Abstract
In contrast to wild-type mouse mammary tumor virus (MMTV), the MMTV mutants with specific deletions in the U3 region of their long terminal repeats cause T-cell lymphomas. In 30% of T-cell lymphomas arising in BALB/c mice infected with MLA-MMTV, a leukemogenic MMTV mutant, we have found that MMTV proviruses were integrated into a short region of the Notch1 genome, so that truncated Notch1 transcripts encoding the transmembrane and the cytoplasmic domains of Notch1 protein could be expressed. Thus, Notch1 is a major target of provirus insertional mutagenesis in these T-cell lymphomas.
Mouse mammary tumor virus (MMTV) is associated primarily with induction of mammary adenocarcinomas. Wnt, fibroblast growth factor, and Notch gene families have been identified as the major cellular proto-oncogenes activated by integrated MMTV proviruses in mammary tumors (reviewed in references 4, 11, 15, and 35). However, in mice with active MMTVs (such as the DBA/2 and GR strains), variants of MMTV provirus with specific deletions of about 350 to 500 nucleotides in the U3 region of the long terminal repeat (LTR) often associate with spontaneously developed T-cell lymphomas (9, 20, 22, 23, 36). Moreover, type B leukemogenic virus (TBLV), a MMTV variant with similar LTR alteration, induces T-cell lymphomas in mice after a very short (ca. 50-day) latency period (2, 3). We have previously cloned the rearranged LTRs from extra MMTV proviruses present in two DBA/2 mouse lymphoma cell lines, MLA and DL-8, and showed that these rearranged LTRs exhibit marked transcriptional activities in T-cell lines (36) compared with the wild-type LTRs. Hsu et al. also demonstrated that MMTV provirus in T-cell lymphomas lack a negative regulatory element in the LTR (14). These results suggested that these MMTV proviruses acquire a selective advantage in lymphocytes by specific LTR alterations. Furthermore, by using an infectious MMTV provirus clone (31), we have constructed pathogenic MMTV proviruses with these rearranged LTRs and demonstrated that these MMTV variants do induce T-cell lymphomas in adult BALB/c mice after an average latency period of 30 weeks, but they no longer induce mammary tumors (37). These results provided direct evidence that the small deletion of specific LTR sequences is necessary and sufficient to convert target tissue of MMTV transformation.
Notch family genes encode transmembrane receptor proteins mediating signals which regulate various cell fate decisions that involve cell-cell interactions (reviewed in reference 1). To date, four members of this family have been identified in the mouse. The extracellular domain of the Notch protein contains signal peptide, 29 epidermal growth factor (EGF)-like repeats (the binding site of the Delta-Serrate-Lag2 family of ligand proteins), and 3 Notch/lin-12 repeats. The intracellular domain contains a RAM domain (34) and Cdc10/ankyrin repeats, both of which can bind the RBP-J/CBF-1 transcription factor. Recent studies showed that ligand binding induces proteolytic cleavage of Notch protein and that the cleaved intracellular form of Notch makes a complex with RBP-J/CBF-1, translocates into the nucleus, and regulates transcription of target genes (7, 32; reviewed in reference 5). Loss of the extracellular domain of the Notch protein causes constitutive activation of the protein and thus is known to be associated with tumorigenesis. MMTV or intracisternal type A particle provirus integration at the Notch4/int3 locus leads to expression of a truncated Notch4/int3 protein which causes mouse mammary tumors (11, 17, 18). Similar MMTV provirus-mediated activation of Notch1 has been reported in mouse mammary tumors (8). On the other hand, in line with the cell fate regulatory function of Notch1 during T-cell development (25, 30), an association of Notch1 rearrangement with T-cell lymphoma induction has been reported: in human T-cell acute lymphoblastic leukemia, the chromosomal translocation t(7;9) joins a portion of Notch1/Tan1 to the T-cell receptor β locus (10); transplantation with bone marrow cells expressing activated Notch1 allele led to exclusive development of T-cell neoplasms in mice (27); and the Notch1 gene is known to be a target of provirus insertions in T-cell lymphomas arising in Moloney murine leukemia virus (Mo-MuLV)-infected MMTVD/myc transgenic mice, a transgenic mouse bred on a CD1 background which overexpresses the c-myc gene under the control of TBLV LTR (12, 13, 26). In this case, collaboration of c-myc and Notch1 for oncogenesis was suggested, because frequent integration of Mo-MuLV proviruses in the Notch1 allele was observed in lymphomas arising in myc transgenic mice but not in nontransgenic littermates (12).
