Abstract
Ribonucleotide reductase (RNR) is an essential enzyme for the de novo synthesis of both cellular and viral DNA and catalyzes the conversion of ribonucleoside diphosphates into the corresponding deoxyribonucleoside diphosphates. The enzyme consists of two nonidentical subunits, termed R1 and R2, whose expression is very low in resting cells and maximal in S-phase cells. Here we show that murine cytomegalovirus (MCMV) replication depends on ribonucleotide reduction since it is prevented by the RNR inhibitor hydroxyurea. MCMV infection of quiescent fibroblasts markedly induces both mRNA and protein corresponding to the cellular R2 subunit, whereas expression of the cellular R1 subunit does not appear to be up-regulated. The increase in R2 gene expression is due to an increase in gene transcription, since the activity of a reporter gene driven by the mouse R2 promoter is induced following virus infection. Cotransfection experiments revealed that expression of the viral immediate-early 1 protein was sufficient to mediate the increase in R2 promoter activity. It was found that the viral gene M45, encoding a putative homologue of the R1 subunit, is expressed 24 and 48 h after infection. Meanwhile, we observed an expansion of the deoxyribonucleoside triphosphate pool between 24 and 48 h after infection; however, neither CDP reduction nor viral replication was inhibited by treatment with 10 mM thymidine. These findings indicate the induction of an RNR activity with an altered allosteric regulation compared to the mouse RNR following MCMV infection and suggest that the virus R1 homologue may complex with the induced cellular R2 protein to reconstitute a new RNR activity.
The replication of both cellular and DNA virus genomes requires a balanced supply of deoxyribonucleoside triphosphates (dNTPs). In eukaryotic cells, conversion of ribonucleoside diphosphates to the corresponding deoxyribonucleoside diphosphates is catalyzed by ribonucleotide reductase (RNR), the rate-limiting enzyme in DNA precursor biosynthesis (56, 60, 61). Ribonucleotide reduction is the first of a series of metabolic reactions leading to DNA synthesis and as such is controlled at several levels. The same enzyme reduces all four ribonucleotides, and both substrate specificity and overall activity are tightly controlled by binding of NTP allosteric effectors. Substrate specificity is controlled by binding of ATP or dATP (CDP/UDP reduction), dTTP (GDP reduction), or dGTP (ADP reduction) to a specificity site in the R1 protein, while overall activity is controlled by binding ATP (activation) or dATP (inactivation) to an activity site (39). The activity of RNR is cell cycle regulated and is very low or not detectable in resting cells and maximal in S-phase cells (56, 61). This is controlled both at the level of transcription and by regulation of protein stability (6, 13, 22, 24).
Three RNR classes have been characterized based on the mechanism for generation of the protein radical, metal cofactor requirement, and subunit composition (39). Human cells, like most eukaryotic cells, contain a class Ia RNR. This form also exists in some prokaryotes, e.g., the well-studied nrdA/nrdB encoded enzyme of Escherichia coli. Class Ia has an α2β2 form of RNR consisting of two homodimeric subunits, proteins R1 (α2) and R2 (β2). The R1 protein is the business end of the enzyme and contains the active site and the binding sites for allosteric effectors. The R2 protein is a radical storage device containing an iron center-generated tyrosyl free radical.
Among the Herpesviridae family, several alpha- and gammaherpesviruses, including herpes simplex virus type 1 (HSV-1), HSV-2, varicella-zoster virus, Epstein-Barr virus, pseudorabies virus, and equine herpesviruses 1, 3, and 4, induce a novel, distinct RNR activity (4, 17, 19, 35, 43). The viral enzyme may be required for virus growth in nondividing cells and for viral pathogenesis and reactivation from latency in infected hosts (12, 20, 28, 29, 34, 37). The HSV-1 RNR enzyme is the most extensively characterized and, like the mammalian and E. coli enzymes, belongs to class Ia. However, it differs from the cellular enzyme in that it completely lacks allosteric regulation as well as most of the residues involved in effector binding in the E. coli and mammalian enzymes at both the activity and specificity sites (16, 42). Therefore, CDP reduction by the HSV RNR is not inhibited by dTTP or dATP, as it is for the mammalian RNR. Furthermore, the N-terminal end of the HSV R1 protein contains a transmembrane helical segment followed by a Ser/Thr protein kinase (18).
Analysis of the protein-coding content of the human and murine cytomegalovirus (HCMV and MCMV) genomes reveals the presence of an open reading frame (ORF), termed UL45 and M45, respectively (14, 55), which shows homology to the R1 subunit of other herpesviruses. For instance, sequence alignment of UL45 or M45 to that of HSV-1 R1, chosen as a representative of herpesvirus R1 proteins, reveals a 25 and a 22% amino acid identity, respectively. However, since the putative HCMV and MCMV R1 subunit lacks certain amino acid residues that are believed to be critical for enzymatic function and are highly conserved among the R1 proteins of other class Ia RNRs, it is not clear whether it acts as an enzyme subunit. One such structural element is the redox-active dithiol on the flexible C-terminal tail of other class Ia R1 proteins, where the CMV R1 has only one cysteine residue.
Like other betaherpesviruses, such as human herpesvirus 6 (HHV-6) and HHV-7 CMV genomes do not carry an ORF for the R2 subunit. It follows that these viruses do not express a functional RNR enzyme.
HCMV and MCMV efficiently replicate in vitro in growth-arrested fibroblasts (21, 44). Since the dNTP concentrations are very low in nondividing cells and limit viral replication, it is still unknown how HCMV and MCMV ensure a sufficient supply of dNTPs to their polymerase in the absence of a functional RNR enzyme. To solve this paradox, one may hypothesize that during their evolution CMV have acquired the ability to force a quiescent cell to express the R1 and R2 subunits of the cellular RNR. Alternatively, the virally encoded R1 subunit may complex with the virus-induced cellular R2 subunit to reconstitute a functional enzyme. A third possible explanation would be salvage of the neccessary deoxynucleosides.
This paper addresses these questions by evaluating the expression and activity of the cellular RNR in quiescent cells during MCMV infection.
MATERIALS AND METHODS
Cells and culture conditions.
NIH 3T3 murine fibroblasts were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% calf serum (Gibco-BRL). Quiescent NIH 3T3 cells (arrested in the G0/G1 phase) were obtained by culturing the subconfluent cultures for 48 h in DMEM medium plus 0.5% calf serum (low-serum medium). Flow cytometry at this time demonstrated that more than 90% of the cells arrested in G0/G1.
Virus preparation and infections.
MCMV (mouse salivary gland virus, strain Smith; ATCC VR.194) was purchased from the American Type Culture Collection (Rockville, Md.). Virus stocks were first produced in salivary glands of BALB/c mice and then propagated in vitro by infecting C57BL/6 mouse embryo fibroblasts (C57BL/6-MEF) at a virus-to-cell ratio of 0.01. Cells were incubated in DMEM supplemented with 2% heat-inactivated calf serum, and virus was harvested by sonication, depending on the cytopathology, at about 1 week postinfection and clarified by centrifugation. Mock-infected fluid was prepared from C57BL/6-MEF by the procedure used to prepare MCMV. A virus stock solution containing approximately 107 PFU/ml (as determined by plaque assay on the B6MEF cell line, an embryonic fibroblast cell line derived from C57BL/6 mice and immortalized through several culture passages) was used in all experiments.
