Abstract
Herpes simplex virus type 1 (HSV-1) glycoprotein D (gD) is an essential component of the entry apparatus that is responsible for viral penetration and subsequent cell-cell spread. To test the hypothesis that gD may serve distinguishable functions in entry of free virus and cell-cell spread, mutants were selected for growth on US11cl19.3 cells, which are resistant to both processes due to the lack of a functional gD receptor, and then tested for their ability to enter as free virus and to spread from cell to cell. Unlike their wild-type parent, HSV-1(F), the variants that emerged from this selection, which were named SP mutants, are all capable of forming macroscopic plaques on the resistant cells. This ability is caused by a marked increase in cell-cell spread without a concomitant increase in efficiency of entry of free virus. gD substitutions that arose within these mutants are sufficient to mediate cell-cell spread in US11cl19.3 cells but are insufficient to overcome the restriction to entry of free virions. These results suggest that mutations in gD (i) are sufficient but not necessary to overcome the block to cell-cell spread exhibited by US11cl19.3 cells and (ii) are insufficient to mediate entry of free virus in the same cells.
Herpes simplex virus (HSV) can enter a naive host cell by either of two distinct methods. Extracellular virions present during primary infection can enter cells in exposed tissue by entry of free virus. Once a cell is initially infected, subsequent infections can begin by lateral spread from the initially infected cell to adjacent uninfected neighbors by cell-cell spread. Both types of entry are important for sustained viral infection. The ability of a virus to spread from cell to cell provides a powerful advantage in vivo since it is able to avoid the strong humoral immune response elicited to extracellular HSV virions. The essential roles of HSV glycoproteins in entry of free virus and cell-cell spread remain poorly characterized.
HSV entry requires a complicated cascade of virus-cell interactions. Initially, an interaction between cellular heparan sulfate proteoglycan and viral glycoprotein C (gC) and/or gB results in attachment of the virion to the cell surface (15, 16, 20, 47, 56). Fusion between the viral membrane and the cellular plasma membrane is not triggered until a secondary interaction occurs between gD and a cellular receptor (12, 17, 21, 54). The fusion event itself requires the coordinated function of the viral glycoproteins gB, gD, gH, and gL. These four proteins play essential roles in both entry of free virus and cell-cell spread (3, 11, 43). These two processes are thus closely related. However, entry of free virus and cell-cell spread can be distinguished in HSV and in related alphaherpesviruses by their differing dependence on specific viral genes and by their differing sensitivities to mutations in the viral genome. Deletion of the viral glycoproteins gE and gI provides a clear distinction between entry of free virus and cell-cell spread since it significantly reduces the efficiency of cell-cell spread in cell lines that closely parallel cells infected in vivo while having no effect on the efficiency of those deletion viruses to enter as free virions (8, 9). In addition, removal of gC reduces the entry of free virus, presumably by decreasing viral attachment or binding, but enhances cell-cell spread (14, 20, 24). The role of the four essential mediators of fusion may also differ in entry of free virus and cell-cell spread. Pseudorabies virus requires gD for entry of free virus but not for cell-cell spread (30, 31, 37), and antibodies against HSV-1 gL can block syncytium formation without affecting the entry of free virus (29). Finally, although wild-type bovine herpesvirus 1, like HSV, requires gD for entry of free virus and cell-cell spread, a point mutation in the bovine herpesvirus gH ectodomain is sufficient to restore to a gD-null virus the capability for cell-cell spread but not the ability to enter as free virus (45, 46).
Two different types of cell lines have been described that fail to support efficient entry of HSV and related alphaherpesviruses due to interference with the function of gD. Some cell lines that express gD constitutively have been found to resist entry, presumably by sequestering receptors (2, 4, 7, 33). Other cell lines have been described that are resistant to entry, evidently due to the lack of a functional receptor for gD (39–41, 47, 49, 50). In either case, the hypothesis that resistance to HSV entry results from the inability of the cells to present functional receptors to the virus is supported by the observation that susceptibility to infection can be partially restored to these cells by expression of proteins that have gD receptor function (13, 25, 41, 48, 51). Selection of mutant viruses that show enhanced growth on these resistant cell types has resulted in the identification of substitution mutations in gD that confer on the mutant the ability to partially overcome the resistance to infection (2, 4, 6, 39). At least some of these mutations alter the interaction of gD with its receptors and are thought to confer the ability to use an alternate receptor. For example, mutations at amino acid 27 of HSV-1 gD increase its rate of complex formation with HveC, prevent its association with HveA and 3-O-sulfated HS, and allow it to interact with HveB (6, 19, 48, 52, 54).
US11cl19.3 is a Syrian hamster kidney cell clone that is highly resistant to HSV entry and cell-cell spread but fully susceptible to other events in HSV infection (39–41). Stable expression of a functional gD receptor in these cells suffices to restore plaque formation to wild-type HSV, suggesting that these cells lack a functional gD receptor and that a gD receptor interaction is a necessary component of cell-cell spread by wild-type virus (41). Virus that enters these cells completes a normal cycle of growth, making them ideally suited to selection and characterization of viral mutants that can overcome the block to entry. We hypothesize that such mutations will fall within two general types: (i) mutations which allow the virus to utilize an alternate gD receptor, and/or (ii) mutations which eliminate the necessity of a gD receptor in viral entry. Furthermore, if the necessity or specificity of gD-receptor interactions in the entry of free virus and cell-cell spread differs, mutants should arise that uncouple these processes. Here we report selection and characterization of HSV-1(F) variants that are capable of forming plaques on US11cl19.3 cells and that carry substitution mutations in the coding sequence for gD. The ability of these mutants to spread from cell to cell is clearly enhanced without concomitant enhancement of entry of free virus. This suggests that the functions of gD in the process of entry of free virus may be distinguishable from its role in cell-cell spread.
MATERIALS AND METHODS
Cells and viruses.
Vero, VD-60 (gift of David Johnson), BHK(TK−), 100-33, and US11cl19.3 cells have been previously described (1, 21, 22, 34, 39, 41, 42) and were maintained in Dulbecco's modified Eagle's medium-high glucose containing 5% fetal bovine serum. The properties of the wild-type strain HSV-1(F) have been described previously (10). The viruses R5001 and HSV-1(U10) (gift of Gabriella Campadelli-Fiume) have also been described (4, 10, 18, 24, 34–36, 39).
Selection of mutant viruses.
