Abstract
The 5′ leader of Rous sarcoma virus (RSV) genomic RNA and of retroviruses in general is long and contains stable secondary structures that are critical in the early and late steps of virus replication such as RNA dimerization and packaging and in the process of reverse transcription. The initiation of RSV Gag translation has been reported to be 5′ cap dependent and controlled by three short open reading frames located in the 380-nucleotide leader upstream of the Gag start codon. Translation of RSV Gag would thus differ from that prevailing in other retroviruses such as murine leukemia virus, reticuloendotheliosis virus type A, and simian immunodeficiency virus, in which an internal ribosome entry segment (IRES) in the 5′ end of the genomic RNA directs efficient Gag expression despite stable 5′ secondary structures. This prompted us to investigate whether RSV Gag translation might be controlled by an IRES-dependent mechanism. The results show that the 5′ leaders of RSV and v-Src RNA exhibit IRES properties, since these viral elements can promote efficient translation of monocistronic RNAs in conditions inhibiting 5′ cap-dependent translation. When inserted between two cistrons in a canonical bicistronic construct, both the RSV and v-Src leaders promote expression of the 3′ cistron. A genetic analysis of the RSV leader allowed the identification of two nonoverlapping 5′ and 3′ leader domains with IRES activity. In addition, the v-Src leader was found to contain unique 3′ sequences promoting an efficient reinitiation of translation. Taken together, these data lead us to propose a new model for RSV translation.
Most eucaryotic mRNAs use the 5′ proximal AUG codon as the site for translation initiation. According to the scanning model, the 40S ribosomal subunit binds to the methylated 5′ cap structure of an mRNA and then scans in the 5′-to-3′ direction until an initiation codon is recognized (39, 42). Genetic elements within the 5′ untranslated region control ribosome access to the downstream major coding region as follows. (i) The consensus sequences surrounding the initiator codon [(A/G)CC AUG G] significantly modulate the efficiency of translation initiation (39). (ii) Stable secondary structures (ΔG < −50 kcal/mol) will inhibit translation initiation by halting ribosome scanning (35, 37). (iii) Small open reading frames (ORFs) upstream of the major coding region (uORFs) attenuate translation initiation at downstream AUGs. Two classes of inhibitory uORFs have been described. In the first class, attenuation occurs because of the intrinsic inefficiency of the reinitiation mechanism. In the second class, attenuation is sequence dependent (13, 14, 30, 31, 44, 62–64). These uORFs seem to act in cis on the translating ribosome, and therefore it has been proposed that the nascent peptide translated from the uORF specifically interacts with a component of the translation apparatus to stall the ribosome at the uORF stop codon (for a review, see reference 41). As a result, ribosomes cease scanning and do not arrive at AUG codons downstream of the uORF. However, when the initiation codon of the uORF is located in a poor initiation context, the majority of the scanning ribosomes ignores the uORF and initiates translation at a downstream AUG. This process is known as leaky scanning (34, 38).
In picornaviruses, translation initiation is promoted by a small domain located within the 5′ region of the viral RNA, designated the internal ribosome entry segment (IRES) (49). In these viruses, the IRES directs the ribosomes to the translation initiation site in a cap-independent manner. When inserted between two genes in a canonical bicistronic construct, the IRES directs expression of the 3′ cistron independently from that of the 5′ one. IRESs have also been identified in retroviruses such as murine leukemia virus (MLV) (Friend MLV and Moloney MLV) (3, 12, 60), Harvey murine sarcoma virus (HaMSV) (4), avian reticuloendotheliosis virus REV-A (40) and simian immunodeficiency virus (SIV) (47). In these viruses, the 5′ leader is formed of stable secondary structures that are required for several steps of virus replication such as genomic RNA dimerization and packaging and for the process of reverse transcription. These stable secondary structures are thought to strongly interfere with ribosome scanning, and IRES-dependent translation might provide a direct way for the ribosomes to gain access to the initiation codon of Gag.
As is the case for MLV and SIV, the 5′ leader of Rous sarcoma virus (RSV) is formed of several stable secondary structures necessary for genomic RNA packaging and reverse transcription (12). In addition, it contains three conserved uORFs (26). Mutations of the initiation codon of these greatly perturbs gene expression and viral replication (17, 45, 46, 54). For example, mutations of uORF 1 and/or uORF 3 cause a strong reduction of RNA packaging (17). Although no direct evidence exists for the translation of these sequences in vivo, their translational properties have been proposed to be implicated in the regulation of viral replication. Although conflicting data have been obtained with respect to the impact of these uORFs on Gag translation, upstream AUG 1 has been found to be a major ribosome binding site on RSV RNA (10, 11, 52, 53, 54). In vitro, its translation generates the predicted 7-amino-acid peptide (27), thus allowing two models for Gag translation to be proposed: one based on reinitiation (29) and the other based on shunt (11).
In view of the discovery of IRESs in MLV, REV-A and SIV, we wanted to reexamine the mechanism of RSV translation. Data presented here show that the 5′ leader of RSV genomic RNA contains an IRES and therefore suggest that synthesis of Gag occurs through a cap-independent mechanism. In addition, our results show that the 5′ leader of v-Src RNA contains a unique 3′ region, downstream of the region corresponding to the RSV 5′ leader, which favors efficient translation reinitiation.
MATERIALS AND METHODS
General methods.
Standard procedures were used for restriction nuclease digestion and plasmid DNA construction. A variant of Escherichia coli HB101, strain 1035 (recA mutant), was used for plasmid DNA amplification. The details of the plasmid constructions are given below. Numbering is with respect to the RNA cap site (position +1) unless otherwise stated.
