Abstract
Chinese hamster ovary (CHO) cells are resistant to infections by gibbon ape leukemia virus (GALV) and amphotropic murine leukemia virus (A-MLV) unless they are pretreated with tunicamycin, an inhibitor of N-linked glycosylation. These viruses use the related sodium-phosphate symporters Pit1 and Pit2, respectively, as receptors in nonhamster cells, and evidence has suggested that the corresponding transporters of CHO cells may be masked by tunicamycin-sensitive secreted inhibitors. Although the E36 line of Chinese hamster cells was reported to secrete the putative Pit2 inhibitor and to be sensitive to the inhibitory CHO factors, E36 cells are highly susceptible to both GALV and A-MLV in the absence of tunicamycin. Moreover, expression of E36 Pit2 in CHO cells conferred tunicamycin-independent susceptibilities to both viruses. Based on the latter results, it was suggested that E36 Pit2 must functionally differ from the endogenous Pit2 of CHO cells. To test these ideas, we analyzed the receptor properties of CHO Pit1 and Pit2 in CHO cells. Surprisingly, and counterintuitively, transfection of a CHO Pit2 expression vector into CHO cells conferred strong susceptibility to both GALV and A-MLV, and similar overexpression of CHO Pit1 conferred susceptibility to GALV. Thus, CHO Pit2 is a promiscuous functional receptor for both viruses, and CHO Pit1 is a functional receptor for GALV. Similarly, we found that the natural resistance of Mus dunni tail fibroblasts to subgroup C feline leukemia viruses (FeLV-C) was eliminated simply by overexpression of the endogenous FeLV-C receptor homologue. These results demonstrate a novel and simple method to unmask latent retroviral receptor activities that occur in some cells. Specifically, resistances to retroviruses that are caused by subthreshold levels of receptor expression or by stoichiometrically limited masking or interference mechanisms can be efficiently overcome simply by overexpressing the endogenous receptors in the same cells.
In most cells, gibbon ape leukemia virus (GALV) and amphotropic murine leukemia virus (A-MLV) use the related Na+-dependent phosphate symporters Pit1 and Pit2, respectively, as receptors for infection (10, 17, 20, 37). Both Pit1 and Pit2 are multiple-membrane-spanning proteins with five presumptive extracellular loops (ECLs). Pit1 and Pit2 cDNAs from a variety of species, including human, mouse, rat, and hamster, have been isolated and extensively characterized (3, 8, 17, 20, 27, 34, 35, 37). While all Pit2 proteins that have been analyzed mediate A-MLV infections, with some mediating GALV infections as well (34, 35), not all Pit1 proteins are able to mediate GALV infections. For example, the resistance of mouse cells to GALV infection, with the exception of that described for the Japanese feral mouse M. m. molossinus (34), is attributed to the inability of mouse Pit1 to function as a GALV receptor (9, 27). Chimera studies of mouse Pit1 and human Pit1 have identified a 9-amino-acid sequence (region A) of Pit1 ECL 4 as critical for GALV receptor function (9, 27). Similarly, the resistances of many other cells to particular retroviruses are caused by mutations at key sites in the receptors (1, 36). In other cases, however, cellular resistances to entry of retroviruses are caused by endogenously inherited interfering envelope glycoproteins (16; reviewed in reference 32) or possibly by other receptor blocking mechanisms (18, 19).
Chinese hamster ovary (CHO) cells are resistant to GALV and A-MLV unless they are pretreated with tunicamycin, an inhibitor of N-linked glycosylation (18, 19). Previous studies have suggested that cells from Chinese hamsters secrete unidentified tunicamycin-sensitive inhibitors that specifically block GALV and A-MLV infections in hamster cells but do not block these infections in nonhamster cells (18, 19). CHO cells are also resistant to ecotropic MLVs unless tunicamycin is present (19). However, a variant of Friend ecotropic MLV that causes neural degeneration can infect untreated CHO cells (15). Tunicamycin is also required for infections of Mus dunni fibroblasts with Moloney ecotropic MLV (6) and for human immunodeficiency virus type 2 infections of some primate cell lines (30). Thus, a tunicamycin requirement for retroviral infections occurs with different viruses and cell lines and can, as was reported in one case (15), be overcome by viral envelope glycoprotein mutants.