US11cl19.3 cells are resistant to both entry of free HSV and cell-cell spread. Escape mutants of HSV-1 capable of productively infecting these cells in culture were selected by repeated passage of resistant cells following high-multiplicity infection. Replicate cultures of 2 × 106 resistant cells were each exposed to 2 × 108 PFU (as measured on Vero cells) of HSV-1(F). Cultures were maintained in growth medium supplemented with fetal bovine serum and passaged in a 1:5 dilution every 3 to 5 days. At each passage, 1 ml of medium was frozen in autoclaved milk to maintain a passage history during selection. This process was repeated until 100% cytopathic effect was achieved or until passage 10 was reached, at which point a freeze-thaw lysate was made from each flask. These lysates were then serially diluted and plated on Vero cells. If virus was present at any dilution, it was plaque purified to homogeneity.
Plaque assays.
Subconfluent cultures of Vero, 100-33, or US11cl19.3 were exposed to virus at 37°C for 90 min and subsequently incubated for 48 h in growth medium containing 0.01% pooled human immune globulin. The cultures were then fixed in methanol at −20°C for 10 min. Virus plaques were detected by immunoassay using monoclonal antibody directed against HSV-1 gD (Goodwin Cancer Research Labs) as previously described (39).
Infectious-center assays.
Infectious-center assays were performed in triplicate as previously described (40, 41). Duplicate cultures of 100-33 and US11cl19.3 cells were exposed to serial dilutions of each virus and incubated to allow entry and initiation of infection with each virus tested. After 90 min, residual virus was inactivated or removed by exposure of the cells to low-pH buffer. Infection was allowed to proceed at 37°C in the presence of neutralizing anti-HSV antibodies. At 4 h after infection, the cells from one of the cultures from each set of duplicates were detached by trypsinization and seeded onto a monolayer of susceptible Vero cells. All cultures were then incubated in the presence of neutralizing antibody. At 48 h after infection, cultures were fixed and immunostained to allow visualization of plaques as described previously (39). Initial serial dilutions, which were repeated in triplicate, were seeded to allow visualization of 20 to 200 plaques per well. The number of infectious centers (i.e., plaques nucleated on Vero cells) is a direct reflection of the number of viral particles that were able to initiate infection following entry of free virus. The number of infectious centers formed on US11cl19.3 cells compared to the number of infectious centers formed on 100-33 cells directly reflects the efficiency with which virus can enter the resistant cells as free virus and controls for the possibility that mutations may affect steps in the viral life cycle other than entry. The number of plaques formed on the remaining duplicate culture compared to the number of infectious centers formed on the Vero monolayer directly reflects the efficiency with which virus that successfully entered as free virus could subsequently spread from cell to cell.
Immunoblotting.
Proteins from Vero or 100-33 cells infected for 18 h were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels using standard methods (44), electrically blotted to nitrocellulose membranes, and probed with a 1:2,000 dilution of anti-gD monoclonal antibody (Goodwin Institute) as previously described (38).
Sequencing.
The genomic sequence corresponding to gD was PCR amplified for sequencing with the following primer pairs: gDN(L) (CGTCATAGTGGGCCTCCA) plus gDN(R) (GTCACCCCCTGCTGGTAG), and gDC(L) (CCGTCAGCCTGCCTCTC) plus gDC(R) (CCCCCGCACCCATTAAG). Genome-length viral DNA from HSV-1(F) or each of the SP mutants was included as a template for PCR amplification. Amplification products were separated on agarose gels and purified using glass fines. Both strands of purified PCR products were subjected to automated sequencing at the University of Iowa DNA Sequencing Facility, using the amplification primers as sequencing primers. This entire process was repeated to ensure that no errors had been incorporated during PCR.
Plasmid constructions.
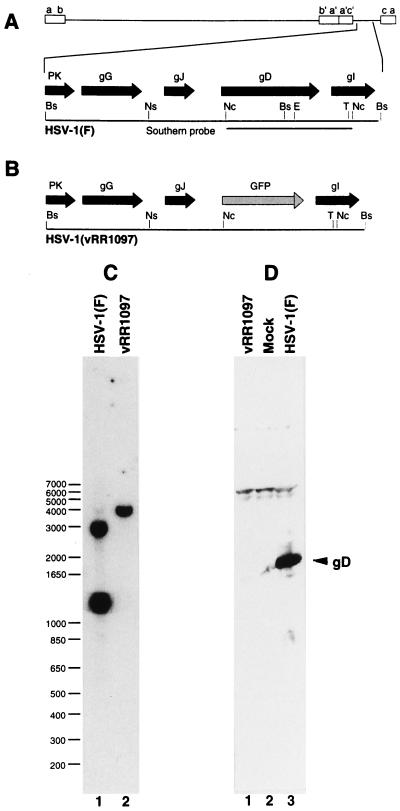
Plasmid pRR1097, used for construction of the gD deletion virus HSV-1(vRR1097), was constructed in several steps. HSV-1(F) DNA was digested to completion with Tth111I restriction endonuclease, treated with Klenow enzyme and deoxynucleoside triphosphates (dNTPs) to produce blunt ends, and then digested with NsiI. The 2.44-kb Tth111I-NsiI fragment from this digestion was purified and ligated into the PstI and SmaI sites of pGEM-3Z (Promega) to generate pRR1074. This plasmid was subsequently mutagenized using a unique site elimination mutagenesis protocol (Clontech) with a mutagenic primer (5′-AGCCCCCCCCATGGCGGAACGCACCA-3′) to introduce an NcoI site overlapping the initiation codon of the gD coding sequence, producing plasmid pRR1075. Sequences coding for residues 1 to 300 of pre-gD were excised from pRR1075 by sequential treatment with EcoNI, Klenow enzyme and dNTPs, and NcoI and then replaced by ligation with a green fluorescent protein (GFP)-containing insert derived from phGFPS65T (Clontech) by sequential treatment with XbaI, Klenow enzyme and dNTPs, and NcoI, generating pRR1097. Plasmid pRR1004, used for generation of radiolabeled probe for Southern blotting, was generated by isolating the 1.54-kb NcoI restriction fragment of HSV-1(F) carrying most of the gD gene, treating with Klenow enzyme and dNTPs to generate blunt ends, and ligating into SmaI-cut pGEM-3Z.
Recombinant virus construction.
To construct vRR1097, HSV-1(F) genomic DNA and pRR1097 were cotransfected into 10-cm2 cultures of VD-60 cells using 12 μl of LipofectAmine (Gibco-BRL) as recommended by the manufacturer. After the cotransfected culture had achieved 100% cytopathic effect, a viral stock was prepared and serial dilutions were plated on VD-60 cells. After 48 h, well-isolated fluorescent plaques were picked and brought through cycles of plaque purification until a homogeneous population of fluorescent plaques was obtained.
Purification of viral DNA.