Currently, two common provirus integration sites, Tblvi1 (24) and c-myc (28) have been found in TBLV-induced T-cell lymphomas, but the gene(s) activated at the Tblvi1 locus is unidentified. In addition, the cellular proto-oncogenes activated by other leukemogenic MMTV in T-cell lymphomas remain unknown. We report here that Notch1 is a major target of provirus insertional mutagenesis in the T-cell lymphoma arising in a leukemogenic MMTV-infected BALB/c mouse. Provirus integrations led to the generation of truncated Notch1 transcripts which encode the transmembrane and the cytoplasmic domains of Notch1 protein.
Rearrangement of the Notch1 gene in T-cell lymphomas developed in MLA-MMTV-infected BALB/c mice.
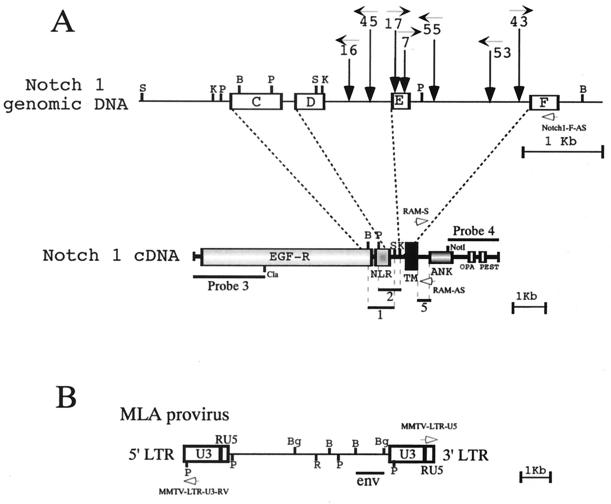
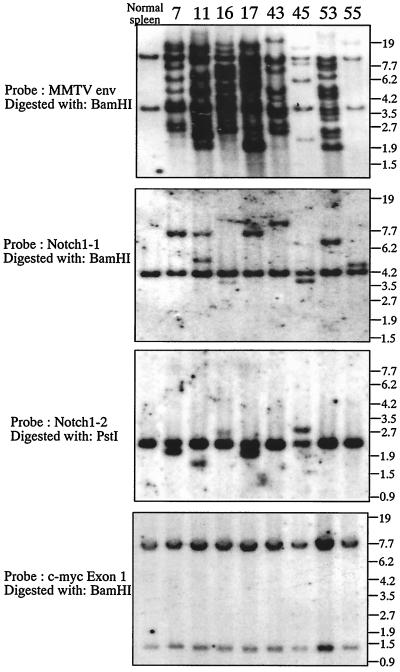
We have analyzed by Southern blotting whether rearrangement of the Notch1 gene occurred in 73 T-cell lymphomas developed in BALB/c mice which were infected with a strain of leukemogenic MMTV, MLA-MMTV (37). The following five Notch1 cDNA fragments were prepared from pmNotch1 (19), the pBluescript II SK(+) containing the entire coding region of mouse Notch1 cDNA and used as probes for Southern and Northern blot analyses: probe 1, a 0.6-kb BamHI-SacI fragment encoding from the C terminus of the EGF-like repeats to a point located between Notch/lin-12 repeats and the transmembrane domain; probe 2, a 0.5-kb PstI-KpnI fragment encoding from the Notch/lin-12 repeats to the N terminus of the transmembrane domain; probe 3, a 2.0-kb ClaI fragment encoding from the N terminus to the middle of the EGF repeats; probe 4, a 1.5-kb NotI fragment encoding from the cdc10/ankyrin repeat to the C terminus; and probe 5, a 0.3-kb fragment encoding the RAM domain (16) which was amplified by PCR using the sense primer RAM-S (5′-CGGCGCCAGCATGGCCAGCTCTGG-3′, corresponding to nucleotide positions 5329 to 5352 of Notch1) and the antisense primer RAM-AS (5′-CTGAGGCGGTGTTGGGGCCAT-3′, corresponding to nucleotide positions 5620 to 5640). In addition, a 1.2-kb BamHI-BglII fragment encoding env of MMTV provirus (31, 37) was used to analyze MMTV provirus integration. (The positions of these probes are shown schematically [see Fig. 2].) Southern blotting with Notch1 probes 1 and 2 revealed that rearrangement of the Notch1 genome occurred in 34% (25 of 73) of the T-cell lymphomas screened. Eight tumors giving rise to rearranged Notch1 genomic DNA fragments which were equimolar relative to the germ line Notch1 genomic DNA fragments were selected for further analysis. The results of Southern blot analysis of these eight tumors and normal spleen cells (used as a control) are shown in Fig. 1. The blot with MMTV env probe showed that DNAs of all lymphomas contained several newly acquired host-virus junction fragments, which were unique for each tumor, indicating that each tumor originated from a single or a few clones which were infected with MMTV (Fig. 1, top panel). The rearrangement of the Notch1 genome in these