Abstract
Infected-cell polypeptide 4 (ICP4) of herpes simplex virus type 1 (HSV-1) activates the expression of many HSV genes during infection. It functions along with the cellular general transcription factors to increase the transcription rates of genes. In this study, an HSV late promoter consisting of only a TATA box and an INR element was immobilized on a magnetic resin and incubated with nuclear extracts or purified TFIID in the presence and absence of ICP4. Analysis of the complexes formed on these promoters revealed that ICP4 increased the formation of transcription preinitiation complexes (PICs) in a TATA box-dependent manner, as determined by the presence of ICP4, TFIID, TFIIB, and polymerase II on the promoter. With both nuclear extract and purified TFIID, it was determined that ICP4 helped TFIID bind to the promoter and the TATA box. These observations differed from those for the activator Gal4-VP16. As previously observed by others, Gal4-VP16 also increased the formation of PICs without helping TFIID bind to the promoter, suggesting that ICP4 and VP16 differ in their mechanism of activation and that ICP4 functions to facilitate PIC formation at an earlier step in the formation of PICs. We also observed that the DNA binding activity of ICP4 was not sufficient to help TFIID bind to the promoter and that the region of ICP4 that was responsible for this activity is located between residues 30 and 274. Taken together these results demonstrate that a specific region of ICP4 helps TFIID bind to the TATA box and that this in turn facilitates the formation of transcription PICs.
Infected-cell polypeptide 4 (ICP4) of herpes simplex virus type 1 (HSV-1) is one of five immediate-early proteins synthesized during productive infection (27). ICP4 is required for the efficient transcription of viral early and late genes (55) and is therefore required for viral growth (12, 17, 42). ICP4 functions to increase the rates of transcription of viral genes in the context of viral infection (23) and activates gene expression in transient assays (13, 20, 22, 39) and transcription in reconstituted in vitro transcription reactions (24). In vitro, it has been shown that ICP4 increases the rate of formation of transcription initiation complexes (24) and that it can activate transcription with a relatively simple set of general transcription factors (GTFs) (6, 7). However, it is not known how ICP4 affects the formation of transcription initiation complexes.
Several studies have shown that transcriptional activators can physically promote the formation of transcription preinitiation complexes (PICs) in vitro by facilitating the recruitment of one or more GTFs to target promoters (2, 11, 34). The transcription of protein-encoding genes into pre-mRNAs necessitates the formation of a transcription PIC on promoters before elongation by RNA polymerase II (Pol II) can proceed. Therefore, the formation of PICs is a major control point for gene expression. PICs can be formed on promoter templates in vitro from the individual assembly of the GTFs TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and RNA Pol II (3) or from the assembly of preformed complexes containing these factors (30, 33). In most cases, the recognition of promoters is mediated by TFIID through the binding of the TATA binding protein (TBP) subunit to TATA box elements and/or recognition of non-TATA box cis elements by TBP-associated factors (TAFs) (4, 18, 19, 29, 35, 36, 38, 43, 49, 52–54, 58). Once a competent TFIID-promoter complex is formed, then the remainder of PIC components can be assembled.
In an attempt to elucidate the mechanism(s) by which transcription factors help in the formation of PICs, a number of studies have reported that activators can physically interact with GTFs or Pol II-associated factors. In several cases it has been shown that a single activator can interact with more than one GTF or with more than one subunit of a single GTF (21, 26, 44). The ability of activators to physically interact with GTFs has been suggested to be part of the mechanistic basis that allows them to recruit or stabilize GTFs on promoters or on PIC subcomplexes (10, 11, 34, 51).
One well-studied activator is the HSV protein VP16, which functions in the viral life cycle to activate viral immediate-early genes (5). The acidic activation domain of VP16 has been postulated to enhance PIC formation by a number of mechanisms. One mechanism involves interaction with TFIIB (11, 34). In these studies, VP16 did not facilitate TFIID binding to the TATA box but rather enhanced TFIIB binding to the complex, which in turn promoted greater recruitment of Pol II. Another study reported that VP16 interacts with TFIIA and that this interaction facilitated PIC formation (31, 32). It has been proposed that ICP4 facilitates PIC formation (24), but, unlike many activators studied thus far, ICP4 does not require TFIIA or cofactors found in the USA fraction (6, 24) to activate transcription. Therefore, it is likely that ICP4 and VP16 affect the formation of PICs in different ways. In this study, we have used promoters immobilized on a magnetic resin to directly show that ICP4 can physically promote PIC formation and that it does so by enhancing the binding of TFIID to the TATA box.
MATERIALS AND METHODS
Transcription factors and proteins.
Human GTFs were extracted from HeLa cell nuclei as described previously (16). Wild-type (wt) and mutant ICP4 molecules were purified from nuclei of Vero cells infected with wt HSV-1 or HSV-1 mutated in the ICP4 locus as previously described (28, 45). The viral and cellular sources of the mutant ICP4 molecules n208, d8-10, and X25 have been previously described (15, 48, 57). Hemagglutinin (HA) epitope-tagged human TFIID (HA-TFIID) was purified from an Hela cell line expressing HA-tagged TBP as described previously (58). Recombinant Gal4-VP16 expressed in Escherichia coli was purified as described previously (9).
