Abstract
The alpha/beta interferon (IFN-α/β) system represents one of the first lines of defense against virus infections. As a result, most viruses encode IFN antagonistic factors which enhance viral replication in their hosts. We have previously shown that a recombinant influenza A virus lacking the NS1 gene (delNS1) only replicates efficiently in IFN-α/β-deficient systems. Consistent with this observation, we found that infection of tissue culture cells with delNS1 virus, but not with wild-type influenza A virus, induced high levels of mRNA synthesis from IFN-α/β genes, including IFN-β. It is known that transactivation of the IFN-β promoter depends on NF-κB and several other transcription factors. Interestingly, cells infected with delNS1 virus showed high levels of NF-κB activation compared with those infected with wild-type virus. Expression of dominant-negative inhibitors of the NF-κB pathway during delNS1 virus infection prevented the transactivation of the IFN-β promoter, demonstrating a functional link between NF-κB activation and IFN-α/β synthesis in delNS1 virus-infected cells. Moreover, expression of the NS1 protein prevented virus- and/or double-stranded RNA (dsRNA)-mediated activation of the NF-κB pathway and of IFN-β synthesis. This inhibitory property of the NS1 protein of influenza A virus was dependent on its ability to bind dsRNA, supporting a model in which binding of NS1 to dsRNA generated during influenza virus infection prevents the activation of the IFN system. NS1-mediated inhibition of the NF-κB pathway may thus play a key role in the pathogenesis of influenza A virus.
Influenza A virus is a negative-strand RNA virus belonging to the Orthomyxoviridae family. The virus genome consists of eight RNA segments which encode 10 proteins. Among these proteins, NS1 is the only nonstructural protein. It is expressed to high levels in virus-infected cells, and it was shown to be able to bind to dsRNA (26). Previous studies suggested that binding of dsRNA by the NS1 protein prevented the activation of the interferon (IFN)-inducible dsRNA-dependent protein kinase (PKR) (38, 56). In addition, other regulatory functions of the NS1 protein during viral replication have been suggested, such as inhibition of host mRNA polyadenylation (42), inhibition of nuclear export of polyadenylated host mRNA (8), inhibition of mRNA splicing (18, 37), stimulation of translation of viral mRNA (2, 10, 13, 14), and modulation of viral RNA transcription and replication (40, 53).
A recombinant influenza A/PR8/34 virus lacking the NS1 gene (delNS1 virus) has been generated (20). This virus appears to efficiently replicate only in substrates or hosts with deficiencies in the alpha/beta IFN (IFN-α/β) system, such as 6-day-old eggs (55), STAT1−/− mice (20), or PKR−/− mice (4). These observations suggest that the NS1 protein may play a critical role in inhibiting IFN responses during viral replication.
IFNs are among the first line of host defenses against virus infections (for a review, see reference 51). There are two types of IFNs, (IFN-α/β), which includes IFN-α and IFN-β, and IFN-γ. IFN-α/β is usually induced within hours after viral infection. Once it is synthesized, it functions in both autocrine and paracrine fashions to prevent the replication and spread of viruses. Induction of IFN-α/β production upon viral infection requires multiple regulatory factors. These factors act mainly at the transcriptional level, inducing the synthesis of mRNAs from the IFN-α/β genes. The transcriptional regulation of the IFN-β promoter has been well studied (1, 48, 58, 59, 63). Critical transcription factors which have been shown to be involved in regulating IFN-β transcription include IRF-3, AP1, and NF-κB. NF-κB comprises a family of transcription factors that play an essential role in the regulation of many physiological responses, ranging from immune and inflammatory responses to cell differentiation and apoptosis (23). Under normal conditions, NF-κB is bound to its inhibitor, IκB, resulting in the cytoplasmic retention of NF-κB. Most of the known inducers of NF-κB act through the recently identified IκB kinase (IKK) complex (12). Activated IKKs phosphorylate IκB, which is subsequently ubiquitinated and undergoes 26S proteosome-mediated degradation. NF-κB is therefore released and enters the nucleus, where it stimulates transcription from genes containing NF-κB-binding sequence elements in their promoters (for a recent review, see reference 31).
It has been shown that nuclear NF-κB activity is induced by exposure to a wide variety of bacterial and viral infections. Subsequently, activated NF-κB contributes to the stimulation of synthesis of IFN-α/β. Because of the importance of IFN-α/β in host antiviral responses, many viruses have evolved different strategies to subvert the IFN system. For example, several negative-strand RNA viruses have been shown to encode inhibitors of the IFN signaling pathway, such as the C proteins of Sendai virus (21, 22, 24, 33), the V proteins of SV5 and PIV2 (11, 64), the NS1 and NS2 proteins of bovine respiratory syncytial virus (50), the VP35 protein of Ebola virus (3), and the NSs proteins of Rift valley fever and bunyamwera viruses (25, 60). In this report, we demonstrate that the NS1 protein of influenza A virus has the ability to prevent NF-κB activation, resulting in the inhibition of IFN-α/β production in virus-infected cells.
MATERIALS AND METHODS
Viruses and cells.
Influenza A/PR/8/34 (PR8) virus (H1N1), Newcastle disease virus (NDV), and Sendai viruses were propagated in 10-day-old embryonated chicken eggs at 37°C. Influenza X-31 virus, a reassortant of influenza A/HK/8/68 and A/PR/8/34 viruses, was supplied by Evans Biological, Ltd., Liverpool, England. The delNS1 virus is a PR8-derived virus in which the NS1 gene is deleted (20). This virus was grown in 7-day-old embryonated chicken eggs, as described previously (55). The NS1(1–126) virus is isogenic with delNS1 virus. Its NS gene has a deletion of 19 nucleotides after nucleotide position 378, resulting in an NS1 gene encoding the first 126 amino acids of the NS1 protein of PR8 virus, followed by the amino acids T, S, and V. Viral titers were obtained by plaque assay on MDCK cells in the presence of 2 μg of trypsin per ml. MDCK cells were maintained in minimal essential medium with 10% fetal calf serum (FCS) and antibiotics. Mouse embryonic fibroblasts (MEFs) and 293 and Vero cells were maintained in Dulbecco's modified Eagle's medium with 10% FCS plus antibiotics.
Virus infections.
