Abstract
Imbalances in protein homeostasis may result in pathologic states highly relevant to human brain development. The major protein degradation machineries in the cell are the ubiquitin-proteasome system (UPS) and autophagy, both of which are frequently associated with the pathogenesis of neurodevelopmental disorders (NDD). One of the main proteasome-associated deubiquitinating enzymes (DUB), USP14, mediates the removal of ubiquitin from protein substrates at the proteasome, and acts as a cellular hub between the UPS and autophagy. Here, we identified biallelic USP14 variants in four individuals from three unrelated families: one fetus, a newborn with a syndromic NDD, and two siblings affected by a progressive neurological disease. Specifically, the two siblings from the latter family carried two compound heterozygous variants (c.8T>C p.(Leu3Pro) and c.988C>T p.(Arg330*), while the fetus had a homozygous frameshift c.899_902del p.(Lys300Serfs*24) variant and the newborn patient harbored a homozygous frameshift c.233_236del p.(Leu78Glnfs*11) variant. To determine the functional consequences of USP14 biallelic variants on UPS function, we assessed patient-derived cells as well as CRISPR-Cas-generated surrogate cells and provided evidence of alterations of USP14 N-terminal methionine excision (NEM) as well as changes in proteasome, autophagy and mitophagy activities. Taken together, our results suggest a key role for USP14 in early human neurodevelopment.
Keywords: Neurodevelopmental disorders, ubiquitin-proteasome system, USP14, loss-of-function variants, N-terminal methionine excision
INTRODUCTION
A tight regulation of protein homeostasis is crucial to vertebrate neurodevelopment. In eukaryotic cells, the ubiquitin proteasome system (UPS) and autophagy participate in maintaining the highly dynamic pool of intracellular proteins by preferentially eliminating damaged or mutant proteins 1. Ubiquitination is a central post-translational modification (PTM) process that mediates the covalent tagging of protein substrates destined for recognition and subsequent degradation by the proteasome and autophagy. Depending in part on ubiquitin chain topology, ubiquitin-modified substrates may indeed be targeted to the 26S proteasome or LC3-positive autophagosomes. Importantly, once proteasome substrates have been committed to degradation, ubiquitin must be removed to facilitate substrate unfolding and subsequent translocation into the 20S catalytic core particle 2. There exist three proteasome-associated deubiquitinating enzymes (DUB) capable of trimming ubiquitin chains from protein substrates prior to their degradation, namely: the two cysteine proteases USP14 3 (Ub-specific protease 14, Ubp6 in Saccharomyces cerevisiae) and UCH37/UCHL5 4, as well as the metalloprotease PSMD14 (Rpn11) 5,6. In addition to cleaving ubiquitin from protein substrates, USP14 and its yeast ortholog Ubp6 have been shown to regulate proteasome activity via both their catalytic domains and their N-terminal ubiquitin-like (UBL) domains 7. Moreover, USP14 exhibits multiple regulatory roles such as the modulation of GABAergic synapses 8 as well as negative feedback between UPS and autophagy 9. USP14 is thus a cellular pivot orchestrating the crosstalk between these two proteolytic pathways.
Proper functioning of the UPS is essential to neurodevelopment, as several critical signaling pathways involved in early development of the central nervous system (CNS) such as synaptogenesis are regulated by ubiquitination 10. Several DUBs have been associated with impaired neurodevelopment, an observation that is in line with the emerging view that UPS dysfunction is a major pathophysiological mechanism of neurodevelopmental disorders (NDD) 11. These include notably Hao-Fountain syndrome (MIM: 602519), which is caused by USP7 dominant variants 12. Variants of the USP9X (MIM: 300072) and USP27X (MIM: 300975) genes are also a common cause of either X-linked dominant (MIM: 300968) 13 or recessive NDD (MIM: 300919, XLID105) 14,15. Similarly, variants in genes coding for DUBs such as EIF3F (MIM: 618295) 16, STAMBP (MIM: 614261) 17, ALG13 (MIM: 300884) 18, or OTUD6B (MIM: 617452) 19 have been associated with NDD. In this study, we provide evidence that biallelic USP14 variants are responsible for severe neurodevelopmental syndrome and explore the functional consequences of USP14 impairment.
MATERIALS AND METHODS
Ethics statement
Human genetic studies conducted in research laboratories were approved by local ethics committees from participating centers. Written informed consent was obtained for all individuals of the study by their parents prior to testing. All affected individuals underwent clinical examination by at least one expert clinical geneticist of each center.
Exome Sequencing
For exome sequencing (ES), DNA was extracted from peripheral blood mononuclear cells (PBMC). We performed ES within diagnostic or research settings according to manufacturer’s instructions, generated and aligned to human genome hg19 assembly. ES data analysis was performed following GATK’s best practices (v3.4).
Generation of CRISPR/Cas9 USP14 knockout clones
A guide RNA sequence (CTCTACTCCGGTGAGCCCTGCGGTG) targeting the first exon of the USP14 gene was cloned into the pSpCas9(BB)-2A-Puro (PX459, V2.0, Addgene, #62988) using the BbsI restriction sites. The Cas9 expression vector containing the guide RNA was introduced into SH-SY5Y cells using the JetPRIME transfection reagent (Polyplus-transfection® SA) following the manufacturer’s recommendations. At 24-hour post-transfection, cells were plated at low density in 96-well plates in the presence of puromycin (10 μg/ml). Resistant clones were further cultivated and assessed for USP14 exon 1 damages by PCR amplification prior to Sanger sequencing.
Cell culture
Peripheral blood mononuclear (PBMC) collected from patients carrying UPS14 variants (Individuals 1 and 2) and related healthy individuals were subjected to T cell expansion by seeding them on irradiated feeder cells in the presence of IL-2 (50 U/ml) and L-PHA (1 mg/ml) for 3–4 weeks following the procedure of Fonteneau et al. 20. The SH-SY5Y neuroblastoma cell line was a laboratory stock cultivated in the presence of DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. After obtaining informed consent, skin biopsies were obtained from individual of Family 1 following standard clinical procedure. Fibroblast cell lines were cultured in DMEM medium supplemented with 10% FBS, 1 mM sodium pyruvate, 4 mM glutamine and 1% penicillin/streptomycin.
SDS-PAGE and western-blotting
Cells were lysed in standard RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP40, 0.1% SDS) and proteins were quantified by BCA following the manufacturer’s instructions. Ten to twenty micrograms of protein lysates were separated by 10, 12, or 15% SDS-PAGE and subsequently blotted (200V, 400 mA, 1 h) onto PVDF membranes (Millipore®). Primary antibodies used in this study were directed against USP14 (Santa Cruz Biotechnology Inc, clone F4), α4 proteasome subunit (i.e.; PSMA7, Enzo Life Sciences, clone MCP34), BNIP3L (Cell Signaling Technology®, 12396), LC3B (Cell Signaling Technology®, 3868), β5 (i.e.; PSMB5) proteasome subunit (Abcam, ab3330), Rpt1 (i.e. PSMC2) proteasome subunit (Enzo Life Sciences, BML-PW8315), α-tubulin (Abcam, DM1A) and GAPDH (Cell Signaling Technology®, clone 14C10). After overnight incubation, membranes were incubated with anti-mouse and -rabbit conjugated secondary antibodies (1/5.000) for 1h at room temperature. Proteins were visualized using Clarity™ Western ECL Substrate (Biorad).
In-plate peptidase activity assay
Cell were lysed in ice-cold homogenization TSDG buffer (10 mM Tris pH 7.0, 10 mM NaCl, 25 mM KCl, 1.1 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 2 mM ATP, 1 mM NaN3, 20 % glycerol) and proteins were extracted using freeze/thawing in liquid nitrogen. Protein quantification of the soluble lysates was determined by Bradford. Proteasome chymotrypsin-like activity was measured on 96-well opaque microtiter plates (Greiner) by incubating ten micrograms of TSDG-generated whole-cell lysates with 0.1 mM of the Suc-Leu-Leu-Val-Tyr-AMC proteasome fluorogenic peptide (Bachem) in quadruplicates in a final volume of 100 μl. AMC release was measured over a 3-h period of time using a fluorescence plate reader at 360/460 nm (NanoQuant Plate™, Tecan).
