Abstract
The beet yellows closterovirus leader proteinase (L-Pro) possesses a C-terminal proteinase domain and a nonproteolytic N-terminal domain. It was found that although L-Pro is not essential for basal-level replication, deletion of its N-terminal domain resulted in a 1,000-fold reduction in RNA accumulation. Mutagenic analysis of the N-terminal domain revealed its structural flexibility except for the 54-codon-long, 5′-terminal element in the corresponding open reading frame that is critical for efficient RNA amplification at both RNA and protein levels.
Viral proteases belong to several structural prototypes; some are unique, whereas others share structural motifs with cellular enzymes (4). In particular, papain-like cysteine proteinases are found in diverse families of positive-strand RNA viruses infecting plants, fungi, and animals (9, 13–15, 23, 25, 30). One class of these proteinases, exemplified by nsP2 of animal alphaviruses, is responsible for processing the nonstructural polyprotein and is intimately involved in RNA replication (31). Similarly, papain-like proteinases encoded by plant tymoviruses and related viruses are involved in the processing of replication-associated polyproteins (6, 17). Proteinases of another class that typically cleave only in cis at their C termini are called leader proteinases (L-proteinases). Examples of these are found in animal arteriviruses (30) and aphthoviruses (15, 28), as well as in plant potyviruses (7) and fungal hypoviruses (25). In addition to autocatalytic processing, several L-proteinases were reported to function in various processes of virus-host interaction (8, 15, 20, 21, 28, 32).
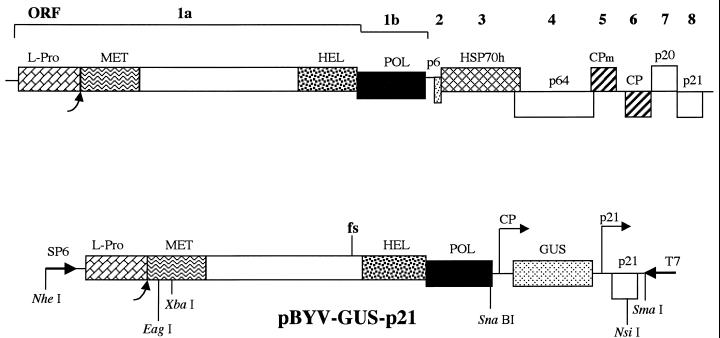
Members of the Closteroviridae family of positive-strand RNA viruses possess 15- to 20-kb genomes encapsidated into filamentous virions (5). Computer-assisted analysis revealed that closteroviruses belong to a Sindbis virus-like superfamily (23). Although the gene content varies among closteroviruses, two genome blocks are conserved among all members (11, 33). The first, 5′-terminal block is represented by open reading frames (ORFs) 1a and 1b, the latter of which encodes RNA polymerase (1, 18, 22). In beet yellows virus (BYV), a prototype closterovirus, ORF 1a codes for a polyprotein that possesses a papain-like L-proteinase (L-Pro), a putative methyltransferase domain, an RNA helicase domain, and a large interdomain region which is unique to closteroviruses (Fig. 1). The second, quintuple, gene block encompasses ORFs encoding proteins responsible for virus assembly (2) and cell-to-cell movement (3, 27).
FIG. 1.
Genomic map of BYV (top) and diagram of the cDNA clone of the mini-BYV genome, pBYV-GUS-p21, tagged by insertion of the GUS gene (bottom). Boxes represent BYV ORFs 1a to 8 encoding L-Pro, replication-associated proteins possessing putative methyltransferase (MET), RNA helicase (HEL), and RNA polymerase (POL) domains, 6-kDa protein (p6), HSP70 homolog (HSP70h), 64-kDa protein (p64), minor capsid protein (CPm), major capsid protein (CP), 20-kDa protein (p20), and 21-kDa protein (p21). Each curved arrow designates the self-processing site for the BYV L-Pro. Arrows marked CP and p21 on the pBYV-GUS-p21 map show approximate positions of the 5′ termini of the subgenomic RNAs expressing GUS and p21 and driven by the CP and p21 promoters, respectively. fs, the frameshift mutation inactivating BYV replicase (26). Selected restriction endonuclease sites are shown below the pBYV-GUS-p21 diagram. Arrows marked SP6 and T7 show positions and orientations of the corresponding RNA polymerase promoters.