Construction of promoter plasmids.
A PCR fragment containing the HSV-1 glycoprotein C (gC) late gene promoter from nucleotide −35 to +70 relative to the mRNA start site that contained a BglII restriction site at its 3′ end was subcloned into the EcoR1 site of pUC18. This subcloned promoter fragment is referred to as gC-wt. The TATA box in the gC-wt fragment was deleted by digestion with restriction enzymes BamH1 and BspE1, which surround the TATA box, followed by filling in with T4 polymerase and religation. This mutation (gC-ΔTA) results in a 12-bp deletion from −35 to −26 of the gC promoter. The INR element in gC-wt was mutated by PCR mutagenesis using sense and antisense oligonucleotides each of which contained a SacI restriction site in place of the −2 to +4 region. The resulting mutant is referred to as gC-ΔINR. The adenovirus E4 promoter fragment containing five DNA binding sites for the GAL4 transcription factor subcloned in pGEM was described previously (34). This promoter fragment is referred to as G5E4T-wt.
Preparation of immobilized promoters.
Plasmids containing the HSV-1 gC promoter fragments or the adenovirus E4T promoter fragment were cut with HindIII, and the ends were filled in with Klenow polymerase (New England Biolabs) in the presence of either deoxyadenosine or deoxycytosine-14-biotin derivatives (Gibco-BRL). After biotinylation of the 5′ end, the gC promoter templates were cut at their 3′ ends with BglII, whereas the G5E4T template was cut with PvuII. The promoter fragments were resolved from the vector fragments by polyacrylamide gel electrophoresis (PAGE). The approximately 200-bp gC and 500-bp G5E4T promoter fragments (for a schematic representation, see Fig. 1) were excised from the gels, electroeluted, ethanol precipitated, and resolubilized in water. The purified biotinylated promoters were than immobilized on a magnetic resin conjugated with streptavidin according to the manufacturer instructions (Dynal) at 100 fmol of fragment per 25 μg of beads. The immobilized promoter templates were kept in Tris-EDTA buffer, pH 7.4, at 4°C for 2 to 3 months.
FIG. 1.
Graphical representation of promoter fragments used in this study. Shown is the HSV-1 gC promoter from position −35 to +70 bp relative to the transcription start site (gC wt). Black box, gC TATA box (TATAAATT); hatched box, gC initiator element (CCCTCACTACC). Two PstI sites (p) used to elute protein complexes bound to gC promoter fragments are shown. TATA box-deleted (gC Δ TA) and INR-mutated (gC Δ INR) versions of gC-wt are also shown, as is the adenovirus E4 promoter [(G)5E4T] with five 5′ DNA binding sites for the yeast GAL4 transcription factor (gray boxes) and with its TATA box (black box; TATATATA) as described previously (34). Black circles, added biotin moieties.
Assembly of transcription PICs on immobilized promoters.
HeLa cell nuclear extract (150 μg of total protein as estimated by the Bradford assay) was added to 300 fmol of immobilized promoters, in the presence or absence of purified ICP4 (approximately 900 fmol or 300 ng, as estimated by Coomassie blue staining) in 75 to 300 μl of binding buffer (20 mM HEPES [pH 7.9], 10% glycerol, 60 mM KCl, 6 mM MgCl2, 1.0 mM dithiothreitol [DTT], 0.1 mM EDTA, 0.01% NP-40). PIC assembly in the absence or presence of Gal4-VP16 was always done in 300-μl volumes with approximately 7,200 fmol of Gal4-VP16 (180 ng as estimated by Coomassie blue staining). The samples were then incubated for 90 min at 30°C on a rotating sample mixer (Dynal). After incubation, the immobilized templates were concentrated with a magnetic concentrator (Dynal), resuspended in 0.5 ml of binding buffer containing 0.04% Sarkosyl, reconcentrated, resuspended in 0.5 ml of binding buffer supplemented to 100 mM KCl and 10 mM MgCl2, and finally reconcentrated. Proteins bound to gC templates were eluted by suspending the beads in 15 μl of New England Biolabs restriction buffer 3 containing 100 U of PstI and 0.01% NP-40 and incubating them at 37°C for 10 min. The supernatant, separated from the beads by magnetic concentration, was mixed with sodium dodecyl sulfate (SDS)-PAGE sample buffer. Complexes bound to G5E4T were eluted directly in SDS-PAGE sample buffer. The proteins in the samples were resolved on 4 to 15% polyacrylamide gradient gels (Bio-Rad) and then transferred to polyvinylidene difluoride (PVDF) membranes (Hybond-P; Amersham Life Science) for Western blot analysis.
Binding of immunopurified HA-TFIID to immobilized promoters.
