Abstract
Norwalk virus (NV), responsible for outbreaks of acute gastroenteritis, comprises the species of the genus Norwalk-like viruses in the family Caliciviridae. Although the study of the molecular biology of NV has been hampered by a lack of culture systems or small experimental animal models, virus-like particles (VLPs) generated with recombinant baculoviruses harboring the capsid protein gene of NV provide a useful tool for investigating NV-cell interactions. In this study, the attachment of the recombinant VLPs derived from the Ueno virus (UEV), a strain belonging to the genogroup II NVs, to mammalian and insect cells was examined. Kinetic analyses of the binding of the recombinant VLPs of the UEV (rUEVs) to Caco-2 cells demonstrated that the binding was specific and occurred in a dose-dependent manner. Approximately 7.5% of the prebound rUEVs were internalized into the Caco-2 cells. Enzymatic and chemical modification of Caco-2 cell surface molecules suggested that the binding was directly mediated by a protein-protein interaction. A virus overlay protein-binding assay (VOPBA) indicated that rUEVs appeared to bind to a 105-kDa molecule, designated as the NV attachment (NORVA) protein. Furthermore, the assay indicated that its native conformational structure was indispensable for the binding activity. In Caco-2 cells, the NORVA protein was detected when VOPBA was carried out with the VLPs from Seto and Funabashi viruses, which are serologically different NVs from UEV, used as probes. The binding of rUEVs to NORVA protein was also observed in six mammalian cell lines other than Caco-2. These data suggest that the attachment of NV to mammalian cells is mediated by NORVA protein, which is ubiquitously expressed in the mammalian cells. The present study is the first report on the role of the cellular molecule in the binding of recombinant VLPs of NV.
Norwalk virus (NVs) has been recognized as the causative agent of acute gastroenteritis in both developing and developed countries (16, 34). In 1972, the 27-nm NV strain 68 (NV/68), the prototype strain of NV, was discovered by immune electron microscopy in fecal specimens collected from cases of a gastroenteritis outbreak in Norwalk, Ohio (36). NVs form a group of noncultivatable human caliciviruses responsible for both sporadic cases and epidemic outbreaks of acute gastroenteritis (19, 28, 47, 48, 50). Currently, NVs are classified into genogroups I and II according to nucleotide and amino acid sequences that are based on the following: a part of the RNA-dependent RNA polymerase in the first open reading frame (ORF1), the capsid protein of ORF2, and ORF3 (2, 64), which encodes a minor structural protein (18).
In spite of extensive research, the study of the molecular biology of NV has been severely hampered by a lack of cell culture and suitable small animal models. While chimpanzees inoculated with NV developed serologic responses, they usually underwent an asymptomatic infection (35, 69). Therefore, physicochemical studies of NV have only been carried out with virions obtained from clinical stool specimens of the patients. Although the amount of virus in such stool specimens is usually low, the complete nucleotide sequence has been determined for several strains of the virus (14, 25, 32, 43, 56). Recent studies showed that in vitro translation of Southampton virus (SAV) RNA with rabbit reticulocyte lysate yielded a 200-kDa precursor protein, which was cotranslationally cleaved into three major products with molecular masses of 113, 48, and 41 kDa. It was demonstrated that the 3C-like protease encoded in the SAV genome has a specificity similar to picornavirus 3C proteases and that the nonstructural precursor polyprotein of SAV is cleaved into at least six smaller products (44, 45, 56).
Several recombinant baculoviruses harboring the gene encoding the ORF2 product were constructed, and the proteins were expressed in insect cells (14, 20, 24, 30, 31). An approximately 58-kDa capsid protein appeared to be self-assembled into virus-like particles (VLPs). Though these VLPs are artificial products, they have been proven to be morphologically and antigenically similar to the native virions (21, 31, 51, 52). Hyperimmune sera against VLPs were subsequently prepared in rabbits, and the enzyme-linked immunosorbent assay was established to detect NV antigens in the stool specimens (24, 29, 49). Furthermore, VLPs are useful for various in vitro experiments in which they could be substituted for native virions. Such experiments include cryoelectron microscopy and image processing, performed to construct the three-dimensional structure of the virion, and X-ray crystallographic studies of the structure of the NV/68 capsid (51, 52). The recombinant NV/68 (rNV) is composed of 180 capsid proteins that form an icosahedron 38 nm in diameter. The rNV was indeed immunogenic and was put to use in oral immunization in order to evaluate its ability to stimulate mucosal immunity (3–5, 46).
Since no permissive cell lines for the growth of NV have yet been established, the VLPs of NV are useful for the study of virus-cell interaction. The binding of rNV to various mammalian cell lines has been analyzed, and it was shown that differentiated Caco-2 cells bound to rNV more efficiently than did other mammalian cell lines from different species (66). The binding of rNV to these cells was blocked by serologically homologous rNV particles, but not by heterologous particles like rotaviruses. A blocking assay was performed with a monoclonal antibody that abolished rNV's binding to Caco-2 cells, and this assay suggested that the virus attachment site within the capsid protein located the amino acid residues at 300 to 384 (66). However, the cellular counterpart for the binding has not yet been determined.
In the present study, recombinant VLPs of the Ueno virus (rUEVs), which is one of the genogroup II NVs, were used to characterize the binding properties between the VLPs and mammalian cells. The virus overlay protein-binding assay (VOPBA) showed that the binding of rUEVs to Caco-2 cells was mediated by a 105-kDa single molecule. This protein was designated the NV attachment (NORVA) protein, and it was found that the NORVA protein was capable of binding to at least two other VLPs generated from the Seto virus (SEV) and the Funabashi virus (FUV), which are serologically distinct from UEV. Interestingly, the NORVA protein was also detected in six mammalian cells other than Caco-2 cells with rUEVs as the probe. These observations suggest that the attachment of the NV to various mammalian cells is mediated by the NORVA protein, which is ubiquitously expressed in the mammalian cells.
MATERIALS AND METHODS
Cells.
Seven mammalian and two insect cell lines were used. Caco-2 (human colon), 293T (human embryonic kidney), HeLa (human cervix), HepG2 (human liver), CV-1 (African green monkey kidney), CHO-K1 (Chinese hamster ovary), and N-MuLi (mouse liver) were grown at 37°C with GIT medium (Wako Pure Chemical Industries, Ltd., Osaka, Japan) containing 3% fetal bovine serum (FBS). Sf9 cells (Riken Cell Bank, Tsukuba, Japan), derived from the insect Spodoptera frugiperda (60), were grown at 27°C with TC-100 (GIBCO BRL, Gaithersburg, Md.) containing 10% FBS. Tn5 cells, an insect cell line from Trichoplusia ni (Invitrogen, San Diego, Calif.), were grown at 27°C with Ex-CELL 400 (JRH Biosciences, Lenexa, Kans.).
Viruses.
UEV was collected from the stool specimen of a female student who had severe diarrhea at the end of January 1994 in Mie Prefecture, Japan. The suspected food was a school lunch, but the causative material had not been identified. SEV was isolated in an outbreak of acute gastroenteritis associated with a school excursion in Aichi Prefecture, Japan, in 1989. FUV was obtained from a patient with gastroenteritis in a mass food poisoning, which occurred in association with a school excursion held in May in 1996 in Chiba Prefecture, Japan. The nucleotide sequences of ORF2 of SEV and FUV indicated that these viruses are genetically close to NV/68 and SAV, respectively.
Recombinant VLPs.