lymphomas is shown in the second and third panels of Fig. 1; in addition to the 4.2-kb BamHI fragment derived from the unaffected Notch1 allele, Notch1 probe 1 hybridized with an additional BamHI fragment which is unique for each tumor. Similarly, in addition to the 2.2-kb PstI fragments derived from the Notch1 unaffected allele, Notch1 probe 2 hybridized with an additional PstI fragment in five tumors. To confirm the location and orientation of the MLA-MMTV provirus inserted in each tumor, PCR analysis was performed. According to the expected orientation of the provirus, one of the following sense primers from the MLA-MMTV LTR sequence (shown in Fig. 2B) was used in combination with an antisense primer from the Notch1 exon F sequence (named Notch1-F-AS, 5′-GAAGGCGGCCGCTGCCACGTACATGAGGTG-3′, corresponding to nucleotide positions 5251 to 5280 of Notch1 cDNA [Fig. 2A]); MMTV-LTR-U5 (5′-CCCGTCTCCGCTCGTCACTTATC-3′, corresponding to the sense strand of the LTR U5 sequence) and MMTV-LTR-U3-RV (5′-GTGTAGGACACTCTCGGGAGT-3′, corresponding to the antisense strand of the LTR U3 sequence). The MMTV-LTR-U5 and MMTV-LTR-U3-RV primers were used when provirus was integrated in the same and in the opposite transcriptional orientations, respectively, as the Notch1 gene. The length of the PCR products amplified from tumor DNAs was then determined (data not shown). Based on these Southern blotting and PCR results and restriction maps of the unaffected Notch1 allele, the integration site and the orientation of MMTV provirus in the rearranged Notch1 allele was determined in each tumor (Fig. 2A). The provirus integrated into lymphoma 55, however, seems to have a deletion in the 3′ part of provirus including the env gene (data not shown).
FIG. 2.
Schematic representation of the proviruses integrated within the Notch1 gene in MLA-MMTV-induced T-cell lymphomas. (A) Rearranged regions of the Notch1 genome are shown together with a schematic representation of the full-length Notch1 cDNA. Exons and introns are indicated by open boxes and solid lines, respectively. Consistent with previous reports (8, 12, 13), the exons are labeled C, D, E, and F. The vertical arrows indicate the sites of provirus integration. The horizontal arrow with a number shows the transcriptional orientation of provirus in each tumor. The structural features of Notch1 cDNA are indicated: EGF-R (EGF-like repeats), NLR (Notch/lin-12 repeats), TM (transmembrane domain), RAM (RAM23 homologous domain), ANK (Cdc10/ankyrin repeats), OPA (opa repeats), and PEST (PEST sequence motif). The probes used in Southern and Northern analyses are indicated by bars. The positions of the Notch1-F-AS, RAM-S, and RAM-AS primers are indicated by open arrows. (B) The structure of MLA-MMTV provirus is schematically presented. The positions of the MMTV env probe and the MMTV-LTR-U5 and the MMTV-LTR-U3-RV primers are indicated by a bar and open arrows, respectively. Restriction sites: B, BamHI; K, KpnI; P, PstI; S, SacI; Bg, BglII; R, EcoRI.