Surprisingly, E36 cells, which were also derived from a Chinese hamster, are susceptible to both GALV and A-MLV in the absence of tunicamycin (5), despite secreting Pit2 inhibitors that inhibit A-MLV infection of CHO cells (18). Moreover, expression of E36 Pit2 in CHO cells confers tunicamycin-independent susceptibility to both of these viruses (35). Therefore, it was inferred that E36 Pit2 is a promiscuous receptor for both GALV and A-MLV and that it must differ from the endogenous CHO Pit2 in its sequence and in its tunicamycin dependency. Subsequently, Chaudry et. al. (3) isolated a cDNA encoding CHO Pit2 and confirmed that the encoded protein differs substantially from E36 Pit2, consistent with the hypothesis that these differences might be responsible for the natural resistance of CHO cells to GALV and A-MLV. These workers also isolated a cDNA encoding CHO Pit1. Based on sequences in the critical ECL 4 region A, they inferred that CHO Pit1 was unlikely to be active as a GALV receptor and they suggested that GALV infection of CHO cells was probably mediated solely by CHO Pit2 (3). In this paper we report the independent isolation of cDNAs for CHO Pit1 and Pit2 and the surprising observation that both of the encoded transporters are active tunicamycin-independent receptors when they are overexpressed within CHO cells. This implies that the endogenous receptors are latent and can be unmasked simply by overexpressing them in the cells from which they were derived. Evidence supporting the generality of this insight was obtained using mouse fibroblasts, which are naturally resistant to subgroup C feline leukemia viruses (FeLV-C). Overexpression of the FeLV-C receptor (FLVCR) homologue isolated from M. dunni tail fibroblasts (MDTF) resulted in strong susceptibility of these cells to FeLV-C.
Isolation of CHO Pit1 and Pit2 cDNAs.
We first endeavored to clone Pit2 cDNA from CHO cells. This proved to be difficult. Initially, we used a CHO cell 5′-stretch lambda gt10 cDNA library (Stratagene, La Jolla, Calif.) with a 32P-labeled nick-translated hybridization probe derived from full-length rat Pit2 cDNA. The hybridizations were performed in stringent conditions at 42°C in a solution containing 50% formamide, 1% sodium dodecyl sulfate, 1 M sodium chloride, and 10% dextran sulfate. Of 12 positive phages that were plaque purified and sequenced, all contained Pit1 rather than Pit2 sequences. Eventually, we succeeded in isolating a Pit2 cDNA clone by PCR using primers that were complementary to the rat Pit2 coding region (upstream primer, 5′-ATGGCCATGGATGAGTATTTGTGG-3′; downstream primer, 5′-TCACACATATGGAAGGATCCCATAC-3′). The CHO Pit2 cDNA was cloned into the vector pcDNA3.1 (Invitrogen, Carlsbad, Calif.) and sequenced at the Microbiology and Molecular Immunology Core Facility on a PE/ABD 377 DNA sequencer using dye terminator cycle chemistry (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The predicted CHO Pit2 protein is 98% identical to the previously reported E36 Pit2 (comparison not shown). Interestingly, our CHO Pit2 sequence has a closer identity to E36 Pit2 than the CHO Pit2 protein reported by Chaudry et al. (3) and differs from the previously reported sequence by 7 amino acids. Despite these differences, the presumptive ECLs of these CHO Pit2 proteins are identical.
As described above, this investigation also generated CHO Pit1 cDNA clones. These clones encoded almost the entire Pit1 protein, excluding the region between amino acids 1 and 92 that comprises the intracellular amino terminus through the first extracellular loop (ECL 1). Our sequence is identical to the CHO Pit1 sequence reported by Chaudry et al. (3), including sequence identity in all ECLs (data not shown). CHO Pit1 differs considerably from E36 Pit1 in region A of ECL 4 (3). This region is highly variable among the Pit1 proteins of different species (3, 33, 34) and has been shown to be critical for GALV receptor functions (9, 27).