Generation of a recombinant baculovirus harboring the gene for the capsid protein of UEV (Ac[UEV]) will be described elsewhere (unpublished data). In brief, viral RNA was extracted with Trizol (GIBCO BRL), and the cDNA synthesis was done by using oligo(dT)33 (61). Amplification was carried out with a forward primer, G2F1 (5′-GTGGGAGGGCGATCGCAATCT-3'), and oligo(dT)33. Approximately-2.5-kb PCR products were cloned into a TA cloning vector, pCR2.1 (Invitrogen), to generate pCR[UEV]. The plasmids were then digested with BamHI, and the resultant 1.7-kb fragment was purified and ligated with a transfer vector, pVL1392 (PharMingen, San Diego, Calif.), to produce pVL[UEV]. Sf9 cells were cotransfected with 0.5 μg of linearized wild-type Autographa californica nuclear polyhedrosis virus DNA (Pharmingen) and 1 μg of pVL[UEV], mediated by Lipofectin, to generate Ac[UEV]. Two other recombinant baculoviruses containing the capsid gene of SEV (Ac[SEV]) (41) and FUV (Ac[FUV]) (unpublished data) were prepared in the same manner. VLPs were prepared by infecting subconfluent Tn5 insect cells with the recombinant baculovirus at a multiplicity of infection (MOI) of 5 to 10 PFU per cell in 250-ml tissue culture flasks (Becton Dickinson Labware, Rutherford, N.J.). The culture medium was harvested at 6 days after infection, centrifuged at 1,000 × g for 10 min to remove the cell debris, and further centrifuged at 10,000 × g for 30 min to remove the baculoviruses. The VLPs in the supernatant were concentrated by centrifugation at 100,000 × g for 2 h at 4°C in an SW28 rotor (Beckman Instruments, Inc., Palo Alto, Calif.). The pellet was resuspended in a solution containing CsCl (1.9 g/4.5 ml) and centrifuged at 120,000 × g for 20 h at 10°C in an SW50.1 rotor (Beckman). Peak fractions containing the VLPs were pooled, diluted with phosphate-buffered saline [PBS(−); pH 7.5], and centrifuged at 200,000 × g for 2 h at 4°C in an SW50.1 rotor. The pellet was resuspended in PBS(−), and the 38-nm VLPs with the native virion size were separated from the smaller 23-nm VLPs by 5 to 30% sucrose gradient centrifugation at 80,000 × g for 2.5 h at 4°C in an SW41 rotor (Beckman). The fractions containing the 38-nm VLPs were diluted with PBS(−) and concentrated by centrifugation at 200,000 × g for 2 h at 4°C in an SW50.1 rotor. The VLPs were resuspended in PBS(−) and used for the binding assay. Protease inhibitors (leupeptin at 5 μg/ml and pepstatin at 13 μg/ml) were included throughout the purification procedures. The VLP preparations were examined by electron microscopy and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined by using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) with bovine serum albumin (BSA) as the protein standard.
Preparation of 35S-labeled rUEVs.
Tn5 cells in a 250-ml flask were infected with Ac[UEV] and incubated for 24 h at 27°C. The cells were washed once with methionine-free Grace's medium (GIBCO BRL) and incubated with 10 ml of the same medium for 30 min at 27°C. Then the UEV capsid proteins were metabolically radiolabeled with 30 μCi of [35S]methionine (Trans 35S-label; ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) for 12 to 20 h at 27°C. At the end of the labeling, 10 ml of Ex-CELL 400 insect medium (JRH Biosciences) was added. The cultures were incubated and harvested 7 days after infection, and the 35S-labeled rUEVs were purified as described above. The culture medium contained penicillin (50 μg/ml), streptomycin (50 μg/ml) (GIBCO BRL), and Fungizone (2.5 μg/ml) (Bristol-Myers Squibb K. K., Tokyo, Japan).
Preparation of 125I-labeled rUEVs.
Purified rUEVs (100 μg) were incubated for 30 min at 4°C with 18.5 MBq of Na125I (NEN Life Science Products, Inc., Boston, Mass.) and Iodo-Gen Iodination Reagent (Pierce, Rockford, Ill.) according to the instructions provided by the manufacturer. The reaction was terminated by adding cold NaI at a final concentration of 1 mM, and free Na125I was removed by passing the solution through a Sephadex G-25 medium column (Pharmacia Biotech AB, Uppsala, Sweden). The 125I-labeled rUEVs were spun down by centrifugation through 1.0 ml of 25% sucrose cushion at 200,000 × g for 2 h at 4°C in an SW50.1 rotor. The pellet was resuspended in PBS(−), and the 38-nm rUEVs were separated from the smaller 23-nm rUEVs by a 5 to 30% sucrose gradient and concentrated by centrifugation as described above. The final preparation was suspended in PBS(−) and stored at 4°C until use.
Binding assays.
Dose-response experiments were performed essentially according to the methods described previously (64). The confluent monolayers of differentiated Caco-2 cells in a 48-well plate (1.5 × 105 cells) (Becton Dickinson Labware, Franklin Lakes, N.J.) were washed three times with cold serum-free Dulbecco's modified Eagle's medium (DMEM) (GIBCO BRL) and chilled to 4°C. Increasing amounts of the 35S-labeled rUEVs (from 0 to 150 μg) in 80 μl of cold serum-free DMEM were added to the duplicate wells and incubated for 1 h at 4°C with gentle agitation. Incubation for 1 h was sufficient for the binding to reach a plateau. The internalization of the rUEVs into the cells was prevented by incubating the cells at 4°C. The binding was terminated by washing the cells three times with cold serum-free DMEM. The cells were then solubilized with radioimmunoprecipitation assay (RIPA) buffer (0.15 M NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 0.01% aprotinin, 10 mM Tris-HCl [pH 7.2]. The binding was tested in the absence of unlabeled rUEVs to determine the total (specific plus nonspecific) binding. The cells were preincubated with a 20-fold excess of unlabeled rUEVs for 1 h at 4°C to measure the nonspecific binding. The specific binding of each concentration of rUEVs was calculated by subtracting the nonspecific binding from the total binding. The number of cells per well was measured by counting the trypsinized cells in duplicate wells. Assays were done in duplicate or triplicate.
Scatchard plot analysis was performed to calculate the number of rUEVs bound to the cells (55). This method determines the binding parameters by using the number of radiolabeled ligand molecules at a binding site. It represents a plot of r/c versus r for different ligand concentrations, where r is the amount of rUEVs bound per 105 cells and c is the concentration of unbound rUEVs (in micrograms per 105 cells). The intercept of the x axis is the value of the saturated quantity (in micrograms) of rUEVs bound to the cell surface. The intercept of the x axis of the Scatchard plot was multiplied by the number present in 1 μg of particles (5.77 × 1010) and then divided by the number of cells (105) to determine the number of binding sites per cell.
Internalization of rUEVs into Caco-2 cells.
