Abstract
Recombinant adeno-associated virus type 2 (AAV2) can be produced in adenovirus-infected cells by cotransfecting a plasmid containing the recombinant AAV2 genome, which is generally comprised of the viral terminal repeats flanking a transgene, together with a second plasmid expressing the AAV2 rep and cap genes. However, recombinant viruses generally replicate inefficiently, often producing 100-fold fewer virus particles per cell than can be obtained after transfection with a plasmid containing a wild-type AAV2 genome. We demonstrate that this defect is due, at least in part, to the presence of a positive-acting cis element between nucleotides 194 and 1882 of AAV2. Recombinant AAV2 genomes lacking this region accumulated 14-fold less double-stranded, monomer-length replicative-form DNA than did wild-type AAV2. In addition, we demonstrate that a minimum genome size of 3.5 kb is required for efficient production of single-stranded viral DNA. Relatively small recombinant genomes (2,992 and 3,445 bp) accumulated three- to eightfold less single-stranded DNA per monomer-length replicative-form DNA molecule than wild-type AAV2. In contrast, recombinant AAV2 with larger genomes (3,555 to 4,712 bp) accumulated similar amounts of single-stranded DNA per monomer-length replicative-form DNA compared to wild-type AAV2. Analysis of two recombinant AAV2 genomes less than 3.5 kb in size indicated that they were deficient in the production of the extended form of monomer-length replicative-form DNA, which is thought to be the immediate precursor to single-stranded AAV2 DNA.
Adeno-associated virus type 2 (AAV2) is a human parvovirus of the dependovirus subgroup. Unlike other parvoviruses, dependoviruses generally require coinfection with a helper virus, such as adenovirus or herpesvirus, to initiate a lytic infection (4). In the absence of a helper virus, AAV2 integrates into the host chromosome and remains latent until it is activated by an adenovirus or herpesvirus infection or other stress, such as DNA damage (46, 66, 67). After adenovirus or herpesvirus infection, AAV2 excises from the chromosome and replicates its genome as linear, double-stranded DNA molecules called replicative forms (51). Repeated sequences at the ends of AAV2 DNA serve as origins of replication and packaging elements. Like other parvoviruses, AAV2 packages one genome-length, single-stranded DNA.
AAV2 contains only two major open reading frames, rep and cap, named for their roles in DNA replication and encapsidation (19, 48, 56). Rep78 and Rep68 are generated from transcripts that derive from the P5 promoter, and they differ in their C termini due to alternative splicing of the P5 transcripts (7, 58). Rep78 and Rep68 are DNA helicases that also have a single-stranded DNA endonuclease activity (5, 23, 65, 69), and either protein is sufficient to support AAV2 DNA replication (21). Rep78 and Rep68 are also required for site-specific integration of AAV DNA into the host cell genome and for excision (2, 49, 52). Rep52 and Rep40 are translated from transcripts generated from a promoter, P19, located within the rep gene. Rep52 and Rep40 are translated from the same open reading frame as Rep78 and Rep68 and vary in their C termini due to the same alternative splicing. Rep52 and Rep40 lack the DNA binding and endonuclease domains of the larger Rep proteins (62) but retain a functional helicase domain (50). Rep52 and Rep40 are not required for the replication of double-stranded DNA, but they are required for the efficient production of single-stranded AAV2 DNA (9). The cap gene encodes three structural proteins: VP1, VP2, and VP3.
Most AAV2-based vectors contain a transgene flanked by the AAV2 terminal repeats (36, 45). AAV2 vectors are propagated by cotransfecting a plasmid carrying the recombinant viral molecule plus a plasmid expressing the AAV2 rep and cap genes into mammalian cells. The cells are either infected with adenovirus or cotransfected with a third plasmid expressing the adenovirus helper genes required to produce AAV2. The recombinant AAV2 (rAAV2) construct and complementing vector can be configured to share no sequence homology (44) so that contaminating pseudo-wild-type virus can arise only via nonhomologous recombination. Unfortunately, however, AAV2 vectors generally replicate less efficiently than wild-type AAV2, producing only 103 to 104 recombinant particles per cell.
Several improvements to the original method for vector production have been reported. Most have involved improved methods for providing the rep and cap gene products in trans, including (i) optimizing the complementing plasmid-to-AAV2 vector ratio (12, 16); (ii) expressing rep and cap from heterologous promoters (31, 61); (iii) reducing the level of Rep78 and -68 by altering their initiation codons from AUG to ACG (31); (iv) producing rep- and cap-expressing cell lines (20, 24, 32, 39, 68); and (v) expressing rep and cap from herpes simplex virus amplicons (13, 25). These modifications improve the yield of recombinant AAV2 but at levels that fall significantly short of the yield obtained after transfection with a plasmid containing wild-type AAV2 DNA.
We demonstrate here that the inefficient replication of recombinant molecules is due in part to the presence of a cis-acting DNA replication element in the wild-type AAV2 genome that is deleted in most recombinant constructs. We also show that efficient production of single-stranded DNA requires an rAAV2 genome that is 3.5 kb in size or greater. Small genomes are deficient in the accumulation of the extended form of monomer-length replicative-form DNA, the apparent precursor to single-stranded AAV2 DNA.
MATERIALS AND METHODS
Plasmids.
To facilitate the cloning of pGET015, we synthesized a 238-bp mega-cloning site that contained all of the restriction endonuclease sites necessary for the cloning of pGET015. The mega-cloning site was created by using six oligonucleotides: Mega1 (5′-GTACATCTAGACTCGAGGACACTCTCTATGATCAGACTTTTTGTCGGCCGGCCTAGTCTT-3′), Mega2 (5′-AGGGTCG ACCAATTGTTAATTAAGACAATAAGTCTACGTAGAAGACTAGGCCG GCCGACA-3′), Mega3 (5′-AATTAACAATTGGTCGACCCTAGGATCCGGCGCGCCGCGGCCGCCTGCAGCTTAAGACCG-3′), Mega4 (5′-CTATGCA TGTTAACGGGCCCGCTAGCACGCGTAAGCTTACCGGTCTTAAGCTG CAGGCGG-3′), Mega5 (5′-GGGCCCGTTAACATGCATAGATCTTAATTAACCAGCGATCGCTTGGACTAGTCCA-3′), and Mega6 (5′-TCGAATCTAG ACCAGCGCTGGCCTGGACTAGTCCAAGCGATCG-3′).
The oligonucleotides (300 pmol of each in 10 mM Tris-HCl [pH 7.5]–5 mM MgCl2–7.5 mM dithiothreitol) were denatured at 95°C for 10 min, and the mixture was allowed to cool slowly until it was below 35°C. Deoxyribonucleotides (dATP, dGTP, dCTP, and dTTP; 10 mM each) and DNA polymerase I (Klenow fragment; 20 U) were added to the mixture (total volume of 80 μl), which was then incubated at 37°C for 1 h. The products of the reaction were separated on a 10% polyacrylamide gel, and DNA approximately 200 bp in length was isolated. This DNA was amplified by PCR using Vent DNA polymerase (New England Biolabs) and the outer two oligonucleotides (Mega1 and Mega6; 100 nM each) as primers. The PCR product was amplified a second time using Taq DNA polymerase (Boehringer Mannheim), to add an A to the 3′ end of the DNA, and the product was then cloned into a T-tailed vector, pT7-Blue (Novagen). The resulting clone, pT7B-MCS, was sequenced to verify that the insert was as expected.
pGET015 was constructed in six subsequent steps: (i) the insert from pT7B-MCS was cloned as an XbaI fragment into the XbaI sites of psub201(−) (44) to produce pGET001; (ii) the simian virus 40 (SV40) polyadenylation site was cloned as an HpaI-to-BglII fragment from pCDM8 (Invitrogen) into pGET001 to produce pGET003; (iii) the SV40 early promoter was isolated from pSVβgal (Promega) as an EcoRI-to-AvrII fragment and cloned into the MfeI and AvrII sites of pGET001 to produce pGET005; (iv) the enhanced green fluorescent protein (EGFP) gene was isolated as a BamHI-to-NotI fragment from pEGFP-N1 (Clontech) and cloned into the same sites in pGET005 to generate pGET007; (v) the puromycin-N-acetyltransferase gene, which confers resistance to puromycin, was isolated as a 730-bp HindIII-to-NheI fragment from pBABE-PURO (37) and cloned into the same sites in pGET007 to produce pGET011; and (vi) the internal ribosomal entry site (IRES) from pCIN4 (from E. S. Rees, Glaxo Wellcome) was cloned as a PstI fragment into pGET007 to generate pGET015.
p015-AAV was constructed by inserting a PvuII fragment from pGT620(+) that contains a wild-type AAV2 genome into the SwaI site of pGET015. Plasmid pGT620(+) was made by cloning the wild-type AAV2 genome as an MscI fragment from pSM620 (43) into psub201(−), so that the wild-type AAV2 genome is flanked by terminal repeats from psub201(−). Therefore, all four terminal repeats in p015-AAV are identical. To create psubλ-AAV2, a 4,378-bp EagI-AvrII segment of λ DNA (sequence positions 19944 to 24322) was cloned into the SpeI and EagI sites in p015-AAV in place of the EGFP-puromycin resistance expression cassette. A series of deletions of various lengths within the λ insert of psubλ-AAV2 were constructed by digesting with the appropriate restriction enzymes, filling in the ends with T4 DNA polymerase as necessary, and religating. The restriction enzymes used were HpaI and RsrII for psubλdl339, HpaI and BglII for psubλdl522, EagI and BlpI for psubλdl797, BlpI and HpaI for psubλdl1157, and BlpI and RsrII for psubλdl1495.
