Abstract
Human papillomaviruses (HPV) are epitheliotropic viruses, with some types suggested to be associated with skin cancer. In this study, swab samples collected from five different sites on the skin of renal transplant recipients, dialysis patients, and age- and sex-matched healthy controls were analyzed for HPV DNA by a newly designed PCR test. Most individuals were found to have asymptomatic HPV infections; more specifically, 94% of the renal transplant patients, 82% of the dialysis patients, and 80% of the healthy controls were positive for HPV DNA. The multiplicity of the HPVs detected was astounding: 20 previously described and 30 putatively new types were identified by cloning and sequencing of 33 samples from 13 individuals. These results demonstrate that normal human skin harbors an array of papillomaviruses, most of them previously unknown.
To date, 85 different genotypes of the human papillomaviruses (HPV) have been fully characterized. In addition to being the causative agents of common skin warts, there is evidence that certain types of HPV play a role in the pathogenesis of skin cancer associated with the rare hereditary disease epidermodysplasia verruciformis (EV) (22, 27), and they are therefore designated EV-associated HPV.
Renal transplant recipients given immunosuppressive therapy for long periods of time have an increased incidence of cutaneous neoplasia (2, 10, 29). Also, more than 90% of kidney recipients develop skin warts and 40% develop skin cancer within 15 years of transplantation, a 50- to 100-fold increase compared to the general population (7), and EV-associated HPV types have been found in skin tumors from such patients (5, 12, 16, 21, 28). Interestingly, EV HPV types have also been detected in hairs plucked from normal skin of 94% of renal transplant recipients (8) and 67% of healthy controls (9), and in another study (1), 35% of biopsy specimens of normal skin obtained during cosmetic surgery were positive for HPV DNA.
In a recent report (20), we presented a PCR test that holds promise as a potent tool for exploring HPV both in skin tumors and in normal skin. In the present investigation, we used the test to study the presence of HPV at various sites on normal skin of both renal patients who were or were not on immunosuppression and matched healthy controls, and there is a serendipitous aspect to the results.
MATERIALS AND METHODS
Subjects.
We studied 52 of about 130 renal transplant recipients being followed up at the outpatient clinic of nephrology of Malmö University Hospital, Malmö, Sweden. The median time since transplantation was 5 years and 2 months, and the range was 2 months to 26 years. Also included in the study were 28 of the 67 patients being treated at the Dialysis Unit; the median length of dialysis treatment was 2 years and 5 months (range, 1 month to 14 years). All of the transplant and dialysis patients were randomly selected, and a sex- and age-matched healthy control was recruited for each of the patients. The age range of the individuals in the three groups was 21 to 80 years, with a median of 54 years. History of skin cancer as well as other forms of cancer was assessed by a questionnaire.
In addition, three healthy volunteers (a female, 26 years old; a male, 38 years old; and a male, 55 years old) were studied longitudinally, using samples that were collected daily for 1 week, thereafter once a week for 2 months, and then once a month for 5 months.
The project was approved by the Committee on Ethics of Lund University (LU-183-98).
Samples.
Samples from the renal transplant recipients, dialysis patients, and healthy controls were collected with prewetted (0.9% NaCl solution) cotton-tipped swabs (Bio Hospital, Kopparberg, Sweden) that were drawn back and forth five times over the skin within an area of 5 by 10 cm and then suspended in 1 ml of 0.9% NaCl solution. These samples were taken at five different sites: the forehead, the volar aspects of the left and right forearms, and the anterior aspects of the left and right thighs. For the longitudinal study, samples were collected from the same five sites, as well as the abdomen. Furthermore, for an environmental survey, samples were taken (with cotton-tipped swabs, as described above) from the floor and laboratory bench of the PCR setup room, and in a room used for preparation of cloned material. Samples were also collected from the floors of the kitchen, bedroom, living room, and bathroom of the apartment of one of the volunteers in the longitudinal study. All samples from renal transplant recipients, dialysis patients, and healthy controls were kept at 4°C for a maximum of 72 h before being analyzed and were then frozen at −20°C until tested. For the longitudinal study, the samples were kept frozen until tested.
PCR.
