Abstract
The infection of CD4+ host cells by human immunodeficiency virus type 1 (HIV-1) is initiated by a temporal progression of interactions between specific cell surface receptors and the viral envelope protein, gp120. These interactions produce a number of intermediate structures with distinct conformational, functional, and antigenic features that may provide important targets for therapeutic and vaccination strategies against HIV infection. One such intermediate, the gp120-CD4 complex, arises from the interaction of gp120 with the CD4 receptor and enables interactions with specific coreceptors needed for viral entry. gp120-CD4 complexes are thus promising targets for anti-HIV vaccines and therapies. The development of such strategies would be greatly facilitated by a means to produce the gp120-CD4 complexes in a wide variety of contexts. Accordingly, we have developed single-chain polypeptide analogues that accurately replicate structural, functional, and antigenic features of the gp120-CD4 complex. One analogue (FLSC) consists of full-length HIV-1BaL gp120 and the D1D2 domains of CD4 joined by a 20-amino-acid linker. The second analogue (TcSC) contains a truncated form of the gp120 lacking portions of the C1, C5, V1, and V2 domains. Both molecules exhibited increased exposure of epitopes in the gp120 coreceptor-binding site but did not present epitopes of either gp120 or CD4 responsible for complex formation. Further, the FLSC and TcSC analogues bound specifically to CCR5 (R5) and blocked R5 virus infection. Thus, these single-chain chimeric molecules represent the first generation of soluble recombinant proteins that mimic the gp120-CD4 complex intermediate that arises during HIV replication.
The fusion of human immunodeficiency virus type 1 (HIV-1) with CD4+ target cells involves an orchestrated appearance of intermediate structures comprised of the viral envelope protein, gp120, the CD4 receptor, and certain seven-transmembrane domain chemokine receptors or coreceptors (2, 9). These intermediates facilitate critical early steps in HIV replication and present structural and antigenic features that are highly conserved among virus strains (19, 27, 38). Accordingly, HIV envelope intermediates are now considered to be promising targets for the development of new therapeutic and anti-HIV vaccine strategies.
One such intermediate, the gp120-CD4 complex, is formed during the attachment of HIV gp120 to the primary host cell receptor, CD4 (38). The functional role of the complex is to induce structural rearrangements that expose a conserved, high-affinity coreceptor-binding site on the gp120 moiety (34, 36). Subsequent attachment of this site to a coreceptor produces a gp120-CD4-coreceptor tricomplex that triggers structural alterations in the viral transmembrane protein, gp41, leading directly to the fusion of viral and host cell membranes (6, 29).
The potential utility of gp120-CD4 complexes in vaccine development has been clearly shown by several studies in which soluble complexes were used to generate antibodies to cryptic gp120 epitopes and broadly neutralizing humoral responses against HIV (5, 8, 12, 17). More recently, complexes presented in the context of cell-cell fusion were also shown to produce neutralizing responses effective against HIV isolates from various geographic clades (20). Other studies have shown that gp120-CD4 complexes present conserved epitopes in the coreceptor binding domain that are recognized by neutralizing human monoclonal antibodies (MAbs) (35, 37). Collectively, these studies suggest that gp120-CD4 complexes should be considered as candidates for subunit vaccine immunogens.
The exposure of the coreceptor-binding domain on gp120-CD4 complexes allows these molecules to also be used for screening panels of compounds for candidates that might inhibit infection at the level of tricomplex formation. Furthermore, these complexes could be used directly as the basis for strategies to competitively block HIV-coreceptor interactions and inhibit viral entry. In this context, the complexes could be considered as analogues of the chemokines that act as natural coreceptor ligands. Such chemokines prevent HIV-1 entry by directly interfering with envelope interactions (2, 9).
Unfortunately, the widespread development of therapeutic and vaccination strategies based on gp120-CD4 complexes is currently hindered by the need to produce and chemically link two polypeptides. Consequently, gp120-CD4 complexes have only been considered as soluble subunits that, although capable of eliciting neutralizing humoral immunity, are unlikely to stimulate a cytotoxic-T-lymphocyte (CTL) response. Envelope-specific CTLs could be generated by DNA- or vector-based vaccines that mediate the coordinated expression of gp120 and CD4. However, a coordinated expression of the CD4 gene and HIV env, which is under the control of the viral rev regulatory sequences, might be difficult to achieve. Instability of the complex structure may also hinder their capacity to block coreceptor interactions with HIV or to screen for other compounds that block viral entry. Although we have shown that stabilization of the soluble complex structure can be achieved by covalent cross-linking, such treatment is not optimal as it may partially obscure critical neutralizing epitopes and alters the antigenic properties of the molecules.
In order to overcome these problems, we have developed single-chain polypeptide molecules that faithfully duplicate the structural, functional, and antigenic properties of the gp120-CD4 complex. The present study describes the construction and characterization of such molecules.
MATERIALS AND METHODS
Cell lines and antibodies.
The 293 cell line is an adenovirus transformed kidney line that was obtained from the NIH AIDS Reagent Repository (Bethesda, Md.). L1.2 cells that express the chemokine receptors CCR5 (R5) and CXCR4 were a generous gift of Lijun Wu (LeukoSite, Cambridge, Mass.) (36). The wild-type L1.2, a murine B-cell lymphoma, was obtained from Eugene Butcher (Stanford, Calif.). U373/CD4/MAGI cells lines that express chemokine receptors CCR5 and CXCR4 were constructed by Mike Emerman (35) and were obtained from the NIH AIDS Reagent Repository. MAb F240 was a generous gift of Lisa Cavacini (Harvard University) (4). MAbs A32, C11 (21a), 48d, and 17b (33) were provided by James Robinson (Tulane University). MAb IgG1b12 (3) was provided by Dennis Burton (San Diego, Calif.). MAb 205-469 (33) was provided by Michael Fung (Tanox, Inc., Houston, Tex.). The anti-myc MAb, 9E10 (11), was obtained from Immunotech (Marseille, France).
Plasmids.
The full-length codon-optimized envelope gene derived from HIV-1BaL was constructed synthetically (Midland Certified Reagents, Midland, Tex.) using codons most frequently used in mammalian cells. The synthetic gene was then subcloned into pUC12 to create pMR1W1-9. The human CD4 sequence used was derived from T4-pMV7 (21) (NIH AIDS Reagent Repository).
Construction of pEF6-FLSC and pEF6-TcSC.