For RNA and protein level determinations, transfection assays, and enzyme assays, quiescent NIH 3T3 cells were infected with MCMV at a multiplicity of infection (MOI) of 5 PFU/cell unless otherwise stated. Mock-infected control cultures were exposed to an equal volume of mock-infecting fluid. Virus adsorptions were carried out for 1 h at 37°C, and 0 h postinfection (p.i.) was defined as the time immediately following this period. At the end of the adsorption, the low-serum medium removed from the cells before infection was returned to the plates to avoid any cellular stimulation that could have resulted from the addition of fresh serum growth factors.
Inactivation of virus by UV light.
MCMV stock or mock-infecting fluid in an uncovered 60-mm-diameter dish was placed in a UV linker (Pbi International) and irradiated with one pulse of UV light at 0.6 J/cm2. Preliminary experiments demonstrated that under these conditions no MCMV gene expression could be demonstrated in UV-irradiated MCMV-infected NIH 3T3 cells (44). The virus stock or the mock-infecting fluid was irradiated just prior to use and then placed on ice. To minimize light exposure and prevent light-induced repair mechanisms, irradiated stocks were kept covered with aluminum foil and infections were performed in the absence of fluorescent lights.
Plasmids.
pET28a(+)R2(+) contains the human R2 cDNA cloned into the E. coli expression vector pET28a(+) (Novagen). p3I contains a fragment (nucleotides 113 to 2825) of the human R1 cDNA. pGL3R2 1.5 contains a 1,517-bp PvuII-to-PvuII fragment of the mouse R2 promoter (nucleotides-1500 to +17 relative to the major transcription start) linked to the luciferase coding region of pGL3 (Promega) (13). pGL3R1 5.7 contains the mouse R1 promoter linked to the luciferase coding region of pGL3 (38). pCMVCAT contains a 1.2kb PstI-NdeI segment from HindIII fragment L of MCMV DNA, positioned upstream from the bacterial chloramphenicol acetyltransferase (CAT) reporter gene of pSVOCAT. The viral genomic segment contains the immediate-early (IE) enhancer and the IE1/3 promoter of MCMV (32).
pIE100/1 and pIE3 contain MCMV genome fragments which encode the pIE1 and pIE3 proteins, respectively. Their expression is driven by the MCMV IE enhancer and the IE1/3 promoter (50).
Transient-transfection and reporter gene assays.
All plasmids were purified by cesium chloride centrifugation. For transient gene expression assay, the day before transfection cells were plated in growth medium at a density of 2 × 105 cells/60-mm-diameter dish. The medium was changed 4 h before transfection. The cells were transfected by the calcium phosphate procedure, and the amount of DNA of each transfection was standardized to 12 μg with carrier DNA (the inert pBluescript SK plasmid) (Stratagene). The DNA-calcium precipitates were added to the culture medium, and the cells were incubated for 18 h. Thereafter, the transfectants were washed twice with medium and incubated for 48 h in DMEM supplemented with 0.5% calf serum. To measure the luciferase activity, the cells were washed twice with phosphate-buffered saline (PBS), scraped from the plates in PBS containing 1 mM EDTA, and collected by centrifugation. The pellets were resuspended in 100 μl of reporter lysis buffer (Promega), and soluble proteins were recovered after centrifugation. Supernatants were quantified for protein concentration, and aliquots were assayed with 100 μl of luciferine substrate (Promega) in a 1600CA Tri-Carb liquid scintillation analyzer (Packard). Reporter gene activity was normalized to the amount of plasmid DNA introduced into recipient cells by DNA dot blot analysis as described by Abken and Reifenrath (1).
Preparation of RNA and Northern analysis.
At the indicated times, cells were rinsed twice with ice-cold PBS and total cellular RNA was isolated by homogenization in 4 M guanidium isothiocyanate and centrifugation through a 5.7 M cesium chloride cushion, as described by Chirgwin et al. (15).
Total RNA (30 μg) was fractionated on a 1% agarose–2.2 M formaldehyde gel and then blotted onto nitrocellulose membrane (Hybond C-Super; Amersham). The filters were baked for 2 h at 80°C and prehybridized for 4 h at 42°C in 50% formamide–750 mM NaCl–48.5 mM Na2HPO4–5 mM EDTA (pH 7.4)–2× Denhardt's solution–0.1% sodium dodecyl sulfate (SDS)–200 μg of denatured salmon sperm DNA per ml. The hybridizations were carried out at 42°C overnight with denatured probes at 106 cpm/ml. The filters were then washed twice for 30 min at room temperature with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) –0.1% SDS and twice for 30 min at 42°C with 0.5× SSC–0.1% SDS. After autoradiography, the hybridization signals were quantitated by densitometric scanning.
Northern blot analysis was performed with random-primed radiolabeled probes corresponding to (i) a 1.8-kb BamHI segment of human R1 cDNA, (ii) a 703-kb EcoRI-EcoRV fragment of human R2 cDNA, and (iii) the mouse glyceraldehyde-3-phosphate dehydrogenase (G3PDH) full-length cDNA. The full-length M45 gene was obtained by PCR amplification of MCMV DNA and completely sequenced.
RT-PCR analysis of MCMV M45 transcription.
Reverse transcriptase PCR (RT-PCR) was employed to analyze the transcription of MCMV M45 following MCMV. Total cellular RNA isolated and purified as described above was treated with RNase-free DNase, repurified, and quantitated spectrophotometrically. A 2-μg quantity of RNA was retrotranscribed at 42°C for 60 min in PCR buffer (1.5 mM MgCl2) containing 5 μM random primers, 0.5 mM each dNTP, and 100 U of Moloney murine leukemia virus reverse transcriptase (Ambion) in a final volume of 20 μl. The resulting cDNAs were amplified with the following primers for MCMV M45: upstream primer, 5′ ATG GCT CGC ATC CGC CGC TAC-3′; downstream primer, 5′ GGC CGA GTA GAA CTG AGC GCG-3′. The following primers were used for β-actin: upstream primer, 5′ TGG AAT CCT GTG GCA TCC ATG AAA-3′; downstream primer, 5′ TAA AAC GCA GCT CAG TAA CAG TCC-3′. Amplification was performed at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for a total of 30 cycles, and the products were analyzed by agarose gel electrophoresis (2% agarose).
Expression of recombinant R2 in E. coli and generation of a rabbit antiserum.
The human R2 subunit was expressed in E. coli BL21(DE3) transformed with pET28a(+)R2(+) as a fusion protein tagged with six residues of histidine and an 11-amino-acid sequence from the T7 capsid protein. Purification of the recombinant R2 and rabbit immunization were performed as described elsewhere (41). The sera were obtained after bleeding at 1 week after the fourth immunization and precipitated with ammonium sulfate at 45% saturation. The precipitates were then resuspended in PBS and further purified on a protein A affinity column (Pharmacia) as specified by the manufacturer.
Preparation of protein extracts and immunoblotting.
Whole-cell extracts were prepared by resuspending pelletted cells in lysis buffer containing 125 mM Tris-HCl (pH 6.8), 3% SDS, 20 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 4 μg of leupeptin per ml, 4 μg of aprotinin per ml, and 1 μg of pepstatin per ml. After a brief sonication, soluble proteins were collected by centrifugation at 15,000 × g. Supernatants were quantified for protein concentration with a Dc protein assay kit (Bio-Rad Laboratories) and stored at −70°C in 10% glycerol. For immunoblotting, after SDS-polyacrylamide gel electrophoresis (PAGE) the proteins were transferred to Immobilon-P membranes (Millipore). The filters were then blocked in 5% nonfat dry milk in 10 mM Tris-HCl (pH 7.5)–100 mM NaCl–0.1% Tween 20 and immunostained with the anti-R1 monoclonal antibody AD203 (49), the anti-R2 polyclonal antibodies, the anti-MCMV IE1 polyclonal antibodies (27), or the anti-actin mouse monoclonal antibody (Boehringer). Immune complexes were then detected by means of sheep anti-mouse immunogloblin or goat anti-rabbit immunoglobulin antibodies, both conjugated to horseradish peroxidase (Amersham), and visualized by using enhanced chemioluminescence (Super Signal; Pierce) as specified by the manufacturer.