RNA extraction and RT-qPCR
Total RNA was isolated from resting T cells using the kit from Analytic Jena AG following the manufacturer’s instructions. Isolated RNA (100–500 ng) was next reverse transcribed using the M-MLV reverse transcriptase (Promega) following the manufacturer’s recommendations. Quantitative PCR was performed using the Premix Ex Taq™ (probe qPCR purchased from TaKaRa) and in duplicates to determine the mRNA levels of six IFN-stimulated genes (ISG) using FAM-tagged TaqMan™ Gene Expression Assays obtained from Thermo Fisher Scientific according to the manufacturer’s instructions. TaqMan™ probes used in this study for ISG quantification included IFI27, IFI44L, IFIT1, ISG15, RSAD2 and IFI44. The cycle threshold (Ct) values for target genes were converted to values of relative expression using the relative quantification (RQ) method (2-ΔΔCt). Target gene expression was calculated relative to Ct values for the GAPDH control housekeeping gene. A type I IFN score representing the median fold change of the six ISG relative to a single calibrator control was calculated for each sample following the procedure of Rice et al. 21.
Plasmid construction and transfection
The cDNA for USP14 was amplified by RT-PCR from total RNA isolated from SH-SY5Y neuroblastoma cells and cloned into the pcDNA3.1/myc-HIS expression vector (Invitrogen). The single amino acid substitutions Leu3Pro and Arg330Term were introduced by site-directed mutagenesis and inverse PCR, respectively. SH-SY5Y wild-type and USP14 knockout cells were transfected using JetPRIME reagent (Polyplus-transfection® SA) according to the instructions of the manufacturer.
Purification of USP14 and proteasome
Human USP14 wild-type (WT) or Leu3Pro (L3P) was subcloned into the pET SUMO TA vector (Invitrogen), then transferred into the pTXB1 vector (NEB) for N-terminal His-sumo and C-terminal intein/chitin binding domain (CBD) tagging. The constructs were expressed in E. coli Rosetta 2 (DE3) cells (Novagen), and purified using Ni-NTA agarose resin (Qiagen) according to the manufacturer’s instructions. The His-SUMO tag was cleaved using the His-Ulp1 SUMO protease, and the resulting USP14-intein-CBD protein was retrieved on Ni-NTA resin. USP14-intein-CBD, WT or L3P, was further purified on chitin beads, and released from the beads in untagged form by DTT cleavage. Human proteasomes were affinity-purified as previously described28 from a stably-transformed HEK293T cell line in which proteasome subunit Rpn11 is HTBH-tagged. This tag induces biotinylation, and proteasomes were thus isolated from the lysate using NeutrAvidin agarose resin (Thermo Scientific). The proteasome was eluted from the resin by TEV cleavage, which removes the HTBH tag. Ubiquitin-vinylsulfone (Ub-VS)-treated proteasome was prepared by on-column incubation with 2.5 μM Ub-VS prior to TEV cleavage.
In vitro de-ubiquitination and degradation assays
To compare the deubiquitinating activity of the proteasome-free form of WT or L3P USP14, each variant was tested at 1.5 μM by incubating with subsaturating amounts of ubiquitin-amido methyl coumarin (Ub-AMC; 1.5 μM) in an assay buffer of 50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, and 1 mg/mL ovalbumin. To measure proteasome-bound USP14 activity for kinetic analysis, 1 nM Ub-VS-treated human proteasomes was reconstituted with graded concentrations of recombinant USP14 and 1 μM Ub-AMC. Ub-AMC cleavage was monitored in real time by measuring fluorescence at Ex365/Em460 with an Envision Plate reader (Perkin Elmer) equipped with an appropriate mirror (e.g., LANCE/DELFIA, 400 nm). To perform in vitro de-ubiquitination and degradation assays with ubiquitin conjugates as the physiological substrate, 4 nM human proteasome was incubated with polyubiquitinated N-terminal cyclin B1 (UbnNCB1; ~120 nM final; HA-tagged) or polyubiquitinated full length cyclin B1 (UbnCCNB1; ~40 nM final) in proteasome assay buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM ATP). Purified recombinant USP14 WT or L3P was reconstituted with proteasome for 5 min before initiating the reaction. Reactions were quenched by adding 5×SDS–PAGE sample buffer, boiled for 5 min, then subjected to SDS–PAGE and immunoblotting analysis using anti-HA-HRP (Roche) or anti-cyclin B1 (Neomarkers) antibody.
Mass spectrometry (MS) analysis
In-gel protein digestion with trypsin was performed on SDS-PAGE loaded with 10 μg of whole-cell lysates generated from fibroblasts of one heathy donor as well as individuals carrying single or biallelic USP14 variants. The resulting digested peptides were extracted from the gel, and a mixture of heavy-labeled peptides containing 20 fmol of both wild-type (PLYSVTVK) and Leu3Pro (PPYSVTVK) USP14 peptides was added. Both peptides were labeled with a heavy valine [13C(5)15N(1), +6 Da]. Subsequently, peptides were then desalted and analyzed on a Q-Exactive MS instrument (Thermo Scientific) using a PRM (parallel reaction monitoring) method to monitor the abundance of the labeled light and heavy peptides used for quantification. Specifically, the heavy-labeled standards were employed for quantifying endogenous wild-type and Leu3Pro peptides in all samples through an Absolute QUAntification (AQUA) MS-based method.
RESULTS
Clinical features of individuals with biallelic USP14 variants
Here, we describe the phenotypic features of four individuals from three unrelated families (Table 1, Figure S1). We identified two USP14 variants in two affected individuals of Family 1 and submitted our data to GeneMatcher platform 22 from which patients 3 and 4 were further enlisted. The two patients from Family 1 showed a progressive neurological disease. The first neurological signs and symptoms were reported at the age of 2 for both siblings, with no specific scale employed for assessing neurological functions. Cognitive functions, assessed using the Wechsler Intelligence Scale for Children (WISC) and the Wechsler Adult Intelligence Scale (WAIS), remained unaffected. Their neurological examination at the age of 16 and 18 years revealed ptosis, nystagmus, tongue fasciculations, proximal upper and lower limb muscular weakness, hand weakness tremor as well as dysmetria. Both individuals presented with a progressive disease, with loss of ability to move (wheelchair), optic neuropathy, multiple tendon retractions, and severe scoliosis requiring surgery.
Table 1.
Clinical features of the patients with point mutations and copy number variants involving USP14.
| Family | Family 1 | Family 1 | Family 2 | Family 3 | Turgut et al., 2021 | Turgut et al., 2021 | Turgut et al., 2021 |
|---|---|---|---|---|---|---|---|
| Individual | P1 | P2 | P3 | P4 | P6 – fetus 1 | P6 – fetus 2 | P6 – fetus 3 |
| Variant type | Compound heterozygous missense + nonsense | Compound heterozygous missense + nonsense | homozygous frameshift | homozygous frameshift | homozygous frameshift | homozygous frameshift | homozygous frameshift |
| Variant (NM_005151.4) | c.8T>C, p.Leu3Pro; c.988C>T, p.Arg330* | c.8T>C, p.Leu3Pro; c.988C>T, p.Arg330* | c.899_902del, p.Lys300Serfs*24 | c.233_236del p.Leu78Glnfs*11 | c.233_236del; p.Leu78Glnfs*11 | c.233_236del; p.Leu78Glnfs*11 | c.233_236del; p.Leu78Glnfs*11 |
| Gender | M | F | M | ND | M | M | M |
| Age at last follow up | 18y | 16y | NA | 2 days | NA | NA | NA |
| Birth term (WG) | 38 | 38 | PTP 29+4 weeks of gestation | Premature birth at 29 weeks of gestation | PTP at 27 weeks of gestation | PTP at 20 weeks of gestation | PTP at 21+4 weeks of gestation |
| Pregnancy complications | – | – | NA | – | polyhydramnios | polyhydramnios | hydramnios |
| Birth weight (grams/SD) | 4020 | 2840 | 1224 | ND | 950 (−0.3SD) | 380 | 460 |
| Birth length (cm/SD) | nd | 48 | 34 | ND | 30 (−2.43 SD) | 25 | 25 |
| OFC at birth (cm/SD) | nd | 34 | 27 | ND | 24.5 (−0.47 SD) | 19 | 19.5 |
| DD/ID | + | + | NA | NA | NA | NA | NA |
| Hypotonia | – | – | NA | NA | NA | NA | NA |
| Speech impairment | + | + | NA | NA | NA | NA | NA |
| Abnormal brain imagery | – | – | + | + | NA | + | + |
| Congenital malformations/additional findings | Severe scoliosis (surgery at 17y) | Severe scoliosis (surgery at 15y) | hypospadias | VSD, Fallot | no | no | no |
| Abnormal extremities | – | – | + | + | + | + | + |
| Ophthalmological findings | Optic neuropathy | Optic neuropathy | NA | NA | NA | NA | NA |
| Dysmorphic features | + | + | + | ND | + | + | + |
M: male, F: female; ND: not determined; SD: standard deviation; OFC: occipital frontal circumference: PTP: premature termination of pregnancy; DD/ID: developmental delay/intellectual disability, VSD: ventricular septal defect.
