Abstract
Generation of neurons through direct reprogramming has emerged as a promising therapeutic approach for treating neurodegenerative diseases. In this study, we present an efficient method for reprogramming retinal glial cells into neurons. By suppressing Notch signaling by disrupting either Rbpj or Notch1/2, we induced mature Müller glial cells to reprogram into bipolar- and amacrine-like neurons. We demonstrate that Rbpj directly activates both Notch effector genes and genes specific to mature Müller glia while indirectly repressing expression of neurogenic basic helix-loop-helix (bHLH) factors. Combined loss of function of Rbpj and Nfia/b/x resulted in conversion of nearly all Müller glia to neurons. Last, inducing Müller glial proliferation by overexpression of dominant-active Yap promotes neurogenesis in both Rbpj- and Nfia/b/x/Rbpj-deficient Müller glia. These findings demonstrate that Notch signaling and NFI factors act in parallel to inhibit neurogenic competence in mammalian Müller glia and help clarify potential strategies for regenerative therapies aimed at treating retinal dystrophies.
Inhibiting Notch signaling and NFI family transcription factors stimulates glia-to-neuron reprogramming in mammalian retina.
INTRODUCTION
Loss of neurons is the key pathological feature of many neurodegenerative diseases such as Parkinson’s disease, Alzheimer’s disease, and retinal dystrophies. Photoreceptors and ganglion cells of the retina, like many other neuronal subtypes in the central nervous system, are susceptible to degeneration, with their loss often resulting in permanent blindness. While there is now no effective regenerative therapy to replace neurons, one of the most potentially promising strategies is through direct reprogramming of endogenous Müller glia (MG) into retinal neurons. MG are progenitor-like radial glial cells that serve as cellular sources for retinal regeneration in cold-blooded vertebrates. Zebrafish MG respond to retinal injury by converting from a resting to an activated state, dividing asymmetrically to produce multipotent neurogenic retinal progenitor cells, and regenerating all types of retinal neurons (1, 2). In mammals, however, MG undergo reactive gliosis and do not spontaneously regenerate neurons following retinal injury.
Previous studies have shown that zebrafish MG rapidly up-regulate neurogenic basic helix-loop-helix (bHLH) factors such as ascl1a following injury (3, 4). In mice, transgenic overexpression of Ascl1 in combination with both histone deacetylase inhibition and other transcription factors (TFs) can induce MG to reprogram into inner retinal-like neurons following injury (5). However, other factors act in parallel with these positive regulators of neurogenesis to repress neurogenic competence. Our previous work showed that the gene expression profile of resting mammalian MG closely resembles retinal progenitor cells. These cells are robustly activated in response to injury; however, their conversion to a neurogenic state is actively inhibited by a dedicated transcriptional regulatory network that restores MG to a resting state (6). Within this network, we identified the nuclear factor I (NFI) family of TFs as negative regulators of injury-induced reprogramming. Simultaneous loss of function of Nfia, Nfib, and Nfix (Nfia/b/x) in mature mouse MG leads a subset of cells to convert to bipolar or amacrine cells (BCs and ACs, respectively) primarily through a process of direct transdifferentiation (6), much like what has been observed following Ascl1 overexpression (5). Therefore, combinatorial approaches that target both positive and negative regulators of neurogenic competence in MG are likely needed for the development of effective reprogramming therapies.
However, major gaps in our understanding of the regulation of neurogenic competence in MG must be bridged before therapeutic applications can be further developed. Current methods for inducing neurogenic competence, whether involving bHLH overexpression or NFI factors loss of function, often require retinal injury to induce MG reprogramming (5, 6). Furthermore, the levels of induced neurogenesis are relatively low, and the modest levels of MG proliferation raise the concerns about potential depletion of MG if the efficiency of neurogenesis is enhanced. Moreover, the range of cell types generated from MG is typically limited to BCs and ACs, although recent studies have also successfully generated retinal ganglion–like cells (7, 8). Therefore, it is imperative to conduct further research aiming at identifying additional candidate regulators, both positive and negative, of neurogenic competence to advance our understanding and develop more effective strategies for efficient glia-to-neuron reprogramming in the retina.
Our previous work has identified transcriptional effectors of the Notch pathway as key candidate regulators of Müller glial quiescence in both zebrafish and mice (6). Notch signaling plays a central role in controlling vertebrate retinal development, where it maintains progenitor status, promotes gliogenesis, and inhibits neurogenesis, particularly photoreceptor specification (9–13). In mature zebrafish MG, it actively represses injury-induced reprogramming (14, 15). Suppression of Notch signaling via gamma-secretase inhibitors efficiently induces injury-independent reprogramming and enhances injury-induced reprogramming (16–20). In mammals, low levels of Notch signaling are detected in mature MG (6, 16), but it remains unclear whether active Notch signaling acts to maintain MG quiescence and represses neurogenic competence in these cells.
In this study, we assessed whether loss of Notch signaling could induce reprogramming of mammalian Müller glial cells. We found that selective deletion of the common Notch transcriptional mediator Rbpj in adult mouse MG induced direct transdifferentiation into bipolar- and amacrine-like cells without injury and that this process was significantly enhanced by N-methyl-d-aspartate (NMDA)–dependent excitotoxic injury. Furthermore, MG-specific loss of function of Notch1 and Notch2 receptors phenocopied the effect of Rbpj deletion. We also showed that simultaneous loss of function of Rbpj and Nfia/b/x led to a conversion of nearly all MG to neuron-like cells and that adeno-associated virus (AAV)–mediated overexpression of dominant-active Yap induced these cells to proliferate. These findings demonstrate that, much like in zebrafish, Notch signaling actively represses neurogenic competence in mammalian MG and imply that efficient endogenous cell reprogramming can be achieved by targeting both negative and positive regulators of neurogenesis.
RESULTS
Suppression of RBPJ-mediated Notch signaling induces MG-derived neurogenesis in the adult mouse retina with and without retinal injury
To comprehensively examine the cellular expression patterns of Notch pathway components in adult retina, we analyzed single-cell RNA sequencing (scRNA-seq) datasets obtained from zebrafish, mice, and humans (6, 21–23). Our scRNA-seq data analysis showed an evolutionary conserved expression of Notch signaling components in MG of all three species. In zebrafish, MG expressed high levels of notch3 and the Notch target genes her6 and id1 (fig. S1A). In mice and humans, in contrast, MG expressed Notch1 and Notch2 and substantially lower levels of Notch3 and Notch4 (fig. S1, B to C). Expression of all key Notch ligands, mediators, and target genes, including Jag1, Rbpj, Numb, Hes1, Hes5, Hey1, Hey2, and Id1/2/3/4, was found in both mouse and human MG. No other retinal cell type, except vascular/endothelial cells, showed comparable expression of Notch signaling components.
Previous studies showed that loss of function of the common Notch transcriptional mediator Rbpj resulted in global inhibition of Notch signaling (24–26) and induced a limited level of astrocyte-to-neuron conversion in the brain following injury (27–29). To assess whether loss of Notch signaling could induce reprogramming of mature mouse MG, we selectively deleted Rbpj in MG using GlastCreER;Rbpjlox/lox;Sun1-GFP mice (Fig. 1A). These transgenic mice express a tamoxifen (TAM)–inducible Cre recombinase under the control of regulatory elements of the MG-specific Glast (Slc1a3) gene. We induced Cre recombination in GlastCreER;Sun1-GFP control and GlastCreER;Rbpjlox/lox;Sun1-GFP mice with five daily intraperitoneal injections of TAM. TAM administration leads to both Cre-dependent removal of exon 4 of Rbpj that results in a null mutation (30) and expression of the Sun1-GFP reporter, thereby allowing lineage tracing of all MG and MG-derived cells (6, 31). We collected uninjured retinas collected for immunohistochemistry (IHC) analysis at multiple time points ranging from 3 weeks to 4 months after TAM injection (Fig. 1B). In previous studies, retinal injury and associated glial activation are required for inducing MG-derived neurogenesis following either Ascl1 overexpression or Nfia/b/x loss of function (5, 6). We also tested whether NMDA-mediated excitotoxic injury, which primarily caused amacrine and retinal ganglion cell death, enhances neurogenesis in Rbpj-deficient MG. We performed intravitreal NMDA injection at 7 days after the final dose of TAM and analyzed retinas analyzed at 3 weeks, 5 weeks, and 4 months following NMDA injury (Fig. 1C).
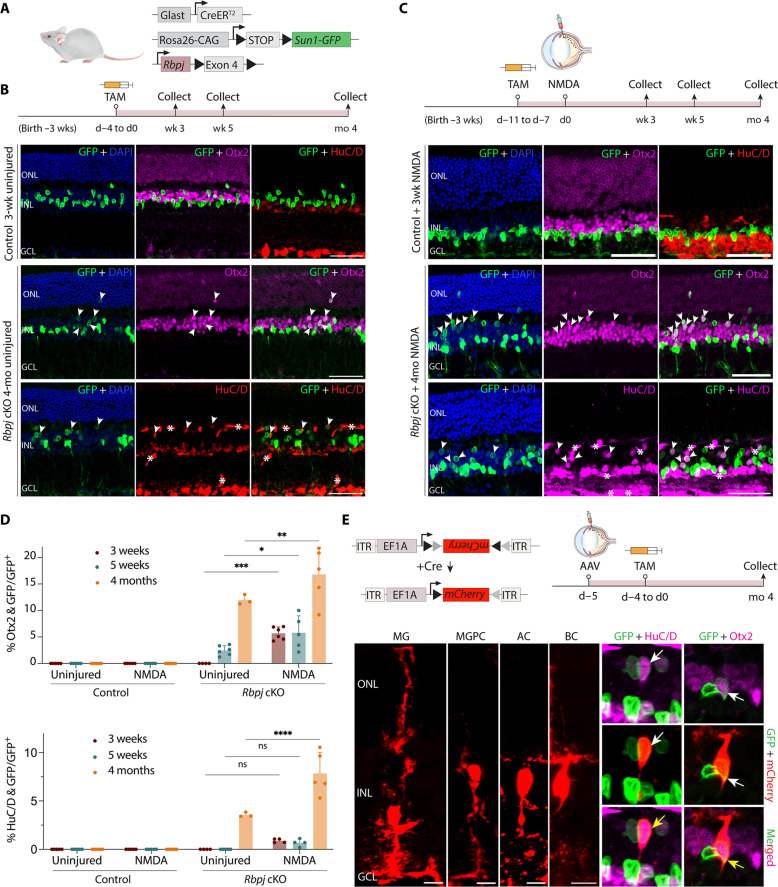
Fig. 1. Selective loss of function of Rbpj induces neurogenesis in adult MG with and without retinal injury.
(A) Schematic of the transgenic constructs used to induce deletion of Rbpj specifically in MG. Cre-mediated removal of Exon 4 of Rbpj leads to premature stop codon formation and disrupts the expression of the RBPJ protein. (B and C) Schematic of the experimental workflow and representative immunostaining for green fluorescent protein (GFP), Otx2, and HuC/D in control and Rbpj-deleted retinas without (B) and with (C) NMDA-induced injury. White arrowheads indicate co-labeled GFP-positive and marker-positive cells. GFP-positive MG-derived neuron-like cells often show relatively lower cellular levels of GFP expression than do MG. Asterisks (*) indicate mouse-on-mouse vascular staining. Scale bars, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole. (D) Quantification of mean percentage ± SD of GFP-positive MG-derived neurons expressing either OTX2 or HuC/D. (E) Morphological characterization of MG-derived neurons in Rbpj-deficient retinas using AAV2.7 m8-Ef1a-Flex-mCherry 4 months after TAM treatment. Yellow arrows indicate co-labeled GFP/mCherry/marker-positive cells. Scale bars, 10 μm. Significance was determined via two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Each data point was calculated from an individual retina. TAM, tamoxifen; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; MG, Müller glia; MGPC, MG-derived progenitor cell; AC, amacrine cell; BC, bipolar cell; wk/wks, week/weeks; d, day; mo, month.