All specimens were tested without previous DNA extraction. The final volume of 25 μl of PCR solution contained 5 μl of the sample, 0.75 μM each primer (FAP59 and FAP64) (20), 0.2 mM each deoxynucleoside triphosphate (Boehringer GmbH, Mannheim, Germany), 0.2% bovine serum albumin, 0.625 U of AmpliTaq Gold DNA polymerase, GeneAmp 10× PCR buffer II, and 3.5 mM MgCl2 (Perkin-Elmer, Foster City, Calif.). Forty-five cycles of amplification were performed after denaturation for 10 min at 94°C. Each cycle consisted of 94°C for 90 s, 50°C for 90 s, and 72°C for 90 s. In each batch of tests, proteinase K-treated human embryonal lung fibroblasts (HEL) and H2O without DNA were included as negative controls. HPV 11 and HPV 20 (both clinical samples) served as positive controls. Moreover, 62 samples that were PCR negative for HPV DNA were analyzed for the presence of the human L1 sequence (13). Human cells contain more than 105 copies of the L1 sequence; thus, this sequence can be used to detect small amounts of human DNA and as an indirect marker to ensure that a sample does not contain any PCR-inhibiting substances. PCR products (5-μl aliquots) were analyzed by electrophoresis in a 2% agarose gel (SeaKem; FMC Bioproducts, Rockland, Maine) containing ethidium bromide (0.02 μg/ml).
Cloning and sequence analysis.
PCR-amplified sample DNA was cloned into the pCR-script SK(+) cloning vector (Stratagene, La Jolla, Calif.). A minimum of 4 and a maximum of 13 clones from each sample were sequenced (Big Dye Terminator cycle sequencing; Perkin-Elmer) and analyzed on a Perkin-Elmer 373A automated sequencer with both forward and reverse primers. The forward and reverse complementary sequences were aligned with MacMolly computer software (version 3.8). The relatively conserved DNA sequence of the L1 open reading frame was used for comparison of the new HPV isolates with previously established HPV types available through the BLAST server (National Center for Biotechnology Information [NCBI]) (http://www.ncbi.nlm.nih.gov/blast/blast.cgi).
An isolate is defined as a new HPV type if the sequence of its L1 gene displays less than 90% homology with the L1 genes of all types that are already known; as a subtype if it shows between 90 and 98% homology with a known HPV type; and as a type variant if it displays greater than 98% homology (Papillomavirus Nomenclature Committee, 1995) (15, 32). This nomenclature was followed to define new HPV type candidates derived from the skin samples.
Sequence alignment and phylogenetic analysis.
Phylogenetic analysis was based on multiple alignment with Clustal X (version 1.8) (24, 31) edited with Genedoc (version 2.4.000) (25). Phylip (version 3.5) (18, 19) was used for neighbor-joining and maximum likelihood analysis. These programs were obtained from the website of the University of Washington Department of Genetics (http://evolution.genetics.washington.edu/phylip/software.html).
The taxonomic system with the papillomavirus supergroups α to ɛ was created by zur Hausen (34) and also proposed by E. M. de Villiers at the taxonomy workshop of the 18th International Papillomavirus Conference in Barcelona, 2000. This system was applied to investigate relatedness between the HPV type candidates and previously established HPV types. Thus far, supergroup α is the largest of the five clades, and it includes mucosal and genital HPV types. Supergroup δ comprises ungulate papillomaviruses, and supergroup ɛ comprises a mix of animal papillomaviruses and cutaneous HPV. Most of the cutaneous HPV are found in supergroups β and γ.
The region of the L1 gene used for the phylogenetic analysis extends from nucleotide 6044 to 6480, relative to the HPV 20 sequence. The following papillomaviruses were included in the analysis: from supergroup β, HPV 5, 8, 9, 12, 14, 15, 17, 19 to 25, 36 to 38, 49, 75, 76, and 80, RTRX7, and Colobus monkey papillomavirus type 2 (CgPV 2); from supergroup γ, HPV 4, 48, 50, 60, and 65; from supergroup δ, bovine papillomavirus types 1 and 2 (BPV 1 and 2), ovine papillomavirus types 1 and 2 (OvPV 1 and 2), European elk papillomavirus (EEPV), and deer papillomavirus (DPV); from supergroup ɛ, HPV 1, 41, and 63, cottontail rabbit papillomavirus (CRPV), and canine oral papillomavirus (COPV); from supergroup α, HPV 13, 44, and 55, and pygmy chimpanzee papillomavirus type 1 (PCPV 1). Also included in the analysis were the HPV type candidates FA1 to FA42. The sequences from previously characterized HPV types were obtained from GenBank (http://www.ncbi.nlm.nih.gov/).
Statistical analysis.
The general chi-square test was used to compare the prevalence of HPV DNA on the forehead, arms, and thighs. Analysis of age-related HPV DNA prevalence was done by the chi-square test for linear trends, and comparison of difference in HPV DNA prevalence between the renal transplant recipients and the healthy control group was accomplished with the chi-square test with Yates' correction.
Nucleotide sequence accession numbers.