DNA constructs.
RSV DNA segments from positions 1 to 379, 1 to 324, 1 to 200, 230 to 379, and 285 to 379 were generated by PCR using plasmid pLAD4 as the template (5), followed by digestion with NheI (PCR-added restriction site). The v-src DNA segments from positions 1 to 473, 1 to 442, and 407 to 473 were generated by PCR using plasmid pSJ2 (5) as the template, followed by digestion with NheI (PCR-added restriction site). The MLV DNA fragments from positions 1 to 620 and 212 to 565 were obtained by digestion with NheI of pMLV-CB63 and pMLV-CB39, respectively (3). To construct plasmids pBi-RSV (1–379), pBi-RSV (1–324), pBi-RSV (1–200), pBi-RSV (230–379), pBi-RSV (285–372), pBi-SRC (1–473), pBi-SRC (1–442), pBi-SRC (407–473), pBi-MLV (1–620), and pBi-MLV (212–651), each fragment described above was inserted between neo and lacZ of plasmid pBi digested by NheI. To construct plasmids pBi-RSV loop (1–379), pBi-RSV loop (1–200), pBi-RSV loop (230–379), pBi-RSV loop (285–379), pBi-SRC loop (1–473), pBi-SRC loop (1–442), pBi-SRC loop (407–473), and pBi-MLV loop (1–620), each fragment described above was inserted between neo and lacZ of plasmid pBi loop digested by NheI. Plasmids pBi and pBi loop were obtained after digestion with BstBI and SmaI, Klenow fragment filling, and subsequent religation of plasmids pMLV-CB63 (3) and pD 891. The pD 891 plasmid was obtained by inserting into the HindIII site of plasmid pMLV-CB63 a sequence with the ability to form a stable stem-loop structure (ΔG = −50 kcal/mol). To construct pBi-RSV (103–379) and pBi-RSV loop (103–379), pCB-65 and pCB-65 loop were digested with BstBI and BstEII, Klenow fragment filled, and subsequently religated. To construct plasmids pM-RSV (1–372) and pM-RSV (1–200) and plasmid pM-SRC (1–475), each previously described fragment was inserted into pCB-63 digested with NheI; subsequently the SmaI-XbaI fragment of each construct, which contains the RSV or v-src 5′ leader and the lacZ gene, was ligated into pMLV-CB28 digested with EcoRV and XbaI (3). In plasmids containing the insert RSV (1–200), the lacZ coding region was fused to the second codon of uORF 3, which led to a poor initiation context (UGC AUG ACG); in all other plasmids containing an RSV insert, the context of the lacZ initiation codon was the same as that of the wild-type AUGgag (AGC AUG G). In plasmids containing a v-Src insert, the context of the lacZ initiation codon was the same as that of the wild-type AUGv-src (ACC AUG G). The pβ-actin-LacZ plasmid contains the lacZ gene under the control of the rat actin promoter. This plasmid was a gift of P. Savatier (Lyon, France). Plasmids pMLP-P2A and pMLP-PR2A contain the poliovirus protease 2A coding sequence derived from poliovirus type 1 (Mahoney strain). In pMLP-P2A, the protease coding sequence was inserted downstream of the adenovirus major late promoter and its tripartite leader. The pMLP-PR2A plasmid contains the insert in the reverse orientation and served as a control. These plasmids were gifts of N. Fouillot (Orsay, France).
Cell culture and DNA transfection.
Murine NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (GIBCO-BRL) containing 10% newborn calf serum at 37°C in the presence of 5% CO2. Avian QT6 cells were grown in Ham's F10 medium (Flow) supplemented with 10% tryptose phosphate broth (Difco), 5% fetal calf serum (Eurobio), 1% chicken serum (Eurobio), 0.2% NaHCO3 at 37°C in the presence of 5% CO2. Transfections were performed on 70% confluent plate by the Fugene method (Roche Inc.) using 1.5 μg of plasmid DNA. Cotransfection experiments were performed with the same method using 1.5 μg of monocistonic plasmid and 1.5 μg of either pMLP-P2A or pMLP-PR2A. Twenty-four hours after transfection, cells were split in three separate aliquots. After 24 additional hours, cells of the first aliquot were fixed and histochemically stained for lacZ expression, cells of the second aliquot were lysed for extraction of cellular proteins, and total RNA was extracted from the third aliquot. Cellular proteins were submitted to a β-galactosidase enzymatic test, and total RNA was analyzed by Northern blot or dot blot analysis using a lacZ probe.
lacZ histochemical staining.
After transfection, cells were fixed with 2% formaldehyde and 0.2% glutaraldehyde, washed twice with phosphate-buffered saline and incubated for 6 h at 37°C in phosphate-buffered saline containing 5-bromo-4-chloro-3-indolyl-β-d-glucoronic acid (X-Gal) (1 mg/ml), ferrocyanure (4 mM), ferrycyanure (4 mM), and MgCl2 (4 mM).
RNA extraction and slot blot and Northern blot analysis.
Extraction of cellular RNAs from transfected cells was performed using the Trizol reagent (Life Technologies) according to the manufacturer's instructions. Northern blot and slot blot analysis were performed as previously described (33). A 32P-labeled probe complementary to the lacZ gene (ClaI-ClaI fragment of pCB 63) was generated using the prim-It room temperature kit (Stratagene).
Protein extraction and enzymatic activities.