CHO Pit2 is an efficient tunicamycin-independent receptor for both GALV and A-MLV.
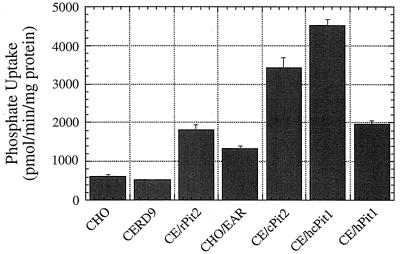
We analyzed the receptor function of CHO Pit1 and Pit2 proteins by expressing them in CHO cells. We first ligated CHO Pit2 cDNA and several Pit1 cDNAs into the retroviral expression vector pSFF and used ping-pong amplification to produce ecotropic host-range virions that encoded these proteins (13). CERD9 cells (31), derived from CHO cells and expressing the mouse receptor for ecotropic MLVs, were subsequently transduced with these virions. As shown in Fig. 1, the transduced cells expressed much higher levels of phosphate transport activity than the control CHO and CERD9 cells that contained only the endogenous phosphate transporters. This implies that the transduced Pit1 and Pit2 proteins were expressed at relatively high levels in these cells. Figure 1 also shows the phosphate transport activity of CHO cells expressing E36 Pit2 (CHO/EAR cells) (35), which were generated by infection of CHO cells with an amphotropic pseudotype virus carrying the E36 Pit2 gene. CHO/EAR cells were generously donated by M. Eiden and C. Wilson (National Institute of Mental Health, Bethesda, Md.).
FIG. 1.
Phosphate uptake of CHO cells expressing Pit1 and Pit2 proteins. CHO cells expressing the mouse ecotropic MLV receptor (CERD9 cells) (31) were transduced with ecotropic virus carrying genes that encode for either rat Pit2 (CE/rPit2) (17), CHO Pit2 (CE/cPit2), human-CHO Pit1 (CE/hcPit1) and human Pit1 (CE/hPit1). CHO/EAR cells are CHO cells expressing E36 Pit2 and were generated by Wilson et al. (35). Phosphate transport was measured using the procedure outlined by Olah et al. (21). The phosphate uptake values are averages of four different replicates in the same experiment. The standard deviations (error bars) are shown.
These CHO cell derivatives were then quantitatively analyzed in the presence and absence of tunicamycin for susceptibility to infection by β-galactosidase-encoding (lacZ) virions pseudotyped with GALV and A-MLV envelope glycoproteins. As shown by the representative results in Table 1, our clone of CHO cells is resistant to GALV and A-MLV but becomes highly susceptible to GALV after pretreatment with tunicamycin. Similarly, tunicamycin caused a weak but somewhat variable susceptibility to A-MLV in the control CHO and CERD9 cells. As determined by multiple independent experiments involving tunicamycin, the control CHO and CERD9 cells were not significantly or reproducibly different in their susceptibilities to these infections (results not shown). In agreement with a previous report (14), CHO cells expressing rat Pit2 (CE/rPit2) (17) were highly susceptible to A-MLVs independently of tunicamycin, suggesting that rat Pit2 is a specific receptor for A-MLVs. Surprisingly, CHO cells expressing CHO Pit2 (CE/cPit2) were also highly susceptible to both GALV and A-MLV in the absence of tunicamycin. Table 1 also shows data from another experiment in which we compared infections of CHO/EAR and CHO/cPit2 cells. CHO/cPit2 cells were generated by transfection of CHO cells with a CHO Pit2 cDNA expression vector. The results show that these cells had very similar properties and confirmed a previous report that CHO/EAR cells are susceptible to both GALV and A-MLV in the absence of tunicamycin (35). Thus, the Pit2 proteins encoded by E36 and CHO cells behave identically when assayed in CHO cells.
TABLE 1.
