Abstract
Melanoma clinical outcomes emerge from incompletely understood genetic mechanisms operating within the tumor and its microenvironment. Here, we used single-cell RNA-based spatial molecular imaging (RNA-SMI) in patient-derived archival tumors to reveal clinically relevant markers of malignancy progression and prognosis. We examined spatial gene expression of 203,472 cells inside benign and malignant melanocytic neoplasms, including melanocytic nevi and primary invasive and metastatic melanomas. Algorithmic cell clustering paired with intratumoral comparative two-dimensional analyses visualized synergistic, spatial gene signatures linking cellular proliferation, metabolism, and malignancy, validated by protein expression. Metastatic niches included up-regulation of CDK2 and FABP5, which independently predicted poor clinical outcome in 473 patients with melanoma via Cox regression analysis. More generally, our work demonstrates a framework for applying single-cell RNA-SMI technology toward identifying gene regulatory landscapes pertinent to cancer progression and patient survival.
Spatial analysis of melanoma microenvironment gene expression at single-cell level identifies markers of malignancy and prognosis.
INTRODUCTION
Melanocytic tumors—both benign and malignant—exhibit a great variety of morphologies and clinical outcomes (1, 2). For example, the melanocytes of nevi (benign melanocytic neoplasms colloquially known as “moles”) initially replicate, quiesce, and remain confined within their tissue microenvironment. The melanocytes of melanoma, in contrast, have the capacity to aggressively proliferate, invade nearby tissues, and metastasize. Although both nevi and melanoma can have similar driver mutations, most commonly in BRAF and NRAS, melanomas are known to harbor a multitude of oncogenic mutations that are not fully understood but differ from the mutations identified in nevi (2). In addition to individual DNA mutations, the clinical fates of patients with melanocytic neoplasms emerge from a combination of incompletely understood molecular, genetic, and cellular mechanisms interacting within the tumor microenvironment.
Unregulated melanocyte cell proliferation (i.e., tumor growth) and inappropriate cell migration (i.e., tissue invasion and metastasis) define melanoma (3–7). Previous studies have defined oscillatory gene expression profiles that promote melanocyte “proliferative” versus “invasive” phenotypes (3, 4, 7). For instance, proliferative melanocytes generally exhibit high levels of transcription factor MITF and other genes (7, 8), although how this phenotype is spatially arranged and modulated within the tumor microenvironment remains poorly understood.
Our understanding of melanocyte biology and gene expression has relied on RNA measurement techniques, e.g., gene expression microarrays, bulk RNA sequencing, and single-cell RNA sequencing (scRNA-seq). These techniques, however, lack spatial information and thus preclude insight into factors such as cell proximity to lymphovasculature (9), tumor-stroma boundary (10), or lymphocytic infiltrate (11). To overcome these limitations, the evolving field of “spatial profiling” (also called “spatial transcriptomics”) (12, 13) holds promise for measuring in situ gene expression within skin and other cancers types (14–18). Realization of the full clinical utility of spatial profiling in the fight against cancer, however, necessitates effective strategies for extracting biologically and clinically useful information from this new class of data.
Here, using a technique enabling in situ simultaneous visualization of ~1000 RNAs at single-cell and subcellular resolution (18), we generated single-cell maps of gene expression within benign, malignant, and metastatic melanocytic neoplasms. We then deconvolute these maps to identify proliferative gene signatures that influence progression to malignancy and melanoma patient survival.
RESULTS
Single-cell RNA-based spatial molecular imaging generates maps of gene expression within benign, malignant, and metastatic melanocytic neoplasms
We visualized in situ gene expression of 203,472 cells using single-cell RNA-based spatial molecular imaging (RNA-SMI) (18), which allows utilization of patient-derived, formalin-fixed, paraffin-embedded (FFPE) specimens (Fig. 1A; further explanatory technical details found in the Supplementary Text).