MEFs in 10-cm-diameter dishes were incubated with viruses in phosphate-buffered saline–0.2% bovine serum albumin (BSA) and infected at a multiplicity of infection (MOI) of 1 at room temperature. One hour after incubation, virus-containing solutions were removed, and cells were incubated in growth medium in a CO2 incubator at 37°C. The same procedure was used for infection of 293 cells, except that the virus inoculum was not removed to prevent cell detachment from the dishes.
Plasmids and cDNAs.
pIFN-CAT was made by inserting the mouse IFN-β promoter (−184 to +19) between NheI and BglII sites of the pCAT 3-enhancer (Promega) in front of the chloramphenicol acetyltransferase (CAT) open reading frame (ORF). The mouse IFN-β promoter was amplified by PCR with primers 5′-GGCCGCTAGCTTGAGAGTTCTTTTATCTTCAGGGCTGTCTC-3′ and 5′-CGCGAGATCTGCAAGCAAGATGAGGTAAAGGCTGTCAAAGGCTGC-3′ and genomic DNA isolated from MEFs as a template. Plasmid pκB-Luc encodes a firefly luciferase reporter gene under the control of an NF-κB-responsive promoter (19). pRL-TK-Luc (Promega), containing a Renilla luciferase reporter gene under the control of the herpes simplex virus thymidine kinase promoter, was used as an internal control of transfection efficiency. pCAGGS-NS1(SAM) was made by inserting the ORF of the NS1 of wild-type PR8 virus between the EcoRI and XhoI sites of pCAGGS (44), as described previously (54). This plasmid contains the NS1 cDNA, in which the splicing acceptor sequence was mutated by a silent point mutation (A to C at nucleotide 541 in the cDNA sense), under the transcriptional control of a chicken β-actin promoter. pCAGGS-NS1-R38A/K41A(SAM), encoding an NS1 protein in which amino acids R38 and K41 were mutated to A, was also previously described (54). pCAGGS-NS1(1–73) was generated by inserting a PCR product corresponding to the sequence encoding the first 73 amino acids of NS1 followed by the hemagglutinin (HA)/TAG epitope between EcoRI and XhoI sites of pCAGGS. This PCR product was generated by using primers NS1EcoRI5′ (5′-GCGCGAATTCAATAATGGATCCAAACACTG-3′) and 5′-GGCCCTCGAATCAGGCATAATCTGGGACATCATAAGGGTACATCCCGGGGGATTCTTCTTTCAGAATCCG-3′ and with pCAGGS-NS1(SAM) as a template. pCAGGS-NS1(1–73,R38A/K41A) was generated with the same set of primers with pCAGGS-NS1-R38A/K41A(SAM) as a template. pLPC-IκB(SA) and pLPC-IKK(KA) are pLPC-derived mammalian expression plasmids encoding a superrepressor form of IκB, IκB(SA), and a dominant-negative form of IKKβ, IKKβ(KA), respectively (6, 47, 62). pT3-NS-IAmut1 contains a mutant NS gene of PR8 virus flanked by the T3 RNA polymerase promoter and BsmI restriction site. The IAmut1 mutation consists of a replacement of amino acids 181 to 185 of the NS1 protein from LIGGL to KQRRS and was generated by site-directed mutagenesis from the pT3/PR8-NS plasmid (15). Specific murine IFN-α (mIFN-α), mIFN-β, murine β-actin (mβ-actin), human IFN-β (hIFN-β), and human β-actin (hβ-actin) probes used in Northern blot analysis were made by random primer labeling (Roche) following the manufacturer's recommendations. DNA fragments used as templates for the probes were made by reverse transcription (RT)-PCR with mRNAs obtained from lipopolysaccharide-treated mouse or human macrophages used as a template. First, mRNAs were reverse transcribed with oligo(dT) as a primer. The following primers were used in subsequent PCRs: mIFN-β, 5′-AATGTCAGGAGCTTCTGGAGC-3′ and 5′-CTCTGATGCTTAAAGGTTGCC; mIFN-α, 5′-AACGCTACACACTGCATCT-3′ and 5′-TGCTCATTGTAATGCTTGG-3′; mβ-actin, 5′-ATGGATGACGATATCGCT-3′ and 5′-ATGAGGTAGTCTGTCAGGT-3′; hIFN-β, 5′-GGCCATGACCAACAAGTGTCTCCTCC-3′ and 5′-GCGCTCAGTTTCGGAGGTAACCTGT-3′; and hβ-actin, 5′-TCCTGTGGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′.
Plasmid transfections.
Transfections of pIFN-CAT were done by using calcium phosphate. pIFN-CAT (0.1 μg) and 1 μg of pLPC, pLPC-IκB(SA), pLPC-IKKβ(KA), pCAGGS-NS1(SAM), pCAGGS-NS1(1–73), or pCAGGS(1–73,R38A/K41A) were used in each transfection. Transfections of pκB-Luc were done with Fugene6 lipofection reagent (Roche) following the manufacturer's instructions. pκB-Luc (0.1 μg) and 1 μg of pLPC, pLPC-IκB(SA), pLPC-IKKβ(KA), pCAGGS-NS1(SAM), pCAGGS-NS1(1–73), or pCAGGS(1–73,R38A/K41A) were used in each transfection. pRL-TK (0.1 μg) was included in these transfections as an internal control to normalize transfection efficiencies. pLPC-IκB(SA), pLPC, pCAGGS, pCAGGS-NS1, and pCAGGS-NS1(1–73) were transfected with Lipofectamine 2000 (Gibco-BRL) in the experiments where Northern analyses were performed. In these cases, 5 μg of each plasmid was used.
Generation of transfectant viruses.
Transfectant viruses were generated by reverse genetics techniques (15, 17, 20). NS1(1–126) virus was rescued in a transfection experiment using a plasmid encoding the NS-IAmut1 RNA segment. After transfection, recombinant viruses were plaque purified three times in MDCK cells covered with agar overlay media at 37°C, and several plaques were used for preparation of viral stocks in 10-day-old embryonated eggs. The genomic RNAs from isolated transfectant viruses were analyzed by RT-PCR using the primers 5′-GGCCTCTAGATAATACGACTCACTATAAGCAAAAGCAGGGTGACAAAG-3′ (complementary to positions 1 to 21 at the 3′ noncoding end of the NS gene) and 5′-GATCGCTCTTCTATTAGTAGAAACAAGGGTGTTTTTTATTAAATAAGCTG-3′ (containing the last 38 nucleotides of the 5′ noncoding end of the NS gene). PCR products were cloned and sequenced. Sequence analysis revealed the presence of two different clonal virus populations. Some viral stocks corresponded to NS-IAmut1 virus, whose NS gene was identical to the transfected NS gene. Some other viral stocks contained a spontaneous deletion of 19 nucleotides in the transfected NS gene, resulting in a virus, NS1(1–126), encoding a truncated NS1 protein. NS1(1–126) virus was further characterized by the assays described in this article.