Susceptibilities of CHO cell derivatives to infection by lacZ pseudotypes of A-MLV and GALV
| Experiment | Target cell type | Titer (CFU/ml) of lacZ pseudotyped of:
|
|||
|---|---|---|---|---|---|
| A-MLV
|
GALV
|
||||
| −Tun | +Tun | −Tun | +Tun | ||
| 1a | CHO | <10 | 4.2 × 102 | <5 | 4.1 × 105 |
| CERD9 | <10 | 1.3 × 102 | <10 | 8.5 × 104 | |
| CE/rPit2 | 3.1 × 106 | 6.9 × 105 | <10 | 5.1 × 104 | |
| CE/cPit2 | 2.0 × 106 | 2.0 × 105 | 6.4 × 104 | 1.6 × 104 | |
| CE/hcPit1b | <10 | 20 | 1.4 × 106 | 6.5 × 105 | |
| CE/hPit1 | <10 | 10 | 1.2 × 106 | 2.9 × 105 | |
| 2c | CHO | <10 | 6.0 × 102 | <10 | 7.0 × 105 |
| CHO/cPit2 | 2.3 × 104 | 4.1 × 104 | 4.0 × 103 | 7.0 × 105 | |
| CHO/EAR | 4.2 × 103 | 1.5 × 104 | 2.3 × 104 | 2.0 × 104 | |
CERD9 cells and CHO cells expressing rat Pit2 (CE/rPit2) (17), CHO Pit2 (CE/cPit2), human-CHO Pit1 (CE/hcPit1), or human Pit1 (CE/hPit1) were tested for susceptibility to lacZ(A-MLV) and lacZ(GALV) pseudotype viruses before treatment with tunicamycin (−Tun) and after treatment with 250 ng of tunicamycin per ml (+Tun).
hcPit1 is a human-CHO Pit1 chimera spliced at amino acid 213 by using an AccI restriction enzyme cleavage site that occurs in both the human and CHO Pit1 cDNAs. It contains presumptive ECL 1 and 2 from hPit1 and the remaining ECLs from CHO Pit1.
The CHO/cPit2 clone used in experiment 2 was made by stable transfection of the CHO Pit2 expression vector (pcDNA3.1-CHOPit2) in CHO cells rather than by the retroviral vector transduction method described in the text and used in experiment 1. Transduction resulted in higher levels of receptor expression and in more stable expression than transfection. CHO/EAR cells express E36 Pit2 and were generated by Wilson et al. (35) by transduction of CHO cells with virions carrying the E36 Pit2 gene.
Producer cells expressing the lacZ(GALV) pseudotype virus were prepared, as previously described (29), by infection of human TE671 cells, containing an integrated MLV vector, MFGnlslacZ (7), with replication-competent GALV (SF strain). lacZ(A-MLV) pseudotype virus was generated by transfecting TELCeB6 cells (kindly provided by Y. Takeuchi and F. L. Cosset) with an FBsalf retroviral expression vector containing the cDNA encoding the A-MLV envelope (4). The TELCeB6 cell line contains a retroviral expression vector expressing Moloney MLV Gag and Pol proteins, and the MFGnlslacZ retroviral vector. The titers of infection are averages of two independent infection studies for experiment 1 and of three independent infection studies for experiment 2.
As shown in Table 1, we also analyzed the receptor function of a human-CHO Pit1 chimera (hcPit1), which contains presumptive ECL regions 1 and 2 of human Pit1 and the remaining sequences of CHO Pit1. CHO cells expressing the hcPit1 chimera (CE/hcPit1) showed specific tunicamycin-independent susceptibility to GALV but not to A-MLV, with GALV titers that were comparable to the titers in CHO cells that express high levels of human Pit1 (CE/hPit1 cells). Similarly, expression of the Pit1 chimera in MDTF resulted in strong susceptibility to GALV (data not shown). These results demonstrate that the critical ECL 4 region A sequence of CHO Pit1 is compatible with GALV receptor function and, more interestingly, that the GALV receptor function occurs in CHO cells and is independent of tunicamycin. This result differs from the inference of Chaudry et al. (3), which was based on mutagenesis of human Pit1. Our result is also consistent with the observation of Miller and Miller (18) that the Pit2 inhibitor(s) secreted by E36 cells does not prevent GALV infections of CHO cells.
Mouse receptor for FeLV-C mediates infections when overexpressed in mouse cells.