Roller bottle (850 cm2) cultures of Vero or VD-60 cells were infected with virus at a multiplicity of infection of 0.5 and incubated until cytopathic effect was evident throughout the culture. Infected cells were washed once with phosphate-buffered saline, scraped into 10 ml of phosphate-buffered saline, and then collected by centrifugation at 800 × g in a clinical centrifuge. Cells were resuspended in 4 ml of viral DNA lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1.5 mM MgCl2), lysed by addition of Nonidet P-40 to 0.1%, and then centrifuged at 5,000 rpm in a Sorvall SS34 rotor for 5 min to pellet nuclei. The supernatant from the centrifugation, containing cytoplasmic virus particles, was adjusted to 5 mM EDTA, 50 mM β-mercaptoethanol, 0.5% SDS, and 100 μg of proteinase K per ml and incubated at 37°C for 30 min to release viral DNA. The suspension was extracted three times with an equal volume of 1:1 water-saturated phenol–chloroform, and then DNA was precipitated from the aqueous phase by addition of 2 volumes of ethanol and incubation at −20°C for 2 h. Precipitated DNA was collected by centrifugation, resuspended in 0.3 ml of TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA) containing 30 μg of RNase A per ml, and incubated for 15 min at 37°C to degrade RNA. Viral DNA was separated from smaller contaminants by pelleting through a linear gradient of 5 to 20% potassium acetate in TE buffer in a Beckman SW40 rotor at 27,000 rpm for 16 h at 20°C. After aspiration of the supernatant, the pelleted viral DNA was resuspended in TE buffer, extracted once with an equal volume of chloroform, ethanol precipitated, and redissolved in distilled water.
Southern analysis.
Radiolabeled probe for Southern hybridization was synthesized by transcription of pRR1004 with T7 RNA polymerase in reaction mixtures containing [α-32P]CTP using protocols provided by Promega Biotec. Viral DNA was digested with BstZ17I, electrophoretically separated on a 1% agarose gel, blotted to a Zeta Probe membrane (Bio-Rad), and probed with sequences corresponding to the Southern probe shown in Fig. 3A.
FIG. 3.
Structure of recombinant vRR1097 genome and gD expression in recombinant virus. (A and B) Sequence arrangement of viral genomes of HSV-1(F) (A) and vRR1097 (B) around the gD locus showing the positions of viral open reading frames (solid arrows), GFP insertion (shaded arrow), and probe used for Southern analysis of recombinant virus genome structure. (C) Autoradiographic image of a Southern blot of BstZ17I-digested viral DNAs from cells infected with HSV-1(F) (lane 1) or the gD deletion virus vRR1097 (lane 2). The sizes of migration standards (in kilobase pairs) are indicated to the left of the panel. (D) Photographic image of a Western blot of SDS-polyacrylamide gel electrophoresis-separated proteins from cells either mock infected (lane 2), or infected with HSV-1(F) (lane 3) or vRR1097 (lane 1). The position of the gD signal is indicated by the arrow. Restriction enzyme abbreviations: Bs, BstZ17I; E, EcoNI; Nc, NcoI; Ns, NsiI; T, Tth111I.
gD complementation assay.
To transfer the gD open reading frames from representative SP mutants, HSV-1(F), and HSV-1(U10) to vRR1097, primers were designed to amplify gD and flanking sequences that contain no part of the protein-coding sequence of neighboring genes, gJ and gI. PCR products of 1.8 kb containing the gD gene were amplified from genome-length viral DNA of wild-type and mutant viruses using the Advantage-HF 2 PCR kit (Clontech) and primers gDflankL (5′-GTGATGTCGGGTCCAAACTC-3′) plus gDflankR (5′-GGGACGGTTCGCAAAAA-3′). The high-fidelity PCR kit is reported to reduce PCR error to one mutation per 40 kb. Each PCR product was gel purified and cotransfected into Vero cells with vRR1097 viral DNA. Subconfluent 10-cm2 cultures of Vero cells were transfected with 200 ng of viral DNA either alone or combined with 300 ng of gel-purified PCR product. Cells were incubated in growth medium for 3 to 5 days to allow the growth of viruses expressing gD. Since Vero cells are not permissive for the production of infectious vRR1097, virus stocks obtained 3 to 5 days posttransfection were highly enriched for recombinants in which the gD locus had been repaired with the mutant gD sequence. To reduce error introduced by variation among individual recombinants, these stocks were amplified without plaque purification on Vero cells. Plate stocks of the individually generated recombinant virus pools were made by freeze-thaw lysis and sonication and amplified once by plating on T75 cultures of Vero cells. Stocks from these cultures were used to infect 100-33, 19.3, and Vero cells in infectious-center assays. Recombinant stocks for each mutant were generated in triplicate from three separate PCRs. Recombinants were not exposed to resistant cells prior to the assay.
RESULTS
Selection of mutant viruses.
Using the selection described for isolation of mutants capable of forming plaques on cells that lack a functional gD receptor, despite the high multiplicity of initial infection, fewer than 25% of cultures produced viable virus after 10 passages on the resistant cells. Because the viruses that emerged from these selection procedures had gained the ability to spread in these cultured cells, the nine resulting isolated were named SP 2, SP 3, SP 4, SP 5, SP 8, SP 9, SP 10, SP 12, and SP 20. The fact that all of these viruses are capable of forming plaques on US11cl19.3 cells suggests that (i) they can enter as free virus and spread from cell to cell with some efficiency in the absence of a gD receptor and (ii) they are not substantially inhibited in other aspects of the viral life cycle. Two other viruses, each previously observed to demonstrate enhanced entry into or spread in cells resistant to gD-mediated events in entry, were also characterized in this study. R5001 is a recombinant virus derived from HSV-1(F) that carries a mutation in the gD coding sequence that substitutes asparagine for the serine at position 140 (39). This mutation is responsible for the ability of this virus to form plaques on resistant US11cl19 cells (39). HSV-1(U10) was derived from HSV-1(F) following selection on cells resistant to infection due to overexpression of HSV-1 gD (4). This virus carries a point mutation in gD that substitutes proline for the leucine at position 25.
Sequencing of mutant gDs.