Transfection of adenovirus-infected cells.
293 cells were propagated in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum (HyClone Laboratories). The 293 cells were seeded at 4 × 105 cells/well in six-well dishes 36 to 40 h before an experiment. The cells were infected at a multiplicity of 5 PFU/cell with Ad5dl312 (26), which contains a deletion in the E1A region of adenovirus type 5. After 1 h, cells were transfected by the calcium phosphate precipitate method. DNA (2 to 4 μg/well) was premixed with 2× HeBS, pH 7.05 (0.280 M NaCl, 50 mM HEPES, 1.5 mM Na2HPO4) (120 μl/well), and then an equal volume of CaTE (0.30 M CaCl2, 1 mM Tris-HCl [pH 7.5], 0.1 mM EDTA) (120 μl/well) was added. The DNA-calcium phosphate precipitate was allowed to form for 2 to 5 min at room temperature before being added to tissue culture medium (2 ml/well). Transfected cells were incubated overnight at 37°C, rinsed two times in phosphate-buffered saline (0.137 M NaCl, 2.7 mM KCl, 1.5 mM KH2PO4), 8.1 mM Na2HPO4), and refed with medium supplemented with 10% fetal bovine serum. In time course experiments, time zero was the point at which cells were refed after the transfection procedure.
Analysis of intracellular AAV2 DNAs.
At various times following transfection, cells (8 × 105 to 106 cells/sample) were resuspended in spent tissue culture medium and then separated from the medium by centrifugation (13,000 rpm for 2 min). The cells were lysed in hypotonic TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and subjected to three rounds of freezing at −80°C and thawing at 37°C, and the cell pellet was separated from the lysate by centrifugation (13,000 rpm for 5 min). The cell pellets were resuspended in lysis buffer (2% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 10 mM Tris-HCl, 1 mM EDTA) at 3 × 106 cell equivalents/ml and incubated for 2 h at 55°C. Cellular DNA was sheared by passing the sample through a syringe fitted with a 25-gauge needle 10 times. Hypotonic TE lysates and tissue culture medium were adjusted to 2% SDS and 0.15 M NaCl. All samples were digested with proteinase K (0.5 mg/ml) for 1 to 2 h at 37°C. An equal number of cell equivalents (104) of each sample was subjected to electrophoresis through a 1% agarose gel in TAE buffer (20 mM Tris-HCl [pH 8.5], 2.5 mM sodium acetate, 0.5 mM EDTA), and the DNA was transferred to a nitrocellulose membrane. The membrane was hybridized overnight at 65°C to a 32P-labeled, random-primed probe DNA in a mixture containing 0.75 M NaCl, 75 mM sodium citrate, 25 mM Na2HPO4, 0.1% polyvinylpyrrolidone 40 (Sigma), 0.1% Ficoll type 400 (Sigma), 0.1% bovine serum albumin, and 0.25 mg of sonicated salmon sperm DNA per ml. The membranes were washed two times in 0.3 M NaCl–0.03 M sodium citrate–0.1% SDS for 30 min at 65°C and two times in 0.03 M NaCl–0.003 M sodium citrate–0.1% SDS for 30 min at 65°C. Bands were quantified using a PhosphorImager (Molecular Dynamics).
For analysis by two-dimensional agarose gel electrophoresis (60), the cell pellet and hypotonic TE lysate fractions containing equal numbers of cell equivalents were combined. One lane containing DNA from both the cell pellet and hypotonic TE lysate was cut off of the first neutral agarose gel (1% agarose in TAE) and fused to a new gel for the second dimension (1% agarose, 50 mM NaCl, 1 mM EDTA), which was presoaked in alkaline running buffer (30 mM NaOH, 1 mM EDTA) for 1 h and then subjected to electrophoresis at 30 V for 48 h. The DNA from the first and second gels was transferred to nitrocellulose and hybridized to a 32P-labeled probe as described above.
RESULTS
Recombinant vGET015 DNA exhibits a cis defect in replication.
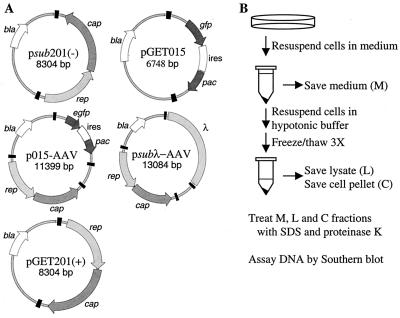
As discussed above, AAV2 vectors do not replicate as efficiently as wild-type AAV2. To determine whether this deficiency results from a problem in providing AAV2 gene products in trans or from a cis-acting defect, we cotransfected adenovirus-infected 293 cells with equal amounts of plasmid carrying a phenotypically wild-type AAV2 genome called psub201(−) (Fig. 1A) (44) and a plasmid with an rAAV2 vector, pGET015 (Fig. 1A). For clarity, we use a “v” prefix to denote the viral DNA that derives from a plasmid, e.g., vGET015 is viral DNA from pGET015. vGET015 encodes a bicistronic mRNA that contains the EGFP gene followed by an IRES followed by a puromycin resistance gene. Expression of the bicistronic message is controlled by the SV40 early promoter. By 60 h following transfection, vGET015 monomer-length replicative-form DNA was approximately 20-fold less abundant than the corresponding vsub201(−) species (data not shown). Additionally, approximately 100-fold less vGET015 single-stranded DNA was produced relative to vsub201(−) (data not shown). Because psub201(−) expresses all of the AAV2 gene products required in trans for replication, it appeared likely that vGET015 exhibits a cis-acting defect.
FIG. 1.
Plasmids and lysis procedure. (A) Maps of plasmids. AAV2 terminal repeats are drawn as black boxes. Open reading frames are designated by arrows and correspond to AAV2 rep, AAV2 cap, EGFP gene (egfp), puromycin-N-acetyltransferase gene (pac), and β-lactamase gene (bla). Additional elements include λ DNA and an IRES (ires). (B) Processing of infected cells. Infected cells were harvested at various times following transfection by resuspension in spent tissue culture medium (M fraction). The cells were removed from the M fraction and subjected to three freeze-thaw cycles in hypotonic buffer; then the cell membrane pellet (C fraction) was separated from the hypotonic lysate (L fraction).
Since vsub201(−) can replicate to high copy number (106 molecules per cell) on its own but vGET015 requires vsub201(−) to replicate, it was possible that the apparent difference in replication efficiencies was due to replication of psub201(−) in cells that did not receive pGET015. To rule out this possibility, we cloned a copy of wild-type AAV2 into pGET015 to create a plasmid called p015-AAV (Fig. 1A). In this way, all transfected cells received exactly equal molar amounts of vGET015 and wild-type AAV2. Since p015-AAV contains four copies of the AAV2 terminal repeat sequence, excision can potentially occur at any combination of these repeats. However, the recombinant and AAV2 genomes are preferentially excised and replicated due to the presence of the 20-bp D sequence, which is located on the inboard side of the 125-bp palindrome within the terminal repeat (63, 64). To ensure that excisions of both wild-type AAV2 and vGET015 from the plasmid were similar, p015-AAV was cloned such that all four terminal repeats are identical and derive from psub201(−). Terminal repeats from psub201(−) lack 13 bp from the end of wild-type AAV2, which may affect their excision (44).