Cytotoxicity assay.
Cells were grown to subconfluence in 24-well plates and then incubated in low-serum medium for 48 h. Thereafter the medium was replaced by low-serum medium containing increasing concentrations of hydroxyurea (HU) (Sigma). After 48 h, cell viability was detemined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method, as previously described (53).
Inhibition of viral replication and DNA synthesis.
Inhibition of viral replication was determined in cells grown to subconfluence in 24-well plates and then incubated in low-serum medium for 48 h. Thereafter they were infected with MCMV at a MOI of 1 PFU/cell. One column per plate was mock infected and served as a cell control. The infected cultures were treated in low-serum medium with increasing concentrations of HU or thymidine (TdR) (Sigma) in duplicate wells. One column per plate was left untreated and served as a virus control. Cultures were incubated until the control cultures displayed an evident cytopathology. Thereafter, the cells and the supernatants from the anti-CMV assay were harvested and disrupted by sonication. The disrupted cells were centrifuged at 500 × g for 10 min, and the supernatant was assayed for infectivity by a standard plaque assay for MCMV on the B6MEF cell line. The number of plaques was plotted as a function of drug concentration, and the concentration producing 50% reduction in plaque formation, i.e., the 50% effective concentration (EC50) was determined.
To evaluate the inhibition of MCMV DNA synthesis, cells were grown to subconfluence in six-well plates and then incubated in low-serum medium for 48 h. Thereafter the cells were infected with MCMV at a MOI of 1 PFU/cell. One well per plate was mock infected and served as a cell control. The infected cultures were treated in low-serum medium with different concentrations of HU or TdR. One well per plate was not treated and served as a virus control. At 48 h p.i., the cells were harvested and total DNA was isolated by resuspending cell pellets in lysis buffer (10 mM Tris-HCl [pH 8.0], 25mM EDTA, 100 mM NaCl, 0.5% SDS, 100 μg of proteinase K per ml) and incubating the mixtures at 50°C for 18 h. The digestion was then followed by phenol-chloroform extraction, ethanol precipitation, and RNase treatment (1 μg of RNase A per ml for 1 h at 37°C). Two-fold dilutions of the DNA samples were then immobilized on a Zeta-Probe hybridization membrane (Bio-Rad). DNA samples were sequentially hybridized with a 32P-labeled 1,104-bp XbaI-AvaI DNA fragment which contains a portion of the fourth exon of the MCMV IE1 gene and with a 32P-labeled mouse G3PDH full-length cDNA. The membranes were autoradiographed, and hybridization signals were quantitated with the Bio-Rad molecular imaging analysis system.
Determination of nucleotide pools in MCMV and mock-infected quiescent NIH 3T3 cells.
Cell cultures with or without 10 mM thymidine were extracted with ice-cold trichloroacetic acid. The extracted nucleotides were separated directly by high-pressure liquid chromatography (NTPs) or first run through a borate affinity column (dNTPs) as described by Hofer et al. (36). The nucleotide pools are given as percentages of the total NTP pool (CTP + UTP + ATP + GTP + dCTP + dTTP + dATP + dGTP) to minimize variations due to small differences in cell numbers in the samples.
RNR assay.
MCMV- or mock-infected quiescent NIH 3T3 cells were extracted as described previously (2). The crude extracts (600 μg of protein) were assayed for reduction of [3H]CDP to [3H]dCDP at 37°C as described previously (23) after the addition of an excess of pure recombinant mouse R2 protein (47) (5 μg) to each assay tube.
RESULTS
Both MCMV replication and DNA synthesis are blocked by an RNR inhibitor.
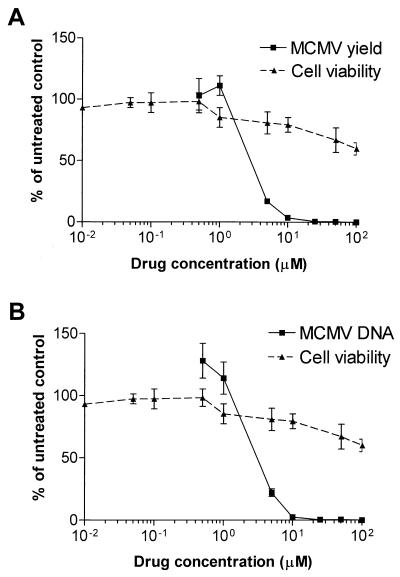
Since the MCMV genome does not encode a functional RNR, we asked whether ribonucleotide reduction is necessary for MCMV replication in quiescent cells. To address this question, we examined the effect of the RNR inhibitor HU on MCMV replication and DNA synthesis. Quiescent cells were infected at a MOI of 1 PFU/cell, and low-serum medium containing HU was added after virus adsorption, to give final concentrations of 0.5 to 100 μM. Culture supernatants collected 4 days after infection were assayed for virus yield on B6MEF cells. As shown in Fig. 1A, HU produced a significant dose-related reduction of MCMV yield at concentrations well below those producing cytotoxic effects. The calculated EC50 and EC90 were 3 and 8 μM, respectively. Cell toxicity assays demonstrated that at these HU concentrations the viability of quiescent mock-infected cells was about 80% and the 50% cytotoxic concentration was >100 μM. This finding indicates that the inhibitory activity of HU on MCMV replication was not due to its generalized cytoxicity.
FIG. 1.
Inhibitory effect of HU on MCMV replication (A) and DNA synthesis (B). Quiescent NIH 3T3 cells were infected at a MOI of 1 PFU/cell and then exposed to increasing HU concentrations for 4 days. (A) Supernatants of cell suspension were assayed for their infectivity by a plaque reduction assay. (B) Total genomic DNA was purified and immobilized on a hybridization membrane by a dot blot apparatus. The same filter was sequentially hybridized with a 32P-labeled viral probe (a 1,104-bp AvaI-XbaI fragment in the fourth exon of the MCMV IE1 gene) and a cell DNA probe (full-length murine G3PDH cDNA). The hybridization signals were quantitated with the Bio-Rad image analysis system. Values are the means of three determinations.
To evaluate the effects of HU on MCMV DNA synthesis, intracellular viral DNA levels were quantified 48 h after infection by dot blot analysis and hybridization with a radiolabeled viral probe. As shown in Fig. 1B, HU treatment resulted in a strong reduction of MCMV DNA levels, with an EC50 of about 3 μM.
These results indicate that MCMV replication and DNA synthesis depend on ribonucleotide reduction in quiescent cells.
Differential regulation of cellular RNR gene expression in quiescent NIH 3T3 cells infected with MCMV.