CAT and luciferase assays.
CAT assays were done as previously described (46). A Dual-Luciferase Reporter Assay system (Promega) was used in luciferase assays according to the manufacturer's recommendations.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were done to determine the activation of NF-κB as previously described (49). Briefly, nuclear extracts were made as follows. Cells were washed with phosphate-buffered saline (PBS) and then resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 60 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.3% NP-40, 1× Complete Protease Inhibitor Cocktail [Roche Molecular Biochemicals]). After 5 min of incubation on ice, nuclei were pelleted by centrifugation at 4°C. The pellet was resuspended in an equal volume of nuclear extract buffer (20 mM Tris-HCl [pH 8.0], 400 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 25% glycerol). After incubation on ice for 10 min, the suspension was vortexed and cleared by centrifugation at 4°C. Aliquots of nuclear extracts containing 2 μg of protein were used in EMSAs. A DNA probe containing a mouse H2 κB binding site was used in these assays (49). Anti-p50 and anti-p65 rabbit polyclonal antibodies used in supershift assays were purchased from Santa Cruz Technologies and Rockland, respectively.
Immunostaining.
MEFs grown on 12-mm-diameter cover glasses (Fisher Scientific) were mock treated, tumor necrosis factor alpha (TNF-α) (10 ng/ml; R&D Systems, Inc.) stimulated, or infected with either PR8 or delNS1 viruses for 6 h at an MOI of 1. Cells were washed with PBS and fixed with ice-cold 100% methanol for 10 min. After blocking for 30 min with 3% bovine serum albumin (BSA) (Sigma), cells were incubated with rabbit anti-p65 antibody (Rockland) diluted 1:200 and mouse anti-NP antibody HT103 (45) diluted 1:500 in 3% BSA–PBS for 40 min. MEFs were washed three times with PBS before incubation with fluorescein isothiocyanate-conjugated antirabbit antibody and Texas red-conjugated antimouse antibody (Boehringer Mannheim) diluted 1:500 in 3% BSA–PBS. Cover glasses were then fixed to slides with mounting medium containing antifading reagent (Molecular Probes). Stained cells were counted for localization of p65 in a blind fashion.
Northern blot analysis.
RNA was isolated from cells mock infected or infected with PR8, delNS1, NS1(1–126) or Sendai viruses at an MOI of 1. Total RNA was extracted with TRIzol reagent (Molecular Research Center) as recommended by the manufacturer. Ten micrograms of total RNA was used for Northern analysis by using Quickhyb hybridization solution (Stratagene). Different mRNAs were detected by hybridization to specific [32P]ATP-labeled probes. RNA loading was controlled by normalization to a β-actin probe.
Western blots.
Dishes (35 mm in diameter) of confluent MDCK cells were mock infected or infected at an MOI of 2 with PR8 or NS1(1–126) viruses. Six hours postinfection (p.i.), cells were lysed in 100 μl of radioimmunoprecipitation assay (RIPA) buffer. Ten microliters of cell lysates was loaded on a sodium dodecyl sulfate (SDS)–15% polyacrylamide gel. Separated proteins were transferred to a membrane and subjected to Western analysis with a rabbit polyclonal anti-NS1 antibody. Goat anti-rabbit immunoglobulin G (IgG) (H+L) peroxidase antibody was used as secondary antibody (Boehringer Mannheim). Western blots were developed with a chemiluminescence reagent (NEN; catalog no. NEL101).
RESULTS
IFN-α/β genes are induced in cells infected with delNS1, but not with PR8 viruses.
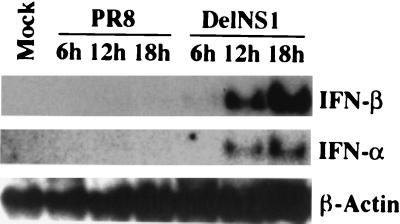
IFN synthesis is one of the early responses of the host against viral infection. Our previous studies showed that an influenza virus with a deletion in the NS1 gene, delNS1 virus, can only replicate efficiently in IFN-deficient systems, suggesting that the NS1 protein of influenza virus inhibits the IFN system of the host (20, 55). We first investigated whether this inhibition is through blocking the synthesis of IFN-α/β. To this end, we infected MEFs with delNS1 or with the parental wild-type PR8 virus at an MOI of 1 and then harvested RNA at 6, 12, and 18 h p.i. Northern blot analysis indicated that both IFN-α and IFN-β genes were induced in cells infected with delNS1 virus (Fig. 1). In contrast, we could not detect induction of these genes following wild-type PR8 virus infection. RT-PCR analysis of a human epithelial cell line, Hec-1b, also showed that IFN-β could be induced in cells infected with delNS1 virus, but not with PR8 virus (54). These results suggest that the NS1 protein directly prevents de novo synthesis of IFN-α/β-specific mRNAs in both human and mouse cells infected with influenza A virus.
FIG. 1.
Induction of IFN-α/β-specific mRNAs by delNS1 virus infection in MEFs. Northern blot analyses of mRNAs corresponding to IFN-α and IFN-β were performed with RNA isolated from mock-infected MEFs or MEFs infected with PR8 or delNS1 viruses at an MOI of 1 for the indicated times. A total of 10 μg of RNA was used. Northern blot detection of mRNA derived from a housekeeping gene (β-actin) is shown as a control.
NF-κB is activated in cells infected with delNS1 virus.