Mutations in gD are sufficient to overcome the entry restriction in US11cl19, BJ, and other resistant cells (2, 4, 6, 39). To ascertain whether and to what extent passage in US11cl19.3 cells provided a similar selection, the region of the viral genome encoding gD from each of the SP mutants was sequenced (Table 1). Observed differences among sequences are shown relative to the parental HSV-1(F) sequence. Mutant SP 2 contains the same substitution found in R5001. Mutants SP 3, SP 4, and SP 20 contain single-point mutations at amino acid 185 which result in substitution of threonine for the alanine. This mutation was shown by Brandimarti et al. to mediate entry into BJ cells, possibly by increasing the affinity for the gD receptor over that for wild-type gD (2). Mutant SP 5 contains a single point mutation at amino acid 22, a site in which no mutation has previously been reported. Surprisingly, viruses carrying mutations at amino acid 25 or 27, which have been reported to exhibit enhanced growth on gD-expressing cells, were not selected on US11cl19.3 cells. Mutants SP 8, SP 9, SP 10, and SP 12 contain no mutations in gD. These results suggest that passage of HSV-1(F) on US11cl19.3 cells selects both for viruses which contain gD mutations and for viruses which are wild type at the gD locus. Only viruses carrying mutations in gD are considered in this study. The properties of the mutants with wild-type gD will be reported elsewhere.
TABLE 1.
gD substitutions
| Virus gD substitution | |
|---|---|
| HSV-1(F) | None |
| U10 | L25P |
| R5001 | S140N |
| SP 2 | S140N |
| SP 3 | A185T |
| SP 4 | A185T |
| SP 5 | L22F |
| SP 8 | None |
| SP 9 | None |
| SP 10 | None |
| SP 12 | None |
| SP 20 | A185T |
Growth properties of mutant viruses.
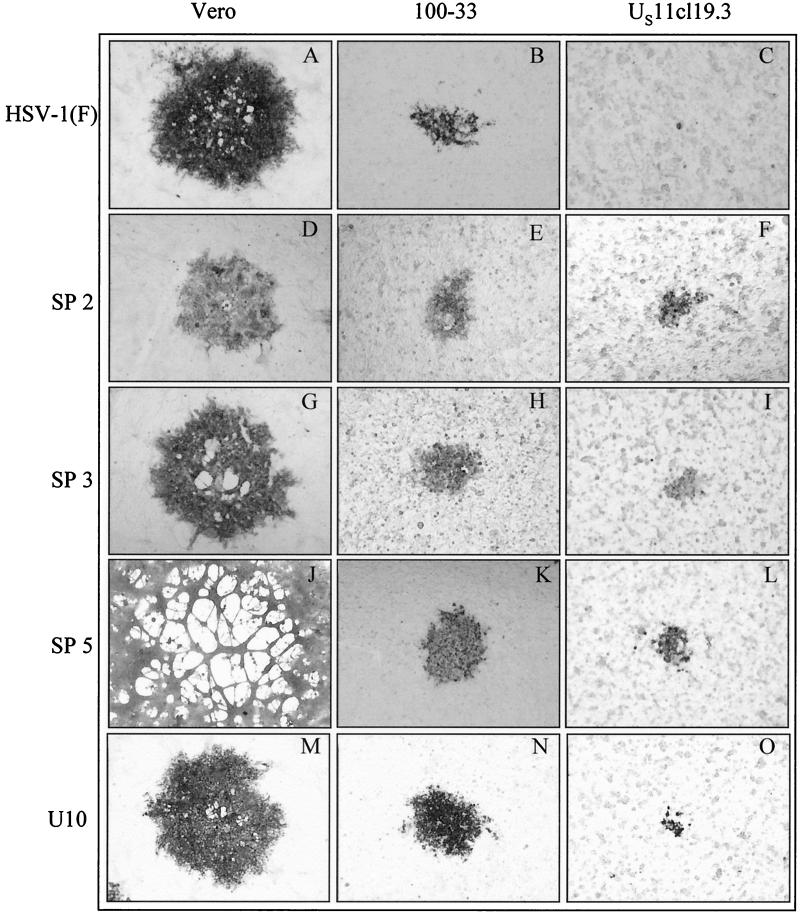
Stocks of all plaque-purified SP mutants and of U10 and R5001 were amplified on Vero cells and plated in serial 10-fold dilution on resistant US11cl19.3 cells and on two susceptible cell lines, Vero and 100-33 (Fig. 1). 100-33 is a clonal cell line that is derived from the same BHK(TK−) cell line as US11cl19.3, is susceptible to wild-type HSV-1 and HSV-2, and is used as a control for BHK(TK−)-specific properties in HSV resistance (39). Three representative SP mutants, as well as U10 and the parental HSV-1(F) are shown. As previously reported (41), HSV-1(F) failed to form plaques on the resistant US11cl19.3 cells (Fig. 1C) and formed wild-type, nonsyncytial plaques on both Vero and 100-33 cells. At least two distinct patterns of cell-cell spread could be distinguished among the mutant viruses. (i) Viruses SP 2 (Fig. 1D to F), U10 (Fig. 1M to O), and SP 4, SP 20, and R5001 (data not shown) formed nonsyncytial plaques on all three cell types. (ii) Viruses SP 3 (Fig. 1G to I) and SP 5 (Fig. 1J to L) demonstrated a cell-type-specific morphology, forming syncytial plaques on Vero cells but nonsyncytial plaques on both 100-33 and US11cl19.3 cells. Within this group, plaque morphology differed between individual mutants on Vero cells. SP 3 (Fig. 1G) showed plaques with a size similar to HSV-1(F) containing relatively small syncytia, whereas SP 5 gave rise to larger-than-wild-type plaques generally made up of a single large syncytium. All mutants showed cell type specificity in the sizes of plaques formed such that the smallest plaques were formed on US11cl19.3 cells and the largest were formed on Vero cells.
FIG. 1.
HSV-1 mutant plaque phenotypes on susceptible and resistant cells. Vero, 100-33, and US11cl19.3 cells were infected for 48 h with the indicated viruses, fixed with cold methanol, and immunostained for the presence of gD. Digital images of representative plaques were obtained under ×40 magnification with a Leitz inverted microscope.
Characterization of entry and cell-cell spread by mutant viruses.