The plasmid pEF6-FLSC, encoding the full-length single chain, was constructed via PCR using the plasmids pMR1W1-9 and T4-pMV7 as templates. The gp120 forward primer was GGG-GGT-ACC-ATG-CCC-ATG-GGG-TCT-CTG-CAA-CCG-CTG-GCC, and the reverse primer was GGG-TCC-GGA-GCC-CGA-GCC-ACC-GCC-ACC-AGA-GGA-TCC-ACG-CTT-CTC-GCG-CTG-CAC-CAC-GCG-GCG-CTT. The CD4 forward primer was GGG-TCC-GGA-GGA-GGT-GGG-TCG-GGT-GGC-GGC - GCG - GCC - GCT - AAG - AAA - GTG - GTG - CTG - GGC - AAA - AAA - GGG - GAT, and the reverse primer was GGG-GTT-TAA-AC-TTA-TTA-CAG-ATC-CTC-TTC-TGA-GAT-GAG-TTT-TTG-TTC-AGC-TAG-CAC-CAC-GAT-GTC-TAT-TTT-GAA-CTC. The resulting PCR product was subcloned into pEF6 (Invitrogen, Carlsbad, Calif.) using KpnI and PmeI restriction sites. To construct the pEF6-TcSC plasmid, the full-length gp120 expressing sequence in pEF6-FLSC was exchanged for a truncated version of the gp120 sequence (DC1DC5DV1V2). The truncated gp120 was generated using GGG-GGT-ACC-ATG-CCC-ATG-GGG - TCT - CTG - CAA - CCG - CTG - GCC - ACC - TTG - TAC - CTG - CTG - GGG - ATG - CTG - GTC - GCT - TCC - TGC - CTC - GGA - AAG - AAC - GTG - ACC - GAG - AAC-TTC-AAC-ATG-TGG as a forward primer and GGG-GGA-TCC-GAT-CTT-CAC-CAC-CTT-GAT-CTT-GTA-CAG-CTC as a reverse primer. The V1 and V2 regions were deleted using CTG-TGC-GTG-ACC-CTG-GGC-GCG-GGC-GAG-ATG-AAG-AAC-TGC-AGC-TTC-AAC-ATC-GGC-GCG-GGC-CGC-CTG-ATC-AGC-TGC as a forward primer and GCA-GCT-GAT-CAG-GCG-GCC - CGC - GCC - GAT - GTT - GAA - GCT - GCA - GTT - CTT - CAT - CTC - GCC - CGC-GCC-CAG-GGT-CAC-GCA-CAG as a reverse primer. All primers were generated by the University of Maryland Baltimore Biopolymer Facility (Baltimore, Md.).
Expression of FLSC and TcSC proteins.
Selected clones of pEF6-FLSC and pEF6-TcSC were transiently transfected into 293 cells using Fugene (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer's protocol. After 48 h, the supernatants were collected and analyzed for protein expression by anti-gp120 and anti-CD4 immunoblot assays.
Immunoblot analysis.
Samples were treated by boiling for 5 minutes in 2% sodium dodecyl sulfate (SDS) and 1% β-mercaptoethanol and then electrophoresed over a 4 to 20% polyacrylamide gradient gel (Owl, Portsmouth, N.H.). Protein was then electrophoretically transferred to nitrocellulose sheets, which were then treated with 10 mM Tris (pH 7.5) containing 140 mM NaCl and 5% nonfat dry milk (BLOTTO) for 1 h at room temperature to prevent nonspecific binding. Transferred protein was detected with either a mixture of murine anti-gp120 MAbs (1); a rabbit anti-human CD4 polyclonal serum, T4-4 (7) (NIH AIDS Reagent Repository); or polyclonal goat anti-human CD4 IgG (R&D Systems, Minneapolis, Minn.) as necessary. Sheets were washed three times with 0.1% Tween in 10 mM Tris (pH 7.5) containing 140 mM NaCl (TBS) and then treated with the appropriate secondary antibody conjugated to either alkaline phosphatase (KPL, Gaithersburg, Md.) or horseradish peroxidase (KPL). All antibodies were suspended in BLOTTO. Bound alkaline phosphatase-conjugated antibodies were visualized with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (KPL), and bound horseradish peroxidase-conjugated antibodies were developed with the ECL-Plus kit (Amersham-Pharmacia Biotech, Piscataway, N.J.). Stained bands were quantified by densitometric analysis using ImageQuant 5.0 (Molecular Dynamics, Sunnyvale, Calif.) on a Storm Fluor-Imager (Molecular Dynamics).
Purification of FLSC and TcSC antigens.
FLSC and TcSC molecules were expressed either from transient transfection of 293 cells or from stable 293 cell lines that had been adapted for growth in 293 serum-free medium (GIBCO-BRL, Gaithersburg, Md.). Single-chain complexes were purified via affinity chromatography using either the anti-gp120 human MAb, A32, coupled to CNBr-activated Sepharose 4B (Amersham-Pharmacia Biotech) or Galantahus nivalis lectin coupled to 4% agarose beads (Sigma, St. Louis, Mo.) as appropriate. Bound protein was eluted from the antibody column with 0.1 M acetic acid (pH 2.5) and from the lectin column with 1 M methyl-α-d-mannopyranoside (13). The eluted material was then dialyzed against phosphate-buffered saline (PBS). Protein concentrations were determined by BCA Assay (Bio-Rad, Hercules, Calif.) according to the manufacturer's protocol.
Gel filtration analysis of FLSC and TcSC molecules.
FLSC and TcSC proteins were analyzed under nondenaturing conditions by gel filtration chromatography on a Superose 6 column (Amersham-Pharmacia Biotech). The column was equilibrated with PBS, calibrated, and then standardized using protein standards ranging from 12.4 to 669 kDa (Sigma). A standard curve of elution volume versus molecular weight was then generated. Regression analyses based on the curve was then used to estimate the molecular weights of test proteins run on the column. Samples of FLSC or TcSC (200 μg) were applied to the column in 0.5 ml of PBS. The column was run at a constant flow rate of approximately 1.0 ml/min, and 5-ml fractions were collected. Uncomplexed BaLgp120 or complexes formed by mixing soluble gp120 with recombinant soluble CD4 (rsCD4) (referred to as soluble gp120-rsCD4 complexes) were also run under identical conditions for comparison. Two hundred micrograms each of these standards was applied to the column in 0.5 ml of PBS. Fractions from runs of BaLgp120, FLSC, and soluble gp120-rsCD4 complexes were analyzed by gp120-capture enzyme-linked immunosorbent assay (ELISA) (22). Fractions from runs of FLSC and TcSC were also analyzed by immunoblot assay with anti-CD4 antibody as described above. The FLSC, TcSC, gp120-rsCD4, and gp120 species were assigned sizes by using the highest point in each peak or shoulder in a chromatogram as the basis for making calculations versus the standard curve.
Gp120-capture ELISA.