Immunostaining data showed that Hes1 protein is enriched in wild-type MG (fig. S2B) and is significantly reduced in Rbpj-deficient MG (fig. S2D), which confirms that loss of Rbpj down-regulates Notch effectors. We detected no MG-derived neurogenesis in the uninjured and NMDA-treated control retinas and all green fluorescent protein (GFP)–positive cells expressed the glial marker Sox9 (Fig. 1C and fig. S2A). However, in Rbpj conditional mutants, we observed that 2.5% of GFP-positive MG-derived cells expressed the pan-bipolar/photoreceptor cell marker Otx2 at 5 weeks, with a progressive increase to 12% at 4 months after induction (Fig. 1, B and D). At this later time point, we also observed that 3.6% of GFP-positive cells expressed the pan-amacrine/retinal ganglion cell marker HuC/D (Fig. 1, B and D). NMDA treatment substantially enhanced levels of MG-derived neurogenesis, with 5.8 and 15% of GFP-positive cells expressing Otx2 at 5 weeks and 4 months, respectively (Fig. 1, C and D). The levels of MG-derived ACs did not change significantly at 3 and 5 weeks in NMDA-treated retinas compared to that in uninjured retinas. However, they were enhanced at 4 months, with 7.8% of GFP-positive cells showing HuC/D expression, in contrast to 3.6% of GFP-positive cells in uninjured retina (Fig. 1, C and D).
Immunostaining analysis of additional cell markers revealed a subset of Rbpj-deficient MG-derived neuron-like cells expressed the glycinergic amacrine-specific marker glycine (fig. S2E) and amacrine/progenitor marker Pax6 (fig. S2F). In addition, these cells also expressed bipolar-specific markers Vsx2 (pan-bipolar), Cabp5 (rod and cone bipolar), Isl1 (rod and cone ON bipolar), and Scgn (cone ON bipolar) (fig. S2, G to J) but did not express the rod bipolar PKCa (fig. S2K). In addition to neurogenesis, we also examined whether loss of Rbpj induces MG-derived proliferation by administering 5-ethynyl-2′-deoxyuridine (EdU) via intraperitoneal injection and drinking water for 7 days at 1 week after TAM. EdU labeling revealed no GFP-positive cells that had incorporated EdU (fig. S2, C and L), nor did these cells exhibit Ki67 staining, indicating that Rbpj-deficient MG did not undergo proliferation. To address the possibility that GFP/HuC/D-positive cells arose from cells that were previously GFP/Otx2-positive, we co-stained for GFP, Otx2, and HuC/D. We observed that GFP-positive MG expressing Otx2 did not co-express HuC/D or vice versa, indicating that these neurons are clearly derived from two different lineages of MG (fig. S3A). Furthermore, expression of the neurogenic bHLH factor Ascl1 was activated in Rbpj-deficient MG but was switched off in GFP-positive induced neurons (fig. S3, B and C). Triple immunostaining for GFP/Lhx2/Otx2 or GFP/Lhx2/HuC/D demonstrated that MG-derived neuron-like cells (GFP/Otx2+ or GFP/HuC/D+) do not express the MG marker Lhx2 (fig. S3, D and E), which suggests that they have lost their glial identity.
The use of nuclear membrane–bound Sun1-GFP reporter allows unambiguous identification of MG-derived neurons; however, the overall cell morphology cannot be observed. To better characterize the neuronal morphology of these induced neuron-like cells, we performed intravitreal injection of Cre-dependent FLEX AAV carrying an mCherry reporter into adult Rbpj conditional knockout (cKO) mice. We then induce Cre recombination with five daily intraperitoneal injections of TAM and harvested the retinas 4 months later. IHC analysis revealed that the MG-derived neuron-like cells retracted their apical glial processes and acquired bipolar- or amacrine-like morphologies, indicated by co-expression of mCherry/GFP with either Otx2 or HuC/D (Fig. 1E). Together, these findings indicate that Notch signaling negatively regulates MG neurogenic competence in mammals and suppressing this pathway induces MG to transdifferentiate into bipolar and amacrine-like cells without injury and this process is significantly enhanced with injury.
Loss of function of Notch1/2 receptors phenocopies neurogenesis observed in Rbpj-deficient MG
While Rbpj is a well-established transcriptional mediator of Notch signaling, previous studies suggest that it may have Notch-independent functions (32–34) and, conversely, that some aspects of Notch signaling may be Rbpj-independent (35, 36). There are four Notch receptors: Notch1 to Notch4, with Notch1 and Notch2 being the two most abundantly expressed in mature MG (fig. S1). To assess whether repression of neurogenic competence in MG was dependent on active Notch signaling, we generated GlastCreER;Notch1lox/lox;Sun1-GFP, GlastCreER;Notch2lox/lox;Sun1-GFP single, and GlastCreER;Notch1lox/lox;Notch2lox/lox;Sun1-GFP double loss-of-function mutants of Notch1 and Notch2 (Fig. 2A) (37, 38). As previously described, we performed five daily intraperitoneal injections of TAM to delete Notch1 and/or Notch2 specifically in MG while simultaneously labeling these cells with Sun1-GFP for lineage tracing (Fig. 2B). Notch1/2 double mutant retinas showed similar levels of MG-derived bipolar-like cells seen in age-matched Rbpj-deficient retinas 4 months after TAM (Fig. 2, C and D). The relative numbers of MG-derived HuC/D-positive amacrine-like cells, however, were significantly lower in Notch1/2 double mutants relative to Rbpj mutants in the absence of injury (Fig. 2D).
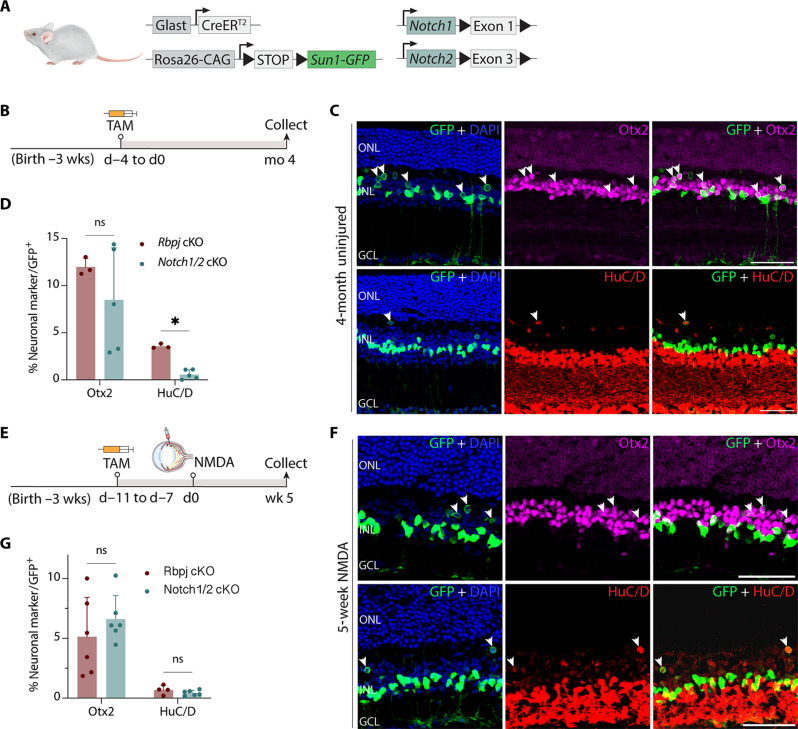
Fig. 2. Notch1/2 deletion phenocopies effects of Rbpj deletion on MG-derived neurogenesis.
(A) Schematic of the transgenic constructs used to induce deletion of Notch1 and Notch2 specifically in MG. Cre-mediated removal of Exon 1 of Notch1 and Exon 3 of Notch2 leads to premature stop codon formation and disrupts the expression of the NOTCH1/2 proteins. (B) Schematic of the experimental workflow for uninjured retinas. (C and D) Representative images immunolabeled for GFP, OTX2, and HuC/D of Notch1/Notch2-deleted retinas after 4 months after TAM injection, and quantification of mean percentage ± SD of GFP-positive MG-derived neurons expressing either Otx2 or HuC/D. White arrowheads indicate co-labeled GFP-positive and marker-positive cells. (E to G) Schematic of the workflow, representative immunostaining, and quantification for NMDA-injured retinas. Significance was determined via Mann-Whitney U test: *P < 0.05. Each data point was calculated from an individual retina. Scale bars, 50 μm. ns, not signifcant.
To compare the level of neurogenesis between Notch1/2 double mutant and Rbpj-deficient retinas after retinal injury, we induced excitotoxic retinal injury with NMDA and analyzed the retina 5 weeks later. We did not observe any induction of neurogenesis in Notch1-deficient MG and observed that only 0.7% of GFP-positive cells expressed Otx2 in Notch2-deficient mice. Likewise, the level of MG-derived neurogenesis in Notch1/2 double mutants is also comparable to those seen with Rbpj mutants following NMDA injury (Fig. 2, E to G). As in Rpbj cKO mice, a subset of Notch1/2-deficient MG-derived cells also expressed the cone bipolar-specific marker Scgn (fig. S3F) but did not express rod bipolar markers such as PKCa (fig. S3G). Expression of neurogenic bHLH factors such as Ascl1 was also activated in Notch1/2-deficient MG (fig. S3H). Sox9 expression was detected in the remaining GFP-positive cells, indicating that they retained glial identity (fig. S3I). These results demonstrate that Notch1/2 loss of function largely phenocopies Rbpj loss of function.
Rbpj directly promotes expression of Notch pathway genes and genes specific to resting MG while indirectly repressing neurogenic bHLH factors
To investigate the early changes after Rbpj loss of function, we conducted scRNA-seq on uninjured whole retinas of GlastCreER;Sun1-GFP control and GlastCreER;Rbpjlox/lox;Sun1-GFP mice at 7 days following TAM injection (Fig. 3A and fig. S4A). We then subsetted the MG population for further downstream analysis. At 7 days after TAM injection, before any neurogenesis is detected, while MG from both samples show broadly similar transcriptional profiles (Fig. 3B), a subset of genes show strongly enriched expression in Rbpj-deficient MG. Genes specific to mature MG (Glul, Aqp4, Apoe, Kcnj10, Mlc1, and Sox9) and Notch pathway genes (Hes1, Hes5, and Id1/2/3) are significantly down-regulated in Rbpj-deficient MG relative to controls. The low level of Hes1 expression that is still observed in Rbpj cKO MG may reflect Notch-independent expression, which has previously been reported in retina (39, 40). In contrast, neurogenic bHLH factors (Ascl1, Neurog2, and Hes6) are strongly up-regulated in Rbpj-deficient MG (Fig. 3, C and D; fig. S4, B and C; and table S1). Expression of cell cycle inhibitors, such as Cdkn1b, Cdkn1c, and Btg2, is up-regulated, consistent with the observation that Rbpj-deficient MG undergo direct conversion into neuron-like cells without proliferation.