Individual 3 from Family 2 is a male fetus from consanguineous parents examined after a premature termination of pregnancy at 29 weeks and 4 days of gestation. Prenatal follow-up detected intrauterine growth restriction. Brain examination showed lissencephaly, corpus callosum agenesis, aqueductal stenosis and ventricular dilation. Hypospadias was also observed and limb examination revealed bilateral clinodactyly, bilateral adducted thumbs, a bilateral unique palmar fold as well as bilateral equinovarus feet.
Individual 4 from Family 3, born to consanguineous parents, died on the second day of life. Autopsy revealed pachygyria, hydrocephalus, and corpus callosum agenesis. He presented with hip dislocation, hepatomegaly, and Fallot tetralogy. Extremities examination showed bilateral post axial hands hexadactyly and multiple congenital contractures.
Genetic variations
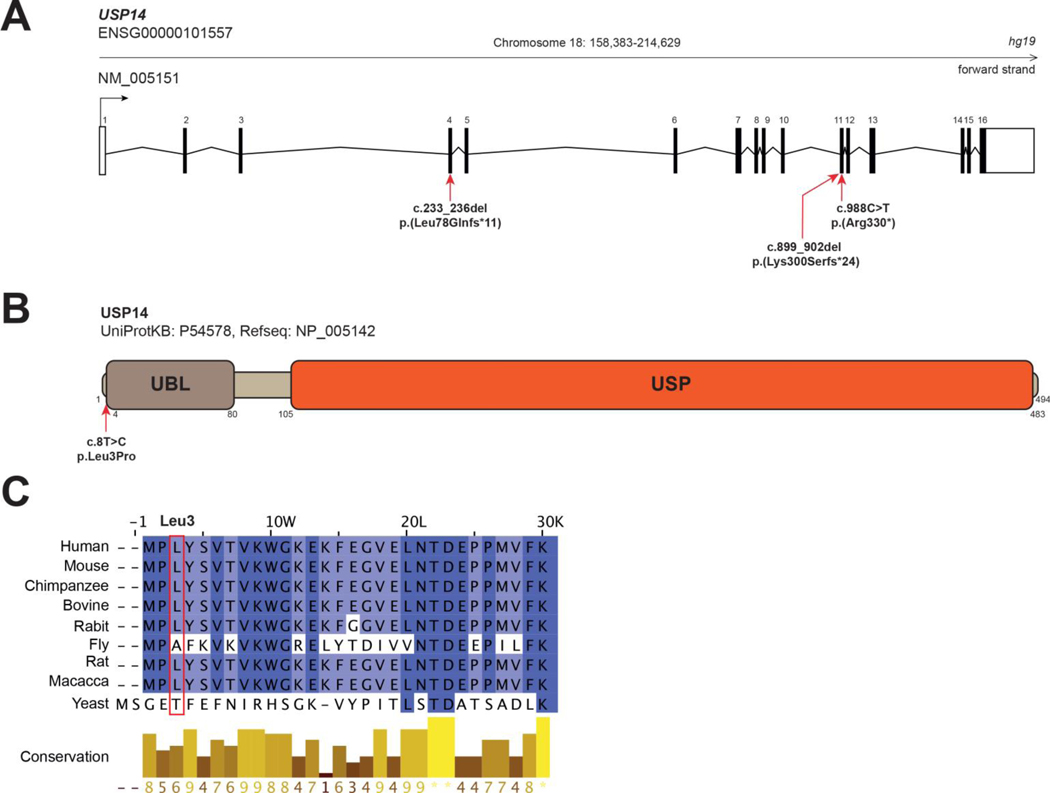
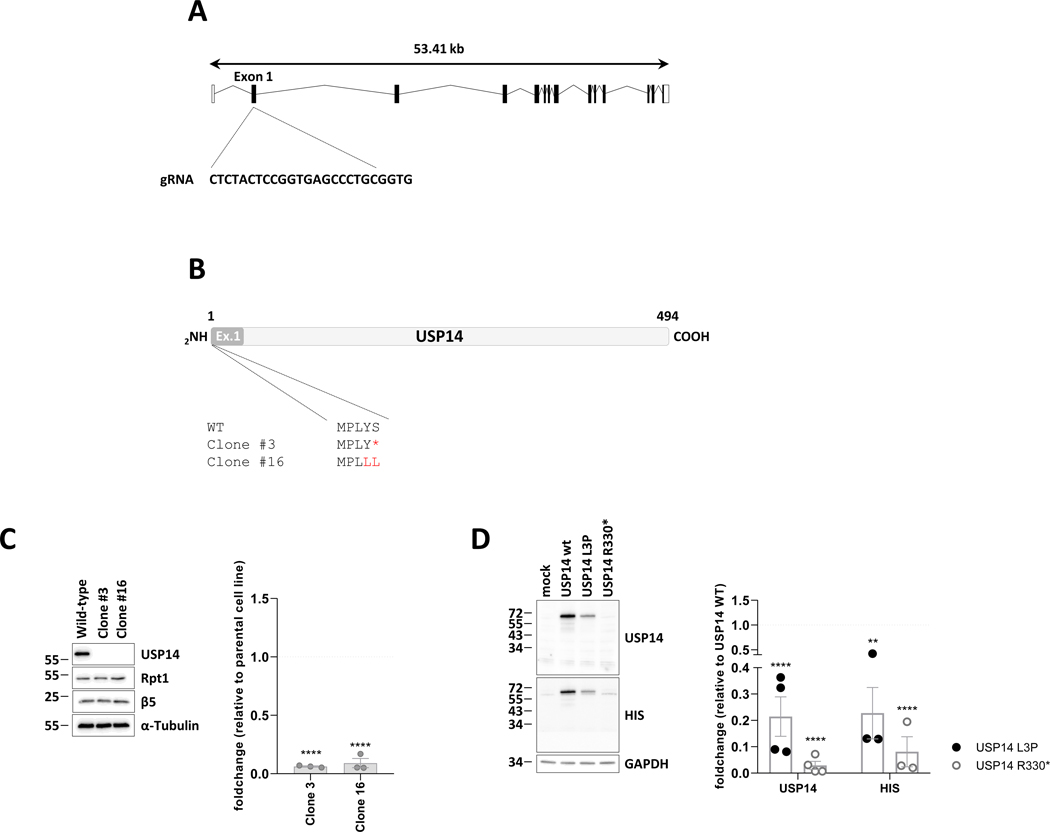
Individuals 1 and 2 carried two heterozygous variants in USP14 (NM_005151.4), namely a c.988C>T, p.(Arg330*) nonsense variant leading to premature protein truncation, and a non-synonymous c.8T>C, p.(Leu3Pro) variant (Figures 1A and S1). Familial segregation study confirmed that each variant was inherited from one of the unaffected parents. Both variants were absent in gnomAD 23 (v3.1 accessed on July 21th, 2023). The c.8T>C, p.(Leu3Pro) variant, located within the first exon of USP14 (Figure 1B), was predicted as deleterious by in silico bioinformatic tool CADD v1.4:.22.4. Importantly, Leu3 is a highly conserved USP14 residue across species and located at the N-terminus of USP14’s UBL domain (Figures 1B and 1C), which is involved in proteasome association 24.
Figxure 1. USP14 variants.
A. Exon structure of longest USP14 transcript, containing 16 exons (NM_005151). Nonsense and frameshift variants identified in the present study are indicated with red arrows. B. Protein structure of USP14 (UniProtKB - P54578). Functional domains (PROSITE annotation rule, UniProtKB/Swiss-Prot format) are indicated. UBL: Ubiquitin-like domain. USP: Ubiquitin specific protease domain. Nonsynonymous variant c.8T>C, (p.Leu3Pro); (individuals 1 and 2) is indicated with a red arrow. C. Multiple sequence alignment of USP14 orthologs from 8 eukaryotic species compared to USP14 human protein sequence. Clustal W multiple sequence alignment program. Conservation scores are mentioned below the alignment. Red boxes identify variant positions.
A homozygous c.899_902del, p.(Lys300Serfs*24) USP14 variant, with a gnomAD frequency ƒ=0.00000796 (2 heterozygous individuals in gnomAD exomes) was found in Individual 3 from Family 2 (Figure S1). Individual 4 from Family 4 carried a homozygous c.233_236del p.(Leu78Glnfs*11) USP14 variant, which was absent in gnomAD. In Families 2 and 3, the variants were detected as heterozygous in each asymptomatic parent (Figure S1). According to the American College of Medical Genetics and Genomics (ACMG) criteria, c.8T>C is classified as variant of unknown significance (VUS, PM2), c.988C>T and c.899_902del c.233_236del as likely pathogenic (PVS1, PM2) and c.233_236del as pathogenic (PVS1, PM2, PP5).