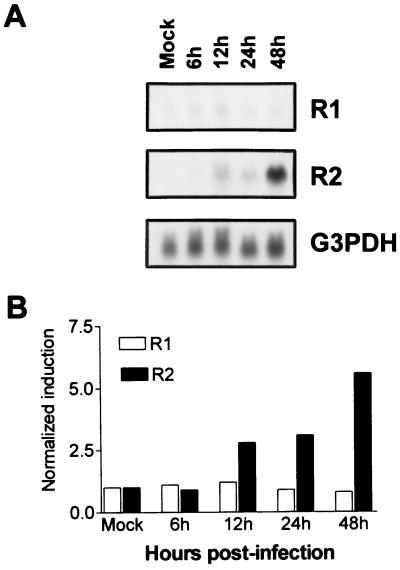
Previous studies demonstrated that MCMV induces the expression of cell nucleotide metabolic enzymes required for virus replication (30, 44). To investigate whether MCMV infection regulates cellular RNR gene expression, serum-depleted NIH 3T3 cells were infected with MCMV (at a MOI of 5 PFU/cell) and total RNA was purified at different times p.i. Both the R1 and R2 mRNA levels were then analyzed by Northern blotting and normalized to the G3PDH mRNA levels. As shown in Fig. 2, the R2 mRNA increased during infection to a maximum level at 48 h p.i. By contrast, the R1 mRNA level was not significantly modified.
FIG. 2.
Steady-state levels of R1 and R2 mRNA during the course of MCMV infection. (A) NIH 3T3 cells were growth arrested in 0.5% calf serum for 48 h and then infected with MCMV (MOI, 5 PFU/cell) or mock infected. Total RNA was isolated at the indicated times after infection and analyzed by Northern blotting. The filter was sequentially hybridized with cellular R1 and R2 and G3PDH radiolabeled probes. (B) The hybridization signals were quantitated with the Bio-Rad image analysis system. The increase in the R1 or R2 mRNA content was calculated by normalizing the amount of radioactivity corresponding to R1 or R2 mRNA to that of G3PDH mRNA (internal control) to correct for differences in RNA loading and recovery. The value at each time point was then normalized to the that observed with mock-infected cells, which was set at 1.
To determine whether viral infection of quiescent cells would lead to a corresponding effect on the expression of the cellular R1 and R2 proteins, cell extracts were prepared at different times p.i. and analyzed by immunoblotting using anti-R1 monoclonal antibodies or anti-R2 polyclonal monospecific antibodies (Fig. 3). As expected, R1 protein was present at very low levels in mock-infected cells and at much higher levels in serum-stimulated cells. As observed at the mRNA level, MCMV infection did not significantly modulate the levels of R1 protein. By contrast, R2 protein was undetectable in mock-infected cells, and its level began to increase at 12 h p.i., and peaked between 24 and 48 h p.i. As expected, serum stimulation induced R2 protein expression in uninfected cells. To verify that the induction detected in infected cells was due to viral gene expression, cells were also infected with UV-inactivated virus and expression of the viral immediate-early protein 1 pIE1 was used as a marker of infectivity. A UV pulse of 0.6 J/cm2 completely abolished pIE1 expression. When the same extract was probed with the anti-R1 or anti-R2 antibodies, no R1 or R2 protein induction was detected, indicating that a potential serum contamination of viral preparations or binding and entry of the inactivated virus particles are not responsible for R2 induction. From these experiments, we conclude that MCMV infection differentially modulates R1 and R2 protein expression and that R2 induction is dependent on virus gene expression.
FIG. 3.
Cellular R1 and R2 protein levels during the course of MCMV infection. NIH 3T3 cells were growth arrested in 0.5% calf serum and then infected with MCMV (MOI, 5 PFU/cell) or UV-irradiated MCMV (MOI, 5 PFU/cell) or mock infected. Total-cell extracts were prepared at the indicated times after infection, fractionated by SDS-PAGE (50 μg of protein/lane), and analyzed by immunoblotting with the anti-R1 monoclonal antibody, with the anti-R2 serum, and with the anti-IE1 serum as described in Materials and Methods. Actin immunodetection with a monoclonal antibody was performed as an internal control. A sample from quiescent cells stimulated with 10% calf serum for 24 h was also run.
Effect of MCMV infection on cell R1 and R2 gene promoters in quiescent NIH 3T3 cells.
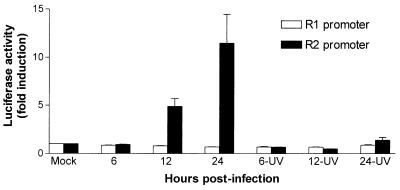
To determine whether the differential modulation of cellular R1 and R2 gene expression by MCMV correlated with a corresponding effect on the respective gene promoters, we analyzed the effects of MCMV infection on the expression of the transiently transfected luciferase reporter gene driven by the R1 or R2 promoters. After transfection, cells were serum starved and then infected with MCMV or UV-inactivated virus. At different times p.i., cell extracts were prepared and assayed for luciferase activity. As shown in Fig. 4, MCMV infection resulted in a time-dependent stimulation of the R2 gene promoter, which began at 12 h p.i. (about 5-fold induction) and reached a maximum level at 24 h p.i. (11-fold induction). By contrast, R1 promoter activity was not affected by MCMV infection. In accord with the Western blot analysis, UV-inactivated virus did not increase luciferase activity driven by the R2 promoter, demonstrating that MCMV-mediated trans-activation requires viral gene expression.
FIG. 4.
Effect of MCMV infection on R1 and R2 promoter activity. DNA (2 μg) from constructs pGL3R1 5.7 or pGL3R2 1.5 was transiently transfected along with carrier DNA [10 μg of pBluescript (SK)] into NIH 3T3 cells, as described in Materials and Methods. After 18 h, the cells were washed and growth arrested in 0.5% calf serum for 48 h. Thereafter, transfectants were infected with MCMV (MOI, 5 PFU/cell) or UV-irradiated MCMV (MOI, 5 PFU/cell) or mock infected. Total cytoplasmic extracts were isolated at the indicated times after infection and assayed for luciferase activity. Reporter gene activity was normalized to the amount of plasmid DNA introduced into recipient cells by DNA dot blot analysis. The resulting luciferase activity is expressed as fold induction relative to basal levels measured in cells transfected with pGL3R1 5.7 or pGL3R2 1.5 and then mock infected, which were set at 1.
Role of MCMV IE proteins in the regulation of R1 and R2 expression.
We have previously demonstrated that promoters of cellular genes involved in DNA precursor metabolism, such as the dihydrofolate reductase (DHFR) and thymidylate synthase (TS) promoters, are trans-activated by MCMV pIE1 but not by pIE3 (30, 44).
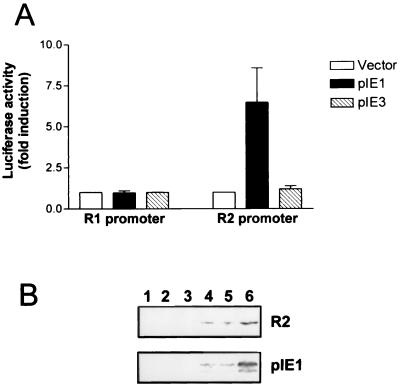
To see whether these IE gene products play a role in regulation of the R1 and R2 promoters, we cotransfected an expression plasmid for pIE1 or pIE3 with the luciferase reporter genes driven by the R1 or R2 promoters into NIH 3T3 cells. To rule out the possibility that the MCMV IE promoter, contained in the IE1 or IE3 expression plasmids, titrates out transcription factors from the target promoters, thereby appearing to regulate them, the amount of MCMV promoter included in the transfection mixtures was kept constant and appropriate amounts of pCMVCAT, which contains the regulatory sequences of the MCMV IE region (the IE enhancer and IE1-3 promoter) linked to the coding region of the irrelevant CAT protein, were included. Figure 5A demonstrates that pIE1 transactivated the R2 promoter (6.5-fold induction) whereas the R1 promoter was not affected. By contrast, pIE3 expression had no effect on R1 or R2 promoter activity. The ability of the IE1 or the IE3 constructs to express functional proteins was verified by cotransfection assays with the pCMVCAT or pE1CAT indicator plasmid, respectively. As previously observed (50), pIE1 expression increased the activity of the MCMV IE enhancer and IE1-3 promoter of pCMVCAT whereas pIE3 expression resulted in trans-activation of the MCMV E1 early promoter of pE1CAT (data not shown).