The BYV L-Pro provides a dual function in viral genome amplification. Autocatalytic cleavage at the C terminus of L-Pro is essential for virus viability, whereas the nonproteolytic, N-terminal domain is required for efficient RNA accumulation (26). This functional profile is reminiscent of that described for the potyvirus leader proteinase HC-Pro (12, 20).
In this study, we expand the functional analysis of L-Pro by using a mini-BYV genome that lacks six virus genes which are superfluous for genome amplification (16, 26). This BYV variant retains ORFs 1a and 1b and a 3′-terminal ORF encoding a 21-kDa protein (p21), which functions as an activator of genome amplification (Fig. 1 and reference 26). To provide a sensitive marker for genome replication and expression, a reporter gene encoding bacterial β-glucuronidase (GUS) was engineered into this BYV variant, creating BYV-GUS-p21 (16).
To further explore structure-function relationships in the L-Pro molecule, we generated 17 mutants (Fig. 2). Analysis of the mutant phenotypes revealed high tolerance to structural changes in most of the N-terminal domain. In contrast, a 54-codon-long, 5′-terminal region of ORF 1a was found to be critical for virus viability. In addition, we demonstrated that although L-Pro is not essential for basal-level genome amplification, its activity increases this level 1,000-fold.
FIG. 2.
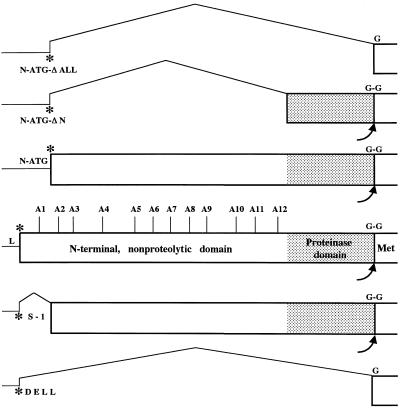
Mutagenic analysis of the function of the N-terminal and proteinase domains of L-Pro in BYV genome amplification. The 5′-terminal part of the BYV genome including the noncoding leader region (L), L-Pro coding region (box), and part of the methyltransferase domain (Met) is shown in the middle. Asterisks, translation start codons; G-G, scissile glycine-glycine bond cleaved by L-Pro. Vertical lines mark sites of the alanine-scanning mutations A1 to A12. N-ATG, mutant in which the original start codon was replaced with ACA and an artificial start codon was introduced at the indicated position; S-1 and DELL, mutants in which in-frame deletions were introduced in the L-Pro coding region; N-ATG-ΔN and N-ATG-ΔALL, mutants in which deletions were introduced into the L-Pro coding region possessing an artificial start codon.
Generation of BYV mutants.
All mutations in the L-Pro coding region were generated using plasmid p5′BYV and site-directed mutagenesis as described elsewhere (24, 26). Each mutation was verified by nucleotide sequencing; the full-length clones of the mutant BYV genomes were engineered by cloning the NheI-EagI fragments of the modified p5′BYV variants into appropriately digested pBYV-GUS-p21 (Fig. 1). The latter plasmid represented the mini-BYV genome in which six viral genes were replaced by a reporter GUS gene (16).