An internalization assay was performed essentially as described previously (6, 64). Caco-2 cell monolayers that were grown to confluency in 24-well plates (4 × 105 cells) were washed three times with cold serum-free DMEM and chilled to 4°C. The plates were divided into two groups. One group was preincubated with 200 μg (20-fold excess) of unlabeled rUEVs for 1 h at 4°C. In the other group, preincubation was carried out without unlabeled rUEVs. In both groups, 10 μg of 35S-labeled rUEVs in serum-free DMEM was added to all of the wells on the plates at final volumes of 300 μl, and the plates of both groups were incubated for 1 h at 4°C with gentle agitation and washed three times with serum-free DMEM. Three wells were selected from each group for assessing the total and nonspecific bindings. The cells were treated with RIPA buffer, and the radioactivity was measured to determine the specific binding (the total binding minus the nonspecific binding). The second three wells from each group were used to determine the efficiency of proteinase K in removing bound rUEVs. The cells were incubated with 500 μl of proteinase K (500 μg/ml) (Sigma Chemical Co., St. Louis, Mo.) for 30 min at 4°C, washed three times, and solubilized with RIPA buffer. Then the radioactivities were measured. Proteinase K was capable of removing approximately 97 to 98% of bound rUEVs from the cell surface under the experimental conditions. The third three wells from each group were used to measure the internalized rUEVs. The cells were transferred to 37°C to allow the bound rUEVs to enter the cell. After 1 h, the cells were washed and treated with proteinase K for 30 min at 4°C, and the radioactivity was measured.
Chemical and enzymatic modification of cell surface molecules.
Caco-2 cells grown in 24-well plates (4 × 105 cells) were washed three times with serum-free DMEM and incubated in the presence of phospholipase C (Sigma) for 1 h at 37°C, sodium periodate (Sigma) for 1 h at 37°C, or proteinase K for 20 min at 37°C in serum-free DMEM. The cells were washed with serum-free DMEM three times to terminate the reactions. Subsequently, 300 μl of purified 35S-labeled rUEVs (10μg) in cold serum-free DMEM was added to triplicate wells, which were then incubated for 1 h at 4°C. The binding was assayed as described above. Assays were done at least twice.
Preparation of cell membranes.
The membrane fraction was prepared by two methods. (i) Cells were harvested, washed twice with PBS(−), and incubated with PBS(−) containing 50 mM EDTA for 10 min at room temperature. The cells were centrifuged for 2 min at 1,000 × g, washed with PBS(−), swollen on ice for 15 min in PBS(−) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma) and 5 mM EDTA, and disrupted with a glass Dounce homogenizer (39). The nuclei and unbroken cells were removed by centrifugation for 5 min at 1,000 × g, and the supernatant was further centrifuged for 1 min at 11,000 × g. The supernatant was stored at −20°C until use. (ii) Cells were harvested, washed twice with PBS(−), and lysed in PBS(−) containing 2 mM PMSF and 1% NP-40 for 1 h at 4°C. Nuclei were removed by centrifugation for 5 min at 1,000 × g, and the supernatant was further centrifuged for 30 min at 11,000 × g. The supernatant was stored as a membrane fraction at −20°C (7).
VOPBA.
The cell membrane proteins were separated by SDS-PAGE under nonreducing conditions as described previously (42). Under these conditions, the sample buffer did not contain 2-mercaptoethanol, and the sample was not boiled. Following SDS-PAGE, the proteins were electroblotted onto a 0.45-μm-pore-diameter nitrocellulose membrane (Bio-Rad) by using the Trans-blot Cell Apparatus (Bio-Rad) at a constant 10 V for 1 h. The membrane was incubated with Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.5], 0.15 M NaCl) containing 5% skim milk for 20 h at 4°C to block nonspecific binding and rinsed with TBS containing 0.5% Tween 20, and then incubated with either 200 μg of unlabeled rUEVs or 50 μg of 125I-labeled rUEVs (2 × 106 cpm) in 10 ml of PBS(−) for 1 h at room temperature on a rocking platform. The membrane was washed three times for total of 1 h with 100 ml of TBS containing 0.5% Tween 20. Unlabeled VLPs were detected by using a polyclonal rabbit anti-VLP immunoglobulin G (IgG) and anti-rabbit Ig-alkaline phosphatase conjugates (Dako A/S, Copenhagen, Denmark). 125I-labeled rUEVs were detected by exposing the membrane to X-OMAT film (Eastman Kodak Company, Rochester, N.Y.).
Competition assay with homologous monomeric capsid protein and heterologous VLPs.
Soluble rUEV capsid proteins were prepared by incubating 200 μg of purified 38-nm rUEVs for 24 h at 4°C in 0.1 M Tris-HCl (pH 9.5), as described previously with slight modifications (67). The disassembly of the rUEVs was examined by 5 to 30% sucrose gradient centrifugation. Caco-2 cells grown in 48-well plates were incubated with 100 μl of serum-free DMEM containing a 20-fold excess amount (200 μg) of the monomeric capsid proteins of rUEVs, rSEV VLPs, or rFUV VLPs for 1 h at 4°C. Subsequently, 10 μg of purified 35S-labeled rUEVs was added to each well for a final volume of 200 μl and incubated for 1 h at 4°C. The binding of the 35S-labeled rUEVs was assayed as described above. Assays were done in duplicate or triplicate.
RESULTS
Purification of self-assembled rUEVs.
The determination of the complete nucleotide sequence and phylogenetic analysis of the ORF2 of UEV indicated that this virus is genetically distinct from any of the previously known NVs, but is a member of genogroup II NVs. From the time course experiment using the recombinant baculovirus Ac[UEV], the expression of the 58-kDa capsid protein in Tn5 cells reached its maximum at 6 days postinfection. The 35S-labeled rUEVs analyzed in CsCl indicated that the buoyant density of the fraction containing both 38- and 23-nm particles was 1.30 g/cm3. Large 38-nm rUEVs were separated from 23-nm rUEVs by sedimentation through 5 to 30% sucrose gradients and exclusively used in the subsequent binding assays. The specific activities of the purified 35S-labeled rUEVs were 3 × 103 to 8 × 103 cpm/μg of protein. Unlabeled rUEVs were prepared in the same manner, and the purified 38-nm particles were labeled with 125I in vitro. The specific activity appeared to be 4 × 104 to 15 × 104 cpm/μg of rUEVs. The molecular weight of the rUEVs in each purification step was monitored by SDS–10% PAGE, and the particle size in the final preparation was confirmed as 38 nm by electron microscopy at a magnification of 100,000.
Binding of rUEVs to differentiated Caco-2 cells.
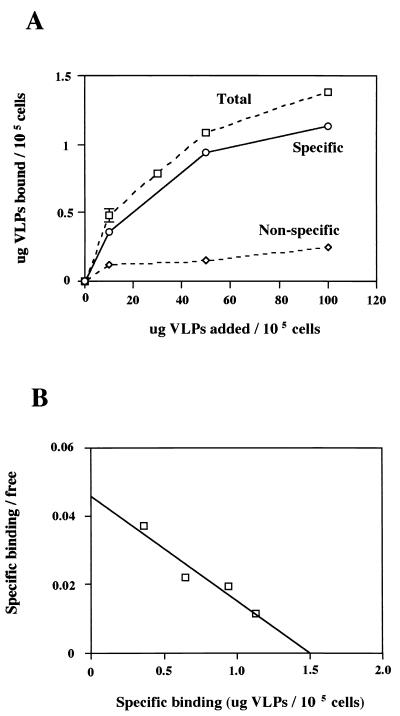
The binding kinetics were examined to study the interaction between rUEVs and differentiated Caco-2 cells. Specific binding should be dose dependent, saturable, and competitively inhibited by a second ligand that binds to the same cell surface molecule. Caco-2 cells were incubated in 48-well plates for at least 1 week after they grew to confluency, during which time typical domes, a marker characteristic for differentiated Caco-2 cells, appeared. Increasing amounts of 35S-labeled rUEVs were added to the wells and incubated for 1 h at 4°C. As shown in Fig. 1A, the binding occurred in a dose-dependent manner and was nearly saturable at a concentration of 100 μg of rUEVs/105 cells. The low value of the nonspecific binding, compared with the total binding at each concentration, indicated that unlabeled rUEVs specifically inhibited the binding.