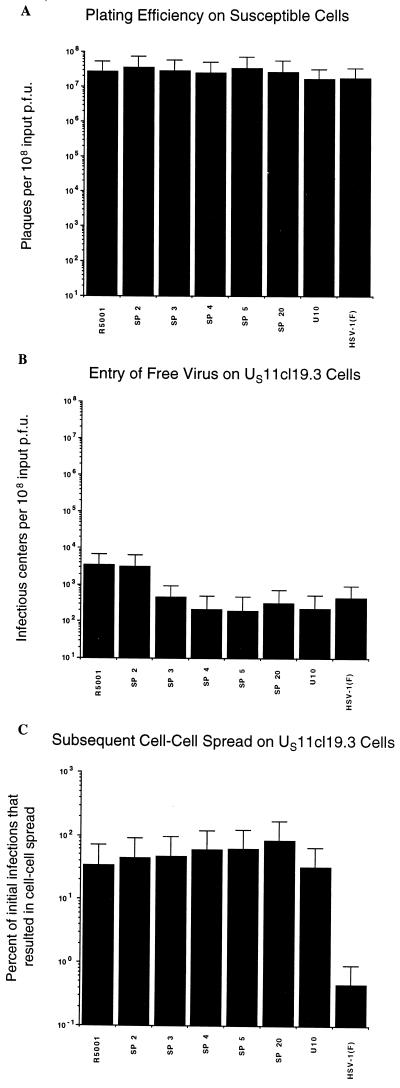
The ability of the mutant viruses to form plaques on the resistant cells suggested that all of these viruses had overcome the US11cl19.3 block to cell-cell spread, at least to some degree. Modified infectious-center assays were performed to quantify their ability to spread and to test for enhancement in their ability to enter as free virus. Shown in Fig. 2A is the control plating efficiency on susceptible 100-33 cells for wild-type HSV-1(F) as well as for SP mutants, R5001, and U10. These data were collected in the same experiments as those from the resistant cells to control for the possibility that mutations affect aspects of viral growth other than entry. Because the production of infectious centers, which is used as a measure of entry of free virus, requires the mutants to complete the viral life cycle, it is critical to demonstrate that no block to viral replication exists other than the block to entry into the resistant cells. As expected from their ability to form plaques on the resistant cells, the behavior of HSV-1 mutants on susceptible cells was not significantly different from that of the wild type, indicating that the mutations neither enhance nor impair any aspect of the viral growth on cells that express functional gD receptors.
FIG. 2.
Efficiencies of initial infection and cell-cell spread by mutant viruses on susceptible and resistant cells. (A) Efficiency of plaque formation on susceptible 100-33 cells. (B) Efficiency of initial entry of free virus on resistant US11cl19.3 cells. (C) Efficiency of subsequent cell-cell spread on resistant US11cl19.3 cells plotted as a percentage of initial infections. Between 20 and 200 plaques were counted for each virus. All assays were conducted in triplicate. Error bars represent the range of values obtained.
As previously reported, the entry of free HSV-1(F) is dramatically hindered in US11cl19.3 cells (Fig. 2B) (39, 41). A 1,000- to 10,000-fold inhibition to entry of free virus is characteristic of wild-type HSV-1(F) on these cells compared to the same process on susceptible 100-33 cells. The ability of two of the mutants, SP2 and R5001, to overcome this block to entry of free virus is slightly enhanced, while the ability of the remaining mutants to do so does not differ significantly from that of wild-type virus. Thus, mutations which result in increased plaque size on cells that lack a functional gD receptor do not correspond to increased ability to enter as free virus.
Previous results have shown that in addition to inhibiting HSV entry, US11cl19.3 cells also block cell-cell spread (41). Shown in Fig. 2C is the percentage of initial infections that led to plaque formation on the resistant cells. Consistent with previous reports of a substantial block to cell-cell spread in US11cl19.3 cells (41), fewer than 1% of the wild-type virions that initially bypassed the block to entry were capable of going on to form plaques by cell-cell spread. In contrast to these results and the results obtained for entry of free virus in Fig. 2B, all of the mutants showed a marked enhancement of cell-cell spread, with the percentage of virus capable of subsequent cell-cell spread approaching 100%. The mutations in these viruses thus improve the efficiency of cell-cell spread on resistant cells to a level typical of susceptible cells but confer little or no ability to overcome the block to entry of free virus exhibited in the same cells.
Characterization of a gD-null deletion recombinant.
To determine if the various mutations in gD are sufficient to confer the mutant phenotype on a wild-type background, recombinant viruses were constructed in which individual point mutations were introduced into HSV-1(F) gD. To simplify and standardize the production of these recombinants, a gD-null virus named vRR1097 was constructed in which the gD gene of HSV-1(F) was replaced with green fluorescent protein (GFP) (Fig. 3A and B). vRR1097 plaques fluoresce under UV illumination, express gC, and are not syncytial (data not shown). Consistent with a loss of essential gD function, vRR1097 is unable to form plaques on Vero, 100-33, or HEp-2 cells but can form plaques on complementing VD-60 cells and, as shown in Fig. 5, is repaired by gD. The structure of the gD locus in the deletion virus was verified by Southern blotting. The gD gene in HSV-1(F) spans two BstZ17I fragments of 2.9 and 1.2 kb (Fig. 3A). In the deletion virus, these two fragments are predicted to be replaced by a single fragment of 4 kb due to the loss of the restriction site that divides the two fragments (Fig. 3B). As predicted, bands of 2.9 and 1.2 kb were seen in DNA from HSV-1(F) but were replaced by a single band of 4 kb in the deletion virus. To test for gD expression from the viral genome, 100-33 cells were either mock infected or infected for 18 h with HSV-1(F) or the deletion virus vRR1097. A strong signal at the position expected for gD was observed in protein from cells infected with wild-type virus (lane 3) but was not detectable in protein from cells infected with the gD deletion recombinant (lane 1). VRR1097 was the parent virus used in the production of all of the gD recombinants reported.
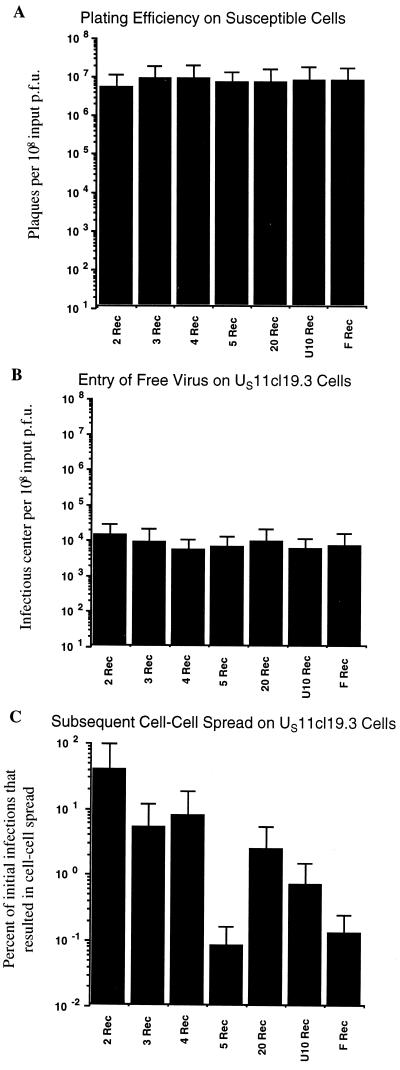
FIG. 5.
Efficiencies of initial infection and cell-cell spread by recombinant viruses on susceptible and resistant cells. (A) Efficiency of plaque formation on susceptible 100-33 cells. (B) Efficiency of initial entry of free virus on resistant US11cl19.3 cells. (C) Efficiency of subsequent cell-cell spread on resistant US11cl19.3 cells plotted as a percentage of initial infections. Between 20 and 200 plaques were counted for each virus. All assays were conducted in triplicate. Error bars represent the range of values obtained.