Adenovirus-infected 293 cells were transfected with p015-AAV and, at various times posttransfection, processed as outlined in Fig. 1B. An equal number of cell equivalents of all samples was subjected to Southern blot analysis using a 32P-labeled probe DNA specific for the AAV2 terminal repeats that detects both wild-type and vGET015 DNAs (Fig. 2A). Most of the progeny virions were in the hypotonic lysate (Fig. 2A, lanes L), as evidenced by the presence of greater than 90% of the single-stranded DNA in this fraction. Most (>70%) of the double-stranded replicating DNA was in the cell pellet (Fig. 2A, lanes C). The amount of double-stranded replicative-form DNAs in the hypotonic lysate varied from one experiment to another, apparently due to slight differences in the lysis procedure. AAV2 packages both plus and minus strands, which could potentially anneal to form a double-stranded DNA. However, this does not appear to be a major source of monomer-length replicative-form DNA in the hypotonic lysate, because the ratio of monomer-length to dimer-length replicative form DNA in the lysate is similar to their ratio in the cell pellet.
FIG. 2.
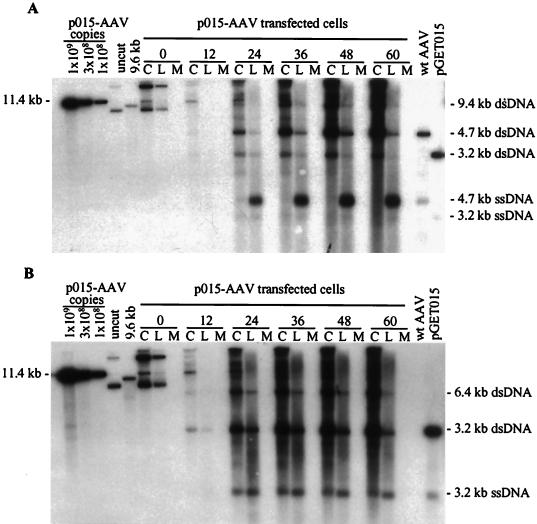
Replication of vGET015 is reduced relative to wild type. Ad5dl312-infected 293 cells were processed at the indicated times after transfection with p015-AAV, and 104 cell equivalents of the cell pellet (lanes C), hypotonic lysate (lanes L), and medium (lanes M) were analyzed by Southern blotting using a probe corresponding to the AAV2 terminal repeats (A) or the EGFP gene (B). To estimate the amounts of AAV2 and vGET015 replicative-form DNAs, known amounts of linearized p015-AAV DNA were included in the analysis. p015-AAV (uncut) was used as a marker for input plasmid. A 9.6-kb PpuMI-SwaI fragment of p015-AAV (9.6 kb) was used as a marker for AAV2 dimer-length replicative-form DNA (dsDNA), which is 9.4 kb. A marker for AAV2 monomer-length replicative-form DNA and single-stranded DNA (ssDNA) was prepared by mixing equal amounts of a 4.7-kb PvuII fragment from psub201(−), which was either left untreated or denatured (wt [wild-type] AAV2). Likewise, markers for vGET015 monomer-length replicative-form DNA and single-stranded DNA were prepared using a 3.2-kb AseI-FspI fragment of pGET015 (pGET015). The AseI-FspI fragment (3,170 bp) is slightly larger than predicted size of vGET015 monomer-length replicative-form DNA (3,112 bp).
By 12 h posttransfection, most of the input plasmid DNA was degraded; residual plasmid was linear (Fig. 2A). This linear form persisted throughout the infection and, as discussed later, appears to be replicating like AAV2 replicative form. By 12 h posttransfection, monomer-length replicative-form DNA of both wild-type AAV2 (4.7 kb) and vGET015 (3.1 kb) was detected. At this time, wild-type monomer-length replicative-form DNA was approximately twofold more abundant than the equivalent vGET015 DNA. By 60 h following transfection, wild-type monomer-length replicative-form DNA was 14-fold more abundant than the vGET015 species. Because both wild-type and vGET015 genomes were delivered to the cell on the same plasmid at equal molar amounts and because wild-type AAV2 provides all of the products necessary for replication in trans, vGET015 was clearly defective in cis.
To better detect vGET015 DNA and to definitively distinguish it from wild-type AAV2 DNA, we analyzed the same samples on a second Southern blot using a 32P-labeled probe corresponding to the EGFP coding region (Fig. 2B). The ratio of dimer to monomer-length replicative-form DNA observed for vGET015 was similar to the ratio observed for wild-type AAV (Fig. 2). However, in comparison to the wild type, the amount of vGET015 single-stranded DNA was reduced relative to the amount of vGET015 monomer-length replicative-form DNA. At 60 h posttransfection, the amount of vGET015 single-stranded DNA was 50-fold less than the amount of wild-type single-stranded DNA present in the same sample. In addition to a defect in the accumulation of vGET015 single-stranded DNA, the distribution of the single-stranded DNA in the cell pellet and hypotonic lysate was altered. Whereas >90% of wild-type single-stranded DNA was in the hypotonic lysate, only 60% of vGET015 single-stranded DNA was in this fraction. Consequently, the amount of single-stranded vGET015 DNA, and presumably vGET015 virions, in the hypotonic lysate was approximately 100-fold less than the amount of wild-type single-stranded DNA. This is noteworthy because the production of a freeze-thaw hypotonic lysate is usually the first step in purification of virus particles. Since the assay does not distinguish between naked and encapsidated single-stranded DNA, we cannot determine whether the vGET015 single-stranded DNA in the cell pellet is a replicating form that may also be present in the wild type or is due to a defect in egress of vGET015 virions from cells.
The terminal repeats of vGET015 are fully functional.
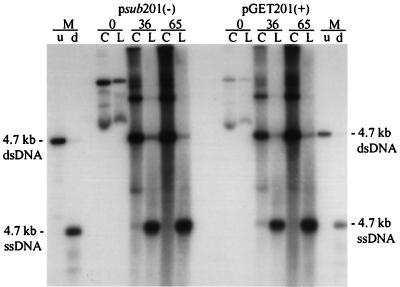
Because the AAV2 terminal repeats are the only cis elements known to be necessary for AAV2 replication and packaging, we wanted to rule out the possibility that the cis defect might result from a lesion in the terminal repeats in pGET015. Accordingly, we cloned the rep and cap genes from psub201(−) between the terminal repeats of pGET015 to create pGET201(+) (Fig. 1A). To ensure that this clone was not merely the original psub201(−), we selected a clone in which the rep and cap genes were inverted relative to their relationship to the terminal repeats in psub201(−). The two orientations of vsub201(−) DNA excise and replicate equally well (44). Adenovirus-infected 293 cells were transfected with either psub201(−) or pGET201(+) and processed at various times after transfection (Fig. 1B). As expected, vGET201(+) excised and replicated as efficiently as wild-type AAV2 (Fig. 3), confirming that the terminal repeats in pGET015 were fully functional.
FIG. 3.
The terminal repeats in pGET015 are fully functional. Ad5dl312-infected 293 cells were processed at the indicated times time after transfection with psub201(−) or pGET201(+), and equal amounts of the cell pellet (lanes C) or hypotonic lysate (lanes L) were analyzed by Southern blotting using a probe corresponding to the AAV2 rep and cap genes. Markers (lanes M) were prepared by loading a 4.7-kb PvuII fragment from psub201(−) and either left untreated (u) or denatured (d). dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
The vGET015 replicon lacks a cis-acting element required for efficient accumulation of monomer-length replicative-form DNA.
It was conceivable that the EGFP-IRES-puromycin resistance cassette in vGET015 contained a cis-acting element that inhibited replication of the recombinant molecule. To test this possibility, we cloned a 4.3-kb DNA fragment from bacteriophage λ in place of the EGFP-IRES-puromycin resistance cassette in p015-AAV to create psubλ-AAV2 (Fig. 1A). Given its prokaryotic origin, this insert is less likely to bind mammalian proteins than the EGFP-IRES-puromycin resistance cassette, and it does not appear to have unusual secondary structure.