FIG. 1.
(A) Dose-response curve of the total, nonspecific, and specific binding of rUEVs. One group of confluent Caco-2 monolayers grown in 48-well plates (1.5 × 105 cells/well) was incubated for 1 h at 4°C with an increasing amount of 35S-labeled rUEVs (0 to 150 μg) in 80 μl to determine the total binding (▫). The other group of plates was preincubated in the presence of a 20-fold excess amount of unlabeled rUEVs to determine nonspecific binding (◊). The specific binding was calculated by subtracting the nonspecific binding from the total binding (○). Each plot represents the mean value (with error bar). Each assay was done in duplicate or triplicate. (B) Scatchard plot analysis of the specific binding of rUEVs to Caco-2 cells. The intercept of the x axis represents the value of the saturated quantity (in micrograms) of the rUEVs specifically bound to 105 cells.
From the Scatchard plot analysis of the specific binding of rUEVs to Caco-2 cells, the number of binding sites per cell was estimated to be 8.6 × 105 (Fig. 1B). This estimated number of binding sites was within the range described in other well-characterized virus-receptor interactions (15, 68). The linear Scatchard plot indicated that only a single class of binding sites was present on the cells and that there was no cooperative binding site in one receptor molecule (55). The Scatchard plot analysis of the specific binding of rUEVs further showed that the Kd value calculated from the slope of the proximate line was 2.9 × 10−8 M.
Internalization of 35S-labeled rUEVs into Caco-2 cells.
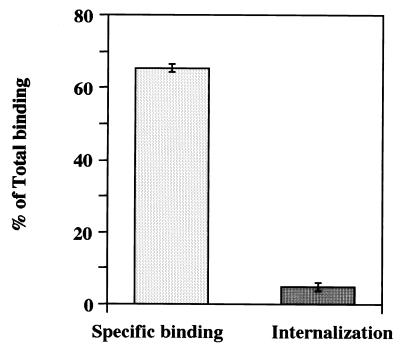
To address whether rUEVs were internalized to Caco-2 cells after the specific binding, an internalization assay was performed. As shown in Fig. 2, the specific binding appeared to represent approximately 65% of the total binding (Fig. 2, left column). This amount is similar to that obtained in the dose-response curve shown in Fig. 1A. The bound rUEVs were internalized by shifting the temperature from 4°C to 37°C. The incubation was carried out for 1 h, and the remaining rUEVs were removed by proteinase K digestion. Approximately 4.9% of the total binding remained associated with the cells after treatment (Fig. 2, right column). This amount is equivalent to approximately 7.5% of the specifically prebound rUEVs. An internalization assay using cells preincubated with 20-fold excess unlabeled rUEVs indicated no radioactivity, demonstrating that the internalization occurred only when rUEVs specifically bound to Caco-2 cells.
FIG. 2.
Internalization of 35S-labeled rUEVs. The Caco-2 monolayers grown in 24-well plates (4 × 105 cells/well) were washed three times with cold DMEM and chilled at 4°C. The plates were divided into two groups to measure the specific binding and internalization. The specific binding was measured as shown in Fig. 1 (left column). To measure the value of internalization, the other group of plates was incubated with 10 μg of 35S-labeled rUEVs per well for 1 h at 4°C with gentle agitation. After washing three times with cold DMEM, the plates were transferred to 37°C for 1 h to allow the bound rUEVs to internalize into the cells. The cells were treated with proteinase K (500 μg/ml) for 30 min at 4°C and solubilized with RIPA buffer, and their radioactivity was measured. The internalization value was determined by subtracting the value of the remaining rUEVs on the cell surface after the proteinase K treatment (right column). Each assay was done in triplicate. Each column represents the mean value (with error bar).
Effects of chemical and enzymatic treatment of Caco-2 cells on binding.
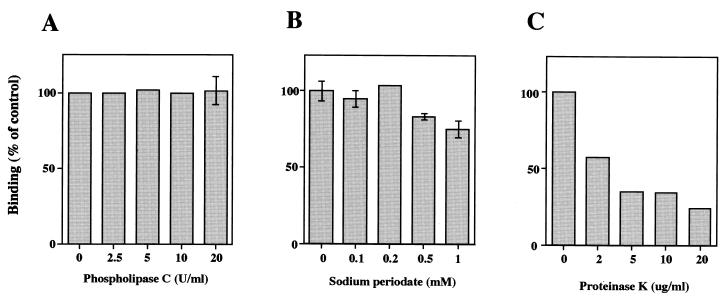
To characterize the cell surface molecule(s) participating in the binding, Caco-2 cells were preincubated with phospholipase C, sodium periodate, or proteinase K. Preincubation of cells with increasing concentrations of phospholipase C or sodium periodate had no effect on the binding of 35S-labeled rUEVs, even in the presence of 20 U of phospholipase C per ml or 1 mM sodium periodate (Fig. 3A and B). In contrast, treatment with proteinase K markedly reduced binding in a dose-dependent manner (Fig. 3C), a finding which suggests that the cellular binding molecule is mainly composed of protein(s). Although the binding decreased during incubation with 0.5 to 1.0 mM sodium periodate, this change may not have been a direct effect of the reagent, but was probably due to cell damage caused by the reagent. In our study, pretreatment of the cells with 0.05 to 0.25% trypsin had no effect on the binding as described for rNV in reference 66 (data not shown). These findings suggest that the binding of rUEVs to Caco-2 cells is mediated by a protein-protein interaction, but not by a protein-phospholipid or protein-carbohydrate interaction.
FIG. 3.
Effect of a chemical modification and enzyme treatments of Caco-2 cells in the binding of rUEVs. Confluent Caco-2 monolayers grown in a 24-well plate (4 × 105 cells/well) were preincubated with 0 to 20 U of phospholipase C per ml in serum-free DMEM at 37°C for 1 h (A), 0 to 1 mM sodium periodate in PBS(−) at 37°C for 1 h (B), and 0 to 20 μg of proteinase K per ml in serum-free DMEM at 37°C for 20 min (C). Subsequently, the cells were washed three times, and purified 35S-labeled rUEVs (10 μg) in 80 μl were added to each well. The incubation was carried out for 1 h at 4°C, and the radioactivity of the bound rUEVs was measured. Assays were done in triplicate plates. Each data point represents the mean value (with error bar).
Detection of membrane proteins for VLP binding.