Cellular proteins were extracted using the β-galactosidase enzyme assay kit (Promega). Protein concentration was determined using the Micro bicinchoninic acid kit (Pierce). Neomycin phosphotransferase (encoded by neo) activity was measured by [γ-32P]ATP phosphate transfer to neomycin (57). β-Galactosidase activity was determined spectrophotometrically (β-galactosidase enzyme assay system; Promega). In order to check for linearity of the assays and to determine the relative activities of neomycin and β-galactosidase in each cell extract, an internal standard curve was generated by serial dilutions of the cell extract giving the strongest Neo or β-galactosidase activity.
RESULTS
Translation mediated by 5′ leader of RSV genomic and v-src RNAs is not inhibited by poliovirus protease 2A.
In poliovirus-infected cells, cap-dependent translation is strongly inhibited by the viral protease 2A (15, 19, 28). Poliovirus protease 2A cleaves the initiation factor eIF4G, a subunit of eIF4F, which bridges together the ribosome and the 5′ cap structure of an mRNA. Intact eIF4G is required in cap-dependent translation mediated by canonical scanning, leaky scanning, reinitiation, and shunt mechanisms. As has been described previously, transient expression of poliovirus protease 2A strongly inhibits cap-dependent translation but not IRES-dependent translation (3, 4, 21). To examine whether gag and v-src translation is cap-dependent, a monocistronic construct with the RSV or v-src 5′ leader sequences upstream of a reporter gene (lacZ) (Fig. 1 and 2) was transfected into NIH 3T3 cells with a plasmid encoding poliovirus protease 2A (pMLP-P2A). In this plasmid, the coding region of the protease 2A is preceded by the adenovirus major late promoter and its tripartite leader, which has been shown to promote efficient translation even in the presence of the poliovirus protease 2A (16). A plasmid containing the protease 2A coding sequence in the reverse orientation was used as a negative control (pMLP-PR2A). The pβ-actin-LacZ plasmid was used as a positive control for cap-dependent translation, and pMLV-CB93 with the MLV IRES was used as a positive control for IRES-dependent translation. Two days after DNA transfection, cells were submitted to histochemical staining for β-galactosidase activity (see Fig. 3). In addition, translation efficiency was estimated from the ratio between the β-galactosidase activity and the concentration of lacZ mRNA (expressed in arbitrary units) (see Fig. 4 for a summary of the results).
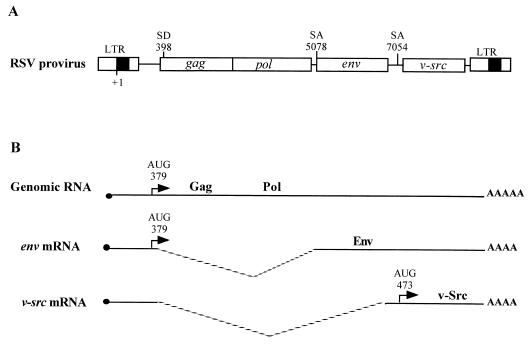
FIG. 1.
Genetic organization and expression of RSV. (A) RSV provirus. LTR, long terminal repeat; SD, splice donor site; SA, splice acceptor site. Numbering is with respect to the RNA cap site (position +1). (B) Viral RNAs (genomic RNA and spliced RNAs).
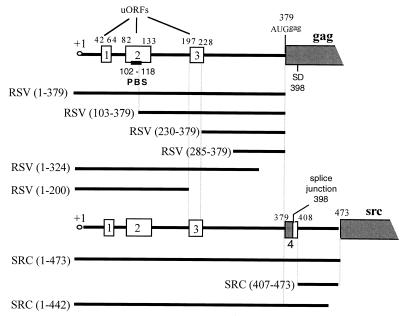
FIG. 2.
Schematic representation of the 5′ leader of RSV genomic and v-src RNAs. The uORFs are represented by rectangles. PBS, primer binding site; SD, splice donor site. Numbering is with respect to the cap site (position +1). The contexts for the initiation codons were as follows: for uORF 1, UUG AUG A; for uORF 2, UGC AUG A; for uORF 3, UCG AUG A; for Gag, AGC AUG G; and for v-src, ACC AUG G. For details of the molecular constructs, see Materials and Methods.
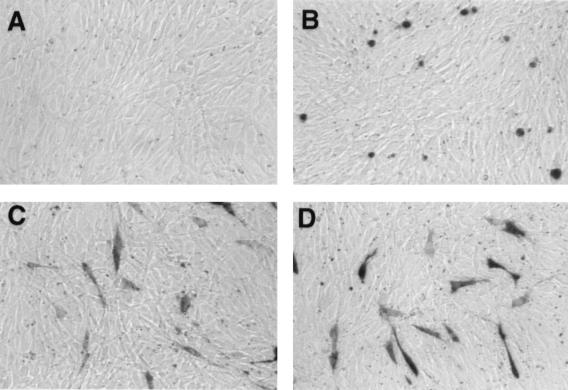
FIG. 3.
Histochemical staining of NIH 3T3 cells for lacZ expression. (A) Cotransfection of pMLP-P2A and pβ-actin-LacZ. The expression of poliovirus protease 2A inhibits lacZ expression. (B) Cotransfection of pMLP-P2A and pM-RSV (1–379). The expression of poliovirus protease 2A does not inhibit lacZ expression, and cells with a round morphology can be visualized. (C) Cotransfection of pMLP-PR2A and pβ-actin-LacZ. (D) Cotransfection of pMLP-PR2A and pM-RSV (1–379).