Between 2 and 8 μl of immunopurified HA-TFIID (4 μl corresponds approximately to 10 fmol of HA-TFIID or approximately to 0.5 ng of HA-TBP as estimated by silver staining) was added to 100 fmol of immobilized promoter templates in binding buffer (20 mM HEPES [pH 7.9], 10% glycerol, 60 mM KCl, 6 mM MgCl2, 1.0 mM DTT, 0.1 mM EDTA, 0.01% NP-40) containing 0.3 μg of bovine serum albumin/μl in the presence or absence of 75 to 300 fmol (approximately 25 to 100 ng) of purified wild-type or mutant ICP4 proteins or with 300 to 2,400 fmol of purified Gal4-VP16 protein (approximately 7.5 to 60 ng, as estimated by Coomassie staining) in 300 μl of binding buffer (20 mM HEPES [pH 7.9], 10% glycerol, 60 mM KCl, 6 mM MgCl2, 1.0 mM DTT, 0.1 mM EDTA, 0.01% NP-40). After incubation at 30°C for 90 min, the samples were washed twice with 0.5 ml of binding buffer. Bound proteins were directly eluted in 20 μl of SDS-PAGE sample buffer, resolved on 4 to 15% gradient gels, and transferred to PVDF (Hybond-P) for Western blot analysis.
Western blot analysis.
Proteins in SDS-PAGE gels were electrophoretically transferred to PVDF membranes and visualized using the SuperSignal West Pico chemiluminescent substrate as described by the manufacturer (Pierce, Rockford, Ill.). The following primary antibody preparations were used to detect the proteins examined in this study. RNA Pol II was detected using the monoclonal 8WG16 antibody (Babco, Richmond, Calif.), which recognizes the large subunit of RNA Pol II. TFIIB was detected using a rabbit polyclonal antibody (C18; Santa Cruz Biotechnology, Santa Cruz, Calif.). HA-tagged TFIID was detected using either monoclonal antibody 12CA5 (Babco), which recognizes the HA epitope-tagged TBP subunit, or a monoclonal antibody that recognizes the TBP subunit (Babco). The TBP-associated factors (TAFs) hCIF150/hTAF150 and hTAF250 were detected using a rabbit polyclonal antibody (generously provided by S. T. Smale) and a monoclonal antibody that recognizes hTAF250 (6B3; Santa Cruz Biotechnology), respectively. Gal4-VP16 was detected using a monoclonal antibody that recognizes the Gal4 DNA binding domain (RKSC1; Santa Cruz Biotechnology). ICP4 was detected using two different rabbit polyclonal antibodies, N15 (45) and an antibody generously provided by Richard Courtney (The Pennsylvania State University Medical Center, Hershey, Pa.). ICP4 and Gal4-VP16 proteins were revealed with antirabbit and antimouse secondary antibodies conjugated to horseradish peroxidase (Sigma), followed by chemiluminescence. Following a reaction with the primary antibodies, GTFs were detected by incubation with either antimouse of antirabbit secondary antibodies conjugated to biotin followed by incubation of avidin-conjugated horseradish peroxidase complexes (Vector; Vectastain ABC kit) prior to chemiluminescence.
RESULTS
Effect of the ICP4 protein on the formation of transcription PICs.
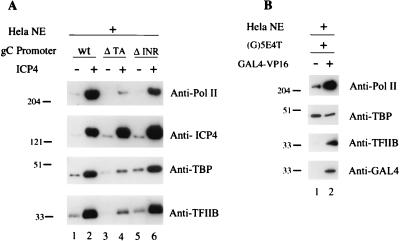
In order to assess the potential role of ICP4 in PIC formation, we examined the interaction of the GTFs with a promoter template immobilized on a magnetic solid support (1, 11, 34) in the presence and absence of ICP4. This procedure allows the rapid isolation and analysis of nucleoprotein complexes formed on immobilized DNA promoter fragments. To assess the role of ICP4 in PIC formation, we used the HSV-1 gC promoter (Fig. 1A), which has been shown to be activated by ICP4 both in vivo and in vitro (24). The only cis-acting elements that affect the level of in vitro transcription of the gC promoter in the presence of ICP4 are a TATA box and an INR element (24) (Fig. 1A). The immobilized gC promoter template was incubated with HeLa nuclear extract in the absence or presence of purified ICP4. Nucleoprotein complexes formed during the incubation were then purified by sequential magnetic separation and washing. Western blot analysis of the proteins bound to the template showed that in the presence of ICP4 a much greater quantity of Pol II was bound to the promoter than when ICP4 was absent from the reaction (Fig. 2A; compare lane 2 with lane 1), suggesting that ICP4 can physically help in the formation of PICs on the gC promoter fragment. The ICP4-mediated increase in Pol II binding was greatly reduced when a gC promoter fragment with a mutated TATA box element was used (Fig. 2A, lane 4). The binding of Pol II was less affected when the gC template containing a mutated INR element was used (Fig. 2A, lane 6).
FIG. 2.