FIG. 5.
The MCMV IE protein pIE1 stimulates cellular R2 expression. (A) Effect of pIE1 on R1 and R2 promoter activity. NIH 3T3 cells were transiently cotransfected with 2 μg of the indicator plasmids pGL3R1 5.7 or pGL3R2 1.5 and 1 μg of the expression vectors for pIE1, pIE3, or CAT respectively. At 18 h after transfection, the cells were washed and then maintained for 48 h in medium containing 0.5% calf serum. Thereafter, total cytoplasmic extracts were isolated and assayed for luciferase activity, as described in Materials and Methods. The resulting luciferase activity is expressed as fold induction relative to basal levels measured in cells cotransfected with pGL3R1 5.7 or pGL3R2 1.5 and the value for the CAT-expressing vector, which was set at 1. (B) Effect of pIE1 expression on cell R2 protein levels. NIH 3T3 cells were transiently transfected with 0.5, 1, and 4 μg of the CAT expression vector (lanes 1, 2, and 3 respectively) or the pIE1 expression vector (lanes 4, 5, and 6 respectively). At 18 h after transfection, the cells were washed and then maintained in medium containing 0.5% calf serum. After 48 h, total-cell extracts were prepared, fractionated by SDS-PAGE (50 μg of protein/lane), and analyzed by immunoblotting with the anti-R2 serum and with the anti-pIE1 serum.
To confirm the ability of pIE1 to induce R2 gene expression, we transfected into NIH 3T3 cells increasing amounts of the pIE1 expression plasmid or pCMVCAT as a control. After transfection the cells were serum starved, and after 48 h cell extracts were analyzed by immunoblotting using anti-R2 and anti-pIE1 polyclonal monospecific antibodies. As shown in Fig. 5B, transfection of the pIE1 plasmid resulted in a dose-dependent expression of pIE1 (lanes 4 to 6) while no signal was observed in the samples transfected with pCMVCAT (lanes 1 to 3). Analysis of the same extracts with the anti-R2 antibodies revealed a dose-dependent induction of cellular R2 (lanes 4 to 6). This was undetectable in the samples from cells transfected with the control plasmid (lanes 1 to 3).
Taken as a whole, these results demonstrate that pIE1 stimulates cellular R2 gene expression in the absence of any other viral product.
An altered RNR activity is induced by MCMV infection.
The finding that the cellular R1 gene is not induced by MCMV infection prompted us to look for RNR activity in infected cells. RNR activity was measured in crude extracts from mock-infected and MCMV-infected cells 24 and 48 h after infection. To make the assay independent of cellular R2 protein, inducible by MCMV infection, all assays were performed in the presence of added, saturating amounts of mouse recombinant R2 protein. We observed that 0.07, 0.18, and 0.20 nmol of dCDP formed per 30 min in the mock-infected, 24-h, and 48-h samples, respectively, using 0.6 μg of protein in each assay. The background value of the assay in the presence of mouse recombinant R2 protein alone (5 μg) was 0.02 nmol/30 min. Although these results show a clear increase in the level of R1 protein after infection, it is not possible from this assay alone to distinguish cellular from virus-induced R1 protein activity. Therefore, we decided to find whether the RNR activity in the infected cells displayed a normal allosteric control.
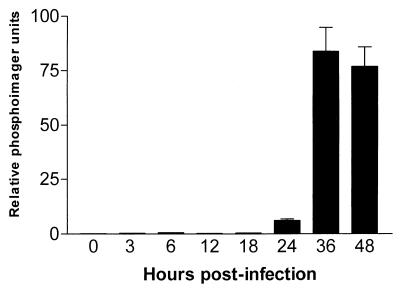
In early studies, measurements of dNTP pools in HSV-infected cells strongly indicated the induction of a new RNR activity with an altered allosteric regulation compared to the mammalian host cell enzyme (16, 42). These findings prompted us to compare the dNTP pools in MCMV- and mock-infected quiescent cells at two time points after infection (Table 1). All four dNTP pools expanded two- to fivefold after MCMV infection, and the most pronounced expansion occurred 48 h after infection. Moreover, a time course analysis of MCMV DNA synthesis revealed a considerable temporal overlap with the dNTP pool expansion since it began at 24 h p.i. and reached maximum levels at 36 and 48 h p.i. as shown in Fig. 6.
TABLE 1.
Distribution of individual dNTPs as a percentage of total NTP content after MCMV infection
| Infection | % of total NTP accounted for by:
|
|||
|---|---|---|---|---|
| dCTP | dTTP | dATP | dGTP | |
| Mock (24 h) | 0.08 | 0.18 | 0.02 | NDa |
| Mock (48 h) | 0.07 | 0.15 | 0.02 | ND |
| MCMV (24 h) | 0.08 | 0.16 | 0.04 | 0.01 |
| MCMV (48 h) | 0.31 | 0.41 | 0.12 | 0.03 |
| Mock (24 h) + TdR (10 mM) | 0.01 | 1.91 | 0.03 | 0.03 |
| MCMV (24 h) + TdR (10 mM) | 0.05 | 2.08 | 0.04 | 0.06 |
| MCMV (48 h) + TdR (10 mM) | 0.18 | 1.58 | 0.06 | 0.05 |
ND, not detectable.
FIG. 6.
Time course of MCMV DNA synthesis. Quiescent NIH 3T3 cells were infected with MCMV (MOI, 5 PFU/cell). Total genomic DNA was purified at the indicated times and immobilized on a hybridization membrane by a dot blot apparatus. The same filter was sequentially hybridized with a 32P-labeled viral probe (a 1,104-bp AvaI-XbaI fragment in the fourth exon of the MCMV IE1 gene) and a cell DNA probe (full-length murine G3PDH cDNA). The hybridization signals were quantitated with the Bio-Rad image analysis system and normalized to the differences in cell DNA. Values are the means of two separate determinations.
When the pool measurements were repeated in cells grown in the presence of 10 mM TdR, a 10-fold increase in the dTTP pool was observed in both mock- and virus-infected cells. As expected from the allosteric regulation of the cell RNR, this expansion was accompanied in the mock-infected cells by a specific drop in the dCTP pool (a 7.6-fold reduction). In strong contrast to the situation in the mock-infected cells, the expanded dTTP pool in the CMV-infected cells did not lead to a decrease in the dCTP pool but, instead, resulted in a 2.5-fold increase. We also measured the virus yield in the supernatants from the cell cultures used for the dNTP pool assay. Significantly, at 48 h p.i., the same yield was obtained from cells infected in the presence of 10 mM TdR (8.3 × 104 PFU/ml) as in its absence (9.4 × 104 PFU/ml). Taken together, these results, along with the earlier results showing no increase of cellular R1 mRNA or protein levels on MCMV infection, indicate the presence of a virus-induced R1 protein which exhibits an altered allosteric regulation in such a way that dTTP does not inhibit CDP reduction. Moreover, our dNTP pool measurements suggest that MCMV infection does not induce the S-phase-specific cellular thymidine kinase 1 since the dTTP pool increased to the same levels in mock- and virus-infected cells in the presence of 10 mM TdR (Table 1). Instead, the expansion of the dTTP pool most probably occurred via the mitochondrial thymidine kinase 2, which is not cell cycle specific.