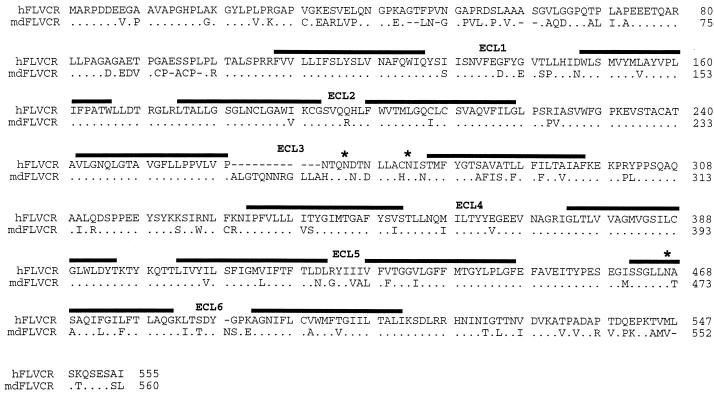
To ascertain the potential generality of the above results for different viruses and cells, we analyzed murine MDTF, which are naturally resistant to FeLV-C infections and become susceptible to the Moloney strain of ecotropic MLV only after treatment with tunicamycin (6). Parental MDTF were tested for susceptibility to lacZ(FeLV-C) and lacZ(RD114) pseudotype viruses before treatment with tunicamycin and after treatment with 250ng of tunicamycin per ml. lacZ(FeLV-C) pseudotype virus was generated, as previously described (28), by transfection of TELCeB6 packaging cells with an FBsalf retroviral expression vector containing the cDNA encoding FeLV-C(Sarma) envelope. lacZ(RD114) pseudotype virus was produced by TELCeB6/RDF-7 helper-free packaging cells (4). The titers of infection were as follows (values are in CFU per milliliter and are averages from three infection experiments): lacZ(FeLV-C) without tunicamycin, <2; lacZ(FeLV-C) with tunicamycin, 2; lacZ(RD114) without tunicamycin, 2.9 × 103; and lacZ(RD114) with tunicamycin, 1.0 × 105. As shown by the data given above, pretreatment of MDTF cells with tunicamycin enhanced infections by RD114 feline endogenous retrovirus approximately 30-fold but did not enhance infections by FeLV-C. These viruses use distinct cell surface receptors (22, 24, 26, 28). To further investigate the MDTF resistance to FeLV-C, we isolated a receptor homologue, MDTF FLVCR (mdFLVCR), from these cells by PCR using primers that were complimentary to the coding region of human FLVCR (hFLVCR) cDNA that was previously isolated by our group (28). The mdFLVCR cDNA was subcloned into the pCDNA3.1V5His-Topo vector (Invitrogen). As shown in Fig. 2, the mdFLVCR protein contains 560 amino acids and is 77% identical to hFLVCR. Interestingly, expression of the mdFLVCR cDNA in MDTF cells conferred strong susceptibility to FeLV-C as shown by the following. MDTF were transiently transfected with the hASCT2, mdFLVCR, or hFLVCR expression constructs and then tested for susceptibility to lacZ(FeLV-C) pseudotype virus. ASCT2 (type 2 neutral amino acid transporter [11]) is the common name for the receptor for RD114 (24, 26). The standard nomenclature for the ASCT2 gene is SLC1A5 (OMIM database, The National Center for Biotechnology Information, National Institutes of Health). The titers of infection were as follows (values are in CFU per milliliter and are averages from three infection experiments): for MDTF/hASCT2, 0; for MDTF/hFLVCR, 2.5 × 103; and for MDTF/mdFLVCR, 1.5 × 103. Thus, mdFLVCR functions as an efficient receptor for FeLV-C when overexpressed in MDTF cells.
FIG. 2.
Comparison of the amino acid sequences of hFLVCR and mdFLVCR. Dots, identical amino acids; dashes, spaces introduced for alignment. ∗, N-linked glycosylation site for hFLVCR. Potential membrane-spanning segments are indicated by a line over the sequence, and the presumptive ECLs are indicated.
Major implications.