FIG. 1.
Southern blot hybridization demonstrating the rearrangement of the Notch1 locus in T-cell lymphomas arising in MLA-MMTV-infected BALB/c mice. The MLA strain of the leukemogenic recombinant MMTV was described previously (36, 37). Southern blotting was performed as described earlier (18, 36). Genomic DNAs (20 μg) from eight representative MLA-MMTV-induced T-cell lymphomas (the identification number of each tumor is shown at the top) and normal BALB/c mouse spleens were digested with restriction endonucleases, Southern blotted, and hybridized. The restriction enzyme and the probe used are indicated on the left side of each panel. Numbers on the right indicate the migration positions of DNA molecular size markers (in kilobases).
Because provirus insertions near the c-myc have been reported in TBLV-induced T-cell lymphomas (28) and collaboration of c-myc and Notch1 has been suggested in the case of Mo-MuLV-induced T-cell lymphomas (12), rearrangement of the c-myc genome in these lymphomas was also analyzed with two probes: a 2.0-kb SacI fragment and a 5.5-kb BamHI fragment spanning exon 1 and exons 2 and 3, respectively, of the mouse c-myc genome. Southern blotting with these probes revealed, however, that no provirus was integrated near the c-myc gene (a 15-kb region covering all the c-myc exons, starting 7.5-kb upstream of exon 1 and ending at 3 kb downstream of exon 3) in these 73 lymphomas (data obtained with the c-myc exon 1 probe are shown in Fig. 1, bottom panel). In addition, probes encoding the RAM domains of the Notch2, -3, and -4 genes which were amplified by PCR (16, 18) failed to reveal rearrangement of these Notch genes (data not shown). Moreover, no rearrangement of the p53 and retinoblastoma (Rb) genes was detected in these tumors (data not shown, a 1.0-kb XhoI-StuI fragment of p53 cDNA and a 1.7-kb BamHI fragment of Rb cDNA were used as probes).
Expression of truncated Notch1 transcripts in lymphomas with Notch1 gene rearrangements.
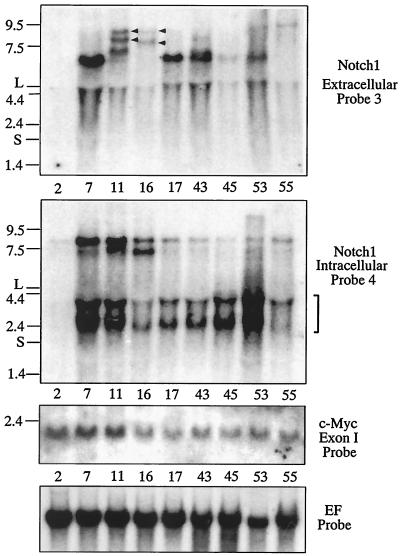
To determine whether provirus insertion affected Notch1 gene expression, a Northern blot analysis was performed with probes encoding the intracellular (probe 4) or the extracellular (probe 3) region of Notch1 protein. The eight MLA-MMTV-induced lymphomas described above, as well as lymphoma 2, an MLA-MMTV- induced T-cell lymphoma with normal Notch1 alleles, were subjected to this analysis (Fig. 3). High levels of expression of 2.5- to 4.0-kb transcripts solely hybridized with the intracellular probe were detected in all of the lymphomas with Notch1 gene rearrangements (Fig. 3, the second panel), suggesting that these truncated RNA encode the intracellular domain of the Notch1 protein. In addition, in some lymphomas (i.e., lymphomas 7, 11, and 16), marked expressions of 7.5- to 8.5-kb transcript were detected with this probe. On the other hand, 7.0- to 7.5-kb transcripts, which solely hybridized with the extracellular probe, were also detected in some lymphomas (lymphoma 7, 11, 17, 43, and 53 in Fig. 3, top panel). In contrast, no Notch1 transcript was detected in lymphoma 2. Basically, these results indicated that provirus insertion commonly induced