The viral M45 gene is expressed in quiescent NIH 3T3 cells infected with MCMV.
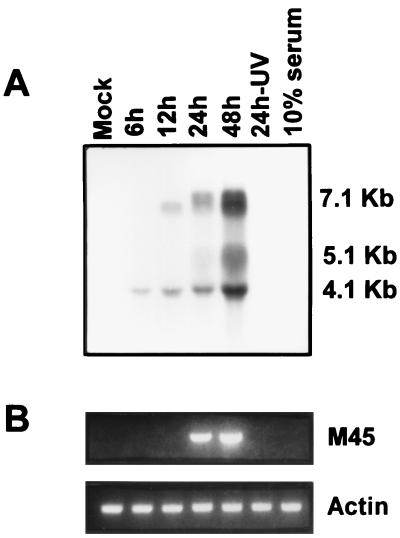
To determine whether the MCMV M45 gene, whose product shows homology to the R1 subunit of other herpesviruses, was expressed during viral replication, total RNA was isolated from mock- or MCMV-infected quiescent NIH 3T3 cells, and analyzed by Northern blotting. As shown in Fig. 7A, a full-length double-stranded M45 probe hybridized to three major viral transcripts of 7.1, 5.1, and 4.1 kb. The absence of any signal with RNA from mock-infected, UV-inactivated MCMV-infected, or serum-treated cells rules out any cross-hybridization with cellular transcripts. To further confirm that the ORF M45 is indeed transcribed, the same RNA samples were treated with DNase and retrotranscribed with random primers and the cDNAs were then amplified by using M45-specific primers. The results (Fig. 7B) clearly show that transcription of MCMV M45 RNA could be detected at 24 h p.i. and persisted up to 48 h p.i. Uninfected cells and cells infected with UV-inactivated MCMV stock or exposed to 10% serum were negative for M45 transcripts. Amplification of β-actin RNA showed that similar amounts of RNA were analyzed.
FIG. 7.
Detection of M45 transcripts in MCMV-infected cells. (A) NIH 3T3 cells were growth arrested in 0.5% calf serum for 48 h and then infected with MCMV (MOI, 5 PFU/cell) or mock infected. As additional controls, cells were also infected with a UV-inactivated MCMV stock or exposed to 10% serum for 24 h. Total RNA was isolated at the indicated times after infection and analyzed by Northern blotting. The filter was hybridized with a radiolabeled full-length M45 probe. (B) The same RNA samples were analyzed by RT-PCR, with M45- and actin-specific primers.
Taken altogether, these results demonstrate that the putative R1 subunit is expressed at times p.i. when either R1 activity, dNTP pool expansion, or viral DNA synthesis occurs, suggesting that it is involved in ribonucleotide reduction during viral replication.
DISCUSSION
An increasing body of evidence indicates that adsorption of the CMV particle to the cell membrane and expression of the IE genes generate an intracellular environment that is more favorable for viral replication (3, 7, 26, 54). Accordingly, it has been demonstrated that CMV infection of quiescent cells induces an “S-phase-like” state by up-regulating cellular enzymes involved in DNA precursor biosynthesis (25, 30, 44, 45, 62) along with an arrest of the host cell cycle, predominantly at the G1/S transition (10, 21, 46, 57, 64). This replicative strategy would provide the viral DNA polymerase with the necessary supply of dNTPs and avoid competitive cellular DNA synthesis and mitosis. We (this study) and others (5) have demonstrated that MCMV and HCMV infection does indeed result in the expansion of all four dNTP pools. However, the mechanism by which CMV fulfills its need for dNTPs in quiescent cells is not fully understood. The complete genome sequencing of many members of the Herpesviridae family demonstrates that alpha- and gammaherpesvirus encode large and small subunits of the RNR. This finding indicates that these viruses do not depend on the host cell for ribonucleotide reduction. By contrast, CMV and other members of the Betaherpesvirinae subfamily, such as HHV-6 and HHV-7, encode only a homologue of the large subunit of RNR. Sun and Conner (59) observed that the HHV-7 R1 homologue encoded by the ORF U28 is not a functional R1 subunit and suggest that betaherpesviruses have no requirement for RNR activity. To address this point, we studied MCMV replication and DNA synthesis in the presence of HU. HU is a selective inhibitor of the R2 protein, acts as a radical scavenger, and increases the rate of iron loss from mammalian R2 proteins. Inhibition of MCMV replication and DNA synthesis by HU in quiescent cells clearly shows that de novo synthesis of dNTPs by an iron-radical RNR is needed and rules out the possibility that the DNA precursors are obtained through the induction of the deoxyribonucleoside salvage pathway. Since CMV does not encode a functional RNR, one way of inducing ribonucleotide reduction in a quiescent cell is to stimulate an unscheduled expression of its R1 and R2 proteins. Here we demonstrated that while MCMV infection does not affect R1 expression, it strongly induces R2 promoter activity and mRNA and protein levels. We reason that R2 stimulation is caused by the infection on the basis of the following pieces of evidence: (i) dependence of the effect on MOI (data not shown); (ii) dependence of the effect on viral infectivity, since UV-inactivated virus cannot trigger any stimulation of the R2 promoter and protein expression; and (iii) ability of a specific MCMV genome fragment encoding the pIE1 to trans-activate the R2 promoter and induce R2 protein expression.
Several reports have shown that CMV infection stimulates the expression of a number of cellular genes important for cell cycle regulation and DNA synthesis. This regulation has been reported to depend on either viral binding to the cell surface (8, 9, 65) or viral IE protein expression (11, 31, 33, 40, 48, 54, 58, 62, 66). The observation that inactivation of MCMV by UV exposure abolished the induction of R2 protein as well as trans-activation of the R2 promoter suggests that virus gene expression, rather than interaction of viral particles with the cell surface, is required to stimulate R2 gene expression. As we have previously observed for the DHFR (44), TS (30) and folylpolyglutamate synthetase (FPGS) (unpublished data) promoters, both MCMV infection and pIE1 transactivated the R2 promoter, and virus-dependent transactivation was observed during the time frame when pIE1 protein was expressed (Fig. 3). Furthermore, transient transfection of a pIE1 expression vector induced R2 protein expression. Taken together, these results indicate that R2 induction by MCMV occurs at least in part via pIE1 expression.
We next asked which R1 protein (cellular or viral) is responsible for ribonucleotide reduction in MCMV-infected cells. Although the cellular R1 protein is barely detectable in both uninfected and infected quiescent cells, we cannot exclude the possibility that these low levels could be sufficient to support MCMV replication. In keeping with the low level of cellular R1 protein in mock-infected cells, its activity, measured in the presence of saturating amounts of recombinant mouse R2 protein, is slightly above the background value (0.07 and 0.02 nmol of dCDP/30 min, respectively). On the other hand, the RNR assay clearly shows an increase of R1 protein activity in MCMV-infected cells (0.18 and 0.2 nmol of dCDP/30 min at 24 and 48 h p.i., respectively), which is in contrast to the uninduced levels of cell R1 mRNA and protein. However, since all the activity values were low, it is not possible from this assay to definitely distinguish a cellular R1 protein from a virus-induced R1 protein.