BaLgp120, gp120-rsCD4 complexes, or single-chain chimeric complexes were captured using a purified polyclonal sheep antibody (International Enzymes, Fallbrook, Calif.) raised against a peptide derived from the C-terminal 15 amino acids of gp120, D7324 (22), adsorbed to the matrix. The D7324 was diluted in PBS to 2 μg/ml and adsorbed to 96-well plates (Maxisorb plates; VWR Scientific, St. Louis, Mo.) by incubating them overnight at room temperature. Plates were treated with BLOTTO in order to prevent nonspecific binding to the wells. After the plates were washed with TBS, test samples were diluted in BLOTTO and 200-μl aliquots were incubated in duplicate D7324-coated wells for 1 h at room temperature. Bound antigen was detected using a pool of inactivated HIV-1+ sera diluted 1:1,000 in BLOTTO followed by goat anti-human IgG labeled with horseradish peroxidase (KPL). Detection was also accomplished using various MAbs, as indicated in the text, followed by the appropriate labeled second antibody. All antibodies were diluted in BLOTTO and incubated for 1 h at room temperature. Plates were washed three times with TBS between each incubation step. The amounts of gp120 sequences present in the test samples were determined based on a standard curve generated with commercial recombinant HIV IIIB gp120 (Bartels, Issaquah, Wash.). In comparative experiments involving BaLgp120-rsCD4 complexes, D7324-coated plates were treated with saturating concentrations of gp120. After the wells were washed, an excess concentration of rsCD4 (1 μg/ml) was then added to the wells and incubated for 1 h to form the complexes. In order to evaluate the TcSC antigen, which lacks the D7324 epitope, an alternate ELISA format using anti-CD4 MAb 45 (Bartels) for capture was developed. The antibody was adsorbed to plastic at 1 μg/ml, and wells were blocked with BLOTTO. Assays were then carried out as described above using the indicated human sera or human MAbs.
Cell surface coreceptor-binding assay.
L1.2 cells expressing coreceptor were grown in sterile media and treated with 5 mM sodium butyrate for 24 h in order to upregulate coreceptor expression. For staining, 105 cells were added to V-bottom culture wells, washed with PBS, and finally pelleted. The cells were then resuspended in 50 μl of PBS containing 0.1% fetal bovine serum (PBS-FBS) and various concentrations of complexes. After 1 h of incubation at 37°C, the cells were pelleted and washed three times with PBS-FBS. Bound material was detected by incubating the cells with a murine anti-human CD4 antibody, MAb 45 (5 μg/ml), for 1 h at 4°C, followed by treatment with phytoerytherin-labeled anti-mouse IgG (Caltag Laboratories, Burlingame, Calif.) for an additional 1 h at 4°C. The cells were then washed three times with PBS-FBS and analyzed by flow cytometry using a FACSCalibur (Becton Dickinson, San Jose, Calif.).
For competition experiments, complexes and competing MAbs were added to the cells at concentrations of 1 and 10 μg/ml, respectively, in PBS-FBS. Cells and protein were incubated together for 1 h at 37°C. Cells were pelleted and washed with PBS-FBS three times. Bound material was detected by flow cytometry with anti-human CD4 MAb 45 (Bartels) as described above.
Virus neutralization assays.
A total of 104 U373/CD4/MAGI cells (35) expressing either CCR5 or CXCR4 were allowed to attach overnight to flat-bottom tissue culture wells. Culture medium was then removed and then replaced with 100 μl of fresh medium containing various concentrations of test protein. An additional 100 μl of medium containing 50 50% tissue culture infective doses of virus was then added. The entire mixture was then incubated at 37°C until syncytia were visible, which typically occurred within 3 to 5 days. Culture wells were then treated with a β-galactosidase chemiluminescent reagent, Galatostar (Tropix, Bedford, Mass.) according to the manufacturer's protocol. Virus infection was then determined as a function of chemiluminescence, quantified using a Victor2 (EG&G Wallac, Gaithersburg, Md.) fluorescence plate reader. Background signal was determined in assays carried out in the absence of virus. Signals obtained for the test assays were then corrected by subtracting the background value. The percent infection was calculated by dividing the corrected relative light units for each experimental well by the corrected light units for control wells containing only cells and virus. The 90% inhibitory dose (ID90) values were determined from plots of the test protein concentration versus the percent inhibition of infection. All test conditions were carried out in triplicate.
RESULTS
Construction of single-chain BaLgp120-CD4 molecules.
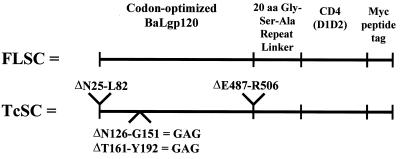
Our strategy for the production of a single-chain gp120-CD4 complex was based on a synthetic, human codon-optimized gene encoding the gp120 of the primary R5 isolate, HIV-1BaL (Fig. 1). This allowed for an efficient, rev-independent expression of envelope in a wide variety of human or mammalian-derived cell lines (15). To construct the single-chain complex, sequences encoding a Gly-Ser-Ala repeat linker were added to the 3′ end of the synthetic gp120 coding sequences. The length of the linker was chosen by spatial analyses of the gp120 structure complexed to soluble CD4 (18). Based on the Swiss PDB Viewer modeling program, we predicted that the 20-amino-acid spacer should provide enough flexibility in the single-chain polypeptide to allow self-association into a complex. The 3′ end of the linker sequence was then attached in frame to an oligonucleotide encoding sequences for the first and second extracellular domains of soluble CD4. The entire single-chain complex gene was completed by adding sequences encoding a short polypeptide tag derived from the c-myc oncogene to the 3′ end. This chimeric recombinant gene, which contained the entire BaL gp120 sequence, was designated full-length single chain (i.e., FLSC). A second construct was designed to produce complexes more closely resembling the molecules used to solve the gp120 crystal structure. This construct, designated truncated single chain (i.e., TcSC), was constructed as before except that a sequence encoding ΔC1ΔC5ΔV1V2 gp120 was used in place of the full-length coding sequence (16, 25, 30, 37, 39) (Fig. 1). The complete single-chain complex genes comprised of these sequences were generated by PCR and inserted into pEF6.
FIG. 1.
Construction of genes encoding single-chain BaLgp120-CD4 molecules. The deletions described for the TcSC construction are numbered according to the BaL gp120 sequence.
Expression and characterization of FLSC and TcSC molecules.
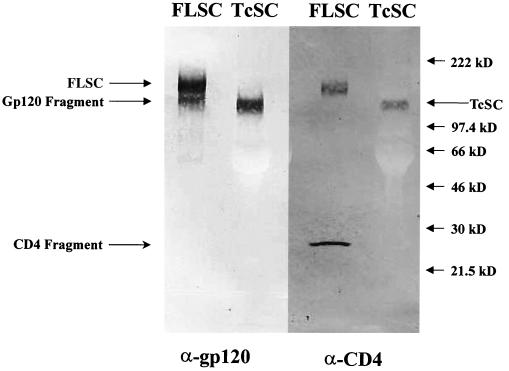
Protein expression by the pEF6-FLSC and pEF6-TcSC plasmids was evaluated by Western immunoblot assays using a mixture of anti-gp120 MAbs (1) or anti-human CD4 polyclonal sera. As shown in Fig. 2, transiently transfected 293 cells expressed a soluble protein with an estimated size of 162 kDa, matching the size predicted for the full-length gp120-CD4 single-chain sequence. As expected, this species was reactive with both anti-gp120 and anti-CD4 antibodies (Fig. 2). In other experiments, we also detected reactivity with anti-myc antibody (data not shown), further confirming the identity of the 162-kDa species as the FLSC. In addition to this high-molecular-mass species, we also observed 120- and 23-kDa bands consistent with the expected size for fragments containing the gp120 and the CD4 D1D2-myc tag moieties, respectively. This indicated that some portion of the single-chain molecules were subjected to proteolysis at or near the gp120-CD4 junction to produce fragments containing either gp120 or CD4 sequences. In contrast to the FLSC, the TcSC was expressed as a single high-molecular-mass protein (Fig. 2) reactive with both anti-CD4 and anti-gp120 antibodies. The apparent molecular mass was approximately 137 kDa, matching the predicted size of the molecule.
