SUMMARY
The human genome has many short tandem repeats, yet the normal functions of these repeats are unclear. The 5′ untranslated region (UTR) of the fragile X messenger ribonucleoprotein 1 (FMR1) gene contains polymorphic CGG repeats, the length of which has differing effects on FMR1 expression and human health, including the neurodevelopmental disorder fragile X syndrome. We deleted the CGG repeats in the FMR1 gene (0CGG) in human stem cells and examined the effects on differentiated neurons. 0CGG neurons have altered subcellular localization of FMR1 mRNA and protein, and differential expression of cellular stress proteins compared with neurons with normal repeats (31CGG). In addition, 0CGG neurons have altered responses to glucocorticoid receptor (GR) activation, including FMR1 mRNA localization, GR chaperone HSP90α expression, GR localization, and cellular stress protein levels. Therefore, the CGG repeats in the FMR1 gene are important for the homeostatic responses of neurons to stress signals.
In brief
Sirois et al. investigated the function of normal CGG repeats in the 5′ UTR of the FMR1 gene in human neurons and showed that neurons without CGG repeats have altered FMR1 mRNA localization and responses to cellular stress, providing evidence for a function of nonpathogenic CGG repeats in FMR1.
Graphical Abstract
INTRODUCTION
Roughly 3% of the human genome is made up of short tandem repeats (STRs) of 2–7 base pairs, which are intrinsically unstable and highly polymorphic.1 To date, more than 50 repeat expansion disorders are known to be caused by expansion of STRs beyond a specific copy number threshold.2–5 In addition, increasing evidence suggests that even repeat length polymorphisms below the disease threshold are correlated with human health, including osteoarthritis and cancer.6–8 However, given their abundance in the human genome, STRs may have important roles in cellular functions9 that have been largely uncharacterized. One particular type of STR is trinucleotide CGG repeats, which have been identified at more than 6,000 sites in the human genome.10 Approximately 93% of these sites are polymorphic, and 30% of those polymorphic sites are located in the 5′ untranslated region (5′ UTR) of genes.10 Despite the importance of the 5′ UTR region in regulating processes such as mRNA translation,11 and evidence that 5′ UTR genetic variants can have deleterious effects,12,13 the normal function of these CGG repeats in most genes remains unknown.
The human Fragile X messenger ribonucleoprotein 1 (FMR1) gene contains a 5′ UTR polymorphic CGG repeat with a modal number of 30 CGG repeats.14 While CGG repeats in the FMR1 gene are conserved in mammals, humans and other great apes have the highest CGG repeats (mean = 30), while other non-human primates (mean = 20) and non-primate mammals (mean = 8) have shorter CGG repeats (Table S1).15 Expansion of FMR1 CGG repeats above 200 copies leads to silencing of the FMR1 gene and fragile X syndrome (FXS), the most prevalent inherited neurodevelopmental disorder that is characterized by intellectual disability.16 Repeats between 55 and 199 CGGs increase FMR1 mRNA expression, but decrease the levels of the protein product (FMRP), and result in neuropsychiatric manifestations, fragile X tremor and ataxia syndrome, or fragile X-associated primary ovarian insufficiency with incomplete penetrance.16 Repeat sizes above 40, but below 55, have been associated with conditions including parkinsonism,17–19 while repeat sizes below 26 are associated with cancer risk.20–22 More recently, several studies have also revealed a correlation between shortened CGG repeat length (<24 repeats), health in later life, and long-term parenting stress.23,24 Specifically, individuals with long-term parenting stress and shorter CGG repeats had poorer health outcomes in areas including physical limitations, cognitive functioning, and mental health symptoms.23,24 Therefore, FMR1 CGG repeat polymorphism has significant effects on human health.
Despite these correlations, the functional significance of normal (30–31) CGG repeats in the human FMR1 gene is unclear. Expression of FMR1 5′ UTR luciferase reporters in vitro shows that both low and high numbers of CGG repeats negatively affected protein translation, but not transcription, compared with the modal number of 30 CGGs.25 Decreased expression of multiple FMR1 mRNA isoforms was observed in luteinized granulosa cells isolated from individuals with fewer than 26 CGG repeats.26 Repeat-associated non-AUG translation from reporter constructs containing normal length repeats helps to control basal and activity-dependent synthesis of FMRP.27 The addition of 24 CGG repeats into the 5′ UTR of non-CGG repeat-containing mRNA can promote its localization to dendrites in primary rat neurons.28 However, direct evidence for functions of a normal number of CGG repeats in the endogenous FMR1 gene is lacking.
To interrogate the function of normal length CGG repeats in FMR1, we generated isogenic CGG-deficient human embryonic stem cell (hESC) lines in which the CGG repeats have been precisely removed (0CGG) from the parental hESCs (31CGG). Removal of CGG repeats leads to an increased abundance of FMR1 mRNA, but a decreased abundance of FMRP protein in dendrites of hESC-derived neurons, with a concomitant decreased abundance of FMRP in the soma. Further, we observe altered expression levels of several cellular stress proteins in neurons, suggesting that these 0CGG neurons may have altered homeostatic response to stress, a feature suggested by human population studies.23,24 In fact, treatment of 0CGG neurons with the glucocorticoid receptor (GR) agonist dexamethasone (DEX) led to differential responses compared with 31CGG neurons, including cellular stress protein levels, FMR1 and FMRP subcellular localization, and the distribution of GR in the soma and nucleus. DEX-treated 0CGG neurons had a decreased abundance of HSP90α, a chaperone protein required for high-affinity steroid binding activity of GR. Our data unveil a role for the CGG repeats in the FMR1 5′ UTR in ensuring the correct localization of both FMR1 mRNA and protein in developing human neurons, and that this correct localization is important for GR signaling and homeostatic response to cellular stress signals. Our findings not only shed light on understanding the function of polymorphic CGG repeats in the FMR1 gene, but also have important implications for designing FXS gene therapy strategies that may remove the entire CGG repeat expansion to reactivate the FMR1 gene.29,30
RESULTS
Removal of CGG repeats from the FMR1 5′ UTR leads to increased localization of FMR1 mRNA in dendrites of immature human neurons
The 5′ UTR region of mRNAs typically contains important regulatory elements,11 including a translation initiation codon, upstream open reading frames, and protein binding sites.31–33 To investigate the function of the FMR1 CGG repeats, we used CRISPR-mediated gene editing to generate two pairs of isogenic male hESC lines in which only the CGG repeats were removed (H1–0CGG and H13–0CGG) from the parental H1 and H13 lines34 that have 31 CGG repeats (Figure S1A; Table S2). Sanger sequencing confirmed removal of the CGG repeats without off-target cutting, and both 0CGG hESC lines were karyotypically normal (Figures S1B–S1D). Both 0CGG and isogenic control hESC lines consistently differentiated into dorsal forebrain neural progenitor cells (NPCs) and immature excitatory neurons (Figure S1F).35 No significant differences in either FMR1 mRNA or FMRP protein levels were detected between 0CGG cells and their respective isogenic controls at the hESC (Figures S1G–S1L), NPC (Figures S1G–S1L), or neuron stages (Figures 1A and 1B), indicating that removal of the CGG repeats from the 5′ UTR does not affect FMR1 expression during early neurogenesis.
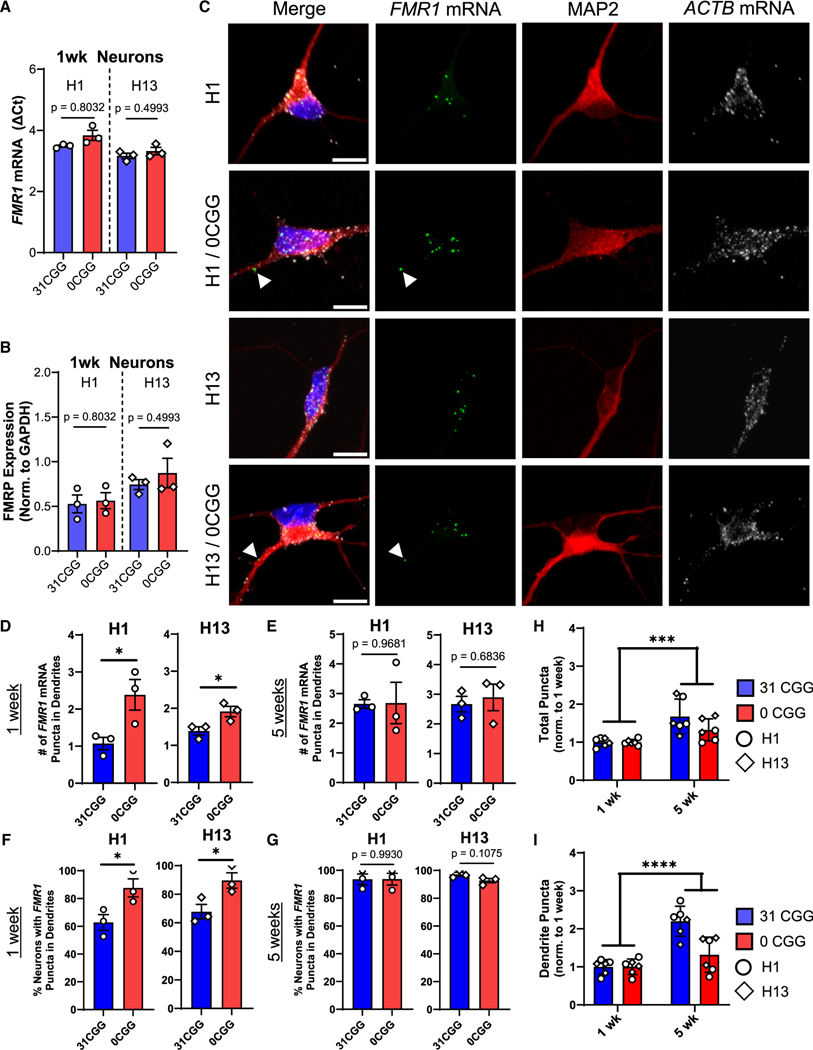
Figure 1. Removal of CGG repeats promotes localization of FMR1 mRNA to dendrites of early post-mitotic neurons.
(A and B) FMR1 mRNA levels (A) and FMRP protein levels (B) in hESC-derived neurons at 1 week of differentiation. See Figure S1J for representative blot images. n = 3 independent batches of differentiation per line.
(c) Representative confocal images showing FMR1 and ACTB mRNA puncta in hESC-derived neurons stained for post-mitotic neuron marker, MAP2 (red). Blue, nuclear staining using DAPI. Scale bar, 10 μm.
(D and E) Quantification of the number of FMR1 mRNA puncta in the dendrites of neurons. n = 3 technical replicates from a single batch of neurons for N = 2 isogenic hESC lines. Each data point represents the average of ≥11 neurons.
(F and G) Percentage of neurons containing at least 1 FMR1 mRNA puncta in their dendrites at 1 week (F) and at 5 weeks (G).
(H and I) Comparison of total FMR1 mRNA puncta (H) and dendritic FMR1 mRNA puncta (I) in 1 week versus 5 week neurons, with all data normalized to the 1-week time point. n = 3 technical replicates from a single batch of neurons for N = 2 isogenic hESC lines. Each data point represents the average of ≥11 neurons. Error bars indicate SEM. (A, B, and D–G): two-tailed Student’s t test *p < 0.05. (H and I) Two-way ANOVA, significant main effect of time; ***p < 0.005, ****p < 0.001.
The 5′ UTR, including the CGG repeats, can modulate the subcellular localization of mRNA within neurons.28,36–38 We thus tested the hypothesis that removal of the CGG repeats from FMR1 would alter the localization of FMR1 mRNA. We compared the localization of FMR1 mRNA in 0CGG and control neurons using single-molecule mRNA fluorescence in situ hybridization (FISH) combined with immunofluorescence of dendritic protein MAP2 in neurons differentiated for 1 week (immature neurons) (Figure 1C) and 5 weeks (mature neurons) (Figure S2). No significant differences were detected in the total number of FMR1 mRNA puncta per neuron between 0CGG and control neurons (Figure S3A), consistent with our qPCR data (Figure 1A). While there were no differences in the number of FMR1 puncta in the soma (Figure S3B) or nucleus (Figure S3C) between 0CGG and control neurons, 0CGG neurons had significantly more FMR1 puncta in their dendrites at 1 week (Figure 1D), but not at 5 weeks (Figure 1E) of differentiation. We next compared the proportion of neurons that contained any FMR1 mRNA puncta in each cellular compartment. Interestingly, while virtually all neurons contained FMR1 mRNA puncta in the soma and nucleus at both ages (Figures S3E and S3F), more 0CGG immature neurons (1 week) contained FMR1 mRNA in their dendrites than did the control neurons (Figure 1F). At 5 weeks, more than 80% of neurons across all genotypes had dendritic FMR1 mRNA puncta (Figures 1G and S3D), and there was an overall increase in FMR1 mRNA puncta throughout the neuron in both genotypes with age (Figures 1H, 1I, S3G, and S3H). Therefore, removal of CGG repeats in the FMR1 5′ UTR leads to increased dendritic localization of FMR1 mRNA in immature neurons.