If we assume that the RNR activity in MCMV-infected cells is the result of cellular R1 and R2 association, the low enzyme activity might reflect the low level of R1 protein in quiescent cells. Alternatively, if the viral R1 associates with the cellular R2, this hybrid RNR might have a different allosteric control and would require different assay conditions. Early studies demonstrated that HSV-1 induced an altered RNR in extracts of infected cells, since pyrimidine nucleotide reduction by the “new” reductase activity was highly resistant to dTTP inhibition (16, 42). Moreover, HSV-1 replication was not affected in cells in which cellular DNA synthesis was inhibited by TdR treatment. In the mammalian cell, TdR is converted to dTTP, which acts as an allosteric inhibitor of the cellular RNR and suppresses CDP reduction (51, 52, 63). dCTP depletion results in blocked DNA synthesis and cell proliferation. As observed with HSV-1, our dNTP pool data support the existence of an altered RNR in the MCMV-infected cells. As expected, measurement of dNTP pools demonstrated that mock-infected cells contain an RNR that is sensitive to dTTP inhibition, since we observed a specific drop in the dCTP pool following the addition of 10 mM TdR whereas in TdR-treated MCMV-infected cells the level of dTTP remained high; also, the level of dCTP increased and MCMV replication was not inhibited. Thus, the MCMV-induced reductase activity allows the virus to override the dTTP-inhibited normal cellular RNR and make dCTP. Taken together, our data demonstrate that MCMV infection stimulates expression of the cellular R2 protein and induces an RNR activity with an altered allosteric regulation compared to the mouse RNR. Whether this latter effect is the consequence of the association of the R2 protein with the product of the M45 gene of MCMV remains to be demonstrated. However, several observations suggest involvement of the M45 gene product in ribonucleotide reduction in the infected cells. First, we have demonstrated that the M45 gene is indeed expressed in MCMV-infected cells. Temporal studies have shown that a significant accumulation of the M45 transcripts can be detected at 24 and 48 h p.i. Moreover, there is a considerable temporal overlap between the increase in M45 mRNA levels, R2 expression, increased R1 activity, expansion of the dNTP pool, and synthesis of the viral DNA. When considered together with the data demonstrating an altered allosteric regulation of RNR following MCMV infection, these findings support the hypothesis that the product of M45 may complex with R2 to form a functional version of the enzyme. Studies are under way to verify this hypothesis.
CMV can replicate in quiescent cells that have shut down their machinery for synthesizing DNA. The reactions catalyzed by RNR and by the enzymes involved in the biosynthesis of thymidylate (dTMP) are highly repressed in cells that are not undergoing DNA synthesis. We have previously demonstrated that MCMV infection of quiescent cells leads to the coordinated stimulation of the cell enzymes FPGS, DHFR, and TS, involved in dTMP synthesis. Here we present evidence that an RNR activity with altered allosteric control is induced in quiescent cells by MCMV infection. The induction of this set of enzymes releases the virus from normal cell control and allows dNTP biosynthesis and viral replication to take place during periods of the cell cycle other than the S phase.
ACKNOWLEDGMENTS
We thank Timothy J. Kinsella for providing plasmid pET28a(+)R2(+) and Nigel Parker for providing plasmid p3I.
This work was supported by grants from the Italian AIDS Research Project (grant 50B.25), from MURST-CNR Biotechnology Program L. 95/95, from A.I.R.C., from the Italian Ministry of Public Health, and from the Swedish Natural Science Research Council.
REFERENCES
- 1.Abken H, Reifenrath B. A procedure to standardize CAT reporter gene assay. Nucleic Acids Res. 1992;20:3527. doi: 10.1093/nar/20.13.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerblom L, Ehrenberg A, Graslund A, Lankinen H, Reichard P, Thelander L. Overproduction of the free radical of ribonucleotide reductase in hydroxyurea-resistant mouse fibroblast 3T6 cells. Proc Natl Acad Sci USA. 1981;78:2159–2163. doi: 10.1073/pnas.78.4.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht T, Boldogh I, Fons M, AbuBakar S, Deng C Z. Cell activation signals and the pathogenesis of human cytomegalovirus. Intervirology. 1990;31:68–75. doi: 10.1159/000150140. [DOI] [PubMed] [Google Scholar]
- 4.Averett D R, Lubbers C, Elion G B, Spector T. Ribonucleotide reductase induced by herpes simplex type 1 virus. Characterization of a distinct enzyme. J Biol Chem. 1983;258:9831–9838. [PubMed] [Google Scholar]
- 5.Biron K K, Fyfe J A, Stanat S C, Leslie L K, Sorrell J A, Lambe C U, Coen D M. A human cytomegalovirus mutant resistant to the nucleoside analog 9-[2-hydroxy-1-(hydroxymethyl)ethoxy]methylguanine (BW B759U) induces reduced levels of BW B759U triphosphate. Proc Natl Acad Sci USA. 1986;83:8769–8773. doi: 10.1073/pnas.83.22.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 7.Boldogh I, AbuBakar S, Albrecht T. Activation of protooncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:961–964. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 8.Boldogh I, AbuBakar S, Millinoff D, Deng C Z, Albrecht T. Cellular oncogenes activation by human cytomegalovirus. Lack of correlation with virus infectivity and immediate early gene expression. Arch Virol. 1991;118:163–177. doi: 10.1007/BF01314027. [DOI] [PubMed] [Google Scholar]
- 9.Boyle K A, Pietropaolo R L, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 11.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 12.Cameron J M, McDougall I, Marsden H S, Preston V G, Ryan D M, Subak-Sharpe J H. Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J Gen Virol. 1988;69:2607–2612. doi: 10.1099/0022-1317-69-10-2607. [DOI] [PubMed] [Google Scholar]
- 13.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–17753. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 14.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 15.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonucleases. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 16.Cohen G H. Ribonucleotide reductase activity of synchronized KB cells infected with herpes simplex virus. J Virol. 1972;9:408–418. doi: 10.1128/jvi.9.3.408-418.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conner J, Marsden H, Clements J B. Ribonucleotide reductase of herpes viruses. Rev Med Virol. 1994;4:25–34. [Google Scholar]
- 18.Cooper J, Conner J, Clements J B. Characterization of the novel protein kinase activity present in the R1 subunit of herpes simplex virus ribonucleotide reductase. J Virol. 1995;69:4979–4985. doi: 10.1128/jvi.69.8.4979-4985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 20.de Wind N, Berns A, Gielkens A, Kimman T. Ribonucleotide reductase-deficient mutants of pseudorabies virus are avirulent for pigs and induce partial protective immunity. J Gen Virol. 1993;74:351–359. doi: 10.1099/0022-1317-74-3-351. [DOI] [PubMed] [Google Scholar]
- 21.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 23.Engstrom Y, Eriksson S, Thelander L, Akerman M. Ribonucleotide reductase from calf thymus. Purification and properties. Biochemistry. 1979;18:2941–2948. doi: 10.1021/bi00581a004. [DOI] [PubMed] [Google Scholar]
- 24.Eriksson S, Graslund A, Skog S, Thelander L, Tribukait B. Cell cycle-dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J Biol Chem. 1984;259:11695–11700. [PubMed] [Google Scholar]