Functional consequences of USP14 variants
Impact of USP14 variants on proteasome activity
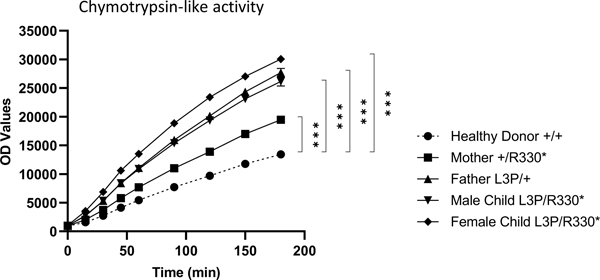
Considering the ability of USP14 to inhibit proteasome activity allosterically 7, we first sought to determine the impact of the p.(Arg330*) and p.(Leu3Pro) USP14 variants on proteasome function. To this end, T cells from both affected siblings as well as their parents were assessed for their capacity to cleave the Suc-LLVY-AMC proteasome reporter substrate using a previously described in-plate activity assay 19,25. As shown in Figure 2, the monoallelic and/or biallelic USP14 variants carried by the family members were all associated with increased proteasome chymotrypsin-like activity when compared to that from a healthy donor control of wild-type genotype. The observation that both p.(Arg330*) and p.(Leu3Pro) USP14 variants suppress the described inhibitory properties of USP14 on proteasomes 26 strongly suggests that such alterations are loss-of-function mutations. Interestingly, the two variants were not equal in their ability to activate proteasomes, with Leu3Pro being more efficient than Arg330* in elevating chymotrypsin-like activity in T cells from the father and the two siblings (Figure 2).
Figure 2. Subjects with UPS14 variants exhibit increased proteasome activity.
Ten micrograms of native cell lysates generated from T cells of a healthy donor (HD) and subjects carrying the p.(Arg330*) and/or p.(Leu3Pro) USP14 variants were incubated at 37°C on a 96-well plate in a final 100 μl volume containing 0.1 mM of the Suc-LLVY-AMC fluorogenic peptide. Fluorescence activity reflecting proteasome chymotrypsin-like activity was measured at 360/460 nm on a microplate reader over a 180-min period of time, as indicated. ***p<0.001 (unpaired two-tailed t test, n=4).
Effects of UPS14 variants on USP14 DUB activity
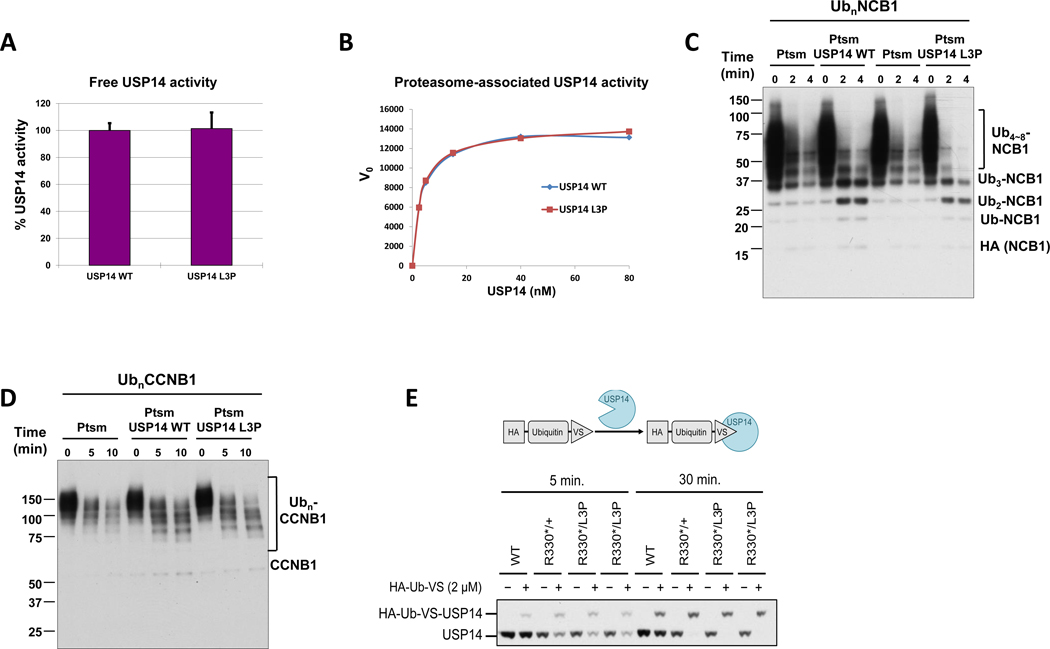
We next examined the impact of the Leu3Pro substitution on the ability of USP14 to remove ubiquitin moieties from protein substrates in vitro. For this purpose, N-terminally SUMO-tagged versions of both wild-type and p.(Leu3Pro) USP14 proteins were expressed in E. coli and subsequently purified following tag cleavage, as previously described 27. As shown in Figure 3A, no discernible differences could be observed between wild-type and p.(Leu3Pro) USP14 in their ability to hydrolyze the standard fluorogenic substrate ubiquitin-AMC. Given that USP14 undergoes an ~800-fold activation upon association with the proteasome 28, we next compared the activity profile of wild-type and p.(Leu3Pro) USP14, when associated with purified 26S proteasomes. Interestingly, the de-ubiquitinating activity of recombinant USP14 in this activated state was also not detectably affected by the Leu3Pro variant. (Figure 3B), suggesting that the p.(Leu3Pro) USP14 variant does not significantly affect the removal of ubiquitin from protein substrates at the proteasome. In agreement with these findings, wild-type and p.(Leu3Pro) USP14 were capable of trimming ubiquitin chains from the NCB1 (an N-terminal fragment derived from cyclin B1; Figure 3C) and CCNB1 (full-length cyclin B1; Figure 3D) poly-ubiquitinated substrates with comparable efficiencies, as determined by Western-blotting. Finally, we investigated the activity state of USP14 in fibroblasts isolated from both affected siblings and their mother by performing a DUB labelling experiment using HA-Ub-VS probe, a strong electrophile that reacts with active cysteine residue of USP14 to form a covalent adduct 28. A shown in Figure 3E, the activity state of the p.(Leu3Pro) USP14 variant in in proband fibroblasts was similar to that of wild-type USP14 in maternal fibroblasts. Altogether, these data indicate that the Leu3Pro substitution does not substantially affect USP14 DUB activity.
Figure 3. Deubiquitinating activity of recombinant USP14-L3P.
A. Ub-AMC hydrolysis assays with free forms of USP14 WT and L3P at 1.5 μM. Error bars represent S.D. from triplicate experiments. B. Kinetic analysis of proteasome-bound USP14 WT and L3P activities with Ub-AMC as substrate. Graded concentrations of recombinant USP14 were reconstituted with 1 nM Ub-VS-treated proteasome, and cleavage of Ub-AMC (1 μM) by USP14 was measured over 80 min in real time. The plots shown represent mean values of initial reaction rates from triplicate experiments. C. In vitro deubiquitination/degradation assays with UbnNCB1, human proteasome (4 nM), and WT or L3P USP14 (80 nM). D. In vitro deubiquitination/degradation assays with UbnCCNB1, human proteasome (7 nM), and WT or L3P USP14 (140 nM). E. The irreversible covalent modifier ubiquitin vinyl sulfone, carrying an HA epitope tag, was added at 2 micromolar to lysates from patient fibroblasts, and the samples incubated at 25°C for either 5 or 30 min as indicated. Samples were then analyzed by SDS-PAGE and immunoblotting, using an antibody to USP14. Each lane was loaded with 25 micrograms of lysate. HA-Ub-VS reacts with the catalytic cysteine of proteasome-activated USP14.