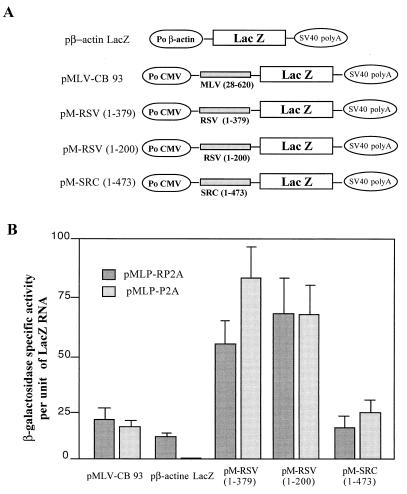
FIG. 4.
Effect of poliovirus protease 2A on translation efficiency of recombinant RSV lacZ RNA. (A) The recombinant plasmids contain the MLV leader (positions 28 to 620) for pMLV-CB 93; the RSV leaders (positions 1 to 379) and (positions 1 to 200) for pM-RSV (1–379) and pM-RSV (1–200), respectively; and the v-src leader (positions 1 to 473) for pM-SRC (1–473). (B) pMLP-P2A encodes the poliovirus protease 2A under the control of the adenovirus late promoter; pMLP-PR2A has the coding sequence of poliovirus protease 2A in the reverse orientation and was used as a control. Cotransfections were performed for each recombinant plasmid with pMLP-P2A or pMLP-PR2A. Forty-eight hours later, lacZ expression was calculated as the ratio between β-galactosidase-specific activity and the amount of recombinant lacZ RNA present in transfected cells (see Materials and Methods). Results are in arbitrary units and are the averages of results from two independent experiments.
Histochemical staining of NIH 3T3 cells for lacZ (Fig. 3) revealed that poliovirus protease 2A drastically inhibited cap-dependent translation since no blue cells were detected when pβ-actin lacZ and pMLP-P2A were coexpressed by DNA transfection (Fig. 3, compare panels A and C). In contrast, efficient β-galactosidase expression occurred after cotransfection of either pMLP-P2A or pMLP-PR2A with pMLV-CB93, pM-RSV (1–379), pM-RSV (1–200), or pM-SRC (1–479) (data shown for pM-RSV (1–379) [Fig. 3, compare panels B and D]). It should also be noted that the expression of poliovirus protease 2A in NIH 3T3 cells caused a round-shaped morphology previously reported to be a proapoptotic phenotype (2, 23) (Fig. 3B). In contrast, cells cotransfected with pMLP-PR2A and any one of the monocistronic lacZ-carrying plasmids exhibited a normal morphology (Fig. 3C and D).
The levels of β-galactosidase activity and of recombinant lacZ RNA in cells cotransfected with the lacZ construct and pMLP-P2A or pMLP-PR2A were monitored, and the values reported are relative levels of enzymatic activity (Fig. 4). In NIH 3T3 cells expressing the pβ-actin lacZ construct and poliovirus protease 2A, the relative level of β-galactosidase activity was drastically lower than that in control cells, confirming that protease 2A strongly inhibited cap-dependent translation (Fig. 4). In contrast, coexpression of protease 2A and pMLV-CB93, pM-RSV (1–379), pM-RSV (1–200), or pM-SRC (1–479) resulted in relative levels of β-galactosidase activity similar to those obtained in the absence of poliovirus protease 2A (Fig. 4). These findings indicate that synthesis of RSV Gag and v-Src does not proceed via a cap-dependent mechanism but most probably uses an IRES. In addition, these results indicate that the AUG of uORF 3 is a strong site for translation initiation and that ribosome recognition at this site is probably cap independent [Fig. 2, pM-RSV (1–200)].
5′ leaders of RSV and v-src RNAs promote efficient translation of downstream cistron in a bicistronic RNA.
In order to further characterize the translation properties of the 5′ leaders of RSV and v-src RNAs, bicistronic plasmids were constructed in which the 5′ leader of RSV genomic or v-src RNAs was inserted between the neo and lacZ genes. In these constructs, a cytomegalovirus promoter allows the production of bicistronic RNAs in cell culture (Fig. 5). Expression of the 5′ cistron (neo) is cap dependent, whereas that of the 3′ cistron (lacZ) can occur only if the intercistronic region contains a functional IRES (3, 4, 12, 40, 49). These bicistronic plasmids were expressed in avian QT6 cells by means of DNA transfection. For each experiment, the relative efficiencies of neo and lacZ translation were calculated as the ratio of the specific enzymatic activity (Neo or β-galactosidase) to the level of cellular bicistronic mRNA (see Materials and Methods).
FIG. 5.
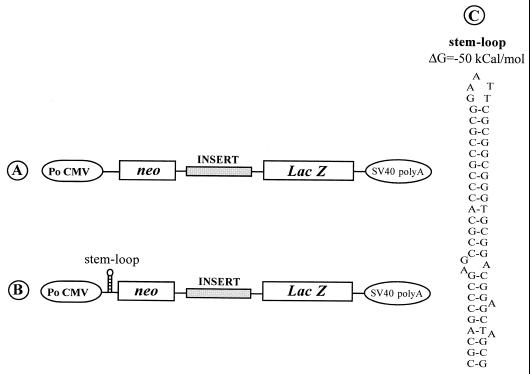
Schematic representation of the bicistronic constructs used for transfection of QT6 cells. Po CMV, cytomegalovirus early promoter; SV40, simian virus 40. (A and B) DNA constructs direct synthesis of bicistronic capped RNAs. However, the presence of a stable stem-loop structure just upstream of neo in the construct shown in panel B is thought to inhibits neo expression (36). (C) Sequence and computer-predicted structure of the stem-loop structure (ΔG = −50 kcal/mol).