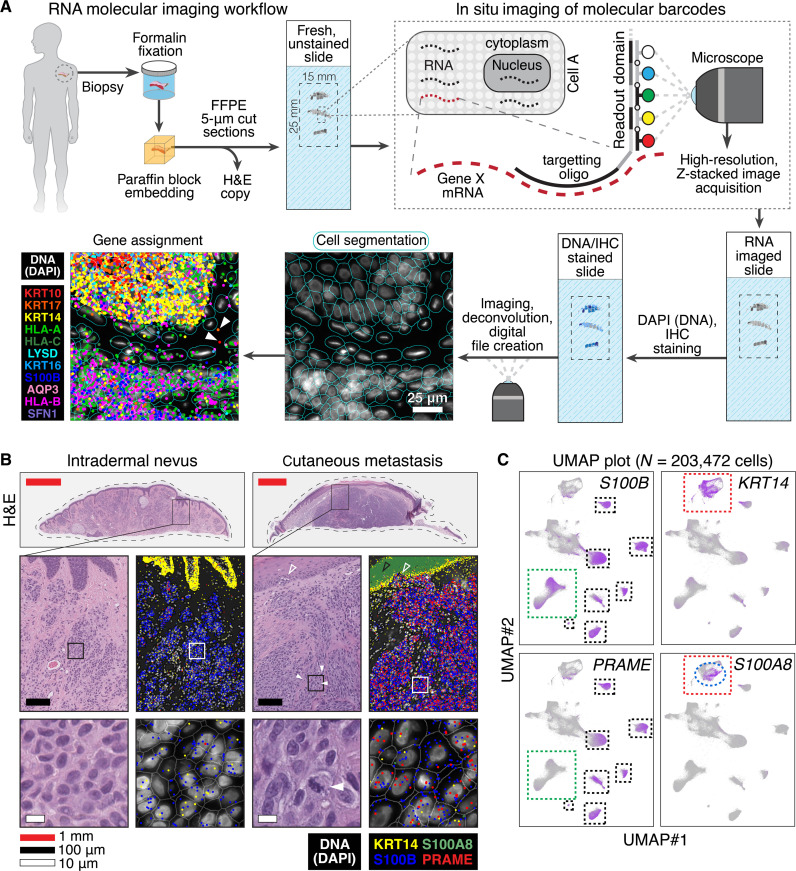
Fig. 1. Single-cell RNA-based SMI generates maps of gene expression within benign, malignant, and metastatic melanocytic neoplasms.
(A) FFPE samples are sectioned and undergo SMI (18). Native RNA is hybridized in situ to gene-specific antisense oligonucleotides fused to multiplexed readout domains, which undergo cyclical reactions with four different fluorophores (Alexa Fluor 488, ATTO 532, Dyomics-605, or Alexa Fluor 647) or null, nonfluorescent domain (represented in white). The readout domain length and number of iterative cycles are plexed to allow detection of ~1000 unique genes. Slides are then stained with DAPI to detect nuclei and other IHC markers, which facilitate automated cell segmentation in silico. Segmented cells and nuclei generate a “map” onto which RNA transcripts are assigned. Bottom left panels show epidermis above melanocyte-containing tissue with 11 differentially colored gene transcripts, with DAPI shown in white. The solid white arrows show a detected RNA transcript outside a cell border, which may represent a z-axis out-of-plane RNA or extracellular RNA. (B) H&E and RNA imaging of an intradermal nevus and cutaneous melanoma metastasis. The solid outlined, rectangular boxes in panels show highlighted insets below. The panels show the expected increased detection of melanoma marker PRAME amongst the S100B melanocytes of metastatic melanoma as well as the expected increase in S100A8 (black open arrow) in epidermal keratinocytes (marked by KRT14) overlying the malignant proliferation. Closed arrows show mitotic cells. Open arrow shows malignant melanocytes juxtaposed to the epidermis. (C) UMAP projections of gene expression amongst 203,472 cells analyzed in 10 patient-derived tumors. S100B expression includes melanocytes, a subset of which express melanoma marker PRAME (black versus green dashed boxes); the red dashed box shows epidermal keratinocytes that express KRT14, a subset of which express S100A8 (blue dashed ellipse).
We first analyzed 10 patient-derived tumors: three intradermal nevi, four primary invasive melanomas, and three cutaneous metastases (fig. S1 and data S1). Metastases had been DNA sequenced during their clinical workup, revealing mutations in KRAS, NRAS, TP53, NF1, and TERT in the respective bulk DNA samples (data S2). The probe set used included ~1000 cancer-associated genes (data S3).
Using machine learning augmented segmentation, we digitally segmented over 63,000 cells per slide, assigning ~70 million gene transcripts to the diagnostic regions of each tumor microenvironment (fig. S2 and data S4). Because we did not find an earlier study examining the skin with this technology, we first confirmed that our dataset accurately recapitulated known spatial gene expression patterns, including superficial-to-deep epidermal keratin and collagen profiles (fig. S3).