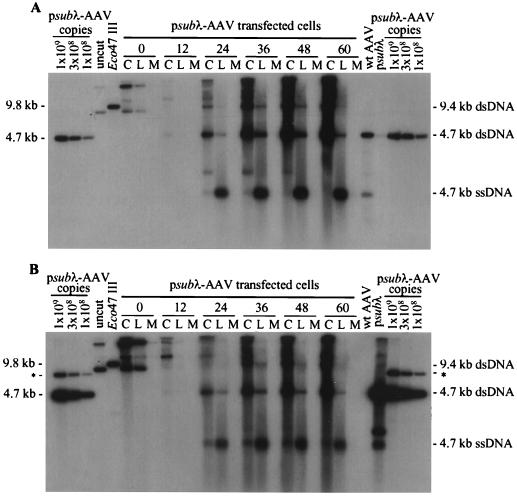
Adenovirus-infected 293 cells were processed (Fig. 1B) at various times following the transfection with psubλ-AAV2. Because vsubλ is similar in size to wild-type AAV2 (4.7 kb), two identical Southern blots were prepared. One was hybridized to a 32P-labeled probe DNA corresponding to the AAV2 rep and cap genes (Fig. 4A), and the other was hybridized to a probe specific for λ DNA (Fig. 4B). To determine the amount of vsubλ monomer-length replicative-form DNA relative to the equivalent wild-type species, we compared the amount of replicative-form DNA obtained from infected cells to a known amount of psubλ-AAV2 that had been digested with SrfI, which cuts twice within each AAV2 terminal repeat (Fig. 1A). As observed for vGET015 (Fig. 2), the amount of vsubλ monomer-length replicative-form DNA was 14-fold reduced in comparison to the wild-type species at 60 h posttransfection (Fig. 4). Since it is unlikely that we introduced negative-acting cis elements with similar inhibitory effects into both vGET015 and vsubλ, this result indicates that a positive element acting in cis is present in wild-type AAV2 but not in vGET015 and vsubλ. Therefore, this cis-acting element must reside between sequence positions 194 and 4498 in AAV2.
FIG. 4.
The AAV2 recombinant vsubλ is deficient in the accumulation of replicative-form DNA. Ad5dl312-infected 293 cells were processed at the indicated times after transfection with psubλ-AAV2 DNA, and 104 cell equivalents of the cell pellet (lanes C), hypotonic lysate (lanes L), and medium (lanes M) were analyzed by Southern blotting using a probe corresponding to the AAV2 rep and cap genes (A) or the λ insert in psubλ-AAV2 (B). To estimate the copy numbers of vsubλ and AAV2 replicative-form DNAs, psubλ-AAV2 was digested with SrfI, which cuts twice in each AAV2 terminal repeat, and 109, 3 × 108, or 108 copies were loaded per lane. Both vsubλ and AAV2 SrfI fragments are approximately 4.6 kb in length. In panel B, the probe to the λ insert also hybridized to a 7.4-kb partial digestion product (∗). As a marker for input plasmid DNA, 108 copies of psubλ-AAV2 was loaded (uncut). To make a marker for AAV2 and vsubλ dimer-length replicative-form DNAs, which are both 9.4 kb, we used a 9.8-kb Eco47III fragment from psubλ-AAV2 (Eco47III). This fragment hybridizes to both the rep and cap probe used in panel A and the λ probe used in panel B. As a marker for AAV2 monomer-length replicative-form (dsRNA) and single-stranded DNA (ssDNA), equal amounts of a 4.7-kb PvuII fragment from psub201(−) that had been denatured or left untreated were combined and loaded into one well (wt [wild-type] AAV2). Likewise, a marker for vsubλ monomer-length replicative-form DNA and single-stranded DNA was made using a 4.6-kb SrfI fragment from psubλ-AAV2 (psubλ).
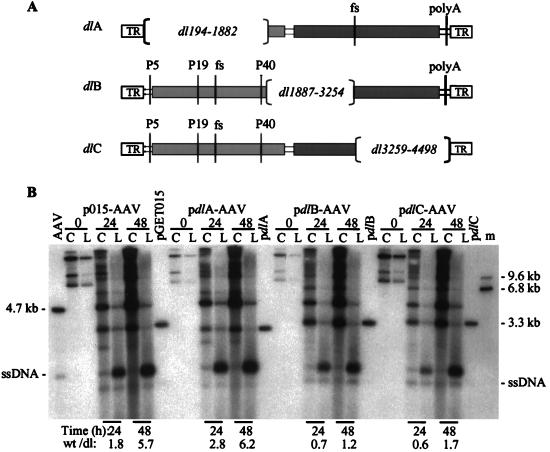
To more precisely localize the element, we introduced deletions into a plasmid containing an AAV2 genome with frameshift mutations that prevent expression of both Rep and Cap proteins. Each deletion removed about one-third of the AAV2 genome (pdlA, pdlB, and pdlC [Fig. 5A]), and as a consequence of the frameshift mutations, none of the deletion constructs express AAV2 gene products (data not shown). We then cloned the mutant sequences in place of the vGET015 genome in p015-AAV to create pdlA-AAV2, pdlB-AAV2, and pdlC-AAV2, respectively. We transfected the set of plasmids into adenovirus-infected 293 cells, processed the cells at various times, and assayed DNA accumulation by Southern blotting using a 32P-labeled probe DNA to the AAV2 terminal repeat. At 48 h posttransfection, the ratio of wild-type to vdlA monomer-length replicative-form DNAs was 6.2, which is similar to the ratio of wild-type to vGET015 monomer-length replicative-form DNAs. In contrast, the ratios of wild-type to vdlB and vdlC monomer-length replicative-form DNAs were 1.0 and 1.7, respectively. Clearly, the element is located within the region deleted in vdlA, i.e., between sequence positions 194 and 1882.
FIG. 5.
Localization of the DNA element responsible for efficient accumulation of monomer-length replicative-form DNA. (A) Map of AAV2 deletion constructs. pdlA contains a large deletion in the rep gene (light gray box) and a frameshift mutation (fs) at the BsiWI site in the cap gene (darker gray box). pdlB and pdlC contain deletions in the capsid gene and frameshift mutations at the BamHI site in the rep gene. Also indicated are the three AAV2 promoters (P5, P19, and P40), polyadenylation site (polyA), and terminal repeats (TR). (B) Ad5dl312-infected 293 cells were processed at the indicated times after transfection with p015-AAV or its mutant derivatives, and equal amounts of the cell pellet (lanes C) or hypotonic lysate (lanes L) were analyzed by Southern blotting using a probe corresponding to the AAV2 terminal repeats. Markers for AAV2 monomer-length replicative-form DNA and single-stranded DNA (ssDNA) were prepared by combining equal amounts of a 4.7-kb PvuII fragment of psub201(−) that had been denatured or left untreated (AAV2). Similar markers were prepared for the rAAV2 genomes using a 3.2-kb AseI-FspI fragment from pGET015 or a 3.0-kb PvuII fragment from a pdlA, a 3.3-kb PvuII fragment from pdlB, or a 3.4-kb PvuII fragment from pdlC. The single-stranded DNA from these markers is not evident, but a longer exposure revealed that for each construct they comigrated with the band below the 4.7-kb single-stranded DNA band. Markers approximately the size of AAV2 dimer-length replicative-form DNA (9.4 kb) and vdlC dimer-length replicative-form DNA (6.8 kb) were prepared using a 9.6-kb PpuMI-SwaI fragment of p015-AAV and NotI-linearized pGET015. These two markers were combined (lane m).
Genome size influences the accumulation of single-stranded DNA.
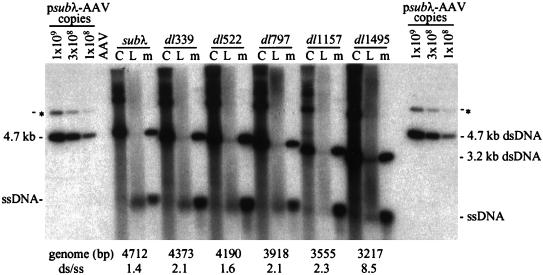
Whereas vGET015 single-stranded DNA was reduced 3.3-fold relative to the amount of its monomer-length replicative-form DNA in comparison to wild-type AAV2 DNA (Fig. 2), the ratio of vsubλ monomer-length replicative-form DNA to vsubλ single-stranded DNA was similar to the wild-type ratio (Fig. 4). Because vsubλ is similar in size to wild-type AAV2 (4.7 kb) and vGET015 is substantially smaller (3.1 kb), we wanted to determine whether the defect in accumulation of vGET015 single-stranded DNA was due to its small size or whether vGET015 contained an inhibitory element. To accomplish this, we made a set of deletions in psubλ-AAV2. The deletions progressively reduced the size of vsubλ from 4,712 to 3,217 bp, which is similar to the size of vGET015. Adenovirus-infected 293 cells were transfected with this set of deletion derivatives and processed 48 h later (Fig. 1B). As noted previously, the amount of monomer-length replicative-form DNA increased as genome length decreased (48). The molar ratio of vsubλ monomer-length replicative-form DNA per vsubλ single-stranded DNA was calculated to be 1.4, similar to the wild-type AAV2 ratio of 1.7 (Fig. 6). Deletion of up to 1,157 bp from the λ DNA in psubλ-AAV2 had only a modest effect on the ratio of monomer-length replicative-form DNA to single-stranded DNA, but deletion of 1,495 bp to give a genome length of 3,217 bp resulted in a fourfold increase in the ratio, to 8.5 (Fig. 6). This is similar to the ratio (5.3) of monomer-length replicative-form DNA to single-stranded DNA observed for vGET015. Therefore, the defect in accumulation of single-stranded DNA is due to the length of the genome rather than the sequence of the insert in vGET015.