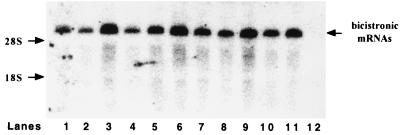
To rule out that a cryptic promoter and/or cryptic splice sites could function during the expression of the bicistronic constructs, RNAs from transfected cells were analyzed by Northern analysis using a lacZ probe. As shown in Fig. 6, lanes 1, 3, 4, 6, 8, and 10, only one major RNA species of the expected size was detected. Thus, it can be concluded that all lacZ RNAs synthesized in transfected QT6 cells were bicistronic.
FIG. 6.
Northern blot analysis of recombinant bicistronic neo-lacZ mRNAs produced in QT6 cells. Total RNA was extracted from QT6 cells 48 h after transfection with the bicistronic construct pBi-RSV (1–379) (lane 1), pBi-RSV loop (1–379) (lane 2), pBi-RSV (103–379) (lane 3), pBi-RSV (230–379) (lane 4), pBi-RSV loop (230–379) (lane 5), pBi-RSV (1–200) (lane 6), pBi-RSV loop (1–200) (lane 7), pBi-SRC (1–473) (lane 8), pBi-SRC loop (1–473) (lane 9), pBi-SRC (407–473) (lane 10), or pBi-SRC loop (407–473) (lane 11) or a negative control (lane 12). RNAs were subjected to electrophoresis in an 0.8% agarose gel and transferred to a nitrocellulose membrane. Hybridization was performed with a 32P-labeled probe corresponding to nucleotides 829 to 3105 of the lacZ cistron (ClaI-ClaI fragment). The 28S and 18S rRNAs were revealed by ethidium bromide staining prior to RNA transfer. Positions of the 18S and 28S rRNA and of the bicistronic neo-lacZ mRNAs are indicated.
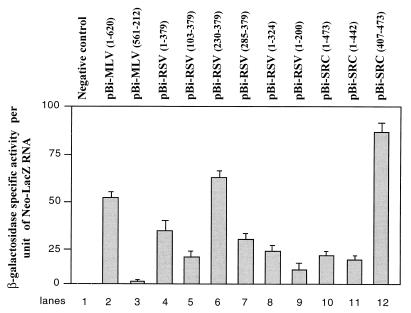
As shown in Fig. 7, the MLV IRES directed a high level of lacZ expression (Fig. 7, lane 2) whereas the MLV IRES in the reverse orientation was inactive (Fig. 7, lane 3). The 5′ leader of RSV and v-src RNAs were active in the canonical bicistronic assay (Fig. 7, lanes 4 and 10). In an attempt to map the regions of the RSV and v-src 5′ leaders responsible for the IRES activity, bicistronic DNAs were constructed and transfected into QT6 cells (Fig. 7, lanes 4 to 9 for RSV RNA and lanes 11 and 12 for v-Src RNA). Clearly two domains of the RSV leader exhibited IRES activity, namely nucleotides (nt) 1 to 200 and nt 285 to 379, corresponding to the 5′ and 3′ ends of the leader (Fig. 7, lanes 7 and 9). Addition of nt 230 to 284 increased by about twofold the IRES activity of the 3′ region of the leader using the canonical bicistronic assays (Fig. 7, lanes 6 and 7). The 5′ domain (nt 1 to 200) had significantly a lower activity than the 3′ domain (nt 285 to 379) (Fig. 7, compare lanes 7 and 9). Finally, a unique domain of the v-src leader (position 407 to 473 [Fig. 2]) was found to direct strong lacZ expression (Fig. 7, lane 11). To rule out the possibility that lacZ expression could be due to translation reinitiation, we examined the influence of a stable 5′ stem-loop structure on the relative expression of neo and lacZ genes.
FIG. 7.
Synthesis of β-galactosidase directed by bicistronic neo-LacZ RNAs in QT6 cells. Transfection of the bicistronic plasmids was performed by the Fugene method (Materials and Methods). Two days later, β-galactosidase-specific activity was measured on cellular extracts and neo-lacZ RNA was quantified by slot blot analysis. Efficiency of lacZ translation for each bicistronic RNA is given as the ratio of β-galactosidase-specific activity per arbitrary unit of bicistronic neo-lacZ mRNA. Results are the averages of the results from two independent experiments.
Influence of 5′ stem-loop structure on the translation of bicistronic neo-lacZ mRNAs.
A stable stem-loop structure (ΔG = −50 kcal/mol) was inserted 30 nucleotides upstream of the neo AUG codon in the bicistronic plasmids (Fig. 5C). This stem-loop structure was designed to block ribosome scanning and thus to impair cap-dependent translation of neo cistron and, as a consequence, of lacZ in situations where reinitiation is driving translation of the second cistron of this bicistronic RNA (37). Northern blot analysis confirmed that the mRNAs synthesized in the transfected QT6 cells were bicistronic (Fig. 6, lanes 2, 5, 7, 9, and 11).