In the DELL (deletion of leader) mutant, the entire region coding for L-Pro was deleted in frame except for the start codon of ORF 1a. This codon was fused with the first glycine codon of the BYV replicase (Fig. 2), resulting in the formation of a replicase that differed from the proteolytically processed, wild-type replicase only by the presence of an N-terminal methionine. The S-1 mutation resulted in the in-frame deletion of ORF 1a codons 2 through 54 (Fig. 2). Mutant N-ATG (new ATG codon) was generated by changing the ORF 1a start codon to ACA (using the mutagenic oligonucleotide 5′-GCTATCGACACACCATTCTTGAACG; changed nucleotides are in boldface), by replacing the G residue downstream from the start codon with C (to disrupt its favorable context), and by engineering a new ORF 1a start codon in place of codon 57 (using the oligonucleotide 5′-CTTCTCTGTCCCGGACATGGTCTTTTTGAACGCG; three nucleotides surrounding ATG were changed to ensure the optimal context for translation). The N-ATG-ΔN mutant was derived from the N-ATG variant via deletion of codons 58 through 442 (original numbering); mutant ORF 1a encoded only the C-terminal, proteinase domain of L-Pro (Fig. 2). In another derivative of the N-ATG mutant, N-ATG-ΔALL, the entire region coding for L-Pro was deleted. In this mutant, the modified ORF 1a produced an unchanged BYV replicase which would be translated from the artificial ATG (Fig. 2). Twelve alanine-scanning mutations (A1 to A12) were introduced throughout the N-terminal domain of L-Pro (Fig. 2). In each of these mutants, three consecutive charged or polar amino acid residues were replaced with three alanine residues (Table 1). The nucleotide sequences of the corresponding mutagenic oligonucleotides are available upon request. The replication-deficient fs variant, harboring a frameshift mutation upstream of the RNA helicase domain, was described previously (26). The corresponding mutant region of fs was cloned into pBYV-GUS-p21 by using unique restriction endonuclease sites XbaI and SnaBI (Fig. 1).
TABLE 1.
GUS activity in BYV variants with alanine-scanning mutations in L-Pro at 4 days after transfection of protoplasts
| BYV variant | Replaced residuesa | Mean GUS activity (% of level in BYV-GUS-p21) ± SD |
|---|---|---|
| A1 | SDD39-41 | 1.1 ± 0.7 |
| A2 | DNP72-74 | 76 ± 12 |
| A3 | NGS92-94 | 128 ± 19 |
| A4 | SKP144-146 | 93 ± 8 |
| A5 | KRK196-198 | 105 ± 17 |
| A6 | SRP228-230 | 97 ± 3 |
| A7 | RRR255-257 | 63 ± 10 |
| A8 | KRK288-290 | 94 ± 14 |
| A9 | KEE316-318 | 90 ± 9 |
| A10 | RRP367-369 | 89 ± 10 |
| A11 | EKK395-397 | 91 ± 12 |
| A12 | SER434-436 | 87 ± 7 |
Three consecutive amino acid residues of L-Pro that were replaced with three alanine residues in each mutant are shown along with their corresponding positions (subscript numbers) in the L-Pro sequence.
Protoplast transfection and analysis of mutant phenotypes.
The mutant BYV-GUS-p21 variants were characterized using transfection of the protoplasts as described previously (12). Each transfection sample contained ∼4 × 106 cells. The capped RNA transcripts were derived using SP6 RNA polymerase (Epicentre) and SmaI-linearized plasmid DNA (Fig. 1). Protoplasts were propagated for 86 h; GUS activity was assayed as described elsewhere (10) and expressed as a percentage of that of the BYV-GUS-p21 variant (positive control). The mock-transfected protoplasts were used as a negative control. Each variant was characterized using at least four independent transfections; means and standard deviations were used to compare GUS activity. The RNA samples were isolated using TRIZOL (Gibco-BRL); Northern hybridization analysis was conducted as described elsewhere (26). The 32P-labeled, single-stranded, negative-polarity RNA probe was generated using T7 RNA polymerase and NsiI-linearized plasmid p3′BYV (Fig. 1 and reference 26). This probe was complementary to the ∼400 3′-terminal nucleotides of the BYV RNA. The radiolabeled hybridization products were detected and quantified using a PhosphorImager (Molecular Dynamics); the means and standard deviations from four independent experiments were used to characterize each variant. In vitro translations were conducted using wheat germ extracts (Promega), [35S]cysteine, and XbaI-linearized variants of p5′BYV exactly as described elsewhere (26).