FIG. 6.
Short recombinant AAV2 molecules are deficient for the accumulation of single-stranded DNA. Ad5dl312-infected 293 cells were transfected with psubλ-AAV2 (subλ) plus a series of deleted derivatives (dl339, dl522, dl797, dl1157, and dl1495), infected cells were harvested 60 h later, and 3 × 104 cell equivalents of the cell pellet (lanes C) or hypotonic lysate (lanes L) were analyzed by Southern blotting using a probe comprised of a λ sequence that is present in all of the deletion constructs. The probe also hybridized to a 7.4-kb partial digestion product (∗). To estimate the amount of replicative-form DNA (dsDNA) and single-stranded DNA (ssDNA) present, 109, 3 × 108, or 108 copies of SrfI-digested psubλ-AAV2 were included in the analysis. Markers (lanes m) for each deletion construct were prepared using a SrfI fragment from the appropriate clone; equal amounts of denatured and untreated marker DNA fragments were combined. Below the autoradiograph, the expected size of each deletion construct is listed, as well as the observed molar ratio of monomer-length replicative-form DNA to single-stranded DNA (ds/ss). For this calculation, we assumed that one monomer-length replicative-form DNA equaled two single-stranded DNA molecules.
All three of the deletion mutants analyzed in Fig. 5 exhibited defects in the accumulation of single-stranded DNA similar to vGET015 and vsubλdl1495. The defect was most dramatic for vdlB, which replicated all double-stranded forms equivalent to wild type but accumulated eightfold less single-stranded DNA than wild type. Therefore, the lower size limit for efficient production of single-stranded DNA is between 3,445 bp (the size of vdlC) and 3,555 bp (the size of vsubλdl1157).
Genome size influences the accumulation of extended monomer-length replicative-form DNA.
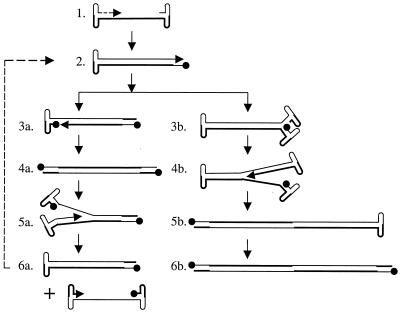
Parvoviruses replicate their DNA (see Fig. 9) by a mechanism known as rolling hairpin replication (8, 51, 55). The terminal palindromes fold back on themselves to form hairpin structures, which serve as primers for subsequent rounds of replication. The hairpin is nicked by Rep78 or -68, and replication by a host DNA polymerase forms a full-length, extended end. Therefore, replicating parvovirus DNAs contain a mixture of extended and covalently closed ends. Replicative forms that are covalently closed on one or both ends are called turnaround forms. Turnaround forms can be distinguished from extended forms by two-dimensional agarose gel electrophoresis (14, 48, 60), where DNA samples are subjected to electrophoresis under neutral conditions in the first dimension and under alkaline conditions in the second dimension. Monomer-length replicative-form DNA that is extended on both ends migrates as a 4.7-kb double-stranded DNA in the first dimension and denatures into 4.7-kb single-stranded DNA under alkaline conditions. Turnaround monomer-length replicative-form DNA, which is covalently closed in one end and extended in the other, comigrates with extended monomer-length replicative-form DNA in the neutral dimension but denatures into 9.4-kb single-stranded DNA under alkaline conditions. Monomer-length replicative-form DNA that is covalently closed on both ends migrates as single-stranded DNA circle in the alkaline dimension (14). It migrates slightly faster than extended monomer-length replicative-form DNA (4.7 kb) in the neutral dimension and slightly slower than 9.4 kb in the alkaline dimension. (For more details on the migration of parvovirus replication intermediates, see reference 60).
FIG. 9.
Replication of AAV2 DNA. AAV2 virions contain a linear single-stranded DNA, which is converted to a double-stranded monomer-length replicative-form DNA that is covalently closed in one end. This is called a turnaround form. Turnaround ends are nicked by Rep78 or -68 (closed circle), which becomes covalently attached to the 5′ end of the DNA (3a). DNA polymerase extends through the terminal repeat to generate a monomer-length replicative-form DNA that has two extended ends (4a). Steps 3a and 4a are often referred to as terminal resolution. Isomerization of the terminal repeat generates a new hairpin, which serves as the primer for DNA synthesis (5a). This may occur at either end. In the absence of encapsidation, both strands are converted to turnaround monomer-length replicative-form DNA, leading to the amplification of monomer-length replicative-form DNA. Capsids bind to displaced strands and package them into viral particles (6a). If in step 2 a new hairpin is formed and DNA synthesis begins prior to resolution of the turnaround end (3b and 4b), the replication complex will proceed through the turnaround end and generate dimer-length replicative-form DNA. Larger concatemers can form by replicating the 5b structure by the same mechanism (not shown). Dimer-length replicative-form DNA (5b and 6b) can be nicked by Rep78 or -68 at the palindrome in the middle of the molecule to form two monomer-length replicative-form DNAs that are turnaround in one or both ends (not shown).
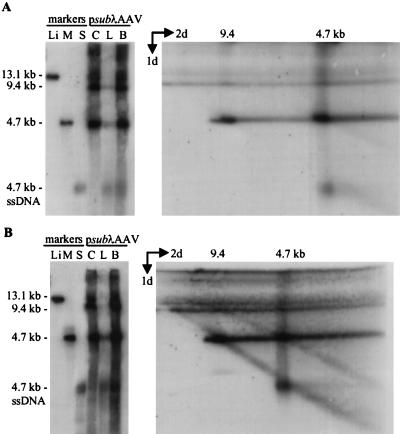
Adenovirus-infected 293 cells were transfected with psubλ-AAV2 and processed 60 h later (Fig. 1B). To detect both single-stranded DNA and double-stranded replicative-form DNA on the same blot, equal amounts of the cell pellet and hypotonic lysate were combined and loaded onto two lanes of a neutral agarose gel. The second lane was cut off and fused to an alkaline agarose gel. After electrophoresis, the DNA was transferred to a nitrocellulose membrane and hybridized to a 32P-labeled probe corresponding to the AAV2 rep and cap genes, which hybridizes only to wild-type DNA (Fig. 7A). As expected, wild-type monomer-length replicative-form DNA separates into three bands under alkaline conditions, which correspond to the extended form (4.7 kb), the turnaround form (9.4 kb), and the covalently closed form, which migrates slightly slower than the 9.4-kb species. The 4.7-kb extended-form band comigrates with wild-type single-stranded DNA in the second dimension; 40% of the wild-type monomer-length replicative-form DNA was turnaround, and 60% was extended DNA (Fig. 7A). A second Southern blot was prepared from the same samples and hybridized to a 32P-labeled probe specific for the λ insert in psubλ-AAV2 (Fig. 7B). Similar to wild type, vsubλ monomer-length replicative-form DNA was comprised of 50% turnaround and 50% extended DNA, indicating that substitution of λ DNA for AAV2 sequences had little effect on the accumulation of extended and turnaround forms. In contrast, when samples from psubλdl1495-transfected cells were subjected to the same analysis, a significant increase in turnaround (70%) relative to extended (30%) DNA was observed (Fig. 8A). Since all of the λ DNA sequence found in psubλdl1495 is also present in the parent vector, psubλ-AAV2, it is likely that this difference is due to the difference in the size of the recombinant genome, rather than due to an inhibitory sequence within the λ insert.
FIG. 7.