These results demonstrate that the resistances of untreated CHO cells to GALV and A-MLV infections and of MDTF to FeLV-C infections are not caused by inherent defects in the endogenous receptors for these viruses. Indeed, although CHO cells are only slightly susceptible to A-MLV infections even after treatment with tunicamycin, overexpression of CHO Pit2 causes substantial tunicamycin-independent susceptibility to both A-MLV and GALV (Table 1). This result is compatible with previous evidence that untreated E36 cells are highly susceptible to GALV and A-MLV infections in the absence of tunicamycin (5), suggesting that E36 cells may express larger amounts of Pit1 and Pit2 than CHO cells or lower concentrations of a masking factor(s) (18). Thus, the E36 and CHO Pit2 proteins function similarly in CHO cells as tunicamycin-independent mediators of GALV and A-MLV infections (Table 1). Similarly, although MDTF are completely resistant to FeLV-C, overexpression of the endogenous mdFLVCR protein in these cells results in strong susceptibility to this infection (see above).
We believe that the simplest explanation for these results that is compatible with previous evidence (18, 19) is that the Pit1 and Pit2 receptors within CHO cells and the FLVCR within MDTF are present in relatively low (subthreshold) quantities and may be additionally inhibited by stoichiometrically limited amounts of masking factors. According to this hypothesis, overexpression of the endogenous receptors would be expected to result in susceptibilities to infections. Previous studies have implied that receptors can be masked by endogenously inherited retrovirus-related envelope glycoproteins by interference mechanisms (reviewed in reference 32) or by other glycoproteins (18) and that these masking glycoproteins can be inactivated by tunicamycin treatment of the cells (18, 19, 25, 30). It is known that processing and folding of retroviral envelope glycoproteins requires N-linked glycosylation (2), which is blocked by tunicamycin. This masking model is clearly consistent with the fact that treatment of CHO cells with tunicamycin induces their susceptibility to GALV and A-MLV infections. However, it is notable that tunicamycin does not induce susceptibility of MDTF to FeLV-C (see above). This result would be compatible with the idea that the putative mask that blocks the mdFLVCR in MDTF might be insensitive to tunicamycin or that both mdFLVCR and its mask might be tunicamycin sensitive. According to the latter explanation, the mask in MDTF might be a retrovirus-related envelope glycoprotein that misfolds in the absence of N-linked glycosylation, but this would not result in tunicamycin-dependent susceptibility to infection because the FLVCR would become inactive in these conditions. This explanation would be compatible with evidence that many but not all glycoproteins misfold in the presence of tunicamycin (2, 12, 23) and that FLVCR contains three consensus sites for N-linked glycosylation (Fig. 2). Although additional studies will be required to test these interpretations, we believe that our results strongly suggest that receptor masking may be more widespread than previously suspected. In addition, the example of mdFLVCR clearly implies that such apparent masking cannot always be reversed by tunicamycin (see above). Finally, our results demonstrate a novel and simple method that may be generally useful for identifying masked or subthreshold quantities of retroviral receptors. In these cases, overexpressing the endogenous receptors within the same cells will result in strong viral susceptibilities.
Nucleotide sequence accession numbers.
The GenBank accession number for the CHO Pit2 cDNA is AF239675 and that for mdFLVCR cDNA is AF239767.
Acknowledgments
We are grateful to Yasuhiro Takeuchi (Wohl Virion Center, University College London, London, United Kingdom) for providing lacZ(GALV) and lacZ(A-MLV) producer cells, TELCeB6 packaging cells, and the FBsalf retroviral expression vector containing the RD114 envelope gene. We are also grateful to Brian J. Willet (Department of Veterinary Pathology, University of Glasgow, Glasgow, United Kingdom) for providing the FBsalf vector containing the FeLV-C(Sarma) envelope cDNA and to Maribeth Eiden and Carolyn Wilson (National Institute of Mental Health, Bethesda, Md.) for providing the CHO/EAR cells. We are grateful to our coworkers Susan Kozak, Mariana Marin, and Emily Platt for encouragement and helpful suggestions.