RNA species, which could encode the intracellular domain of Notch1. Since the provirus had integrated in the direction of Notch1 gene transcription in lymphomas 7, 17, and 43, the proviral 3′ LTR appears to function as a promoter for the Notch1 transcripts of about 2.5 kb detected with the intracellular probe. In the same lymphomas, on the other hand, marked expression of ∼7.0-kb transcripts, which solely hybridized with the extracellular probe, was also detected. Probably, the 5′ LTR of the integrated provirus provided not only enhancer(s) to the Notch1 promoter to achieve high levels of Notch1 transcription but also a transcriptional termination signal. This might be the mechanism generating the Notch1 transcripts of 7.0 kb. In lymphomas 11 and 16, expression of 8- to 9-kb RNA species which hybridized with both extracellular and intracellular probes was detected (Fig. 3, top panel). The mechanism generating these Notch1 transcripts is not clear, but the following is possible: some sequence in the integrated provirus may have provided enhancer(s) for Notch1 promoter, which led to transcription of entire Notch1 gene even in the presence of integrated provirus (assuming that the provirus integrated in the opposite orientation as that in lymphoma 16 could not provide transcriptional termination signal). Then, the Notch1 RNA species encoding both extracellular and intracellular domains of Notch1 were generated by RNA splicing.
FIG. 3.
Northern analyses with extracellular or intracellular Notch1 probes and c-myc probe. Total RNAs were prepared from MLA-MMTV-induced T-cell lymphomas with or without Notch1 gene rearrangement. Total RNAs (10 μg) were separated on a 0.75% formaldehyde-agarose gel and Northern blotted as described earlier (17). The blot was hybridized with Notch1 extracellular (top panel), Notch1 intracellular (second panel), c-myc–exon 1 (third panel), and elongation factor-1α (bottom panel) probes. Numbers and the letters shown on the left indicate the migration positions of RNA molecular size markers (in kilobases) and of the large (L) and small (S) subunits of the rRNA. Positions of the 2.5- to 4.0-kb transcript solely hybridized with the intracellular probe are indicated with a blanket on the right side of the second panel. Arrowheads in the top panel indicate RNA species hybridized with both extracellular and intracellular probes. A 3.5-kb BamHI fragment of the human elongation factor-1α (EF) cDNA was used as a probe for RNA loading control.
Because the collaboration of c-myc and Notch1 in development of T-cell lymphoma was reported (12), we compared levels of c-myc mRNA between T-cell lymphomas with or without Notch1 gene rearrangements. Eight lymphomas with the rearrangements (shown in Fig. 1 and 3) and seven MLA-MMTV-induced lymphomas without (lymphomas 2 and 6 and other lymphomas) then expressed similar levels of c-myc mRNA, suggesting that Notch1 activation was not related to the activation of c-myc expression in MLA-MMTV-induced lymphomas. However, these results did not exclude the possibility that the levels of c-myc expression were generally elevated in these lymphomas.
Structure of the truncated Notch1 transcripts.