For further validation, we examined known melanoma gene expression of melanocytic nevi versus melanoma cutaneous metastases. This confirmed the expected comparative increase in the known melanoma marker PRAME (19) amongst S100B melanocytes of the metastatic melanoma versus nevus as well as the previously identified increased S100A8 in the keratinocytes overlying the melanoma (Fig. 1B) (17).
To visualize gene expression in 203,472 cells simultaneously, we used uniform manifold approximation and projection (UMAP) (Fig. 1C), a dimension reduction technique allowing visualization of clustered patterns of high-dimensional data (20). Consistently, UMAP showed distinct subsets of PRAME expression within S100B-postive cells (i.e., presumptive malignant melanocytes) and the S100A8-expressing subset of KRT14-positive cells (presumptive keratinocytes overlying melanoma).
Collectively, these and other candidate gene UMAP plots and interlesional melanocyte comparisons (fig. S4) showed that our dataset—a series of two-dimensional (2D) maps showing the location of ~70 million mRNA transcripts in 203,472 cells—was amenable to further algorithmic extraction of biologically meaningful gene expression signatures between the different lesion classes examined.
Clustering analyses identify a proliferative gene signature that includes CDK2
We next sought to identify gene signatures that correlated with cell proliferation via algorithmic clustering approaches. We clustered the 203,472 cells into 11 unique groups using unsupervised Leiden clustering, an unbiased clustering algorithm that improves on Louvain clustering to yield better-connected communities (Fig. 2A) (21).
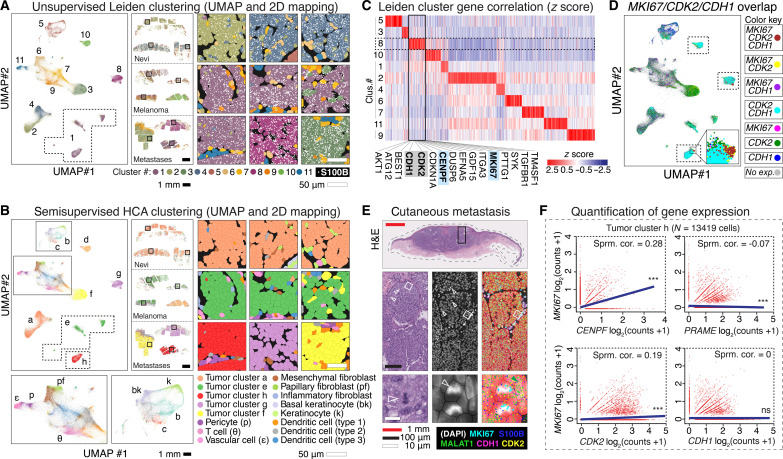
Fig. 2. Clustering analyses identify a proliferative gene signature that includes CDK2.
(A) UMAP projection of 203,472 cells into 11 unsupervised Leiden clusters, 2D mapped onto the nevi, melanoma, and cutaneous metastases samples. Black square insets magnified in right panels, showing portions of melanocytic tumors expressing S100B (white dots). (B) UMAP and 2D mapping with semisupervised clusters using HCA skin reference (solid rectangles magnified below). Black square insets magnified in right panels. (C) Heatmap of normalized gene expression in Leiden clusters. Proliferative markers MKI67 and CENPF localized to cluster 8, which also included melanoma-associated proliferative genes CDK2 and CDH1 (top hits listed below cluster in alphabetical order). (D) UMAP projection with combinatorial overlap of MKI67-, CDK2-, and CDH1-expressing cells; inset shows a portion of cluster h enriched for MKI67, CDK2, and CDH1 coexpression. (E) H&E and RNA-SMI of MKI67, CDK2, CDH1, S100B, and predominately nuclear localized MALAT1 within cutaneous metastasis #1 (predominately composed of tumor cluster h–type cells). Black solid rectangle is magnified in the middle horizontal row; white solid squares magnified in the bottom horizontal row. White open arrows point to mitoses. Green arrow shows predominately nuclear localization of MALAT1. The middle column DAPI panels are overlayed in the right column with semisupervised HCA cell clusters, most of which are tumor cluster h, as shown in (B). Detected RNA transcripts are shown as outlined in the key at bottom right key. Collectively, the panels show expression of MKI67, CDK2, and CDH1 in cells of the mitotically active portion of the tumor. (F) Gene expression correlation of tumor cluster h, showing a positive Spearman correlation (Sprm. cor.) of CENPF and CDK2 with MKI67 (correlation absent with PRAME and CDH1). ***, adjusted P value < 0.0001; ns, P value > 0.05.