Tropism and plaque morphology of recombinant viruses carrying gD mutations.
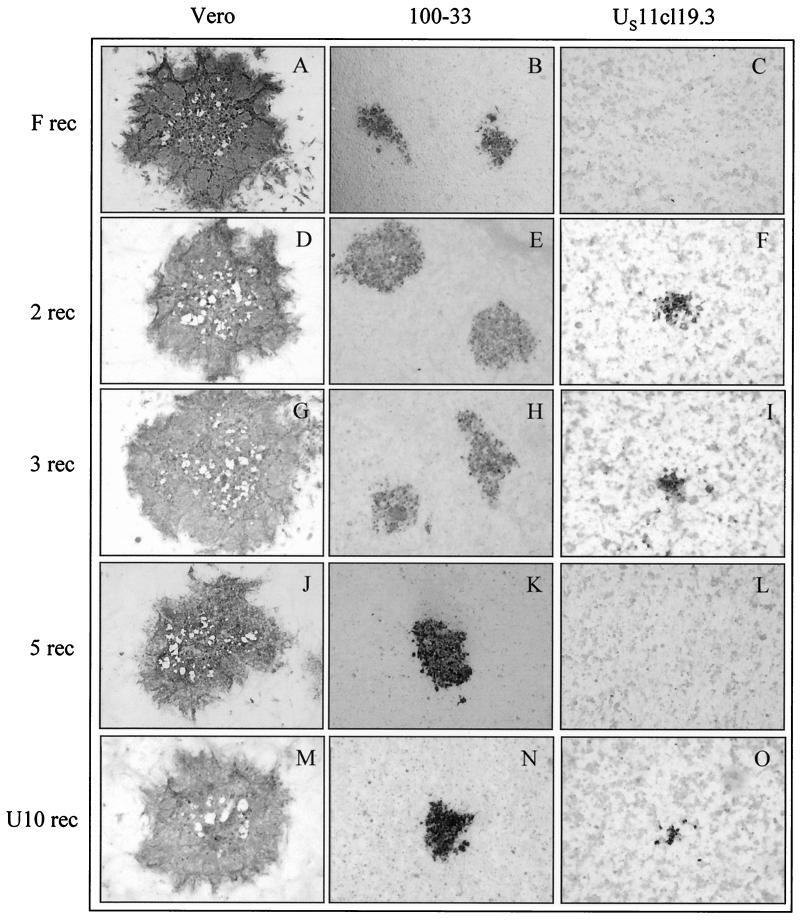
To assess the effect of the individual gD mutations on plaque morphology, gD open reading frames from the indicated mutants were individually introduced into vRR1097. The procedure for production of recombinants (rec mutants), beginning with PCR amplification of the gD coding sequence, was repeated three times to control for the possibility of error in amplification of one of the gD sequences or the introduction of an adventitious mutation in any single recombinant. Thus, three individual recombinants were generated from a common parent for each gD mutation. Each of the recombinant viruses was plated at low multiplicity on Vero, 100-33, and US11cl19.3 cells. Cultures were incubated for 48 h to allow plaque development and were then fixed and immunostained (Fig. 4). The results shown for each gD mutant are ranges of values generated by identical treatment of the three individually derived recombinants. Noteworthy observations include the following. First, in no case did the transfer of mutant gD sequences confer a syncytial phenotype on the resulting recombinants, demonstrating that the syncytial phenotype of SP 3 (compare Fig. 1G and Fig. 4G) and SP 5 (Fig. 1J and 4J) is not due to the observed mutations in the gD gene. Second, although SP 5 was capable of forming plaques on US11cl19.3 (Fig. 1L), 5 rec was not (Fig. 4L), showing that the difference in the gD locus between HSV-1(F) and this virus is not sufficient to confer its cell-cell spread capability on US11cl19.3 cells. However, U10 rec, 2 rec, 3 rec (Fig. 4F, I, and O) and 4 rec and 20 rec (data not shown) could form plaques on US11cl19.3 cells demonstrating that the gD lesions carried by these mutants are sufficient to confer cell-cell spread capability on the resistant cells. These results suggest that SP 5 carries mutations in a gene or genes other than gD that confer cell-cell spread capability on US11cl19.3 cells.
FIG. 4.
gD recombinant plaque phenotypes on susceptible and resistant cells. Vero, 100-33, and US11cl19.3 cells were infected for 48 h with the indicated viruses, fixed with cold methanol, and immunostained for the presence of gD. Digital images of representative plaques were obtained under ×40 magnification with a Leitz inverted microscope.
Efficiency of entry and cell-cell spread by recombinant mutant viruses.
Modified infectious-center assays were done with recombinant mutant viruses just as they were conducted with the original mutants (Fig. 5). All of the recombinant mutants, like the recombinant wild-type repair, spread from cell to cell on susceptible 100-33 cells with equal efficiencies (Fig. 5A), confirming that the gD mutations do not affect cell-cell spread or other aspects of viral replication on susceptible cells. In addition, no recombinant virus demonstrated significant enhancement over the wild-type repair in its ability to enter resistant cells as free virus (Fig. 5B). In contrast, the efficiency of a virus to subsequently spread from cell to cell on resistant US11cl19.3 cells (Fig. 5C) depended strongly on the coding sequence of the gD gene. As expected, the wild-type gD sequence derived from HSV-1(F) was unable to enhance cell-cell spread. The gD sequences from SP 2, SP 3, SP 4, SP 20, and U10 were all sufficient to confer at least a 10-fold enhancement in cell-cell spread on resistant cells. Cell-cell spread of the recombinant carrying the SP2 gD was as efficient as that of the nonrecombinant SP 2 mutant, suggesting that the S140N mutation in gD is sufficient to completely reconstitute the enhanced-spread phenotype of the original mutant. The recombinant SP 3, SP 4, SP 20, and U10 viruses, however, spread less efficiently than the mutants from which the gD sequence was derived, suggesting that each of the original mutants carries other spread-enhancing mutations. Surprisingly, the gD sequence from SP 5, although it carries a substitution in gD, conferred on its recombinant no enhancement of cell-cell spread on the resistant cells.
DISCUSSION
The process of cell-cell spread is poorly understood but is critical in understanding infection and pathogenesis in vivo. The studies that have examined cell-cell spread have generally focused on viral proteins which are not essential for all entry events but tend to play functionally unique roles in cell-cell spread. This research is novel in that we have examined viruses which carry mutations in a glycoprotein that plays an essential role in both entry of free virus and cell-cell spread and demonstrated that these processes are functionally distinguishable with respect to gD.