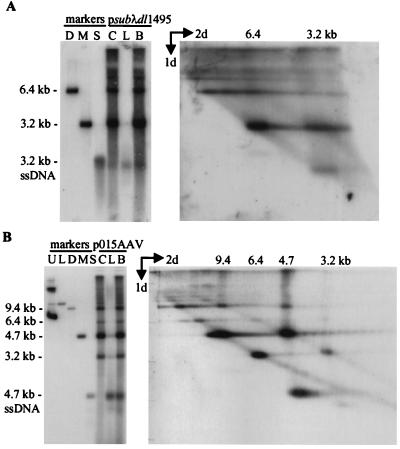
AAV2 recombinant vsubλ accumulates normal proportions of extended and turnaround replicative-form DNAs. Ad5dl312-infected 293 cells were transfected with psubλ-AAV2 and harvested 60 h later; 104 cell equivalents of the cell pellet (lanes C), the hypotonic lysate (lanes L) or the two combined (lanes B) were subjected to electrophoresis in a neutral agarose gel and then analyzed by Southern blotting (left autoradiograms). For two-dimensional electrophoresis (right autoradiograms), a lane identical to lane B was cut off the neutral gel, fused to an alkaline gel, and subjected to electrophoresis. Southern blots were analyzed using a probe corresponding to AAV2 (A) or λ DNA (B). Markers for panel A were ClaI-linearized psubλ-AAV2 (lane Li) and a 4.7-kb PvuII fragment of psub201(−) that was either untreated (lane M) or denatured (lane S). Markers for panel B were ClaI-linearized psubλ-AAV2 (lane Li) and a 4.6-kb SmaI fragment from psubλ-AAV2 that was either untreated (lane M) or denatured (lane S). Markers for the alkaline gel were loaded onto a well at the bottom of the first gel. These markers have been cut off of the autoradiographs, but their positions are identified by size designations above the autoradiograms. For panel A, the markers were a 4.7-kb PvuII fragment of psub201(−) plus a 9.7-kb ClaI-to-BsiWI fragment of psubλ-AAV2. For panel B, the markers were a 4.6-kb SmaI fragment of psubλ-AAV2 plus a 9.7-kb ClaI-BsiWI fragment of psubλ-AAV2. ssDNA, single-stranded.
FIG. 8.
Short recombinant AAV2 genomes are deficient in the accumulation of extended relative to turnaround replicative-form DNA. Ad5dl312-infected 293 cells were transfected with psubλdl1495 (A) or p015-AAV (B), infected cells were harvested 60 h later, and 104 cell equivalents of the cell pellet (lanes C), the hypotonic lysate (lanes L), or a mixture of the two (lanes B) were subjected to electrophoresis in neutral agarose and analyzed by Southern blotting (left autoradiograms). Two-dimensional electrophoresis was carried out as described in the legend to Fig. 7 (right autoradiograms). Southern blots were hybridized to a probe specific for λDNA. Markers for panel A were 6.4-kb SfiI-to-Eco47III fragment of psubλdl1495 (lane D) and a 3.3-kb SmaI fragment of psubλdl1495 that was either untreated (lane M) or denatured (lane S). In panel B, both Southern blots were hybridized to a probe corresponding to the AAV2 terminal repeats. Markers for panel B are undigested p015-AAV (lane U), 11.4-kb p015-AAV NheI-linearized fragment (lane L), a 9.6-kb PpuMI-SwaI fragment from p015-AAV that is similar in size to AAV2 dimer-length replicative-form DNA (lane D), and a 4.7-kb PvuII fragment from psub201(−) that was either left untreated (lane M) or denatured (lane S). Markers for the second dimension were loaded in a well that was originally at the bottom of the first gel. The markers are not shown, but their positions are identified by size designations above the autoradiogram. They were a mixture of a 3.2-kb AseI-FspI fragment of pGET015, a 4.7-kb PvuII fragment from psub201(−), a 6.8-kb NotI-linearized pGET015, and a 9.6-kb PpuMI-SwaI fragment from p015-AAV. ssDNA, single-stranded DNA.
Similar results were obtained with vGET015, which shares no homology with vsubλdl1495 other than the AAV2 terminal repeats but is similar in size (Fig. 8B). Both vGET015 and wild-type AAV2 replicative forms can be visualized in the same blot because of the difference in their sizes. As expected, the majority of the replicative-form DNA was derived from the wild-type genome since vGET015 lacks the cis-acting element needed for efficient accumulation of double-stranded DNA. Approximately 60% of wild-type monomer-length replicative-form DNA was turnaround, and 40% was extended DNA. In this Southern blot, the gel was slightly distorted during transfer, such that the wild-type AAV2 single-stranded DNA appears to be migrating faster than the 4.7-kb extended monomer-length replicative-form DNA. Similar to wild-type monomer-length replicative-form DNA, vGET015 monomer-length replicative-form DNA separates into two bands corresponding to extended (3.2 kb) and turnaround (6.4 kb) DNA. A third band corresponding to vGET015 covalently closed monomer-length replicative-form DNA was not observed. These molecules probably comigrate with vGET015 turnaround monomer-length replicative-form DNA molecules, because the distance separating single-stranded circles and single-stranded linear DNA in the alkaline dimension appears to be inversely proportional to size (Fig. 8B); compare the distance between wild-type covalently closed monomer-length replicative-form and turnaround DNA (9.4 kb) with the distance between vGET015 covalently closed dimer-length replicative-form and turnaround DNA (12.8 kb) and with the distance between wild-type covalently closed and turnaround dimer-length replicative-form DNA (18.8 kb). However, whereas wild-type monomer-length replicative-form DNA was comprised of approximately 60% turnaround and 40% extended DNA, vGET015 monomer-length replicative-form DNA in the same sample was comprised of about 80% turnaround and only 20% extended DNA. As with vsubλdl1495, vGET015 was clearly deficient in the accumulation extended monomer-length replicative-form DNA.
Both vsubλdl1495 and vGET015 were deficient in the accumulation of both extended monomer-length replicative-form DNA and single-stranded DNA, and extended monomer-length replicative-form DNA is presumed to be the immediate precursor to single-stranded DNA (Fig. 9). Consequently, our results suggest that the suboptimal sizes of vsubλdl1495 and vGET015 result in a deficiency in the accumulation extended monomer-length replicative-form DNA, and this in turn leads to a deficiency in the production of single-stranded DNA.
Analysis of wild-type and vGET015 dimer-length replicative-form DNA on the same two-dimensional gel (Fig. 8B) suggests that an additional cis-acting feature of either dimer-length replicative-form DNA or vGET015 may affect the accumulation of extended forms. Similar to monomer-length DNA, AAV2 dimer-length replicative-form DNA separated into a 9.4-kb band, which derives from DNA in the extended form and an 18.8-kb band that derives from DNA that is turnaround in one end. A third band that migrated slower than the 18.8 kb band in the second dimension probably corresponds to DNA that is covalently closed in both ends. Additionally, we observed a 4.7-kb band that probably derives from dimer-length replicative-form DNA that has been nicked in the dimer bridge region. Another band deriving from dimer-length replicative-form DNA is also present but is obscured by the tail of the 18.8-kb band. This band is probably three genomes (14.1 kb) in length and derives from turnaround forms of dimer-length replicative-form DNA that have been nicked in the dimer bridge. About 60% of AAV2 dimer-length replicative-form DNA was turnaround and about 40% was extended form, whereas vGET015 dimer-length replicative-form DNA was >80% turnaround form. Since vGET015 dimer-length replicative-form DNA (6.4 kb) is larger than wild-type monomer-length replicative-form DNA (4.7 kb), this result is not due to the size of the replicating molecule. This difference may be due to a negative-acting cis element in vGET015 but not vsubλ, or due to a difference in the replication of dimer-length relative to monomer-length DNA. For example, dimer-length replicative-form DNA contains two potential replication origins in the middle of the molecule that would be lacking in monomer-length replicative-form DNA.
In addition to wild-type and vGET015 replicative forms, we detected a set of bands that presumably corresponds to linear p015-AAV that is replicating like AAV2 (Fig. 8B). This DNA comigrated with linear p015-AAV (11.4 kb) in the neutral dimension and separated into three bands in the alkaline dimension, similar to extended, turnaround, and circular replicative forms. None of these bands comigrated with the vGET015 or wild-type replicative form in the second dimension, which is consistent with their identity as 11.4-kb linear v015-AAV2.
DISCUSSION
We have identified two elements that are important in the production of recombinant AAV2. First, wild-type AAV2 contains a positive, cis-acting element located between sequence positions 194 and 1882; rAAV2 lacking this region accumulated 14-fold less monomer-length replicative-form DNA than did wild-type AAV2 (Fig. 2 and 4). Second, a genome size of 3.5 kb or greater is required for the efficient production of single-stranded AAV2 DNA. This requirement was best demonstrated using the deletion mutants vdlB and vdlC (Fig. 5A), because they contain the cis-acting element required for efficient accumulation of double-stranded replicative forms. These mutants were both about eightfold deficient in the accumulation of single-stranded DNA (Fig. 5B). Additionally, two recombinant molecules of suboptimal length, vGET015 (3.1 kb) and vsubλdl1495 (3.2 kb), were deficient in the accumulation of extended relative to covalently closed, turnaround replicative form (Fig. 8).