To visualize Leiden clusters within tumor microenvironments, we mapped cell cluster identities onto the nevi, melanoma, and cutaneous metastases samples (Fig. 2A). The S100B-expressing melanocytes of two of three intradermal nevi (nevi #2 and #3) segregated into a single cluster (cluster 4); one melanoma (melanoma #1) and two metastasis samples (cutaneous metastases #2 and #3) clustered independently (clusters 10, 8, and 3, respectively). Three melanomas and one cutaneous metastasis formed a single cluster (cluster 1). These results led us to explore additional clustering methods to group clinically similar tumoral melanocytes to facilitate subsequent comparative analyses.
We improved cell clustering by using the scRNA-seq–derived, Human Cell Atlas (HCA) skin gene expression dataset as a cell type reference (22–24). Considering the expression of all genes, we assigned cells to known cell types (e.g., inflammatory cells, keratinocytes, fibroblasts, endothelial cells, etc.) while simultaneously allowing space for identification of previously undefined cell profiles. This “semisupervised” approach resulted in eight distinct neoplastic melanocyte cell types (clusters a to h) that notably grouped all nevus melanocytes into a single group (cluster a), three of the four melanoma samples into a single group (cluster e), and each metastasis independently (Fig. 2B).
To identify the gene signature that most correlated with cell proliferation, we examined expression of MKI67 (marker of proliferation Ki-67) (25) and CENPF (centromere protein F) (26), which are reliable mRNA proxies for cell cycling used in scRNA analyses (16, 27). Amongst the unsupervised Leiden clusters, cluster 8 had the highest expression of MKI67 and CENPF and contained other genes implicated in melanoma growth e.g., CDH1 (cadherin-1) (8), the MITF transcriptional target CDK2 (cyclin-dependent kinase 2) (28), growth factor signaling pathway component AKT (29), and others (Fig. 2C and data S5).
To examine the global expression of this proliferative gene signature within our dataset, we generated UMAP plots labeled by coexpression of these markers, noting that MKI67-expressing cells represented a subset of cell that expressed other proliferative signature markers, e.g., CDH1 and CDK2 (Fig. 2D). Consistently, we found that MKI67/CDH1/CDK2+ cells were qualitatively enriched in metastatic tumors (e.g., cluster h), particularly within tissue aggregates undergoing active cell cycling (Fig. 2E).
To determine which proliferative signature genes best correlated with the MKI67 subset, we quantified expression within the 13,419 cells comprising metastatic melanoma cluster h. As expected, cell proliferative marker CENPF positively correlated with MKI67, whereas the expression levels of nonproliferative melanoma marker PRAME did not (Fig. 2F). Notably, we found that CDK2 (but not CDH1) showed a positive correlation with MKI67, a finding also observed at the protein level (fig. S5).
To assess the generalizability of these findings, we examined The Cancer Genome Atlas (TCGA) (30) bulk tumor-derived transcriptomic data from 473 patient-derived melanoma samples. These data confirmed the significant correlations of MKI67 with CENPF and CDK2 (fig. S6); we found that the correlation between MKI67 and CDK2 was lower than that of MKI67 and CENPF, which could be explained by the baseline, noncycling level of expression of CDK2 in melanocytes (28) and potential additional CDK functions other than cell proliferation, such as genomic and chromosomal modulation (31). Notably, we found that increased expression of CDK2 was comparatively specific to melanoma versus 16 other cancer types found in The Human Protein Atlas (fig. S7) (32); moreover, CDK2 was the only member of the CDK family to show this melanoma specificity at the transcript level (fig. S7).
Together, these analyses demonstrate how unsupervised and semisupervised clustering algorithms from single-cell spatial data can identify gene expression signatures of candidate cellular processes, including cell proliferation, in melanoma.
Candidate gene analysis reveals differential tumor-stroma boundary expression of CDK2
We next sought to identify differential spatial expression patterns of genes in nevi versus melanoma using a candidate approach. Toward this aim, we chose to examine CDK2 due to its constellation of intriguing features, e.g., its correlation with cell proliferation (Fig. 2), comparative specificity of up-regulation in melanoma versus other cancers (fig. S7), and status as a MITF downstream transcriptional target (28).