The human FMR1 5′ UTR leads to CGG repeat-mediated differences in dendritic mRNA localization of reporter transcripts in mouse neurons
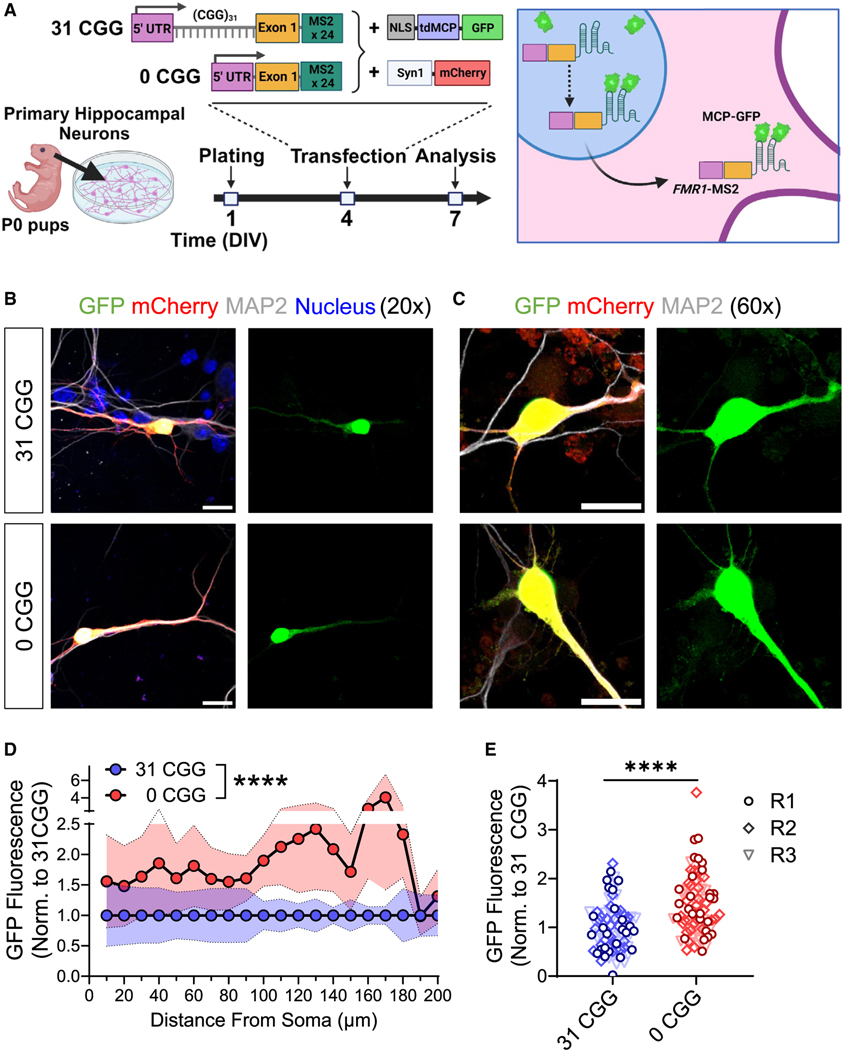
To validate that the CGG repeats in FMR1 mRNA are responsible for the differences in dendritic FMR1 mRNA localization that we observed by FISH, we created MS2 reporters39 containing the human FMR1 5′ UTR and exon 1 sequence (Figure 2A), with either normal repeats (FMR1-MS2–31CGG) or no repeats (FMR1-MS2–0CGG). We co-transfected mouse neurons at 4 days in vitro (DIV) with the MS2 reporter, together with a vector expressing a nucleus-targeted fusion protein of GFP and MS2 binding protein (GFP-MCP) for monitoring MS2 containing mRNA transcripts in the cytosol, and a vector expressing synapsin I promoter-driven mCherry for the visualization of transfected neurons (Syn1-mCherry) (Figures 2A and S4A–S4C). The GFP intensity in the primary dendrites of mCherry+ neurons was higher at 10 and 14 DIV compared with 6 DIV (Figures S4D–S4F), consistent with our observation that FMR1 mRNA puncta in dendrites increased during neuronal maturation (Figure 1D). In addition, FMR1-MS2–0CGG transfected neurons showed higher dendritic GFP intensity compared with FMR1-MS2–31CGG reporter transfected neurons at DIV6 (Figure S4G), but not at DIV 10 or 14 (Figures S4H and S4I). We, therefore, chose DIV 7 to investigate whether CGG repeats affect mRNA localization in immature neurons. Indeed, we observe both greater GFP intensities throughout the length of the primary dendrite and higher levels of total GFP signal in the dendrites of FMR1-MS2–0CGG compared with FMR1-MS2–31CGG transfected neurons (Figures 2B–2E). These results were not due to differences in dendritic length between the two conditions (n = 86–94 cells from N = 3 isolations; p = 0.0815), in overall cellular GFP signal (n = 29–31 cells from N = 3 isolations; p = 0.7383), or in the cellular composition of the primary neuron cultures (Figure S4B). Taken together, our findings in both mouse and human neurons suggest that the CGG repeats in the 5′ UTR of FMR1 play an important role in controlling the localization of FMR1 mRNA in immature neurons, and that removal of these repeats may result in the premature localization of FMR1 mRNA to dendrites.
Figure 2. Expression of the human FMR1 5′ UTR in mouse neurons recapitulates CGG repeat-mediated differences in dendritic mRNA localization.
(A) Schematic illustration of transfection of primary mouse hippocampal neurons with MS2 and Syn1-mCherry constructs and timing of experiments.
(B and C) Representative confocal images of MS2-transfected neurons at 20× magnification (B) and 60× magnification (C). Scale bars, 25 μm (B), 20 μm (C). Green, MS2-reporter; red, Synapsin-mCherry reporter; white, MAP2 (post-mitotic neuron label; blue, nuclear staining using Hoechst.
(D) Quantification the GFP fluorescence intensity along the length of primary dendrites of GFP+/mCherry+/MAP2+ neurons. Shaded area indicates SEM.
(E) Summation of the fluorescence intensity in primary dendrites, normalized for dendritic length. Each data point represents single neurons. Data in D and E are from N = 3 independent neuronal isolations/biological replicates (R1–R3; 22–29 neurons per replicate). Values were normalized to 31 CGG condition for each batch of neurons. (D) Two-way ANOVA, significant main effect of genotype ****p < 0.001. (E) Welch’s t test ****p < 0.001.
Localization of mRNA with normal number of CGG repeats is mediated by G-quadruplexes
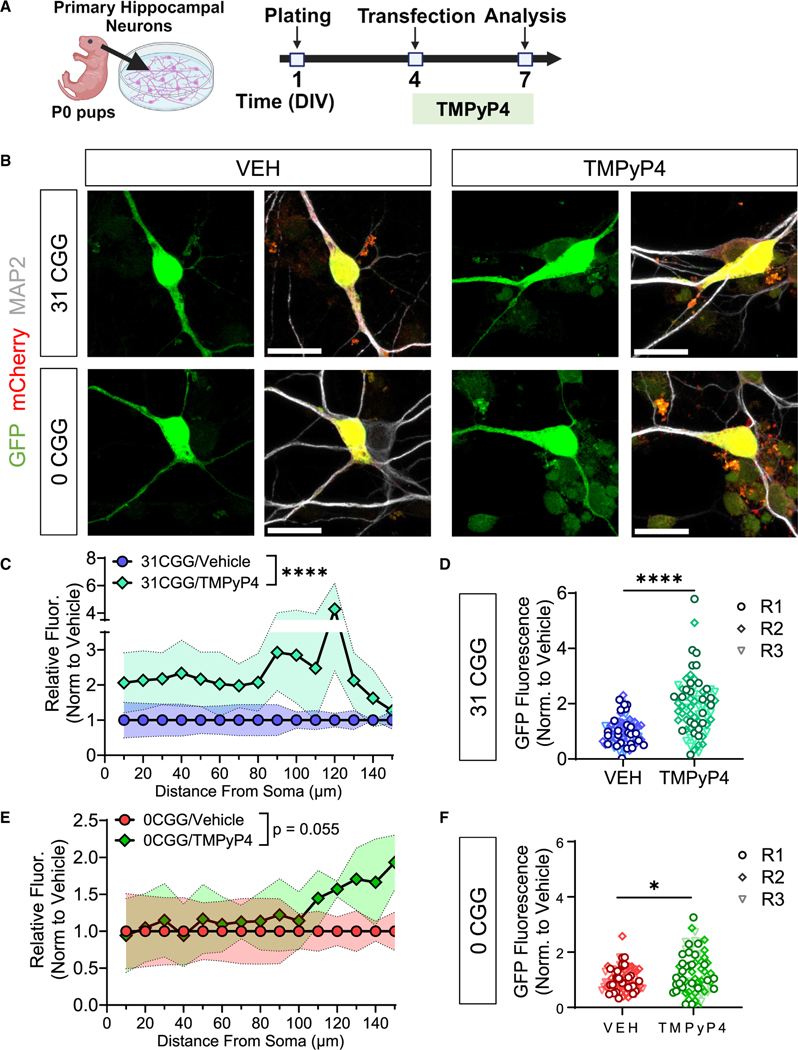
The CGG repeats in the 5′ UTR of FMR1 form secondary structures, including hairpins and G-quadruplexes (GQs),40–42 the latter of which can mediate mRNA localization to neurites.43–45 Secondary structure prediction using QGRS Mapper46 (Figure S5A) and RNAfold47 (Figures S5B and S5C) indicated that the deletion of CGG repeats leads to the absence of multiple GQs in the 5′ UTR of the FMR1 gene. To test the contribution of GQ secondary structure on FMR1 mRNA localization, we used TMPyP4, a cationic porphyrin that destabilizes DNA and RNA GQs (Figures 3A and 3B).48–51 Treatment of FMR1-MS2–31CGG-transfected neurons with TMPyP4 dramatically increases GFP-MCP signal in primary dendrites compared with vehicle (Figures 3C and 3D). In contrast, TMPyP4 only had a modest effect on FMR1-MS2–0CGG neurons (Figures 3E and 3F), and the effect of TMPyP4 on FMR1-MS2–31CGG neurons was greater than the FMR1-MS2–0CGG neurons (Figure S5D). Hence, our results suggest that the retention of FMR1 mRNA to the soma or nucleus in early neuronal development may be mediated by the GQ structure formed by the CGG repeats in its 5′ UTR.
Figure 3. Localization of mRNA with normal length CGG repeats is mediated by GQs.
(A) Schematic illustration of the timing of primary mouse hippocampal neuron transfection and TMPyP4 treatment.
(B) Representative confocal images of MS2-transfected neurons treated with VEH (left) or TMPyP4 (right) at 60× magnification. Scale bar, 20 μm. Green, MS2-reporter; red, Synapsin-mCherry reporter; white, MAP2 (postmitotic neuron marker).
(C and E) Quantification the GFP fluorescence intensity along the length of primary dendrites of GFP+/mCherry+/MAP2+ neurons transfected with 31 CGG (C) or 0 CGG (E) plasmids. Shaded area indicates SEM.
(D and F) Summation of the fluorescence intensity in primary dendrites, normalized for dendritic length in 31 CGG (D) or 0 CGG (F) transfected neurons. Each data point represents single neurons (R1–R3). Data in (C–F) are from N = 3 independent neuronal isolations/biological replicates (18–29 neurons per replicate). Values were normalized to vehicle (VEH) condition for each batch of neurons. (C and E) Two-way ANOVA, significant main effect of treatment ****p < 0.001. (D and F) Welch’s t test; *p < 0.05; ****p < 0.001.
CGG repeat-dependent effects of GR activation on FMR1 mRNA localization
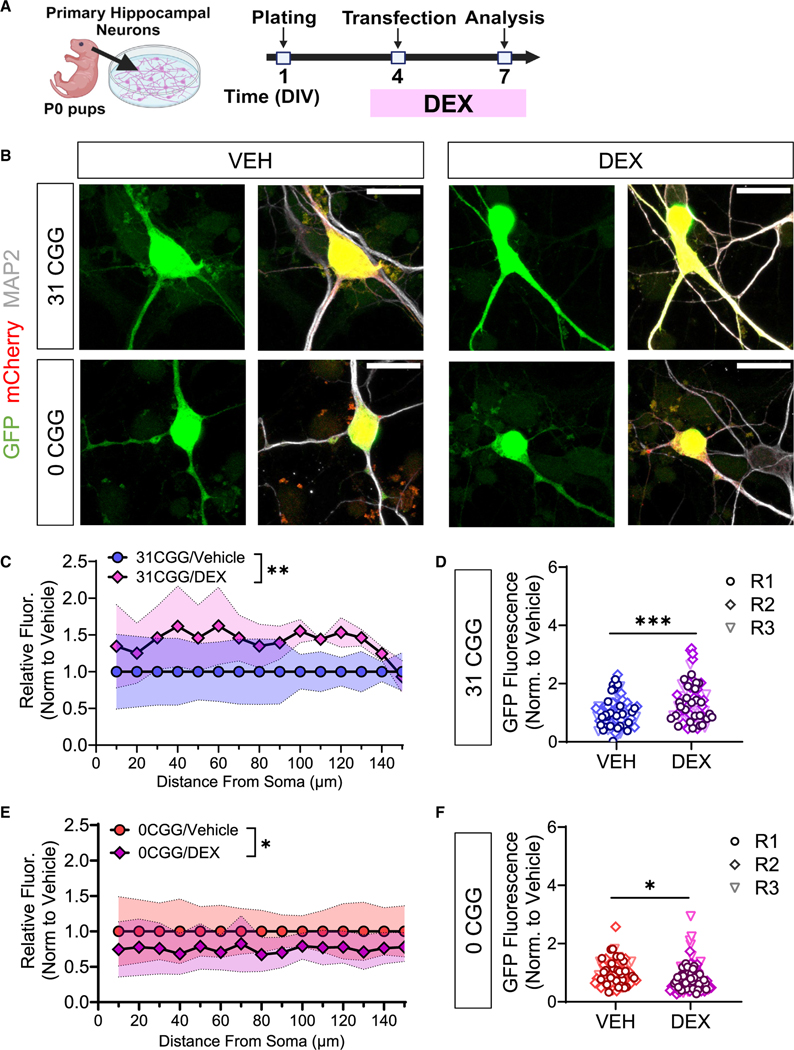
Individuals with fewer FMR1 CGG repeats (<24) exhibit poorer health outcomes in response to certain types of chronic life stress.23,24 During stressful events, corticosteroids are released from the hypothalamus, leading to the activation of both the fast and slow modes of the stress response system.52 The GR is involved in both stress responses, and regulates stress termination.52 To determine whether the CGG repeat-mediated FMR1 mRNA localization is sensitive to cellular stress signals, we treated primary hippocampal neurons transfected with FMR1-MS2 reporters with the GR agonist DEX (Figure 4A). We found that DEX treatment had opposite effects on reporter mRNA localization in FMR1-MS2–31CGG compared with FMR1-MS2–0CGG neurons (Figure 4B): 31CGG repeat RNA had increased localization to dendrites after DEX treatment (Figures 4C and 4D), while 0CGG RNA had decreased localization to dendrites after DEX (Figures 4E and 4F). Therefore, our data support a connection between CGG repeat-mediated FMR1 mRNA localization and GR activation.
Figure 4. CGG repeat-dependent effects of GR activation on FMR1 mRNA localization.
(A) Schematic illustration of the timing of primary mouse hippocampal neuron transfection and DEX treatment.
(B) Representative maximum intensity confocal images of MS2-transfected neurons treated with VEH (left) or DEX (right). Scale bar, 20 μm. Green, MS2-reporter; red, Synapsin-mCherry reporter; white, MAP2 (postmitotic neuron marker).
(C and E) Quantification the GFP fluorescence intensity along the length of primary dendrites of GFP+/mCherry+/MAP2+ neurons transfected with 31 CGG (C) or 0 CGG (E) plasmids. Shaded area indicates SEM.
(D and F) Summation of the fluorescence intensity in primary dendrites, normalized for dendritic length in 31 CGG (D) or 0 CGG (F) transfected neurons. Each data point represents single neurons (R1–R3). Data in (C–F) are from N = 3 independent neuronal isolations/biological replicates (19–30 neurons per replicate). Values were normalized to vehicle (VEH) condition for each batch of neurons. (C and E) Two-way ANOVA, significant min effect of treatment *p < 0.05, **p < 0.01. (D and F) Welch’s t test; *p < 0.05; ***p < 0.005.
Removal of CGG repeats from the FMR1 5′ UTR leads to altered cellular stress and response to GR activation
Mislocalization of mRNA within neurons is detrimental to important cellular processes, such as axon guidance,53 local mRNA translation and protein synthesis,54 long-term potentiation,54 and dendritic morphogenesis.55 Further, mRNA localization has been linked to cellular stress in non-neuronal cell types.56 We, therefore, tested whether the treatment of human 0CGG neurons with DEX impacted cellular stress. We first confirmed that hESC-derived neurons express GR (Figure S6A) and that long-term treatment with various doses of DEX did not have significant effects on viability (Figure S6B). We selected a concentration of DEX (1 μM) that activates the high-affinity GR, but not the low-affinity mineralocorticoid receptor57 and has been previously used on human neural cultures.58,59 The treatment of immature neurons with 1 μM DEX for 24 h upregulated mRNA levels of PTPN11, a previously reported DEX response gene,60,61 in both control and 0CGG neurons (Figure S6C), confirming GR activation. Because our earlier results in hESC-derived neurons were consistent across both hESC lines examined (Figures 1 and S1–S3), we combined these two lines in subsequent analyses.
