Abstract
The transmembrane subunit (TM) of the envelope glycoprotein (Env) of the oncovirus avian sarcoma/leukosis virus (ASLV) contains an internal fusion peptide flanked by two cysteines (C9 and C45). These cysteines, as well as an analogous pair in the Ebola virus GP glycoprotein, are predicted to be joined by a disulfide bond. To examine the importance of these cysteines, we mutated C9 and C45 in the ASLV subtype A Env (EnvA), individually and together, to serine. All of the mutant EnvAs formed trimers that were composed of the proteolytically processed surface (SU) and TM subunits. All mutant EnvAs were incorporated into murine leukemia virus pseudotyped virions and bound receptor with wild-type affinity. Nonetheless, all mutant EnvAs were significantly impaired (∼1,000-fold) in their ability to support infectivity. They were also significantly impaired in their ability to mediate cell-cell fusion. Our data are consistent with a model in which the internal fusion peptide of ASLV-A EnvA exists as a loop that is stabilized by a disulfide bond at its base and in which this stabilized loop serves an important function during virus-cell fusion. The fusion peptide of the Ebola virus GP glycoprotein may conform to a similar structure.
Avian sarcoma/leukosis virus, subtype A (ASLV-A), is an enveloped avian retrovirus that contains a single glycoprotein, EnvA, that is present in virions as trimers of two disulfide-bonded subunits, SU (gp85) and TM (gp37) (12). SU binds to the target cell receptor, Tva (16); TM contains a hydrophobic sequence, termed the fusion peptide, that functions in virus-cell fusion (3, 11) and the membrane anchor. The protein is initially synthesized as a single chain precursor, pr95, which is processed into SU and TM by a furin-like enzyme present in the Golgi complex (5). EnvA and its single target cell receptor, Tva, appear to be sufficient to initiate the fusion process (1, 2, 10).
For many viral fusion proteins (most notably the influenza virus hemagglutinin and the human immunodeficiency virus Env glycoprotein), the maturation processing event positions the fusion peptide at the N terminus of the fusion-mediating subunit; for others, the fusion peptide remains internal. ASLV is a member of the latter group. Whereas N-terminal fusion peptides have been subjected to considerable scrutiny (reviewed in reference 4), less is known about internal fusion peptides. We have recently provided evidence consistent with a model in which the fusion peptide of ASLV EnvA (residues 22 to 37 of TM) exists as a looped structure (3). Other internal fusion peptides can also be modeled as looped structures (3) and, indeed, the internal fusion peptide of the tick-borne encephalitis virus envelope glycoprotein, E, exists as a loop in its prefusogenic conformation. What is not clear for any internal fusion peptide, however, is whether the loop structure is important for fusion.
A modeling study has suggested that the TM subunit of EnvA and the Ebola virus GP2 adopt similar structures (6). Both contain internal fusion peptides flanked by cysteines (C9 and C45 for ASLV TM; C10 and C55 for Ebola virus GP2) that are predicted to be joined in a disulfide bond (6). This prediction was supported by recent crystallographic data on the core of Ebola virus GP2 (13a, 15). However, it has not yet been formally shown whether the cysteines in question form a disulfide bond and, if so, whether said disulfide bond is important for the structure and/or function(s) of EnvA or Ebola virus GP. A disulfide bond at the base of the looped fusion peptide region might be important for proper folding and processing of the glycoprotein, for its assembly into trimers, and/or for its fusion activity. The concept that a disulfide bond might be needed to stabilize an internal fusion peptide has been suggested, but not verified, for tick-borne encephalitis virus (9) and for viral hemorrhagic septicemia virus (7).
In the present study, we investigated the role of C9 and C45 of the TM domain of the ASLV-A envelope glycoprotein (EnvA) by mutating each of these residues, individually and together, to serine. Following expression by transient transfection in 293T cells (3), each mutant EnvA was evaluated for trimerization, proteolytic processing, incorporation into pseudotyped virions, receptor binding, infectivity, and its ability to mediate cell-cell fusion.