Influence of USP14 dysfunction on autophagy/mitophagy
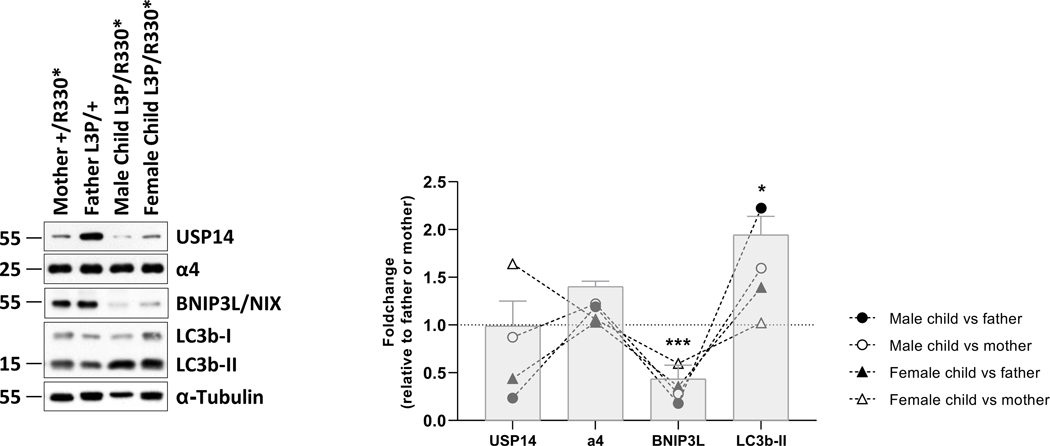
Western-blot analysis of the T-cell samples from the siblings and their parents from family 1 further revealed that the p.(Arg330*) variant was associated with reduced amounts of USP14 full-length protein in the mother as well as male and female children (Figure 4). It is noteworthy that we were unable to detect the USP14(1–229) truncated protein, which would theoretically arise from the p.(Arg330*) variant and run at the expected size of 36.7 kDa. The inability of the p.(Arg330*) variant to generate a protein product was likely due to the significant reduction in mRNA expression of the c.(988C>T) mutant allele compared to that of the c.(988C) wild-type allele (Figure S2A). This observation also suggests that USP14 mRNAs carrying the c.(988C>T) mutation were promptly targeted for nonsense-mediated mRNA decay (NMD), preventing their translation into proteins. Despite comparable amounts of total USP14 mRNA (Figure S2B), the protein levels of the p.(Leu3Pro) USP14 variant were also slightly diminished in the father’s T cells compared to those of wild-type USP14 in T cells from a healthy donor (Figure S3A and B). This suggests that this variant may exhibit a higher protein turnover rate than its wild-type counterpart, even though proteasome inhibition by bortezomib treatment, although promoting the accumulation of ubiquitin-modified proteins failed to stabilize all forms of USP14 (Figures S3A and B).
Figure 4. Patients with UPS14 biallelic variants are associated with alterations of the autophagy/mitophagy pathways.
Left panel: ten to twenty micrograms of RIPA cell lysates obtained from T cells of subjects with USP14 variants were separated by SDS-PAGE prior to western-blotting using antibodies specific for USP14, α4, BNIP3L/NIX, LC3B and α-tubulin (loading control), as indicated. Shown are representative western-blots of three independent experiments performed. Right panel: Shown is the quantification of the Western-blots by densitometry. Data are presented as foldchanges in the affected male and female children vs their father and/or mother whose densitometric measurements were set to 1 (gridline), as indicated. Columns indicate the foldchange mean values ± SEM calculated from the normalizations of three independent biological replicates. Statistical significance was assessed by unpaired Student’s test (*p<0.05 and ***p<0.001).
Noteworthy, the expression of α4 proteasome subunits did not substantially vary across the four samples, indicating that the increased chymotrypsin-like activity triggered by the USP14 variants did not occur as a consequence of increased proteasome amounts. Because USP14 is a well-known modulator of autophagy 29, we next monitored the steady-state expression level of the autophagy marker LC3B-II in both parental and progenitor T cells. As shown in Figure 4, our Western-blot analysis of these samples uncovered that both affected siblings carrying biallelic UPS14 variants exhibited increased levels of LC3b-II, as compared to their parents, suggesting elevated autophagosome formation in the cells of affected individuals. The dysregulated autophagy associated with USP14 biallelic variants was further accompanied by a severe loss of the mitophagy receptor BNIP3L/NIX (Figure 4), indicating that mitophagy was also perturbed in these samples. Further investigations on these specimens confirmed that USP14 compound heterozygosity led to increased mitochondrial autophagy flux, as evidenced by decreased expression of the mitochondrial proteins BNIP3 and PINK1 in both affected siblings, as compared to their parents (Figures S4A and B).
Introduction of tagged USP14 variants into USP14−/− SH-SY5Y cells
We next investigated whether the p.(Arg330*) nonsense variant gives rise to a truncated USP14 protein devoid of active site but still carrying the N-terminal ubiquitin-like (UBL) domain required for proteasome binding. Herein, we first generated USP14 knockout SH-SY5Y cells by a CRISPR/Cas9-based strategy prior to rescue experiments. As shown in Figure 5A and B, the use of guide RNA targeting the first exon of the USP14 gene in SH-SY5Y cells efficiently caused nonsense or frameshift mutations, which completely abolished USP14 expression in two different puromycin-resistant clones (Figure 5C). The loss of USP14 in these cells was not accompanied by a change of proteasome amounts and/or composition, as evidenced by their Rpt1 and β5 expression levels, which were quite similar to those of the wild-type SH-SY5Y cell line (Figure 5C). Most importantly, introducing a HIS-tagged version of the p.(Arg330*) variant in USP14−/− clone #16 failed to produce a USP14 truncated protein with an expected size of 36,7 kDa, as determined by Western-blotting using antibodies specific for USP14 and HIS (Figure 5D). By contrast, transfection of these cells with HIS-tagged p.(Leu3Pro) USP14 was followed by a parallel rise of USP14 whose expression was, however, much weaker than that of wild-type USP14 (Figure 5D, S5A and B). Importantly, the observed decreased expression of the p.(Leu3Pro) USP14 variant in these cells did not stem from variations in transfection efficiencies, as co-administration of p.(Leu3Pro) USP14 with EGFP did not lead to lower EGFP expression levels compared to cells transfected with both wild-type USP14 and EGFP (Figure S5C). On the contrary, EGFP levels were even higher when introduced together with p.(Leu3Pro) USP14 (Figure S5C), suggesting a stabilizing effect of this variant on EGFP protein turnover. Altogether, these data indicate that both p.(Arg330*) and p.(Leu3Pro) USP14 variants negatively impact on USP14 steady-state expression in SH-SY5Y cells.
Figure 5. The p.(Arg330*) nonsense mutation fails to give rise to a USP14 truncated variant in USP14−/− SH-SY5Y cells.
A. Schematic representation of the USP14 gene as well as the region in exon 1 targeted by the specific guide RNA used for gene deletion by CRISPR/Cas9. B. Primary structures generated from the sequencing results of USP14 PCR amplicons obtained for wild-type SHSY-5Y cells and clones #3 and #16, as indicated. C. Left panel: wild-type SHSY-5Y cells and clones #3 and #16 were subjected to SDS-PAGE and western-blotting using antibodies specific for USP14, Rpt1 and Beta5, as indicated. Equal protein loading was ensured by using an antibody directed to α-tubulin. Right panel: Shown is the quantification of the Western-blots by densitometry. Data are presented as foldchanges (n=3) in clones #3 and #16 vs the SHSY-5Y parental cell line whose densitometric measurements were set to 1 (gridline), as indicated. Statistical significance was assessed by unpaired Student’s test (****p<0.0001). D. Left panel: SHSY-5Y clone #16 was subjected to a 24-h transfection with expression vectors coding for HIS-tagged wild-type UPS14 as well as the p.(Arg330*) or p.(Leu3Pro) USP14 variants. Overexpressed proteins were quantified by SDS-PAGE/western-blotting using anti-USP14, anti-HIS and anti-GAPDH (loading control) antibodies, as indicated. Right panel: Shown is the quantification of the Western-blots by densitometry. Data are presented as foldchanges of the USP14 and HIS signals in SHSY-5Y cells transected with the p.(Leu3Pro) (black circles) and p;(Arg330*) (white circles) USP14 variants relative to SHSY-5Y cells transected with wild-type USP14 whose densitometric measurements were set to 1 (gridline), as indicated. Statistical significance was assessed by unpaired Student’s test (**p<0.01 and ****p<0.0001).