The 5′-terminal region of ORF 1a is critical for RNA replication and L-Pro function.
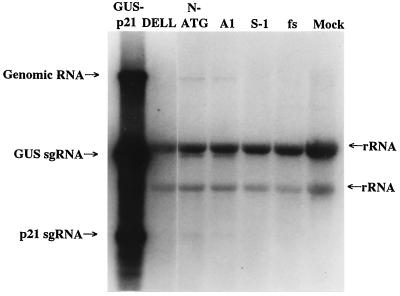
To determine the role that each of the L-Pro domains plays in BYV RNA amplification, a series of mutations was introduced into the region of ORF 1a encoding L-Pro. The previously generated cDNA clone encompassing a mini-BYV genome containing the GUS ORF was used for this purpose (Fig. 1 and reference 16). The capped RNA transcripts derived from linearized pBYV-GUS-p21 variants were transfected into tobacco protoplasts. The GUS assays were used as a sensitive surrogate marker for quantification of the levels of genome amplification. In our previous work we demonstrated that cleavage between L-Pro and the remainder of the ORF 1a product is essential for virus viability, whereas the N-terminal, nonproteolytic domain functions as an activator of genome amplification (26). However, it was not known if release of the mature replicase is the only function of L-Pro that is essential for RNA replication, and if the proteinase domain provides any additional activity required for efficient RNA accumulation. To address these questions, we generated a mutant called DELL, in which the complete L-Pro ORF except for the start codon was deleted such that the translation of mutant RNA would result in production of mature, unchanged replicase (Fig. 2). Protoplast transfection experiments revealed that the DELL variant produced no detectable GUS activity (Table 2) and accumulated no virus-specific RNA (Fig. 3, lane DELL). In fact, this mutant was indistinguishable from the replication-deficient fs mutant expressing nonfunctional replicase (Table 2; Fig. 3, lane fs; reference 26).
TABLE 2.
Comparison between GUS activity and accumulation of genomic RNA in BYV variants at 4 days after transfection of protoplasts
| BYV variant | % of level in BYV-GUS-p21 (mean ± SD)
|
|
|---|---|---|
| GUS activity | RNA accumulation | |
| Mock | >0.001 | UDa |
| fs | >0.001 | UD |
| DELL | >0.001 | UD |
| S-1 | >0.001 | UD |
| N-ATG | 2.5 ± 1.5 | 2.3 ± 0.6 |
| N-ATG-ΔN | 0.1 ± 0.003 | UD |
| N-ATG-ΔALL | 0.1 ± 0.007 | UD |
| A1 | 1.1 ± 0.7 | 1.7 ± 0.7 |
UD, undetectable.
FIG. 3.
Northern analysis of the RNA accumulation in protoplasts transfected with parental and mutant BYV variants. Lane GUS-p21, parental BYV-GUS-p21 variant. Other lanes represent the mutants marked at the top and mock-transfected protoplasts (lane Mock). Arrows mark the positions of genomic RNA and subgenomic RNAs (sgRNAs) encoding GUS and p21, as well as of background bands corresponding to plant rRNAs (26). The membrane was overexposed to visualize low levels of BYV RNA accumulation detected in N-ATG and A1 mutants.
One possible interpretation of the inability of the DELL variant to replicate is that the function of the proteolytic domain is not limited to a single autocatalytic cleavage at the C terminus of L-Pro but may also involve cleavage(s) elsewhere in the replicase. An alternative explanation would be that the noncatalytic, N-terminal domain is indispensable for virus viability. However, we have shown earlier that a mutant, called 1-4, lacking most of the N-terminal domain and retaining its very N-terminal, 54-amino-acid-long peptide was viable but accumulated ∼5 times less RNA than the wild-type virus (26). Thus, complete loss of viability in the DELL mutant could be attributed to loss of either the 54-codon-long RNA region or a region encoding the proteinase domain.