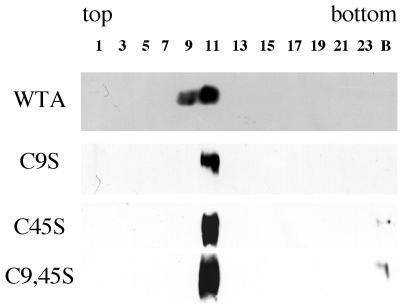
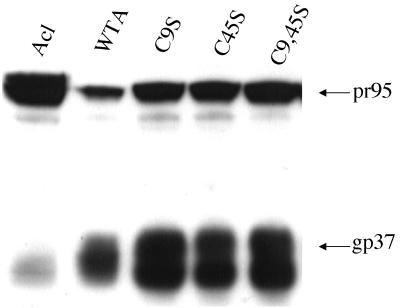
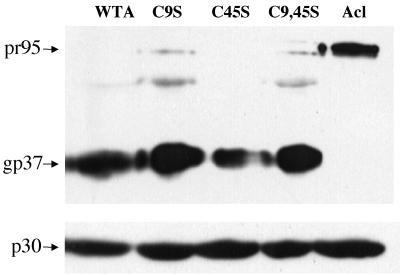
As seen in Fig. 1, all mutant EnvAs sedimented on sucrose gradients at the same position as the wild-type trimer. As seen in Fig. 2, all three mutant EnvAs were processed from the precursor (pr95) into SU (gp85) and TM (gp37) as well as was wild-type EnvA. As seen in Table 1, each of the mutant EnvAs bound receptor with equal or higher affinity than wild-type EnvA. As seen in Fig. 3, processed forms of all EnvAs were incorporated into pseudotyped virions to levels approximately equivalent to those of wild-type EnvA. Hence, none of the mutations impaired either the proteolytic processing, trimer formation, receptor binding, or virion incorporation properties of EnvA.
FIG. 1.
Trimerization of mutant EnvAs. 293T cells were transfected with pCB6-EnvA DNA by the calcium phosphate method and induced with 10 mM sodium butyrate 24 h later. Sixteen to 18 h posttransfection, cells were harvested, lysed in 1% NP-40 buffer containing protease inhibitors (3), and subjected to sucrose density centrifugation in 10 to 30% sucrose in n-octyl glucoside buffer as described previously (3). Fractions (500 μl each) were collected and processed for Western blot analysis with the anti-Ngp37 antibody which recognizes both TM and the full-length EnvA precursor, pr95 (10). The gp37 band is shown. The gradient density increases from left to right. C9S, C45S, and C9,45S contain serines in place of the cysteine at the numbered site(s); numbering is from the initial residue of the mature TM subunit. The numbered lanes contain samples from the specified gradient fractions. B denotes a sample from the bottom of the centrifuge tube. WTA, wild-type EnvA.
FIG. 2.
Processing of mutant EnvAs. Cell lysates were prepared and processed for Western blot analysis as described in the legend to Fig. 1. Both the uncleaved pr95 (top) and cleaved gp37 (bottom) are indicated. Samples are labeled as in the legend to Fig. 1. Acl is described in Table 1, footnote b.
TABLE 1.
Receptor binding and infectivity of cysteine mutantsa
| Mutant | Assay
|
|
|---|---|---|
| Receptor binding (Kd) | Infectivity (% wild type) | |
| Wild type | 1.7 × 10−8 | 100.00 |
| C9S | 1.6 × 10−8 | 0.16 |
| C45S | 0.7 × 10−8 | 0.09 |
| C9,45S | 1.6 × 10−8 | 0.09 |
| Aclb | NDc | 0.02 |
The apparent Kd for binding of s47 to EnvA-expressing cells and infectivity mediated by EnvA pseudotyped MLV virions were measured as previously described (3). For the infectivity assay, the results from four independent experiments, normalized to wild-type infectivity within each experiment, were averaged. The standard deviation for the infectivity results ranged from ±0.01% for C9,45S to ±0.09% for C9S. ND, not determined.
Acl is a mutant EnvA in which the cleavage site has been mutated (8). Although it forms trimers, is expressed at high levels at the cell surface, and is well incorporated into virions, this EnvA is poorly cleaved and does not support infection.
Although the Kd valve for the Acl mutant was not determined, immunoprecipitation studies indicate that Acl retains wild-type receptor binding (data not shown).
FIG. 3.
Incorporation of mutant EnvAs into MLV pseudotyped virions. EnvA-MLV pseudotyped virions were prepared and concentrated as previously described (3). Virions were diluted into sodium dodecyl sulfate sample buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with the anti-Ngp37 antibody. Parallel blots were probed with an antibody recognizing the MLV capsid (p30). Samples are labeled as described in the legend to Fig. 1.