We visualized CDK2 expression in the melanocytic neoplasms of our dataset, and unexpectedly, we found CDK2 expressed in benign melanocytes at the dermal-epidermal junction and in superficial dermal melanocytic nests of nevi (fig. S8). The deep dermal expression of CDK2, however, vastly differed between nevi and melanoma: In intradermal nevi, CDK2 expression was gradually lost with dermal depth, whereas CDK2 expression was retained at comparatively high level at the deep tumor-stromal boundary in primary melanoma (Fig. 3A).
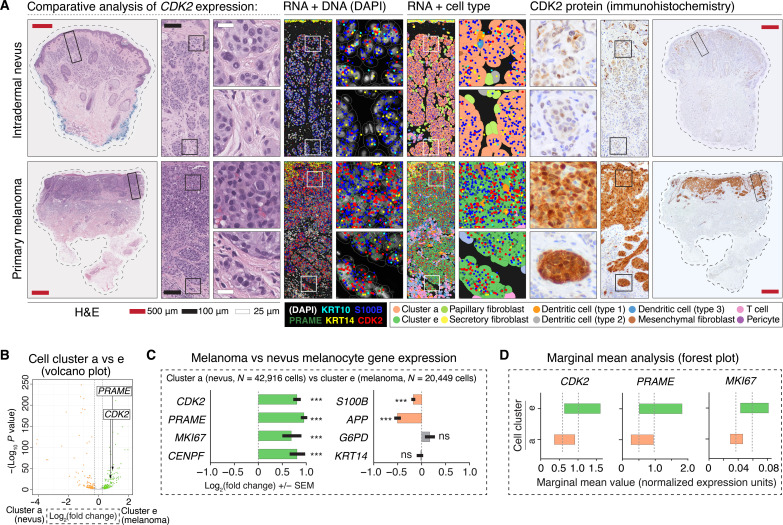
Fig. 3. Candidate gene analysis reveals differential tumor-stroma boundary expression of CDK2.
(A) Panels show the comparative loss of CDK2 expression in dermal melanocytes of intradermal nevi versus retained expression of CDK2 in dermally invasive primary melanoma. Left H&E panels have solid rectangular insets subsequently magnified in the rightward panels. H&E stains show characteristic, bland appearing nested melanocytes of an intradermal nevus versus the hyperchromatic, crowded, and pleomorphic cells of primary melanoma. RNA + DNA panels show comparatively increased expression of CDK2 and PRAME in S100B-expressing malignant melanoma melanocytes. RNA + cell type panels show semisupervised HCA clustering of the cells of the tumor, with annotation of intradermal nevus-like melanocytes (cluster a) and malignant melanoma melanocytes (cluster e). Right panels show CDK2 protein expression, which was notable for comparatively increased expression at the deep, tumor-stroma border in malignant melanoma versus nevus. (B) Volcano plot showing quantification of differential gene expression between nevus (cluster a) and melanoma (cluster e) melanocytes. Each dot on this plot represents a gene. The location of PRAME and CDK2 within the plot is shown. Orange dots represent genes with higher expression in nevus (cluster a), whereas green dots represent genes with higher expression in melanoma (cluster e). Gray dots represent genes that failed to reach the FDR threshold of 0.05 (Benjamini-Hochberg correction). (C) Bar graphs highlighting select genes from volcano plot shown in (B). ***P value < 0.0001; ns, P value > 0.05. Error bars show SEM. (D) Forest plots showing marginal means (gray dashed line) with a 95% confidence interval. Note that graphics shown in (B) to (D) are different ways to visualize the same quantification of differential expression of PRAME, CDK2, MKI67, and other genes between clusters a and e.
To validate these findings at the protein level, we used a patient-derived tumor panel consisting of 59 nevi and 69 primary melanomas (fig. S9) and characterized their expression patterns (figs. S10 to S12). We found that comparatively high CDK2 expression was at least focally retained amongst ~80% of primary melanoma with ≥0.8-mm tumor thickness (fig. S10, patterns #4 and #5), a depth of invasion that correlates with comparatively poor clinical outcomes (33), as well as in melanoma metastases (34). In contrast, all intradermal nevi of the panel with comparable depths exhibited decreased CDK2 expression at the deep tumor-stroma boundary.