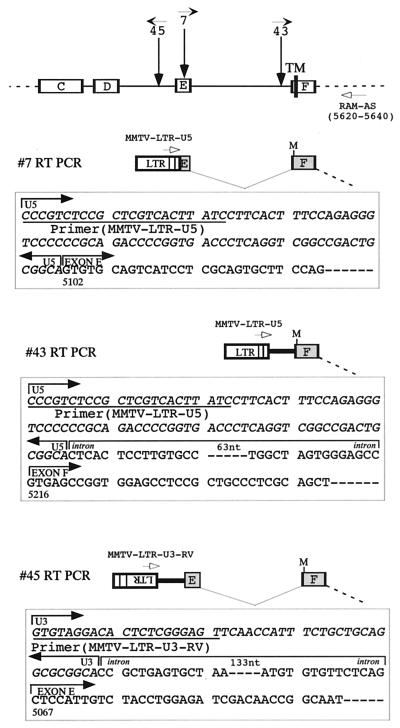
To define the structure of the 2.5-kb Notch1 transcripts solely hybridized with the intracellular probe, and possibly their mode of production as well, lymphomas 7, 43, and 45 were chosen, and the 5′ regions of their Notch1 transcripts were analyzed by reverse transcription-PCR (RT-PCR) (Fig. 4). Single-stranded cDNAs were synthesized from total RNAs from these lymphomas using the SuperScript Preamplification System for First Strand cDNA Synthesis (Gibco-BRL, Gaithersburg, Md.). In the case of lymphomas 7 and 43, where the provirus is integrated in the same transcriptional orientation to Notch1, the cDNA fragment containing sequences of the U5 region of MMTV LTR and the Notch1 RAM domain was amplified by RT-PCR with the single-stranded cDNAs synthesized and a set of primers (sense primer MMTV-LTR-U5 and antisense primer RAM-AS). In case of lymphoma 45, however, in which the provirus is integrated in an orientation opposite to that of Notch1, we assumed that truncated transcripts originated from a cryptic promoter within the provirus and that sequences in the reverse strand in the MMTV LTR U3 might be transcribed and constitute a part of the MMTV-Notch1 chimeric transcript. As expected, RT-PCR with MMTV-LTR-U3-RV and RAM-AS primers generated a 0.78-kb product which hybridized with Notch1 probe 5 (data not shown). The PCR products thus obtained were blunted and phosphorylated with T4 DNA polymerase (NEB) and with T4 polynucleotide kinase (NEB), respectively, and then cloned into the EcoRV site of pBluescript II KS(+) by blunt-end ligation. Sequence analysis revealed that the RT-PCR product obtained from lymphoma 7 was a 0.62-kb DNA fragment whose sequence corresponded to a chimeric virus-Notch1 RNA that started in the LTR (from MMTV-LTR-U5 primer) and was followed by the Notch1 cDNA sequence that started and ended at nucleotide positions 5102 (middle of exon E) and 5640 (position of the RAM-AS primer), respectively. On the other hand, the RT-PCR product from lymphoma 43 was a 0.56-kb DNA fragment corresponding to a chimeric virus-Notch1 RNA that started in the LTR and was followed by 93 nucleotides of the Notch1 intron sequence adjacent to exon F and the Notch1 cDNA sequence that started and ended at nucleotide positions 5216 (first nucleotide of exon F) and 5640, respectively. Furthermore, the RT-PCR product from lymphoma 45 was a 0.78-kb DNA fragment corresponding to a chimeric virus-Notch1 RNA that started in the MMTV-LTR-U3 and was followed by 161 nucleotides of the Notch1 intron sequence adjacent to exon E and Notch1 cDNA sequence that started and ended at nucleotide positions 5067 (first nucleotide of exon E) and 5640, respectively. In all of cases, chimeric virus-Notch1 RNAs used the ATG codon at nucleotide position 5257 (located just N-terminal to the transmembrane domain in exon F) of Notch1 cDNA as an initiation methionine to generate the truncated Notch1 proteins consisting of the transmembrane and intracellular domains. In lymphomas in which the provirus is integrated in an orientation opposite to that of Notch1, however, structure of the 5′ part of the MMTV-Notch1 chimeric transcripts hybridizing with the intracellular probe remains unknown, and their generation mechanism is not clear. In this regard, Girard et al. (12) have analyzed the structure of similar Notch1 transcripts in lymphomas in which Mo-MuLV had integrated in the opposite orientation to Notch1 and reported following two modes of generation for these transcripts: (i) the enhancer in the 5′ LTR of the integrated provirus activated a cryptic promoter within the intronic Notch1 sequence to yield the truncated transcripts and (ii) some distinct truncated transcripts originated from a cryptic promoter within the provirus itself. In our MLA-MMTV-induced tumor cases, the second mode may be operative in lymphoma 45. However, the first mode is also possible because we previously identified a T-lymphocyte-specific enhancer in the U3 region of the MLA LTR (at nucleotide positions 536 to 557 of the U3 sequence [36]). In any case, these results further substantiated the notion that Notch proteins lacking the extracellular domain are tumorigenic. However, the molecular mechanism(s) by which these truncated Notch proteins induce tumors remains to be elucidated.