Bacteriophage φX174 has a lower limit (78.5% of genome length) for the production of infectious virus particles (1), similar to the limit observed here for the efficient accumulation of AAV2 single-stranded DNA (75% of wild-type length). φX174 is similar to parvoviruses in that it packages a single-stranded circular DNA into an icosahedral particle, it replicates its DNA by a mechanism similar to that of parvoviruses, and φX174 gene A protein nicks a single strand of DNA and becomes covalently attached to the 5′ end in a fashion analogous to the AAV2 large Rep proteins. The lower limit on φX174 genome length is thought to be due instability of the virions rather than a defect in either the production or packaging of single-stranded DNA (1). As yet, we cannot rule out the possibility that less than optimal-length vectors package single-stranded DNA as efficiently as wild-type AAV2 but are unstable so that single-stranded DNA is quickly degraded or is converted back to double-stranded monomer-length replicative-form DNA. This hypothesis, however, leaves open the question as to why parvovirus particles, which are markedly stable when empty or full, would be particularly unstable when only partially full. Although we have not measured the stability of vGET015 virions directly, the infectivity of vGET015 particles (DNA per infectious unit) was similar to that of wild-type AAV2, suggesting that vGET015 virions are not particularly unstable (G. Tullis and T. Shenk, unpublished data).
Accumulation of single-stranded AAV2 DNA is believed to require sequestration of displaced single-stranded DNA by encapsidation, because little or no single-stranded DNA is made in the absence of capsids (19, 56). Therefore, the amount of single-stranded DNA produced is indicative of the amount of virus produced. Dong et al. (15) found that rAAV DNAs <4.1 kb in length were suboptimal for the production of recombinant virus, which they interpreted as a packaging defect. Our results, however, indicate that the deficiency in production of rAAV from shorter than optimal genomes is actually due to aberrant DNA replication. In our experiments, the amount of double-stranded replicative-form DNA was increased and the amount of single-stranded DNA produced was decreased for shorter than optimal genomes (Fig. 6). Senapathy et al. (48) have made a similar observation using large deletion mutants that derived from naturally occurring defective interfering particles (40 to 50% of wild-type length). These deletion mutants accumulated double-stranded replicative forms but were defective in cis for the accumulation of single-stranded DNA. The lower limit for efficient production of single-stranded DNA may, therefore, be linked to the time it takes a replication fork to proceed from one hairpin to the other.
Current models of parvovirus replication suggest that extended monomer-length replicative-form DNA is the immediate precursor to single-stranded DNA (Fig. 9). Consequently, it seems likely that the defect in the accumulation of single-stranded DNA by small rAAV2 genomes results from a deficiency in the accumulation of extended replicative-form DNA, rather than the other way around. As the replication fork reaches the opposite hairpin, it may induce the isomerization of the hairpin and the initiation of a subsequent round of DNA replication by unwinding the DNA helix. If this occurs before resolution of the turnaround end can occur (Fig. 9, steps 3a and 4a), it would result in the accumulation of more dimer-length replicative-form DNA and higher concatemers (Fig. 9, steps 3b to 6b). A dimer-length replicative-form DNA is then processed into two monomer-length turnaround DNAs by a mechanism similar to terminal resolution. Therefore, the smaller the genome, the more quickly the DNA replication complex will transverse the genome, and this will increase the probability that reinitiation will occur before resolution of the turnaround end (Fig. 9, step 3b versus step 3a). This would generate more dimer-length replicative-form DNA and larger concatemers. We did not observe an increase in the ratio of vGET015 dimer-length to monomer-length replicative-form DNA compared to AAV2 ratio (Fig. 2). Perhaps the processing of dimer-length into monomer-length replicative-form DNA reduces this effect so that we failed to detect it. We did, however, consistently observe an increase in higher-order concatemers of vGET015 replicative-form DNA, up to eight genomes in length (25 kb), in contrast to wild-type AAV2, which is limited to a length of four genomes (18.8 kb) or less (Fig. 2 and 8). With even smaller (0.7- to 2.4-kb) AAV2 DNAs, a ladder of concatemers was observed (G. Tullis, J. LaBonte, and T. Shenk, unpublished data). Thus, the number of concatemers was inversely proportional to the size of the rAAV2 genome, consistent with the above model.
Alternatively, as the replication fork reaches the opposite hairpin, it might displace the 3′ hairpin before the encapsidation reaction is completed (Fig. 9, steps 5a and 6a). The displaced single-stranded DNA may then be converted to a turnaround monomer-length replicative-form DNA molecule, resulting in a surplus of turnaround monomer-length replicative-form DNA relative to extended forms. Thus, the deficiency in extended monomer-length replicative-form DNA might be a result of inefficient or aborted encapsidation. We do not favor this model because the complete absence of encapsidation due to mutations in the capsid gene did not result in abnormal ratios of extended and turnaround DNAs in another parvovirus, minute virus of mice (MVM) (59).
Our first model assumes that terminal resolution is independent of elongation and therefore may occur while elongation is proceeding. Ni et al. (38) have proposed that the two may be linked. This hypothesis is based on their failure to observe DNA on two-dimensional agarose gels with the mobility predicted for intermediates that have been nicked in the terminal palindrome during elongation. A similar observation has been reported for MVM DNA (60). Due to the absence of the predicted DNA intermediates, Ni et al. (38) suggested that terminal resolution occurs only after the completion of the elongation step. This may be due to either the physical state of the DNA during DNA polymerization or masking of the Rep78 binding site by protein factors associated with DNA synthesis. Reduction of genome length may, therefore, disrupt the coordination between completion of elongation and terminal resolution.
Astell and coworkers have identified a tripartite, cis-acting element in MVM DNA that is functionally similar to the element that we have identified in AAV2 (6, 53, 54). This element was mapped to sequence positions 4489 to 4695, which is 213 bp upstream of the MVM polyadenylation site. This is on the opposite end of the genome from the AAV2 element, which we have mapped to between sequence positions 194 to 1882 (Fig. 5). Several cellular factors that bind to MVM DNA within the cis-acting element have been identified (54). However, none of their binding sites is evident in the AAV2 domain containing the cis-acting element, suggesting that these elements might function through the binding of different factors.
What might bind to the AAV2 cis-acting element that enhances the accumulation of double-stranded replicative-form DNA? The AAV2 domain between sequence positions 194 and 1882 includes all three AAV2 promoters (Fig. 5A), and consequently it contains numerous binding sites for cellular factors, which may play a dual role in transcription and replication. The SV40 origin, for example, contains six Sp1 binding sites that enhance both DNA replication and transcription (18). Alternatively, the virus-encoded Rep78 and -68 proteins might play a role. These proteins are essential for DNA replication (19, 56), for repression of the P5 and P19 promoters (3, 22, 28, 29, 30, 41, 57), and for transactivation of the P19 and P40 promoters (30, 33, 41, 42, 57). In addition to the binding sites in the AAV2 terminal repeats (10, 23, 35, 40), Rep binds to a high-affinity binding site between the TATA box and the transcription initiation site in P5 and to two lower-affinity sites upstream of P19 (11, 34).
Senapathy and Carter (47) observed that the 3′ end of the capsid gene, which is located between sequence positions 2944 and 4680, tends to be preferentially retained in defective interfering genomes. However, the cis-acting element that we have identified maps to the other end of the genome. The retention of the capsid gene in defective particles may be due to recombination hot spots in this region that increase the probability that the deletion junction will occur within this region of the genome.
Kestler et al. (27) identified in the cap open reading frames of MVM and the closely related virus H1 an approximately 800-bp cis element that is required for the efficient production of recombinant virus. Unlike the AAV2 element, this domain is not required for the efficient accumulation of MVM or H1 replicative-form DNA but is presumably required for either the production or the encapsidation of single-stranded DNA. The inclusion of this region of MVM recombinants resulted in up to a 50-fold increase in the yield.
Using a monoclonal antibody that recognizes full capsids, Grimm et al. (17) found that wild-type AAV2 particles are approximately 50% empty and 50% full, whereas greater than 80% of recombinant AAV2 particles are empty, leading to the suggestion that the encapsidation of recombinant genomes may be inefficient. Our data suggest that the excess empty capsids in rAAV2 stocks probably result from a DNA replication defect rather than encapsidation. If such a packaging element exists, it has only a modest (<2-fold) effect on single-stranded DNA production, because vsubλ accumulated single-stranded DNA and monomer-length replicative-form DNA in a ratio similar to wild-type AAV2 (Fig. 4).