FIG. 2.
Expression of single-chain complexes. 293 cells were transiently transfected with pEF6-FLSC or pEF6-TcSC. After 48 h, supernatant was collected and subjected to SDS-PAGE electrophoresis as described in Materials and Methods and then immunoblotted with either a mixture of anti-gp120 MAbs or anti-CD4 polyclonal sera. Uncleaved FLSC, cleaved gp120 and CD4 fragments, and TcSC are indicated.
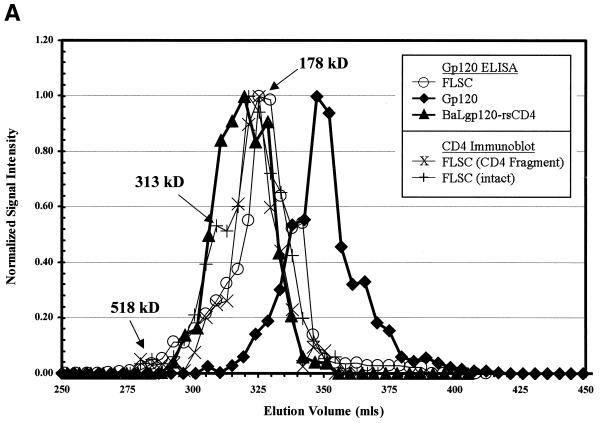
The characteristics of the native single-chain complexes were examined further by gel filtration chromatography on a Superose 6 column under nondenaturing conditions. Column fractions were analyzed by anti-gp120 ELISA and anti-CD4 immunoblot assays, and the collected data were used to generate chromatographic profiles for the single-chain molecules. Densitometric analysis of the immunoblot assays was used to determine the relative positions and amounts of the intact FLSC versus the CD4 fragments. As shown in Fig. 3A, the majority of the intact FLSC eluted as a protein peak of approximately 178 kDa, in agreement with the size expected for a monomer of the polypeptide. Accordingly, the apparent size of the major peak was somewhat smaller than the peak obtained with soluble gp120-rsCD4 complexes (227 kDa), which contain a larger (D1D4) CD4 fragment. The major peak also exhibited a shoulder of higher-molecular-mass, approximately 313 kDa. In addition, a single minor FLSC peak could be distinguished by the ELISA and immunoblot assays corresponding to approximately 518 kDa, roughly matching the expected size of an FLSC trimer. A minor shoulder of similar size was also evident within the gp120-rsCD4 complex standard peak. Notably, fractions containing the two FLSC peaks also contained the cleaved CD4 fragment (Fig. 3A). However, no such anti-CD4 reactive protein was detected in fractions corresponding to the position expected for the free CD4 fragment. Notably, no FLSC protein was detected by the gp120 ELISA in fractions matching the peak position of the BaLgp120 standard (97 kDa; Fig. 3A). Thus, the elution profile of the FLSC indicated that the gp120 and CD4 fragments are extensively incorporated into gp120-CD4 complexes of the same size as the intact FLSC species.
FIG. 3.
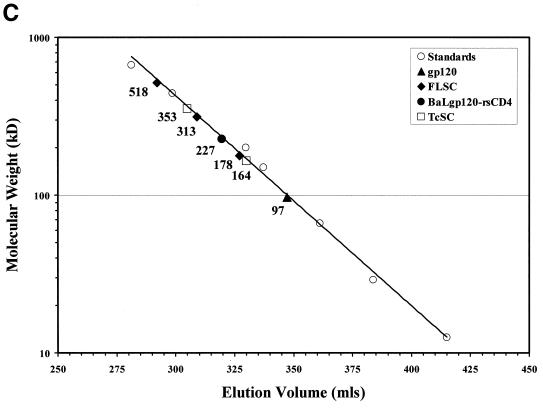
Gel filtration analysis of native FLSC and TcSC polypeptides. (A) Chromatographic profiles of FLSC, gp120-rsCD4 complexes, and BaLgp120 on Superose 6. Chromatography was carried out as described in Materials and Methods. Fractions were collected from the column and analyzed by gp120 capture ELISA using pooled HIV+ sera and by immunoblot assay for CD4 sequences using polyclonal goat anti-CD4 IgG as described in Materials and Methods. In order to clearly compare chromatographic profiles from the different assays, ELISA and immunoblot readings were normalized by assigning the highest signal a value of 1 and then adjusting the rest of the data accordingly. The molecular masses corresponding to distinguishable FLSC species were calculated as described in Materials and Methods and below and are indicated with arrows. (B) Chromatographic profile of TcSC on Superose 6. Chromatography and anti-gp120 immunoblot analyses of column fractions were performed as described in Materials and Methods. (C) Molecular-mass estimations for FLSC and TcSC. Elution volumes for the column protein standards (○) were plotted versus their molecular masses. The volumes shown are averages of measurements made in two separate runs on the same column. Solid symbols represent the elution volume corresponding to the highest normalized signal calculated for each test protein shown in the other panels. The molecular masses estimated from the standard curve are shown. The FLSC, TcSC, gp120-rsCD4, and gp120 species were assigned sizes by using the highest point in each peak or shoulder in the chromatogram as the basis for making calculations versus the standard curve. For the TcSC, estimated molecular masses for the largest and smallest forms of the molecule evident in the gel filtration profile are shown.
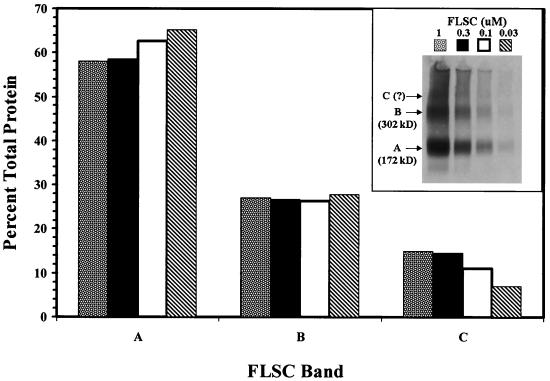
To examine the structural properties of the native FLSC in greater detail, different concentrations (1 to 0.03 μM) of the same protein preparation examined above were covalently cross-linked in PBS in order to fix any multimeric structures existing in solution. The cross-linked material was then analyzed by immunoblot assay with anti-CD4 antibody. As shown in Fig. 4, a major protein band (inset, band A) of 172 kDa was consistently visible, along with two minor bands of higher molecular mass. One of the minor bands (inset, band B) had an apparent size of approximately 302 kDa, while the other (inset, band C) failed to migrate far enough into the gel to allow an accurate assignment of size. The appearance and proportions of the different protein bands were not dependent on the FLSC concentration prior to cross-linking.
FIG. 4.