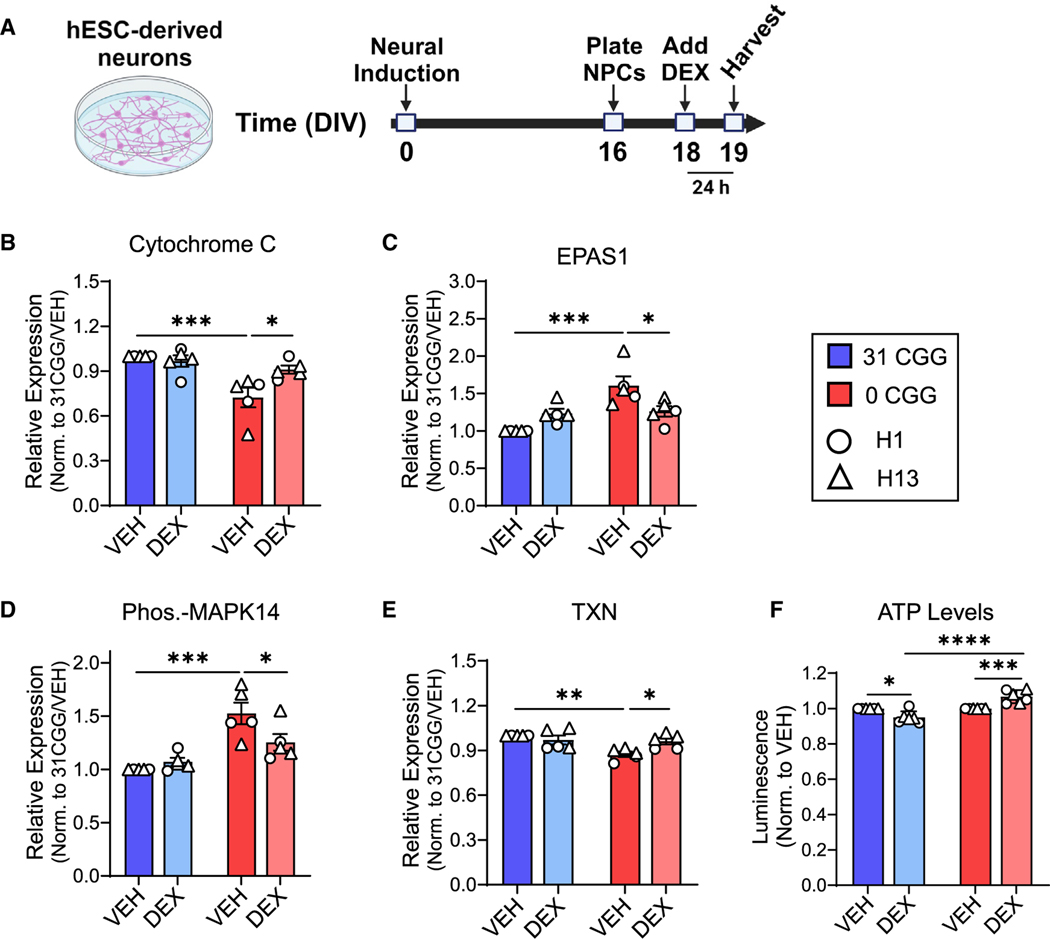
Because stress or GR signaling affects multiple cellular stress pathways,62–64 we analyzed 26 different cellular stress proteins in DEX-treated neurons using protein arrays (Figures 5A and S6D; Table S3). Differences in cellular stress proteins were already apparent at the baseline (VEH) level, with six proteins showing significantly lower levels and seven proteins showing significantly higher levels in 0CGG neurons compared with control neurons (31CGG-VEH vs. 0CGG-VEH) (Figures 5B–5E, S6E, and S6G). Interestingly, among the 13 proteins differentially expressed in 0CGG neurons at baseline, the expression of four proteins (cytochrome c, EPAS1 (HIF2α), phosphorylated p38α/MAPK14, and thioredoxin [TXN]) returned to control levels after DEX treatment (significant difference in 0CGG-DEX vs. 0CGG-VEH, no difference in 0CGG-DEX vs. 31CGG-VEH) (Figures 5B–5E). The expression of an additional five proteins (ADAMTS1, BCL2, CA9, COX2, and phosphorylated HSPB1) was partially rescued by DEX (no significant difference in 0CGG-DEX vs. 31CGG-VEH) (Figure S6E). Interestingly, phospho-TP53 was partially rescued in 0CGG neurons, but also significantly increased by DEX treatment in control neurons. FABP1 was differentially expressed in response to DEX in control neurons, but not in CGG-deficient neurons, whereas HIF1α was significantly increased in DEX-treated control neurons compared with DEX-treated CGG-deficient neurons (Figure S6F). Three proteins that were altered at baseline (CDKN1A/p21, CDKN1B/p27, and phospho-JNK/MAPK8) did not respond to DEX in either genotype (Figure S6G). Therefore, out of 26 stress proteins analyzed by arrays, 15 proteins were affected by either CGG repeats or by DEX treatment (see Table S3). Additionally, examination of cytochrome c, EPAS1, and TXN levels in neurons treated with DEX for 1 week showed that longer DEX treatment had a similar effect as 24 h treatment, with all three proteins partially or fully returning to control levels after DEX treatment (Figures S6H–S6J). These data support a potential role of FMR1 CGG repeats in regulating the cellular stress of neurons in response to GR activation.
Figure 5. Removal of CGG repeats from the FMR1 5’ UTR leads to altered cellular stress and response to GR activation.
(A) Schematic showing the timing of DEX treatment in hESC-derived neurons.
(B–E) Quantification of total protein levels in VEH-treated versus DEX-treated neurons: cytochrome c (B), EPAS1 (C), phosphorylated-MAPK14 (D), and TXN (E). Data are from n = 5 independent neuronal differentiations from N = 2 cell lines.
(F) ATP levels in DEX-treated neurons. (Left) H1 and H1–0CGG. (Right) H13 and H13–0CGG. Data shown are from n = 3 independent neuronal differentiations per line. DEX data point for each batch of cells was normalized to matched VEH control. Error bars indicate SEM. (B–F) Two-way ANOVA with Tukey’s multiple comparison’s test; *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
FMRP is involved in mitochondrial function during neuronal development,65,66 and several mitochondrial stress proteins were altered in 0CGG neurons (cytochrome c, TXN, cyclo-oxygenase 2). This led us to measure adenosine triphosphate (ATP) levels in control and CGG-deficient neurons in response to DEX. We observed that cellular ATP levels decreased in control neurons but increased in 0CGG neurons, in response to DEX (Figure 5F) (31CGG-DEX vs. 0CGG-DEX). Taken together, these results indicate that the removal of the CGG repeats from the FMR1 5′ UTR affects cellular stress pathways and leads to differential responses to GR activation.
Removal of CGG repeats from the FMR1 5′ UTR affects GR subcellular localization after DEX treatment
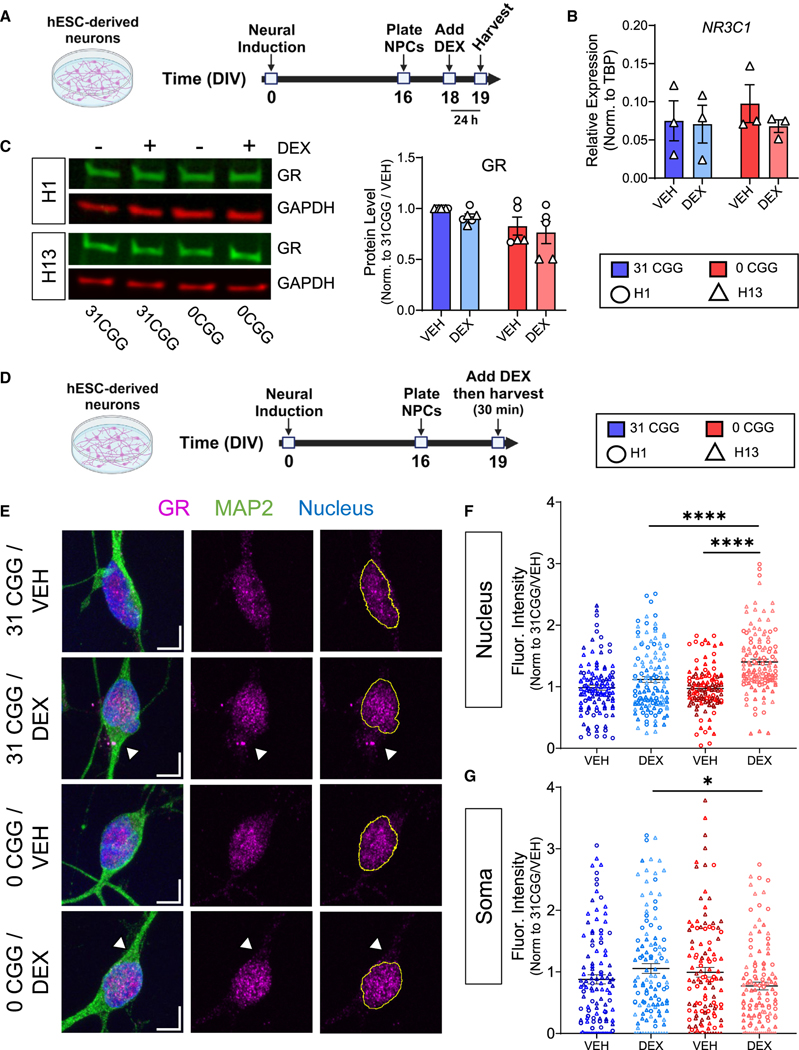
Altered response to GR activation can result from multiple mechanisms, including downregulation of the transcriptionally active GR alpha isoform (GRα), upregulation of non-transcriptionally active GR isoforms (GRβ, GRγ, and GR-A), or impaired GR nuclear translocation.67–69 We first tested the hypothesis that 0CGG neurons had altered levels of GR mRNA (NR3C1) or protein, which could be responsible for the altered DEX response. No differences in total NR3C1 mRNA or GR protein levels were detected between 0CGG and control neurons at baseline or after DEX treatment (Figures 6A–6C).
Figure 6. Removal of CGG repeats from the FMR1 5’ UTR affects GR subcellular localization after DEX treatment.
(A) Schematic showing the timing of DEX treatment in hESC-derived neurons.
(B) qRT-PCR data showing NR3C1 mRNA levels in H13 and H13–0CGG neurons. n = 3 independent differentiations. Error bars indicate SEM.
(C) GR protein levels in DEX-treated hESC-derived neurons. (Left) Representative western blot images from H1 and H13 neurons. (Right) Quantification of GR protein levels. n = 5–6 independent batches of differentiation from N = 2 cell lines. Data shown are normalized to 31 CGG-VEH condition. Error bars indicate SEM.
(D) Schematic showing the timing of acute DEX treatment of hESC-derived neurons.
(E) Representative confocal images of the GR receptor expression in MAP2+ neurons. Scale bars, 5 μm. Arrowheads demonstrate differences in soma GR signal in DEX-treated 31CGG and 0CGG neurons. Magenta, GR; green, MAP2 (postmitotic neuron marker); blue, nuclear staining using Hoechst.
(F and G) Quantification of GR fluorescent signal in nucleus (F) and soma (G). n = 111–125 individual neurons from N = 2 cell lines. (B and C) Two-way ANOVA. (F) Brown-Forsythe and Welch ANOVA test followed by Games-Howell’s multiple comparison’s test, ****p < 0.001. (G) One-way ANOVA followed by Tukey’s multiple comparison’s test, *p < 0.05.
GR is predominantly localized to the cytosol when not bound by a ligand, but rapidly translocates to the nucleus following binding by its agonists cortisol or DEX, where it regulates transcription, before eventually being exported back into the cytosol.70 Shuttling of GR between the nucleus and cytoplasm, and the balance between nuclear import and export, ultimately determine its subcellular localization.71 We hypothesized that GR localization or translocation may be altered in 0CGG neurons. Therefore, we assessed the localization of GR in the nucleus and soma of human neurons treated with vehicle or DEX for 30 min (Figures 6D and 6E). Although baseline levels of GR in the soma and nucleus did not differ between control and 0CGG neurons (Figures 6F and 6G), 0CGG neurons had more GR in the nucleus after DEX treatment, whereas control neurons showed no difference (Figure 6F). Conversely, DEX-treated 0CGG neurons had lower GR levels in the soma compared with DEX-treated control neurons (Figure 6G). These results indicate an increased nuclear GR localization after DEX treatment in 0CGG neurons, providing potential mechanistic insight into the altered stress response of these neurons.
Removal of CGG repeats from the FMR1 5′ UTR leads to altered FMRP localization and decreased levels of GR chaperone protein HSP90α after DEX treatment
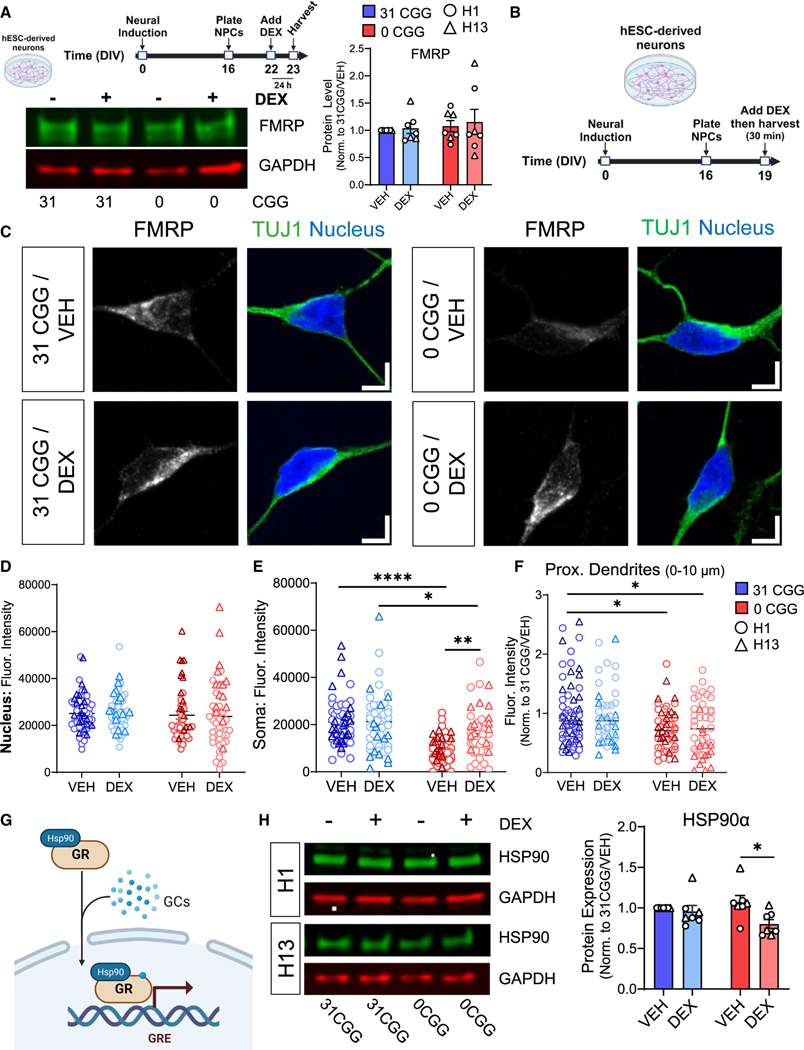
FMRP is an RNA binding protein that binds to many mRNAs in human neurons, thereby affecting their translation and function.35,66,72 The mRNA encoding GR, NR3C1, is predicted to be an FMRP target in human neurons (Figure S6K),35 as well as in mouse and human brain tissue.73–75 We reasoned that the altered GR localization after DEX treatment could be a result of aberrant FMRP protein localization resulting from mislocalization of FMR1 mRNA, because total levels of FMRP are not significantly different between 0CGG and control neurons at baseline or after DEX treatment (Figure 7A). We, thus, defined the subcellular localization of FMRP in human neurons at baseline and after acute DEX treatment (Figures 7B and 7C). There were no differences in the amount of nuclear FMRP in any of the conditions examined (Figure 7D). Interestingly, 0CGG neurons had significantly less FMRP in the soma (Figures 7E and S7A) and in proximal dendrites (Figures 7F and S7B) compared with control neurons, opposite to the localization of human FMR1 MS2 reporters (Figure 2). Additionally, in response to DEX, FMRP localization did not change in control neurons, but was significantly increased in the soma of 0CGG neurons (Figures 7E and S7A) (0CGG-VEH vs. 0CGG DEX). Therefore, the removal of CGG repeats from FMR1 affects the localization of both FMR1 mRNA and its protein product FMRP at baseline, and in response to GR activation.