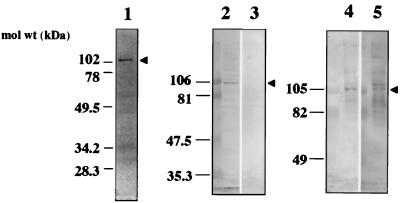
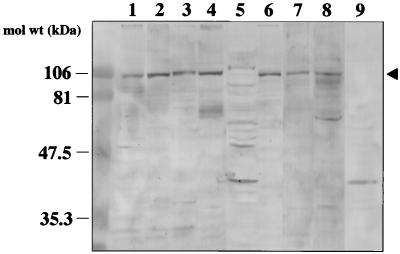
To identify the membrane protein(s) participating in the binding, VOPBA was performed. This method has been used to detect the receptor molecules of various viruses, including reovirus type 3, encephalomyocarditis virus, Sendai virus, and mouse hepatitis virus (9, 10, 17, 33). Approximately 10 to 20 μg of membrane proteins was prepared from Caco-2 cells by Dounce homogenization and separated by SDS-PAGE under nonreducing conditions. The proteins were transferred onto a nitrocellulose membrane and then probed with rUEVs. A single band of 105 kDa was observed as a major band when 125I-labeled rUEVs were used in the assay (Fig. 4, lane 1). Preincubation of the nitrocellulose membrane with a 20-fold excess amount of unlabeled rUEVs reduced the intensity of the band to approximately 30%, suggesting that the reaction was specific (data not shown). The inhibition caused by the homologous rUEVs may explain the similar degree of inhibition observed in the dose-response experiment as shown in Fig. 1A. The same band was also observed when unlabeled rUEVs were used as the probe and a rabbit anti-rUEV antibody was used as the detector antibody (Fig. 4, lanes 2 and 4). No band was observed when the membrane protein was denatured by boiling for 15 min in the presence of 5% 2-mercaptoethanol (Fig. 4, lane 3). Interestingly, the 105-kDa protein band was detectable when soluble monomeric rUEV capsid proteins were used (Fig. 4, lane 5). Though the band with higher molecular mass (110 kDa) appeared when the monomeric rUEVs were used, it is likely that the region located inside of the VLP was exposed and bound nonspecifically to the cellular protein. These results indicate that the 105-kDa protein is a cellular protein participating in the specific binding of rUEVs, and this protein was designated as a NORVA protein. The results also indicate that the assembly of the 58-kDa capsid proteins to form an icosahedron is not necessary for the interaction, whereas the conformational structure of the NORVA protein is indispensable for binding.
FIG. 4.
VOPBA in the detection of the binding membrane proteins. A membrane fraction (10 to 20 μg of protein) of Caco-2 cells was prepared by homogenization, separated by SDS-PAGE under nondenaturing conditions, and electroblotted onto nitrocellulose filter paper (lanes 1, 2, 4, and 5). The membrane protein was denatured by boiling in the presence of 2-mercaptoethanol for 15 min prior to SDS-PAGE and blotting (lane 3). The filter paper was incubated with 2 × 106 cpm (50 μg) of 125I-labeled rUEVs, and the signal was detected by exposing the membrane to X-OMAT film (lane 1). The filter paper was incubated with 200 μg of intact 38-nm rUEVs (lanes 2, 3, and 4) or disrupted monomeric rUEV capsid proteins, prepared by incubating 38-nm rUEVs at high pH (lane 5), followed by detection with anti-rUEV rabbit serum. Arrowheads indicate the positions of the NORVA protein. mol wt, molecular mass.
Detection of NORVA proteins in various mammalian cell lines.
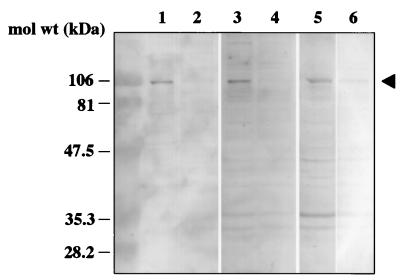
To determine whether the binding of rUEVs to NORVA protein is specific to Caco-2 cells, VOPBA was performed with eight other cell lines from various species. The cell membrane fraction was prepared by either homogenization (Caco-2, HeLa, HepG2, N-MuLi, and CV-1) or treatment with 1% NP-40 (293T, CHO, Tn5, and Sf9) as described in Materials and Methods. Unlabeled rUEVs were used as the probe. As shown in Fig. 5, NORVA protein was detected in six mammalian cell lines, as in Caco-2 cells (Fig. 5, lanes 1 to 4 and 6 to 8), whereas no band corresponding to a 105-kDa NORVA protein was detected in either of the insect cell lines. Instead, an approximately 42-kDa molecule was detected as a major band in insect cells (Fig. 5, lanes 5 and 9). Mammalian cells thus share NORVA protein for binding, but this protein is not present in insect cells. NORVA protein seems to be a ubiquitous molecule expressed in mammalian cells, because it was detected in cells not only from humans, but also from mice, hamsters, and monkeys.
FIG. 5.
Detection of NORVA membrane proteins from various cells. The membrane fraction was prepared by homogenization (Caco-2, HeLa, HepG2, N-MuLi, and CV-1) or treatment with detergent (1% NP-40 [293T, CHO, Tn5, and Sf9]). Approximately 20 μg of each cell membrane lysate was used for VOPBA. Lanes 1 to 9 contained Caco-2, 293T, N-MuLi, CHO, Tn5, CV-1, HeLa, HepG2, and Sf9, respectively. The membrane was incubated with 200 μg of unlabeled rUEVs. The NORVA protein was indicated by an arrowhead.
Inhibition of binding with monomeric capsid proteins and serologically different VLPs.
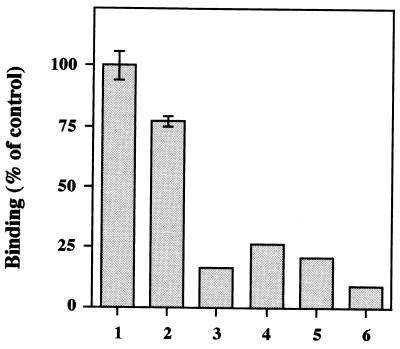
To determine whether the monomeric capsid protein and other serologically different VLPs block the binding of rUEVs to the Caco-2 cell, competition experiments were performed. Because the tertiary structure of rUEV itself was not essential for binding, the monomeric capsid proteins (MCPs) were prepared and used as a competitor. rSEV and rFUV were also used as competitors. Incubation with a 20-fold excess amount of intact rUEVs inhibited 83.7% of the total binding of 35S-labeled rUEVs (Fig. 6, column 3). This value is similar to that observed in the dose-dependent curve, shown in Fig. 1A. The homologous MCPs markedly inhibited the binding of the rUEVs (Fig. 6, column 4), indicating that the attachment sites of rUEVs are not concave structures like the canyons observed in picornaviruses (27, 54). Interestingly, heterologous monomeric rSEV and rFUV extensively inhibited the binding of 35S-labeled rUEVs (Fig. 6, columns 5 and 6). These competition experiments indicated that the MCPs can bind to the same molecule as intact 38-nm rUEVs do, and serologically different VLPs also bind to the same Caco-2 cellular molecule.
FIG. 6.
Competition assay with homologous monomeric capsid proteins or heterologous VLPs (rSEV and rFUV). Caco-2 cells were grown in a 48-well plate, and the binding of 35S-labeled rUEVs (10 μg) without competitors was examined (column 1). The cells were preincubated for 1 h at 4°C with 1% BSA (column 2), 20-fold excess (200 μg) of intact 38-nm rUEVs (column 3), homologous monomeric capsid proteins (column 4), rSEV (column 5), or rFUV (column 6). Subsequently, purified 35S-labeled rUEVs (10 μg) were added and incubated for 1 h at 4°C. The cells were washed five times and solubilized, and the radioactivity was measured.
Detection of NORVA proteins by probing with serologically different VLPs.
To determine the role of NORVA protein in the binding of rSEV and rFUV, VOPBA was performed with differentiated Caco-2 cells. High-titer rabbit sera to rSEV and rFUV were used as the detector antibodies. As shown in Fig. 7, a 105-kDa band corresponding to NORVA protein was clearly observed when 38-nm VLPs of rSEV and rFUV were used as the probe (Fig. 7, lanes 1, 3, and 5). Like rUEVs, rSEV and rFUV did not bind to the denatured NORVA protein (Fig. 7, lanes 2, 4, and 6). These findings indicate that NORVA protein is a common cellular binding protein for at least three serologically different NVs.