To test the role of the 5′-proximal region of ORF 1a in RNA amplification, we generated a mutant (S-1) in which codons 2 to 54 were deleted in frame to result in expression of the truncated L-Pro possessing most of the N-terminal domain and a complete proteinase domain (Fig. 2). Unexpectedly, the S-1 mutant was nonviable (Table 2; Fig. 3, lane S-1). This result could be due to the indispensability of the short N-terminal peptide for L-Pro function or to a critical role played by the deleted RNA region (e.g., in RNA folding or interaction with the replicase).
To distinguish between these two possibilities, we generated a double-point mutant, N-ATG, in which the original start codon of ORF 1a was replaced with the ACA and an artificial start codon was engineered in place of codon 57 of ORF 1a (Fig. 2). As expected, in vitro translation of the N-ATG RNA yielded a truncated L-Pro. This mutant product accumulated in vitro to a level similar to that of the nonmutant L-Pro, indicating that no significant changes in translational and proteolytic activity occurred due to the transfer of the start codon to the downstream location (data not shown).
In protoplast transfection experiments, the N-ATG variant was viable, although it produced only 2.5% of the GUS activity of the parental variant (Table 2). Northern hybridization analysis yielded similar results (Table 2; Fig. 3, lane N-ATG), once again indicating that the GUS activity accurately reflects RNA accumulation. The phenotype exhibited by the N-ATG mutant is indicative of a major defect in RNA amplification. Comparison of the phenotypes of the S-1 and N-ATG mutants suggests that the 5′-terminal, 54-codon-long region of ORF 1a provides a dual function. At the RNA level, this region is indispensable for virus viability, likely due to its role in overall RNA folding or its function as a cis-replicational signal. At the protein level, the peptide encoded in this region plays an important role in the L-Pro function in accumulation of viral RNA.
Roles played by each of the L-Pro domains in RNA accumulation.
The viability of the N-ATG variant allowed us to revisit the problem of the relative functional importance of the N-terminal and proteinase domains for BYV RNA accumulation. To this end, we engineered two deletion mutants based on the N-ATG variant. In mutant N-ATG-ΔN, an artificial start codon was fused with the proteinase domain to result in expression of L-Pro variant lacking all of its N-terminal domain but possessing a proteinase domain (Fig. 2). In vitro translation experiments using the mutant mRNA revealed formation of the expected ∼16-kDa proteinase domain that efficiently released itself from the downstream protein product (not shown). This result was in agreement with our previous work demonstrating that the N-terminal domain is not required for the proteolytic activity of the C-terminal domain (26).
In mutant N-ATG-ΔALL, the same artificial start codon was placed immediately upstream of the first codon of the putative methyltransferase domain. This mutant was designed to express intact replicase in the absence of L-Pro expression (Fig. 2). Protoplast transfection experiments demonstrated that the N-ATG-ΔN variant was viable, although it produced only ∼0.1% of the GUS activity found in parental variant (Table 2). This result emphasized the importance of the N-terminal domain for RNA amplification: in its absence, only a low, basal level of viral RNA was produced. The level of GUS activity in protoplasts transfected with the N-ATG-ΔALL variant was indistinguishable from that found in N-ATG-ΔN variant (Table 2). This result can be interpreted to mean that in the absence of a need for the proteolytic release of the replicase N terminus, the proteinase domain provides no other activity in genome amplification. Alternatively, strong debilitation of genome amplification after deletion of the N-terminal domain could itself be a rate-limiting event masking the need in a proteinase domain.
It should be emphasized that although the GUS activity measured in N-ATG-ΔN and N-ATG-ΔALL variants was only 0.1% of that found in parental BYV-GUS-p21, it was ∼100-fold higher than the background GUS activity detected in the replication-deficient fs variant. This result confirmed that low GUS activity detected in the N-ATG-ΔN and N-ATG-ΔALL mutants was due to amplification and transcription of the viral RNA rather than to direct translation of the input RNA transcripts.