Figure 7. Removal of CGG repeats from the FMR1 5′ UTR leads to decreased levels of GR chaperone protein HSP90α following DEX treatment.
(A) Total FMRP levels in neurons treated with DEX for 24 h. (Top left) A schematic diagram showing the timeline for DEX treatment. (Bottom left) Representative western blots from H13 and H13–0CGG neurons treated with DEX. (Right) Quantification of FMRP protein levels from Western blots. n = 7 technical replicates from N = 2 isogenic pairs of cells (three independent batches of differentiation and DEX treatment per line). Error bars indicate SEM.
(B) Schematic showing the timing of DEX treatment in hESC-derived neurons.
(C) Representative confocal images showing FMRP localization in TUJ1+ (green) neurons. (Left) 31 CGG. (Right) 0 CGG. Blue, nuclear staining using Hoechst. Scale bar, 5 μm.
(D–F) Quantification of FMRP signal in the nucleus (D), soma (E), and proximal dendrites (first 10 μm proximal to soma, F). Data shown in (F) are normalized to 31 CGG-VEH condition. (D and E) n = 48–57 neurons from N = 2 cell lines. (F) n = 44–60 neurons from N = 2 cell lines.
(G) Schematic illustrating the role of chaperone protein HSP90α in GR nuclear translocation.
(H) HSP90α protein levels in DEX-treated hESC-derived neurons. (Left) Representative western blots from H1 and H13 neurons treated with DEX. (Right) Quantification of GR protein levels. n = 7 independent batches of differentiation from N = 2 cell lines. Error bars indicate SEM. (A, D–F, and H) Two-way ANOVA followed by Tukey’s multiple comparison’s test, *p < 0.05, **p < 0.01, ****p < 0.001.
As GR mRNA or protein levels (Figures 6A and 6B) did not differ at baseline or in response to DEX between 0CGG and control neurons, we hypothesized that the differential response to GR activation in 0CGG neurons is due to misregulation of FMRP targets involved in GR response, and examined other FMRP targets that could be mediating this difference. GR translocation is a highly regulated, multi-stage process involving numerous chaperone proteins that stabilize the structure of GR before and after ligand binding, followed by the formation of additional protein complexes that allow GR to be imported into the nucleus through the nuclear pore complex.76,77 Interestingly, many of the proteins involved in this process are mRNA targets of FMRP (Figure S7C).35,74,75,78 We quantified the levels of six proteins known to be involved in GR translocation and that are, except for HSP70, FMRP targets (Figures S7C and S7D). There were no significant differences between 0CGG and control neurons in the levels of importin β1 (KPNB1),79 HSP90β (HSP90AB1),80 dynein cytoplasmic heavy chain 1 (DYNC1H1),81 importin 7 (IPO7),79 or HSP7082 at baseline or in response to DEX (Figures S7E–S7H). There were also no baseline expression differences in HSP90α (HSP90AA180,82 (Figure 7H) (31CGG-VEH vs. 0CGG-VEH). However, DEX treatment significantly decreased HSP90α levels in 0CGG neurons, but not control neurons (Figure 7H) (0 CGG-VEH vs. 0 CGG DEX; 31 CGG-VEH vs. 31 CGG-DEX). 0CGG neurons exposed to 7-day DEX treatments also exhibited decreased HSP90α levels, but instead had decreased GR levels in the nucleus (Figures S7I and S7J), which may indicate that less GR can be activated by additional DEX exposure due to the decreased levels of HSP90α chaperone. Therefore, FMR1 CGG repeats have a significant effect on the levels of GR chaperone HSP90α in response to DEX.
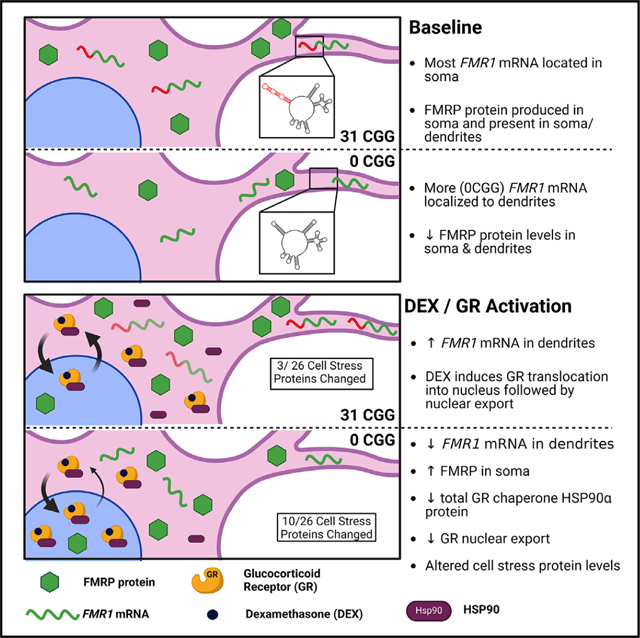
Based on our findings, we propose the following model: In immature neurons, FMR1 mRNA translation mainly occurs in the soma, and then FMRP protein is transported into dendrites. Normal length CGG repeats in the FMR1 5′ UTR may ensure the correct localization of FMR1 mRNA and, therefore, FMRP protein,83 which is important for temporal and spatial regulation of specific FMRP mRNA targets (such as HSP90AA1/HSP90α). Regulation of HSP90α levels, in turn, allows for proper shuttling of GR into and out of the nucleus in response to events that trigger GR activation, such as environmental stress. In the absence of CGG repeats, FMR1 mRNA is prematurely localized to dendrites, decreasing FMRP protein production and expression of proteins in cellular stress pathways. Upon GR activation, 0CGG neurons fail to export FMR1 mRNA to dendrites, leading to increased FMRP protein in the soma where it represses translation of its mRNA target HSP90AA1. Decreases in HSP90α lead to increased retention of GR in the nucleus, contributing to the difference in response to DEX treatment in 0CGG neurons. This model may help to explain the observation that patients with shorter CGG repeats in the FMR1 gene have adverse effects of long-term life stress compared with individuals with normal range CGG repeat lengths.
DISCUSSION
The function of STRs in the human genome remains largely unknown, despite their abundance and disease association.1–4,9 Here, we used human stem cell-derived neurons and mouse primary neurons to demonstrate that the CGG repeats in the FMR1 gene are important for temporal and spatial localization of FMR1 mRNA and protein, which in turn is important in the homeostatic response of neurons to cellular stress signals. Our study provides direct evidence for a role of normal length CGG repeats in the FMR1 gene in neurons. Our results not only improve our understanding of the FMR1 gene in normal development, but also provide critical insight for designing CRISPR-based gene therapy approaches for FXS, in which the expanded CGG repeats are removed or contracted in order to reactivate the FMR1 gene.29,30,84
Removal of CGG repeats from the FMR1 5′ UTR did not affect levels of FMR1 mRNA or protein in hESCs, NPCs, or immature neurons. These results are consistent with normal FMR1 mRNA or protein levels in rare patients lacking CGG repeats due to de novo deletions within the 5′ UTR,85–87 but are inconsistent with studies examining the effects of FMR1 CGG repeat length on translation using luciferase reporters.25 The discrepancy could be due to differences in methodology, use of different cell types, or maturation state of the neurons.
Since CGG repeats or 5′ UTR sequences in other genes affect mRNA localization,28,36–38,45 we hypothesized that removal of CGG repeats causes mislocalization of FMR1 mRNA. Mislocalization of mRNA within neurons can negatively affect cellular processes such as axon guidance,53 local mRNA translation and protein synthesis,54 long-term potentiation,54 and dendritic morphogenesis.55 Here, FMR1 transcripts preferentially localized to dendrites in the absence of CGG repeats in hESC-derived immature neurons, as well as mouse neurons expressing the human FMR1 5′ UTR. The FMRP protein level was decreased in the soma and proximal dendrites in human neurons lacking CGG repeats. Although 3′ UTRs are known to regulate mRNA localization, 5′ UTRs can also modulate, and are sometimes necessary for, the correct subcellular localization of some mRNAs in neurons.36–38 In addition, the differences between our results and published studies further support the complexity of regulation for FMR1 mRNA. Muslimov et al.28 showed that 24 CGG repeats added to the 5′ UTR of the ACTA1 mRNA promoted its localization to the dendrites of rat sympathetic neurons as compared with the addition of 24 CCC repeats. Rodriguez et al.27 constructed a reporter containing both the 5′ UTR and 3′ UTR regions of the FMR1 transcript with either 0CGG or 20CGG repeats and showed that both exhibited similar levels of dendritic localization in primary rat hippocampal neurons. The results of these studies suggest that the length of CGG repeat, the 3′ UTR of FMR1, and the cell type may influence the localization of FMR1 mRNA. We, therefore, believe that published findings and our own are complimentary and illustrate that mRNA localization is highly gene and context specific. Our results suggest that the CGG repeats in the 5′ UTR of FMR1 play a role in the proper localization of FMR1 mRNA and protein during early neuronal development.
The development-dependent mislocalization of FMR1 mRNA in 0CGG neurons suggests that the 5′ UTR is only one of multiple mechanisms by which developing neurons control the localization of FMR1 mRNA transcripts. Neurons may employ developmental stage-specific mechanisms to localize FMR1 mRNA, with the 5′ UTR being more important in immature neurons and less important later in development. Alternatively, removal of the CGG repeats could affect the splicing of FMR1 mRNA,88 leading to increased production of mRNA transcripts that are exported to dendrites during early neuronal development, similar to the brain-derived neurotrophic factor (BDNF) gene.36,89 Future studies should also evaluate the role of candidate mRNA binding proteins5,90,91 in the transport and splicing of 0CGG versus control FMR1 mRNA in both immature and mature neurons. Additionally, mislocalization of FMR1 mRNA in older 0CGG neurons may still occur, but require specific stimuli, such as electrical or chemical stimulation,92 or exposure to specific environmental stressors.
One mechanism by which the CGG repeats in the 5′ UTR may modulate the localization of FMR1 mRNA is through the GQs formed by the CGG repeats. FMRP binds to GQs in its target mRNAs,93,94 and at least one other neuronal mRNA is dependent upon GQs in its 5′ UTR for proper dendritic localization.45 Treatment of neurons with TMPyP4, a compound that destabilizes the GQs formed by the FMR1 CGG repeats,95–97 increases the dendritic export of control length CGG repeats, indicating that the structure formed by the CGG repeats is likely important for its localization. However, since TMPyP4 destabilizes many mRNA GQs,49,50 future studies should replicate these findings using an approach that specifically targets the FMR1 CGG repeats.
The function of the CGG repeats in the FMR1 5′ UTR warrants investigation, given their connection with multiple facets of human health.17–22 The molecular biology of the CGG repeats in the FMR1 5′ UTR is established in the context of the premutation (>55 repeats) and full mutation (>200 repeats),16,98,99 yet few studies have examined the cellular and molecular function of these repeats in the low (6–23 repeats), normal (24–40 repeats), or gray zone (41–54 repeats) ranges.25,27,28,91,95 FMR1 CGG repeat length is correlated with long-term health outcomes in individuals with higher life stress, including associations with cognitive functioning, depression, anxiety, and balance.23,24 To model the potential interaction between FMR1 CGG repeat length and life stress in vitro, we tried to mimic the events that occur following life stress in vivo by treating neurons with DEX at a concentration known to activate GR, but not the mineralocorticoid receptor, for a sustained period. This approach has been successfully applied to in vitro models of post-traumatic stress disorder,100,101 and several studies have shown an association between GR102 or cortisol103,104 and FXS. Interestingly, stress or DEX treatment differentially affected the localization of FMR1-MS2 RNA containing no CGG repeats compared with normal repeats, indicating a connection between FMR1 mRNA localization, CGG repeats, and GR activation. Our results also establish a link between normal length FMR1 CGG repeats, and cellular stress response, as CGG-deficient neurons exhibit changes in multiple cellular stress proteins at baseline and in response to DEX treatment.
How does mislocalization of FMR1 mRNA lead to altered GR localization? FMRP binds hundreds of target mRNAs in neuronal cells, many of which have been implicated in cellular stress.35,65,66,74,105 Although FMR1 mRNA and protein tend to be localized to similar compartments in neurons, mGluR activation in rat hippocampal neurons decreases the levels of FMRP protein, but not mRNA, in synapses,92 suggesting that their localization can be uncoupled in response to certain stimuli. Although it is unclear how GR activation changes dendritic FMR1 mRNA localization, potential mechanisms that can be explored include altered formation of RNA or stress granules,106 binding of GR to FMR1 mRNA directly,107 or modulation of local protein synthesis by GR via its regulation of BDNF.108–110
Based on our findings, we propose that the mislocalization of FMRP protein in 0CGG neurons causes misregulation of at least some FMRP target mRNAs involved in GR signaling. This model is supported by our finding that DEX treatment decreased total levels of HSP90α (encoded by HSP90AA1), which is required for GR nuclear translocation and is one of the key factors that regulates GR chromatin binding in the nucleus.111,112 Our results showing decreased GR in the cytoplasm of DEX-treated 0CGG neurons is consistent with findings in human podocytes with short hairpin RNA knockdown of HSP90.113 We believe that increased FMRP in the soma of 0CGG neurons in response to DEX leads to increased binding of FMRP to HSP90AA1 mRNA and its subsequent translational repression.
In summary, we demonstrate that CGG repeats in the 5′ UTR of FMR1 at normal, non-disease-associated lengths, regulate FMR1 localization, cellular stress responses, and GR signaling during early neuronal development. Our results suggest potential mechanisms by which FMR1 CGG repeat length polymorphism in the low, normal, and gray zones could lead to the clinically-relevant differences seen across individuals in the context of life stress. Given the ever-increasing number of polymorphic repeats in non-coding regions and disease-associated repeat expansion genes being identified, our results also highlight the importance of studying the molecular biology and function of repeats, both in the normal and disease contexts.
Limitations of the study
This study is not without limitations. Human neuron studies were performed in predominantly excitatory neurons; therefore, whether localization of endogenous FMR1 mRNA in stem cell-derived inhibitory neurons behaves similarly is not known. This study only used male hESC lines, since they only have one copy of the FMR1 gene. Future studies should examine the consequences of removing CGG repeats from either one or both copies of FMR1 in neurons derived from female hPSC lines. Future studies should explore how GR activation via DEX leads to the altered subcellular localization of FMR1 mRNA and protein in 0CGG neurons, which remains unclear based on the findings presented here. The correlation between low CGG repeat lengths and response to chronic life stress led us to assess how 0CGG neurons may respond to cellular stress signals. We acknowledge that it is not possible to model life stress in a dish. Therefore, we studied one molecular aspect of life stress, GR activation, and whether it had any relevance to CGG repeats in FMR1. Finally, we do not know whether there are functional outcomes of the transient FMR1 mislocalization phenotype caused by removal of CGG repeats, so further characterization of neurons lacking FMR1 CGG repeats is needed to determine whether short-term mislocalization of FMR1 mRNA and protein has long-term consequences.