HPV type candidate sequences FA14 to FA43 have been submitted to GenBank with the following accession numbers: FA14, AF217656; FA15, AF217657; FA16.1, AF217658; FA16.2, AF217659; FA17, AF217660; FA18, AF217661; FA19, AF217662; FA20, AF217663; FA21, AF217664; FA22, AF217665; FA23.1, AF217666; FA23.2, AF217667; FA24, AF217668; FA25, FA217670; FA26, AF217671; FA27, AF217672; FA28, AF217673; FA29, AF217674; FA30, AF217675; FA31, AF217676; FA32, AF217677; FA33, AF217678; FA34, AF217679; FA35, AF217680; FA36, AF217681; FA37, AF217682; FA38, AF217683; FA39, AF217684; FA40, AF217685; FA41, AF217686; FA42, AF217687; and FA43, AF252606.
RESULTS
HPV DNA was detected in at least one of the samples from 94% (49 of 52) of the renal transplant recipients, 82% (23 of 28) of the dialysis patients, and 80% (64 of 80) of the healthy controls (Table 1). A history of skin cancer (basal or squamous cell carcinoma) was common in the renal transplant recipient group (11.5% [6 of 52]), whereas no cases of skin cancer had been noted in the dialysis patients or the healthy controls. Five of the six renal transplant recipients with a history of skin cancer were positive for HPV DNA.
TABLE 1.
Prevalence of HPV in skin samples from renal transplant recipients, dialysis patients, and healthy controls
| Group | HPV prevalencea
|
|||||
|---|---|---|---|---|---|---|
| Total | Left arm | Right arm | Forehead | Left thigh | Right thigh | |
| Renal transplant recipients (n = 52) | 49 (94) | 30 (58) | 30 (58) | 45 (86) | 32 (61) | 30 (58) |
| Dialysis patients (n = 28) | 23 (82) | 14 (50) | 12 (43) | 22 (79) | 16 (57) | 14 (50) |
| Healthy controls (n = 80) | 64 (80) | 38 (48) | 39 (49) | 61 (76) | 30 (38) | 35 (44) |
| All subjects (n = 160) | 136 (85) | 82 (51) | 81 (51) | 128 (80) | 78 (49) | 79 (49) |
Number (percentage) of individuals with at least one sample positive for HPV DNA.
There was a significant difference between the renal transplant recipients and the dialysis and healthy control groups with regard to the prevalence of HPV DNA (P < 0.05). Also, HPV DNA was significantly more prevalent in the forehead samples than in samples from arms and thighs (P < 0.001).
HPV type determination.
PCR products of the samples from one male and one female from each of the three groups of subjects (i.e., the transplant patients, the dialysis patients, and the healthy controls) were selected for cloning. Five of these individuals had HPV DNA-positive samples from the forehead, arms, and thighs, whereas one of the healthy controls had one HPV DNA-positive sample only, from the forehead. Five clones from each sample were DNA sequenced and analyzed (Table 2).
TABLE 2.
HPV DNA in skin samples from two renal transplant recipients, two dialysis patients, and two healthy controls, based on analysis of five clones from each sample
| Subject (age, in yrs) | HPV type or type candidatea (no. of clones) in samples from the:
|
||||
|---|---|---|---|---|---|
| Left arm | Right arm | Forehead | Left thigh | Right thigh | |
| Renal transplant recipients | |||||
| Female (40) | HPV FA12 (3) | HPV 49 (2) | HPV 49 (1) | HPV 5 (2) | HPV 49 (2) |
| HPV JC9710 (1) | HPV FA1.1 (2) | FA14 (3) | HPV FA1.1 (1) | HPV 58 (1) | |
| FA28 (1) | FA29 (1) | FA28 (1) | FA14 (1) | HPV FA1.1 (2) | |
| FA28 (1) | |||||
| Male (72) | FA16.2 (2) | HPV 8 (1) | HPV 8 (1) | FA16.2 (1) | HPV FA9 (2) |
| FA30 (1) | HPV FA5 (3) | HPV FA13 (1) | FA23.1 (1) | FA35 (2) | |
| FA31 (1) | FA32 (1) | FA30 (1) | FA30 (1) | FA41 (1) | |
| *b (1) | FA33 (1) | FA32 (2) | |||
| FA34 (1) | |||||
| Dialysis patients | |||||
| Female (35) | FA24 (5) | HPV23 (4) | HPV 17 (1) | HPV 76 (4) | HPV23 (1) |
| FA24 (1) | HPV23 (1) | HPV vs92-1 (1) | HPV 76 (1) | ||
| HPV 76 (2) | FA23.2 (2) | ||||
| FA24 (1) | FA24 (1) | ||||
| Male (71) | HPV 19 (1) | HPV 19 (1) | HPV 19 (2) | HPV 19 (1) | HPV 49 (1) |
| HPV 49 (1) | HPV 49 (1) | HPV 49 (2) | HPV 49 (2) | HPV vs73-1 (1) | |
| FA22 (3) | FA22 (2) | FA26 (1) | FA22 (1) | FA22 (2) | |
| FA25 (1) | FA27 (1) | FA27 (1) | |||
| Healthy controls | |||||
| Female (36) | FA36 (5) | ||||
| Male (72) | HPV 47 (2) | HPV 47 (4) | HPV 20 (1) | HPV 20 (1) | HPV 23 (1) |
| FA38 (1) | FA39 (1) | HPV 47 (3) | HPV 47 (3) | HPV 47 (4) | |
| FA39 (2) | FA42 (1) | FA40 (1) | |||
Boldface indicates new HPV type candidates.