FIG. 7.
Detection of membrane protein(s) with three serologically different VLPs. Approximately 20 μg of the detergent-solubilized Caco-2 membrane fraction, either nondenatured (lanes 1, 3, and 5) or denatured (lanes 2, 4, and 6), was loaded into each lane and transferred to nitrocellulose membranes. VOPBA was carried out with 200 μg of unlabeled rUEVs (lanes 1 and 2), rSEV (lanes 3 and 4), and rFUV (lanes 5 and 6). The NORVA protein is indicated by an arrowhead. mol wt, molecular mass.
DISCUSSION
Infection of the virus starts with the attachment of the virions to the cell surface receptor molecules with either a high-affinity interaction (13, 22, 40, 59, 63) or low-affinity interactions (12, 37). The interaction event is generally highly specific, in that only cells permissive for viral replication possess receptor molecules on the surface membranes (53, 65). Although the prototype virus of NV was found more than 30 years ago, little is known about the mechanism by which the virus enters into its sensitive cells and initiates the replication. This is because neither cell culture systems that allow in vitro growth nor small animal models have been developed. In the present study, rUEVs were used to elucidate the mechanism of the early stages of infection. The experiments were based on the assumption that the recombinant VLPs behave as authentic native virions, at least during the early virus-cell interactions (66).
The following three findings suggest that rUEVs bind specifically to a single class of cellular molecule, the NORVA protein, which possesses receptor-like properties. (i) The dose-response experiment indicated that the specific binding of rUEVs to differentiated Caco-2 cells was nearly saturable at a concentration of 100 μg of rUEVs/105 cells (Fig. 1A). The experiment also indicated that the amount of particles necessary for saturable binding, 1.5 μg of rUEVs/105 cells (Fig. 1B), was of the same order as that of rNV (2.9 μg of VLPs/105 cells), as previously described (66). (ii) The number of cellular binding sites per cell for rUEVs was calculated to be 8.6 × 105 on Caco-2 cells, and this number is within the range of 104 to 106 as described by other well-characterized virus-receptor interactions (15, 68). (iii) The linear Scatchard plots shown in Fig. 1B demonstrated that a single class of molecule is involved in the binding of rUEVs to Caco-2 cells. The Scatchard analysis also revealed that the dissociation constant (Kd) of the Caco-2 binding protein–rUEV complex was approximately 2.9 × 10−8 M. This value is within the range of 10−8 to 10−9 M described for virion-receptor interactions in various viruses (15, 68).
The internalization assay demonstrated that approximately 7.5% of the specifically bound 35S-labeled rUEVs penetrated Caco-2 cells when the incubation was shifted from 4°C to 37°C. This rate of internalization was low compared with that of rotavirus, where 15 and 50 to 70% of the bound particles were shown to be internalized (6, 66). We do not know whether this low rate itself could fully explain the incapability of NV to grow in cultured cells at the moment.
VOPBA has been used to identify receptor proteins for various viruses, such as encephalomyocarditis virus (33), Sendai virus (17), mouse hepatitis virus (9), and cytomegalovirus (1, 62). This assay is sensitive enough to detect the binding of less than 100 μg of protein in a crude membrane preparation or to detect 5 μg of purified glycophorin (17). A single 105-kDa band was observed when 20 μg of the membrane fraction was probed with either 125I-labeled or unlabeled rUEVs (Fig. 4). It should be noted that NORVA protein was detected not only in Caco-2 and other human cell lines, but also in monkey, hamster, and mouse cells. This result indicates that NORVA protein is a ubiquitous cellular molecule of animal cells and therefore has an important similar biological function(s).
It is well known that human rhinovirus consists of two groups: the major receptor group and the minor receptor group. Although the structural and physicochemical properties are closely related, the major group shares ICAM-1, an Ig-like protein, as the receptor (11, 22, 23), whereas the minor group uses low-density lipoprotein receptor-related protein (26). Decay-accelerating factor, a complement regulator molecule, can serve as an attachment protein for a variety of enteroviruses, including echoviruses, coxsackie B viruses, coxsackievirus A21, and enterovirus 70 (8, 38, 57, 58). This evidence illustrates that serologically different strains of the same genus choose the same receptor. In competition assays, an rSEV and rFUV that were serologically different NVs from rUEV blocked the binding of rUEVs to Caco-2 cells (Fig. 6). Therefore, rSEV and rFUV share the cellular binding protein with rUEVs, and, indeed, the VOPBA indicated that rSEV and rFUV bind to a 105-kDa protein corresponding to NORVA protein (Fig. 7). Furthermore, a preliminary experiment demonstrated that several VLPs, serologically different from rUEVs, rSEV, and rFUV, also bound to NORVA protein (data not shown). Therefore, we hypothesized that the binding molecules on the different animal cell lines are the same.
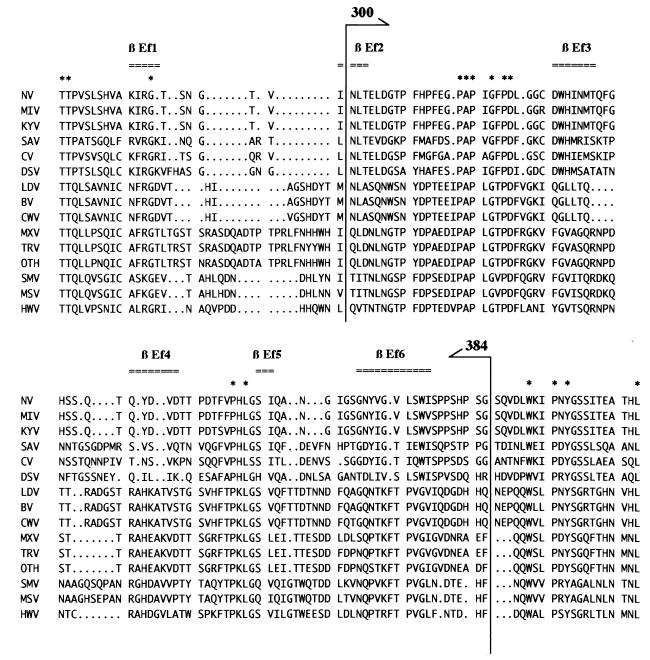
One monoclonal antibody (NV8812) raised against the rNV was shown to be capable of blocking the binding of rNV to human and animal cell lines (66). By immunoprecipitation with truncated and cleaved forms of the capsid protein, the binding site of the monoclonal antibody was localized to the C-terminal 300 to 384 residues of the rNV capsid, indicating that these regions are involved in cell-binding activity. The recent study of the X-ray crystallographic structure of rNV demonstrated that these residues form the P2 subdomain, which is the most exposed region of the rNV capsid protein (51). A comparison of the nucleotide sequence between genetically distinct NVs indicates that the P2 subdomain is highly variable and that this region may contain the determinant of strain specificity. However, VOPBA indicated that the same 105-kDa protein on the different animal cell lines was recognized by serologically different VLPs (Fig. 7). Therefore, it is highly likely that conserved amino acid residues in the variable sequence may function as the virion attachment site. Candidate sequences were found in the alignment shown in Fig. 8, where 315 to 322 and 354 to 356 residues are highly conserved among the genetically different NVs. The proline residues are most frequent. We hypothesized that the cell surface protein interacts with the hydrophobic portion of the capsid protein. This contrasts with the virus-cell interaction elucidated in the poliovirus and human rhinovirus, where marked depressions (canyons) encircling the vertices of the icosahedrons have been shown to bind specific cellular receptors (27, 54).