STAR★METHODS
RESOURCES AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Xinyu Zhao (xinyu.zhao@wisc.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact. Plasmids generated in this study have been deposited to Addgene. Human 0CGG hESC lines generated in this study have been deposited to WiCell.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. Protein array data have been reported in Table S3.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Generation of 0CGG hESC lines
H1–0CGG and H13–0CGG hESC lines were generated using CRISPR/Cas9-based genome editing using published methods.114 Briefly, dissociated single hESCs were electroporated with lentiCRISPRv1 (Addgene #49535)117 containing sgRNA and donor plasmid. Donor plasmids were constructed by modifying the OCT4–2A-eGFP-PGK-Puro plasmid (Addgene #31938)115 resulting in FMR1 5′UTR donor plasmids containing either 31CGG or 0CGG. Cells were selected with 0.5 μg/mL of puromycin 24–72 h post electroporation. Approximately 2 weeks later, colonies were picked for expansion and screening. Primers spanning the CGG repeat within the 5′UTR of the FMR1 gene were used to screen colonies by conventional PCR. Putative positive colonies were then sequenced by Sanger sequencing to confirm homology-directed repair. To confirm a lack of off-target effects from genome editing, primers were designed to sequence the top 5 predicted off-target sites (Benchling) by PCR and Sanger sequencing. hESCs were also karyotyped (WiCell, Madison, WI) to confirm a lack of large insertions or deletions. Finally, all hESC lines were confirmed mycoplasma negative at several passages during the experiments (WiCell, Madison, WI). See Table S2 for primer and RNA sequences.
Neuronal differentiation
hESCs were differentiated into forebrain glutamatergic neurons using a published protocol.35 Briefly, 5 days after passaging hESCs were maintained in neural induction medium (NIM) containing dual SMAD and Wnt inhibitors (XLSB) for 10 days until neuroepithelium formed, at which point cells were passaged at high density using TrypLE Express onto Matrigel-coated plates in neural stem cell (NSC) media containing ROCK inhibitor. Cells were kept in NSC media for 7 days, at which point neural rosettes formed. Neural progenitor cells (NPCs) were harvested for analysis or plated for terminal differentiation onto Matrigel-coated plastic dishes or polyornithine/laminin-coated coverslips in NSC media containing ROCK inhibitor and Compound E. The next day, half the media was replaced with neural differentiation medium (NDM) containing Compound E. Half media changes (NDM only) were performed every 3–4 days thereafter.
Mouse husbandry and isolation of mouse hippocampal neurons
We performed all procedures involving live mice in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the protocols approved by the University of Wisconsin-Madison Animal Care and Use Committee. The C57BL/6J mice (JAX stock #000664) were purchased from Jackson Laboratory and maintained in house. Mice were housed in groups and maintained on a 12-h light/dark cycle with food and water available ad libitum. Mouse hippocampal neurons were isolated from WT P0 neonate mice and were grown as dispersed cultures as described previously.65,118
METHOD DETAILS
CGG repeat analysis
Genomic DNA (gDNA) was isolated from H1 and H13 hESCs as follows: Cells were incubated in lysis buffer (100 mM NaCl, 10 nM Tris HCl, 25 mM EDTA, and 0.5% SDS) and Proteinase K (20 mg/mL) at 55°C overnight. Phase separation of gDNA was achieved by adding phenol, vortexing, and collecting aqueous layer, followed by adding chloroform, vortexing, collection of aqueous layer, and precipitation with 100% ethanol. gDNA quality was confirmed by NanoDrop (260:280 value ≥ 1.89). CGG repeat analysis of gDNA (50–100 ng/μL) was performed by Rush University Medical Center Molecular Diagnostics Laboratory using a published protocol.119
qRT-PCR
RNA was isolated from cells using the Direct-zol RNA microprep kit (Zymo). Reverse transcription was carried out on 500 ng of RNA using the Prime Script RT kit (Takara). To quantify mRNA levels using real-time PCR, first-strand cDNA was amplified with gene-specific primers and universal SYBR Green PCR Supermix (Bio-Rad, #172–5124) using the Step-1 Real-Time PCR System (Applied Biosystems). GAPDH was used as a housekeeping gene for quantification. See Table S2 for primer sequences.
Western Blotting
Cell lysates were prepared by lysing cells on ice in cold RIPA buffer containing protease inhibitors for 30 min (50 mM Tris HCl, 150 mM NaCl, 1% NP-40, 0.5% SDC, 0.1% SDS) followed by centrifugation. Protein concentration was determined using the Bradford Assay (Bio-Rad). 50 μg of protein was loaded per lane. Western blots were run using Mini-PROTEAN TGX gels (4–20%, Bio-Rad) followed by standard transfer onto nitrocellulose membranes. Membranes were blocked in 5% milk in TBS-T for 1 h at room temperature, followed by incubation in blocking buffer containing primary antibodies overnight at 4°C. Membranes were washed, incubated in blocking buffer containing secondary antibodies for 1 h at room temperature, washed again, then imaged using the LI-COR Odyssey imaging system. IRDye secondary antibodies were used at a concentration of 1:10,000 (LI-COR). Western blots were quantified using Image Studio Lite (LI-COR).
| Primary antibodies and dilutions | ||
|---|---|---|
|
| ||
| Marker | Antibody information | Application |
| FMRP | ThermoFisher MA5–15499 (mouse) | WB (1:1000; Figure S1) |
| FMRP | Millipore-Sigma MAB2160 (mouse) | WB (1:1000; Figure S1; Figure 7A) |
| FMRP | Santa Cruz sc101048 (mouse) | ICC (1:100; Figure 7C) |
| GAPDH | ThermoFisher TAB1001 (rabbit) | WB (1:5000) |
| GAPDH | ThermoFisher MA5–15738 (mouse) | WB (1:5000) |
| TUJ1 | Biolegend 802001 (rabbit) | ICC (1:10000) |
| MAP2 | Sigma M1406 (mouse) | ICC (1:500) |
| GFAP | Agilent Z0334 (rabbit) | ICC (1:1000) |
| Glucocorticoid Receptor (GR) | Cell Signaling 3660 (rabbit) | ICC (1:100) |
| Glucocorticoid Receptor (GR) | Proteintech 24050–1-AP (rabbit) | WB (1:5000) |
| DYNC1H1 | Proteintech 12345–1-AP (rabbit) | WB (1:1000) |
| HSP90 | Proteintech 13171–1-AP (rabbit) | WB (1:5000) |
| HSP90AB1 | Proteintech 11405–1-AP (rabbit) | WB (1:5000) |
| KPNB1 | Proteintech 10077–1-AP (rabbit) | WB (1:2000) |
| IPO7 | Proteintech 28289–1-AP (rabbit) | WB (1:1000) |
| HSP70 | Proteintech 10995–1-AP (rabbit) | WB (1:5000) |
| Cytochrome c | Proteintech 10993–1-AP (rabbit) | WB (1:1000) |
| EPAS1 (HIF2α) | Proteintech 26422–1-AP (rabbit) | WB (1:1000) |
| TXN | Proteintech 14999–1-AP (rabbit) | WB (1:2000) |
ICC
NPCs were plated onto poly-ornithine and laminin-coated glass coverslips at a density of 100,000 cells/coverslip, or onto 8 well glass chamber slides (Nunc Lab-Tek II, ThermoFisher Scientific) at a density of 150,000 cells/well. Neurons were fixed in 4% PFA for 10 min at room temperature, blocked and permeabilized in blocking buffer (5% NGS and 0.2% Triton X-100 in PBS), then incubated in blocking buffer containing primary antibodies overnight at 4°C. Cells were then washed 3 × 5 min, incubated in blocking buffer containing secondary antibodies for 1 h at room temperature, washed 2 × 5 min, then incubated with DAPI or Hoechst for 5 min at room temperature. Following another wash (1 × 5 min), cells were mounted using PVA-DABCO. Slides were allowed to dry overnight at room temperature, protected from light, then were stored at 4°C until imaging.
RNA FISH plus Immunocytochemistry (ICC)
NPCs were plated onto poly-ornithine and laminin-coated glass coverslips at a density of 75,000 cells/coverslip. RNA FISH was performed with the ViewRNA ISH Cell Assay (ThermoFisher) using commercially available probes for FMR1 and ACTB mRNAs (ThermoFisher). The manufacturer’s protocol was followed with the following modification: protease digest (Step 4) was omitted (i.e., hybridization with probes (Step 5) occurred immediately following permeabilization (Step 3)). Following the label probe hybridization and wash steps (Steps 11 and 12), neurons were incubated in blocking buffer (5% NGS in PBS-T) for 1 h at room temperature, then incubated in blocking buffer containing primary antibody (mouse anti-MAP2, M1406, Sigma, 1:500) overnight at 4°C. Coverslips were washed then incubated in blocking buffer containing secondary antibody (Alexa Fluor 568 Goat Anti-Mouse, Invitrogen, 1:500) for 1 h at room temperature. Coverslips were washed briefly, incubated with DAPI for 5 min at room temperature, washed again, then mounted onto glass slides using PVA-DABCO.
Confocal Microscopy & image analysis (RNA FISH)
Confocal z series were acquired with a 60x oil immersion objective (NA 1.4) on an A1R-HD (Nikon) system. Images were acquired at 2048 x 2048 resolution at 1/8 frames per second, 0.15 μm interval. Eight to ten randomly chosen areas were imaged per coverslip and three coverslips were imaged per cell line. Images were analyzed using Fiji (ImageJ) software.120 z series images were reconstructed using the 3D Viewer Plugin. For each cell analyzed, 360° rotation images were recorded for each channel, as well as each combination of channels (red/green, green/blue, etc.). Total FMR1 mRNA puncta colocalizing with MAP2 staining were manually counted for each neuron, followed by quantification of puncta within the nucleus, soma, and in the neuronal processes.
Human FMR1 5′ UTR MS2 reporter system
Phage-ubc-nls-ha-tdMCP-gfp (MCP-GFP) plasmid was obtained from Addgene (Addgene #40649).116 The MS2-FMR1 plasmids were constructed by replacing the CMV promoter of phage-cmv-cfp-24xMS2 (Addgene #40651)116 with EFSns promoter, and replacing ECFP with human FMR1 5′ UTR and exon 1 containing either 31 CGG or 0 CGG repeats. Human FMR1 insertions were generated from cDNA that was amplified from H1 hESCs. pcDNA3 plasmid co-transfected with MCP-GFP served as a Nuclear localization positive control, and unmodified phage-cmv-cfp-24xMS2 (Addgene #40651) plasmid co-transfected with MCP-GFP served as a Cytoplasmic localization positive control (Figure S4).
Primary hippocampal neuron culture and transfection for MS2 experiments
Mouse hippocampal neurons were transfected as described previously.65,118 Briefly, neurons were transfected with plasmids using Lipofectamine 2000 (Thermo Fisher Scientific)) on DIV 4 as they were undergoing dendritic morphogenesis. 1uM TMPyP4 (Sigma 613560), 1uM dexamethasone (DEX; Sigma D4902), or vehicle (DMSO) were added to the cultures 24 h prior to transfection, then added to transfected neurons 6 h after transfection. Neurons were fixed with 4% paraformaldehyde at 72 h after transfection, unless started otherwise (Figure S4), then stained for MAP2 using the ICC protocol described above.
MS2 quantification
MS2 transfected-neurons were imaged on a Zeiss Apotome microscope with 20x objective. Intensity quantification of MS2-GFP in neurites was done using Fiji/ImageJ.120 For the time course (Figure S4), 13–16 neurons were imaged across two coverslips (1 batch). For all other experiments, 3 independent experiments were performed (18–30 neurons per batch). Only Map2+/Syn-mCherry+/MS2-GFP+ cells were chosen for analysis. Syn-mCherry was used for dendrite tracing using the SNT plugin in Fiji.121 GFP fluorescence intensity in traced primary dendrites was measured every 0.4 μm using the Plot Profile function in SNT using the following settings (Shape: None.; Radius: 0; Integration Metric: N/A; with Spatially calibrated distances selected). Background subtraction was performed by averaging the fluorescence intensity of 2–3 traced dendrites of Map2+/Syn-mCherry-/MS2-GFP- cells. Because TMPyP4-treated cells had increased GFP fluorescence overall (MS2+ and MS2-), sections of dendrites overlapping with other cells were omitted from analysis. Fluorescence intensity was quantified by examining the fluorescence intensity value at 10 μm intervals along the dendrite. The average intensity at each interval across primary dendrites was calculated for each neuron. Additionally, we quantified the total fluorescence intensity in primary dendrites by normalizing the total GFP signal in each dendrite (after background subtraction) to its total length [Σ(intensity measurements along length of dendrite) ÷ dendritic length in μm]. The number of MAP2+ cells and total number of nuclei per field (Figure S4B) was quantified in 5 fields per condition using the Cell Counter plugin in Fiji/ImageJ. Representative high-resolution maximum intensity projection images shown in figures were acquired on the Nikon A1R-HD confocal microscope as described above using 60x immersion objective and 0.3 μm z-interval distance.
Dendritic length quantification
Dendritic length of lentivirus- Syn-mCherry infected primary hippocampal neurons was measured using published methods.65,105 Briefly, morphological analysis of neurons that were positive for both Syn-mCherry and MS2-GFP was carried out using Neurolucida software (MBF Biosciences) using the same cells and images as "MS2 Quantification" (above).
RNA structure prediction
RNA structure of the human FMR1 gene with 31 CGG repeats or 0 CGG repeats was predicted using RNA fold with default parameters (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi)47 and GQRS Mapper (https://bioinformatics.ramapo.edu/QGRS/index.php).46 FMR1 mRNA sequence was accessed using Ensembl genome browser (https://www.ensembl.org).122 Ensembl transcript ENST00000370475.9 (FMR1–205) was used for RNA structure prediction.