- 25.Estes J E, Huang E-S. Stimulation of cellular thymidine kinase by human cytomegalovirus. J Virol. 1977;24:13–21. doi: 10.1128/jvi.24.1.13-21.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortunato E A, McElroy A K, Sanchez I, Spector D H. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 2000;8:111–119. doi: 10.1016/s0966-842x(00)01699-1. [DOI] [PubMed] [Google Scholar]
- 27.Gariglio M, Foresta P, Sacchi C, Lembo M, Hertel L, Landolfo S. Suppression of high mobility group protein T160 expression impairs mouse cytomegalovirus replication. J Gen Virol. 1997;78:665–670. doi: 10.1099/0022-1317-78-3-665. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein D J, Weller S K. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988;166:41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gribaudo G, Riera L, Lembo D, De Andrea M, Gariglio M, Rudge T L, Johnson L F, Landolfo S. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J Virol. 2000;74:4979–4987. doi: 10.1128/jvi.74.11.4979-4987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gribaudo G, Ravaglia S, Guandalini L, Cavallo R, Gariglio M, Landolfo S. The murine cytomegalovirus immediate early 1 protein stimulates NF-κB activity by transactivating the NF-κB p105/p50 promoter. Virus Res. 1996;45:15–27. doi: 10.1016/0168-1702(96)01356-1. [DOI] [PubMed] [Google Scholar]
- 32.Gribaudo G, Ravaglia S, Gaboli M, Gariglio M, Cavallo R, Landolfo S. Interferon-α inhibits the murine cytomegalovirus immediate-early gene expression by down-regulating NF-κB activity. Virology. 1995;211:251–260. doi: 10.1006/viro.1995.1398. [DOI] [PubMed] [Google Scholar]
- 33.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heineman T C, Cohen J I. Deletion of the varicella-zoster virus large subunit of ribonucleotide reductase impairs growth of virus in vitro. J Virol. 1994;68:3317–3323. doi: 10.1128/jvi.68.5.3317-3323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry B E, Glaser R, Hewetson J, O'Callaghan D J. Expression of altered ribonucleotide reductase activity associated with the replication of the Epstein-Barr virus. Virology. 1978;89:262–271. doi: 10.1016/0042-6822(78)90058-2. [DOI] [PubMed] [Google Scholar]
- 36.Hofer A, Ekanem J T, Thelander L. Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J Biol Chem. 1998;273:34098–34104. doi: 10.1074/jbc.273.51.34098. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson J G, Leib D A, Goldstein D J, Bogard C L, Schaffer P A, Weller S K, Coen D M. A herpes simplex virus ribonucleotide reductase deletion mutant is defective for productive acute and reactivatable latent infections of mice and for replication in mouse cells. Virology. 1989;173:276–283. doi: 10.1016/0042-6822(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 38.Johansson E, Hjortsberg K, Thelander L. Two YY-1-binding proximal elements regulate the promoter strength of the TATA-less mouse ribonucleotide reductase R1 gene. J Biol Chem. 1998;273:29816–29821. doi: 10.1074/jbc.273.45.29816. [DOI] [PubMed] [Google Scholar]
- 39.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 40.Koszinowski U H, Keil G M, Volkmer H, Fibi M R, Ebeling-Keil A, Munch K. The 89,000-Mr murine cytomegalovirus immediate-early protein activates gene transcription. J Virol. 1986;58:59–66. doi: 10.1128/jvi.58.1.59-66.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo M-L, Kinsella T J. Overexpression of a hexa-histidine and T7 peptide tagged human ribonucleotide reductase small subunit, R2 in Escherichia coli and the generation of human R2 antibodies. Int J Oncol. 1997;10:515–520. doi: 10.3892/ijo.10.3.515. [DOI] [PubMed] [Google Scholar]
- 42.Langelier Y, Buttin G. Characterization of ribonucleotide reductase induction in BHK-21/C13 Syrian hamster cell line upon infection by herpes simplex virus (HSV) J Gen Virol. 1981;57:21–31. doi: 10.1099/0022-1317-57-1-21. [DOI] [PubMed] [Google Scholar]
- 43.Lankinen H, Graslund A, Thelander L. Induction of a new ribonucleotide reductase after infection of mouse L cells with pseudorabies virus. J Virol. 1982;41:893–900. doi: 10.1128/jvi.41.3.893-900.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lembo D, Angeretti A, Gariglio M, Landolfo S. Murine cytomegalovirus induces expression and enzyme activity of dihydrofolate reductase in quiescent cells. J Gen Virol. 1998;78:2803–2808. doi: 10.1099/0022-1317-79-11-2803. [DOI] [PubMed] [Google Scholar]
- 45.Lembo D, Gribaudo G, Cavallo R, Riera L, Angeretti A, Hertel L, Landolfo S. Human cytomegalovirus stimulates cellular dihydrofolate reductase activity in quiescent cells. Intervirology. 1999;42:30–36. doi: 10.1159/000024957. [DOI] [PubMed] [Google Scholar]
- 46.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mann G J, Graslund A, Ochiai E, Ingemarson R, Thelander L. Purification and characterization of recombinant mouse and herpes simplex virus ribonucleotide reductase R2 subunit. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 48.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Clifford Azizkhan J. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClarty G A, Chan A K, Engstrom Y, Wright J A, Thelander L. Elevated expression of M1 and M2 components and drug-induced posttranscriptional modulation of ribonucleotide reductase in a hydroxyurea-resistant mouse cell line. Biochemistry. 1987;26:8004–8011. doi: 10.1021/bi00398a068. [DOI] [PubMed] [Google Scholar]
- 50.Messerle M, Buhler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris N R, Fisher G A. Studies concerning the inhibition of cellular reproduction by deoxyribonucleosides. I. Inhibition of the synthesis of deoxycytidine by a phosphorylated derivative of thymidine. Biochim Biophys Acta. 1963;68:84–92. doi: 10.1016/0006-3002(60)90777-0. [DOI] [PubMed] [Google Scholar]
- 52.Morris N R, Reichard P, Fisher G A. Studies concerning the inhibition of cellular reproduction by deoxyribonucleosides. II. Inhibition of the synthesis of deoxycytidine by thymidine, deoxyadenosine and deoxyguanosine. Biochim Biophys Acta. 1963;68:93–99. [Google Scholar]
- 53.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Hederwijin P, Desmyter J, De Clerq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 54.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E-S. The human cytomegalovirus IE1–72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 57.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schickedanz J, Philipson L, Ansorge W, Pepperkork R, Klein R, Koszinowski U H. The 89,000-Mr murine cytomegalovirus immediate-early protein stimulates c-fos expression and cellular DNA synthesis. J Virol. 1988;62:3341–3347. doi: 10.1128/jvi.62.9.3341-3347.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y, Conner J. The U28 ORF of human herpesvirus-7 does not encode a functional ribonucleotide reductase R1 subunit. J Gen Virol. 1999;80:2713–2718. doi: 10.1099/0022-1317-80-10-2713. [DOI] [PubMed] [Google Scholar]
- 60.Thelander L, Gräslund A. Ribonucleotide reductase in mammalian cells. Metal Ions Biol Syst. 1994;30:109. [Google Scholar]
- 61.Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 62.Wade M, Kowalik T F, Mudryj M, Huang E-S, Clifford Azizkhan J. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whittle E D. Effect of thymidine on deoxyribonucleic acid synthesis and cytidine metabolism in rat thymus cells. Biochim Biophys Acta. 1966;114:44–60. [Google Scholar]
- 64.Wiebusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yurochko A D, Hwang E-S, Rasmussen L, Keay S, Pereira L, Huang E-S. The human cytomegalovirus UL55 (gB) and UL 75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yurochko A D, Kowalik T F, Huong S M, Huang E-S. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoter. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]