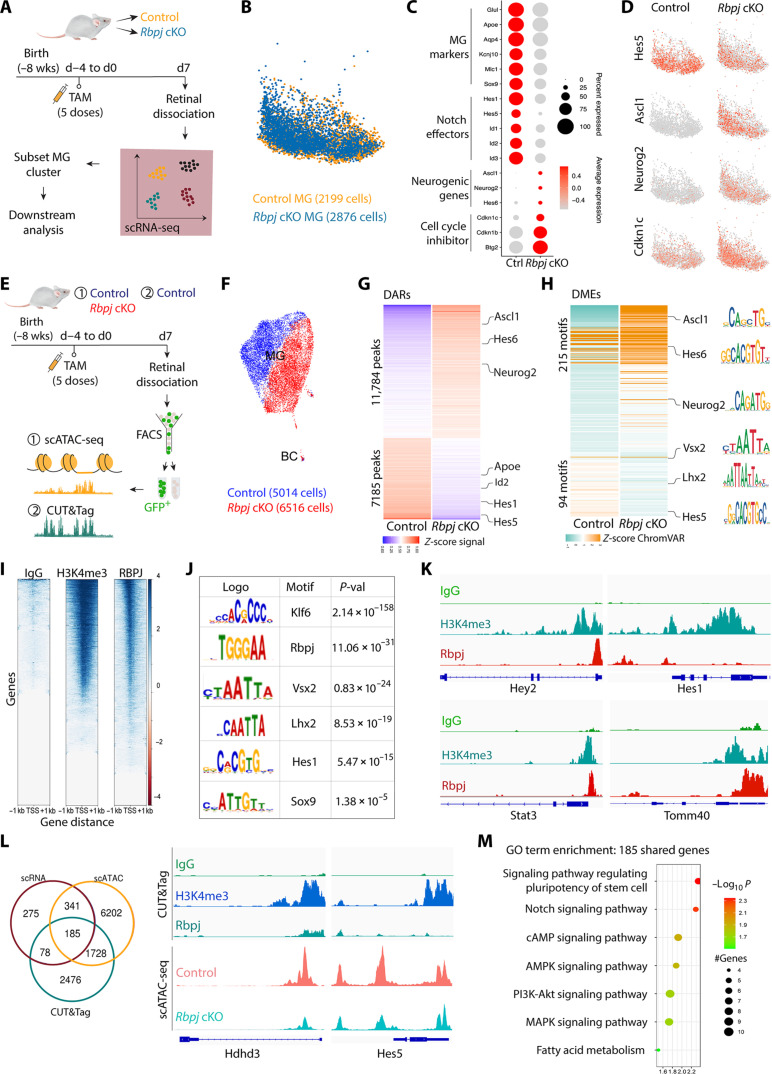
Fig. 3. Rbpj deletion in MG down-regulates Notch target genes and activates expression of neurogenic genes.
(A) Schematic of the experimental workflow used to generate scRNA-Seq data from whole retinas of control and Rbpj cKO mice. (B) Uniform manifold approximation and projection (UMAP) of scRNA-seq data showing the clustering of control and Rbpj-deficient MG subsetted from whole retina cell populations. (C) Dot plot showing down-regulation of genes specific to resting MG and Notch-regulated genes, and up-regulation of neurogenic bHLH factors as well as cell cycle inhibitors in Rbpj-deficient MG relative to controls. (D) Feature plots highlighting differential expression of Hes5, Ascl1, Neurog2, and Cdkn1c in control and Rbpj-deficient MG. (E) Schematic of the experimental workflow used to generate scATAC-seq data from control and Rbpj cKO MG and CUT&Tag data from control MG. (F) UMAP plot showing global differences in chromatin accessibility observed by scATAC-seq in control and Rbpj-deficient MG. (G) Differentially accessible chromatin regions (DARs) and nearby genes between Rbpj-deficient and control MG. (H) Differential TF motif enrichments (DMEs) in the DARs between Rbpj-deficient and control MG. (I) CUT&Tag analysis of IgG, H3K4me3, and RBPJ from fluorescence-activated cell sorting (FACS)–isolated GFP-positive MG from GlastCreER;Sun1GFP retinas. (J) TF motifs enriched in RBPJ CUT&Tag samples relative to control. (K) Representative genome tracks for CUT&Tag analysis. (L) Integrative analysis of scRNA-seq, scATAC-seq, and CUT&Tag to identify genes regulated by RBPJ. (M) Gene Ontology (GO) enrichment analysis for genes shared among the three datasets. AMPK, AMP-activated protein kinase; cAMP, cyclic adenosine 3′,5′-monophosphate; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase.
To examine the changes in chromatin accessibility caused by Rbpj deletion, we performed Single-cell ATAC sequencing (scATAC-seq) on fluorescence-activated cell sorting (FACS)–isolated GFP-positive cells from uninjured retinas of GlastCreER;Sun1-GFP control and GlastCreER;Rbpjlox/lox;Sun1-GFP mice at 7 days following TAM injection (Fig. 3E). Clustering analysis showed a much clearer separation of control and Rbpj-deficient MG cells than was seen using scRNA-seq, indicating that Rbpj loss of function leads to global changes in chromatin accessibility (Fig. 3F and fig. S4E). We then compared control and Rbpj-deleted MG and identified 18,969 differentially accessible chromatin regions (DARs) between the two samples. Furthermore, we also observed that changes in chromatin accessibility broadly mirror changes in transcription, with decreased accessibility at Notch pathway genes such as Hes1, Hes5, Hey2, and Id1/2/3/4 and increased accessibility at neurogenic bHLH genes such as Ascl1, Neurog2, and Hes6 (Fig. 3G and table S1). We next examined TF motif enrichment in these DARs. As expected, we observed substantial motif enrichments for neurogenic TFs, including Ascl1, Neurog2, and Hes6, and reduction in accessibility associated with TF motifs for MG-enriched genes, such as Rbpj, Lhx2, Hes5, and Vsx2, in Rbpj-deficient MG (Fig. 3H and fig. S4, E and D).
To identify genomic binding targets of Rbpj, we next performed CUT&Tag on FACS-isolated GFP-positive adult MG from GlastCreER;Sun1-GFP mice at 7 days following TAM injection (Fig. 3, E and I). Rbpj-binding genomic regions were significantly enriched for consensus motifs for Rbpj and MG-enriched TFs such as Klf6, Lhx2, Hes1, Hes5, and Sox8/9 (Fig. 3J). As expected, Rbpj binds directly to cis-regulatory sites associated with Notch pathway genes, including Hes1, Hes5, Hey2, and Id1/2/3/4 as well as other TFs such as Tcf7l2 that are known to promote gliogenesis (Fig. 3K, fig. S4F, and table S2) (41). We found that Rbpj also directly binds to regulatory sites associated with genes that promote cell cycle inhibition, including Cdkn1c and Btg2 (table S2). We next sought to comprehensively identify genes directly regulated by Rbpj by integrating differential gene expression, DARs, and Rbpj-binding regions from scRNA-seq, scATAC-seq, and CUT&Tag data, respectively. We found that 185 genes were shared among the three datasets, indicating that the expression of these genes is directly controlled by Rbpj (Fig. 3L). Gene Ontology (GO) enrichment analysis showed that these genes are enriched for Notch, Akt, and mitogen-activated protein kinase signaling, as well as fatty acid metabolism (Fig. 3M). Together, our findings revealed that Rbpj directly activates Notch effector genes and genes specific to mature MG while also indirectly represses the expression of neurogenic bHLH factors.
Combined loss of function of Nfia/b/x and Rbpj leads to conversion of the majority of MG to neuron-like cells
We previously showed that the NFI factors inhibit neurogenic competence in mature MG (6). Additional analysis of Nfia/b/x-deficient MG demonstrates that Notch pathway genes such as Hes1, Hes5, and Hey2 remained expressed in Nfia/b/x-deficient MG (fig. S5, A to C). Furthermore, the expression and motif activity of Nfia/b/x were largely unaffected in Rbpj-deficient MG (fig. S4, C and D, and table S1). This raises the question whether Notch signaling and NFI factors act in parallel to inhibit neurogenic competence in adult MG. To address this, we used the gamma-secretase inhibitor N-[N-(3, 5-difluorophenacetyl)-l-alanyl]-s-phenylglycinet-butyl ester (DAPT) to inhibit Notch signaling in combination with our previously described MG-specific TAM-inducible mouse model of Nfia/b/x loss of function (GlastCreER;Nfia/b/xlox/lox; Sun1-GFP) (6). Briefly, adult mice were fed TAM diet for 3 weeks, followed by intravitreal injection of DAPT, then NMDA, and lastly a second dose of DAPT (fig. S5D). We then collected retinas 3 weeks later for immunostaining analysis. Quantification of HuC/D/GFP-positive cells revealed over twofold increase in the number of MG-derived neuron-like cells in DAPT-treated retinas compared to that in vehicle control (fig. S5D). This finding supports our hypothesis that Nfia/b/x and Rbpj act in parallel to repress MG neurogenic competence.
To better assess potential combinatorial roles for Nfia/b/x and Rbpj in repressing neurogenic competence, we generated GlastCreER;Nfia/b/xlox/lox;Rbpjlox/lox;Sun1-GFP mice (Fig. 4A). This transgenic mouse model allows a more specific inhibition of the Notch signaling pathway in MG than global inhibition of gamma-secretase and also serves as cross-validation of the DAPT treatment. After five daily intraperitoneal injections of TAM to delete Nfia/b/x and Rbpj in MG, we induced retinal injury with injection of NMDA in the right eye while leaving the left eye uninjured. We then harvested the retinas for immunostaining analysis 3 weeks later to examine the level of MG-derived neurogenesis (Fig. 4B). We observed that combined loss of function of Nfia/b/x and Rbpj leads to a robust and synergistic increase in neurogenesis relative to both Nfia/b/x- (6) and Rbpj-deficient MG (Fig. 1). In uninjured Nfia/b/x;Rbpj-deficient retinas, we found that 45.3% of GFP-positive cells expressed either Otx2 or HuC/D at 4 weeks following TAM induction, with many of these cells undergoing radial migration into the outer nuclear layer (ONL; Fig. 4, C and D). NMDA-mediated excitotoxic injury induces even higher levels of glia-to-neuron conversion in Nfia/b/x;Rbpj-deficient MG. At 3 weeks after injury, we observe that the vast majority of Sun1-GFP–positive cell nuclei are radially displaced away from their typical location in the inner nuclear layer, with many now present in the ONL (Fig. 4C). We observe that 52.6% of all GFP-positive MG-derived cells are strongly immunopositive for Otx2, indicating that they are likely BCs, while 19.9% are HuC/D-positive ACs (Fig. 4, C and D).
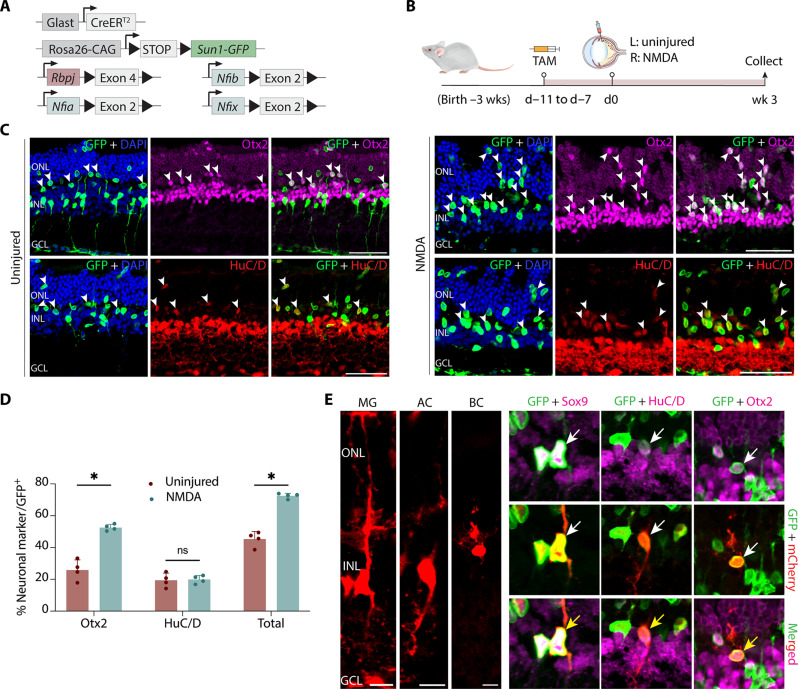
Fig. 4. Simultaneous disruption of Nfia/b/x and Rbpj convert nearly all adult MG into retinal neurons.