We previously reported that US11cl19.3 cells manifest a block to HSV at a point after attachment but prior to penetration and that this restriction precludes plaque formation. This cellular restriction is mediated against entry of free HSV virions as well as particles passing from an infected cell to an adjacent uninfected cell via cell-cell spread. Failure to express a gD receptor or interference with the function of that receptor constitutes at least part of this restriction, since the introduction and stable expression of a known gD receptor, HveA, restores the ability of HSV-1(F) to form plaques. In some resistant cells that stably express HveA, cell-cell spread was enhanced to a much greater degree than was entry of free virus, suggesting that gD-receptor interactions in entry of free virus and cell-cell spread may be functionally distinguishable. Here we support and extend these studies to show that HSV-1 mutants capable of forming plaques on US11cl19.3 cells are competent in cell-cell spread while remaining as impaired as wild-type HSV-1 in entry of free virus.
Entry of free virus and cell-cell spread may differ in such a way that gD-receptor interactions that suffice to mediate the fusion event required for cell-cell spread are not sufficient to mediate the entry of free virus. At least two distinct explanations for this difference can be proposed. (i) Low-affinity gD-receptor interactions may be sufficient to promote fusion when the local concentration of gD and receptor are very high, as in regions of apposition between the membranes of an infected cell and an uninfected neighbor. (ii) Promotion of fusion in these mutant viruses may require additional factors that are present on membranes at the site of apposition between adjacent cells but are not present in the envelope of free virus particles. These secondary factors could be cellular molecules or nonstructural viral proteins.
Four of the mutant viruses isolated in this study (SP 2, SP 3, SP 4, and SP 20) and two derived from other studies (R5001 and U10) carry mutations in the coding sequence of gD that are sufficient to confer the enhanced cell-cell spread phenotype on recombinant viruses. The L25P substitution found in U10 and the A185T substitution found in SP3, SP 4, and SP 20 have previously been shown to enhance the infection of cells resistant to infection due to cellular expression of HSV-1 gD (2). Although the kinds of gD mutations that confer enhanced infectivity on gD-expressing cells clearly overlap with those that enhance cell-cell spread on US11cl19.3 cells, enough mutants have now been isolated to support the suggestion that the two restrictions are distinct. The mutation in U10 gD is in a region of the protein known to contain determinants of receptor-binding specificity, suggesting that this gD mutation enhances cell-cell spread on the resistant cells by altering the receptor binding affinity or specificity, allowing the use of an alternative receptor. In fact, the L25P gD mutation, but not the S140N and A185T mutations, has recently been reported to confer the ability upon HSV-1 to utilize HveB as a cellular receptor (23). Interestingly, US11cl19.3 cells are resistant to entry of free virus and cell-cell spread of HSV-2, which has also been reported to utilize HveB as a gD receptor.
The S140N substitution seen in HSV-1(R5001) and SP 2, which provides the most efficient restoration of cell-cell spread on US11cl19.3 cells, has never been isolated from similar selections on gD-expressing cell lines and the S140N mutants were the least efficient among the mutants tested in overcoming the gD-mediated restriction of BJ cells (2). Amino acid 140 is located within functional region 2 of gD, which, like functional regions 1, 3 and 4, is essential for the process of entry of free virus (5). Within the context of properly folded gD, however, some sequences within functional regions 2 and 3 must be adjacent, since they form a discontinuous epitope that is recognized by neutralizing monoclonal antibodies (26, 53). Several lines of evidence suggest that functional region 3 plays a role in gD functions separate from receptor binding. Specific mutations in functional region 3 abolish the ability of gD to mediate the entry of free virus without affecting receptor affinity (28, 55). Similarly, a truncated, soluble gD lacking region 3 sequences binds well to gD receptors but cannot efficiently block infection (53), and group 1a and II monoclonal antibodies, which bind to region 3 sequences, do not block gD-HveA interaction but still neutralize HveA-mediated entry (27). The mutation present in gD of HSV-1(R5001) and SP 2 provides a link between two essential domains of gD, at least one of which appears to function in a process other than receptor binding. This mutation may therefore overcome US11cl19.3 resistance in a manner unrelated to receptor binding.
Two of the mutants selected in this study (SP 3 and SP 5) have, in addition to mutations in gD, mutations that cause cell-type-dependent syncytium formation. These viruses are capable of forming syncytia on Vero cells but not on susceptible or resistant BHK-derived cells. In neither of these mutants is the mutation in gD sufficient to completely reconstitute the enhancement of cell-cell spread on resistant cells. Indeed, the gD substitution in SP 5 was completely ineffective by itself in promoting cell-cell spread. It seems likely that the mutations conferring the syncytial phenotype on Vero cells also contribute to the cell-cell spread phenotype of these viruses and, in the case of SP 5, may be the primary mediator of spread on the resistant cells. The ability of these mutants to spread on the resistant cells without causing concomitant syncytium formation suggests that a mechanism associated with but separate from syncytium formation can mediate cell-cell spread in the US11cl19.3 cells. A similar observation was reported by Pertel and Spear, who also selected HSV-1 variants that carried lesions in syn loci but were nonsyncytial on the cells in which they were selected (32). Two nonexclusive mechanisms can be envisioned by which these mutations outside of gD might enhance cell-cell spread on cells that are resistant because they lack a functional gD receptor: (i) alterations in proteins that functionally interact with gD might change the receptor binding specificity of gD such that an alternative receptor for cell-cell spread can be used, and (ii) changes in proteins that functionally interact with the fusion apparatus might aberrantly regulate fusion such that weak or absent gD-receptor interactions suffice for cell-cell spread.
The varied properties of the mutants described in this report suggest that HSV-1 can adapt to the absence of a wild-type gD receptor in multiple ways. As expected, mutations in gD that would be expected to alter receptor binding specificity can contribute to enhanced viral spread. Also as expected, mutations outside of gD that alter the regulation of fusion can operate alone or in combination with a mutated gD to enhance spread. Moreover, the restriction of these adaptations to effects on cell-cell spread suggest that this process is regulated differently from entry of free virus. Specifically, both gD and non-gD fusion regulators evidently participate in cell-cell spread in a manner distinct from their participation in entry of free virus.
ACKNOWLEDGMENTS
This work was funded by Research Project grant RPG-97-070-01-VM from the American Cancer Society. D.A.R. was supported by grant AI07533 from NIAID and NCI for training in molecular virology and viral pathogenesis.
We thank David Johnson for the gift of VD-60 cells, Gabriela Campadelli-Fiume for providing HSV-1(U10), and Bernard Roizman for providing viruses and plasmids. We are grateful to other members of our laboratory for invaluable discussions and for critical reading of the manuscript.