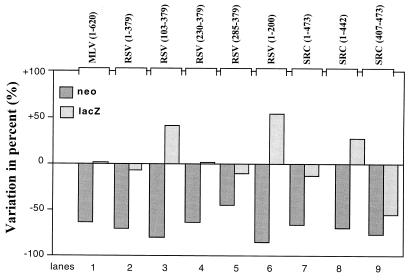
As shown in Fig. 8, the presence of the stable stem-loop structure reduced neo expression by 50 to 80% with respect to the control. In contrast, lacZ expression directed by pBi-MLV loop (1–620), pBi-RSV loop (1–379), pBi-RSV loop (230–379), pBi-RSV loop (285–379), and pBi-SRC loop (1–473) was not inhibited. Interestingly, the expression of lacZ was even enhanced with pBi-RSV loop (103–379) (Fig. 8, lane 3) and pBi-RSV loop (1–200) (Fig. 8, lane 6). This confirms that in the corresponding RNAs, translation of the lacZ cistron is independent from that of the neo cistron and that the intercistronic region contains a functional IRES. However, in pBi-SRC loop (403–476) bicistronic RNA, the presence of the stem-loop decreased lacZ translation efficiency by 70% with respect to the control (Fig. 8, lane 9). This shows that in these bicistronic RNAs, the translations of neo and lacZ do not occur independently, indicating that the 5′ leader of v-Src RNA from positions 403 to 476 does not contain an IRES but instead allows efficient reinitiation.
FIG. 8.
Influence of stable 5′ stem-loop structure on neo and lacZ expression. QT6 cells were transfected with a bicistronic plasmid allowing synthesis of a bicistronic RNA with or without the potential to form a very stable secondary structure (ΔG = −50 kcal/mol) 5′ to the neo cistron. The inserts used as the intercistronic spacers are indicated at the top of the figure. Forty-eight hours later, the percent variation of Neo and β-galactosidase enzymatic activities in the presence or absence of the 5′ stem-loop structure was calculated (activity with the stem-loop − activity without the stem loop)/activity without the stem-loop × 100).
DISCUSSION
The 5′ leader of avian sarcoma and leukosis virus (ASLV) and of other related oncoviruses is long and contains several stable secondary structures that are involved in key functions of the viral life cycle such as dimerization and packaging of the genomic RNA and in the process of reverse transcription (8, 11, 65). The 5′ leaders of several MLV oncoviruses and SIV each contain an IRES (4, 5, 40, 47, 60). This suggests that to initiate translation of the viral RNA, ribosomes do not scan the 5′ leader but are directly recruited at or near the Gag initiation codon. This is in agreement with the observation that stable secondary structures do not allow efficient ribosome scanning (35, 37). In addition to stable secondary structures, the 5′ leader of RSV RNA contains three conserved, short ORFs (uORFs) which seem to constitute an additional barrier for efficient translation initiation mediated by any of the known cap-dependent mechanisms (scanning, leaky scanning, and reinitiation). This prompted us to reexamine the mechanism of translation initiation of RSV RNA.
As first indicated by the results obtained with monocistronic RNAs, the 5′ leaders of RSV and v-src RNAs most probably both contain an IRES that drives translation, since expression of the poliovirus protease 2A, which cleaves the initiation factor eIF4G, thus impairing cap-dependent translation, had no effect on β-galactosidase synthesis (Fig. 3 and 4). In the monocistronic RSV lacZ RNA, which contains the 5′ leader from position 1 to 200, lacZ translation uses the initiation codon of uORF 3, which is in a poor consensus context (UGC AUG ACC). Therefore, if lacZ expression were to occur by a canonical ribosome scanning, the AUG of uORF 3 would have probably been poorly recognized by the ribosomes and thus lacZ would have been poorly expressed. On the contrary, pM-RSV (1–200) was well expressed in the presence or absence of protease 2A (Fig. 4) and thus most probably was expressed by an IRES-directed mechanism. These findings were confirmed by investigating the translation of canonical bicistronic neo-lacZ RNAs in which neo expression is 5′ cap dependent whereas that of lacZ must rely on either reinitiation or the presence of an IRES (Fig. 5). As shown in Fig. 7 and 8, both the RSV and v-src leaders are able to drive translation of the 3′ cistron of bicistronic RNA independently from that of the 5′ cistron, a property of IRESs (49). Interestingly, the first 200 nt 5′ of the RSV leader display a low level of IRES activity in avian cells (Fig. 7, lane 9) whereas the last 150 nt (positions 230 to 379) 3′ of the leader direct a strong IRES-dependent translation (Fig. 7, lane 6). Also, site-directed mutagenesis suggests that the AUG of Gag is probably an integral part of the RSV IRES. The AUG of Gag was changed to a CUG known to be functional for the synthesis of MLV GlycoGag (55, 60) and although the CUG was in an optimal context (AGC CUG G in place of AGC AUG G) (38), this mutation completely inhibited the RSV IRES activity (data not shown). This bipartite IRES in the RSV 5′ leader may ensure a high level of RSV and more generally of ASLV expression in infected cells. Possible interactions between the 5′ and 3′ domains of this bipartite IRES, as suggested by the scheme shown in Fig. 9, are presently under investigation.
FIG. 9.
Schematic representation of RSV 5′ leader structure upon ribosome binding to uORF 1. Numbering is with respect to the cap site (position +1). PBS, primer binding site. In this model, nucleotides from positions 1 to 77 are engaged in ribosome binding at uORF 1 (10, 11). Structures described in other models (26) and conserved in this model are indicated; the L3 stem-loop is implicated in RSV RNA dimerization (5, 20), and the US-IR stem-loop is implicated in the initiation of reverse transcription (43). The ΔG of the structure is estimated to be −148 kcal/mol. Note that the extended secondary structure with stem-loops and bulges (from the PBS to the AUG of Gag) corresponds to the bipartite IRES.