Immunoblot analyses of native FLSC. Four concentrations of purified FLSC (1 to 0.03 μM) were covalently cross-linked as previously described (8, 10) by treatment with 1.5 μM bis(sulfosuccinimidyl) suberate (BS3; Pierce, Rockford, Ill.) for 30 min at room temperature. The cross-linked material was then subjected to immunoblot analyses with anti-CD4 antibody as described in Materials and Methods. The visible bands (inset) are shown with arrows; sizes for bands A (172 kDa) and B (302 kDa) were calculated relative to protein standards run in a parallel lane. The relative proportion of protein represented by each band was estimated by densitometric analyses and plotted. Total FLSC protein was determined by adding the densitometry values for the three bands in each lane.
In comparison to the FLSC, the chromatographic profile of the TcSC was significantly more complex. Under nondenaturing conditions the protein eluted as a broad series of peaks ranging from 166 to 353 kDa (Fig. 3B). Such a profile indicated that the shorter TcSC polypeptide forms multiple higher-order structures upon expression and/or purification.
Antigenic properties of FLSC and TcSC molecules.
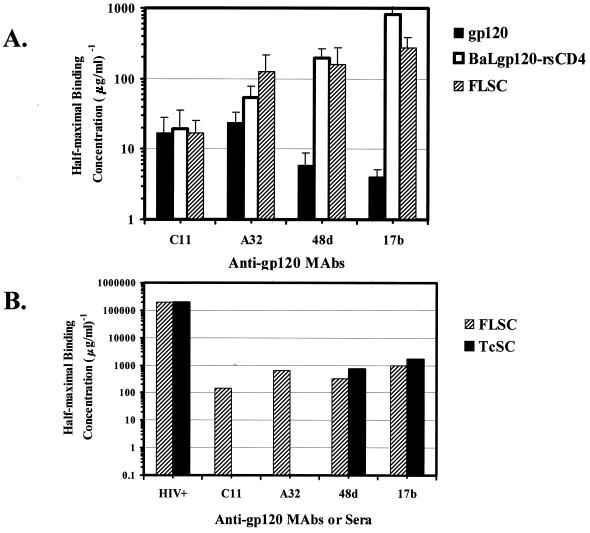
The binding of gp120 to CD4 causes conformational changes in the molecule leading to the exposure of the coreceptor-binding domain. Therefore, antibodies directed against epitopes in this domain should react strongly with properly folded single-chain complexes. To determine if this was the case, purified FLSC and TcSC were subjected to immunochemical analyses by antigen capture ELISA. A sheep anti-gp120 antibody, D7324 (22), was adsorbed to the solid phase in order to capture the FLSC, which was then analyzed using a panel of human MAbs (A32, 17b, and 48d) previously shown to preferentially bind gp120 after engagement of CD4 (21a, 33). Two of the antibodies, 17b and 48d, bind within the coreceptor attachment site that is induced by CD4 binding (31, 34, 36). Antibody C11 (21a), which recognizes a conserved epitope in the C1-C5 region of free gp120, was also tested for comparison.
As shown in Fig. 5A, all of the antibodies reacted strongly with the FLSC. However, the half-maximal binding concentrations of antibodies 17b, 48d, and A32 were consistently higher with FLSC versus gp120 alone and equivalent to what was observed with soluble, noncovalent BaLgp120-rsCD4 complexes. The higher immunoreactivity of FLSC was specific to the antibodies directed against the CD4-induced epitopes, as there was no significant difference in the half-maximal binding concentrations of antibody C11 with FLSC versus free gp120.
FIG. 5.
Exposure of CD4-induced epitopes on single-chain complexes. (A) Reciprocal half-maximal binding concentrations of human anti-gp120 MAbs with purified FLSC. An antigen capture format with antibody D7324 was used as described in Materials and Methods. Antibodies 17b, 48d, and A32, which recognize CD4-induced epitopes on gp120, were tested with FLSC or with BaLgp120 either alone or complexed with rsCD4. C11, an anti-gp120 MAb that is not complex dependent, was used as a control. Threefold dilutions of each antibody were tested starting at 10 μg/ml. Average values derived from three separate experiments are shown. Standard errors are shown with bars. (B) Reciprocal half-maximal binding concentrations of human anti-gp120 MAbs in TcSC versus FLSC ELISAs. Single-chain complexes were captured from transfected 293 cell conditioned medium onto plastic via the anti-CD4 MAb 45, as described in Materials and Methods, and probed with the same antibodies as in panel A. Since different amounts of the FLSC and TcSC were present in the conditioned medium, comparisons of ELISAs were made by normalizing half-maximal binding concentrations based on a conversion factor that equalized the signal obtained with the pooled HIV+ human sera. The level of FLSC reactivity with the HIV+ human sera was 6.6-fold higher than with the TcSC.
Since the D7324 epitope sequences are deleted from the TcSC construct, immunochemical analyses of the TcSC protein were performed using an alternative ELISA format in which anti-CD4 MAb 45 was to capture antigen via the C-terminal CD4 sequences. As shown in Fig. 5B, the level of 17b and 48d reactivity with TcSC was equivalent to what was observed with FLSC analyzed in parallel. As expected, antibodies C11 and A32 did not react with TcSC since the bulk of their respective epitopes were deleted from the TcSC construct.
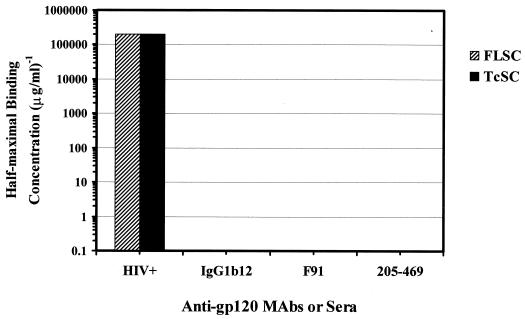
The binding of gp120 and CD4 sequences in the single-chain polypeptides should also block the exposure of epitopes in the CD4 binding site on gp120. To evaluate whether such binding had occurred, the FLSC and TcSC were evaluated using the MAb 45 capture format and a series of MAbs (IgG1b12, F91, and 205-469) directed against the CD4 binding domain (CD4bd) on gp120. As shown in Fig. 6, none of the antibodies reacted with either single-chain complex, although positive reactivity was observed with pooled HIV+ sera tested in parallel. This suggested an interaction between CD4 sequences and the CD4bd present within the single chain complexes.
FIG. 6.
Absence of exposed CD4bd epitopes on the single-chain complexes FLSC and TcSC. Reciprocal half-maximal binding concentrations of HIV+ sera and MAbs IgG1b12, F91, or 205-469 in FLSC and TcSC ELISAs. Single-chain complexes were captured from transfected 293 cell conditioned medium onto plastic via the anti-CD4 MAb 45, as described in Materials and Methods. Threefold dilutions of each antibody were tested starting at 10 μg/ml. Since different amounts of the FLSC and TcSC were captured from the conditioned medium, comparisons of ELISAs were made by normalizing half-maximal binding concentrations based on a conversion factor that equalized the signal obtained with the HIV+ human sera. The level of FLSC reactivity with the HIV+ human sera was 6.6-fold higher than with the TcSC.
