, 3229–3233. 10.1039/D0OB00265H. [DOI] [PubMed] [Google Scholar]
- Boyman O.; Sprent J. The Role of Interleukin-2 during Homeostasis and Activation of the Immune System. Nat. Rev. Immunol. 2012, 12, 180–190. 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- Yuan Y.; Kolios A. G. A.; Liu Y.; Zhang B.; Li H.; Tsokos G. C.; Zhang X. Therapeutic Potential of Interleukin-2 in Autoimmune Diseases. Trends Mol. Med. 2022, 28, 596–612. 10.1016/j.molmed.2022.04.010. [DOI] [PubMed] [Google Scholar]
- Klatzmann D.; Abbas A. K. The Promise of Low-Dose Interleukin-2 Therapy for Autoimmune and Inflammatory Diseases. Nat. Rev. Immunol. 2015, 15, 283–294. 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- Charych D. H.; Hoch U.; Langowski J. L.; Lee S. R.; Addepalli M. K.; Kirk P. B.; Sheng D.; Liu X.; Sims P. W.; VanderVeen L. A.; et al. NKTR-214, an Engineered Cytokine with Biased IL2 Receptor Binding, Increased Tumor Exposure, and Marked Efficacy in Mouse Tumor Models. Clin. Cancer Res. 2016, 22, 680–690. 10.1158/1078-0432.CCR-15-1631. [DOI] [PubMed] [Google Scholar]
- Karakus U.; Sahin D.; Mittl P. R. E.; Mooij P.; Koopman G.; Boyman O. Receptor-Gated IL-2 Delivery by an Anti-Human IL-2 Antibody Activates Regulatory T Cells in Three Different Species. Sci. Transl. Med. 2020, 12, eabb9283 10.1126/scitranslmed.abb9283. [DOI] [PubMed] [Google Scholar]
- Hernandez R.; Põder J.; LaPorte K. M.; Malek T. R. Engineering IL-2 for Immunotherapy of Autoimmunity and Cancer. Nat. Rev. Immunol. 2022, 22, 614–628. 10.1038/s41577-022-00680-w. [DOI] [PubMed] [Google Scholar]
- Rao B. M.; Driver I.; Lauffenburger D. A.; Wittrup K. D. High-Affinity CD25-Binding IL-2 Mutants Potently Stimulate Persistent T Cell Growth. Biochemistry 2005, 44, 10696–10701. 10.1021/bi050436x. [DOI] [PubMed] [Google Scholar]
- Levin A. M.; Bates D. L.; Ring A. M.; Krieg C.; Lin J. T.; Su L.; Moraga I.; Raeber M. E.; Bowman G. R.; Novick P.; et al. Exploiting a Natural Conformational Switch to Engineer an Interleukin-2 ‘Superkine.’. Nature 2012, 484, 529–533. 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Sun J.; Yuan Y.; Ji D.; Sun Y.; Liu Y.; Li S.; Zhu X.; Wu X.; Hu J.; et al. Proximity-Enabled Covalent Binding of IL-2 to IL-2Rα Selectively Activates Regulatory T Cells and Suppresses Autoimmunity. Sig. Transduct. Target. Ther. 2023, 8, 28. 10.1038/s41392-022-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodei L.; Herrmann K.; Schöder H.; Scott A. M.; Lewis J. S. Radiotheranostics in Oncology: Current Challenges and Emerging Opportunities. Nat. Rev. Clin. Oncol. 2022, 19, 534–550. 10.1038/s41571-022-00652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouros G.; Bodei L.; McDevitt M. R.; Nedrow J. R. Radiopharmaceutical Therapy in Cancer: Clinical Advances and Challenges. Nat. Rev. Drug Discovery 2020, 19, 589–608. 10.1038/s41573-020-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon A.; Degoul F. Antibody Pretargeting Based on Bioorthogonal Click Chemistry for Cancer Imaging and Targeted Radionuclide Therapy. Bioconjugate Chem. 2020, 31, 159–173. 10.1021/acs.bioconjchem.9b00761. [DOI] [PubMed] [Google Scholar]
- Kelly J. M.; Amor-Coarasa A.; Ponnala S.; Nikolopoulou A.; Williams C.; DiMagno S. G.; Babich J. W. Albumin-Binding PSMA Ligands: Implications for Expanding the Therapeutic Window. J. Nucl. Med. 2019, 60, 656–663. 10.2967/jnumed.118.221150. [DOI] [PubMed] [Google Scholar]
- Klauser P. C.; Chopra S.; Cao L.; Bobba K. N.; Yu B.; Seo Y.; Chan E.; Flavell R. R.; Evans M. J.; Wang L. Covalent Proteins as Targeted Radionuclide Therapies Enhance Antitumor Effects. ACS Cent. Sci. 2023, 9, 1241–1251. 10.1021/acscentsci.3c00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S.; Crocker P. R.; Paulson J. C. Siglec-Mediated Regulation of Immune Cell Function in Disease. Nat. Rev. Immunol. 2014, 14, 653–666. 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker P. R.; Paulson J. C.; Varki A. Siglecs and Their Roles in the Immune System. Nat. Rev. Immunol. 2007, 7, 255–266. 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- van de Wall S.; Santegoets K. C. M.; van Houtum E. J. H.; Büll C.; Adema G. J. Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol. 2020, 41, 274–285. 10.1016/j.it.2020.02.001. [DOI] [PubMed] [Google Scholar]
- Murugesan G.; Weigle B.; Crocker P. R. Siglec and Anti-Siglec Therapies. Curr. Opin. Chem. Biol. 2021, 62, 34–42. 10.1016/j.cbpa.2021.01.001. [DOI] [PubMed] [Google Scholar]
- Angata T.; Nycholat C. M.; Macauley M. S. Therapeutic Targeting of Siglecs Using Antibody- and Glycan-Based Approaches. Trends Pharmacol. Sci. 2015, 36, 645–660. 10.1016/j.tips.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H.; Woods E. C.; Vukojicic P.; Bertozzi C. R. Precision Glycocalyx Editing as a Strategy for Cancer Immunotherapy. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 10304–10309. 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.; Xu L.; Chen L.; Zhang Z.; Zhang Y.; Wu W.; Li C.; Liu S.; Xiang S.; Dai S.; et al. Epitope-Directed Antibody Elicitation by Genetically Encoded Chemical Cross-Linking Reactivity in the Antigen. ACS Cent. Sci. 2023, 9, 1229–1240. 10.1021/acscentsci.3c00265. [DOI] [PMC free article] [PubMed] [Google Scholar]