Although both IFN-α and IFN-β genes were induced in MEFs infected by delNS1 virus, we focused our efforts on investigating the activation of molecules involved in signaling pathways leading to IFN-β induction during delNS1 virus infection. Many transcription factors have been defined in the regulation of IFN-β expression, among which NF-κB and IRF-3 play a key role. Previously, we have demonstrated that delNS1 virus infection, but not PR8 virus infection, results in the activation of IRF-3 (54). We now tested whether NF-κB was also activated in cells infected with delNS1 virus. MEFs were infected with either wild-type PR8 virus or mutant delNS1 virus at an MOI of 1. At 2 and 6 h p.i., nuclear extracts were made and subjected to EMSA with a probe specific for NF-κB. delNS1 virus infection clearly activated NF-κB at 6 h p.i. (Fig. 2A). However, there was no significant difference in the levels of activated NF-κB between mock-infected and PR8-infected cells. The activation of NF-κB by delNS1 virus infection was maintained at 16 h p.i. (data not shown). delNS1-mediated activation of NF-κB was comparable in this assay to TNF-α-, dsRNA-, or NDV-mediated activation of NF-κB (Fig. 2A). These results suggest that the NS1 protein prevents NF-κB activation in influenza A virus-infected cells.
FIG. 2.
Activation of NF-κB by delNS1 virus infection in MEFs. (A) Detection of activated NF-κB by EMSA. MEFs were untreated (UT), mock infected, or infected with PR8, delNS1, or NDV viruses at an MOI of 1 for the indicated times. Nuclear extracts were subjected to EMSA with a DNA probe specific for NF-κB. TNF-α- and dsRNA-treated cells were included as positive controls. (B) Supershift of NF-κB complexes. Anti-p65 and anti-p50 antibodies (Ab) were used to shift the NF-κB complexes in delNS1- or NDV-infected MEFs at 6 h p.i.
NF-κB normally consists of two subunits, p50 and p65. To test the presence of such polypeptides in activated NF-κB by delNS1 virus infection, we performed a supershift assay (Fig. 2B). Both anti-p50 and anti-p65 antibodies were able to supershift activated NF-κB in response to delNS1 virus infection, demonstrating that both subunits were present in the NF-κB complex.
We also determined activation of NF-κB by immunostaining of virus-infected MEFs by using an antibody specific for the p65 subunit of NF-κB. Inactive NF-κB is retained in the cytoplasm by binding to its inhibitor IκB. Nuclear translocation of NF-κB is then indicative of its activation. MEFs infected with delNS1, but not with PR8 virus, showed significant nuclear translocation of p65 (data not shown). In fact, we found that more than 85% of delNS1 virus-infected cells showed nuclear translocation of NF-κB at 6 h p.i., while only about 10% of PR8 virus-infected cells showed NF-κB translocation. These results demonstrate that expression of NS1 is required to prevent NF-κB activation in cells infected with influenza A virus.
NF-κB is essential for the induction of the IFN-β gene by delNS1 virus infection.
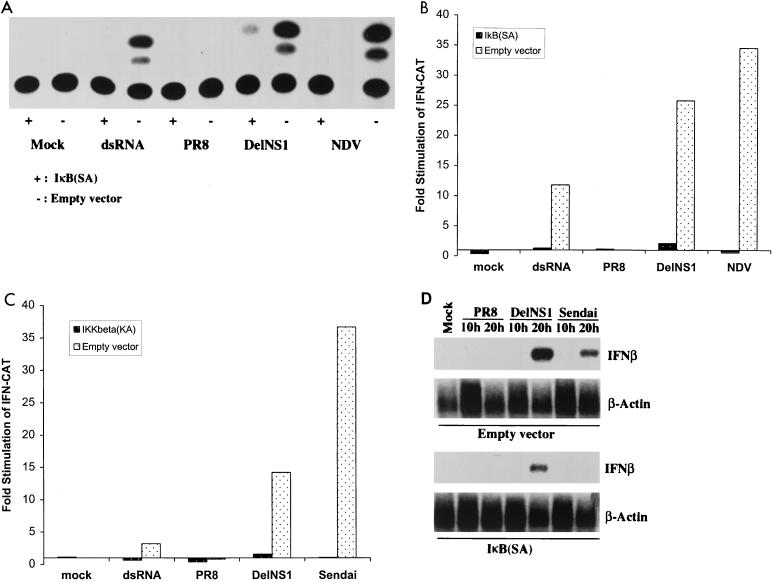
NF-κB has been shown to bind to the positive regulatory domain II of the IFN-β promoter and plays an essential role in regulating IFN-β transcription (32). We then investigated the role of NF-κB activation in IFN-β induction following delNS1 virus infection. Transfections were performed in 293 cells with a reporter gene, pIFN-CAT, which encodes CAT under the control of the mouse IFN-β promoter. Consistent with our Northern analysis in MEFs, infection with delNS1, but not with PR8 virus, activates this reporter gene (Fig. 3A, B, and C). To test the importance of NF-κB in the activation of the IFN-β promoter during delNS1 virus infection, we used a plasmid encoding a superrepressor form of IκB, IκB(SA), which encodes a human IκBα protein containing point mutations at its phosphorylation sites, S32 and S36 (6). Thus, this mutant form of IκBα binds to NF-κB, but cannot be phosphorylated and degraded, therefore inhibiting NF-κB activation. Cotransfection of pLPC-IκB(SA) with pIFN-CAT significantly attenuated the activation of the CAT reporter gene by delNS1 virus infection (Fig. 3A and B), demonstrating that NF-κB is essential for the activation of the IFN-β promoter. pLPC-IκB(SA) transfection also inhibited dsRNA- or NDV-induced CAT activity (Fig. 3A and B), in agreement with previous data suggesting that NF-κB plays a critical role for the regulation of the IFN-β promoter by different inducers (32, 41).
FIG. 3.
NF-κB is essential for delNS1 virus-induced IFN-β gene expression. (A) Monolayers of 293 cells were cotransfected with pIFN-CAT (encoding a CAT reporter gene under a mouse IFN-β promoter) and a plasmid expressing a superrepressor form of IκB, IκB(SA), or empty vector. One day posttransfection, cells were mock treated or transfected with 50 μg of dsRNA or infected with PR8, delNS1, or NDV viruses at an MOI of 1. One day later, CAT assays were performed with 5 μl of cell extracts. (B) Quantitative analysis of the results shown in Fig. 3A. (C) Monolayers of 293 cells were cotransfected with pIFN-CAT and a plasmid expressing a dominant-negative form of IκB kinase, IKKβ(KA), or empty vector. One day posttransfection, cells were treated as in panel A. Quantitative results are indicated. (D) 293 cells were transfected with a plasmid expressing IκB(SA) or empty vector. One day posttransfection, cells were infected with PR8, delNS1, or Sendai viruses at an MOI of 1 for the indicated times. RNAs were extracted and subjected to Northern blot analysis with probes specific for IFN-β and β-actin mRNAs.