Analyses of FLSC and TcSC function.
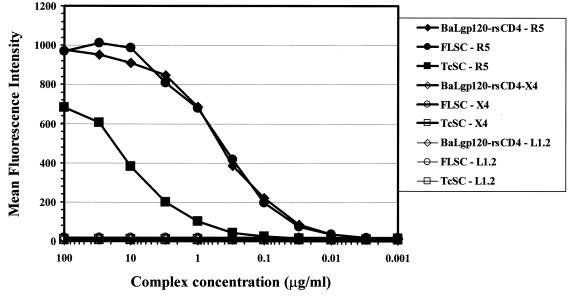
The formation of the gp120-CD4 complex normally exposes the envelope domains that interact with an appropriate coreceptor (33, 35). Therefore, a correctly folded single-chain complex containing the R5 BaL envelope should bind specifically to R5. To test for this property, purified FLSC and TcSC proteins were incubated with either L1.2 cells expressing either R5 or CXCR4 (X4) (35). Parental cells not expressing coreceptor were also tested as controls. As shown in Fig. 7, both single-chain forms bound only to the R5-expressing cells. Maximal binding was observed with FLSC at concentrations (10 μg/ml) equivalent to what was observed with soluble BaLgp120-rsCD4 complexes tested as controls. In comparison, approximately 10-fold-higher concentrations of the TcSC were required to approach saturation binding.
FIG. 7.
Specific interaction of single-chain and BaLgp120-rsCD4 complexes with the CCR5 coreceptor. Various concentrations of complexes were incubated with either L1.2 cells expressing CCR5 (R5), L1.2 cells expressing CXCR4 (X4), or parental L1.2 cells (L1.2). Bound complexes were detected by flow cytometry using 5 μg of MAb 45 per ml as described in Materials and Methods. The values shown are from a representative experiment repeated three times with the same results.
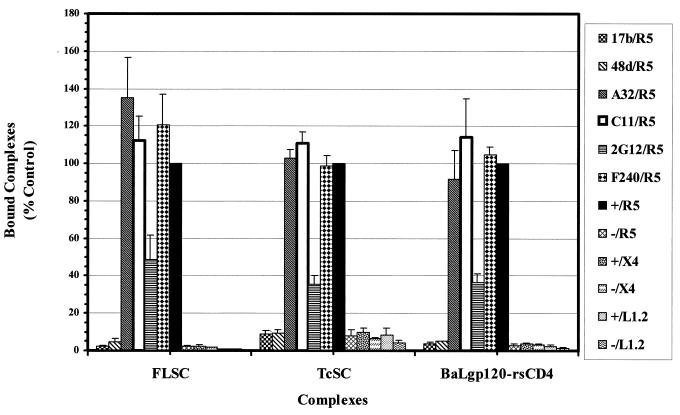
To verify that the expected coreceptor-binding domain on gp120 mediated the binding to R5, the single-chain complexes were retested for cell surface binding after treatment with anti-coreceptor domain antibodies. As shown in Fig. 8, 17b and 48d strongly inhibited the binding of both single-chain complexes to the cells. In the presence of these antibodies, the binding signal on R5-expressing cells was the same as the background binding seen with L1.2-X4 and L1.2 parental cells. Interestingly, 2G12, a potent neutralizing antibody, also reduced the interaction of all complex forms with R5. In comparison, anti-gp120 antibodies recognizing epitopes outside the coreceptor binding domain, C11, A32, and an anti-gp41 antibody, F240, all failed to reduce the binding of FLSC or TcSC to the R5-expressing L1.2 cells.
FIG. 8.
Inhibition of FLSC and TcSC interaction by antibodies against the coreceptor binding site on gp120. FLSC, TcSC, or BaLgp120-rsCD4 complexes (1 μg/ml) were incubated with L1.2 cells expressing CCR5 (R5), CXCR4 (X4), or no coreceptor (L1.2) and 10 μg of the indicated antibodies per ml. Complex binding was then determined by flow cytometry with 5 μg of MAb 45 per ml as described in Materials and Methods. Control assays with each cell type were carried out with each type of complex in the absence of antibody and are designated with a “+.” Results are presented as the percent binding relative to the mean fluorescence intensity obtained in the matched control assay. Background measurements obtained with untreated cells are designated with a “−.” Average values derived from three separate experiments are shown. Standard errors are shown with bars.
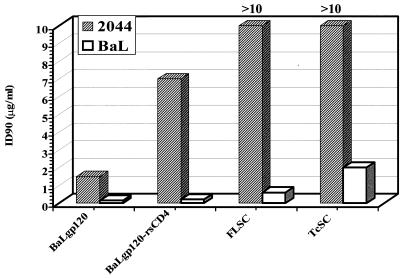
The single-chain complexes were further examined for coreceptor-binding specificity by testing their ability to neutralize R5 and X4 viruses. As shown in Fig. 9, both FLSC and TcSC potently and selectively neutralized the R5 HIV-1BaL isolate, while there was only a slight inhibition (ID90 > 10 μg/ml) of the X4 isolate. In comparison, uncomplexed BaLgp120 inhibited entry of both HIV-1BaL and X4 (HIV-12044) viruses as expected due to its direct interactions with CD4.
FIG. 9.
Neutralization of HIV-12044 and HIV-1BaL by FLSC and TcSC. BaLgp120, BaLgp120-rsCD4 complexes, FLSC, and TcSC were examined for neutralization of HIV-1 infection, as described in Materials and Methods, using U373 cells that express CD4, either R5 or X4, and β-galactosidase governed by the HIV-1LTR promoter. An X4-specific isolate, HIV-12044, and the R5-specific isolate, HIV-1BaL, were tested in parallel. An ID90 for FLSC and TcSC against HIV-12044 was not achieved with the maximum concentrations tested and is therefore presented as >10 μg/ml.
DISCUSSION
The gp120-CD4 complex represents a critical intermediate structure in early HIV replication that is essential for virus entry. The formation of this complex requires an interaction between one gp120 molecule and a single CD4 receptor and can be accomplished with soluble molecules. Thus, a chimeric polypeptide containing gp120 and CD4 sequences separated by a flexible linker could be expected to fold into a structure duplicating the gp120-CD4 complex. We describe here the construction and characterization of two such chimeras: one having full-length gp120 and the other containing a truncated HIV envelope closely resembling the molecules used to resolve the crystal structure of gp120 bound to CD4 (18).
Both single-chain forms exhibited the expected antigenic properties of the gp120-CD4 complex, being strongly reactive with MAbs to the coreceptor-binding site. Further, the levels of reactivity with such antibodies were significantly higher than those observed with free gp120 (Fig. 5), in which the coreceptor binding site is partially inaccessible. Such results strongly indicated that the gp120 and CD4 sequences in the single-chain molecules form stable interactions. In accordance, antibodies to the CD4bd, which would be occluded in the gp120-CD4 complex, failed to recognize either the FLSC or TcSC in antigen capture ELISA (Fig. 6).