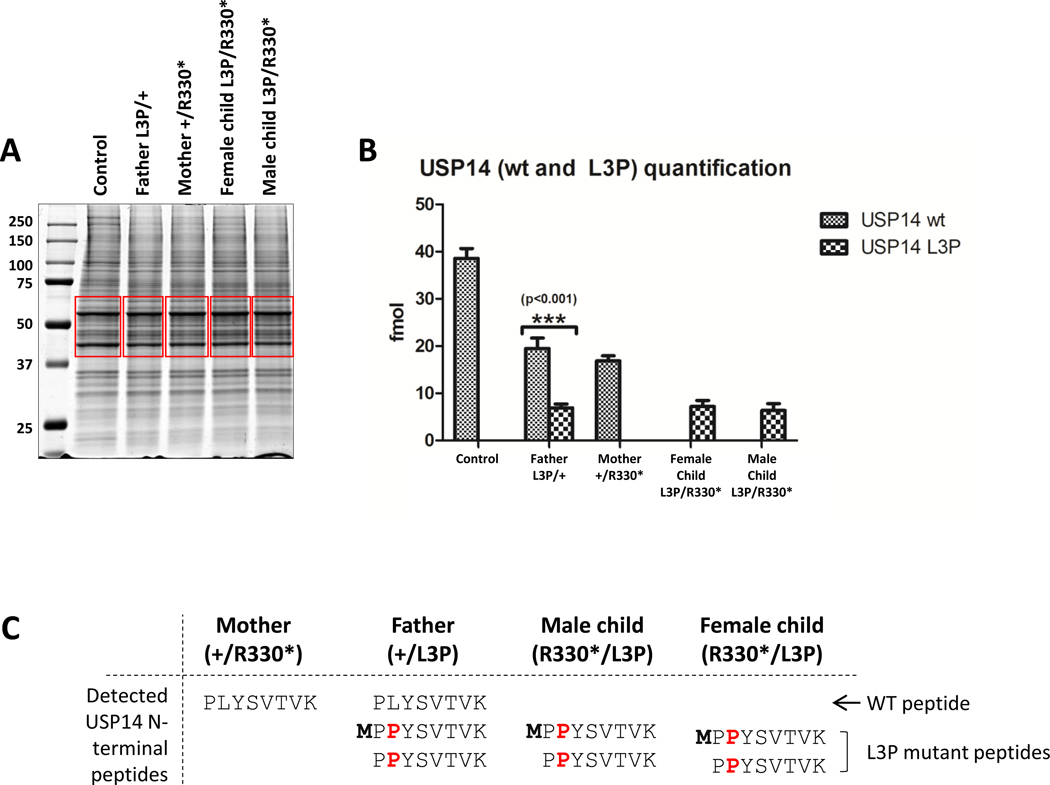
Mass spectrometry analysis of USP14 protein N-termini in patient-derived T cells
To further ascertain the impact of the p.(Arg330*) and p.(Leu3Pro) variants on USP14 steady-state expression level, we next conducted a mass spectrometry (MS) analysis of the proteomes from both the siblings and their parents. To this end, whole-cell lysates from primary fibroblasts from one unrelated heathy individual and the four family members were separated by SDS-PAGE prior to Coomassie staining. As shown in Figure 6A, gel sections containing the USP14 bands were extracted and subjected to in-gel digestion using trypsin. Interestingly, MS analysis of the resulting products showed that individuals that carry one copy of wild-type USP14 express 50% (≈20 fmol) of the protein that is found in healthy individual (Figure 6B). As expected, the p.(Leu3Pro) USP14 mutant peptide was only found in individuals carrying the variant (father and both siblings). In line with our Western-blot analysis (Figure 5D), the p.(Leu3Pro) USP14 variant was weaker expressed than its wild-type counterpart (Figure 6B).
Figure 6. Mass spectral analysis of whole-cell lysates derived from patient cells reveals decreased expression and N-terminal methionine processing of the Leu3Pro (L3P) USP14 variant.
A. Coomassie-stained SDS-PAGE of protein lysates from subjects carrying the p.(Arg330*) and/or p.(Leu3Pro) USP14 variants, as indicated. Gel slices excised for trypsin digestion and subsequent mass spectrometry analysis are indicated in red. B. Quantification of wild-type (wt) and Leu3Pro (L3P) USP14 peptides by MS using a PRM (parallel reaction monitoring) method. C. Table recapitulating the identification of N-terminal methionine untrimmed and trimmed USP14 wild-type (WT)- and Leu3Pro-derived peptides found in USP14 immuno-precipitates of each sample, as indicated.
Considering the previously documented deleterious effects of proline residues at position +3 on N-terminal methionine excision (NME) of nascent proteins by methionine aminopeptidase (MAP) 30, we next proceeded to investigate whether the p.(Leu3Pro) variant had an impact on this process. Herein, USP14 immuno-precipitates from primary fibroblast cell lines of all family members were subjected to trypsin digestion prior to MS analysis. As summarized in Figure 6C, there were no detectable remnants of N-terminal methionine on USP14 peptides derived from the wild-type allele, suggesting complete N-terminal methionine removal of the newly synthesized USP14 protein under normal conditions. In contrast, a portion of the USP14 p.(Leu3Pro) mutant peptides in all three family members harboring this variant retained N-terminal methionine (Figure 6C). These findings suggest that the Leu3Pro missense variant partially impairs NME of USP14, and this defect appears to be linked to reduced expression of the p.(Leu3Pro) USP14 at the protein level. To further validate this point, we disrupted the accessibility of the N-terminal methionine to MAP by tagging USP14 N-terminally with a double HA epitope. As shown in Figure S6, the Leu3Pro substitution had no discernable effect on the steady-state expression of the HA-USP14 fusion protein, confirming that this variant was deleterious only when the initiator methionine is be subjected to NME. Importantly, both c.(233_236del) and c.(899_902del) USP14 homozygous deletions identified in P3 and P4 failed to supply stable truncated USP14 proteins in these cells, as determined by Western-blotting (Figure S6). These data confirm that the four USP14 genomic alterations identified in the three families were loss-of-function variants.
Impact of UPS14 dysfunction on type I interferon (IFN) status in patient T cells
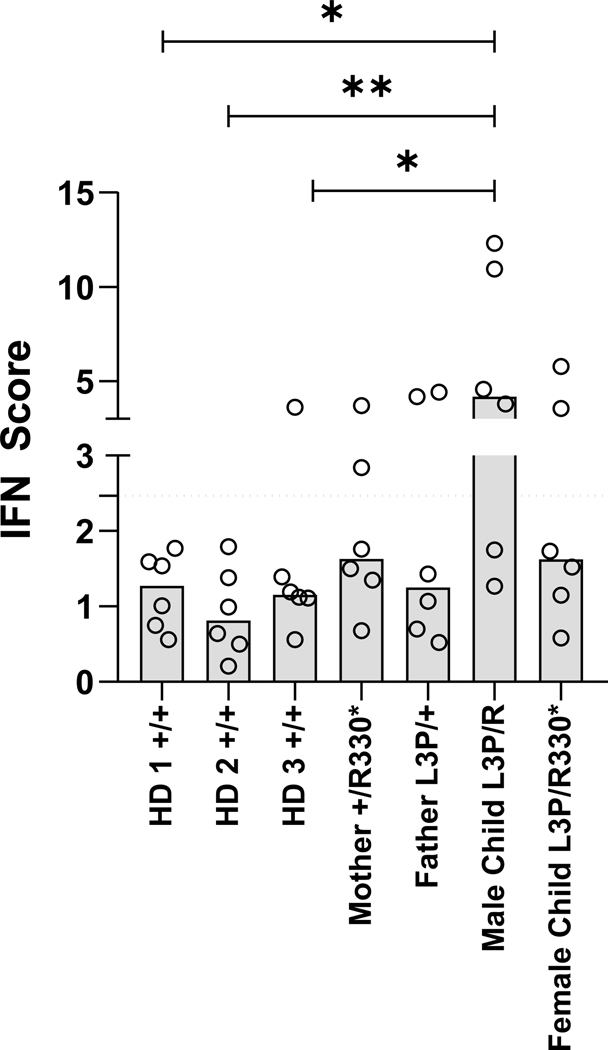
Since perturbations of proteasome function typically trigger sterile type I interferon (IFN) responses in patients 31, T cells of the four family members were next assessed for their content of IFN-stimulated gene (ISG) transcripts using qPCR. As shown in Figure 7, the IFN scores exhibited by the affected female sibling or her parents were not significantly higher than those displayed by any of the three healthy donor controls of wild-type genotype. The male sibling showed a slight increased type I IFN score (Figure 7)., which may potentially indicate proteotoxic stress not necessarily related to USP14 loss of function.
Figure 7. UPS14 loss-of-function is not consistently associated with sterile type I IFN responses.
Interferon (IFN) scores of whole-blood samples collected and stabilized in PAXgene RNA tubes from three unrelated healthy individuals (HD) and subjects carrying the p.(Arg330*) and/or p.(Leu3Pro) USP14 variants, as indicated. Scores were calculated as the median of the relative quantification (RQ) of seven ISG (i.e., IFIT1, IFI27, IFI44L, ISG15, RSAD2 and SIGLEC1) over a single calibrator control, as measured by RT-qPCR (see Materials & Methods). Statistical significance was assessed by paired ratio t test (*p<0.05 and **p<0.01).
Altogether these data indicate that both p.(Arg330*) and p.(Leu3Pro) USP14 variants are deleterious for USP14 function (possibly with some differences with p.(Arg330*) being a complete loss-of-function mutation, while p.(Leu3Pro) a hypomorphic allele) and their biallelic expression affects autophagy and mitophagy processes without triggering autoinflammation. By adding all these functional studies showing damaging effect on the gene or gene product (ACMG criterion PS3), the new classification for p.(Leu3Pro) is likely pathogenic. On the other hand, it was not possible to investigate the functional impact of the two frame-shift variants because suitable biological samples could not be obtained from Individuals 3 and 4.