Effects of ICP4 and Gal4-VP16 on the formation of PICs. (A) Effect of ICP4 on PIC formation on immobilized HSV-1 gC promoter templates. Immobilized wt, TATA box-deleted (Δ TA), and INR-mutated (Δ INR) gC promoters (300 fmol) were incubated with HeLa nuclear extract (NE; 150 μg of total protein) in the absence or presence of purified ICP4 (300 ng) in a total volume of 75 μl for 90 min at 30°C. After incubation at 30°C, nucleoprotein complexes bound to immobilized promoters were isolated as described in Materials and Methods. The presence of ICP4, TFIIB, TFIID, and RNA Pol II was assessed by Western blot analysis. (B) Effect of Gal4-VP16 on PIC formation on immobilized G5E4T promoter templates. Approximately 7,200 fmol of purified Gal4-VP16 (180 ng) and 300 fmol of immobilized G5E4T were used as described for panel A but in a 300-μl volume and with direct elution of the promoter-bound proteins in SDS-PAGE sample buffer. The presence of Gal4-VP16, TFIIB, TFIID, and RNA Pol II was assessed by Western blot analysis.
The effects of ICP4 on the binding of TFIID and TFIIB were also monitored in the same reactions. Western blot analysis with anti-TBP and anti-TFIIB antibodies showed that the TATA box-dependent increase of Pol II binding observed in the presence of ICP4 was also observed for TBP and TFIIB (Fig. 2A). Western blots performed in similar experiments also revealed that another component of TFIID, CIF/TAF150, was also recruited by ICP4 (data not shown). The observed TATA box-dependent increase in the binding of Pol II, TFIID, and TFIIB in the presence of ICP4 strongly suggests that ICP4 can promote the formation of PICs on the promoter.
For comparison the effects of the VP16 activation domain on the formation of PICs was determined using nuclear extract, a purified Gal4-VP16 protein, and an immobilized promoter that contains Gal4 binding sites upstream of the adenovirus E4 TATA box. The results depicted in Fig. 2B are consistent with those previously published (34) and show that the VP16 activation domain promotes Pol II and TFIIB binding with little or no effect on the binding of TFIID. This is different from the effects of ICP4 on the binding of these factors in that ICP4 also promoted the binding of TFIID to the gC promoter.
Effect of ICP4 on the formation of TFIID-gC promoter complexes.
The experiments described above suggest that ICP4 facilitates the formation of PICs by directly helping TFIID to bind to the promoter. To examine this possibility more thoroughly, we determined the effects of ICP4 on the binding of purified TFIID using this system. For this purpose, a HA epitope-tagged TFIID (HA-TFIID) was purified to near homogeneity from an HeLa cell line expressing an HA-tagged TBP (58). Figure 3A depicts a silver-stained SDS-PAGE gel of the TFIID used in these experiments showing TBP and a number of the TAFs known to be associated with TFIID.
FIG. 3.
Effect of ICP4 on the binding of purified HA-TFIID to immobilized gC promoters. (A) Silver-stained SDS-PAGE gel of HA-tagged TFIID purified from Hela cells expressing HA-tagged TBP. TAF subunits, according to their estimated molecular weights, and TBP are shown on the left side of the gel. (B) Effect of ICP4 concentration on the binding of HA-TFIID to immobilized gC promoter templates. HA-TFIID (4 μl; (approximately 10 fmol) was incubated with 100 fmol of immobilized gC promoter in the presence of 75 to 1,200 fmol of purified ICP4 protein or in the absence of ICP4 in a total volume of 300 μl. After incubation at 30°C, nucleoprotein complexes bound to immobilized promoter templates were isolated and analyzed as described in Materials and Methods. (C) Effect of the HA-TFIID concentration on its recruitment to immobilized gC promoter templates in the presence of ICP4. HA-TFIID (from 2 to 8 μl) was incubated with 100 fmol of immobilized gC templates in the presence of approximately 300 fmol of ICP4 or in the absence of ICP4 in a total volume of 300 μl. gC-bound ICP4, TAF250, TAF150, and HA-TFIID were isolated and analyzed as described in Materials and Methods.
In order to assess if the level of HA-TFIID recruitment can be influenced by the amount of ICP4 bound to immobilized gC promoters, we examined the binding capacity of HA-TFIID from a fixed concentration in the absence or presence of increasing concentrations of ICP4. HA-TFIID (10 fmol) was incubated with 100 fmol of immobilized gC promoter in the presence of 75 to 1,200 fmol of purified ICP4 protein (approximately 25 to 400 ng of ICP4) or in the absence of ICP4. Western blot analysis of the proteins bound to the immobilized templates was performed to detect the ICP4 and HA-TFIID complexes (Fig. 3B). The results of this experiment show that the amount of recruited HA-TFIID increased as a function of increased amounts of bound ICP4. An overexposure from the filter used for this experiment allows the visualization of bound ICP4 protein at the lowest concentration used (Fig. 3B, lane 2, and data not shown). It is interesting to note that the immobilized promoters became saturated when about three ICP4 molecules were added per template molecule. When additional ICP4 was added, there was no additional increase in the amount of bound TFIID, suggesting that the saturating levels of ICP4 precluded the recruitment of TFIID or that all of the TFIID binding sites were occupied. Interestingly, the same dependence on ICP4 concentration is often seen for activated transcription in vitro, implying a squelching mechanism.