As does the RSV 5′ leader (positions 1 to 379), the v-src leader contains a unique sequence (positions 407 to 473) (Fig. 2) with a stop codon in-frame with the gag start codon. This small v-src sequence was found to promote a high level of translation reinitiation (Fig. 7, lane 12, and Fig. 8, lane 9). These findings would thus favor the notion that translation of v-src is mediated by the IRES of the RSV 5′ leader followed by a reinitiation mechanism between the AUGgag and AUGv-src. In agreement with this, a previous work has shown that mutating the stop codon downstream of the AUGgag resulted in the production of a v-Src fusion protein (32).
In the light of the present data, previous studies on the RSV should be reinterpreted. We present an RSV translation model (Fig. 9) that could explain how mutating the initiation codon of uORF 1 or uORF 3 can strongly affect RSV RNA translation and packaging (17, 45, 46). Our model implies that the ribosome is binding to and stalling at uORF 1 (10, 11) due to stable secondary structures located downstream of uORF 1 which are required for internal translation initiation of Gag and RNA packaging. In fact, a computer-predicted structure of the RSV 5′ leader where a ribosome is bound to uORF 1 highlights an extended stable structure with a ΔG of −148 kcal/mol (Fig. 9). In this model the RSV IRES maps to a Y-like structure, since the long stem and the two short upper stem-loops each appear to contribute to the IRES activity (Fig. 7, lanes 5 to 8, and Fig. 9). Also, this model structure suggests that translation reinitiation at uORF 3 or gag is very unlikely due to the predicted stable RNA structures (Fig. 9) (12, 25, 37). In addition, ribosome stalling at the level of uORF 1 (10) might require an interaction between the uORF-encoded peptide and the translation apparatus which has been shown to block ribosome dissociation at the stop codon (6, 7, 41, 61). In agreement with this notion, the RSV uORF 1 encoded peptide (MAGPLIP) displays similarities with the inhibitory peptide encoded by the uORF present in the mammalian S-adenosylmethionine decarboxylase RNA (MAGDIS) (31). Mutating the AUG codon of uORF 1 might allow the preinitiation complexes to scan downstream of uORF 1 and thus unwind some RNA structures engaged in IRES activity and RNA packaging. Consistent with this hypothesis, after translation termination, the 40S subunits can resume scanning but progression is easily inhibited by secondary structures (25); in contrast, preinitiation complexes can unfold more-stable secondary structures (ΔG = −30kcal/mol) (35, 37).
The existence of an IRES in the 5′ leader of RSV RNA and other retroviruses such as MLV (3, 61), REV-A (40) and SIV (47) should direct ribosome recruitment at or near the Gag translation start site, thus bypassing the secondary structures which are required for the process of reverse transcription. Furthermore, colocalization of the RSV IRES with the dimerization and packaging sequences favors the notion that the full-length retroviral RNA could be used both as a messenger RNA and as pregenomic RNA (5, 11), as in the case of poliovirus RNA (22). It has recently been shown that uORF 3 is located in stem-loop O3, which is capable of promoting encapsidation of an heterologous RNA (1). In addition, a previous report shows that uORF 3 is critical for RNA encapsidation, probably through its translational properties (18). The present findings suggest that uORF 3 is not translated after reinitiation from uORF 1 but uses an IRES-dependent mechanism. This, however, has major consequences because IRES-dependent translation does follow the same rules as classical cap-dependent translation. In that respect, trans-acting factors are thought to play a major role in internal initiation. In particular, it has been shown that Gag acts as a translational repressor of RSV RNA which would be capable of sorting viral RNA for translation or encapsidation (5, 59). It is therefore tempting to speculate that, by binding to the RSV IRES, Gag can regulate its own translation and, therefore, virus assembly.
Interestingly IRES-dependent translation might well provide some advantages to the virus since it occurs independently of the initiation factor eIF4E which recognizes the 5′ cap structure and regulates cellular protein synthesis (48, 50, 51). In that respect, the retroviral IRES-directed translation, which avoids eIF4E regulation, may represent a strategy to favor viral protein synthesis, especially during the G2/M phase of the cell cycle, when cap-dependent translation is significantly decreased (9, 56, 58).
ACKNOWLEDGMENTS
We thank Christelle Daudé for technical assistance, Nathalie Fouillot for the gift of plasmids pMLP-P2A and pMLP-PR2A, Pierre Savatier for the gift of plasmid pβ-Actin-LacZ, and Edmund Derrington for a critical reading of the manuscript.
This work was supported by grants from ANRS and MGEN. Clarence Deffaud is supported by an ARC fellowship.