IκB phosphorylation appears to be mediated by the recently identified IKKs. The IKK complex consists of two kinases, IKKα and IKKβ, and other regulatory subunits. Knockout studies demonstrated that IKKβ is especially important in mediating NF-κB activation by a variety of different inducers, including dsRNA (9). To test the possible involvement of IKKβ in the activation of NF-κB and induction of IFN-β by delNS1 virus, we used a plasmid encoding a dominant-negative form of IKKβ, IKKβ(KA). The point mutation K44 to A renders this kinase inactive, and it now functions as a dominant-negative mutant of wild-type IKKβ. Cotransfection of pLPC-IKKβ(KA) with pIFN-CAT significantly inhibited the delNS1 virus-mediated activation of the CAT reporter gene (Fig. 3C), suggesting that IKKβ is critically involved in the activation of the IFN-β promoter. pLPC-IKKβ(KA) transfection also blocked induction of the IFN-β promoter by dsRNA or by Sendai virus (Fig. 3C), suggesting that IKKβ is important in mediating IFN induction by different viruses. These data are consistent with a recent report using vesicular stomatitis virus infections of IKKβ−/− MEFs (9).
Next, we tested whether inhibition of NF-κB resulted in inhibition of the activation of the endogenous IFN-β gene by delNS1 virus infection. Consistent with our previous experiments, infection with delNS1, but not with PR8 virus, induces IFN-β mRNA production in 293 cells (Fig. 3D). Expression of IκB(SA) significantly inhibited delNS1 virus-induced IFN-β mRNA expression, demonstrating that activation of NF-κB is required for the induction of IFN-β gene transcription by delNS1 virus infection. As expected, IκB(SA) expression also inhibited Sendai virus-induced IFN-β mRNA synthesis (Fig. 3D).
The dsRNA binding domain of NS1 is sufficient for the inhibition of NF-κB activation and IFN-β induction.
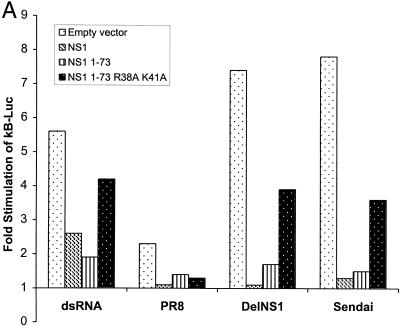
To determine how NS1 blocks the activation of NF-κB, we used an NF-κB reporter gene, pκB-Luc, which encodes a luciferase reporter gene under the control of two NF-κB binding sites. As expected, infection with delNS1 virus induced the expression of the reporter gene activity by approximately sevenfold in 293 cells, while PR8 virus infection only moderately affected reporter gene activity (Fig. 4A). These results are consistent with the EMSA and NF-κB translocation assays in MEFs (Fig. 2). Cotransfection of pκB-Luc with an expression plasmid for the NS1 protein, pCAGGS-NS1(SAM), almost completely inhibited delNS1 virus-induced NF-κB activation (Fig. 4A), demonstrating that NS1 is critically involved in suppressing NF-κB activation in influenza A virus-infected cells. NS1 expression could also inhibit dsRNA or Sendai virus-induced NF-κB activation (Fig. 4A), demonstrating that the NS1 protein is able to prevent NF-κB activation in the absence of expression of any other influenza A virus protein.
FIG. 4.
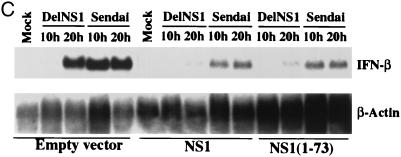
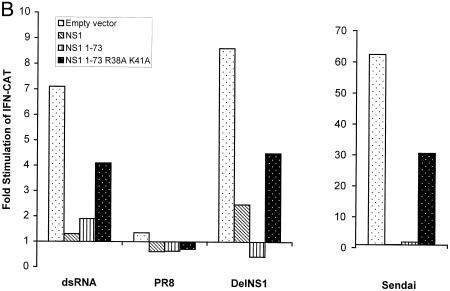
The dsRNA binding domain of the NS1 protein is sufficient to prevent NF-κB activation and IFN-β induction. (A) 293 cells were transfected with a plasmid containing a reporter gene under the control of an NF-κB-responsive promoter, pκB-Luc. In addition, cells were cotransfected with plasmids expressing NS1, NS1(1–73), NS1(1–73,R38A/K41A), or empty vector. One day posttransfection, cells were transfected with 10 μg of dsRNA or infected with PR8, delNS1, or Sendai viruses at an MOI of 1. Two days posttransfection, luciferase activity was determined. In all transfections, pRL-TK-Luc, encoding a Renilla luciferase under the control of a constitutive promoter, was cotransfected, and Renilla luciferase activity was used as an internal control to normalize the results. (B) 293 cells were cotransfected with pIFN-CAT and plasmids expressing NS1, NS1(1–73), or NS1(1–73,R38A/K41A) proteins or empty vector. One day posttransfection, cells were transfected with 10 μg of dsRNA or infected with PR8 or delNS1 viruses at an MOI of 1 or infected with Sendai virus at an MOI of 10. Two days posttransfection, CAT assays were performed, and the results were quantified. (C) 293 cells were transfected with plasmids expressing NS1 or NS1(1–73) proteins or empty vector. One day posttransfection, cells were infected with delNS1 or Sendai viruses at an MOI of 1 for the indicated time points. RNA was extracted and subjected to Northern blot analysis with probes specific for IFN-β and β-actin mRNAs.