We next examined the binding capacities of various concentrations of HA-TFIID in the absence or presence of a fixed amount of purified ICP4. HA-TFIID (from 2 to 8 μl, or approximately 5 to 20 fmol) was incubated with 100 fmol of immobilized gC promoter in the absence or presence of 300 fmol of purified ICP4 (approximately 100 ng of ICP4). The concentration of ICP4 used allowed most of the gC templates to be occupied (Fig. 3B). The presence of ICP4 and HA-TFIID bound to the gC promoter after elution from the magnetic resin was revealed by Western blot analysis using anti-ICP4, anti-HA (anti-HA-TBP), anti-CIF150/TAF150, and anti-TAF250 antibodies. TAF250 and TAF150 are stable components of TFIID. This experiment shows that gC promoter templates bound with ICP4 (Fig. 3C, lanes 2, 4, and 6) contained many more HA-TFIID complexes than templates not bound by ICP4. In addition, the amount of bound TFIID in the sample containing the lowest concentration of TFIID in the presence of ICP4 was considerably greater than the amount of bound TFIID in the sample containing the highest concentration of TFIID in the absence of ICP4 (compare lanes 5 and 2). Thus, the presence of ICP4 allowed more TFIID to bind to the promoter from a solution of lower TFIID concentration. The same effects were observed for the known components of TFIID that do not directly bind to the TATA box.
The previous experiments demonstrated that ICP4 can recruit TFIID to the promoter in the absence of other proteins. The activation domain of VP16 has been reported to recruit TFIID-TFIIA complexes and TFIIB to the promoter. It has not been reported to recruit TFIID on its own. Therefore, it serves as a good factor to contrast ICP4 to in the same experiment. Accordingly, we compared the abilities of ICP4 and Gal4-VP16 (VP16 amino acids 413 to 490) to recruit TFIID to immobilized G5E4T promoter fragments. This promoter consists of an adenovirus E4 TATA box and five tandem Gal4 binding sites upstream of the TATA box. HA-TFIID (4 μl) was incubated with 100 fmol of immobilized G5E4T template in the absence of ICP4 or in the presence of approximately 25 to 100 ng (75 to 300 fmol) of purified ICP4 protein or with approximately 7.5 to 60 ng of purified Gal4-VP16 protein (300 to 2,400 fmol). Bound ICP4, Gal4-VP16, and TFIID were detected by Western blot analysis using ICP4, Gal4, and TBP antibodies, respectively. The results of this experiment show that ICP4 can efficiently recruit TFIID (Fig. 4, lanes 1 to 4) to the G5E4T promoter template, as well as to the gC promoter template (Fig. 4, lanes 9 to 13), while the transcriptional activator Gal4-VP16 was not effective in recruiting TFIID (Fig. 5, lanes 1 and 5 to 8).
FIG. 4.
Comparison of the effect of ICP4 and Gal4-VP16 on the recruitment of HA-TFIID to immobilized promoters. HA-TFIID (4 μl) was incubated with 100 fmol of immobilized G5E4T or gC promoter templates in the presence of 75 to 300 fmol of purified ICP4 (approximately 25 to 100 ng) on in the absence of ICP4 or with 300 to 2,400 fmol of purified Gal4-VP16 (approximately 7.5 to 60 ng) in a total volume of 300 μl. As a control for the DNA binding specificity of the Gal4-VP16 protein, this protein (at the highest concentration used only) was incubated with HA-TFIID and immobilized gC promoters (lane 13). After incubation at 30°C, nucleoprotein complexes bound to immobilized promoters were isolated and analyzed as described in Materials and Methods.
FIG. 5.
Effect of wt and mutant ICP4 proteins on the recruitment of HA-TFIID complexes to gC promoters. (A) Primary structures of wt and mutant ICP4 proteins. The primary sequence of ICP4 is shown along with a summary of the regions of ICP4 that are similar to those of varicella-zoster virus IE140, with the rectangles indicating similarity (solid rectangles indicate the greatest amount of similarity) (37). Also shown are some of the regions of ICP4 that contribute to various activities, as deduced from genetic and biochemical studies (7, 14, 15, 25, 40, 41, 47, 50, 57). (B) Effect of wt (KOS), n208, and d8-10 ICP4 proteins on HA-TFIID recruitment. HA-TFIID (4 μl) was incubated with 100 fmol of immobilized gC templates in the presence of 75 to 300 fmol (approximately 25 to 75 ng) of purified wt, n208, and d8-10 ICP4 proteins or in the absence of ICP4 in a total volume of 300 μl. As a control for nonspecific binding, HA-TFIID and ICP4 (300 fmol only) were incubated with magnetic resin devoid of immobilized promoter (lanes 5, 9, and 13). After incubation at 30°C, nucleoprotein complexes bound to immobilized promoters were isolated and analyzed as described in Materials and Methods. The presence of ICP4 and HA-TFIID was assessed by Western blot analysis with anti-ICP4 (N15) and anti-HA (anti-HA-TBP) antibodies, respectively. (C) Same as panel B, except that wt and X25 ICP4 proteins were used. The anti-ICP4 antibody used in this experiment (provided by Richard Courtney) allows the detection of the X25 protein.