This clone contained a sequence of human DNA.
The two renal transplant recipients harbored 9 and 13 different HPV types or type candidates, respectively, and the two dialysis patients carried 6 and 7 HPV types or type candidates, respectively. One new HPV type candidate (FA36) was detected from the healthy control who had only an HPV DNA-positive forehead sample, whereas seven different HPV types or type candidates were isolated from the second healthy control person.
PCR products from forehead specimens from another six individuals (again, two from each of the three groups) were also cloned and sequenced (Table 3). New clones from each of these six specimens were analyzed until at least 3 clones with one and the same HPV type or type candidate had been found; in that way, 4 to 13 clones from each sample were analyzed. Five of the six samples contained more than one HPV type or type candidate. The sample from one of the renal transplant recipients contained six different HPV types: HPV 8 and FA2.1, and the four HPV type candidates FA14 to FA17. Two different HPV candidates (FA18 and FA19) and HPV 5 and 49 were isolated from the samples from the second renal transplant recipient and a dialysis patient, whereas the sample from the second dialysis patient contained only one new HPV type candidate (FA20). Each of the forehead samples from the two healthy controls contained four different HPV types or type candidates.
TABLE 3.
HPV findings for six forehead samples
| Subject (age, in yrs) | HPV type or candidatea | No. of clones |
|---|---|---|
| Renal transplant recipients | ||
| Female (40) | HPV 8 | 1 |
| HPV FA 2.1 | 1 | |
| FA14 | 7 | |
| FA15 | 1 | |
| FA16.1 | 1 | |
| FA17 | 2 | |
| Male (56) | FA18 | 4 |
| FA19 | 1 | |
| Dialysis patients | ||
| Female (77) | FA20 | 4 |
| Male (54) | HPV 49 | 3 |
| HPV 5 | 2 | |
| Healthy controls | ||
| Female (54) | HPV 8 | 1 |
| HPV 20 | 3 | |
| HPV 38 | 1 | |
| FA21 | 1 | |
| Female (32) | HPV 23 | 6 |
| HPV 47 | 1 | |
| FA23.1 | 1 | |
| FA24 | 2 |
New clones from each sample were analyzed until at least three clones with the same HPV type or type candidate had been found. The new HPV candidates are designated FA14 to FA24 and are boldfaced.
Altogether, 33 samples from 13 individuals were subjected to HPV type determination, which revealed 20 previously described HPV types or type candidates (HPV 5, 8, 17, 19, 20, 23, 38, 47, 49, 58, and 76, HPV vs73-1, HPV vs92-1, HPV JC9710, HPV FA1.1, FA2.1, FA5, FA9, FA12, and FA13) and 30 new HPV type candidates. The 30 putative HPV types and their closest relatives are presented in Table 4. Furthermore, for two of these new HPV type candidates, we identified two subtypes each: FA16.1 and FA16.2 (97.88% homology), and FA23.1 and FA23.2 (97.87% homology). The fraction of HPV-positive samples was observed to increase with the age of the individual when all three groups of subjects were investigated together (P < 0.01) (Table 5). No difference was seen between the HPV prevalences in males and females.
TABLE 4.
Thirty new HPV candidates and two subtypes isolated from human skin samples and an environmental samplea
| HPV candidate | Sourceb | Size of fragment analyzedc (bp) | Closest related HPV type or candidate | Nucleotide sequence homology (%) |
|---|---|---|---|---|
| FA14 | RTR | 434 | HPV 8 | 72 |
| FA15 | RTR | 443 | HPV FA4 | 68 |
| FA16.1d | RTR | 437 | HPV 23 | 81 |
| FA16.2d | RTR | 437 | HPV 23 | 81 |
| FA17 | RTR | 437 | HPV RTRX7 | 78 |
| FA18 | RTR | 434 | HPV 8 | 71 |
| FA19 | RTR | 215 | PCPV 1 | 61 |
| FA20 | D | 431 | HPV FA11 | 72 |
| FA21 | HC | 431 | HPV 4 | 67 |
| FA22 | D | 440 | HPV FA7 | 81 |
| FA23.1d | HC, RTR | 434 | HPV 8 | 71 |
| FA23.2d | D | 434 | HPV 8 | 71 |
| FA24 | HC, D | 440 | HPV FA4 | 70 |
| FA25 | D | 434 | HPV 5 | 72 |
| FA26 | D | 434 | HPV 8 | 72 |
| FA27 | D | 434 | HPV 50 | 82 |
| FA28 | RTR | 437 | HPV FA13 | 68 |
| FA29 | RTR | 440 | HPV FA2.1 | 67 |
| FA30 | RTR | 440 | HPV FA13 | 76 |
| FA31 | RTR | 215 | HPV 48 | 63 |
| FA32 | RTR | 437 | HPV FA11 | 65 |
| FA33 | RTR | 215 | HPV 48 | 59 |
| FA34 | RTR | 440 | HPV 50 | 64 |
| FA35 | RTR | 440 | HPV FA13 | 64 |
| FA36 | HC | 218 | HPV 5 NAf3 | 77 |
| FA37 | Floor | 437 | HPV 5 | 77 |
| FA38 | HC | 221 | HPV FA2.3 | 62 |
| FA39 | HC | 440 | HPV FA7 | 82 |
| FA40 | HC | 437 | HPV 38 | 74 |
| FA41 | RTR | 440 | HPV 4 | 70 |
| FA42 | HC | 437 | HPV FA7 | 80 |
| FA43 | HC | 446 | HPV FA6 | 81 |
HPV candidates are presented together with the previously characterized HPV types to which they show the closest DNA sequence homology.