The NS1 protein has been shown to contain two important domains, a dsRNA binding domain at the N terminus, and an effector domain at the C terminus. The effector domain refers to sequences required for NS1-mediated inhibition of mRNA splicing, polyadenylation, and transport (34). dsRNA generated during viral infection has been suggested to trigger the antiviral signaling pathways in virus-infected cells. Therefore, we tested whether the dsRNA binding domain of the NS1 protein is sufficient to inhibit NF-κB activation. We generated plasmids expressing two NS1 mutants, pCAGGS-NS1(1–73), encoding the minimal dsRNA binding domain of NS1 (36), and pCAGGS-NS1(1–73,R38A/K41A), encoding the same NS1 domain containing a double mutation responsible for the attenuation of its dsRNA binding activity (57). Expression of NS1(1–73) protein resulted in inhibition of NF-κB activation during delNS1 or Sendai virus infection or during dsRNA treatment, while this inhibition was significantly reduced when the dsRNA mutant NS1 protein NS1(1–73,R38A/K41A) was used (Fig. 4A).
We also investigated whether the dsRNA binding domain of the NS1 is sufficient to inhibit IFN-β induction by virus infection. Cotransfection of plasmids expressing full-length NS1 or NS1(1–73) with pIFN-CAT significantly inhibited delNS1 virus-induced activation of the CAT reporter gene under the control of the IFN-β promoter. This inhibitory effect was reduced when the dsRNA binding mutant NS1(1–73,R38A/K41A) was expressed (Fig. 4B). NS1(1–73) also inhibited dsRNA or Sendai virus-induced IFN promoter activity (Fig. 4B), demonstrating that the dsRNA binding domain of the NS1 protein prevents the activation of the IFN promoter, as well as the activation of NF-κB, in the absence of expression of any other influenza A virus protein. Consistent with that, we found that expression of NS1(1–73) is sufficient to inhibit virus-induced endogenous IFN-β mRNA synthesis in 293 cells (Fig. 4C).
Infection with a recombinant influenza A virus expressing a truncated NS1 protein of 129 amino acids, NS1(1–126) virus, prevents the activation of NF-κB and the induction of IFN-β.
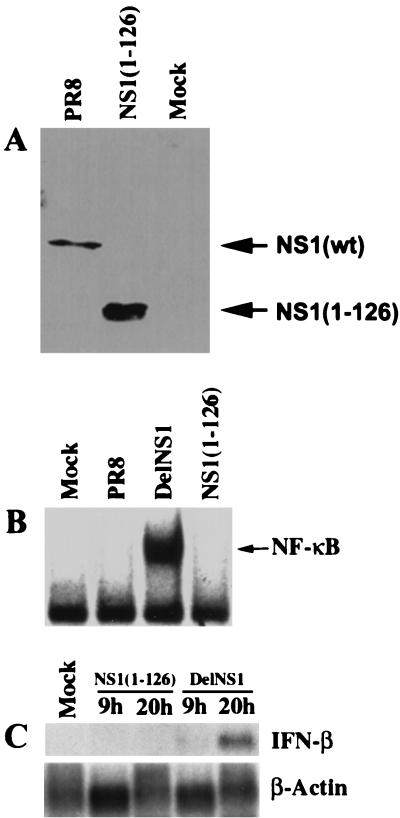
To confirm that the dsRNA binding domain of the NS1 protein is sufficient to inhibit NF-κB activation and IFN-β induction during influenza A virus infection, we used a recombinant influenza virus, NS1(1–126), which expresses a truncated form of the NS1 protein (Fig. 5A). This truncated NS1 protein has an intact dsRNA binding domain, but lacks the carboxy-terminal effector domain. We investigated by EMSA whether NS1(1–126) virus infection activates NF-κB. As expected, NS1(1–126) virus behaved the same as the wild-type PR8 virus (Fig. 5B). Thus, NS1(1–126) virus infection prevented NF-κB activation. Consistent with that, NS1(1–126) virus infection also prevented IFN-β mRNA expression as measured by Northern blotting (Fig. 5C). These results demonstrate that the dsRNA binding domain of the NS1 protein of influenza A virus is sufficient to suppress both NF-κB activation and IFN-β induction.
FIG. 5.
NS1(1–126) virus infection prevents the activation of NF-κB and the induction of IFN-β. (A) MDCK cells were infected with PR8 or NS1(1–126) viruses at an MOI of 1. Six hours p.i., cell extracts were made. Cell extracts (10 μl) were subjected to Western analysis with an anti-NS1 antibody. wt, wild type. (B) 293 cells were mock infected or infected with delNS1, PR8, or NS1(1–126) viruses at an MOI of 1. Six hours p.i., nuclear extracts were made and subjected to EMSA with a probe specific for activated NF-κB. (C) 293 cells were infected with either NS1(1–126) or delNS1 viruses. RNA was extracted at the indicated times and subjected to Northern blot analysis with probes specific for IFN-β and β-actin mRNAs.
DISCUSSION
The results presented here highlight a previously unknown effect of the NS1 protein of influenza A virus: the inhibition of activation of NF-κB. Several products might contribute to the activation of NF-κB during wild-type influenza A virus infection, including dsRNA, which is presumed to be generated during virus infections. In addition, overexpression of different influenza virus proteins, such as NP, M, and HA, was shown to result in the activation of NF-κB and the transcription of NF-κB-responsive reporter genes (16). Despite this observation involving the expression of individual influenza virus proteins, we found only a marginal activation of NF-κB in wild-type influenza virus-infected cells. Significantly, our results demonstrate that the NS1 protein of influenza A virus prevents the activation of NF-κB during virus infection. In fact, expression of the NS1 protein efficiently prevented the dsRNA-, Sendai virus-, and NDV-mediated activation of NF-κB. Moreover, infection with the influenza A virus NS1 knockout virus (delNS1) resulted in uncontrolled NF-κB activation. Downstream genes activated by NF-κB include genes involved in stimulation of T-cell proliferation, such as the interleukin 2 (29, 52), major histocompatibility complex class I (30), and B7 (66) genes, among others. In addition, NF-κB participates in the transcriptional activation of the IFN-β gene (28, 35), which leads to the expression of antiviral genes. Therefore, activation of NF-κB plays an important role in the inhibition of virus replication by stimulating both innate and adaptive immune responses in the host. In this study, we demonstrate that one possible mechanism by which viruses can inhibit antiviral defense mechanisms of the host is by preventing NF-κB activation.