(A) Schematic of the transgenic constructs used to induce loss of function of Rbpj and Nfi/a/b/x specifically in MG. (B and C) Schematic of experimental pipeline and representative images of uninjured and NMDA-damaged retinas immunolabeled for GFP, Otx2, and HuC/D. White arrowheads indicate GFP-positive MG-derived neurons expressing neuronal markers Otx2 or HuC/D. (D) Quantification of mean percentage ± SD of GFP-positive MG-derived neurons expressing either Otx2 or HuC/D in uninjured and NMDA-treated retina. (E) Morphological characterization of MG-derived neurons in Nfia/b/x;Rbpj-deficient retinas using AAV2.7 m8-Ef1a-Flex-mCherry 4 weeks after TAM treatment. Yellow arrows indicate co-labeled GFP/mCherry/marker-positive cells. Scale bars, 10 μm. Significance was determined via unpaired t test: *P < 0.05. Scale bars, 50 μm.
To better characterize the neuron-like cells derived from Nfia/b/x;Rbpj-deficient MG, we performed morphological analysis using a similar experimental paradigm used for Rbpj cKO mice (Fig. 1E). We harvested retinas 4 weeks after TAM for IHC analysis, which revealed the MG-derived cells retracted their apical glial processes and acquired bipolar- or amacrine-like morphologies (Fig. 4E). Similar to what is seen in Rpbj-deficient retinas, a subset of Nfia/b/x;Rbpj-deficient MG-derived cells also expressed bipolar-specific markers Scgn, Vsx2, and CaBP5 (fig. S6, A to C) but did not express the rod bipolar marker PKCa (fig. S6D). A subset of induced neurons expresses amacrine-marker Pax6 (fig. S6E). Triple immunostaining for GFP/Lhx2/Otx2 or GFP/Lhx2/HuC/D demonstrated that MG-derived neuron-like cells (GFP/Otx2+ or GFP/HuC/D+) do not express the MG marker Lhx2, which suggests that they have lost their glial identity (fig. S6, G and H). In line with this massive reduction in the number of MG due to direct transdifferentiation, we also observe widespread disruptions in retinal lamination, with whorls and rosettes formed throughout the ONL (Fig. 4C). This disorganization of the outer limiting membrane mimics the effects observed by selective loss of a large fraction of MG (42–45). In addition to these MG-derived bipolar- and amacrine-like cells, we also observed small numbers of GFP-positive cells expressing Nrl or Recoverin in the ONL, indicating that we also are generating some rod-like photoreceptors (fig. S5, J and K).
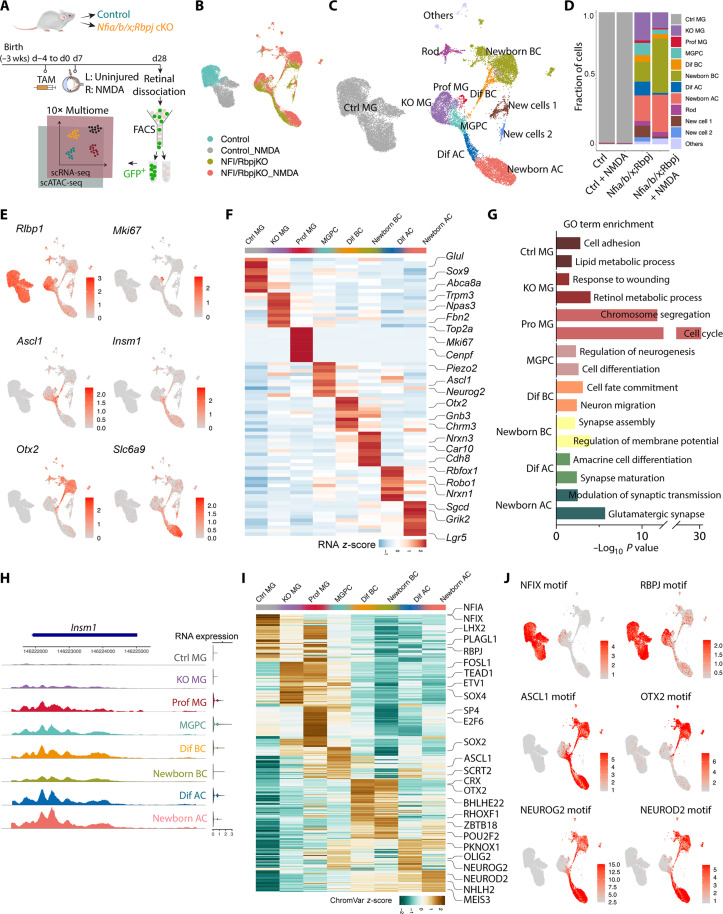
To better understand the molecular mechanism underlying MG reprogramming in the setting of a deficiency in both Nfia/b/x and Rbpj, we conducted single-nucleus multiomic (combined RNA-seq and ATAC-seq) analysis of FACS-isolated GFP-positive cells from control GlastCreER;Sun1-GFP and GlastCreER;Nfia/b/xlox/lox; Rbpjlox/lox;Sun1-GFP mice. Both uninjured and NMDA-treated retinas were profiled 4 weeks following TAM induction (Fig. 5A and table S3). The combined uniform manifold approximation and projection (UMAP) plot of these scRNA-seq and scATAC-seq data show a clear separation of control and Nfia/b/x;Rbpj-deficient MG clusters, suggesting that deleting Nfia/b/x and Rbpj markedly alters gene expression globally (Fig. 5B). Cell annotation based on known marker genes revealed no MG-derived neurons in uninjured and NMDA-treated control samples (Fig. 5D). In contrast, Nfia/b/x;Rbpj-deficient MG generated several distinct cell populations, including proliferative MG, neurogenic MG-derived progenitor cells (MGPCs), and amacrine and bipolar-like cells, as well as a small number of rod photoreceptors (Fig. 5C). Unexpectedly, we also observed two additional clusters of cells that selectively expressed Col25a1 and Alk, respectively (fig. S6A). While the Col25a1-positive cluster expressed a subset of photoreceptor/bipolar (Crx, Gnb3, and Tacr3)–and retinal pigment epithelium (Rgr, Lrat, and Rrh)–specific markers, neither of these clearly corresponded to any cell type in the wild-type retina. These cells were barely detected in the NMDA-treated retina where substantially higher levels of neurogenesis were observed, implying that they may represent transitional rather than stable cell states (Fig. 5, C and D).
Fig. 5. Integrated snRNA/scATAC-seq analysis of control and Nfia/b/x;Rbpj-deficient adult MG and their progeny.
(A) Schematic of the multiomic snRNA/ATAC-seq experimental pipeline. (B) UMAP plot of Multiome (scRNA/ATAC-seq) datasets showing the clustering of control and Nfia/b/x;Rbpj-deficient MG from uninjured and NMDA-treated retinas. (C) UMAP plot showing the identity of cell clusters determined by marker gene expression. (D) Stacked barplots represent the proportion of cells in each cluster across different sample groups. (E) Feature plots highlighting the cluster of MG (Rlbp1), proliferating MG (Mki67), neurogenic MGPC (Ascl1 and Insm1), BCs (Otx2), and ACs (Slc6a9). (F) Heatmap showing the expression of top 10 differentially expressed genes for each cell cluster. (G) GO enrichment for each cell cluster. The x axis indicates the −log10(P value) of the GO term. (H) Representative ATAC peaks and RNA expression level of Insm1 across different cell clusters. (I) Heatmap showing activity of top 50 differential motifs across different cell clusters. (J) Feature plots showing activity of indicated motifs. Prof MG, proliferating MG; Dif BC, differentiating BCs; Dif AC, differentiating ACs.
Our multiomic analysis showed that a substantial number of neuron-like cells are derived from MG lacking Nfia/b/x and Rbpj (Fig. 5D). NMDA-induced injury further enhanced the level of MG-derived neurogenesis, which is consistent with our IHC data (Figs. 4D and 5D). However, a small subset of MG-derived cells expressed the proliferation marker, Mki67 (Fig. 5, E and F), which was also detected in our IHC analysis (fig. S6G). MGPCs selectively expressed neurogenic TFs such as Ascl1 and Insm1 (Fig. 5, E and G). This MGPC cluster forms clear differentiation trajectories connecting Rlbp1-expressing MG to Otx2/Cabp5-positive bipolar and Slc6a9/Elavl3-positive ACs (Fig. 5, E and F, and fig. S7, A and B). While subsets of MGPCs express certain genes that are enriched in either the BC trajectory (e.g. Otx2, Crx) or the AC trajectory (e.g., Pax6, Neurog2, and Olig2), it is not clear whether these represent cells that show any bias toward one or the other trajectory. GO enrichment analysis revealed that genes in the proliferating MG cluster were enriched for cell cycle regulation, genes in the MGPC cluster for cell differentiation, and genes in the induced bipolar and amacrine for membrane potential and synapse regulation (Fig. 5G). While no clear differentiation trajectory connected neurogenic MG and rod photoreceptors, a subset of cells in the bipolar differentiation trajectory expressed TFs that promote rod specification (Prdm1) or differentiation (Crx) (46–48) or genes that are known to be selectively expressed in photoreceptor precursors (Wnt5b/Ankrd33b) (fig. S7B and table S3) (49). The origin of the Col25a1 and Alk-positive cell clusters, however, remains less clear.
Many other known cell-type–specific genes were selectively up-regulated in these cells as they differentiated, including Car10/Lhx4/Trpm1 in BCs and Rbfox3/Gad2/Neurod2 in ACs (Fig. 5F and table S3). These transcriptional changes were reflected in changes in the chromatin accessibility observed in the ATAC-seq analysis. As in Rbpj-deficient MG, chromatin accessibility at genes specifically expressed in resting Müller glial genes was decreased, while accessibility for neurogenic TFs such as Insm1 and Neurod2 and the rod photoreceptor-specific factor Nrl was increased and maintained in differentiating neurons (Fig. 5H and fig. S7, C and D). Analysis of open chromatin regions revealed significant enrichment for motifs for neurogenic TFs such as Ascl1, Neurog2, and Neurod2 and reduction in accessibility associated with TF motifs for MG genes—including Nfia, Nfix, Rbpj, and Lhx2—in the Nfia/b/x;Rbpj-deficient MG (Fig. 5, I and J). Chromatin accessibility patterns were highly dynamic as reprogrammed MG transitions from MGPCs to differentiating and mature BCs, on the one hand, and to differentiating and mature ACs, on the other (fig. S7C and table S3). For instance, the Ascl1 target motif is highly accessible in MGPCs and shows lower accessibility in differentiating BCs and even lower accessibility in both mature BCs and the AC trajectory. Otx2 and Vsx2 motifs, on the other hand, show accessibility in differentiating and mature BC and low/absent accessibility in the AC trajectory. This closely resembles patterns observed in developing retina (41).
In summary, because no detectable levels of neurogenesis were observed in uninjured Nfia/b/x-deficient retina (6) and only 5.8% of Rbpj-deficient MG undergo conversion to neuron-like cells at this age (Fig. 1), these findings indicate that Nfia/b/x and Rbpj act synergistically to repress neurogenic competence in MG.