Expression of cellular stress proteins
NPCs were plated for terminal differentiation as described above, then treated with vehicle (DMSO) or 1 μM DEX starting 2 days after plating (Day 18). 24 h after treatment, cells were harvested in PBS, pelleted, and stored at −80°C until needed. Cellular stress proteins were analyzed using the Human Cell Stress Proteome Profiler Array (R&D Systems ARY018) according to manufacturer’s instructions. Briefly, cell pellets were lysed in Lysis Buffer 6 (R&D Systems) containing 10 μg/mL each of aprotinin (Sigma A6279), leupeptin (Tocris 1167), and pepstatin (Tocris 1190) for 30 min at 4°C, centrifuged, and the supernatant collected. Bradford Assay was used to determine the total protein concentration, as described above. Membranes were blocked with Array Buffer 6 (R&D Systems) for 1 h at room temperature. During this time, cell lysates (400 μg of total protein) were incubated with Array Buffer 4 (R&D Systems) and Detection Antibody Cocktail (R&D Systems) for 1 h at room temperature. Membranes were then incubated with protein lysates on a shaker overnight at 4°C, washed 3 × 10 min with 1x Wash Buffer (R&D Systems) at room temperature, incubated with Streptavidin-HRP diluted in Array Buffer 6 for 30 min at room temperature, then again washed 3 x 10 min. To image membranes, Chemi Reagent Mix was added to each membrane for 1 min, then membranes were imaged on the c600 Phosphorimaging System (Azure Biosystems). A list of the cellular stress markers included on the assay can be found below. Array images were quantified using densitometry on Fiji/ImageJ software. Each protein array contained n = 2 technical replicates for each protein/antibody, which were averaged for each sample.
| Proteins included on human cellular stress array | ||
|---|---|---|
| ADAMTS1 | EPAS1 (HIF2α) | Phosphorylated-p38α (MAPK14) (T180/Y182) |
| BCL2 | Phosphoylated-HSPB1 (HSP27) (S78/S82) | Phosphorylated-p53 (TP53) (S46) |
| CA9 | HSPD1 (HSP60) | PON1 |
| CITED2 | HSPA1A (HSP70) | PON2 |
| COX2 | IDO1 | PON3 |
| Cytochrome c (CYCS) | Phosphorylated-Pan JNK (MAPK8) (T183/Y185) | TXN (Thioredoxin) |
| DKK4 | NFKB1 (NFκB) | SIRT2 |
| FABP1 | CDKN1A (p21/CIP1) | SOD2 |
| HIF1A (HIF1α) | CDKN1B (p27) | |
To examine the expression of cellular stress proteins following 1 week of DEX treatment, NPCs were plated for terminal differentiation at Day 16 onto Matrigel-coated plates, then treated with 1 μM DEX on Days 22, 25, and 28. Neurons were harvested at Day 29 by removing media, then collected in 1 mL PBS, centrifuged, supernatant removed, and cell pellets stored at −80°C until lysate preparation. Expression of Cytochrome c, EPAS1, and TXN were then assessed by Western Blot as described above (see Western Blotting).
Cell viability and ATP levels
ATP levels were measured using the Cell Titer GLO 2.0 assay (Promega) according to manufacturer’s instructions and our publication.66 Briefly, NPCs were plated for terminal differentiation as described above onto Matrigel-coated 96 well tissue culture plates (25,000 cells/well). Cells were treated with vehicle or 1 μM DEX at Day 18 (same as for Cell Stress Array above) for 24 h. Cells were placed at room temperature to equilibrate for 30 min, then Cell Titer Glo reagent was added equal to the volume of medium in each well. The plate was placed on an orbital shaker for 2 min to induce cell lysis, then incubated, protected from light, for 10 min at room temperature. Luminescence was recorded using the pre-programmed CellTiter-GLO settings on a GloMax plate reader (Promega).
Microscopy & image analysis (GR localization)
NPCs were plated for terminal differentiation onto glass chamber slides as described above. Four days after plating for terminal differentiation (Day 19), cells were treated with vehicle or 1 μM DEX for 30 min (Figure 6) or for 72 h (Figure S7I) prior to fixation (4% PFA for 10 min). ICC was performed as described above for GR and MAP2. Slides were imaged on the Nikon ECLIPSE Ti2-E widefield fluorescence microscope, equipped with a Multi-wavelength LED Light Engine. Multiplane 2048x2048 images were obtained using 100x oil immersion objective (NA 1.45) with 0.3 μm Z-interval. 10 to 19 images (different fields within each well) were acquired for each replicate. MAP2 and Hoechst channels were used to define soma and nuclear compartments, respectively, and fluorescence intensity of GR in each compartment was measured in ImageJ after subtracting background pixel intensity. 1 to 3 neurons were quantified in each image. For GR representative images shown in Figure 6E, single plane images were acquired with a 100x oil immersion objective using the A1R system as described above.
Microscopy & image analysis (FMRP localization)
NPCs were plated for terminal differentiation onto glass chamber slides as described above. Four days after plating for terminal differentiation (Day 19), cells were treated with vehicle or 1 μM DEX for 30 min prior to fixation (4% PFA for 10 min). ICC was performed as described above for FMRP and TUJ1. Confocal z series were acquired with a 60x oil immersion objective (NA 1.4) on a Nikon A1R-HD system. Images were acquired at 1024x1024 resolution at 1/8 frames per second, 0.15 μm interval. Eight to eleven randomly chosen areas were imaged for each replicate (2 separate batches of differentiation and treatment). TUJ1 and Hoechst channels were used to define soma, nucleus, and dendrites, and fluorescence intensity of FMRP in each compartment was measured in ImageJ after subtracting background pixel intensity. 1 to 3 neurons were quantified in each image. FMRP signal in proximal dendrites was quantified using SNT plugin (see MS2 quantification section above).
Characterization of hESC cultures
NPCs were plated for terminal differentiation onto poly-o/laminin-coated 96 well imaging plates (CellVis, P96–1.5P) at a density of 25,000 cells per well. Neurons were fixed at 1 week post-plating and ICC for MAP2 and GFAP was performed using the same ICC procedure as above with the following exception: instead of mounting in PVA-DABCO, neurons were kept in PBS and immediately imaged on the Nano (Molecular Devices) high content imaging system using a 20x objective. The number of cells in each field was manually quantified using the Cell Counter plugin in Fiji/ImageJ. For each condition, 4–6 wells (technical replicates) were imaged, and 2–3 fields were quantified per well.
Digital droplet PCR (ddPCR)
DEX-treated neurons were harvested in Trizol and RNA was isolated using the Direct-zol RNA Micro Prep Kit (Zymo). cDNA was synthesized from 500 ng of RNA using the Prime Script RT kit (Takara). Digital droplet PCR was performed using a published method123 using the Bio-Rad QX200 system. Gene-specific primers/probes were obtained from IDT and used at 20x. PCR reaction mixture (consisting of ddPCR Super Mix for Probes (no dUTP), gene of interest primer/probe, housekeeping gene (TBP) primer/probe, and 1 ng/μL cDNA template) was partitioned into 10,000 to 20,000 droplets, then parallel PCR amplification carried out on the Bio-Rad C1000. PCR signals were quantified using QX Manager software (Bio-Rad), and thresholds were manually adjusted for each reaction. Two-color PCR reaction was utilized to normalize gene of interest expression to the housekeeping gene TBP prior to comparison between conditions.
| ddPCR assays | ||
|---|---|---|
|
| ||
| Gene Symbol | Dye-Quencher method (probe) | Assay |
| PTPN11 | FAM/ZEN/IBFQ | Hs.PT.58.39503117 |
| NR3C1 | FAM/ZEN/IBFQ | Hs.PT.58.27480377 |
| TBP (housekeeping) | HEX/ZEN/IBFQ | Hs.PT.58v.39858774 |
QUANTIFICATION AND STATISTICAL ANALYSIS
Data collection timing and blinding
Data collection was carried out for a predetermined period of time, as dictated by literature or core facility-based standards. All cell counting, tracing, quantification and behavioral analyses were performed by experimenters who were blind to the identity and treatments of the samples.
Statistical analysis
Power analysis was used to pre-determine sample sizes, and our sample sizes are similar to those reported in publications (see citations within each procedure). Data distribution was assumed to be normal but this was not formally tested. Statistical comparisons were performed using Prism software (GraphPad). For data with equal variances, student’s t-tests were used for comparisons of two conditions, and one-way or two-way ANOVA was used for comparisons across multiple conditions and/or variables. Multiple comparisons correction was done using Sidak’s or Tukey’s post-hoc test. For data that was normalized and/or that did not have equal variances, t test with Welch’s correction was used for comparisons of two conditions, and Brown-Forsythe and Welch ANOVA tests were used for comparisons across multiple conditions/variables. Multiple comparisons correction was done using Games-Howell post-hoc test. Any outliers were identified using ROUT method (Q = 1%) and removed from analysis. Probabilities of p < 0.05 were considered as significant.
Graphs/plots for all figures were generated using Prism. Schematic drawings were created using Power Point or BioRender. Predicted RNA structures were generated using RNAFold.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Anti-FMRP | Thermo Fisher Scientific | Cat# MA5–15499; RRID: AB_10979450 |
| Anti-FMRP | Millipore-Sigma | Cat# MAB2016; RRID: AB_2283007 |
| Anti-FMRP | Santa Cruz Biotechnology | Cat# sc-101048; RRID: AB_1122951 |
| Anti-GAPDH | Thermo Fisher Scientific | Cat# TAB1001; RRID: AB_10709707 |
| Anti-GAPDH | Thermo Fisher Scientific | Cat# MA5–15738; RRID: AB_10977387 |
| Anti-TUJ1 | Biolegend | Cat# 802001; RRID: AB_2564645 |
| Anti-MAP2 | Sigma-Aldrich | Cat# M1406; RRID: AB_477171 |
| Anti-GFAP | Agilent | Cat# Z0334; RRID: AB_10013382 |
| Anti-Glucocorticoid Receptor | Cell Signaling | Cat# 3660; RRID: AB_11179215 |
| Anti-DYNC1H1 | Proteintech | Cat# 12345–1-AP; RRID: AB_2261765 |
| Anti-HSP90 | Proteintech | Cat# 13171–1-AP; RRID: AB_2120924 |
| Anti-HSP90AB1 | Proteintech | Cat# 11405–1-AP; RRID: AB_2121207 |
| Anti-KPNB1 | Proteintech | Cat# 10077–1-AP; RRID: AB_2133977 |
| Anti-IPO7 | Proteintech | Cat# 28289–1-AP; RRID: AB_2881106 |
| HSP70 | Proteintech | Cat# 10995–1-AP; RRID: AB_2264230 |
| Cytochrome c | Proteintech | Cat# 10993–1-AP; RRID: AB_2090467 |
| EPAS1 (HIF2α) | Proteintech | Cat# 26422–1-AP; RRID: AB_2880510 |
| TXN | Proteintech | Cat# 14999–1-AP; RRID: AB_2272597 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dexamethasone | Sigma-Aldrich | D4902; CAS 50–02-2 |
| TMPyP4 | Sigma-Aldrich | Cat# 613560; CAS 36951–72-1 |
| DMEM/F12 | Thermo Fisher Scientific | Cat# 11330032 |
| Neurobasal | Thermo Fisher Scientific | Cat# 21103049 |
| Knockout Serum Replacement (KOSR) | Thermo Fisher Scientific | Cat# 10828028 |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | Cat # 15240062 |
| Dispase II | Thermo Fisher Scientific | Cat# 17105041 |
| Collagenase, Type IV | Thermo Fisher Scientific | Cat# 17104019 |
| StemPro™ Accutase | Thermo Fisher Scientific | Cat# A1110501 |
| TrypLE Express Enzyme | Thermo Fisher Scientific | Cat# 12605010 |
| 0.5% Trypsin | Thermo Fisher Scientific | Cat# 15400054 |
| B-27 (without vitamin A) | Thermo Fisher Scientific | Cat# 12587020 |
| B-27 | Thermo Fisher Scientific | Cat# 17504044 |
| Glutamax | Thermo Fisher Scientific | Cat# 35050061 |
| L-glutamine | Thermo Fisher Scientific | Cat# 25030081 |
| Laminin Mouse Protein, Natural | Thermo Fisher Scientific | Cat# 23017015 |
| MEM NEAA | Thermo Fisher Scientific | Cat# 11140050 |
| SB431542 | Biogems | Cat# 3014193 |
| Puromycin | InvivoGen | Cat# ant-pr-1 |
| LDN-193189 | Selleck Chemicals | Cat# S2618; CAS: 1062368–24-4 |
| XAV 939 | Tocris | Cat# 3748; CAS: 284028–89-3 |
| Y-27632 | Tocris | Cat# 1254; CAS:129830–38-2 |
| Matrigel | Corning | Cat# 354248 |
| Glucose | Dot Scientific | Cat# DSG32040 |
| Recombinant Human/Murine/Rat BDNF | Peprotech | Cat# 450–02 |
| Recombinant Human GDNF | Peprotech | Cat# 450–10 |
| DMEM, High Glucose | Sigma-Aldrich | Cat# D5796 |
| L-ascorbic acid | Sigma-Aldrich | Cat# A8960; CAS: 1713265–25-8 |
| Cyclic AMP (cAMP) | Sigma-Aldrich | Cat# D0260; CAS: 241–059-4 |
| γ-Secretase Inhibitor XXI, Compound E | Sigma-Aldrich | Cat# 565790; CAS: 209986–17-4 |
| 2-mercapto-ethanol | Sigma-Aldrich | Cat# M6250; CAS: 60–24-2 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D2650; CAS: 67–68-5 |
| Poly-L-ornithine | Sigma-Aldrich | Cat# P4638; CAS: 27378–49-0 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668027 |
| Aprotinin | Sigma-Aldrich | Cat# A6279; CAS: 9087–70-1 |
| Leupeptin | Tocris | Cat# 1167; CAS: 103476–89-7 |
| Pepstatin | Tocris | Cat# 1190; CAS: 26305–03-3 |
| DAPI (4’, 6-diamidine-2’-phenylindole dihydrochloride) | Millipore Sigma | Cat# 10236276001; CAS 28718–90-3 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat# 62249; CAS 23491–53-3 |
|
| ||
| Critical commercial assays | ||
|
| ||
| CellTiter-Glo® 2.0 Cell Viability Assay | Promega | Cat# G9242 |
| ViewRNA ISH Cell Assay | Thermo Fisher Scientific | Cat# QVC0001 |
| Human Cell Stress Proteome Profiler Array Kit | R & D Systems | Cat# ARY018 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| H9 (WA09) | WiCell | RRID: CVCL_9773 |
| H1 (WA01) | WiCell | RRID: CVCL_9771 |
| H13 (WA13) | WiCell | RRID: CVCL_9774 |
| H1–0CGG | This paper | N/A |
| H13–0CGG | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | Jackson Laboratory | RRID: IMSR_JAX:000664 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for Cloning Cas9/sgRNA and Donor Vectors for Genome Editing | This manuscript | See Table S2 |
| Primers for Screening Gene Edited Clones | This manuscript | See Table S2 |
| Primers for CRISPR Off-Target Analysis | This manuscript | See Table S2 |
| Primers for qRT-PCR | This manuscript | See Table S2 |
| PTPN11 PrimeTime qPCR Assay for ddPCR | IDT | Cat# Hs.PT.58.39503117 |
| NR3C1 PrimeTime qPCR Assay for ddPCR | IDT | Cat# Hs.PT.58.27480377 |
| TBP PrimeTime qPCR Assay for ddPCR | IDT | Cat# Hs.PT.58v.39858774 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pLentiCRISPRv1 | Li et al. 2020.114 | RRID: Addgene #49535 |
| pLentiCRISPRv1–49535-sgFmr1_CGG5–2 | This manuscript | N/A |
| OCT4–2A-eGFP-PGK-Puro | Hockemeyer et al., 2011115 | RRID: Addgene #31938 |
| pDonor-FMR1-dCGG0 | This manuscript | N/A |
| pDonor-FMR1-CGG-BsmBI | This manuscript | N/A |
| pLV-Syn1-mCherry | This manuscript | N/A |
| p40651-EFSNS-FMR1_e1–24xms2 | This manuscript | N/A |
| p40651-EFSNS-FMR1_e1ΔCGG-24xms2 | This manuscript | N/A |
| P40651- phage-cmv-cfp-24xMS2 | Wu et al. 2012116 | RRID: Addgene_40651 |
| p40649-phage-ubc-nls-ha-tdMCP-gfp | Wu et al. 2012116 | RRID: Addgene_40649 |
|
| ||
| Software and algorithms | ||
|
| ||
| Prism (v10) | GraphPad |
https://www.graphpad.com; RRID: SCR_002798 |
| Benchling | Benchling |
https://www.benchling.com/; RRID: SCR_013955 |
| Fiji | National Institute of Health |
https://imagej.net/software/fiji/; RRID: SCR_002285 |
| SNT (plugin) | National Institute of Health | https://imagej.net/plugins/snt/ |
| NIS-Elements | Nikon |
https://www.nikoninstruments.com/Products/Software; RRID: SCR_014329 |
| ImageStudio (v5.2) | Li-Cor |
https://www.licor.com/bio/products/software/image_studio/; RRID: SCR_015795 |
| Neurolucida | MBF Biosciences |
http://www.mbfbioscience.com/neurolucida; RRID:SCR_001775 |
| RNA Fold | Vienna RNA Web Services |
http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi; RRID:SCR_008550 |
| QGRS Mapper | Ramapo College of New Jersey | https://bioinformatics.ramapo.edu/QGRS/index.php |
| QX Manager | Bio-Rad |
https://www.bio-rad.com/en-us/life-science/digital-pcr/qx-software; RRID: SCR_019707 |
| BioRender | BioRender |
http://biorender.com/; RRID: SCR_018361 |
Highlights.