Activation of NF-κB is mediated by phosphorylation of its inhibitor, IκB, by the IKK kinase complex (IKKα, IKKβ, and IKKγ) (12). In turn, IKKβ can become activated by PKR (9, 65). It is well established that binding to dsRNA results in the activation of PKR (61). Thus, it is possible that binding of dsRNA by the NS1 protein (26) prevents dsRNA from activating constitutive levels of endogenous PKR, therefore preventing NF-κB activation. Our observations are in agreement with this hypothesis. Thus, mutant forms of the NS1 protein containing only the dsRNA binding domain, NS1(1–73) and NS1(1–126), are competent in preventing NF-κB activation when expressed from plasmids in transfected cells or when expressed by a recombinant influenza A virus. In contrast, a mutant NS1 affected in its dsRNA binding ability, NS1(1–73,R38A/K41A), was not able to efficiently prevent NF-κB activation. On the other hand, expression of NS1(1–73) did not affect the activation by TNF or by overexpression of p65 of an NF-κB-responsive promoter (data not shown), suggesting that the dsRNA binding domain of NS1 is specifically involved in inhibiting virus and/or dsRNA-induced NF-κB activation. These results are consistent with a model in which synthesis of NS1 prevents the dsRNA-mediated activation of PKR, inhibiting the PKR-mediated stimulation of the NF-κB pathway. Specifically, dsRNA generated during influenza virus infection might be sequestered by the NS1 protein and might not be accessible for activation of PKR. On the other hand, the NS1 protein might be targeted to interact with PKR by virtue of its dsRNA binding properties, resulting in PKR inhibition. In fact, infection with delNS1 virus or with NS1 temperature-sensitive influenza A viruses, but not with wild-type virus, resulted in PKR activation (4, 27). Interestingly, transfection of a plasmid expressing a kinase dominant-negative form of PKR (PKR K296R) did not prevent the activation of the IFN-β promoter in delNS1- or Sendai virus-infected cells (data not shown). These results are in agreement with recent observations demonstrating that overexpression of catalytically inactive PKR results in stimulation of IKK, most likely through protein-protein interactions (5, 9). Therefore, experiments using enzymatically inactive dominant-negative mutants of PKR do not rule out the possibility that PKR is responsible for NF-κB activation in delNS1 virus-infected cells. Nevertheless, we cannot exclude that an unknown dsRNA-activated kinase different from PKR is the major target of the NS1-mediated inhibition of the NF-κB pathway.
NF-κB is an essential positive regulator for the activation of the IFN-β gene (35). Stimulation of the synthesis of IFN-β is presumed to initiate the IFN-α/β cascade (39). Previous studies also suggested that NF-κB might be important for the activation of IFN-α genes (9). Consistent with this, we observed that IFN-α/β synthesis was stimulated in delNS1 virus-infected cells (Fig. 1). In contrast, we were unable to detect significant levels of IFN-α/β mRNA in wild-type PR8 virus-infected cells by Northern blot analysis. These results suggest that the NS1 of influenza A virus serves as a virus-encoded IFN antagonist by inhibiting the synthesis of IFN-α/β. In addition to NF-κB, other transcription factors such as AP-1, IRF3, and IRF7 have also been implicated in the activation of the IFN-β gene during viral infections (58). Previous studies in our laboratory demonstrated that the NS1 protein also inhibits the activation of IRF-3 in virus-infected cells (54), further supporting a critical role of the NS1 protein in the inhibition of the IFN-α/β system during influenza virus infection (Fig. 6).
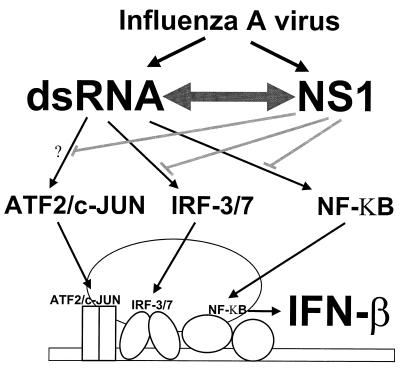
FIG. 6.
Model for the mechanism of inhibition of IFN-β induction by the NS1 protein of influenza A virus. Influenza virus infection results in the generation of dsRNA, which in turn activates transcription factors AP-1 (ATF2/c-JUN), IRFs (IRF-3/7), and NF-κB. Cooperation between these transcription factors upon binding to the IFN-β promoter facilitates the recruitment of the RNA polymerase II machinery (enhancesome formation) and stimulates the synthesis of IFN-β mRNA. Expression of NS1 protein during influenza virus infection prevents the dsRNA-mediated activation of IRF-3 (54) and of NF-κB (this study), therefore inhibiting IFN-β production. This inhibitory effect is dependent on the ability of NS1 to bind dsRNA. Therefore, activation of ATF2/c-JUN might also be prevented during influenza A virus infection by the NS1 protein. In addition, it should be noted that PKR, a dsRNA-activated kinase which plays an important role in different IFN pathways, both as an inducer of IFN synthesis, as well as an inhibitor of translation whose levels are transcriptionally increased in response to IFN and IRF-1 activation (43, 61), has also been found to be inhibited by the NS1 protein during influenza A virus infections (4, 27). Inhibition of IRFs and NF-κB activation by the NS1 protein most likely involves inhibition of PKR and/or uncharacterized upstream kinases activated by dsRNA.
Coevolution of viruses and hosts has resulted in the establishment of complex interactions which modulate virus pathogenicity and host disease. The IFN-α/β system serves as a potent first line of defense against virus infections. Activation of the synthesis of IFN during viral infection results in the transcriptional activation of many host genes involved in antiviral defense mechanisms. However, most viruses have responded to this antiviral system by encoding IFN antagonists. The NS1 of influenza A virus appears to target the synthesis of IFN-α/β by virtue of its dsRNA-binding properties. In this respect, it appears to have a functional role analogous to that of the E3L protein of vaccinia virus, a DNA virus (7). The presence of analogous proteins performing similar functions in vaccinia and influenza A viruses underscores the significance of the role of these proteins in the replication of viruses within the host.
ACKNOWLEDGMENTS
X.W. and M.L. contributed equally to this work.
We acknowledge members of the A.G.-S. and P.P. laboratories for critical discussions. We are also grateful to Otto Haller for critical reading of the manuscript and to Thorsten Wolff for providing pT3-NS-IAmut1 plasmid.