FIG. 8.
Amino acid sequence alignment of the P2 subdomain of the NV capsid protein. Alignments of the amino acid sequence of the capsid protein were generated by the GCG Pileup program, and the regions corresponding to the P2 subdomain were extracted according to the three-dimensional structure of the rNV (51). The β-strand in the P2 subdomain is indicated at the top of the figure. Consensus amino acid sequences are shown by asterisks. The region corresponding to amino acid residues 300 to 384 of NV is indicated by arrows. GenBank accession numbers for the alignment are as follows: NV, M87661; MIV, unpublished; KYV, L23828; SAV, L07418; CV, AB022679; DSV, U04469; LDV, X86557; BV, X76716; CWV, U46500; MXV, U22498; TRV, U02030; OTH, L23830; SMV, U70059; MSV, X81879; and HWV, U07611.
Although the data indicate that NORVA protein is expressed on various mammalian cell surfaces and may function as the cellular receptor of NV, more specific data about the NORVA binding protein are needed to determine its role as a receptor.
ACKNOWLEDGMENTS
We thank Y. Matsuura for helpful discussion and suggestions. We also thank T. Mizoguchi for secretarial work. We thank N. Sakurai (Mie Prefectural Institute of Public Health, Mie, Japan), S. Kobayashi (Aichi Prefectural Institute of Public Health, Aichi, Japan), and K. Shinozaki (Chiba Prefectural Institute of Public Health, Chiba, Japan) for providing UEV, SEV, and FUV.
This work was supported in part by Health Sciences Research grants, including grants for Research on Emerging and Re-emerging Infectious Diseases, Research on Environmental Health, Research on Pharmaceutical and Medical Safety, and Research on Health Sciences focusing on Drug Innovation from the Ministry of Health and Welfare, Japan.
REFERENCES
- 1.Adlish J D, Lahijani R S, St. Jeor S C. Identification of a putative cell receptor for human cytomegalovirus. Virology. 1990;176:337–345. doi: 10.1016/0042-6822(90)90003-a. [DOI] [PubMed] [Google Scholar]
- 2.Ando T, Mulders M N, Lewis D C, Estes M K, Monroe S S, Glass R I. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch Virol. 1994;135:217–226. doi: 10.1007/BF01309781. [DOI] [PubMed] [Google Scholar]
- 3.Ball J M, Estes M K, Hardy M E, Conner M E, Opekun A R, Graham D Y. Recombinant Norwalk virus-like particles as an oral vaccine. Arch Virol Suppl. 1996;12:251–262. doi: 10.1007/978-3-7091-6553-9_26. [DOI] [PubMed] [Google Scholar]
- 4.Ball J M, Graham D Y, Opekun A R, Gilger M A, Guerrero R A, Estes M K. Recombinant Norwalk virus-like particles given orally to volunteers: phase I study. Gastroenterology. 1999;117:40–48. doi: 10.1016/s0016-5085(99)70548-2. [DOI] [PubMed] [Google Scholar]
- 5.Ball J M, Hardy M E, Atmar R L, Conner M E, Estes M K. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J Virol. 1998;72:1345–1353. doi: 10.1128/jvi.72.2.1345-1353.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass D M, Baylor M R, Chen C, Mackow E M, Bremont M, Greenberg H B. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J Clin Investig. 1992;90:2313–2320. doi: 10.1172/JCI116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass D M, Mackow E R, Greenberg H B. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology. 1991;183:602–610. doi: 10.1016/0042-6822(91)90989-o. [DOI] [PubMed] [Google Scholar]
- 8.Bergelson J M, Chan M, Solomon K R, St. John N F, Lin H, Finberg R W. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc Natl Acad Sci USA. 1994;91:6245–6249. doi: 10.1073/pnas.91.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle J F, Weismiller D G, Holmes K V. Genetic resistance to mouse hepatitis virus correlates with absence of virus-binding activity on target tissues. J Virol. 1987;61:185–189. doi: 10.1128/jvi.61.1.185-189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Co M S, Gaulton G N, Fields B N, Greene M I. Isolation and biochemical characterization of the mammalian reovirus type 3 cell-surface receptor. Proc Natl Acad Sci USA. 1985;82:1494–1498. doi: 10.1073/pnas.82.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colonno R J, Callahan P L, Long W J. Isolation of a monoclonal antibody that blocks attachment of the major group of human rhinoviruses. J Virol. 1986;57:7–12. doi: 10.1128/jvi.57.1.7-12.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 13.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 14.Dingle K E, Lambden P R, Caul E O, Clarke I N. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- 15.Epstein R L, Powers M L, Rogart R B, Weiner H L. Binding of 125I-labeled reovirus to cell surface receptors. Virology. 1984;133:46–55. doi: 10.1016/0042-6822(84)90424-0. [DOI] [PubMed] [Google Scholar]
- 16.Estes M K, Atmar R L, Hardy M E. Norwalk and related diarrhea viruses. In: Richmann D D, Whitley R J, Hayden F G, editors. Clinical virology. New York, N.Y: Churchill Livingstone, Inc.; 1997. pp. 1073–1095. [Google Scholar]
- 17.Gershoni J M, Lapidot M, Zakai N, Loyter A. Protein blot analysis of virus receptors: identification and characterization of the Sendai virus receptor. Biochim Biophys Acta. 1986;856:19–26. doi: 10.1016/0005-2736(86)90004-0. [DOI] [PubMed] [Google Scholar]
- 18.Glass P J, White L J, Ball J M, Leparc-Goffart I, Hardy M E, Estes M K. Norwalk virus open reading frame 3 encodes a minor structural protein. J Virol. 2000;74:6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green K Y. The role of human caliciviruses in epidemic gastroenteritis. Arch Virol Suppl. 1997;13:153–165. doi: 10.1007/978-3-7091-6534-8_15. [DOI] [PubMed] [Google Scholar]
- 20.Green K Y, Kapikian A Z, Valdesuso J, Sosnovtsev S, Treanor J J, Lew J F. Expression and self-assembly of recombinant capsid protein from the antigenically distinct Hawaii human calicivirus. J Clin Microbiol. 1997;35:1909–1914. doi: 10.1128/jcm.35.7.1909-1914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green K Y, Lew J F, Jiang X, Kapikian A Z, Estes M K. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J Clin Microbiol. 1993;31:2185–2191. doi: 10.1128/jcm.31.8.2185-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 23.Greve J M, Rossmann M G. Interaction of rhinovirus with its receptor, ICAM-1. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. pp. 195–213. [Google Scholar]
- 24.Hale A D, Crawford S E, Ciarlet M, Green J, Gallimore C, Brown D W G, Jiang X, Estes M K. Expression and self-assembly of Grimsby virus: antigenic distinction from Norwalk and Mexico viruses. Clin Diagn Lab Immunol. 1999;6:142–145. doi: 10.1128/cdli.6.1.142-145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy M E, Estes M K. Completion of the Norwalk virus genome sequence. Virus Genes. 1996;12:287–290. doi: 10.1007/BF00284649. [DOI] [PubMed] [Google Scholar]