Effect of wt and mutant ICP4 proteins on TFIID recruitment.
In order to map the domain(s) of the ICP4 protein responsible for the recruitment of TFIID, we compared the abilities of different ICP4 mutants (Fig. 5A) to that of wt ICP4 to recruit HA-TFIID to immobilized gC promoter templates. HA-TFIID (4 μl) was incubated with 100 fmol of immobilized template in the absence of ICP4 or in the presence of 25 to 100 ng (75 to 300 fmol) of purified ICP4 or with the approximate molar equivalent of purified ICP4 mutant proteins. As a control, wt and mutant ICP4 proteins (at the highest concentration used only) were incubated with HA-TFIID and magnetic resin alone. Consistent with the known properties of the ICP4 proteins (15, 47, 48), all of the ICP4 molecules used in this experiment bound to the immobilized template (Fig. 5). In addition, all of the proteins tested, with the exception of X25, were able to recruit TFIID to the template. Therefore, the DNA binding property of ICP4 is not sufficient to recruit TFIID. Consistent with this result are our previous observations that X25 is the only ICP4 protein used in this experiment that completely lacks the ability to activate transcription (7, 15, 46, 47).
Specificity of TFIID-ICP4 promoter interactions.
Both ICP4 and TFIID are site-specific DNA binding proteins that will also bind nonspecifically to DNA. The promoter used in these experiments contains a TFIID binding site, the TATA box, but does not contain a recognized ICP4 binding site. It is presumed that the observed binding of ICP4 in these experiments is a function of nonspecific DNA-protein interactions. Therefore, it is important to determine if ICP4 can promote TFIID on the TATA box from where gC transcription is initiated. Accordingly, we conducted an experiment to challenge bound complexes with an excess of nonspecific competitor DNA. Four microliters of HA-TFIID and 100 fmol of immobilized promoter were incubated in the presence and absence of ICP4 (300 fmol) for the indicated times (Fig. 6). In some of the reactions a 300-fold mass excess (4.5 μg) of double-stranded poly(dG-dC) was added at the indicated times.
FIG. 6.
Effect of the gC TATA box on ICP4-mediated recruitment of HA-TFIID to immobilized gC promoter templates. HA-TFIID (4 μl) was incubated with 100 fmol of immobilized wt or TATA box-mutated (ΔTA) gC promoter in the absence or presence of purified ICP4 (300 fmol) and in the absence or presence of poly(dG-dC) (30 ng/μl) in a total volume of 150 μl. Poly(dG-dC) was added either at time zero (lane 3) or following 30 min of incubation at 30°C (lanes 8 to 11). After incubation at 30°C for 30 min (lanes 1 to 3) or for 90 min (lanes 4 to 11), nucleoprotein complexes bound to immobilized promoters were isolated and analyzed as described in Materials and Methods.
Following a 30-min incubation, ICP4 greatly enhanced the binding of TFIID (Fig. 6, lanes 1 and 2). Both TFIID and ICP4 binding was completely competed when the poly(dG-dC) was added for the duration of the 30-min incubation (lane 3). Following a 90-min incubation, ICP4 also enhanced TFIID binding, and there was only a marginal increase in the amount of TFIID bound relative to that for the 30-min incubation (lanes 4 and 5), suggesting that most of the binding occurred in the first 30 min. However, there was only a marginal decrease in the amount of TFIID binding in the presence and absence of ICP4 when the template with the mutated TATA box was used (lanes 6 and 7), suggesting that much of the TFIID binding and recruitment under these condition was not to the TATA box. When the excess poly(dG-dC) was added after a 30-min incubation and incubation was continued for an additional hour, the amounts of bound ICP4 and TFIID declined; however there was a significant enhancement of TFIID binding in the presence of ICP4 (lanes 8 and 9). This enhancement was not seen under these conditions when the TATA box was mutated (lanes 10 and 11). Taken together, these results demonstrate that ICP4 promotes the formation of TFIID-ICP4 complexes on TATA boxes. Therefore, this experiment suggests that ICP4 can recruit TFIID to DNA templates independently of cis-acting sites and that some of the recruited complexes require the presence of the TATA box; these complexes are resistant to competitive challenge by a 300-fold mass excess of nonspecific DNA.
DISCUSSION
In order to help understand the mechanism(s) by which the HSV-1 ICP4 protein activates transcription, we have assessed if this protein can physically promote the formation of transcriptional PICs on an immobilized HSV-1 promoter. Promoter elements that were immobilized on magnetic resin were used to assemble and isolate PICs formed from the GTFs present in HeLa nuclear extracts in the absence or presence of purified ICP4 proteins. We show in this study that the ICP4 protein can facilitate the formation of transcription PICs on immobilized gC promoters in a manner that is dependent on the presence of the TATA box. This facilitation appears to involve the recruitment of the TFIID complex to the promoter.