This work was supported by NIH grants CA25810 and CA83835 and by The Wellcome Trust.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassin R H, Ruscetti S, Ali I, Haapala D K, Rein A. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology. 1982;123:139–151. doi: 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 3.Chaudry G J, Farrell K B, Ting Y T, Schmitz C, Lie Y S, Petropoulos C J, Eiden M V. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J Virol. 1999;73:2916–2920. doi: 10.1128/jvi.73.4.2916-2920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eglitis M A, Eiden M V, Wilson C A. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J Virol. 1993;67:5472–5477. doi: 10.1128/jvi.67.9.5472-5477.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiden M V, Farrell K, Wilson C A. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferry N, Duplessis O, Houssin D, Danos O, Heard J M. Retroviral mediated gene transfer to hepatocytes in vivo. Proc Natl Acad Sci USA. 1991;88:8377–8381. doi: 10.1073/pnas.88.19.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johann S V, Van Zeijl M, Cekleniak J, O'Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly variable between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kekuda R, Prasad P D, Fei Y J, Torres-Zamorano V, Sinha S, Yang-Feng T L, Leibach F H, Ganapathy V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 12.Kornfield R, Kornfield S. Assembly of aspargine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 13.Kozak S L, Kabat D. Ping-pong amplification of a retroviral vector achieves high-level gene expression: human growth hormone production. J Virol. 1990;64:3500–3508. doi: 10.1128/jvi.64.7.3500-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda M, Hanson C A, Hoffman P M, Ruscetti S K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDougall A S, Terry A, Tzavaras T, Cheney C, Rojko J, Neil J C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for role in natural resistance to subgroup B feline leukemia viruses. J Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D G, Miller A D. Inhibitors of retrovirus infection are secreted by several hamster cell lines and are also present in hamster sera. J Virol. 1993;67:5346–5352. doi: 10.1128/jvi.67.9.5346-5352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robbins T. Characterization of the human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Diff. 1990;1:119–127. [PubMed] [Google Scholar]
- 21.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 22.Quigley J G, Burns C C, Anderson M M, Lynch E D, Sabo K M, Overbaugh J, Abkowitz J L. Cloning of the cellular receptor for feline leukemia virus subgroup C(FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95:1093–1099. [PubMed] [Google Scholar]
- 23.Rademacher T W, Parekh R B, Dwek R A. Glycobiology. Annu Rev Biochem. 1988;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 24.Rasko J E, Battini J L, Gottschalk R J, Mazo I, Miller A D. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rein A, Schultz A M, Bader J P, Bassin R H. Inhibitors of glycosylation reverse retroviral interference. Virology. 1982;119:185–192. doi: 10.1016/0042-6822(82)90075-7. [DOI] [PubMed] [Google Scholar]
- 26.Tailor C S, Nouri A, Zhao Y, Takeuchi Y, Kabat D. A sodium dependent neutral amino acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol. 1999;73:4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K L. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma associated virus, and GALV infection. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tailor C S, Willet B J, Kabat D. A putative cell surface receptor for anemia-inducing subgroup C feline leukemia virus is a member of a transporter superfamily. J Virol. 1999;73:6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi Y, Simpson G, Vile R G, Weiss R A, Collins M K L. Retroviral pseudotypes produced by rescue of a Moloney murine leukemia virus vector by C-type, but not D-type, retroviruses. Virology. 1992;186:792–794. doi: 10.1016/0042-6822(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 30.Talbot S J, Weiss R A, Schulz T F. Reduced glycosylation of human cell lines increases susceptibility to CD4-independent infection by human immunodeficiency virus type 2 (LAV-2/B) J Virol. 1995;69:3399–3406. doi: 10.1128/jvi.69.6.3399-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Paul R, Burgeson R E, Keene D R, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991;65:6468–6477. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss R A. Cellular receptors and viral glycoproteins involved in retroviral entry. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 1–108. [Google Scholar]
- 33.Weiss R A, Tailor C S. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 34.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukemia virus receptor from susceptible and non-susceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retroviruses. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeijl M V, Johann S V, Cross E, Cunningham J, Eddy R, Shows T B, O'Hara B. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]