The normal CGG repeats in the 5′ UTR of human FMR1 regulate FMR1 mRNA localization
0CGG human neurons have altered expression levels of stress and metabolic proteins
DEX-treated 0CGG neurons have reduced GR chaperone HSP90α levels and altered GR translocation
FMR1 CGG repeats are important for homeostatic responses to cellular stress in neurons
ACKNOWLEDGMENTS
We thank Dr. Yina Xing, Ryan Risgaard, and Bradley Levesque for technical assistance; Dr. Karla Knobel at the Waisman IDD Models Core for services; and Drs. Paul Hagerman, Marsha Mailick, Albee Messing, Su-Chun Zhang, and Qiang Chang for their guidance and input on this project. This work was supported by grants from the National Institutes of Health (R01MH118827, R01NS105200, R01MH116582, and R01MH136152 to X.Z.; a diversity supplement to R01MH118827 and R36MH136790 for S.O.S.; U54HD090256 and P50HD105353 to the Waisman Center), DOD IIRA grant W81XWH-22-1-0621 (to X.Z. and A.B.); Brain Research Foundation, UW Vilas Mid-Career Award, Kellett Mid-Career Award, Wisconsin Alumni Research Foundation, Jenni and Kyle Professorship, and Eagles Autism Foundation (to X.Z.); Simons Foundation Autism Research Initiative pilot grant (to X.Z. and A.M.M.S.); Brain Research Foundation BRFSG-2023–11 (to A.M.M.S.); Science and Medicine Graduate Research scholarship (to S.O.S.); postdoctoral fellowships from FRAXA (to C.L.S and M.S.); and postdoctoral fellowships from the UW Stem Cell and Regenerative Medicine Center and the Autism Science Foundation (to C.L.S.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114330.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Fan Y, Shen S, Yang J, Yao D, Li M, Mao C, Wang Y, Hao X, Ma D, Li J, et al. (2022). GIPC1 CGG Repeat Expansion Is Associated with Movement Disorders. Ann. Neurol. 91, 704–715. 10.1002/ana.26325. [DOI] [PubMed] [Google Scholar]
- 3.Erwin GS, Gursoy G, Al-Abri R, Suriyaprakash A, Dolzhenko E, Zhu K, Hoerner CR, White SM, Ramirez L, Vadlakonda A, et al. (2023). Recurrent repeat expansions in human cancer genomes. Nature 613, 96–102. 10.1038/s41586-022-05515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rafehi H, Read J, Szmulewicz DJ, Davies KC, Snell P, Fearnley LG, Scott L, Thomsen M, Gillies G, Pope K, et al. (2023). An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA27B/ATX-FGF14. Am. J. Hum. Genet. 110, 1018. 10.1016/j.ajhg.2023.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik I, Kelley CP, Wang ET, and Todd PK (2021). Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607. 10.1038/s41580-021-00382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fytili P, Giannatou E, Papanikolaou V, Stripeli F, Karachalios T, Malizos K, and Tsezou A. (2005). Association of repeat polymorphisms in the estrogen receptors alpha, beta, and androgen receptor genes with knee osteoarthritis. Clin. Genet. 68, 268–277. 10.1111/j.1399-0004.2005.00495.x. [DOI] [PubMed] [Google Scholar]
- 7.Auer RL, Dighiero G, Goldin LR, Syndercombe-Court D, Jones C, McElwaine S, Newland AC, Fegan CD, Caporaso N, and Cotter FE (2007). Trinucleotide repeat dynamic mutation identifying susceptibility in familial and sporadic chronic lymphocytic leukaemia. Br. J. Haematol. 136, 73–79. 10.1111/j.1365-2141.2006.06388.x. [DOI] [PubMed] [Google Scholar]
- 8.Thion MS, Tézenas du Montcel S, Golmard JL, Vacher S, Barjhoux L, Sornin V, Cazeneuve C, Bièche I, Sinilnikova O, Stoppa-Lyonnet D, et al. (2016). CAG repeat size in Huntingtin alleles is associated with cancer prognosis. Eur. J. Hum. Genet. 24, 1310–1315. 10.1038/ejhg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright SE, and Todd PK (2023). Native functions of short tandem repeats. Elife 12, e84043. 10.7554/eLife.84043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annear DJ, Vandeweyer G, Elinck E, Sanchis-Juan A, French CE, Raymond L, and Kooy RF (2021). Abundancy of polymorphic CGG repeats in the human genome suggest a broad involvement in neurological disease. Sci. Rep. 11, 2515. 10.1038/s41598-021-82050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steri M, Idda ML, Whalen MB, and Orrù V. (2018). Genetic variants in mRNA untranslated regions. Wiley Interdiscip. Rev. RNA 9, e1474. 10.1002/wrna.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiffin N, Karczewski KJ, Zhang X, Chothani S, Smith MJ, Evans DG, Roberts AM, Quaife NM, Schafer S, Rackham O, et al. (2020). Characterising the loss-of-function impact of 5’ untranslated region variants in 15,708 individuals. Nat. Commun. 11, 2523. 10.1038/s41467-019-10717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boivin M, and Charlet-Berguerand N. (2022). Trinucleotide CGG Repeat Diseases: An Expanding Field of Polyglycine Proteins? Front. Genet. 13, 843014. 10.3389/fgene.2022.843014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peprah E. (2012). Fragile X syndrome: the FMR1 CGG repeat distribution among world populations. Ann. Hum. Genet. 76, 178–191. 10.1111/j.1469-1809.2011.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichler EE, Kunst CB, Lugenbeel KA, Ryder OA, Davison D, Warren ST, and Nelson DL (1995). Evolution of the cryptic FMR1 CGG repeat. Nat. Genet. 11, 301–308. 10.1038/ng1195-301. [DOI] [PubMed] [Google Scholar]
- 16.Hunter JE, Berry-Kravis E, Hipp H, and Todd PK (1993). FMR1 Disorders. In GeneReviews(®), Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, and Amemiya A, eds. (University of Washington, Seattle: ). [Google Scholar]
- 17.Debrey SM, Leehey MA, Klepitskaya O, Filley CM, Shah RC, Kluger B, Berry-Kravis E, Spector E, Tassone F, and Hall DA (2016). Clinical Phenotype of Adult Fragile X Gray Zone Allele Carriers: a Case Series. Cerebellum 15, 623–631. 10.1007/s12311-016-0809-6. [DOI] [PubMed] [Google Scholar]
- 18.Loesch DZ, Tassone F, Mellick GD, Horne M, Rubio JP, Bui MQ, Francis D, and Storey E. (2018). Evidence for the role of FMR1 gray zone alleles as a risk factor for parkinsonism in females. Mov. Disord. 33, 1178–1181. 10.1002/mds.27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall DA, Nag S, Ouyang B, Bennett DA, Liu Y, Ali A, Zhou L, and Berry-Kravis E. (2020). Fragile X Gray Zone Alleles Are Associated With Signs of Parkinsonism and Earlier Death. Mov. Disord. 35, 1448–1456. 10.1002/mds.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weghofer A, Tea MK, Barad DH, Kim A, Singer CF, Wagner K, and Gleicher N. (2012). BRCA1/2 mutations appear embryo-lethal unless rescued by low (CGG n<26) FMR1 sub-genotypes: explanation for the "BRCA paradox"? PLoS One 7, e44753. 10.1371/journal.pone.0044753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laitman Y, Ries-Levavi L, Berkensdadt M, Korach J, Perri T, Pras E, and Friedman E. (2014). FMR1 CGG allele length in Israeli BRCA1/BRCA2 mutation carriers and the general population display distinct distribution patterns. Genet. Res. 96, e11. 10.1017/s0016672314000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamsheck HC, Petty EM, Hong J, Baker MW, Brilliant MH, and Mailick MR (2017). Is Low FMR1 CGG Repeat Length in Males Correlated with Family History of BRCA-Associated Cancers? An Exploratory Analysis of Medical Records. J. Genet. Couns. 26, 1401–1410. 10.1007/s10897-017-0116-5. [DOI] [PubMed] [Google Scholar]
- 23.Mailick M, Hong J, Greenberg J, Dawalt LS, Baker MW, and Rathouz PJ (2017). FMR1 genotype interacts with parenting stress to shape health and functional abilities in older age. Am. J. Med. Genet. B Neuropsychiatr. Genet. 174, 399–412. 10.1002/ajmg.b.32529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailick MR, Hong J, DaWalt LS, Greenberg JS, Movaghar A, Baker MW, Rathouz PJ, and Brilliant MH (2020). FMR1 Low Zone CGG Repeats: Phenotypic Associations in the Context of Parenting Stress. Front. Pediatr. 8, 223. 10.3389/fped.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LS, Tassone F, Sahota P, and Hagerman PJ (2003). The (CGG)n repeat element within the 5’ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 12, 3067–3074. 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Barad DH, Darmon SK, Kushnir VA, Wu YG, Lazzaroni-Tealdi E, Zhang L, Albertini DF, and Gleicher N. (2018). Reduced RNA expression of the FMR1 gene in women with low (CGGn<26) repeats. PLoS One 13, e0209309. 10.1371/journal.pone.0209309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez CM, Wright SE, Kearse MG, Haenfler JM, Flores BN, Liu Y, Ifrim MF, Glineburg MR, Krans A, Jafar-Nejad P, et al. (2020). A native function for RAN translation and CGG repeats in regulating fragile X protein synthesis. Nat. Neurosci. 23, 386–397. 10.1038/s41593-020-0590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muslimov IA, Patel MV, Rose A, and Tiedge H. (2011). Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J. Cell Biol. 194, 441–457. 10.1083/jcb.201010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie N, Gong H, Suhl JA, Chopra P, Wang T, and Warren ST (2016). Reactivation of FMR1 by CRISPR/Cas9-Mediated Deletion of the Expanded CGG-Repeat of the Fragile X Chromosome. PLoS One 11, e0165499. 10.1371/journal.pone.0165499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park CY, Halevy T, Lee DR, Sung JJ, Lee JS, Yanuka O, Benvenisty N, and Kim DW (2015). Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 13, 234–241. 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- 31.Renz PF, Valdivia-Francia F, and Sendoel A. (2020). Some like it translated: small ORFs in the 5’UTR. Exp. Cell Res. 396, 112229. 10.1016/j.yexcr.2020.112229. [DOI] [PubMed] [Google Scholar]
- 32.Soukarieh O, Meguerditchian C, Proust C, Aïssi D, Eyries M, Goyenvalle A, and Trégouët DA (2022). Common and Rare 5’UTR Variants Altering Upstream Open Reading Frames in Cardiovascular Genomics. Front. Cardiovasc. Med. 9, 841032. 10.3389/fcvm.2022.841032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stripecke R, Oliveira CC, McCarthy JE, and Hentze MW (1994). Proteins binding to 5’ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell Biol. 14, 5898–5909. 10.1128/mcb.14.9.5898-5909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Shin J, Risgaard RD, Parries MJ, Wang J, Chasman D, Liu S, Roy S, Bhattacharyya A, and Zhao X. (2020). Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 30, 361–374. 10.1101/gr.251405.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baj G, Leone E, Chao MV, and Tongiorgi E. (2011). Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc. Natl. Acad. Sci. USA 108, 16813–16818. 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal R, Agbas A, Bao X, Hui D, Leary C, Hunt J, Naniwadekar A, Michaelis ML, Kumar KN, and Michaelis EK (2003). Selective dendrite-targeting of mRNAs of NR1 splice variants without exon 5: identification of a cis-acting sequence and isolation of sequence-binding proteins. Brain Res. 994, 1–18. 10.1016/j.brainres.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 38.Bi J, Tsai NP, Lin YP, Loh HH, and Wei LN (2006). Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc. Natl. Acad. Sci. USA 103, 19919–19924. 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, and Long RM (1998). Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445. 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 40.Handa V, Saha T, and Usdin K. (2003). The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 31, 6243–6248. 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Napiera1a M, and Krzyzosiak WJ (1997). CUG repeats present in myotonin kinase RNA form metastable "slippery" hairpins. J. Biol. Chem. 272, 31079–31085. 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 42.Zumwalt M, Ludwig A, Hagerman PJ, and Dieckmann T. (2007). Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 4, 93–100. 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, and Moine H. (2011). G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 12, 697–704. 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imperatore JA, McAninch DS, Valdez-Sinon AN, Bassell GJ, and Mihailescu MR (2020). FUS Recognizes G Quadruplex Structures Within Neuronal mRNAs. Front. Mol. Biosci. 7, 6. 10.3389/fmolb.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maltby CJ, Schofield JPR, Houghton SD, O’Kelly I, Vargas-Caballero M, Deinhardt K, and Coldwell MJ (2020). A 5’ UTR GGN repeat controls localisation and translation of a potassium leak channel mRNA through G-quadruplex formation. Nucleic Acids Res. 48, 9822–9839. 10.1093/nar/gkaa699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikin O, D’Antonio L, and Bagga PS (2006). QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–W682. 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, and Hofacker IL (2011). ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martino L, Pagano B, Fotticchia I, Neidle S, and Giancola C. (2009). Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B 113, 14779–14786. 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 49.Morris MJ, Wingate KL, Silwal J, Leeper TC, and Basu S. (2012). The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 40, 4137–4145. 10.1093/nar/gkr1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamiri B, Reddy K, Macgregor RB Jr., and Pearson CE (2014). TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 289, 4653–4659. 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haldar S, Zhang Y, Xia Y, Islam B, Liu S, Gervasio FL, Mulholland AJ, Waller ZAE, Wei D, and Haider S. (2022). Mechanistic Insights into the Ligand-Induced Unfolding of an RNA G-Quadruplex. J. Am. Chem. Soc. 144, 935–950. 10.1021/jacs.1c11248. [DOI] [PubMed] [Google Scholar]
- 52.de Kloet ER, Joëls M, and Holsboer F. (2005). Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 6, 463–475. 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 53.Yao J, Sasaki Y, Wen Z, Bassell GJ, and Zheng JQ (2006). An essential role for beta-actin mRNA localization and translation in Ca2+--dependent growth cone guidance. Nat. Neurosci. 9, 1265–1273. 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 54.Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, and Mayford M. (2002). Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36, 507–519. 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 55.Brechbiel JL, and Gavis ER (2008). Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr. Biol. 18, 745–750. 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hochberg-Laufer H, Schwed-Gross A, Neugebauer KM, and Shav-Tal Y. (2019). Uncoupling of nucleo-cytoplasmic RNA export and localization during stress. Nucleic Acids Res. 47, 4778–4797. 10.1093/nar/gkz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anacker C, Cattaneo A, Luoni A, Musaelyan K, Zunszain PA, Milanesi E, Rybka J, Berry A, Cirulli F, Thuret S, et al. (2013). Glucocorticoid-related molecular signaling pathways regulating hippocampal neurogenesis. Neuropsychopharmacology 38, 872–883. 10.1038/npp.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moors M, Bose R, Johansson-Haque K, Edoff K, Okret S, and Ceccatelli S. (2012). Dickkopf 1 mediates glucocorticoid-induced changes in human neural progenitor cell proliferation and differentiation. Toxicol. Sci. 125, 488–495. 10.1093/toxsci/kfr304. [DOI] [PubMed] [Google Scholar]