Alanine-scanning mutagenesis of the N-terminal domain.
To further examine the functional significance of the different regions in the N-terminal L-Pro domain, we generated 12 alanine-scanning mutations, designated A1 to A12 and located through the entire domain's length (Fig. 2). In each of these mutants, three adjacent codons specifying charged or polar amino acid residues were replaced with alanine codons (Table 1). These mutations were expected to affect the L-Pro function in RNA amplification by disrupting the electrostatic and/or hydrophilic interactions within the L-Pro molecule or between L-Pro and its putative protein partners. Surprisingly, the effects of 11 out of 12 alanine-scanning mutations on GUS accumulation were relatively weak. The levels of GUS activity detected in protoplasts were from 63 to 128% of that found in the parental variant (Table 1). Statistical analysis of the data revealed that these mutants were not significantly different from the nonmutant variant (P > 0.1), except for mutant A7 (P < 0.001). In contrast, mutant A1 accumulated only ∼1% of the GUS activity found in a nonmutant variant (Table 1). This result was also confirmed by Northern hybridization analysis (Fig. 3, lane A1). Since A1 was the only alanine-scanning mutation located within the limits of the N-terminal, 54-residue-long peptide, this result further emphasized the particular significance of this N-terminal region in L-Pro function. It is also possible that A1 mutation affected replication due to disturbance in the overall folding of the 5′-terminal RNA region.
Tagged mini-BYV variant as a model system.
In this work, we used GUS activity as a surrogate marker of BYV genome amplification. Since GUS activity is a final result of viral genome replication, transcription of a subgenomic RNA, and its translation, we wished to investigate whether the level of GUS activity is an accurate measure of genome amplification. More specifically, we determined whether the mutations in L-Pro could selectively affect the processes of transcription or translation without affecting genomic RNA accumulation. Northern hybridization analyses demonstrated that the GUS-negative fs, DELL, and S-1 mutants failed to accumulate any detectable viral RNA, indicating that each of these mutations blocked accumulation of viral RNA. Comparative analyses of the relative levels of GUS activity and RNA accumulation for mutants A1 and N-ATG revealed similarly low levels of replication between the two types of assay. It should be noted that the sensitivity of GUS assays is much higher than that of Northern analysis. Quantification of RNA levels lower than 1% of the wild-type level was impractical due to the background signal. On the other hand, the high signal-to-background ratio of the GUS assays allowed confident measurements of enzymatic activity at levels of 0.001% of the wild-type level. These results established the GUS-tagged mini-BYV genome as an adequate model with which to study amplification of BYV RNA. An additional benefit of using the mini-BYV variant is the relative ease of manipulation of the truncated genome. A similar minimal replicon was engineered recently for another closterovirus, citrus tristeza virus (29).
Structure-function relationships in the L-Pro molecule.
The GUS-tagged mini-BYV was used to reveal the roles played by each of two major domains of BYV L-Pro in genome amplification. As we demonstrated previously, the cleavage mediated by the C-terminal proteinase domain is essential for virus viability, whereas the N-terminal L-Pro domain acts as an activator of RNA amplification (26). However, it was not known if L-Pro is essential for RNA replication, nor were the specific roles played by each of the L-Pro domains understood. The data presented in this work demonstrate that mutant N-ATG-ΔALL, expressing none of the L-Pro domains, is capable of replicating in tobacco protoplasts, albeit to a very low level. The results indicate that L-Pro is not necessary for basal-level replication. On the other hand, a 1,000-fold decrease in RNA accumulation exhibited by the L-Pro null mutant stresses the importance of L-Pro for efficient amplification of the closterovirus genome.