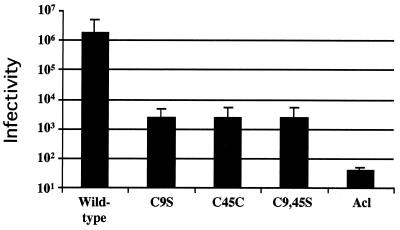
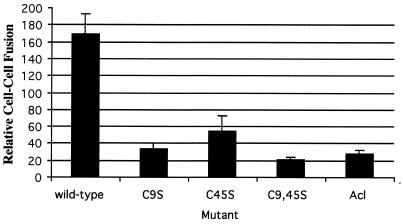
We next used the murine leukemia virus (MLV) pseudotype system to asses the ability of each mutant EnvA to support infection of Tva-expressing cells (3). In this triple-transfection, single-round infection system, virions produced by MLV gag(pHIT60) (13) incorporate ASLV EnvAs expressed at the cell surface (pCB6/EnvA) (8) and package DNA for the expression of β-galactosidase (pHIT111) (13). Infection is scored by counting blue cells. As seen in Table 1 and Fig. 4, all three mutant EnvAs showed a significant (∼1,000-fold) decrease in their ability to support infection compared to that of wild-type EnvA. To determine if the defect in infectivity was at the level of membrane fusion, we used a β-galactosidase reporter gene cell-cell fusion assay (11, 14). As seen in Fig. 5, all three mutant EnvAs were significantly impaired in their ability to mediate cell-cell fusion, suggesting that the defect in infectivity is at the level of virus-cell fusion.
FIG. 4.
Infectivity of MLV pseudotyped mutant EnvA viruses. EnvA pseudotyped viruses were prepared and titered on PG950 cells as described previously (3). Results are an average of three independent experiments. Samples are labeled as in the legend to Fig. 1. Wild-type, wild-type EnvA.
FIG. 5.
Cell-cell fusion mediated by mutant EnvAs. Cell-cell fusion assays were performed as described previously (11), except that modified vaccinia virus Ankara-infected PG950 cells were incubated overnight in medium containing rifampin (100 μg/ml), and the cell-cell fusion medium contained both rifampin (100 μg/ml) and AraC (150 μg/ml). It should be noted that at no time did the pH of any of these media dip below pH 7.2. The data from triplicate samples from a representative experiment were averaged. Samples are labeled as in the legend to Fig. 1. Wild-type, wild-type EnvA.
In the present study, we mutated C9 and C45, the two cysteines in the TM subunit of EnvA that flank its internal fusion peptide. The cysteines were changed singly and together to serine. We found that neither C9 nor C45 was required for the formation of native EnvA structure, as measured by the ability of all mutant EnvAs (C9S, C45S, and C9,45S) to trimerize, to be processed into SU and TM subunits, and to be incorporated into pseudotyped virions. In addition, all three mutant EnvAs bound receptor with equal or higher affinity than wild-type EnvA. However, both cysteines proved to be critical for the ability of EnvA to support infection and cell-cell fusion. Because our infectivity assay is a single-round infectivity assay designed to measure early events in entry, and because our cell-cell fusion assay is a measure of membrane fusion, we infer that both C9 and C45 are required for virus-cell fusion.
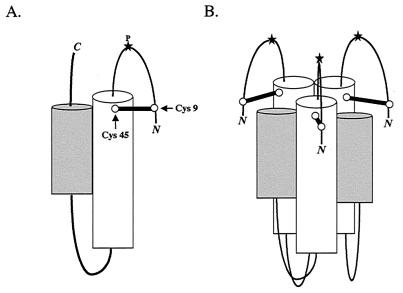
In Fig. 6, we present a model for a fusion-active conformation of ASLV TM. The core monomer (Fig. 6A) and six-helix bundle (Fig. 6B) formed by the gp37 trimer are based on crystal structures of the core fragment of the Ebola virus GP2 (13a, 15), which has been predicted to be similar to the core of the ASLV-A EnvA TM subunit (6). The C-terminal and N-terminal residues (including the internal fusion peptide), which are not present in the Ebola virus GP2 crystal structure, are depicted extending toward the common fused membrane. The N-terminal sequence is shown as a looped structure with the internal fusion peptide at its center, stabilized by a disulfide bond between C9 and C45. Analysis of the central proline within the fusion peptide has suggested that it may perform a function similar to that of the N-terminal residues of the influenza virus hemagglutinin fusion peptide in initial target membrane interaction (3). Accordingly, we have placed the proline at the apices of the fusion peptide loop. We predict that the Ebola virus GP2 fusion peptide region will adopt a similar structure.
FIG. 6.