More precise localization of the cis-acting element that we have identified within sequence positions 194 to 1882 should facilitate the construction of rAAV2 vectors that replicate more efficiently than current vectors.
ACKNOWLEDGMENTS
We thank J. LaBonte for help in the construction of pGET015.
This work was supported by grant CA38965 from the National Cancer Institute.
REFERENCES
- 1.Aoyama A, Hayashi M. Effects of genome size on bacteriophage φX174 DNA packaging in vitro. J Biol Chem. 1985;260:11033–11038. [PubMed] [Google Scholar]
- 2.Balague C, Kalla M, Zhang W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the Rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, editor. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Raven Press; 1996. pp. 2173–2197. [Google Scholar]
- 5.Brister J R, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunstein J, Astell C R. Analysis of the internal replication sequence indicates that there are three elements required for efficient replication of minute virus of mice minigenomes. J Virol. 1997;71:9087–9095. doi: 10.1128/jvi.71.12.9087-9095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassinotti P, Weitz M, Tratschin J D. Organization of the adeno-associated virus (AAV2) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology. 1988;167:176–184. [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. Palindromic base sequences and replication of eukaryotic chromosome ends. Nature (London) 1974;250:467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- 9.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 10.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini J A, Yang L, Safer B, Kotin R M. Determination of adeno-associated virus Rep68 and Rep78 binding site by random sequence oligonucleotide selection. J Virol. 1995;69:7334–7338. doi: 10.1128/jvi.69.11.7334-7338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chirico J, Trempe J P. Optimization of packaging of adeno-associated virus gene therapy vectors using plasmid transfections. J Virol Methods. 1998;76:31–41. doi: 10.1016/s0166-0934(98)00120-7. [DOI] [PubMed] [Google Scholar]
- 13.Conway J E, Zolotukhin S, Muzyczka N, Hayward G S, Bryne B J. Recombinant adeno-associated virus type 2 replication and packaging is entirely supported by a herpes simplex virus type 1 amplicon expressing Rep and Cap. J Virol. 1997;71:8780–8789. doi: 10.1128/jvi.71.11.8780-8789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Tattersall P. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J Y, Fan P D, Frizzell R A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 16.Fan P D, Dong J Y. Replication of rep-cap genes is essential for the high-efficiency production of recombinant AAV2. Hum Gene Ther. 1997;8:87–98. doi: 10.1089/hum.1997.8.1-87. [DOI] [PubMed] [Google Scholar]
- 17.Grimm D, Kern A, Pawlita M, Ferrari F K, Samulski R J, Kleinschmidt J A. Titration of AAV2–2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV2–2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 18.Guo Z-S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holscher C, Horer M, Kleinschmidt J A, Zentgraf H, Burkle A, Heilbronn R. Cell lines inducibly expressing the adeno-associated virus (AAV2) rep gene: requirements for productive replication of rep-negative AAV2 mutants. J Virol. 1994;68:7169–7177. doi: 10.1128/jvi.68.11.7169-7177.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holscher C, Kleinschmidt J A, Burkle A. High-level expression of adeno-associated virus (AAV2) Rep78 or Rep68 protein is sufficient for infectious particle formation by a rep-negative mutant. J Virol. 1995;69:6880–6885. doi: 10.1128/jvi.69.11.6880-6885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im D-S, Muzyczka N. The AAV2 origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 24.Inoue N, Russell D W. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston K M, Jacoby D, Pechan P A, Fraefel C, Borghesani P, Schuback D, Dunn R J, Smith F I, Breakefeild X O. HSV/AAV2 hybrid amplicon vectors extend transgene expression in human glioma cells. Hum Gen Ther. 1997;8:359–370. doi: 10.1089/hum.1997.8.3-359. [DOI] [PubMed] [Google Scholar]
- 26.Jones N, Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 27.Kestler J, Neeb B, Struyf S, Van Damme J, Cotmore S F, D'Abramo A, Tattersall P, Rommelaere J, Dinsart C, Cornelis J J. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum Gene Ther. 1999;10:1619–1632. doi: 10.1089/10430349950017626. [DOI] [PubMed] [Google Scholar]
- 28.Kyöstiö S R M, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV2) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV2 p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyöstiö S R, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV2 Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Samulski R J, Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X L, Clark K R, Johnson P R. Production of recombinant adeno-associated virus vectors using a packaging cell line and a hybrid recombinant adenovirus. Gene Ther. 1999;6:293–299. doi: 10.1038/sj.gt.3300807. [DOI] [PubMed] [Google Scholar]
- 33.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarty D M, Pereira D J, Zolothkhin I, Zhou Z, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monahan P E, Samulski R J. AAV2 vectors: is clinical success on the horizon? Gene Ther. 2000;7:24–30. doi: 10.1038/sj.gt.3301109. [DOI] [PubMed] [Google Scholar]
- 37.Morgenstern J P, Land H. Advanced mammalian gene transfer high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1992;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogasawara Y, Mizukami H, Urabe M, Kume A, Kanegae Y, Saito I, Monahan J, Ozawa K. Highly regulated expression of adeno-associated virus large Rep proteins in stable 293 cell lines using the Cre/loxP switching system. J Gen Virol. 1999;80:2477–2480. doi: 10.1099/0022-1317-80-9-2477. [DOI] [PubMed] [Google Scholar]
- 40.Owens R A, Weitzman M D, Kyöstiö S R M, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV2) Rep protein acts as both a repressor and an activator to regulate AAV2 transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira D J, Muzyczka N. The adeno-associated virus type 2 p40 promoter requires a proximal Sp1 interaction and a p19 CArG-like element to facilitate Rep transactivation. J Virol. 1997;71:4300–4309. doi: 10.1128/jvi.71.6.4300-4309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samulski R J, Berns K I, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samulski R J, Chang L S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlehofer J R, Ehrbar M, zur Hausen H. Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology. 1986;152:110–117. doi: 10.1016/0042-6822(86)90376-4. [DOI] [PubMed] [Google Scholar]
- 47.Senapathy P, Carter B J. Molecular cloning of adeno-associated virus variant genomes and generation of infectious virus by recombination in mammalian cells. J Biol Chem. 1984;259:4661–4666. [PubMed] [Google Scholar]
- 48.Senapathy P, Tratschin J D, Carter B J. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep- mutants by a wild-type genome or an ori- mutant and correction of terminal palindrome deletions. J Mol Biol. 1984;179:1–20. doi: 10.1016/0022-2836(84)90303-6. [DOI] [PubMed] [Google Scholar]
- 49.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 50.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straus S E, Sebring E, Rose J A. Concatemers of alternating plus and minus strands are intermediates in adeno-associated virus DNA synthesis. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam P, Astell C R. Replication of minute virus of mice minigenomes: novel replication elements required for MVM DNA replication. Virology. 1993;193:812–824. doi: 10.1006/viro.1993.1190. [DOI] [PubMed] [Google Scholar]
- 54.Tam P, Astell C R. Multiple cellular factors bind to cis-regulatory elements found inboard of the 5′ palindrome of minute virus of mice. J Virol. 1994;68:2840–2848. doi: 10.1128/jvi.68.5.2840-2848.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tattersall P, Ward D C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature (London) 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 56.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tratschin J D, Tal J, Carter B J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral rep gene product. Mol Cell Biol. 1986;6:2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trempe J P, Carter B J. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J Virol. 1988;62:3356–3363. doi: 10.1128/jvi.62.9.3356-3363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tullis G E, Burger L R, Pintel D J. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol. 1993;67:131–141. doi: 10.1128/jvi.67.1.131-141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tullis G, Schoborg R V, Pintel D J. Characterization of the temporal accumulation of minute virus of mice replicative intermediates. J Gen Virol. 1994;75:1633–1646. doi: 10.1099/0022-1317-75-7-1633. [DOI] [PubMed] [Google Scholar]
- 61.Vincent K A, Piraino S T, Wadsworth S C. Analysis of recombinant adeno-associated virus packaging and requirements for rep and cap gene products. J Virol. 1997;71:1897–1905. doi: 10.1128/jvi.71.3.1897-1905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71:2722–2730. doi: 10.1128/jvi.71.4.2722-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X S, Ponnazhagan S, Srivastava A. Rescue and replication signals of the adeno-associated virus type 2 genome. J Mol Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 64.Wang X S, Ponnazhagan S, Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wonderling R S, Kyostio S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yakinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 67.Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X, Zolotukhin I, Im D S, Muzyczka N. Biochemical characterization of adeno-associated virus rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]