AAV-mediated overexpression of constitutively active Yap5SA promotes proliferation in Nfia/b/x- and Rbpj-deficient retina
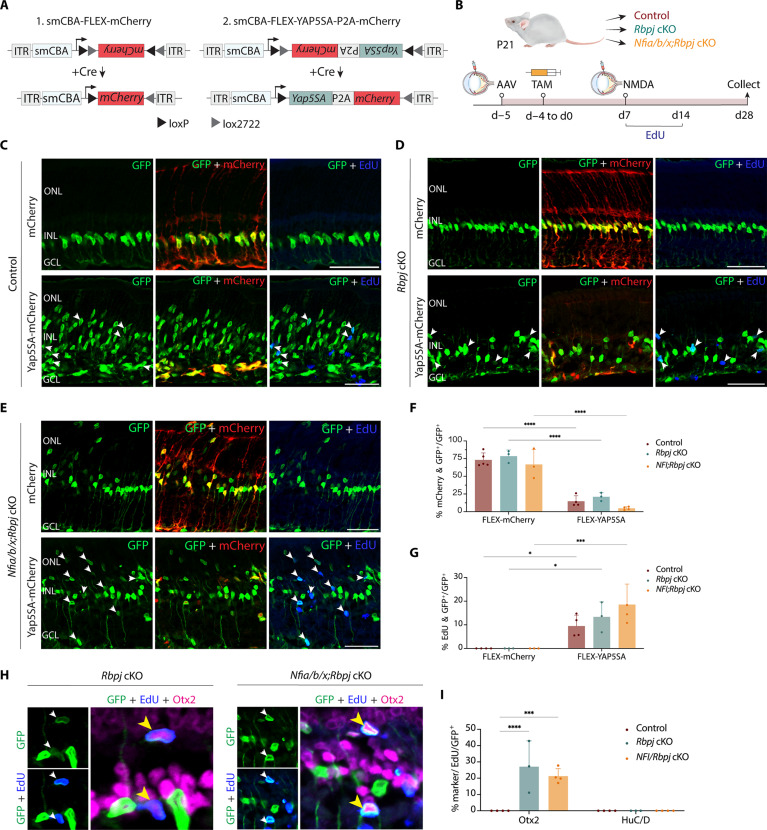
While loss of function of Rbpj induces MG-derived neurogenesis, we detected no proliferation in Rbpj-deficient MG. Moreover, we observed that Rbpj loss of function activates expression of cell cycle inhibitors such as Cdkn1b/c and Btg2 (Fig. 3). Likewise, only a limited level of MG proliferation was induced by Nfia/b/x- or combined Nfia/b/x;Rbpj deletion (Fig. 5). This process of direct transdifferentiation to neuron-like cells depletes endogenous MG and severely limits any potential clinical application of Notch pathway inhibition. However, recent studies show that Hippo signaling represses MG proliferation (50, 51) and that transgenic mice overexpressing the dominant-active Yap5SA mutant induce Müller glial proliferation (52). To determine whether overexpression of Yap5SA could induce MG to proliferate, we intravitreally injected adult wild-type control, Rbpj, and Nfia/b/x;Rbpj cKO mice with Cre-inducible FLEX AAV constructs that overexpressed stably either Yap5SA-P2A-mCherry or mCherry alone (Fig. 6A). We then induced Cre activation with five daily doses of TAM intraperitoneal injection followed by retinal injury with NMDA. To label proliferating cells, we administered EdU to mice via drinking water and daily intraperitoneal injections for 7 days. We then harvested retinas at 1 and 3 weeks after NMDA injection for immunostaining to detect MG-derived proliferation and neurogenesis (fig. S8A and Fig. 6B).
Fig. 6. Overexpression of dominant active YapS5A stimulates proliferation and enhanced neurogenesis in both Rbpj- and Nfia/b/x/Rbpj-deficient adult MG.
(A) A schematic of Cre-inducible control and Yap5SA FLEX constructs used in the study. (B) Schematic of experimental pipeline. (C to E) Representative immunostaining of GFP, mCherry, and EdU on retinal sections from control, Rbpj cKO, and Nfia/b/x;Rbpj cKO with mCherry control and Yap5SA AAVs. White arrowheads indicate co-labeled GFP-positive and EdU-positive cells. (F) Quantification of AAV transduction efficiency in MG. (G) Quantification of MG co-labeled with EdU. (H) Representative immunostaining of EdU-positive MG-derived neurons expressing BC marker Otx2. Yellow arrowheads indicate EdU/GFP/Otx2 triple-positive cells. (I) Quantification of mean percentage ± SD of EdU-positive MG-derived neurons expressing neuronal markers: Otx2 and HuC/D. Significance was determined via two-way ANOVA with Tukey’s test: *P < 0.05, ***P < 0.001, ****P < 0.0001. Each data point was calculated from an individual retina. Scale bars, 50 μm.
We observed selective MG-specific expression of mCherry in both constructs, as evidenced by colocalization of Sun1-GFP and mCherry (Fig. 6, C to E). Little to no EdU/GFP-positive cells were detected in retinas injected with AAV-flex-mCherry, although induction of mCherry is highly efficient (Fig. 6, C to F). However, in retinas injected with AAV-flex-Yap5SA-P2A-mCherry, we observed a significant fraction of GFP/EdU-positive cells across all three sample groups (Fig. 6, C to E and G; 9.5% of control, 13.4% of Rbpj-deficient, and 14.6% of Nfia/b/x;Rbpj-deficient MG co-labeled with EdU). While some mCherry expression colocalized with Sun1-GFP–positive cells in the ganglion cell layer, immunostaining for GFP, mCherry, EdU, and Pax2 (astrocyte marker) revealed no GFP/Pax2-positive cells co-labeled with EdU nor mCherry, indicating that the proliferating cells are MG, not astrocytes (fig. S8, B to D). We also observed that a significant fraction of EdU/GFP+ cells exhibited faint or no mCherry fluorescent signal. This suggests that YAP5SA overexpression induces MG to undergo multiple cell divisions, subsequently leading to the dilution of AAV mCherry reporter (Fig. 6, C to E). In Nfia/b/x;Rbpj cKO retinas, which had significant MG depletion (Fig. 4C), we detected an increase in the number of Sox9/GFP+ cells and Sox2/GFP+ cells with YAP5SA overexpression (fig. S9, A and B).
To determine whether YAP5SA-induced proliferating MG cells give rise to neurons, we performed co-immunostaining with the neuronal markers Otx2 and HuC/D. No neurons derived from MG were observed in the control retinas. While the relative fraction of Otx2/GFP-positive cells was similar among retinas infected with AAV-flex-Yap5SA-P2A-mCherry and AAV-flex-mCherry, we observed that 27.1 and 18.4% of MG-derived cells expressing Otx2 were co-labeled with EdU in Rbpj- and Nfia/b/x;Rbpj-deficient retinas, respectively (Fig. 6, H and I, and fig. S9C). This indicates that they were derived from proliferating MG. In addition, co-immunostaining for Ki67 and Otx2 or HuC/D in YAP5SA-overexpressed Nfia/b/x;Rbpj-deficient retinas revealed no colocalization of Ki67 with either neuronal marker. This suggests that these MG-derived neuron-like cells are not actively proliferating (fig. S9, D and E). We did not detect any HuC/D/GFP-positive cells co-labeled with EdU in any of the three genotypes (Fig. 6I). This observation suggests that proliferating MG induced by YAP5SA overexpression are, for reasons that remain unclear, prevented from differentiating into ACs. Overall, our data revealed that AAV-mediated overexpression of YAP5SA can be used to effectively replenish MG that were depleted due to high levels of non-proliferative transdifferentiation into neuron-like cells that results from the loss of Nfia/b/x and Rbpj.
DISCUSSION
In this study, we identified Rbpj and Notch signaling as critical negative regulators of neurogenic competence in mature mammalian MG. Using genetic lineage analysis, IHC, and scRNA-seq and ATAC-seq, we unambiguously demonstrate that both Rbpj and Notch1/2-deficient MG cells generate retinal neuron-like cells in adult mouse retinas. We show that loss of function of Rbpj induces direct transdifferentiation of MG into bipolar- and amacrine-like cells in the absence of injury and that retinal injury further enhances this effect. We also found that loss of function of Notch1/2 phenocopies Rbpj loss of function. CUT&Tag analysis revealed that Rbpj regulates expression of both Notch pathway components and mature MG-specific genes in MG. Furthermore, we report that combined deletion of Rbpj and Nfia/b/x factors results in robust levels of MG-derived neurogenesis, indicating that Nfia/b/x and Rbpj act in parallel pathways to independently inhibit neurogenesis in mature MG. We also conducted comprehensive multiomic analyses to characterize neurogenic glia and glia-derived neuron-like cells. Last, we show that AAV-mediated expression of constitutively active Yap strongly induces proliferation and neurogenesis in both Rbpj-deficient and Nfia/b/x;Rbpj-deficient MG. It is unclear the exact extent to which these induced bipolar and amacrine neurons resemble their endogenous counterparts. However, our findings confirm that neurogenic competence is actively repressed in mammalian MG and demonstrate that disrupting these negative regulators has the potential for developing regenerative therapies designed to treat retinal dystrophies. The near-complete induction of neurogenesis seen in Nfia/b/x;Rbpj-deficient MG also demonstrates that there is no clear intrinsic barrier to glial reprogramming in retina, further strengthening the feasibility of reprogramming as a viable therapeutic strategy for retinal degeneration.
Our findings align with previous studies in zebrafish indicating that Notch signaling maintains MG quiescence and is transiently down-regulated following injury (6, 17, 19, 20). However, while chemical inhibition of Notch signaling is sufficient to induce MG reprogramming in zebrafish (14, 16), it is not effective in mice (16). Although DAPT-mediated Notch inhibition enhanced neurogenesis in Nfia/b/x-deficient MG, this effect was modest compared to the robust effect observed in the Nfia/b/x;Rbpj conditional mutant. This discrepancy may be attributed to difficulties in efficiently inhibiting Notch signaling using small molecules and the fact that Nfia/b/x and possibly other factors act in a redundant, parallel, manner to repress neurogenic competence in mammalian MG. Notably, loss of function of both Rbpj and Notch1/2 induces robust levels of MG-derived neurogenesis in the absence of acute injury, in sharp contrast to Nfia/b/x loss of function and also Ascl1 overexpression, where injury is essential for inducing neurogenesis (5, 6). To date, only simultaneous overexpression of either Ascl1 and Atoh1, or ikzf1 and Ikzf4 has been reported to induce injury-independent MG-derived neurogenesis (7, 53). Injury-induced transition to a reactive state is essential for inducing neurogenic competence in zebrafish by down-regulating genes that maintain Müller glial quiescence (6, 54), and our findings imply that inhibition of Notch signaling is an essential component of this process.
During retinal development, dynamic Notch signaling regulates the balance between retinal progenitor maintenance and the differentiation of retinal neurons and MG (9, 12, 55), with Notch pathway inhibition resulting in precocious differentiation of retinal progenitors. Here, we find that inhibition of the Notch pathway leads to a direct conversion of MG into retinal interneurons in the absence of detectable proliferation, which contrasts with the limited induction of proliferation seen in Nfia/b/x-deficient MG (6). This can be reversed by viral misexpression of dominant-active Yap5SA, which induces robust glial proliferation in all genotypes examined and rescues the loss of MG seen in Nfia/b/x;Rbpj conditional mutants. Previous studies that have used constitutive transgenic models to overexpress Yap5SA in MG do not observe tumor formation or defects in retinal lamination (56), implying that excess glia may simply undergo apoptosis and that potential safety concerns may be overestimated. Moreover, any glial proliferation induced by non-replicative AAV vectors will result in progressive dilution with each round of division, further limiting these risks. However, a full evaluation of the long-term consequences of sustained Yap5SA overexpression in MG remains to be evaluated, as do the effects of sustained Yap activity on differentiation of MG-derived neurons.