The single-chain molecules also exhibited functional properties characteristic of a gp120-CD4 complex. Both TcSC and FLSC, containing the R5 HIV-1BaL envelope sequence, bound specifically to R5-expressing cells in a dose-dependent manner. In each case the binding was completely blocked by 17b and 48d MAbs directed against the coreceptor-binding domain, confirming that the interaction with R5 was mediated by the gp120 moiety of the complexes. In agreement with these data, the single-chain chimeras mediated selective antiviral effects consistent with their coreceptor preference. Both FLSC and TcSC selectively neutralized the R5-specific isolate, HIV-1BaL, in a coreceptor-specific neutralization assay. It is noteworthy that the soluble BaLgp120-rsCD4 complexes were less-specific inhibitors and neutralized the X4 HIV-12044 isolate in the 6- to 7-μg/ml range, whereas the single-chain complexes did not. Such neutralization was likely due to more extensive or irreversible dissociation of the gp120-rsCD4 complexes during the extended incubation periods of the neutralization assay, allowing free gp120 to interfere in CD4 interactions. A greater stability of the single-chain complexes may have limited such effects. However, at high concentrations weak neutralization of the X4 virus was also mediated by the FLSC (data not shown). The complexes of gp120 and CD4 fragments present in the FLSC preparation (see below) could explain this effect, as they are capable of dissociating in a manner analogous to the gp120-rsCD4 complexes.
A major consideration for the single-chain chimeras is whether their antigenic and functional properties arise from multimeric, interchain binding (gp120 in one molecule interacts with CD4 sequences in another) or from monomeric intrachain interactions. The latter would give rise to bona fide single-chain gp120-CD4 complexes. This question was addressed by gel filtration, SDS-PAGE, and immunoblot analyses of the single-chain chimeras. These analyses revealed that the FLSC and TcSC had significantly different physical features.
The majority of the intact FLSC behaved as a monomer, as it exhibited similar sizes under nondenaturing (178 kDa; Fig. 3) and denaturing (162 kDa; Fig. 2) conditions. These sizes were in agreement with predictions of a monomeric molecular mass based on the composition of the molecule. Further, immunoblot analyses (Fig. 4) showed that FLSC behaves primarily as a 172-kDa protein even after covalent cross-linking of the native material. In view of its antigenic and functional properties the intact FLSC seems capable of folding into an intrachain complex. Thus, as predicted, the 20-amino-acid linker positioned between the gp120 and CD4 sequences seems to be sufficient to allow the CD4bd of gp120 to engage the CD4 moiety.
However, it is also clear that the FLSC is not exclusively monomeric, since larger forms of the molecule were detected under nondenaturing conditions. The gel filtration experiments suggested the presence of minor species of 313 and 518 kDa, while the immunoblot analyses of cross-linked FLSC showed two minor high-molecular-mass forms (Fig. 4, inset). The smaller of these exhibited a molecular mass of approximately 302 kDa, possibly representing an FLSC dimer. This species probably corresponds to the 313-kDa shoulder of the major FLSC peak evident in the gel filtration chromatogram. Although the larger cross-linked species (Fig. 4) could not be assigned an accurate molecular mass by immunoblot analyses, it is likely to be the same as the 518-kDa species that was also fractionated by the gel filtration column. In either case, we cannot conclude that these oligomers reflect interchain complex formation, since we have detected analogous high-molecular-mass molecules resembling dimers and trimers in preparations of cross-linked BaLgp120 and rsCD4 (Fig. 3A and data not shown). Similarly, oligomeric molecules were also evident in preparations of cross-linked IIIB gp120 and rsCD4 complexes (10). Therefore, limited oligomerization appears to be a general characteristic of gp120-CD4 complexes and is not unique to the FLSC. Whether these oligomers arise via interactions between sequences in gp120, CD4, or both is as yet unclear.
Unlike the TcSC, the FLSC was sensitive to proteolysis that generated fragments containing either gp120 or CD4 sequences. However, this action did not grossly disrupt the relevant structure of the FLSC. The fragments coeluted from the gel filtration column and were not recovered separately (Fig. 3A), indicating that they were almost entirely incorporated into gp120-CD4 complexes. These complexes, which are untethered and almost certainly exist as monomers, were physically indistinguishable from the intact 178-kDa FLSC under nondenaturing conditions. Notably, the complexed fragments coeluted with the larger species of intact FLSC, indicating that they form oligomers in the same manner as the intact single chain.
The position of the cleavage site that separates the fragments is probably located within the C-terminal gp120 sequences present only in the FLSC, since the shorter TcSC did not exhibit degradation. Notably, these sequences encompass the gp120-gp41 junction normally cleaved by the furin protease (14). Cleavage of the FLSC at the natural furin site would be consistent with the behavior of the FLSC fragments, as it would have minimal impact on the structures of the gp120 and CD4 moieties and their capacity to interact. In accordance with this, we have recently determined that mutations in the putative furin site in fact dramatically reduce proteolytic degradation of the FLSC (data not shown).
In contrast to the FLSC, the TcSC eluted from the gel filtration column (Fig. 3B) as a very broad peak representative of a collection of oligomers. This behavior indicates that the TcSC exists primarily as variably sized chains of polypeptides joined by interactions between gp120 sequences and CD4 sequences in separate molecules. Since the TcSC was created by deleting 20 C-terminal amino acids from gp120, the distance between the CD4 core structure and the CD4bd of gp120 was significantly shortened. This may have hindered the ability of the TcSC to achieve an intramolecular gp120-CD4 interaction and favored the formation of interchain complexes. Nevertheless, the TcSC also exhibited the antigenic and functional features of a gp120-CD4 complex. However, the maximal binding of the TcSC to cell surfaces required approximately 10-fold higher concentrations compared to the FLSC. It is possible that because of intermolecular interactions involving multiple TcSC molecules, a smaller proportion of the total protein expressed a coreceptor binding site capable of interacting with surface coreceptors. Alternatively, the deletion of the V1 and/or V2 regions in the TcSC may decrease the relative affinity of the BaL envelope for R5. Further modification of the TcSC to elongate the linker between the gp120 and CD4 moieties might allow it to form intrachain complexes. Whether the multimeric nature of the TcSC represents a disadvantage remains an open question, since studies with other multimeric molecules suggest they are more potent immunogens than their monomeric counterparts (9, 16, 20). Studies to address this question are ongoing.
The availability of single-chain analogues of the gp120-CD4 complex should greatly facilitate the identification of molecules that interact with the coreceptor binding domain. Folded single-chain complexes can be expressed at high levels in culture and conveniently purified in a single immunoaffinity step using either lectin or antibodies specific for CD4-induced epitopes. Because they reliably present the coreceptor binding site, such molecules can now be used as convenient tools for identifying and evaluating the therapeutic potential of MAbs or other compounds that might interfere with HIV-1 coreceptor interactions.