The recruitment of TFIID is particularly interesting since it is generally thought that the binding of this factor to the TATA box is part of the initial steps in the formation of PICs. Therefore, it is likely that the recruitment of TFIID may facilitate subsequent steps in complex formation. The recruitment of TFIID is most likely a direct effect since ICP4 was able to recruit purified TFIID to the promoter in the absence of other proteins. This is in contrast to the situation seen for the cytomegalovirus E2 protein, which has been proposed to counteract the effect of the mammalian Dr1 repressor on TBP-promoter interactions (8).
The effect of ICP4 on PIC formation also differs from that of the VP16 activation domain. The studies with the VP16 activation domain have resulted in two proposed mechanisms. One mechanism appears to involve the recruitment of TFIIB to preformed TFIID-promoter complexes followed by a post-TFIIB recruitment step that is as yet uncharacterized (11, 34, 56). Another mechanism involves the recruitment of TFIIA-TFIID complexes (31). Neither of these mechanisms involves the sole recruitment of TFIID to the TATA box. Our experiments examining PIC formation as a function of the VP16 activation domain were performed in parallel with ICP4 and showed that, relative to ICP4, VP16 very poorly, if at all, helped recruit TFIID, despite its ability to recruit TFIIB and RNA Pol II. These studies further suggest that ICP4 intervenes at a very early step in the formation of the transcription initiation complex and that its mechanism of recruitment is fundamentally different from that of other activators such as VP16. It is also important to note that our studies with ICP4 utilize the intact protein isolated from HSV-infected cells undergoing a productive infection. The studies with VP16 (ours included) utilized a chimeric Gal4 construct produced in bacteria. Therefore, it is possible that there are subtle differences between the effects of bona fide VP16 isolated from HSV-infected cells and Gal4-VP16 on the formation of PICs.
Another observation of note in our study is that the facilitation of TFIID binding is a function of amino acid sequences from residues 30 to 274. The ICP4 protein purified from n208 (15)-infected cells contains residues 1 to 774. The X25 mutant was derived from the n208 allele and lacks residues 30 to 274 (47, 48). X25 binds to DNA but does not repress or activate transcription in vivo or in vitro (7, 25, 47, 48). Therefore, the DNA binding activity of ICP4 is not sufficient to facilitate TFIID binding. The requirement for residues 30 to 274 is interesting in that these are the residues previously found to be required for ICP4 to form a tripartite complex on DNA with TBP and TFIIB (50).
The n208 mutant (residues 1 to 774) molecule activates transcription in virus infection (15), transient assays (14), and in vitro (7). However, the magnitude of activation is considerably less than that observed for wt (residues 1 to 1298) ICP4. The carboxy-terminal residues from 775 to 1298 are required for efficient activation and viral growth (7, 15). These residues are also required for ICP4 to interact with TFIID in solution through interaction with TAF250 (7), a component of TFIID (58). More recently we have also found that the residues from 774 to 1298 are also involved in the multimerization of ICP4 on DNA (R. Kuddus and N. DeLuca, unpublished data). These later interactions, while not crucial for the recruitment of TFIID in our present studies, may have significant effects on the level of activation by ICP4. This carboxy-terminal region may affect the ability of ICP4 to bind to DNA in a complex mixture of proteins. Consistent with this is the observation that the n208 protein bound less efficiently to the immobilized template than wt ICP4 in the presence of nuclear extract (data not shown). Additionally the carboxy-terminal region of ICP4 may help stabilize PICs. This could occur simply as a manifestation of the interaction between the carboxy terminus of ICP4 and TAF250 and/or possible as yet unknown conformational changes brought on by this interaction. Studies reported by other groups (11, 34, 56) have shown that the Gal4-VP16 activator facilitates another step in PIC function after facilitation of TFIIB assembly. Like the Gal4-VP16 chimera, the ICP4 protein, after its effect on the recruitment of TFIID, may facilitate another step that is necessary to stabilize PIC formation or function. Resolution of this question will require the construction of site-directed mutations in the carboxy-terminal domain and an analysis that separates TFIID interaction activity from activity responsible for multimerization on DNA, as well as an assessment of the activation function of the mutant proteins.
ICP4 can activate the transcription of relatively simple promoters in vitro with a relatively simple set of transcription factors (6). However, it is a large and structurally complex protein, and it would not be surprising if multiple activities and interactions work in concert to produce the high transcription rates seen in HSV infection, and hence the abundant and rapid synthesis of HSV proteins and virions. This study demonstrates that one activity of ICP4 which is consistent with its role in the activation of transcription is the facilitation of TFIID binding to the TATA box and possibly the recruitment of the remainder of the transcription PIC, as suggested by the increase in TFIIB and Pol II binding.