RTR, renal transplant recipient; D, dialysis patient; HC, healthy control.
Primer sequences not included.
FA16.1 and FA16.2, as well as FA23.1 and FA23.2, represent subtypes.
TABLE 5.
Fraction of HPV-positive samplesa from all 160 subjects,b categorized according to the ages of the individuals
| Age (yrs) | HPV DNA prevalencec |
|---|---|
| 20–30 | 38 (23/60) |
| 31–40 | 44 (53/120) |
| 41–50 | 45 (58/130) |
| 51–60 | 56 (130/230) |
| 61–70 | 71 (85/120 |
| 71–80 | 73 (102/140) |
Five samples were analyzed per individual.
Comprising 52 renal transplant recipients, 28 dialysis patients, and 80 healthy controls.
Expressed as a percentage (number of positive samples/total samples).
Sixty-two samples that tested negative for HPV DNA were all PCR positive for human DNA, indicating that no PCR-inhibiting substances were present.
Longitudinal study.
Considering all three volunteers over the 7-month study period, skin swab specimens collected from the foreheads showed a higher prevalence of HPV DNA than specimens from the other sampling sites (arms and thighs). At least one sample was positive for HPV DNA on 19 of the 20 (95%) different sampling occasions for the 38-year-old male, on 14 of the 20 (70%) occasions for the female, and on 15 of 17 (88%) occasions for the 55-year-old male. The total prevalence of positive samples was 23 of 120 (19%) for the female, 59 of 120 (49%) for the younger male, and 27 of 102 (26%) for the older male. Detailed results for the younger male are given in Table 6.
TABLE 6.
HPV DNA in 120 swab samples collected according to the indicated time schedule from six different sites on a healthy 38-year-old male
| Sampling frequency | Date (1999) | Presence of HPV DNA in the:
|
|||||
|---|---|---|---|---|---|---|---|
| Left arm | Right arm | Abdomen | Left thigh | Right thigh | Forehead | ||
| Once a day | May 6 | + | + | + | + | + | + |
| May 7 | + | + | + | + | |||
| May 8 | + | + | + | ||||
| May 9 | |||||||
| May 10 | + | + | + | ||||
| May 11 | + | + | |||||
| May 12 | + | + | + | ||||
| Once a week | May 19 | + | + | ||||
| May 27 | + | + | |||||
| June 2 | + | + | + | + | + | + | |
| June 9 | + | + | |||||
| June 16 | + | + | |||||
| June 24 | + | + | + | ||||
| July 1 | + | + | + | ||||
| July 8 | + | + | |||||
| Once a month | August 4 | + | + | ||||
| September 7 | + | + | |||||
| October 5 | + | + | + | + | + | + | |
| November 7 | + | + | + | ||||
| December 2 | + | + | + | ||||
Five samples from the female were cloned, and two clones from each sample were sequenced. The specimens had been collected on four different occasions from four different sites: on day 1, from the abdomen; on day 3, from the forehead; after 1 month, from the right arm; and after 5 months, from the abdomen and right thigh. Only one new HPV type candidate, FA43, was found in the five samples.
Environmental survey.
HPV DNA was detected in the samples from the floor of the PCR setup room (HPV type candidate FA37, not found in any of the clinical samples), from the laboratory bench (HPV 23 and FA14), and from the floor of the room used for work with cloned material (sample not sequenced). The samples collected from the floors of the kitchen and bedroom in the apartment of one of the volunteers were HPV DNA positive, whereas the bathroom and living room samples were negative. All four of these samples were PCR positive for human DNA.
Phylogenetic analysis.