DISCUSSION
Here, we describe four individuals, including a fetal patient, with a phenotypic spectrum involving both neurodevelopmental and neuromuscular features caused by USP14 biallelic pathogenic variants, including a fetal patient. Recently, the c.233_236del; p.Leu78Glnfs*11 homozygous USP14 variant has been associated with cerebral malformations with an arthrogryposis multiplex congenita (AMC) phenotype in three fetuses from a consanguineous family 32. In our cohort, Individual 3 carrying the c.899_902del, p.Lys300Serfs*24 variant presented with lissencephaly, corpus callosum agenesis, aqueductal stenosis, ventricular dilation and limbs contractures. Individual 4 carrying the c.233_236del p.(Leu78Glnfs*11) variant at homozygous state also presented with pachygyria, hydrocephalus, corpus callosum anomalies, and AMC. Introduction of these genomic alterations into SH-SY5Y USP14−/− cells by plasmid-driven expression failed to generate the corresponding truncated proteins (Figure S6), thereby confirming that the two biallelic USP14 deletions were loss-of-function variants associated with CNS malformations.
The two individuals carrying compound heterozygous p.(Arg330*) and p.(Leu3Pro) USP14 variants showed a progressive neurological disease (Table 1). Our data revealed that both alterations were associated with reduced USP14 protein expression, although to a much lesser extent for the p.(Leu3Pro) variant (Figures 4, 5 and 6). Indeed, the 36.7 kDa truncated form of USP14 theoretically produced by p.(Arg330*) variant was not detected in patient-derived T cells or fibroblasts using SDS-PAGE/Western blotting or MS, respectively. Furthermore, this variant could not be ectopically expressed as HIS- or HA-tagged USP14(1–329) fusion protein in USP14−/− SH-SY5Y cells (Figures 5D and S6), most likely as a result of a failure of the c(988C>T) transcript to escape NMD (Figure S2).
Conversely, the p.(Leu3Pro) USP14 variant was detected at the protein level in patient-derived T cells and fibroblasts. However, its steady-state expression was lower than wild-type USP14 (Figures 5D and 6B) and the reduced amounts of p.(Leu3Pro) USP14 were associated with reduced NME (Figure 6C). This observation is fully in line with previous studies showing that NME by MAP is favored by proline residues at position P2 – directly after the initiator methionine (P1), but not at P3 30,33. NME is a highly conserved limited proteolysis process that occurs co-translationally and whose functional significance remains poorly understood. It has been proposed that NME might serve various functional purposes such as in methionine recycling, post-translational modification, the latter being closely linked to protein folding and/or turnover, particularly through N-terminal acetylation 34 As such, it is tempting to speculate that p.(Leu3Pro) USP14 mutants are less stable than their wild-type counterparts because of inefficient NME. Supporting this notion, rendering p.(Leu3Pro) USP14 insensitive to NME by tagging it at the N-terminus with a double HA epitope led to protein expression levels comparable to those of HA-tagged wild-type USP14 (Figure S6). However, how perturbed NME alters the turnover of the p.(Leu3Pro) USP14 variant remains unclear, as we were unable to rescue its expression following proteasome inhibition (Figures S2A and S2B). These observations suggest that untrimmed p.(Leu3Pro) USP14 may not be targeted for proteasomal degradation or exhibit a half-life longer than the 4-hour bortezomib treatment used in these assays. Another factor contributing to the reduced levels of p.(Leu3Pro) USP14 might be a decreased expression of the corresponding c.(8T>C) mutant mRNA. Regrettably, quantifying the expression of this transcript by allele-specific RT-qPCR proved challenging due to GC-rich regions surrounding the mutation. Interestingly, the p.(Leu3Pro) USP14 variant seemed to exhibit slightly higher activity than wild-type USP14 in cellulo (Figure 3E), suggesting that aberrant DUB function might participate in the pathogenesis of this variant as well.
Interestingly, to the best of our knowledge, this study represents one of the very few reporting impaired NME as a potential underlying factor in diseases caused by genetic variants. Indeed, aside from a study by Sheikh et al. documenting the effects of p.(Ala2Val) variant on methyl CpG-binding protein 2 (MeCP2) NME in Rett syndrome (RTT) 35, our literature research failed to identify any other studies reporting similar pathogenic mechanisms.
In addition to their impact on USP14 protein expression, USP14 variant exerted effects on proteasomes whose chymotrypsin-like activity was increased individuals carrying single and/or biallelic variants (Figure 2). The downregulation of proteasome activity by USP14 is a well-documented process relying on USP14’s ubiquitin-like (UBL) domain which targets USP14 to 26S complexes 24,36. Because the Leu3Pro variant is located within the UBL domain of USP14 (Figure 1), it is conceivable that the ability of the p.(Leu3Pro) variant to activate proteasomes may reflect its inability to associate with them. The observation that wild-type and p.(Leu3Pro) USP14 reach comparable active states in the presence of purified 26S proteasomes in vitro (Figure 3B) rather suggests that this effect might be due to a reduced availability of USP14 for binding to proteasomes. It is noteworthy that USP14 is subjected to post-translational modification, in particular by the AKT kinase, which phosphorylates Ser432 within the catalytic domain of USP14 (Xu et al 2015). Modification at this site has been estimated to enhance the deubiquitinating activity of proteasome-bound USP14 by approximately 2-fold. Whether this modification affects the expressivity of the mutations described in this study remains, however, to be determined. Despite exhibiting similar affinities for the proteasome, these forms of USP14 may potentially have distinct effects on ATPase alignment and/or 20S gating mechanisms. In any case, the effects exerted by the p.(Arg330*) or p.(Leu3Pro) monoallelic variants on proteasome activity are unlikely to reach the threshold for disease pathogenesis, since these effects are observed in the T cells from the siblings’ parents who are clinically unaffected.
An additional driver of these phenotypes might be a dysregulation of the autophagy and mitophagy processes which was detected in the T cells from both male and female affected children (Figure 4). The molecular mechanisms by which the combined –but not single– expression of the p.(Arg330*) and p.(Leu3Pro) USP14 variants promote autophagy in patients’ T cells are unclear, but this observation is consistent with previous studies showing that USP14 inhibition stimulates autophagosome formation 37. It is indeed argued that UPS14 typically counteracts autophagy by de-ubiquitinating K63-linked ubiquitin chains from Beclin-1, a process required for activating the vacuolar protein sorting (Vps) 34 kinase 38. Whether Beclin-1 ubiquitination is specifically affected by the two p.(Arg330*) and p.(Leu3Pro) USP14 recessive alleles in these patients remains to be formally demonstrated.
Interestingly, recent studies have revealed that the induction of mitophagy by USP14 gene silencing or inhibition can effectively rescue mitochondrial dysfunction in a Drosophila model of Parkinson’s disease 39. Remarkably, this was accompanied by improved locomotion behavior and a notable delay in the progression neurodegeneration. These compelling observations suggest that while USP14 deficiency is associated with early-onset NDD, it paradoxically exerts beneficial effects in the context of late-onset neurodegenerative diseases. One plausible explanation for these contrasting results is that a major contributing factor of neurodegeneration is a decline in proteasome function, which can be effectively corrected by suppressing USP14, although future studies are needed to fully clarify this point.
All the experimental studies performed indicated that p.(Arg330*) behaves as a nearly complete loss-of-function mutation; conversely, the p.(Leu3Pro) variant is possibly a hypomorphic allele, partly affecting some of the USP14 functions. This could explain the different phenotypes observed in the individuals reported in this study: indeed, affected subjects from family 1 (compound heterozygotes for p.Arg330* and p.Leu3Pro) have a milder clinical presentation compared to the severe phenotype characterizing patients 3 and 4 (who were homozygous for complete loss-of-function variants).
The molecular mechanisms underlying the impact of USP14 loss-of-function variants on the pathogenesis of NDD beyond alterations in proteasome-mediated protein breakdown remain unclear. It is plausible that USP14 deficiency could induce perturbations in CNS function and/or development by influencing the expression of specific regulators, such as PRNP, as previously described 40. In this regard, subsequent studies are warranted to unravel the proteome of neuronal tissues carrying the p.(Arg330*) and p.(Leu3Pro) biallelic USP14 variants. Surprisingly, unlike other proteasome components 31, USP14 dysfunction was not associated with spontaneous and sterile type I IFN responses (Figure 7). To the best of our knowledge, this is the first proteasome element for which genomic alterations do not lead to type I IFN gene signatures. This might be attributed to the fact that only a fraction of USP14 is bound to the 26S complex. However, further investigations are required to address this hypothesis.