While both genetic and chemical inhibition of Notch signaling strongly induce photoreceptor generation in the developing retina (10, 57–59), we do not detect substantial levels of MG-derived photoreceptor generation in adult Rbpj cKO or Notch1/2 cKO mice. However, we do observe limited numbers of Nrl- and Recoverin-positive photoreceptor-like cells in the ONL of Rbpj/Nfia/b/x-deficient retinas. This relative lack of MG-derived photoreceptors is consistent with previous findings from overexpression of Ascl1 alone or in combination with other TFs, as well as from Nfia/b/x-deficient MG (5–8). This is unexpected, given that over 80% of neurons in the mouse retina are rod photoreceptors and comprise the overwhelming majority of late-born cells. It implies that photoreceptor generation is actively repressed in neurogenic MG, but the underlying reasons remain unclear. It is likely that a combination of additional treatments that can both promote photoreceptor specification and inhibit these negative regulators will be needed for the development of successful regenerative therapies for photoreceptor dystrophies.
Our findings highlight the importance of targeting active negative regulators of neurogenic competence for effective induction of glia-to-neuron reprogramming. Induced pluripotent stem cells clearly demonstrated the power of TF overexpression as an effective tool for cell conversion (60), and most studies aimed at inducing glia-to-neuron conversion in retina (5, 7, 8) and brain (61, 62) have used similar gain of function approaches. Although powerful, high and persistent expression of TFs that promote cell-type specification can potentially inhibit terminal differentiation of converted neurons, or generate unwanted neuronal subtypes. This could potentially be avoided by instead blocking endogenous inhibitors of neurogenesis. Identifying effective methods of doing so, however, has proven challenging. Overexpression of microRNAs that block expression of gliogenic factors can induce fibroblast to neuron conversion in vitro (63–65) and can enhance the efficiency of glia-to-neuron reprogramming in both brain and retina, but this approach requires overexpression of other TFs to induce neurogenesis in vivo (66–69). Claims of efficient glia-to-neuron conversion in brain and retina by knockdown of the glial-enriched RNA binding protein Ptbp1 have not been replicated (22, 70–76) and instead likely reflect technical artifacts resulting from use of AAV-based minipromoter constructs (77, 78). Nevertheless, our utilization of rigorous cell lineage analysis and multiomic analysis to show that combined Rbpj and Nfia/b/x loss of function leads to conversion of over 70% of retinal MG into neurons demonstrates the promise of targeting negative regulatory factors for effective in vivo glia-to-neuron conversion. Because these genes are also expressed in many different glial cell types in the brain, this strategy may hold broader potential for developing cell-based therapies for treating neurodegenerative disorders.
Several important questions remain unanswered in this study, however. First, it is unclear whether the induced bipolar and amacrine neurons from Rbpj- or Nfia/b/x;Rbpj-deficient MG are capable of integrating into endogenous retinal circuitry. Second, while their gene expression pattern and morphology closely resemble those of BCs and ACs, further physiological analysis is essential to fully characterize these neurons. Last, it remains to be determined whether MG, in which neurogenic competence has been restored by a combination of Notch inhibition and Nfia/b/x loss of function, can be guided by additional fate-determining factors to efficiently generate functional photoreceptors or retinal ganglion cells that are lost in blinding diseases.
MATERIALS AND METHODS
Mice
Mice were raised and housed in a climate-controlled pathogen-free facility on a 14-hour/10-hour light/dark cycle. Mice used in this study were GlastCreER;Sun1-GFP, which were generated by crossing the GlastCreER and Sun1-GFP lines developed by J. Nathans at Johns Hopkins (31, 79) and were obtained from his group. GlastCreER;Rbpjlox/lox;Sun1-GFP mice were generated by crossing GlastCreER;Sun1GFP with conditional Rbpjlox/lox mice (Jackson Laboratory, no. 034200).
GlastCreER;Notch1lox/lox;Notch2lox/lox;Sun1-GFP mice were obtained by crossing GlastCreER;Notch1lox/lox;Sun1-GFP and GlastCreER;Notch2lox/lox;Sun1-GFP mice. GlastCreER;Nfia/b/xlox/lox;Rbpjlox/lox;Sun1-GFP quadruple cKO mice were generated by crossing GlastCreER;Nfia/b/xlox/lox;Sun1-GFP and GlastCreER;Rbpjlox/lox;Sun1-GFP mice. Maintenance and experimental procedures performed on mice were in accordance with the protocol approved by the Institutional Animal Care and Use Committee at the Johns Hopkins School of Medicine.
Intraperitoneal TAM injection
To induce Cre recombination, animals at ~3 weeks of age were intraperitoneally injected with TAM (Sigma-Aldrich, no. H6278-50mg) in corn oil (Sigma-Aldrich, no. C8267-500ML) at 1.5 mg per dose for five consecutive days.
NMDA treatment
Adult mice were anesthetized with isoflurane inhalation. Two microliters of 100 mM NMDA in phosphate-buffered saline (PBS) were intravitreally injected using a microsyringe with a 33-gauge blunt-ended needle.
EdU treatment
At 1 week after TAM induction, mice were administered EdU (Abcam, no. ab146186) via intraperitoneal injections [150 μl of PBS (5 mg/ml)] and drinking water (0.3 mg/ml) for seven consecutive days.
DAPT treatment
Adult GlastCreER;Nfia/b/xlox/lox; Sun1-GFP mice were anesthetized with isoflurane inhalation and intravitreally injected with 200 μM (2 μl per injection) of DAPT (MedChemExpress, no. HY-13027) in dimethyl sulfoxide using a microsyringe with a 33-gauge blunt-ended needle.
Cloning and production of AAV
The Addgene (no. 50462) construct containing an EF1a promoter was replaced with a smCBA promoter. The P2A ribosomal self-cleaving peptide is used to simultaneously express Yap5SA 5′ to the mCherry reporter as a single polypeptide, which is then cleaved to generate the TF and mCherry. The coding sequence of Yap5SA was synthesized by GeneWiz. AAV constructs were packaged into AAV2.7m8 by Boston Children’s Hospital Viral Core.
FLEX-AAV delivery
GlastCreER;Sun1-GFP and GlastCreER;Rbpjlox/lox;Sun1-GFP mice (~3 weeks old) were anesthetized with isoflurane inhalation and intravitreally injected with AAV-FLEX constructs using a microsyringe with a 33-gauge blunt-ended needle. One day following AAV transduction, five daily doses of TAM (1.5 mg per dose in corn oil) were administered to activate Cre recombinase. Titer and injection volume for each construct are listed in table S4.
IHC and imaging
Collection and immunohistochemical analysis of retinas were performed as described previously (6). Briefly, mouse eye globes were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, no. 15710) for 4 hours at 4°C. Retinas were dissected in 1× PBS and incubated in 30% sucrose overnight at 4°C. Retinas were then embedded in frozen section medium (VWR, no. 95057-838), cryosectioned at 16-μm thickness, and stored at −20°C. Sections were dried for 30 min in a 37°C incubator and washed three times for 5 min with 0.1% Triton X-100 in PBS (PBST). EdU labeling was performed by using a Click-iT EdU kit (Thermo Fisher Scientific, nos. C10340 and C10636) following the manufacturer’s instructions. Sections were then incubated in 10% horse serum (Thermo Fisher Scientific, no. 26050070), 0.4% Triton X-100 in 1× PBS (blocking buffer) for 2 hours at room temperature (RT) and then incubated with primary antibodies in the blocking buffer overnight at 4°C. Primary antibodies used are listed in table S5.
Sections were washed four times for 5 min with PBST to remove excess primary antibodies and were incubated in secondary antibodies in blocking buffer for 2 hours at RT. Sections were then counterstained with 4′,6-diamidino-2-phenylindole in PBST, washed four times for 5 min in PBST and mounted with ProLong Gold Antifade Mountant (Invitrogen, no. P36935) under coverslips (VWR, no. 48404-453), air-dried, and stored at 4°C. Fluorescent images were captured using a Zeiss LSM 700 confocal microscope. Secondary antibodies used are listed in table S5.
Cell quantification and statistical analysis
Otx2/GFP-positive and HuC/D/GFP-positive cells were counted and divided by the total number of GFP-positive cells from a single random whole section per retina. For cell proliferation quantification, EdU/GFP-positive cells were counted and divided by the total number of GFP-positive cells from a single random whole section per retina. EdU-positive MG-derived neurons were quantified by the number of EdU/GFP/Otx2-positive cells divided by the total GFP/Otx2-positive cells. AAV infection efficiency was calculated by the number of mCherry/GFP-positive cells divided by the total number of GFP-positive cells. Each data point in the bar graphs was calculated from an individual retina. All cell quantification data were graphed and analyzed using GraphPad Prism 10. Two-way analysis of variance (ANOVA) were used for analysis between three or more samples of multiple groups. All results are presented as means ± SD.
Retinal cell dissociation
Retinas were dissected in fresh ice-cold PBS, and retinal cells were dissociated using an optimized protocol as previously described (80). Each sample contains a minimum of four retinas from four animals of both sex. Dissociated cells were resuspended in ice-cold Hibernate Buffer A GlutamaxBuffer containing Hibernate A (BrainBits, no. HALF500), B-27 supplement (Thermo Fisher Scientific, no. 17504044), and GlutaMAX (Thermo Fisher Scientific, no. 35050061).
CUT&Tag library preparation
CUT&Tag was performed using the CUTANA CUT&Tag kit following the manufacturer’s instructions. Adult GlastCreER;Sun1-GFP mice were induced with TAM injection 1 week before retinal dissociation for CUT&Tag experiment. Nuclei were isolated from ~100,000 FACS-isolated GFP-positive cells in Nuclear Extraction Buffer for 10 min on ice. Primary antibodies used were rabbit anti–immunoglobulin G (IgG; negative control), rabbit anti-H3K27me3 (positive control), and rabbit anti-Rbpj. Libraries were sequenced on NextSeq Mid150 with 130 million reads per library.
CUT&Tag data analysis
To analyze the CUT&Tag data, Fastq files were trimmed for adapters by Cutadapt and mapped to the mm10 genome by Bowtie2 (81) using the parameters: --local --very-sensitive -no-mixed --no-discordant -I 10 -X 700. Bam files were generated and multimapping reads were removed by SAMtools (82), and duplicate reads were removed by Picard. Pileups were generated as bigwig files using deepTools (83) function “bamCoverage” with count-per-million normalization and visualized in IGV. Replicates from each biological group, i.e., IgG, H3K4me3, and Rpbj, were merged before peak calling using SAMtools. MACS2 (84) was used to call peaks following a standard procedure. The heatmaps showing peak distribution for each merged dataset of IgG, H3K4me3, and Rbpj were generated using the deepTools function “plotHeatmap.” Motif-enrichment analysis was performed using MEME function “Enriched motif” (85); only the top 5000 peaks ranked by the P-adjusted values on the merged data for each group were used for this analysis.
scRNA-seq library preparation
scRNA-seq was prepared on dissociated retinal cells using the 10x Genomics Chromium Single Cell 3′ Reagents Kit v3.1 (10x Genomics, Pleasanton, CA). Libraries were constructed following the manufacturer’s instructions and were sequenced using Illumina NextSeq. Sequencing data were processed through the Cell Ranger 7.0.1 pipeline (10x Genomics) using default parameters.
scATAC-seq library preparation
scATAC-seq were prepared on FACS-isolated cells using the 10x Genomics Chromium NextGEM SingleCell ATAC Reagent Kits v1.1 (10x Genomics, Pleasanton, CA). Briefly, cells were spun down at 500g for 5 min and resuspended in 100 μl of ice-cold 0.1× Lysis Buffer and lysed by pipette-mixing four times and incubated on ice for 4 min total. Cells were washed with 0.5 ml of ice-cold Wash Buffer and spun down at 500g for 5 min at 4°C. Nuclei pellets were resuspended in 10 to 15 μl of Nuclei Buffer and counted using Trypan blue. Resuspended cell nuclei (10,000 to 15,000) were used for transposition and loaded into the 10x Genomics Chromium Single Cell system. ATAC libraries were amplified with 10 polymerase chain reaction (PCR) cycles and were sequenced on Illumina NovaSeq with ~200 million reads per library. Sequencing data were processed through the Cell Ranger ATAC 1.1.0 pipeline (10x Genomics) using default parameters.