ACKNOWLEDGMENTS
We thank Michael Carrozza and Dool-Bboon Kim for helpful discussions and technical assistance. We also acknowledge The National Cell Culture Center in Minneapolis, Minnesota, as a source of grown HeLa cells.
This work was supported by NIH grant AI30612 to N.D. and by a postdoctoral fellowship from the Medical Research Council of Canada to B.G.
REFERENCES
- 1.Arias J A, Dynan W S. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J Biol Chem. 1989;264:3223–3229. [PubMed] [Google Scholar]
- 2.Arnosti D N, Merino A, Reinberg D, Schaffner W. Oct-2 facilitates functional preinitiation complex assembly and is continuously required at the promoter for multiple rounds of transcription. EMBO J. 1993;12:157–166. doi: 10.1002/j.1460-2075.1993.tb05641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 5.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 6.Carrozza M J, DeLuca N. The high mobility group protein 1 is a coactivator of herpes simplex virus ICP4 in vitro. J Virol. 1998;72:6752–6757. doi: 10.1128/jvi.72.8.6752-6757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrozza M J, DeLuca N A. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caswell R, Bryant L, Sinclair J. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J Virol. 1996;70:4028–4037. doi: 10.1128/jvi.70.6.4028-4037.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 11.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLuca N A, Schaffer P A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5:1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca N A, Schaffer P A. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 1987;15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca N A, Schaffer P A. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J Virol. 1988;62:732–743. doi: 10.1128/jvi.62.3.732-743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon R A, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emami K H, Jain A, Smale S T. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emanuel P A, Gilmour D S. Transcription factor TFIID recognizes DNA sequences downstream of the TATA element in the Hsp70 heat shock gene. Proc Natl Acad Sci USA. 1993;90:8449–8453. doi: 10.1073/pnas.90.18.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisberg J V, Chen J L, Ricciardi R P. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol Cell Biol. 1995;15:6283–6290. doi: 10.1128/mcb.15.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godowski P J, Knipe D M. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci USA. 1986;83:256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu B, DeLuca N. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J Virol. 1994;68:7953–7965. doi: 10.1128/jvi.68.12.7953-7965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu B, Kuddus R, DeLuca N A. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol Cell Biol. 1995;15:3618–3626. doi: 10.1128/mcb.15.7.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guermah M, Malik S, Roeder R G. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol Cell Biol. 1998;18:3234–3244. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imbalzano A N, Shepard A A, DeLuca N A. Functional relevance of specific interactions between herpes simplex virus type 1 ICP4 and sequences from the promoter-regulatory domain of the viral thymidine kinase gene. J Virol. 1990;64:2620–2631. doi: 10.1128/jvi.64.6.2620-2631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufmann J, Smale S T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi N, Horn P J, Sullivan S M, Triezenberg S J, Boyer T G, Berk A J. DA-complex assembly activity required for VP16C transcriptional activation. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 35.Martinez E, Chiang C M, Ge H, Roeder R G. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez E, Zhou Q, L'Etoile N D, Oelgeschlager T, Berk A J, Roeder R G. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci USA. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeoch D J, Dolan A, Donald S, Brauer D H. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 39.O'Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson T, Everett R D. Mutational dissection of the HSV-1 immediate-early protein Vmw175 involved in transcriptional transactivation and repression. Virology. 1988;166:186–196. doi: 10.1016/0042-6822(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 41.Paterson T, Everett R D. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 1988;16:11005–11025. doi: 10.1093/nar/16.23.11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 44.Sauer F, Hansen S K, Tjian R. DNA template and activator-coactivator requirements for transcriptional synergism by Drosophila bicoid. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 45.Shepard A A, DeLuca N A. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J Virol. 1991;65:299–307. doi: 10.1128/jvi.65.1.299-307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard A A, DeLuca N A. Intragenic complementation among partial peptides of herpes simplex virus regulatory protein ICP4. J Virol. 1989;63:1203–1211. doi: 10.1128/jvi.63.3.1203-1211.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepard A A, Imbalzano A N, DeLuca N A. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J Virol. 1989;63:3714–3728. doi: 10.1128/jvi.63.9.3714-3728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shepard A A, Tolentino P, DeLuca N A. trans-Dominant inhibition of herpes simplex virus transcriptional regulatory protein ICP4 by heterodimer formation. J Virol. 1990;64:3916–3926. doi: 10.1128/jvi.64.8.3916-3926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 50.Smith C A, Bates P, Rivera-Gonzalez R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steger G, Ham J, Lefebvre O, Yaniv M. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sypes M A, Gilmour D S. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 1994;22:807–814. doi: 10.1093/nar/22.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 54.Verrijzer C P, Yokomori K, Chen J L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 55.Watson R J, Clements J B. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature. 1980;285:329–330. doi: 10.1038/285329a0. [DOI] [PubMed] [Google Scholar]
- 56.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia K, Knipe D M, DeLuca N A. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J Virol. 1996;70:1050–1060. doi: 10.1128/jvi.70.2.1050-1060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]