Model of a fusion-active state of the ASLV TM trimer. The core monomer and six-helix bundle formed by the gp37 trimer were modeled after the crystal structures of the core fragment of Ebola virus GP2 (13a, 15). (A) The gp37 monomer. The predicted N-terminal (white) and C-terminal (gray) α-helices pack against each other in an antiparallel orientation, placing the N-terminal loop (containing the fusion peptide) and the C-terminal portions of the protein near each other, extending toward the common fused membrane. A star at the apex of the loop (P) denotes the predicted position of the proline within the fusion peptide. The loop containing the proline is shown stabilized by a disulfide bond between C9 (Cys9) and C45 (Cys45). The disulfide-bonded cysteines are represented by a dumbbell and marked by arrows. The N and C termini are marked. (B) The gp37 trimer. By analogy to other trimeric viral fusion proteins, the N-terminal helices are expected to pack against each other, forming a threefold symmetry axis down the center of the resulting coil (13a, 15) with a given C-terminal helix resting in the groove made by its own and a neighboring N-terminal helix. The C-terminal residues extending from these helices have been omitted for clarity.
The fact that neither of the single mutations (C9S or C45S) impaired the formation of the native EnvA structure was somewhat surprising, as they could have led to aberrant disulfide bond formation. Furthermore, some mutations within the fusion peptide have caused a significant portion of EnvA to be misfolded (3, 11), and it appears that a residue with high reverse turn probability (the wild-type Pro is the best known) is required in the middle of the fusion peptide for optimal processing of the EnvA percursor, pr95, into SU and TM (3). Based upon these results, we predicted that the fusion peptide region exists as a looped structure in the EnvA precursor (3). If a disulfide bond was needed to stabilize a looped fusion peptide in the EnvA precursor, to afford optimal presentation of the SU-TM cleavage site, then it was formally possible that all of our cysteine mutants would have been impaired in SU-TM processing. However, none of these concerns proved to be the case. Perhaps the fusion peptide is packed in the precursor form of EnvA well enough (as a loop with a reverse turn about the central Pro) so that a disulfide bond at its base is not needed at this stage of the protein's life cycle. Alternatively, a disulfide bond between C9 and C45 may form only after the proteolytic processing event.
Our results suggest that a disulfide bond between C9 and C45 is important for EnvA function during virus-cell fusion. In a current model of this process (hhtp://www.people.virginia.edu/∼jag6n/whitelab.html), the interaction of the fusion peptide with the target membrane helps to bring the viral and target membranes close together as the fusion protein is undergoing a series of conformational rearrangements. It may be that a disulfide bond between C9 and C45 is needed so that proper interaction between the fusion peptide and the target membrane can be maintained as the protein changes conformation at the fusion site. Experiments are underway to further define the defects exhibited by the mutant EnvAs C9S, C45S, and C9,45S.
Gallaher predicted that there is a disulfide bond between C9 and C45 based on a modeling study of the fusion mediating subunit of ASLV EnvA TM and the homologous subunit of the Ebola virus glycoprotein (6). Crystal structures of the core fragment of GP2 supported this prediction (13a, 15). Our mutational analysis of ASLV EnvA complements these predictions and provides the first evidence that these cysteines are important for the fusion process. Given the similarity in the predicted structures of the Ebola virus GP2 and the ASLV TM, we predict that C10 and C55 of GP2 will also be important for Ebola virus-mediated fusion. It will be interesting to determine whether the cysteines flanking other internal fusion peptides are also important for fusion.
Acknowledgments
We thank Michael Burdick for help with preparing cDNAs for the expression of the mutant EnvAs.
This work was supported by NIH grant A122470 to J.M.W.
REFERENCES
- 1.Damico R, Bates P. Soluble receptor-induced retroviral infection of receptor-deficient cells. J Virol. 2000;74:6469–6475. doi: 10.1128/jvi.74.14.6469-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delos S E, Gilbert J M, White J M. The central proline of an internal viral fusion peptide serves two important roles. J Virol. 2000;74:1686–1693. doi: 10.1128/jvi.74.4.1686-1693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 5.Einfeld D A, Hunter E. Mutational analysis of the oligomer assembly domain in the transmembrane subunit of the Rous sarcoma virus glycoprotein. J Virol. 1997;71:2383–2389. doi: 10.1128/jvi.71.3.2383-2389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 7.Gaudin Y, de Kinkelin P, Benmansour A. Mutations in the glycoprotein of viral haemorrhagic septicaemia virus that affect virulence for fish and the pH threshold for membrane fusion. J Gen Virol. 1999;80:1221–1229. doi: 10.1099/0022-1317-80-5-1221. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinz F, Stiasny K, Puschner-Auer G, Holzmann H, Allison S, Mandl C, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez L D, Peters R R, Delos S E, Young J A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez L D, White J M. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter E, Hill E, Hardwick M, Brown A, Schwartz D E, Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J Virol. 1983;46:920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Malashkevich V N, Schneider B J, McNally M L, Millhollen M A, Pang J X, Kim P S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoprotein analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissenhorn W, Carff A, Lee K, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 16.Young J A, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]