Single-cell Multiome ATAC and GEX sequencing library preparation
scATAC-seq and scRNA-seq were prepared on FACS-isolated GFP-positive cells using the 10X Genomic Chromium Next GEM Single Cell Multiome ATAC and Gene Expression kit following the manufacturer’s instructions. Briefly, cells were spun down at 500g for 5 min and resuspended in 100 μl of ice-cold 0.1× Lysis Buffer and lysed by pipette-mixing four times and incubated on ice for 4 min total. Cells were washed with 0.5 ml of ice-cold Wash Buffer and spun down at 500g for 5 min at 4°C. Nuclei pellets were resuspended in 10 to 15 μl of Nuclei Buffer and counted using Trypan blue. Resuspended cell nuclei (10,000 to 15,000) were used for transposition and loaded into the 10x Genomics Chromium Single Cell system. ATAC libraries were amplified with 10 PCR cycles and were sequenced on Illumina NovaSeq with ~200 million reads per library. RNA libraries were amplified from cDNA with 14 PCR cycles and were sequenced on Illumina NovaSeq 6000.
scRNA-seq data analysis
scRNA-seq of the control and Rbpj cKO mice was analyzed using the Seurat package(86). Briefly, the gene expression data were jointly normalized by function “NormalizeData,” the principal components analysis dimensionality reduction, Louvain clustering, and UMAP visualization were performed on the top 30 principal components. MG cells then were subset from other cell types by the function “subset,” and dimensionality reduction and visualization were performed a second time on the top 30 principal components. Expression of key marker genes within each cell cluster was used to confirm the appropriate assignment of cell types.
scATAC-seq data analysis
scATAC-seq of the control and Rbpj KO mice were analyzed using the Signac package (87). Briefly, MG were subset from other cell types, dimensionality reduction was performed using functions “FindClusters” and “FindNeighbors.” UMAP visualization was performed on the top 30 principal components. Enhanced accessibility of marker gene bodies and enrichment of marker TF motifs within each MG cell were used to confirm the appropriate assignments. The scATAC-seq peak calling was performed using MACS2, and the DARs were obtained using function “FindMarker” comparing between the control and Rbpj cKO groups. The high-confidence peaks were used for chromatin accessibility heatmap, and depth-normalized pileups from MACS2 were used for genome-browser visualization in IGV (88). The scATAC-seq motif enrichment was performed using the function “chromVAR.” The footprinting information for sets of motifs was obtained and visualized using the Signac functions “Footprint” and “PlotFootprint,” respectively.
Multiomic data analysis
For the data processing, raw scRNA-seq and scATAC-seq data were processed with the Cell Ranger software for formatting reads, demultiplexing samples, genomic alignment, and generating the cell-by-gene count matrix. The cell-by-gene count matrix is the final output from the Cell Ranger pipeline and was used for all downstream analysis. Then, Seurat (86) and Signac (87) packages were used to create Seurat objects for each sample with the cell-by-gene count matrix with the function “CreateSeuratObject.” After visual checking the violin plot of the total counts for each cell, cells with nCount_ATAC > 100,000 and nCount_RNA > 30,000, nCount_ATAC < 1000 and nCount_RNA < 1000, nFeature_RNA < 500, nucleosome_signal > 1.2, and TSS.enrichment < 2 were filtered out. In addition, cells with a mitochondrial fraction of >15% were also removed.
For the data analysis, the weighted nearest neighbor method was used to jointly analyze a single-cell dataset measuring both DNA accessibility and gene expression in the same cells using Signac and Seurat function “FindMultiModalNeighbors” (89). The “SCTransform” and “RunTFIDF” functions were used to normalize the data for the scRNA-seq and scATAC-seq analysis, respectively. The two-dimensional UMAP was generated using the first 30 dimensions for both the scRNA-seq and scATAC-seq. Cluster-based differential gene expression and DARs between cell types were performed using the Seurat and Signac function “FindAllMarkers.” The average gene expression and peak of each cell type were obtained using the Seurat and Signac function “AverageExpression.” The TF motif activity per cell was performed using Signac function “RunChromVAR.” The average gene expression, average peak, and average motif score were visualized in the heatmaps using Seurat and Signac function “Doheatmap.”
GO analysis
To obtain the significantly enriched GO terms (biological process), the Database for Annotation Visualization and Integrated Discovery (90) was applied under the cutoff of P < 0.01.
Acknowledgments
We thank P. Lyu, L. Orzolek, T. Creamer, and members of the Hopkins Single-Cell and Transcriptomic Core for technical assistance. We thank A. Fischer, J. Nathans, and A. Kolodkin, and members of the Blackshaw lab for comments on this manuscript.
Funding: This work was supported by an award from the National Eye Institute (R01EY031685) to S.B., a Stein Innovation Award from Research to Prevent Blindness to S.B., and a Young Investigator grant from Alcon Research Institute to T.H.
Author contributions: Conceptualization: N.L., S.B., and T.H. Methodology: N.L., S.B., and T.H. Investigation: N.L., I.P., Y.K., S.B., and T.H. Validation: N.L. Formal Analysis: T.H., N.L., T-D.V., and R.P. Supervision: S.B. and T.H. Funding acquisition: S.B. and T.H. Writing—original draft: N.L., S.B., and T.H. Writing—review and editing: N.L., T-D.V, S.B., and T.H.
Competing interests: S.B. is a cofounder and shareholder in CDI Labs LLC and has received financial support from Genentech. S.B. and T.H. are inventors on patent application PCT/US2023/071472 submitted by Johns Hopkins University that covers the use of the findings described here in reprogramming retinal glia into neurons for cell replacement therapy. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All scRNA-seq, snRNA-seq, and scATAC-seq data described in this study are available at Gene Expression Omnibus under accession number GSE246574.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S4 to S5
Legends for tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Wan J., Goldman D., Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 40, 41–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahne M., Nagashima M., Hyde D. R., Hitchcock P. F., Reprogramming Müller glia to regenerate retinal neurons. Annu. Rev. Vis. Sci. 6, 171–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorsuch R. A., Lahne M., Yarka C. E., Petravick M. E., Li J., Hyde D. R., Sox2 regulates Müller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a. Exp. Eye Res. 161, 174–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fausett B. V., Gumerson J. D., Goldman D., The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J. Neurosci. 28, 1109–1117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorstad N. L., Wilken M. S., Grimes W. N., Wohl S. G., VandenBosch L. S., Yoshimatsu T., Wong R. O., Rieke F., Reh T. A., Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature 548, 103–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoang T., Wang J., Boyd P., Wang F., Santiago C., Jiang L., Yoo S., Lahne M., Todd L. J., Jia M., Saez C., Keuthan C., Palazzo I., Squires N., Campbell W. A., Rajaii F., Parayil T., Trinh V., Kim D. W., Wang G., Campbell L. J., Ash J., Fischer A. J., Hyde D. R., Qian J., Blackshaw S., Gene regulatory networks controlling vertebrate retinal regeneration. Science 370, eabb8598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd L., Hooper M. J., Haugan A. K., Finkbeiner C., Jorstad N., Radulovich N., Wong C. K., Donaldson P. C., Jenkins W., Chen Q., Rieke F., Reh T. A., Efficient stimulation of retinal regeneration from Müller glia in adult mice using combinations of proneural bHLH transcription factors. Cell Rep. 37, 109857 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd L., Jenkins W., Finkbeiner C., Hooper M. J., Donaldson P. C., Pavlou M., Wohlschlegel J., Ingram N., Rieke F., Reh T. A., Mu X., Reprogramming Müller glia to regenerate ganglion-like cells in adult mouse retina with developmental transcription factors. Sci. Adv. 8, eabq7219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jadhav A. P., Cho S.-H., Cepko C. L., Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc. Natl. Acad. Sci. U.S.A. 103, 18998–19003 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jadhav A. P., Mason H. A., Cepko C. L., Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–923 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Mills E. A., Goldman D., The regulation of notch signaling in retinal development and regeneration. Curr. Pathobiol. Rep. 5, 323–331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson B. R., Hartman B. H., Georgi S. A., Lan M. S., Reh T. A., Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 304, 479–498 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer K. A., Riesenberg A. N., Brown N. L., Notch signaling differentially regulates Atoh7 and Neurog2 in the distal mouse retina. Development 141, 3243–3254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M.-S., Wan J., Goldman D., Tgfb3 collaborates with PP2A and notch signaling pathways to inhibit retina regeneration. eLife 9, e55137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan J., Goldman D., Opposing actions of Fgf8a on notch signaling distinguish two muller glial cell populations that contribute to retina growth and regeneration. Cell Rep. 19, 849–862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsaeidi F., Macpherson P., Mills E. A., Jui J., Flannery J. G., Goldman D., Notch suppression collaborates with Ascl1 and Lin28 to unleash a regenerative response in fish retina, but not in mice. J. Neurosci. 38, 2246–2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahu A., Devi S., Jui J., Goldman D., Notch signaling via Hey1 and Id2b regulates Müller glia’s regenerative response to retinal injury. Glia 69, 2882–2898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell L. J., Hobgood J. S., Jia M., Boyd P., Hipp R. I., Hyde D. R., Notch3 and DeltaB maintain Müller glia quiescence and act as negative regulators of regeneration in the light-damaged zebrafish retina. Glia 69, 546–566 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Fogerty J., Song P., Boyd P., Grabinski S. E., Hoang T., Reich A., Cianciolo L. T., Blackshaw S., Mumm J. S., Hyde D. R., Perkins B. D., Notch inhibition promotes regeneration and immunosuppression supports cone survival in a zebrafish model of inherited retinal dystrophy. J. Neurosci. 42, 5144–5158 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell L. J., Levendusky J. L., Steines S. A., Hyde D. R., Retinal regeneration requires dynamic Notch signaling. Neural Regen. Res. 17, 1199–1209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y., Shiau F., Yi W., Lu S., Wu Q., Pearson J. D., Kallman A., Zhong S., Hoang T., Zuo Z., Zhao F., Zhang M., Tsai N., Zhuo Y., He S., Zhang J., Stein-O’Brien G. L., Sherman T. D., Duan X., Fertig E. J., Goff L. A., Zack D. J., Handa J. T., Xue T., Bremner R., Blackshaw S., Wang X., Clark B. S., Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell 53, 473–491.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang T., Kim D. W., Appel H., Pannullo N. A., Leavey P., Ozawa M., Zheng S., Yu M., Peachey N. S., Blackshaw S., Genetic loss of function of Ptbp1 does not induce glia-to-neuron conversion in retina. Cell Rep. 39, 110849 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu P., Iribarne M., Serjanov D., Zhai Y., Hoang T., Campbell L. J., Boyd P., Palazzo I., Nagashima M., Silva N. J., Others, common and divergent gene regulatory networks control injury-induced and developmental neurogenesis in zebrafish retina. Nat. Commun. 14, 8477 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riesenberg A. N., Liu Z., Kopan R., Brown N. L., Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J. Neurosci. 29, 12865–12877 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. J. Collins, “Structure-function analysis of the Notch signaling CSL-KyoT2 and SPOC-NCoR corepressor complexes: Understanding how corepressor assembly is regulated at Notch target genes,” thesis, University of Cincinnati, Cincinnati, OH (2014). [Google Scholar]