REFERENCES
- 1.Banks J D, Linial M L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barco A, Feduchi E, Carrasco L. A stable HeLa cell line that inducibly expresses poliovirus 2Apro: effects on cellular and viral gene expression. J Virol. 2000;74:2383–2392. doi: 10.1128/jvi.74.5.2383-2392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlioz C, Torrent C, Darlix J-L. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieth E, Gabus C, Darlix J-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbin A, Darlix J-L. Functions of the 5′ leader of murine leukemia virus genomic RNA in virion structure, viral replication and pathogenesis, and MLV-derived vectors. Biochimie. 1996;78:632–638. doi: 10.1016/s0300-9084(96)80009-5. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis S, Bruynooghe G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 10.Darlix J-L, Spahr P F, Bromley P A, Jaton J C. In vitro, the major ribosome binding site on Rous sarcoma virus RNA does not contain the nucleotide sequence coding for the N-terminal amino acids of the gag gene product. J Virol. 1979;29:597–611. doi: 10.1128/jvi.29.2.597-611.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlix J L, Zuker M, Spahr P F. Structure-function relationship of Rous sarcoma virus leader RNA. Nucleic Acids Res. 1982;10:5183–5196. doi: 10.1093/nar/10.17.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deffaud C, Darlix J-L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J Virol. 2000;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degnin C R, Schleiss M R, Cao J, Geballe A P. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delbecq P, Werner M, Feller A, Filipkowski R K, Messenguy F, Pierard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaney M A, Vakharia V N, Lloyd R E, Ehrenfeld E, Grubman M J. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol. 1988;62:4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolph P J, Huang J T, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 20.Fosse P, Motte N, Roumier A, Gabus C, Muriaux D, Darlix J-L, Paoletti J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996;35:16601–16609. doi: 10.1021/bi9613786. [DOI] [PubMed] [Google Scholar]
- 21.Fouillot N, Tlouzeau S, Rossignol J M, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstaub D, Gradi A, Bercovitch Z, Grosmann Z, Nophar Y, Luria S, Sonenberg N, Kahana C. Poliovirus 2A protease induces apoptotic cell death. Mol Cell Biol. 2000;20:1271–1277. doi: 10.1128/mcb.20.4.1271-1277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackett P B, Dalton M W, Johnson D P, Petersen R B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett P B, Petersen R B, Hensel C H, Albericio F, Gunderson S I, Palmenberg A C, Barany G. Synthesis in vitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J Mol Biol. 1986;190:45–57. doi: 10.1016/0022-2836(86)90074-4. [DOI] [PubMed] [Google Scholar]
- 28.Hellen C U, Facke M, Krausslich H G, Lee C K, Wimmer E. Characterization of poliovirus 2A proteinase by mutational analysis: residues required for autocatalytic activity are essential for induction of cleavage of eukaryotic initiation factor 4F polypeptide p220. J Virol. 1991;65:4226–4231. doi: 10.1128/jvi.65.8.4226-4231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hensel C H, Petersen R B, Hackett P B. Effects of alterations in the leader sequence of Rous sarcoma virus RNA on initiation of translation. J Virol. 1989;63:4986–4990. doi: 10.1128/jvi.63.11.4986-4990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill J R, Morris D R. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. Regulation by the 5′ transcript leader. J Biol Chem. 1992;267:21886–21893. [PubMed] [Google Scholar]
- 31.Hill J R, Morris D R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- 32.Hughes S, Mellstrom K, Kosik E, Tamanoi F, Brugge J. Mutation of a termination codon affects src initiation. Mol Cell Biol. 1984;4:1738–1746. doi: 10.1128/mcb.4.9.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khandjian E W, Meric C. A procedure for Northern blot analysis of native RNA. Anal Biochem. 1986;159:227–232. doi: 10.1016/0003-2697(86)90332-5. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc Natl Acad Sci USA. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. . (Erratum, 92:7134.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 39.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Lastra M, Gabus C, Darlix J-L. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum Gene Ther. 1997;8:1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 41.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller J T, Ge Z, Morris S, Das K, Leis J. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J Virol. 1997;71:7648–7656. doi: 10.1128/jvi.71.10.7648-7656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mize G J, Ruan H, Low J J, Morris D R. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- 45.Moustakas A, Sonstegard T S, Hackett P B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moustakas A, Sonstegard T S, Hackett P B. Effects of the open reading frames in the Rous sarcoma virus leader RNA on translation. J Virol. 1993;67:4350–4357. doi: 10.1128/jvi.67.7.4350-4357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohlmann T, Lopez-Lastra M, Darlix J-L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J Biol Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 48.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 49.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 50.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen R B, Hackett P B. Characterization of ribosome binding on Rous sarcoma virus RNA in vitro. J Virol. 1985;56:683–690. doi: 10.1128/jvi.56.3.683-690.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen R B, Hensel C H, Hackett P B. Identification of a ribosome-binding site for a leader peptide encoded by Rous sarcoma virus RNA. J Virol. 1984;51:722–729. doi: 10.1128/jvi.51.3.722-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen R B, Moustakas A, Hackett P B. A mutation in the short 5′-proximal open reading frame on Rous sarcoma virus RNA alters virus production. J Virol. 1989;63:4787–4796. doi: 10.1128/jvi.63.11.4787-4796.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prats A, DeBilly G, Wang P, Darlix J-L. A CUG initiation codon is used for the synthesis of a cell surface antigen coded by MuLV. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 56.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 57.Ramesh N, Osborne W R. Assay of neomycin phosphotransferase activity in cell extracts. Anal Biochem. 1991;193:316–318. doi: 10.1016/0003-2697(91)90028-r. [DOI] [PubMed] [Google Scholar]
- 58.Sonenberg N. Remarks on the mechanism of ribosome binding to eukaryotic mRNAs. Gene Expr. 1993;3:317–323. [PMC free article] [PubMed] [Google Scholar]
- 59.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 60.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Fang P, Sachs M S. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol. 1998;18:7528–36. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Gaba A, Sachs M S. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J Biol Chem. 1999;274:37565–37574. doi: 10.1074/jbc.274.53.37565. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Sachs M S. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J Biol Chem. 1997;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss R T N, Varmus H, Coffin J. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. [Google Scholar]