FIG. 4.
Analysis of the 5′ end of the 2.5-kb truncated Notch1 transcripts in lymphomas 7, 43, and 45 by RT-PCR. At the top, the Notch1 genome is shown as in Fig. 2A. The PCR products generated with the RAM-AS in combination with MMTV-LTR-U5 or MMTV-LTR-U3-RV primers (indicated by open arrows) were cloned and sequenced. Regions of interest are shown in boxes. Sequence homology between the PCR products and the MMTV LTR (21; GenBank accession no. K00556) or the Notch1 cDNA (6; no. Z11886) was analyzed. In this report, all nucleotide positions of the Notch1 cDNA and MMTV are cited from these databases. Nucleotides of the MMTV LTR and Notch1 sequences are indicated by italics and plain letters, respectively. “M” indicates the potential initiating methionine codon corresponding to nucleotide position 5257 of the Notch1 cDNA. TM, transmembrane domain. Sequencing was performed using Thermo Sequenase (Amersham) Kit and a 373 SEQUENCER (Applied Biosystems).
Recently, Sorensen et al. reported that Sint1, a common integration site in SL3-3 MuLV-induced mouse T-cell lymphomas, harbors a putative proto-oncogene of the Septin gene family (33). Interestingly, this locus was suggested to be identical to an MMTV integration site in a T-cell lymphoma, Pad3 (29). Southern blot analysis with Sint1 genomic probe revealed, however, that no MMTV provirus was integrated in the 14-kb HindIII genomic fragment of this locus (in which SL3-3 proviruses were integrated [33]) in 73 MLA-MMTV-induced T-cell lymphomas screened (S.-I. Yanagawa, A. B. Sorensen, and F. S. Pedersen, unpublished results). Because Notch1 rearrangements were observed in only 30% of these lymphomas, clearly, further screenings for proviral common integration sites is necessary.
Acknowledgments
We thank T. Oikawa (Sasaki Institute) and N. Tsuchida (Tokyo Medical and Dental University) for c-myc and the p53 plasmid, respectively. We also thank A. B. Sorensen and F. S. Pedersen for the Sint1 genomic probe.
This work was supported by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan to S.-I.Y.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Ball J K, Dekaban G A. Characterization of early molecular events associated with thymic lymphoma induction following infection with a thymotropic type-B retrovirus. Virology. 1987;161:357–365. doi: 10.1016/0042-6822(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 3.Ball J K, Diggelmann H, Dekaban G A, Grossi G F, Semmler R, Waight P A, Fletcher R F. Alteration on the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J Virol. 1988;62:2985–2993. doi: 10.1128/jvi.62.8.2985-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan R. MMTV induced mutations in mouse mammary tumors: their potential relevance to human breast cancer. Breast Cancer Res Treatment. 1996;39:33–44. doi: 10.1007/BF01806076. [DOI] [PubMed] [Google Scholar]
- 5.Chan Y-M, Jan Y N. Roles for proteolysis and trafficking in Notch maturation and signal transduction. Cell. 1998;94:423–426. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- 6.Del Amo F F, Gendron-Maguire M, Swiatek P J, Jenkins N A, Copeland N G, Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15:259–264. doi: 10.1006/geno.1993.1055. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Anneart W, Cupers P, Safting P, Craessaerts K, Munn L S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, Goate A, Kopan R. A presenilin-1-like dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 8.Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 9.Dudley J, Risser R. Amplification and novel locations of mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984;49:92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 11.Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- 12.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in the thymomas of MMTVD/myc transgenic mice suggest a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 13.Girard L, Jolicoeur P. A full-length Notch1 allele is dispensable for transformation associated with a provirally activated truncated Notch1 allele in MuLV-infected MMTVD/myc transgenic mice. Oncogene. 1998;16:517–522. doi: 10.1038/sj.onc.1201562. [DOI] [PubMed] [Google Scholar]