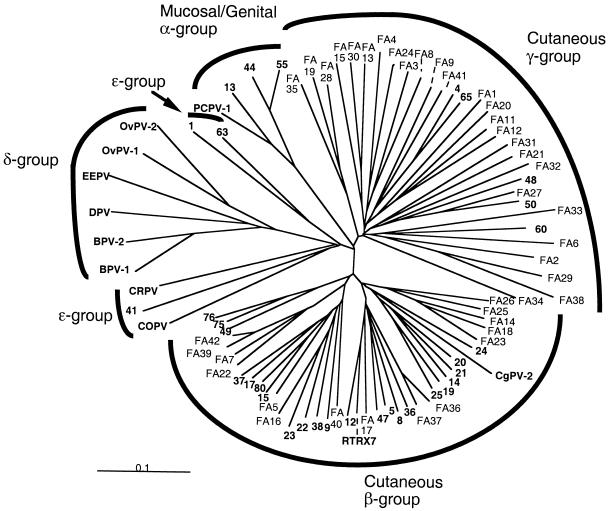
The phylogenetic tree in Fig. 1 shows that the previously characterized HPV types fall into six assemblages: the five supergroups α, β, γ, δ, and ɛ, with supergroup ɛ divided into two subordinate groups. Thirteen of the FA HPV candidates were found to belong to supergroup β and 16 to supergroup γ, the latter comprising only five previously fully characterized HPV types. Trees obtained using the neighbor-joining and maximum-likelihood algorithms were similar.
FIG. 1.
Phylogenetic analysis of the HPV type candidates FA1 to FA42, the established papillomaviruses (indicated in boldface) of the supergroups β, γ, ɛ, and δ, and four HPV types of supergroup α based on neighbor-joining evaluation of a segment of the L1 gene. Only 4 out of the total of about 70 papillomavirus types of the supergroup α were used in the analysis.
DISCUSSION
Our results disclose what has been indicated in previous publications (1, 4, 5, 8, 16, 20), namely, that the human skin harbors a very large spectrum of HPV genotypes, most of them previously unknown. Moreover, there is reason to believe that a further substantial number of skin papillomavirus types remain to be detected, because our limited study of 33 samples from only 13 individuals revealed 20 previously described HPV types and 30 novel HPV type candidates. Fifteen of the putative HPV types FA1 to FA42 were found to belong to supergroup β, which thereby increased in size by 62%. Interestingly, we noted that 26 of the putative HPV types belonged to the small supergroup γ; this represents a more than fivefold expansion of supergroup γ, which previously included only five fully characterized HPV types. However, the possibility that some of the FA HPV type candidates we detected have also been found by other researchers (5, 28) cannot be ruled out, because we analyzed a different segment of the L1 gene than they did.
We used a simple sampling method in which a saline-soaked cotton-tipped swab was gently drawn over a small area of skin, and we observed that more than 75% of the samples from the foreheads of healthy individuals were positive for HPV DNA. Also, cloning and DNA sequencing of single PCR products revealed as many as six different HPV types or type candidates in a single sample. Thus, it seems logical to assume that we would have found even more types of HPV if we had analyzed additional clones from the same PCR products.
Many different HPV types and type candidates were found at all of the analyzed skin sites, although prevalence was greatest on the forehead. Other investigators have used more invasively collected samples, such as plucked hairs (8, 9) and skin biopsy specimens (1, 4, 11, 12), but the analytical methods they applied gave HPV harvests that were lower compared to ours.
We also observed that greater age and immunosuppression were correlated with higher prevalences of HPV in the skin samples, but that might simply reflect a difference in the quantity of HPV DNA present rather than a true difference in the prevalence of HPV infection.
Generally, we found certain types or HPV type candidates on several skin sites within one individual as well as longitudinally on the same person.
Skin cancers in EV and immunocompromised patients occur predominantly on parts of the body exposed to UV radiation, indicating that UV light plays a key role in the development of such disease (14, 17, 23). In our study, HPV was found more frequently on the forehead than on the arms or thighs. If the production of HPV in infected skin is balanced by a local immune response, it is conceivable that the local photoimmunosuppression can occur even at low doses of UVB (3, 33), and it would thus be possible that the higher HPV prevalence we observed on foreheads was due to immunosuppression on sun-exposed sites, in both the immunosuppressed patients and the control population.
The ubiquitousness of skin papillomaviruses revealed in our study puts the supposed role of these agents in the natural history of certain skin cancers to a severe test. Obviously, HPV DNA found in a skin tumor might merely be a passenger that has no relevance to the genesis of the malignancy. Therefore, methods such as in situ hybridization tests and measurement of the expression of certain viral genes and their interaction with host cell functions will probably be needed to provide essential information about the involvement of papillomaviruses in skin cancer.