The synthetic genes for the single-chain complexes should also vastly improve the versatility of vaccine approaches to target novel conformational epitopes on gp120. The single-chain gene can be introduced into vaccine delivery vehicles such as attenuated vaccinia virus (14, 23), Semliki Forest virus (14, 24), or Salmonella sp. (26, 28) to provide an efficient and reliable means for the expression of properly associated and folded gp120 and CD4 sequences. Thus, both humoral and cellular responses to complex-dependent epitopes could be generated. Introducing envelope genes derived from viruses that use alternative coreceptors could further expand the potential of these single-chain molecules.
ACKNOWLEDGMENTS
We thank Lijun Wu for the generous contribution of the transfected L1.2 cells and Robert Powell and Megan Vorthman for assistance in the purification of the complexes.
This work was supported by grants NHLBI R01 03-5-20064, NIAID R21 03-5-21326 to A.L.D., NIAID P01 A143046 to A.L.D., D. H., and G.K.L., and NIAID R01 A138112 to G.K.L.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claassen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retrovir. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Burton D R, Pyati J, Koduri R, Thornton G B, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 4.Cavacini L A, Emes C L, Wisnewski A V, Power J, Lewis G, Montefiori D, Posner M R. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res Hum Retrovir. 1998;14:1271–1280. doi: 10.1089/aid.1998.14.1271. [DOI] [PubMed] [Google Scholar]
- 5.Celada F, Cambiaggi C, Maccari J, Burastero S, Gregory T, Patzer E, Porter J, McDanal C, Matthews T. Antibody raised against soluble CD4-gp120 complex recognizes the CD4 moiety and blocks membrane fusion without inhibiting CD4-gp120 binding. J Exp Med. 1990;172:1143–1150. doi: 10.1084/jem.172.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;1998:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 7.Deen K C, McDougal J S, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon P J, Axel R, Sweet R W. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature. 1988;331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 8.DeVico A, Silver A, Thornton A M, Sarngadharan M G, Pal R. Covalently crosslinked complexes of human immunodeficiency virus type 1 (HIV-1) gp120 and CD4 receptor elicit a neutralizing immune response that includes antibodies selective for primary virus isolates. Virology. 1996;218:258–263. doi: 10.1006/viro.1996.0188. [DOI] [PubMed] [Google Scholar]
- 9.DeVico A L, Gallo R C. HIV coreceptors and chemokines as therapeutic targets against HIV/AIDS. AIDS Rev 1999. 1999;1:4–14. [Google Scholar]
- 10.DeVico A L, Rahman R, Welch J, Crowley R, Lusso P, Sarngadharan M G, Pal R. Monoclonal antibodies raised against covalently crosslinked complexes of human immunodeficiency virus type 1 gp120 and CD4 receptor identify a novel complex-dependent epitope on gp120. Virology. 1995;211:583–588. doi: 10.1006/viro.1995.1441. [DOI] [PubMed] [Google Scholar]
- 11.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershoni J M, Denisova G, Raviv D, Smorodinsky N I, Buyaner D. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J. 1993;7:1185–1187. doi: 10.1096/fasebj.7.12.7690724. [DOI] [PubMed] [Google Scholar]
- 13.Gilljam G. Envelope glycoproteins of HIV-1, HIV-2, and SIV purified with Galanthus nivalis agglutinin induce strong immune responses. AIDS Res Hum Retrovir. 1993;9:431–438. doi: 10.1089/aid.1993.9.431. [DOI] [PubMed] [Google Scholar]
- 14.Girard M, Habel A, Chanel C. New prospects for the development of a vaccine against human immunodeficiency virus type 1. An overview. C R Acad Sci III. 1999;322:959–966. doi: 10.1016/s0764-4469(00)87193-0. [DOI] [PubMed] [Google Scholar]
- 15.Haas J, Park E C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 16.Jeffs S A, McKeating J, Lewis S, Craft H, Biram D, Stephens P E, Brady R L. Antigenicity of truncated forms of the human immunodeficiency virus type 1 envelope glycoprotein. J Gen Virol. 1996;77:1403–1410. doi: 10.1099/0022-1317-77-7-1403. [DOI] [PubMed] [Google Scholar]
- 17.Kang C-Y, Hariharan K, Nara P L, Sodroski J, Moore J P. Immunization with a soluble CD4-gp120 complex preferentially induces neutralizing anti-human immunodeficiency virus type 1 antibodies directed to conformation-dependent epitopes of gp120. J Virol. 1994;68:5854–5862. doi: 10.1128/jvi.68.9.5854-5862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 21.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 21a.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore J P, Jarrett R F. Sensitive ELISA for the gp120 and gp160 surface glycoproteins of HIV-1. AIDS Res Hum Retrovir. 1988;4:369–379. doi: 10.1089/aid.1988.4.369. [DOI] [PubMed] [Google Scholar]
- 23.Moss B. Use of vaccinia virus vectors for development of AIDS vaccines. Aids. 1988;2(Suppl. 1):S103–S105. doi: 10.1097/00002030-198800001-00015. [DOI] [PubMed] [Google Scholar]
- 24.Mossman S P, Bex F, Berglund P, Arthos J, O'Neil S P, Riley D, Maul D H, Bruck C, Momin P, Burny A, Fultz P N, Mullins J I, Liljestrom P, Hoover E A. Protection against lethal simian immunodeficiency virus SIVsmmPBj14 disease by a recombinant Semliki Forest virus gp160 vaccine and by a gp120 subunit vaccine. J Virol. 1996;70:1953–1960. doi: 10.1128/jvi.70.3.1953-1960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard S R, Rosa M D, Rosa J J, Wiley D C. Truncated variants of gp120 bind CD4 with high affinity and suggest a minimum CD4 binding region. EMBO J. 1992;11:585–591. doi: 10.1002/j.1460-2075.1992.tb05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell R, Lewis G, Hone D. Introduction of eukaryotic expression cassettes into animal cells using a bacterial vector delivery system. In: Bran F, Norrby E, Burton D, Meckalanos J, editors. Molecular approaches to the control of infectious diseases. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 183–187. [Google Scholar]
- 27.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 28.Shata M T, Stevceva L, Agwale S, Lewis G K, Hone D M. Recent advances with recombinant bacterial vaccine vectors. Mol Med Today. 2000;6:66–71. doi: 10.1016/s1357-4310(99)01633-0. [DOI] [PubMed] [Google Scholar]
- 29.Sodroski J. HIV-1 entry inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 30.Stamatatos L, Wiskerchen M, Cheng-Mayer C. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res Hum Retrovir. 1998;14:1129–1139. doi: 10.1089/aid.1998.14.1129. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J P, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun N C, Ho D D, Sun C R Y, Liou R-S, Gordon W, Fung M S C, Li S-L, Ting R C, Lee T-H, Chang N T, Chang T W. Generation and characterization of monoclonal antibodies to the putative CD4-binding domain of human immunodeficiency virus type 1 gp120. J Virol. 1989;63:3579–3585. doi: 10.1128/jvi.63.9.3579-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3986. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 35.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 37.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 39.Wyatt R, Sullivan N, Thali M, Repke H, Ho D D, Robinson J, Posner M, Sodroski J. Characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]