- 14.Hsu C-L L, Fabritius C, Dudley J. Mouse mammary tumor virus provirus in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J Virol. 1988;62:4644–4652. doi: 10.1128/jvi.62.12.4644-4652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochem Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 17.Kordon E C, Smith G H, Callahan R, Gallahan D. A novel non-mouse mammary tumor virus activation of the int-3 gene in a spontaneous mouse mammary tumor. J Virol. 1995;69:8066–8069. doi: 10.1128/jvi.69.12.8066-8069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J-S, Haruna T, Ishimoto A, Honjo T, Yanagawa S. Intracisternal type A particle-mediated activation of the Notch4/int3 gene in a mouse mammary tumor: generation of truncated Notch4/int3 mRNAs by retroviral splicing events. J Virol. 1999;73:5166–5171. doi: 10.1128/jvi.73.6.5166-5171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J-S, Ishimoto A, Honjo T, Yanagawa S. Murine leukemia provirus-mediated activation of the Notch1 gene leads to induction of HES-1 in a mouse T lymphoma cell line, DL-3. FEBS Lett. 1999;455:276–280. doi: 10.1016/s0014-5793(99)00901-1. [DOI] [PubMed] [Google Scholar]
- 20.Lee W T-L, Prakash O, Klein D, Sarker N H. Structural alteration in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987;159:39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- 21.Majors J E, Varmus H E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983;70:495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalides R, Wagenaar E. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology. 1986;154:76–84. doi: 10.1016/0042-6822(86)90431-9. [DOI] [PubMed] [Google Scholar]
- 23.Michalides R, Wagenaar E, Hilkins J, Hilgers J, Groner B, Hynes N E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982;43:819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller R E, Baggio L, Kozak C A, Ball J K. A common integration locus in type B retrovirus-induced thymic lymphomas. Virology. 1992;191:628–639. doi: 10.1016/0042-6822(92)90238-k. [DOI] [PubMed] [Google Scholar]
- 25.Osborne B, Miele I. Notch and the immune system. Immunity. 1999;11:653–663. doi: 10.1016/s1074-7613(00)80140-5. [DOI] [PubMed] [Google Scholar]
- 26.Panquette Y, Doyon L, Laperriere A, Hanna Z, Ball J, Sekaly R P, Jolicoueur P. A viral longterminal repeat expressed in CD4+ CD8+ precursors is downregulated in mature peripheral CD4− CD8+ or CD4+ CD8− T cells. Mol Cell Biol. 1992;12:3522–3530. doi: 10.1128/mcb.12.8.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch allele. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajan L, Brroussard D, Lozano M, Lee C G, Kozak C A, Dudley J P. The c-myc locus is a common integration site in type B retrovirus-induced T-cell lymphomas. J Virol. 2000;74:2466–2471. doi: 10.1128/jvi.74.5.2466-2471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajan L, Dudley J P. An MMTV integration site maps near the distal end of mouse chromosome 11. Mamm Genome. 1997;8:295–296. doi: 10.1007/s003359900420. [DOI] [PubMed] [Google Scholar]
- 30.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 31.Shackleford G M, Varmus H E. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroter E H, Kissinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen A B, Lund A H, Ethelberg S, Copeland N G, Jenkins N J, Pedersen F S. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family. J Virol. 2000;74:2161–2168. doi: 10.1128/jvi.74.5.2161-2168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jk/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 35.van Leeuwen F, Nusse R. Oncogene activation and oncogene cooperation in MMTV-induced mouse mammary cancer. Semin Cancer Biol. 1995;6:127–133. doi: 10.1006/scbi.1995.0018. [DOI] [PubMed] [Google Scholar]
- 36.Yanagawa S, Murakami A, Tanaka H. Extra mouse mammary tumor proviruses in DBA/2 mouse lymphomas acquire a selective advantage in lymphocytes by alteration in the U3 region of the long terminal repeat. J Virol. 1990;64:2474–2483. doi: 10.1128/jvi.64.6.2474-2483.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanagawa S, Kakimi K, Tanaka H, Murakami A, Nakagawa Y, Kubo Y, Yamada Y, Hiai H, Kuribayashi K, Masuda T, Ishimoto A. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J Virol. 1993;67:112–118. doi: 10.1128/jvi.67.1.112-118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]