This work was supported by NIH research grants to P.P., A.G.-S., and A.A.B. and by a grant from the Austrian Science Fund to T.M.
REFERENCES
- 1.Algarté M, Nguyen H, Heylbroeck C, Lin R, Hiscott J. IκB-mediated inhibition of virus-induced beta interferon transcription. J Virol. 1999;73:2694–2702. doi: 10.1128/jvi.73.4.2694-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragón T, de La Luna S, Novoa I, Carrasco L, Ortín J, Nieto A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol. 2000;20:6259–6268. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., X. Wang, E. Mühlberger, V. Volchkov, J. Paragas, H.-D. Klenk, A. García-Sastre, and P. Palese. The Ebola virus VP35 protein functions as a type I interferon antagonist. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 4.Bergmann M, García-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet M C, Weil R, Dam E, Hovanessian A G, Meurs E F. PKR stimulates NF-κB irrespective of its kinase function by interacting with the IκB kinase complex. Mol Cell Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Li Y, Krug R M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 10.de la Luna S, Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 13.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami M, Enami K. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J Virol. 2000;74:5556–5561. doi: 10.1128/jvi.74.12.5556-5561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enami M, Palese P. High-efficiency formation of influenza virus transfectants. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flory E, Kunz M, Scheller C, Jassoy C, Stauber R, Rapp U R, Ludwig S. Influenza virus-induced NF-κB-dependent gene expression is mediated by overexpression of viral proteins and involves oxidative radicals and activation of IκB kinase. J Biol Chem. 2000;275:8307–8314. doi: 10.1074/jbc.275.12.8307. [DOI] [PubMed] [Google Scholar]
- 17.Fodor E, Devenish L, Engelhardt O G, Palese P, Brownlee G G, García-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita T, Nolan G P, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 20.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 21.Garcin D, Curran J, Kolakofsky D. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J Virol. 2000;74:8823–8830. doi: 10.1128/jvi.74.19.8823-8830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh S, May M J, Kopp E B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 24.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-α/β-mediated responses. FEBS Lett. 1999;459:205–210. doi: 10.1016/s0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 25.Haller O, Janzen C, Vialat P, Huerre M, Pavlovic J, Bouloy M. Proceedings of the 11th International Conference on Negative Strand Viruses. 2000. High virulence of attenuated Rift valley fever virus in mice lacking a functional type I interferon system; p. 98. Québec City, Canada. [Google Scholar]
- 26.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 27.Hatada E, Saito S, Fukuda R. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiscott J, Alper D, Cohen L, Leblanc J F, Sportza L, Wong A, Xanthoudakis S. Induction of human interferon gene expression is associated with a nuclear factor that interacts with the NF-κB site of the human immunodeficiency virus enhancer. J Virol. 1989;63:2557–2566. doi: 10.1128/jvi.63.6.2557-2566.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoyos B, Ballard D W, Bohnlein E, Siekevitz M, Greene W C. Kappa B-specific DNA binding proteins: role in the regulation of human interleukin-2 gene expression. Science. 1989;244:457–460. doi: 10.1126/science.2497518. [DOI] [PubMed] [Google Scholar]
- 30.Israël A, Le Bail O, Hatat D, Piette J, Kieran M, Logeat F, Wallach D, Fellous M, Kourilsky P. TNF stimulates expression of mouse MHC class I genes by inducing an NF-κB-like enhancer binding activity which displaces constitutive factors. EMBO J. 1989;8:3793–3800. doi: 10.1002/j.1460-2075.1989.tb08556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 32.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krug R M. Unique functions of the NS1 protein. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. Oxford, United Kingdom: Blackwell Science Ltd.; 1998. pp. 82–92. [Google Scholar]
- 35.Lenardo M J, Fan C M, Maniatis T, Baltimore D. The involvement of NF-κB in β-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Lynch P A, Chien C Y, Montelione G T, Krug R M, Berman H M. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Qian X Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 39.Marié I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marión R M, Zürcher T, de la Luna S, Ortín J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 41.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 42.Nemeroff M E, Barabino S M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen H, Lin R, Hiscott J. Activation of multiple growth regulatory genes following inducible expression of IRF-1 or IRF/RelA fusion proteins. Oncogene. 1997;15:1425–1435. doi: 10.1038/sj.onc.1201318. [DOI] [PubMed] [Google Scholar]
- 44.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Percy N, Barclay W S, García-Sastre A, Palese P. Expression of a foreign protein by influenza A virus. J Virol. 1994;68:4486–4492. doi: 10.1128/jvi.68.7.4486-4492.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Régnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 48.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 49.Scheinman R I, Beg A A, Baldwin A S., Jr NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol Cell Biol. 1993;13:6089–6101. doi: 10.1128/mcb.13.10.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlender J, Bossert B, Buchholz U, Conzelmann K-K. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J Virol. 2000;74:8234–8242. doi: 10.1128/jvi.74.18.8234-8242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 52.Serfling E, Barthelmas R, Pfeuffer I, Schenk B, Zarius S, Swoboda R, Mercurio F, Karin M. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. EMBO J. 1989;8:465–473. doi: 10.1002/j.1460-2075.1989.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu K, Handa H, Nakada S, Nagata K. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 1994;22:5047–5053. doi: 10.1093/nar/22.23.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, García-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talon J, Salvatore M, O'Neill R E, Nakaya Y, Zheng H, Muster T, García-Sastre A, Palese P. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan S L, Katze M G. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 57.Wang W, Riedel K, Lynch P, Chien C Y, Montelione G T, Krug R M. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 59.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber F, Bridgen A, Elliott R M. Proceedings of the 11th International Conference on Negative Strand RNA Viruses. 2000. The Bunyamwera virus nonstructural protein NSs is a repressor of the viral polymerase and confers interferon antagonistic function; p. 98. Québec City, Canada. [Google Scholar]
- 61.Williams B R. PKR: a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 62.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 63.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young D F, Didcock L, Goodbourn S, Randall R E. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]
- 65.Zamanian-Daryoush M, Mogensen T H, DiDonato J A, Williams B R. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol Cell Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao J, Freeman G J, Gray G S, Nadler L M, Glimcher L H. A cell type-specific enhancer in the human B7.1 gene regulated by NF-κB. J Exp Med. 1996;183:777–789. doi: 10.1084/jem.183.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]