REFERENCES
- 1.Arsenakis M, Hubenthal-Voss J, Campadelli-Fiume G, Pereira L, Roizman B. Construction and properties of a cell line constitutively expressing the herpes simplex virus glycoprotein B dependent upon functional α4 protein synthesis. J Virol. 1986;60:674–682. doi: 10.1128/jvi.60.2.674-682.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandimarti R, Huang T, Roizman B, Campadelli-Fiume G. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc Natl Acad Sci USA. 1994;91:5406–5410. doi: 10.1073/pnas.91.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entery and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campadelli-Fiume G, Qi S, Avitabile E, Foa-Tomasi L, Brandimarti R, Roizman B. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J Virol. 1990;64:6070–6079. doi: 10.1128/jvi.64.12.6070-6079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang H-Y, Cohen G, Eisenberg R. Identification of functional regions of herpes simplex virus glycoprotein D by using linker insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean H J, Terhune S S, Shieh M T, Susmarski N, Spear P G. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 7.Dean H J, Warner M S, Terhune S S, Johnson R M, Spear P G. Viral determinants of the variable sensitivity of herpes simplex virus strains to gD-mediated interference. J Virol. 1995;69:5171–5176. doi: 10.1128/jvi.69.8.5171-5176.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwell K S, Doering L C, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejercito P M, Kieff E D, Roizman B. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 11.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller A O, Spear P G. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc Natl Acad Sci USA. 1987;84:5454–5458. doi: 10.1073/pnas.84.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of Alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 14.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold B C, Visalli R J, Susmarski N, Brandt C R, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 16.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principle role in adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Highlander S, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoggan M D, Roizman B. The isolation and properties of a variant of herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- 19.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laquerre S, Argnani R, Anderson D B, Zucchini S, Manservigi R, Glorioso J C. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlefield J W, Basilico C. Infection of thymidine kinase-deficient BHK cells with polyoma virus. Nature. 1966;211:250–252. doi: 10.1038/211250a0. [DOI] [PubMed] [Google Scholar]
- 23.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Capmadelli-Fiume G. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manservigi R, Spear P G, Buchan A. Cell fusion induced by herpes simplex is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci USA. 1977;74:3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 26.Muggeridge M I, Wu T-T, Johnson D C, Glorioso J C, Eisenberg R J, Cohen G H. Antigenic and functional analysis of a neutralization site of HSV-1 glycoprotein D. Virology. 1990;174:375–387. doi: 10.1016/0042-6822(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 27.Nicola A V, Ponce De Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Mongomery R I, Spear P G, Eisenberg R J, Cohen G C. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–5601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus gD. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novotny M J, Parish M L, Spear P G. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology. 1996;221:1–13. doi: 10.1006/viro.1996.0347. [DOI] [PubMed] [Google Scholar]
- 30.Peeters B, Bouma A, de Bruin T, Moormann R, Gielkens A, Kimman T. Nontransmissible pseudorabies virus gp50 mutants: a new generation of safe life vaccines. Vaccine. 1994;12:375–380. doi: 10.1016/0264-410x(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 31.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertel P E, Spear P G. Modified entry and syncytium formation by herpes simplex virus type 1 mutants selected for resistance to heparin inhibition. Virology. 1996;226:22–33. doi: 10.1006/viro.1996.0624. [DOI] [PubMed] [Google Scholar]
- 33.Pertel P E, Spear P G. Partial resistance to gD-mediated interference conferred by mutations affecting herpes simplex virus type 1 gC and gK. J Virol. 1997;71:8024–8028. doi: 10.1128/jvi.71.10.8024-8028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pogue-Geile K L, Lee G T-Y, Shapira S K, Spear P G. Fine mapping of mutations in the fusion-inducing MP Strain of herpes simplex type 1. Virology. 1984;136:100–109. doi: 10.1016/0042-6822(84)90251-4. [DOI] [PubMed] [Google Scholar]
- 35.Pogue-Geile K L, Spear P G. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 36.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: a gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 37.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roller R, Roizman B. Herpes simplex virus 1 RNA binding protein US11 negatively regulates the accumulation of a truncated viral RNA. J Virol. 1991;65:5873–5879. doi: 10.1128/jvi.65.11.5873-5879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roller R, Roizman B. A herpes simplex virus-1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J Virol. 1994;68:2830–2839. doi: 10.1128/jvi.68.5.2830-2839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roller R J, Herold B C. Characterization of a BHK(TK−) cell clone resistant to postattachment entry by herpes simplex virus types 1 and 2. J Virol. 1997;71:5805–5813. doi: 10.1128/jvi.71.8.5805-5813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller R J, Rauch D. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK−) cell clones. J Virol. 1998;72:1411–1417. doi: 10.1128/jvi.72.2.1411-1417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roller R J, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Schroder C, Keil G M. Bovine herpesvirus 1 requires glycoprotein H for infectivity and direct spreading and glycoproteins gHW450 and gB for glycoprotein D-independent cell-to-cell spread. J Gen Virol. 1999;80:57–61. doi: 10.1099/0022-1317-80-1-57. [DOI] [PubMed] [Google Scholar]
- 46.Schroder C, Linde G, Fehler F, Keil G M. From essential to beneficial: glycoprotein D loses importance for replication of bovine herpesvirus 1 in cell culture. J Virol. 1997;71:25–33. doi: 10.1128/jvi.71.1.25-33.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shieh M-T, WuDunn D, Montgomery R I, Esko J, Spear P G P G. Cell surface receptors for herpes simplex virus are heparin sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian G, LeBlanc R A, Wardley R C, Fuller A O. Defective entry of herpes simplex virus types 1 and 2 into porcine cells and lack of infection in infant pigs indicate species tropism. J Gen Virol. 1995;76:2375–2379. doi: 10.1099/0022-1317-76-9-2375. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian G, McClain D S, Perez A, Fuller A O. Swine testis cells contain functional heparan sulfate but are defective in entry of herpes simplex virus. J Virol. 1994;68:5667–5676. doi: 10.1128/jvi.68.9.5667-5676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R, Cohen G H, Eisenberg R J, Spear P G. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner S W, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection of herpes simplex virus type 1, herpes simplex virus type 2, and psuedorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 53.Whitbeck C J, Muggeridge M I, Rux A H, Hou W, Krummenacher C, Lou H, Van Geelen A, Eisenberg R J, Cohen G H. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J Virol. 1999;73:9879–9890. doi: 10.1128/jvi.73.12.9879-9890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce De Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.WuDunn D W, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparin sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]