In conclusion, we describe here the clinical consequences of USP14 biallelic loss-of-function variants and underline the importance of a tight regulation of USP14 for both autophagy and UPS-dependent cellular protein degradation, stressing the crucial role of these degradation pathways in human development.
Supplementary Material
Acknowledgments
We are grateful to individuals who participated in the study.
Funding Statement
This research was funded by the European Joint Programme on Rare Diseases (EJP RD) project GENOMIT (Italian Ministry of Health ERP-2019–23671045 and the German Federal Ministry of Education and Research and Horizon2020 01GM1920A) as well as the ERA PerMed project PerMiM (01KU2016A) to D.G. and H.P. This work was further supported by the European Reference Network (ERN), with D.G. and I.M. being members of the ERN EURO-NMD and ERN-RND, respectively. Additionally, B.H.L. received funding from the National Research Foundation of Korea (NRF) of the Ministry of Science and ICT (2022R1A4A2000703 & 2022R1A2C1092638) and D.F. acknowledges funding the National Institutes of Health (1R35GM145246–01). This study was part of the Project “CP21/00017”, funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union (M.A.P.). EK and SM were supported by funding from the German Research Foundation (RTG PRO 2719). EK also received support from COST (European Cooperation in Science and Technology) Action ProteoCure CA20113 for this work.
Footnotes
Ethics Declaration
Written informed consent was obtained for use of medical history, genetic testing report, and photographs (if applicable), as approved by the Institutional Review Board of CHU Nantes.
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
All raw data presented in this paper are available to qualified researchers upon reasonable request.
REFERENCES
- 1.Rock KL, Gramm C, Rothstein L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11(14):1245–1256. doi: 10.1096/FASEBJ.11.14.9409543 [DOI] [PubMed] [Google Scholar]
- 3.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/EMBOJ/20.18.5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T, Naqvi NI, Yang H, Teo TS. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem Biophys Res Commun. 2000;272(1):270–275. doi: 10.1006/BBRC.2000.2767 [DOI] [PubMed] [Google Scholar]
- 5.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/NATURE01071 [DOI] [PubMed] [Google Scholar]
- 6.Verma R, Aravind L, Oania R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/SCIENCE.1075898 [DOI] [PubMed] [Google Scholar]
- 7.Hanna J, Hathaway NA, Tone Y, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. doi: 10.1016/J.CELL.2006.07.038 [DOI] [PubMed] [Google Scholar]
- 8.Lappe-Siefke C, Loebrich S, Hevers W, et al. The ataxia (axJ) mutation causes abnormal GABAA receptor turnover in mice. PLoS Genet. 2009;5(9). doi: 10.1371/JOURNAL.PGEN.1000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim E, Park S, Lee JH, et al. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018;24(3):732–743. doi: 10.1016/J.CELREP.2018.06.058 [DOI] [PubMed] [Google Scholar]
- 10.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412(6845):449–452. doi: 10.1038/35086595 [DOI] [PubMed] [Google Scholar]
- 11.Ebstein F, Küry S, Papendorf JJ, Krüger E. Neurodevelopmental Disorders (NDD) Caused by Genomic Alterations of the Ubiquitin-Proteasome System (UPS): the Possible Contribution of Immune Dysregulation to Disease Pathogenesis. Front Mol Neurosci. 2021;14. doi: 10.3389/fnmol.2021.733012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fountain MD, Oleson DS, Rech ME, et al. Pathogenic variants in USP7 cause a neurodevelopmental disorder with speech delays, altered behavior, and neurologic anomalies. Genet Med. 2019;21(8):1797–1807. doi: 10.1038/S41436-019-0433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homan CC, Kumar R, Nguyen LS, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014;94(3):470–478. doi: 10.1016/J.AJHG.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H, Haas SA, Chelly J, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21(1):133–148. doi: 10.1038/MP.2014.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolly LA, Parnell E, Gardner AE, et al. Missense variant contribution to USP9X-female syndrome. NPJ Genom Med. 2020;5(1):53. doi: 10.1038/s41525-020-00162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin HC, Jones WD, McIntyre R, et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018;362(6419):1161–1164. doi: 10.1126/SCIENCE.AAR6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonell LM, Mirzaa GM, Alcantara D, et al. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat Genet. 2013;45(5):556–562. doi: 10.1038/NG.2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng BG, Eklund EA, Shiryaev SA, et al. Predominant and novel de novo variants in 29 individuals with ALG13 deficiency: Clinical description, biomarker status, biochemical analysis, and treatment suggestions. J Inherit Metab Dis. 2020;43(6):1333–1348. doi: 10.1002/JIMD.12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santiago-Sim T, Burrage LC, Ebstein F, et al. Biallelic Variants in OTUD6B Cause an Intellectual Disability Syndrome Associated with Seizures and Dysmorphic Features. Am J Hum Genet. 2017;100(4). doi: 10.1016/j.ajhg.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonteneau JF, Larsson M, Somersan S, et al. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells. J Immunol Methods. 2001;258(1–2):111–126. doi: 10.1016/S0022-1759(01)00477-X [DOI] [PubMed] [Google Scholar]
- 21.Rice GI, Melki I, Frémond ML, et al. Assessment of Type I Interferon Signaling in Pediatric Inflammatory Disease. J Clin Immunol. 2017;37(2):123–132. doi: 10.1007/S10875-016-0359-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/HUMU.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/NATURE19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Ning S, Yu B, Wang Y. USP14: Structure, Function, and Target Inhibition. Front Pharmacol. 2022;12. doi: 10.3389/FPHAR.2021.801328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Küry S, Besnard T, Ebstein F, et al. De Novo Disruption of the Proteasome Regulatory Subunit PSMD12 Causes a Syndromic Neurodevelopmental Disorder. Am J Hum Genet. 2017;100(2). doi: 10.1016/j.ajhg.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HT, Goldberg AL. The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. J Biol Chem. 2017;292(23):9830–9839. doi: 10.1074/jbc.M116.763128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43(1):1–9. doi: 10.1016/J.PEP.2005.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee BH, Lee MJ, Park S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/NATURE09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai Y, Xia C, Sun Z. The Inhibitory Effect of 6-Gingerol on Ubiquitin-Specific Peptidase 14 Enhances Autophagy-Dependent Ferroptosis and Anti-Tumor in vivo and in vitro. Front Pharmacol. 2020;11:598555. doi: 10.3389/fphar.2020.598555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boissel JP, Kasper TJ, Shah SC, Malone JI, Bunn HF. Amino-terminal processing of proteins: hemoglobin South Florida, a variant with retention of initiator methionine and N alpha-acetylation. Proc Natl Acad Sci U S A. 1985;82(24):8448–8452. doi: 10.1073/pnas.82.24.8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papendorf JJ, Krüger E, Ebstein F. Proteostasis Perturbations and Their Roles in Causing Sterile Inflammation and Autoinflammatory Diseases. Cells. 2022;11(9). doi: 10.3390/CELLS11091422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turgut GT, Altunoglu U, Sivrikoz TS, et al. Functional loss of ubiquitin-specific protease 14 may lead to a novel distal arthrogryposis phenotype. Clin Genet. 2022;101(4):421–428. doi: 10.1111/CGE.14117 [DOI] [PubMed] [Google Scholar]
- 33.Flinta C, Persson B, Jörnvall H, von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986;154(1):193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x [DOI] [PubMed] [Google Scholar]
- 34.Ree R, Varland S, Arnesen T. Spotlight on protein N-terminal acetylation. Exp Mol Med. 2018;50(7):1–13. doi: 10.1038/s12276-018-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheikh TI, de Paz AM, Akhtar S, Ausió J, Vincent JB. MeCP2_E1 N-terminal modifications affect its degradation rate and are disrupted by the Ala2Val Rett mutation. Hum Mol Genet. 2017;26(21):4132–4141. doi: 10.1093/hmg/ddx300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin JY, Muniyappan S, Tran NN, Park H, Lee SB, Lee BH. Deubiquitination Reactions on the Proteasome for Proteasome Versatility. Int J Mol Sci. 2020;21(15):1–16. doi: 10.3390/IJMS21155312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Shan B, Sun H, et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016;30(15):1718–1730. doi: 10.1101/GAD.285122.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia P, Wang S, Du Y, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32(20):2685–2696. doi: 10.1038/EMBOJ.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakraborty J, von Stockum S, Marchesan E, et al. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol Med. 2018;10(11). doi: 10.15252/emmm.201809014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian T, McLean JW, Wilson JA, Wilson SM. Examination of genetic and pharmacological tools to study the proteasomal deubiquitinating enzyme ubiquitin-specific protease 14 in the nervous system. J Neurochem. 2021;156(3):309–323. doi: 10.1111/jnc.15180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data presented in this paper are available to qualified researchers upon reasonable request.