- 26.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blass D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 28.Jaykus L-A, De Leon R, Sobsey M D. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl Environ Microbiol. 1996;62:2074–2080. doi: 10.1128/aem.62.6.2074-2080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Cubitt D, Hu J, Dai X, Treanor J, Matson D O, Pickering L K. Development of an ELISA to detect MX virus, a human calicivirus in the snow Mountain agent genogroup. J Gen Virol. 1995;76:2739–2747. doi: 10.1099/0022-1317-76-11-2739. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Matson D O, Ruiz-Palacios G M, Hu J, Treanor J, Pickering L K. Expression, self-assembly, and antigenicity of a Snow Mountain agent-like calicivirus capsid protein. J Clin Microbiol. 1995;33:1452–1455. doi: 10.1128/jcm.33.6.1452-1455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 33.Jin Y-M, Pardoe I U, Burness A T H, Michalak T I. Identification and characterization of the cell surface 70-kilodalton sialoglycoprotein(s) as a candidate receptor for encephalomyocarditis virus on human nucleated cells. J Virol. 1994;68:7308–7319. doi: 10.1128/jvi.68.11.7308-7319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapikian A Z. Norwalk and Norwalk-like viruses. In: Kapikian A Z, editor. Viral infections of the gastrointestinal tract. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 471–518. [Google Scholar]
- 35.Kapikian A Z, Estes M K, Chanock R M. Norwalk group of viruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 783–810. [Google Scholar]
- 36.Kapikian A Z, Wyatt R G, Dolin R, Thornhill T S, Kalica A R, Chanock R M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10:1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnauchow T M, Tolson D L, Harrison B A, Altman E, Lublin D M, Dimock K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55) J Virol. 1996;70:5143–5152. doi: 10.1128/jvi.70.8.5143-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilpatrick D R, Lipton H L. Predominant binding of Theiler's viruses to a 34-kilodalton receptor protein on susceptible cell lines. J Virol. 1991;65:5244–5249. doi: 10.1128/jvi.65.10.5244-5249.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi S, Sakae K, Suzuki Y, Shinozaki K, Okada M, Ishiko H, Kamata K, Suzuki K, Natori K, Miyamura T, Takeda N. Molecular cloning, expression, and antigenicity of Seto virus belonging to genogroup I Norwalk-like viruses. J Clin Microbiol. 2000;38:3492–3494. doi: 10.1128/jcm.38.9.3492-3494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Lambden P R, Caul E O, Ashley C R, Clarke I N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Clarke I N, Lambden P R. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70:2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B L, Viljoen G J, Clarke I N, Lambden P R. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J Gen Virol. 1999;80:291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- 46.Mason H S, Ball J M, Shi J J, Jiang X, Estes M K, Arntzen C J. Expression of Norwalk virus capsid protein in transgenic tobacco and potato and its oral immunogenicity in mice. Proc Natl Acad Sci USA. 1996;93:5335–5340. doi: 10.1073/pnas.93.11.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morens D M, Zweighaft R M, Vernon T M, Gary G W, Eslien J J, Wood B T, Holman R C, Dolin R. A waterborne outbreak of gastroenteritis with secondary person-to-person spread. Association with a viral agent. Lancet. 1979;i:964–966. doi: 10.1016/s0140-6736(79)91734-3. [DOI] [PubMed] [Google Scholar]
- 48.Morse D L, Guzewich J J, Hanrahan J P, Stricof R, Shayegani M, Deibel R, Grabau J C, Nowak N A, Herrmann J E, Cukor G, et al. Widespread outbreaks of clam- and oyster-associated gastroenteritis. Role of Norwalk virus. J Med Virol. 1986;19:11–18. doi: 10.1056/NEJM198603133141103. [DOI] [PubMed] [Google Scholar]
- 49.Numata K, Nakata S, Jiang X, Estes M K, Chiba S. Epidemiological study of Norwalk virus infections in Japan and Southeast Asia by enzyme-linked immunosorbent assays with Norwalk virus capsid protein produced by the baculovirus expression system. J Clin Microbiol. 1994;32:121–126. doi: 10.1128/jcm.32.1.121-126.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pether J V, Caul E O. An outbreak of food-borne gastroenteritis in two hospitals associated with a Norwalk-like virus. J Hyg. 1983;91:343–350. doi: 10.1017/s0022172400060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad B V, Hardy M E, Dokland T, Bella J, Rossmann M G, Estes M K. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 52.Prasad B V V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren R B, Costantini F, Gorgacz E J, Lee J J, Racaniello V R. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell. 1990;63:353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- 54.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 55.Scatchard G. The attraction of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660–672. [Google Scholar]
- 56.Seah E L, Marshall J A, Wright P J. Open reading frame 1 of the Norwalk-like virus Camberwell completion of sequence and expression in mammalian cells. J Virol. 1999;73:10531–10535. doi: 10.1128/jvi.73.12.10531-10535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shafren D R, Bates R C, Agrez M V, Herd R L, Burns G F, Barry R D. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J Virol. 1995;69:3873–3877. doi: 10.1128/jvi.69.6.3873-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shafren D R, Dorahy D J, Ingham R A, Burns G F, Barry R D. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J Virol. 1997;71:4736–4743. doi: 10.1128/jvi.71.6.4736-4743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 60.Stewart L M D, Posse R D. Baculovirus expression vectors. In: Davidson A J, Elliotts R M, editors. Molecular virology: a practical approach. Oxford, England: IRL Press; 1993. pp. 227–256. [Google Scholar]
- 61.Supanaranond K, Takeda N, Yamazaki S. The complete nucleotide sequence of a variant of Coxsackievirus A24, an agent causing acute hemorrhagic conjunctivitis. Virus Genes. 1992;6:149–158. doi: 10.1007/BF01703064. [DOI] [PubMed] [Google Scholar]
- 62.Taylor H P, Cooper N R. The human cytomegalovirus receptor on fibroblasts is a 30-kilodalton membrane protein. J Virol. 1990;64:2484–2490. doi: 10.1128/jvi.64.6.2484-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomassini J E, Graham D, DeWitt C M, Lineberger D W, Rodkey J A, Colonno R J. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1989;86:4907–4911. doi: 10.1073/pnas.86.13.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Jiang X, Madore H P, Gray J, Desselberger U, Ando T, Seto Y, Oishi I, Lew J F, Green K Y, Estes M K. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–5990. doi: 10.1128/jvi.68.9.5982-5990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe M, Reimann K A, DeLong P A, Liu T, Fisher R A, Letvin N L. Effect of recombinant soluble CD4 in rhesus monkeys infected with simian immunodeficiency virus of macaques. Nature. 1989;337:267–270. doi: 10.1038/337267a0. [DOI] [PubMed] [Google Scholar]
- 66.White L J, Ball J M, Hardy M E, Tanaka T N, Kitamoto N, Estes M K. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White L J, Hardy M E, Estes M K. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71:8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wickham T J, Granados R R, Wood H A, Hammer D A, Shuler M L. General analysis of receptor-mediated viral attachment to cell surfaces. Biophys J. 1990;58:1501–1516. doi: 10.1016/S0006-3495(90)82495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyatt R G, Greenberg H B, Dalgard D W, Allen W P, Sly D L, Thornhill T S, Chanock R M, Kapikian A Z. Experimental infection of chimpanzees with the Norwalk agent of epidemic viral gastroenteritis. J Med Virol. 1978;2:89–96. doi: 10.1002/jmv.1890020203. [DOI] [PubMed] [Google Scholar]