- 59.Lieberman R, Kranzler HR, Levine ES, and Covault J. (2017). Examining FKBP5 mRNA expression in human iPSC-derived neural cells. Psychiatry Res. 247, 172–181. 10.1016/j.psychres.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buurstede JC, van Weert LTCM, Colucci P, Gentenaar M, Viho EMG, Koorneef LL, Schoonderwoerd RA, Lanooij SD, Moustakas I, Balog J, et al. (2022). Hippocampal glucocorticoid target genes associated with enhancement of memory consolidation. Eur. J. Neurosci. 55, 2666–2683. 10.1111/ejn.15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeshima K, Stanford SM, Hammaker D, Sacchetti C, Zeng LF, Ai R, Zhang V, Boyle DL, Aleman Muench GR, Feng GS, et al. (2016). Abnormal PTPN11 enhancer methylation promotes rheumatoid arthritis fibroblast-like synoviocyte aggressiveness and joint inflammation. JCI Insight 1, e86580. 10.1172/jci.insight.86580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bauer A, Tronche F, Wessely O, Kellendonk C, Reichardt HM, Steinlein P, Schütz G, and Beug H. (1999). The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13, 2996–3002. 10.1101/gad.13.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vyas S, Rodrigues AJ, Silva JM, Tronche F, Almeida OFX, Sousa N, and Sotiropoulos I. (2016). Chronic Stress and Glucocorticoids: From Neuronal Plasticity to Neurodegeneration. Neural Plast. 2016, 6391686. 10.1155/2016/6391686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen HJC, Yip T, Lee JK, Juliani J, Sernia C, Hill AF, Lavidis NA, and Spiers JG (2020). Restraint Stress Alters Expression of Glucocorticoid Bioavailability Mediators, Suppresses Nrf2, and Promotes Oxidative Stress in Liver Tissue. Antioxidants 9, 853. 10.3390/antiox9090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen M, Wang F, Li M, Sah N, Stockton ME, Tidei JJ, Gao Y, Korabelnikov T, Kannan S, Vevea JD, et al. (2019). Reduced mitochondrial fusion and Huntingtin levels contribute to impaired dendritic maturation and behavioral deficits in Fmr1-mutant mice. Nat. Neurosci. 22, 386–400. 10.1038/s41593-019-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen M, Sirois CL, Guo Y, Li M, Dong Q, Méndez-Albelo NM, Gao Y, Khullar S, Kissel L, Sandoval SO, et al. (2023). Species-specific FMRP regulation of RACK1 is critical for prenatal cortical development. Neuron 111, 3988–4005.e11. 10.1016/j.neuron.2023.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchi D, and van Eeden FJM (2021). Homeostatic Regulation of Glucocorticoid Receptor Activity by Hypoxia-Inducible Factor 1: From Physiology to Clinic. Cells 10, 3441. 10.3390/cells10123441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spies LML, Verhoog NJD, and Louw A. (2021). Acquired Glucocorticoid Resistance Due to Homologous Glucocorticoid Receptor Downregulation: A Modern Look at an Age-Old Problem. Cells 10, 2529. 10.3390/cells10102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang H, and Wang W. (2023). Molecular mechanisms of glucocorticoid resistance. Eur. J. Clin. Invest. 53, e13901. 10.1111/eci.13901. [DOI] [PubMed] [Google Scholar]
- 70.Nicolaides NC, Chrousos G, and Kino T. (2000). In Glucocorticoid Receptor, Endotext, Feingold KR, Anawalt B, Blackman MR, Boyce A, Chrousos G, Corpas E, de Herder WW, Dhatariya K, and Dungan K, et al. , eds. (MDText.com, Inc.Copyright © 2000–2023, MDText.com, Inc.). [Google Scholar]
- 71.Meijer OC, Buurstede JC, and Schaaf MJM (2019). Corticosteroid Receptors in the Brain: Transcriptional Mechanisms for Specificity and Context-Dependent Effects. Cell. Mol. Neurobiol. 39, 539–549. 10.1007/s10571-018-0625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richter JD, and Zhao X. (2021). The molecular biology of FMRP: new insights into fragile X syndrome. Nat. Rev. Neurosci. 22, 209–222. 10.1038/s41583-021-00432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ascano M Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386. 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, Grossi M, Rage F, and Bardoni B. (2018). HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 46, 6344–6355. 10.1093/nar/gky267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran SS, Jun HI, Bahn JH, Azghadi A, Ramaswami G, Van Nostrand EL, Nguyen TB, Hsiao YHE, Lee C, Pratt GA, et al. (2019). Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25–36. 10.1038/s41593-018-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vandevyver S, Dejager L, and Libert C. (2012). On the trail of the glucocorticoid receptor: into the nucleus and back. Traffic 13, 364–374. 10.1111/j.1600-0854.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- 77.Mazaira GI, Echeverria PC, and Galigniana MD (2020). Nucleocytoplasmic shuttling of the glucocorticoid receptor is influenced by tetratricopeptide repeat-containing proteins. J. Cell Sci. 133, jcs238873. 10.1242/jcs.238873. [DOI] [PubMed] [Google Scholar]
- 78.Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Freedman ND, and Yamamoto KR (2004). Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15, 2276–2286. 10.1091/mbc.e03-11-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morishima Y, Mehta RK, Yoshimura M, Lau M, Southworth DR, Lawrence TS, Pratt WB, Nyati MK, and Osawa Y. (2018). Chaperone Activity and Dimerization Properties of HSP90α and Hsp90β in Glucocorticoid Receptor Activation by the Multiprotein Hsp90/Hsp70-Dependent Chaperone Machinery. Mol. Pharmacol. 94, 984–991. 10.1124/mol.118.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galigniana MD, Radanyi C, Renoir JM, Housley PR, and Pratt WB (2001). Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 276, 14884–14889. 10.1074/jbc.M010809200. [DOI] [PubMed] [Google Scholar]
- 82.Kirschke E, Goswami D, Southworth D, Griffin PR, and Agard DA (2014). Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697. 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mangano GD, Fontana A, Salpietro V, Antona V, Mangano GR, and Nardello R. (2022). Recurrent missense variant in the nuclear export signal of FMR1 associated with FXS-like phenotype including intellectual disability, ASD, facial abnormalities. Eur. J. Med. Genet. 65, 104441. 10.1016/j.ejmg.2022.104441. [DOI] [PubMed] [Google Scholar]
- 84.Lee HG, Imaichi S, Kraeutler E, Aguilar R, Lee YW, Sheridan SD, and Lee JT (2023). Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation. Cell 186, 2593–2609.e18. 10.1016/j.cell.2023.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Graaff E, de Vries BB, Willemsen R, van Hemel JO, Mohkamsing S, Oostra BA, and van den Ouweland AM (1996). The fragile X phenotype in a mosaic male with a deletion showing expression of the FMR1 protein in 28% of the cells. Am. J. Med. Genet. 64, 302–308. . [DOI] [PubMed] [Google Scholar]
- 86.Grønskov K, Hjalgrim H, Bjerager MO, and Brøndum-Nielsen K. (1997). Deletion of all CGG repeats plus flanking sequences in FMR1 does not abolish gene expression. Am. J. Hum. Genet. 61, 961–967. 10.1086/514872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erbs E, Fenger-Grøn J, Jacobsen CM, Lildballe DL, and Rasmussen M. (2021). Spontaneous rescue of a FMR1 repeat expansion and review of deletions in the FMR1 non-coding region. Eur. J. Med. Genet. 64, 104244. 10.1016/j.ejmg.2021.104244. [DOI] [PubMed] [Google Scholar]
- 88.Sittler A, Devys D, Weber C, and Mandel JL (1996). Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet. 5, 95–102. 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 89.Colliva A, and Tongiorgi E. (2021). Distinct role of 5’UTR sequences in dendritic trafficking of BDNF mRNA: additional mechanisms for the BDNF splice variants spatial code. Mol. Brain 14, 10. 10.1186/s13041-020-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suhl JA, Muddashetty RS, Anderson BR, Ifrim MF, Visootsak J, Bassell GJ, and Warren ST (2015). A 3’ untranslated region variant in FMR1 eliminates neuronal activity-dependent translation of FMRP by disrupting binding of the RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 112, E6553–E6561. 10.1073/pnas.1514260112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, et al. (2013). Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters micro-RNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 3, 869–880. 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Antar LN, Afroz R, Dictenberg JB, Carroll RC, and Bassell GJ (2004). Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 24, 2648–2655. 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, and Darnell RB (2001). Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489–499. 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 94.Menon L, and Mihailescu MR (2007). Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 35, 5379–5392. 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rovozzo R, Korza G, Baker MW, Li M, Bhattacharyya A, Barbarese E, and Carson JH (2016). CGG Repeats in the 5’UTR of FMR1 RNA Regulate Translation of Other RNAs Localized in the Same RNA Granules. PLoS One 11, e0168204. 10.1371/journal.pone.0168204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ofer N, Weisman-Shomer P, Shklover J, and Fry M. (2009). The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 37, 2712–2722. 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weisman-Shomer P, Cohen E, Hershco I, Khateb S, Wolfovitz-Barchad O, Hurley LH, and Fry M. (2003). The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res. 31, 3963–3970. 10.1093/nar/gkg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. (2013). CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455. 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bell MV, Hirst MC, Nakahori Y, MacKinnon RN, Roche A, Flint TJ, Jacobs PA, Tommerup N, Tranebjaerg L, Froster-Iskenius U, et al. (1991). Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell 64, 861–866. 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- 100.Breen MS, Bierer LM, Daskalakis NP, Bader HN, Makotkine I, Chattopadhyay M, Xu C, Buxbaum Grice A, Tocheva AS, Flory JD, et al. (2019). Differential transcriptional response following glucocorticoid activation in cultured blood immune cells: a novel approach to PTSD biomarker development. Transl. Psychiatry 9, 201. 10.1038/s41398-019-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seah C, Breen MS, Rusielewicz T, Bader HN, Xu C, Hunter CJ, McCarthy B, Deans PJM, Chattopadhyay M, Goldberg J, et al. (2022). Modeling gene 3 environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression. Nat. Neurosci. 25, 1434–1445. 10.1038/s41593-022-01161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bruno JL, Hong DS, Lightbody AA, Hosseini SMH, Hallmayer J, and Reiss AL (2021). Glucocorticoid regulation and neuroanatomy in fragile x syndrome. J. Psychiatr. Res. 134, 81–88. 10.1016/j.jpsychires.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hessl D, Glaser B, Dyer-Friedman J, Blasey C, Hastie T, Gunnar M, and Reiss AL (2002). Cortisol and behavior in fragile X syndrome. Psychoneuroendocrinology 27, 855–872. 10.1016/s0306-4530(01)00087-7. [DOI] [PubMed] [Google Scholar]
- 104.Hessl D, Glaser B, Dyer-Friedman J, and Reiss AL (2006). Social behavior and cortisol reactivity in children with fragile X syndrome. J. Child Psychol. Psychiatry 47, 602–610. 10.1111/j.1469-7610.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- 105.Guo Y, Shen M, Dong Q, Méndez-Albelo NM, Huang SX, Sirois CL, Le J, Li M, Jarzembowski ED, Schoeller KA, et al. (2023). Elevated levels of FMRP-target MAP1B impair human and mouse neuronal development and mouse social behaviors via autophagy pathway. Nat. Commun. 14, 3801. 10.1038/s41467-023-39337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Silva JM, Rodrigues S, Sampaio-Marques B, Gomes P, Neves-Carvalho A, Dioli C, Soares-Cunha C, Mazuik BF, Takashima A, Ludovico P, et al. (2019). Dysregulation of autophagy and stress granule-related proteins in stress-driven Tau pathology. Cell Death Differ. 26, 1411–1427. 10.1038/s41418-018-0217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parsonnet NV, Lammer NC, Holmes ZE, Batey RT, and Wuttke DS (2019). The glucocorticoid receptor DNA-binding domain recognizes RNA hairpin structures with high affinity. Nucleic Acids Res. 47, 8180–8192. 10.1093/nar/gkz486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suri D, and Vaidya VA (2013). Glucocorticoid regulation of brain-derived neurotrophic factor: relevance to hippocampal structural and functional plasticity. Neuroscience 239, 196–213. 10.1016/j.neuroscience.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 109.Numakawa T, Odaka H, and Adachi N. (2017). Actions of Brain-Derived Neurotrophic Factor and Glucocorticoid Stress in Neurogenesis. Int. J. Mol. Sci. 18, 2312. 10.3390/ijms18112312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Castrén ML, and Castrén E. (2014). BDNF in fragile X syndrome. Neuropharmacology. Neuropharmacology 76, 729–736. 10.1016/j.neuropharm.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 111.Liu J, and DeFranco DB (1999). Chromatin recycling of glucocorticoid receptors: implications for multiple roles of heat shock protein 90. Mol. Endocrinol. 13, 355–365. 10.1210/mend.13.3.0258. [DOI] [PubMed] [Google Scholar]
- 112.Stavreva DA, Müller WG, Hager GL, Smith CL, and McNally JG (2004). Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell Biol. 24, 2682–2697. 10.1128/mcb.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao X, Khurana S, Charkraborty S, Tian Y, Sedor JR, Bruggman LA, and Kao HY (2017). α Actinin 4 (ACTN4) Regulates Glucocorticoid Receptor-mediated Transactivation and Transrepression in Podocytes. J. Biol. Chem. 292, 1637–1647. 10.1074/jbc.M116.755546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li M, Zhao H, Ananiev GE, Musser MT, Ness KH, Maglaque DL, Saha K, Bhattacharyya A, and Zhao X. (2017). Establishment of Reporter Lines for Detecting Fragile X Mental Retardation (FMR1) Gene Reactivation in Human Neural Cells. Stem Cell. 35, 158–169. 10.1002/stem.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734. 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu B, Chao JA, and Singer RH (2012). Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 102, 2936–2944. 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo W, Polich ED, Su J, Gao Y, Christopher DM, Allan AM, Wang M, Wang F, Wang G, and Zhao X. (2015). Fragile X Proteins FMRP and FXR2P Control Synaptic GluA1 Expression and Neuronal Maturation via Distinct Mechanisms. Cell Rep. 11, 1651–1666. 10.1016/j.celrep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mailick MR, Hong J, Movaghar A, DaWalt L, Berry-Kravis EM, Brilliant MH, Boero J, Todd PK, and Hall D. (2021). Mild Neurological Signs in FMR1 Premutation Women in an Unselected Community-Based Cohort. Mov. Disord. 36, 2378–2386. 10.1002/mds.28683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arshadi C, Günther U, Eddison M, Harrington KIS, and Ferreira TA (2021). SNT: a unifying toolbox for quantification of neuronal anatomy. Nat. Methods 18, 374–377. 10.1038/s41592-021-01105-7. [DOI] [PubMed] [Google Scholar]
- 122.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. (2022). Ensembl 2022. Nucleic Acids Res. 50, D988–d995. 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. (2017). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. 10.1126/science.aan3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. Protein array data have been reported in Table S3.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.