The identical phenotypes of the mutants lacking the N-terminal domain only and those lacking both N-terminal and proteinase domains suggested that the proteinase itself plays no specific role in the enhancement of genome amplification. Previous work with potyvirus HC-Pro, which also possesses a C-terminal papain-like proteinase domain, suggested that either this domain itself or the cis cleavage mediated by this domain is indispensable for viral viability (19). Since we were able to generate a viable BYV mutant in which the need for cis cleavage was abolished, we propose that the major function of the proteinase domain is to cleave between the L-Pro and bona fide replicase. However, extreme debilitation of genome amplification in the absence of the N-terminal domain of L-Pro could interfere with our ability to detect possible additional functions provided by the proteinase domain.
Mutation analysis of the N-terminal domain revealed its unexpected structural flexibility. Indeed, 11 out of 12 alanine-scanning mutations introduced into this domain had no major effect on RNA accumulation. Computer analysis suggested that the N-terminal domain of L-Pro possesses a nonglobular, elongated structure, in contrast to the globular proteinase domain (A. R. Mushegian and V. V. Dolja, unpublished data). This type of structure may account for the unusual tolerance of the former domain to mutations. Alternatively, this domain may be required for other than genome amplification phases of the virus life cycle. The only region in which an alanine-scanning mutation was not tolerated was the very N-terminal region of L-Pro. The A1 mutation, which changed amino acids 39 to 41, resulted in a 100-fold reduction in RNA accumulation. A similar level of genome amplification was obtained with the mutant in which the ORF 1a start codon was engineered ∼50 codons downstream from its natural position. In contrast, deletion of the ∼50-codon-long RNA segment completely abolished genome amplification, indicating that this region of ORF 1a functions not only as a coding sequence but also as, or as part of, the cis element required for RNA replication. Understanding of the multiple roles played by the L-Pro-encoding region will permit us to investigate the molecular mechanisms involved in activation of genome amplification mediated by this important part of the BYV genome.
Acknowledgments
We thank Yuka Hagiwara for participation in an initial step of this study, Arcady Mushegian for help with computer analysis of amino acid sequences, and George Rohrman and Theo Dreher for critical reading of the manuscript. We are grateful to Jonathan Reed for excellent technical assistance.
This work was supported by grants from the U.S. Department of Agriculture (NRICGP 97-35303-4515) and National Institutes of Health (R1GM53190B) to V.V.D.
REFERENCES
- 1.Agranovsky A A, Koonin E V, Boyko V P, Maiss E, Frotschl R, Lunina N A, Atabekov J G. Beet yellows closterovirus: complete genome structure and identification of a leader papain-like thiol protease. Virology. 1994;198:311–324. doi: 10.1006/viro.1994.1034. [DOI] [PubMed] [Google Scholar]
- 2.Agranovsky A A, Lesemann D E, Maiss E, Hull R, Atabekov J G. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc Natl Acad Sci USA. 1995;92:2470–2473. doi: 10.1073/pnas.92.7.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzhanova D V, Hagiwara Y, Peremyslov V V, Dolja V V. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology. 2000;268:192–200. doi: 10.1006/viro.1999.0155. [DOI] [PubMed] [Google Scholar]
- 4.Babe L M, Craik C S. Viral proteases: evolution of diverse structural motifs to optimize function. Cell. 1997;91:427–430. doi: 10.1016/s0092-8674(00)80426-2. [DOI] [PubMed] [Google Scholar]
- 5.Bar-Joseph M, Garnsey S M, Gonsalves D. The closteroviruses: a distinct group of elongated plant viruses. Adv Virus Res. 1979;25:93–168. doi: 10.1016/s0065-3527(08)60569-2. [DOI] [PubMed] [Google Scholar]
- 6.Bransom K L, Dreher T W. Identification of the essential cysteine and histidine residues of the turnip yellow mosaic virus protease. Virology. 1994;198:148–154. doi: 10.1006/viro.1994.1017. [DOI] [PubMed] [Google Scholar]