Most of the previously recognized skin HPV types and 15 of the HPV type candidates detected in our study belonged to the so-called EV-associated HPVs. However, we found that these HPV types or type candidates are frequently present on the skin of normal, healthy individuals, and it has also been reported that they occur in skin cancer lesions of renal transplant recipients (4, 5, 11, 16, 21). Therefore, it seems warranted to stop calling them EV-associated HPVs. Preferably, they should be classified according to phylogenetic supergroups (34), or, in a broader sense, simply referred to as skin HPV types.
Papillomaviruses have been found in most vertebrates investigated (30), and it is assumed that they have developed together with various animal species over hundreds of millions of years (6, 26). Therefore, it is not surprising that some papillomaviruses have adapted in such a way that they can efficiently spread between individuals and cause chronic infections without inducing any tissue damage, at least under normal conditions. An inescapable and fascinating question is whether papillomavirus infections have brought any evolutionary benefits to the vertebrate hosts.
ACKNOWLEDGMENTS
We thank Rose-Marie Carlsson and Susanne Brännlund, nurses at the Dialysis Unit, University Hospital, Malmö, for collecting the samples from the dialysis patients. Thanks are also due to Kenneth Persson, Department of Medical Microbiology, Lund University, University Hospital, Malmö, for invaluable help with statistical analysis of the data.
This work was supported by the Cancer Foundation of University Hospital, Malmö, and the Alfred Österlund Foundation.
REFERENCES
- 1.Astori G, Lavergne D, Benton C, Hockmayr B, Egawa K, Garbe C, de Villiers E M. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J Investig Dermatol. 1998;110:752–755. doi: 10.1046/j.1523-1747.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 2.Barr B B, Benton E C, McLaren K, Bunney M H, Smith I W, Blessing K, Hunter J A. Human papillomavirus infection and skin cancer in renal allograft recipients. Lancet. 1989;i:124–129. doi: 10.1016/s0140-6736(89)90412-1. [DOI] [PubMed] [Google Scholar]
- 3.Beissert S, Schwarz T. Mechanisms involved in ultraviolet light-induced immunosuppression. J Investig Dermatol Symp Proc. 1999;4:61–64. doi: 10.1038/sj.jidsp.5640183. [DOI] [PubMed] [Google Scholar]
- 4.Bens G, Wieland U, Hofmann A, Hopfl R, Pfister H. Detection of new human papillomavirus sequences in skin lesions of a renal transplant recipient and characterization of one complete genome related to epidermodysplasia verruciformis-associated types. J Gen Virol. 1998;79:779–787. doi: 10.1099/0022-1317-79-4-779. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout R J, Tieben L M, Smits H L, Bavinck J N, Vermeer B J, ter Schegget J. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol. 1995;33:690–695. doi: 10.1128/jcm.33.3.690-695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard H U. Coevolution of papillomaviruses with human populations. Trends Microbiol. 1994;2:140–143. doi: 10.1016/0966-842x(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 7.Birkeland S A, Storm H H, Lamm L U, Barlow L, Blohme I, Forsberg B, Eklund B, Fjeldborg O, Friedberg M, Frodin L, et al. Cancer risk after renal transplantation in the Nordic countries, 1964–1986. Int J Cancer. 1995;60:183–189. doi: 10.1002/ijc.2910600209. [DOI] [PubMed] [Google Scholar]
- 8.Boxman I L, Berkhout R J, Mulder L H, Wolkers M C, Bouwes Bavinck J N, Vermeer B J, ter Schegget J. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Investig Dermatol. 1997;108:712–715. doi: 10.1111/1523-1747.ep12292090. [DOI] [PubMed] [Google Scholar]
- 9.Boxman I L, Russell A, Mulder L H, Bavinck J N, Schegget J T, Green A. Case-control study in a subtropical Australian population to assess the relation between non-melanoma skin cancer and epidermodysplasia verruciformis human papillomavirus DNA in plucked eyebrow hairs. The Nambour Skin Cancer Prevention Study Group. Int J Cancer. 2000;86:118–121. doi: 10.1002/(sici)1097-0215(20000401)86:1<118::aid-ijc18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Boyle J, MacKie R M, Briggs J D, Junor B J, Aitchison T C. Cancer, warts, and sunshine in renal transplant patients. A case-control study. Lancet. 1984;i:702–705. doi: 10.1016/s0140-6736(84)92221-9. [DOI] [PubMed] [Google Scholar]
- 11.de Jong-Tieben L M, Berkhout R J, Smits H L, Bouwes Bavinck J N, Vermeer B J, van der Woude F J, ter Schegget J. High frequency of detection of epidermodysplasia verruciformis-associated human papillomavirus DNA in biopsies from malignant and premalignant skin lesions from renal transplant recipients. J Investig Dermatol. 1995;105:367–371. doi: 10.1111/1523-1747.ep12320803. [DOI] [PubMed] [Google Scholar]