- 26.Siebel C., Lendahl U., Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Magnusson J. P., Göritz C., Tatarishvili J., Dias D. O., Smith E. M. K., Lindvall O., Kokaia Z., Frisén J., A latent neurogenic program in astrocytes regulated by Notch signaling in the mouse. Science 346, 237–241 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Zamboni M., Llorens-Bobadilla E., Magnusson J. P., Frisén J., A widespread neurogenic potential of neocortical astrocytes is induced by injury. Cell Stem Cell 27, 605–617.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlén M., Meletis K., Göritz C., Darsalia V., Evergren E., Tanigaki K., Amendola M., Barnabé-Heider F., Yeung M. S. Y., Naldini L., Honjo T., Kokaia Z., Shupliakov O., Cassidy R. M., Lindvall O., Frisén J., Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci. 12, 259–267 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Han H., Tanigaki K., Yamamoto N., Kuroda K., Yoshimoto M., Nakahata T., Ikuta K., Honjo T., Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002). [DOI] [PubMed] [Google Scholar]
- 31.de Melo J., Miki K., Rattner A., Smallwood P., Zibetti C., Hirokawa K., Monuki E. S., Campochiaro P. A., Blackshaw S., Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc. Natl. Acad. Sci. U.S.A. 109, 4657–4662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedrich T., Ferrante F., Pioger L., Nist A., Stiewe T., Andrau J.-C., Bartkuhn M., Giaimo B. D., Borggrefe T., Notch-dependent and -independent functions of transcription factor RBPJ. Nucleic Acids Res. 50, 7925–7937 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz-Trelles R., Scimia M. C., Bushway P., Tran D., Monosov A., Monosov E., Peterson K., Rentschler S., Cabrales P., Ruiz-Lozano P., Mercola M., Notch-independent RBPJ controls angiogenesis in the adult heart. Nat. Commun. 7, 12088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hori K., Cholewa-Waclaw J., Nakada Y., Glasgow S. M., Masui T., Henke R. M., Wildner H., Martarelli B., Beres T. M., Epstein J. A., Magnuson M. A., Macdonald R. J., Birchmeier C., Johnson J. E., A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 22, 166–178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turkoz M., Townsend R. R., Kopan R., The notch intracellular domain has an RBPj-independent role during mouse hair follicular development. J. Invest. Dermatol. 136, 1106–1115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinbuck M. P., Winandy S., A review of notch processing with new insights into ligand-independent notch signaling in T-cells. Front. Immunol. 9, 1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X., Klein R., Tian X., Cheng H.-T., Kopan R., Shen J., Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81–94 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Pan Y., Lin M.-H., Tian X., Cheng H.-T., Gridley T., Shen J., Kopan R., γ-Secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell 7, 731–743 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Bosze B., Suarez-Navarro J., Cajias I., Brzezinski Iv J. A., Brown N. L., Notch pathway mutants do not equivalently perturb mouse embryonic retinal development. PLOS Genet. 19, e1010928 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wall D. S., Mears A. J., McNeill B., Mazerolle C., Thurig S., Wang Y., Kageyama R., Wallace V. A., Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184, 101–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyu P., Hoang T., Santiago C. P., Thomas E. D., Timms A. E., Appel H., Gimmen M., Le N., Jiang L., Kim D. W., Chen S., Espinoza D. F., Telger A. E., Weir K., Clark B. S., Cherry T. J., Qian J., Blackshaw S., Gene regulatory networks controlling temporal patterning, neurogenesis, and cell-fate specification in mammalian retina. Cell Rep. 37, 109994 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Melo J., Zibetti C., Clark B. S., Hwang W., Miranda-Angulo A. L., Qian J., Blackshaw S., Lhx2 is an essential factor for retinal gliogenesis and notch signaling. J. Neurosci. 36, 2391–2405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen W., Fruttiger M., Zhu L., Chung S. H., Barnett N. L., Kirk J. K., Lee S., Coorey N. J., Killingsworth M., Sherman L. S., Gillies M. C., Conditional Müllercell ablation causes independent neuronal and vascular pathologies in a novel transgenic model. J. Neurosci. 32, 15715–15727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byrne L. C., Khalid F., Lee T., Zin E. A., Greenberg K. P., Visel M., Schaffer D. V., Flannery J. G., AAV-mediated, optogenetic ablation of Müller Glia leads to structural and functional changes in the mouse retina. PLOS ONE 8, e76075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann B., Sterling J., Song Y., Song D., Fruttiger M., Gillies M., Shen W., Dunaief J. L., Conditional Müller cell ablation leads to retinal iron accumulation. Invest. Ophthalmol. Vis. Sci. 58, 4223–4234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K., Omori Y., Onishi A., Sato S., Kondo M., Furukawa T., Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci. 30, 6515–6526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brzezinski J. A. IV, Lamba D. A., Reh T. A., Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furukawa T., Morrow E. M., Li T., Davis F. C., Cepko C. L., Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 23, 466–470 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Clark B. S., Stein-O’Brien G. L., Shiau F., Cannon G. H., Davis-Marcisak E., Sherman T., Santiago C. P., Hoang T. V., Rajaii F., James-Esposito R. E., Gronostajski R. M., Fertig E. J., Goff L. A., Blackshaw S., Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lourenço R., Brandão A. S., Borbinha J., Gorgulho R., Jacinto A., Yap regulates müller glia reprogramming in damaged zebrafish retinas. Front. Cell Dev. Biol. 9, 667796 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masson C., García-García D., Bitard J., Grellier É.-K., Roger J. E., Perron M., Yap haploinsufficiency leads to Müller cell dysfunction and late-onset cone dystrophy. Cell Death Dis. 11, 631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamon A., García-García D., Ail D., Bitard J., Chesneau A., Dalkara D., Locker M., Roger J. E., Perron M., Linking YAP to Müller glia quiescence exit in the degenerative retina. Cell Rep. 27, 1712–1725.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Boudreau-Pinsonneault C., David L. A., Lourenço Fernandes J. A., Javed A., Fries M., Mattar P., Cayouette M., Direct neuronal reprogramming by temporal identity factors. Proc. Natl. Acad. Sci. U.S.A. 120, e2122168120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas J. L., Ranski A. H., Morgan G. W., Thummel R., Reactive gliosis in the adult zebrafish retina. Exp. Eye Res. 143, 98–109 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Mizeracka K., DeMaso C. R., Cepko C. L., Notch1 is required in newly postmitotic cells to inhibit the rod photoreceptor fate. Development 140, 3188–3197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rueda E. M., Hall B. M., Hill M. C., Swinton P. G., Tong X., Martin J. F., Poché R. A., The hippo pathway blocks mammalian retinal Müller glial cell reprogramming. Cell Rep. 27, 1637–1649.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X., Emerson M. M., Notch signaling represses cone photoreceptor formation through the regulation of retinal progenitor cell states. Sci. Rep. 11, 14525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chew S. H., Martinez C., Chirco K. R., Kandoi S., Lamba D. A., Timed notch inhibition drives photoreceptor fate specification in human retinal organoids. Invest. Ophthalmol. Vis. Sci. 63, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yaron O., Farhy C., Marquardt T., Applebury M., Ashery-Padan R., Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367–1378 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Torper O., Ottosson D. R., Pereira M., Lau S., Cardoso T., Grealish S., Parmar M., In vivo reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep. 12, 474–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y., Li B., Cananzi S., Han C., Wang L.-L., Zou Y., Fu Y.-X., Hon G. C., Zhang C.-L., A single factor elicits multilineage reprogramming of astrocytes in the adult mouse striatum. Proc. Natl. Acad. Sci. U.S.A. 119, e2107339119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Victor M. B., Richner M., Hermanstyne T. O., Ransdell J. L., Sobieski C., Deng P.-Y., Klyachko V. A., Nerbonne J. M., Yoo A. S., Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cates K., McCoy M. J., Kwon J.-S., Liu Y., Abernathy D. G., Zhang B., Liu S., Gontarz P., Kim W. K., Chen S., Kong W., Ho J. N., Burbach K. F., Gabel H. W., Morris S. A., Yoo A. S., Deconstructing stepwise fate conversion of human fibroblasts to neurons by MicroRNAs. Cell Stem Cell 28, 127–140.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R., MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wohl S. G., Reh T. A., miR-124-9-9* potentiates Ascl1-induced reprogramming of cultured Müller glia. Glia 64, 743–762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papadimitriou E., Koutsoudaki P. N., Thanou I., Karagkouni D., Karamitros T., Chroni-Tzartou D., Gaitanou M., Gkemisis C., Margariti M., Xingi E., Tzartos S. J., Hatzigeorgiou A. G., Thomaidou D., A miR-124-mediated post-transcriptional mechanism controlling the cell fate switch of astrocytes to induced neurons. Stem Cell Rep. 18, 915–935 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niu W., Zang T., Zou Y., Fang S., Smith D. K., Bachoo R., Zhang C.-L., In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.di Val Cervo P. R., Romanov R. A., Spigolon G., Masini D., Martín-Montañez E., Toledo E. M., La Manno G., Feyder M., Pifl C., Ng Y.-H., Sánchez S. P., Linnarsson S., Wernig M., Harkany T., Fisone G., Arenas E., Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 35, 444–452 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Xie Y., Zhou J., Chen B., Critical examination of Ptbp1-mediated glia-to-neuron conversion in the mouse retina. Cell Rep. 39, 110960 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoang T., Kim D. W., Appel H., Ozawa M., Zheng S., Kim J., Blackshaw S., Ptbp1 deletion does not induce astrocyte-to-neuron conversion. Nature 618, E1–E7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W., Zheng Q., Huang Q., Ma S., Li M., Repressing PTBP1 fails to convert reactive astrocytes to dopaminergic neurons in a 6-hydroxydopamine mouse model of Parkinson’s disease. eLife 11, e75636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo T., Pan X., Jiang G., Zhang D., Qi J., Shao L., Wang Z., Xu H., Zhao Y., Downregulating PTBP1 fails to convert astrocytes into hippocampal neurons and to alleviate symptoms in Alzheimer’s mouse models. J. Neurosci. 42, 7309–7317 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang G., Yan Z., Wu X., Zhang M., Xu C., Shi L., Yang H., Fang K., Ptbp1 knockdown failed to induce astrocytes to neurons in vivo. Gene Ther. 30, 801–806 (2023). [DOI] [PubMed] [Google Scholar]
- 75.Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., Xiao Q., Wang B., Wu W., Sun Y., Zhou Y., Tang C., Liu F., Wang L., Feng C., Liu M., Li S., Zhang Y., Xu H., Yao H., Shi L., Yang H., Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell 181, 590–603.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Qian H., Kang X., Hu J., Zhang D., Liang Z., Meng F., Zhang X., Xue Y., Maimon R., Dowdy S. F., Devaraj N. K., Zhou Z., Mobley W. C., Cleveland D. W., Fu X.-D., Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 582, 550–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L.-L., Serrano C., Zhong X., Ma S., Zou Y., Zhang C.-L., Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 184, 5465–5481.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le N., Appel H., Pannullo N., Hoang T., Blackshaw S., Ectopic insert-dependent neuronal expression of GFAP promoter-driven AAV constructs in adult mouse retina. Front. Cell Dev. Biol. 10, 914386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mo A., Mukamel E. A., Davis F. P., Luo C., Henry G. L., Picard S., Urich M. A., Nery J. R., Sejnowski T. J., Lister R., Eddy S. R., Ecker J. R., Nathans J., Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fadl B. R., Brodie S. A., Malasky M., Boland J. F., Kelly M. C., Kelley M. W., Boger E., Fariss R., Swaroop A., Campello L., An optimized protocol for retina single-cell RNA sequencing. Mol. Vis. 26, 705–717 (2020). [PMC free article] [PubMed] [Google Scholar]
- 81.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu T., Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 1150, 81–95 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stuart T., Srivastava A., Madad S., Lareau C. A., Satija R., Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ko K. D., Jiang K., Dell’Orso S., Sartorelli V., Integrating single-cell transcriptomes, chromatin accessibility, and multiomics analysis of mesoderm-induced embryonic stem cells. STAR Protoc. 4, 102307 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S4 to S5
Legends for tables S1 to S3
Tables S1 to S3