- 7.Carrington J C, Cary S M, Parks T D, Dougherty W G. A second proteinase encoded by a plant potyvirus genome. EMBO J. 1989;8:365–370. doi: 10.1002/j.1460-2075.1989.tb03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven M G, Pawlyk D M, Choi G H, Nuss D L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J Virol. 1993;67:6513–6521. doi: 10.1128/jvi.67.11.6513-6521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A L M, Plagemann P G W, Gorbalenya A E, Snijder E J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase (GUS) into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolja V V, Karasev A V, Koonin E V. Molecular biology and evolution of closteroviruses: sophisticated build-up of large RNA genomes. Annu Rev Phytopathol. 1994;32:261–285. [Google Scholar]
- 12.Dolja V V, Hong J, Keller K E, Martin R R, Peremyslov V V. Suppression of potyvirus infection by coexpressed closterovirus protein. Virology. 1997;234:243–252. doi: 10.1006/viro.1997.8660. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A E, Koonin E V, Lai M M-C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarne A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagiwara Y, Peremyslov V V, Dolja V V. Regulation of closterovirus gene expression examined by insertion of a self-processing reporter and by Northern hybridization. J Virol. 1999;73:7988–7993. doi: 10.1128/jvi.73.10.7988-7993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadare G, Rozanov M, Haenni A-L. Expression of the turnip yellow mosaic virus proteinase in Escherichia coli and determination of the cleavage site within the 206 kDa protein. J Gen Virol. 1995;76:2853–2857. doi: 10.1099/0022-1317-76-11-2853. [DOI] [PubMed] [Google Scholar]
- 18.Karasev A V, Boyko V P, Gowda S, Nikolaeva O V, Hilf M E, Koonin E V, Niblett C L, Cline K, Gumpf D J, Lee R F, Garnsey S M, Lewandowski D J, Dawson W O. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 19.Kasschau K D, Carrington J C. Requirement for HC-Pro during genome amplification of tobacco etch potyvirus. Virology. 1995;209:268–273. doi: 10.1006/viro.1995.1254. [DOI] [PubMed] [Google Scholar]
- 20.Kasschau K D, Cronin S, Carrington J C. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology. 1997;228:251–262. doi: 10.1006/viro.1996.8368. [DOI] [PubMed] [Google Scholar]
- 21.Kasschau K D, Carrington J C. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 22.Klaassen V A, Boeshore M, Koonin E V, Tian T, Falk B W. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly transmitted, bipartite closterovirus. Virology. 1995;208:99–110. doi: 10.1006/viro.1995.1133. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel T A, Roberts J D, Zakour R. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 25.Nuss D L. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peremyslov V V, Hagiwara Y, Dolja V V. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J Virol. 1998;72:5870–5876. doi: 10.1128/jvi.72.7.5870-5876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peremyslov V V, Hagiwara Y, Dolja V V. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc Natl Acad Sci USA. 1999;96:14771–14776. doi: 10.1073/pnas.96.26.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccone M E, Rieder E, Mason P W, Grubman M J. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J Virol. 1995;69:5376–5382. doi: 10.1128/jvi.69.9.5376-5382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satyanarayana T, Gowda S, Boyko V P, Albiach-Marti M R, Mawassi M, Navas-Castillo J, Karasev A V, Dolja V, Hilf M E, Lewandowski D J, Moreno P, Bar-Joseph M, Garnsey S M, Dawson W O. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci USA. 1999;96:7433–7438. doi: 10.1073/pnas.96.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijder E J, Meulenberg J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 31.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki N, Chen B, Nuss D L. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J Virol. 1999;73:9478–9484. doi: 10.1128/jvi.73.11.9478-9484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H-Y, Ling K-S, Goszczynski D E, McFerson J R, Gonsalves D. Nucleotide sequence and genome organization of grapevine leafroll-associated virus-2 are similar to beet yellows virus, the closterovirus type member. J Gen Virol. 1998;79:1289–1298. doi: 10.1099/0022-1317-79-5-1289. [DOI] [PubMed] [Google Scholar]