- 12.de Jong-Tieben L M, Berkhout R J, ter Schegget J, Vermeer B J, de Fijter J W, Bruijn J A, Westendorp R G, Bouwes Bavinck J N. The prevalence of human papillomavirus DNA in benign keratotic skin lesions of renal transplant recipients with and without a history of skin cancer is equally high: a clinical study to assess risk factors for keratotic skin lesions and skin cancer. Transplantation. 2000;69:44–49. [PubMed] [Google Scholar]
- 13.Deragon J M, Sinnett D, Mitchell G, Potier M, Labuda D. Use of gamma irradiation to eliminate DNA contamination for PCR. Nucleic Acids Res. 1990;18:6149. doi: 10.1093/nar/18.20.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Villiers E M. Human papillomavirus infections in skin cancers. Biomed Pharmacother. 1998;52:26–33. doi: 10.1016/s0753-3322(97)86238-5. [DOI] [PubMed] [Google Scholar]
- 15.de Villiers E M. Human pathogenic papillomavirus types: an update. Curr Top Microbiol Immunol. 1994;186:1–12. doi: 10.1007/978-3-642-78487-3_1. [DOI] [PubMed] [Google Scholar]
- 16.de Villiers E M, Lavergne D, McLaren K, Benton E C. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. Int J Cancer. 1997;73:356–361. doi: 10.1002/(sici)1097-0215(19971104)73:3<356::aid-ijc9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 17.de Villiers E M, Ruhland A, Sekaric P. Human papillomaviruses in non-melanoma skin cancer. Semin Cancer Biol. 1999;9:413–422. doi: 10.1006/scbi.1999.0145. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Numerical methods for inferring evolutionary trees. Q Rev Biol. 1982;57:379–404. [Google Scholar]
- 20.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson B G. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80:2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 21.Höpfl R, Bens G, Wieland U, Petter A, Zelger B, Fritsch P, Pfister H. Human papillomavirus DNA in non-melanoma skin cancers of a renal transplant recipient: detection of a new sequence related to epidermodysplasia verruciformis associated types. J Investig Dermatol. 1997;108:53–56. doi: 10.1111/1523-1747.ep12285630. [DOI] [PubMed] [Google Scholar]
- 22.Jablonska S, Dabrowski J, Jakubowicz K. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res. 1972;32:583–589. [PubMed] [Google Scholar]
- 23.Jablonska S, Majewski S. Epidermodysplasia verruciformis: immunological and clinical aspects. Curr Top Microbiol Immunol. 1994;186:157–175. doi: 10.1007/978-3-642-78487-3_9. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 25.Nicholas K B, Nicholas H B, Jr, Deerfield D W. Genedoc: analysis and visualization of genetic variation. EMBnet News. 1997;4:14. [Google Scholar]
- 26.Ong C K, Chan S Y, Campo M S, Fujinaga K, Mavromara Nazos P, Labropoulou V, Pfister H, Tay S K, ter Meulen J, Villa L L, et al. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67:6424–6431. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orth G, Jablonska S, Jarzabek-Chorzelska M, Obalek S, Rzesa G, Favre M, Croissant O. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 1979;39:1074–1082. [PubMed] [Google Scholar]
- 28.Shamanin V, Glover M, Rausch C, Proby C, Leigh I M, zur Hausen H, de Villiers E M. Specific types of human papillomavirus found in benign proliferations and carcinomas of the skin in immunosuppressed patients. Cancer Res. 1994;54:4610–4613. [PubMed] [Google Scholar]
- 29.Stark L A, Arends M J, McLaren K M, Benton E C, Shahidullah H, Hunter J A, Bird C C. Prevalence of human papillomavirus DNA in cutaneous neoplasms from renal allograft recipients supports a possible viral role in tumour promotion. Br J Cancer. 1994;69:222–229. doi: 10.1038/bjc.1994.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundberg J P. Papillomavirus infections in animals. In: Syrjänen K, Gissmann L, Koss L G, editors. Papillomaviruses and human disease. Berlin, Germany: Springer-Verlag; 1987. pp. 40–103. [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. Improved sensitivity of profile searches through the use of sequence weights and gap excision. CABIOS. 1994;10:19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- 32.Van Ranst M A, Tachezy A R, Burk R D. Taxonomy of the human papillomaviruses. Papillomavirus Rep. 1993;4:61–65. [Google Scholar]
- 33.Vermeer B J, Hurks M. The clinical relevance of immunosuppression by UV irradiation. J Photochem Photobiol B. 1994;24:149–154. doi: 10.1016/1011-1344(94)07033-4. [DOI] [PubMed] [Google Scholar]
- 34.zur Hausen H. Papillomaviruses in human cancers. Proc Assoc Am Physicians. 1999;111:581–587. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]