To quantify the differential gene expression of CDK2 and other primary melanoma-increased genes in our dataset, we analyzed the semisupervised clusters of intradermal nevi (cluster a, N = 42,916 cells) and primary melanoma (cluster e, N = 20,449 cells) (Fig. 3, B and C) and performed differential expression analysis using negative binomial regression. Within our model for each gene, we included patient as a random effect, the expression of the gene in surrounding cells as a fixed effect, and the total counts in each cell as an offset. As an alternative visualization, we show the marginal means estimated from the model for three select genes (Fig. 3D). These data showed that quantitative up-regulation of CDK2 closely resembles that of PRAME, consistent with our prior study using sequencing-based spatial approaches (17).
Overall, this candidate gene approach revealed that comparatively high CDK2 expression at the tumor-stroma boundary is a feature of most primary melanomas, as might be expected from previous bulk RNA microarray-based studies (31). However, CDK2 expression is not a marker of melanocyte malignancy per se given its expression in certain benign melanocytes, e.g., superficial nevus melanocytes. This paradox hinted at the existence of other spatially defined factors that modulate cell phenotype in the benign and malignant tumor environment.
Intratumoral spatial analyses reconcile field-level proliferative and metabolic gene expression signatures
To search for microenvironmental factors that help define benign versus malignant melanocytes, we sought to directly compare benign and malignant CDK2-expressing melanocytes. To reduce potential confounders from intertumoral analyses (e.g., different patient, age, biopsy site, sun exposure, and genetic background), we performed intratumoral analyses in a single tumor having both melanoma and nevus portions.
We confirmed the canonical nevus and melanoma portions of the tumor via histomorphological and immunohistochemical analyses (Fig. 4A and fig. S12). Melanocytes expressed both PRAME and CDK2 as expected of their benign or malignant status (Fig. 4B). Similarly, keratinocytes associated with the malignant portion of the tumor expressed S100A8 (Fig. 4B). Consistently, Leiden unsupervised and semisupervised clustering generated differential cell clusters within the nevus and melanoma portions of the combined tumor (Fig. 3C).
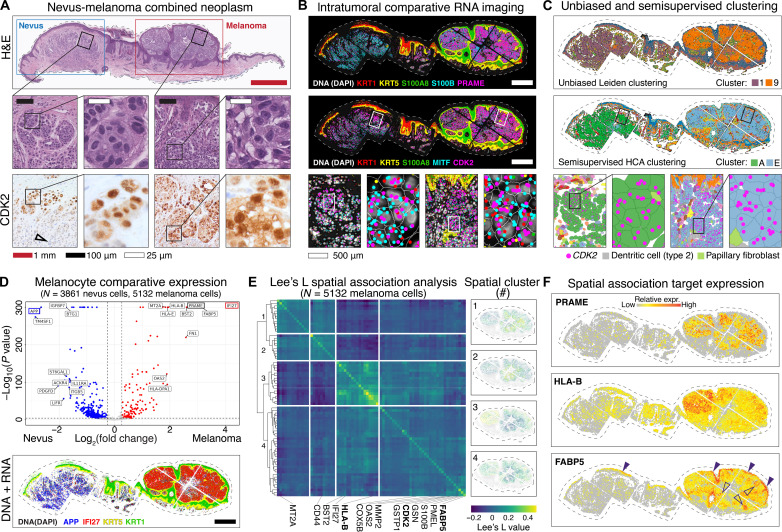
Fig. 4. Intratumoral spatial analyses reconcile field-level proliferative and metabolic gene expression signatures.
(A) Melanocytic tumor with nevus and malignant melanoma portions, with insets shown as indicated by black squares. Bottom row shows nuclear and cytoplasmic CDK2 protein expression in melanocytes. Black open arrow shows loss of CDK2 expression in deep nevus cells. (B) Top panel shows comparatively increased PRAME and S100A8 in melanoma portion of the tumor. The middle panel shows expression of CDK2 and melanocytic marker MITF, progressively magnified as indicated with white bordered rectangular insets. (C) Unsupervised Leiden (top) and semisupervised HCA (middle) clustering generated nevus-type and melanoma-type melanocyte clusters (clusters A and E, respectively). (D) Top panel shows volcano plot of differential gene expression between nevus (cluster A) and melanoma (cluster E) melanocytes. Blue dots represent genes with higher expression in nevus (cluster A), whereas red dots represent genes with higher expression in melanoma (cluster E). Gray dots represent genes that failed to reach the FDR threshold of 0.05 (Benjamini-Hochberg correction). The top hit in each condition outlined in blue (APP) or red (IFI27) were mapped out in the bottom DNA + RNA imaging panel. Scale bar: 500 μm. (E) Heatmap shows results of Lee’s L spatial association analysis of malignant melanocytes (cluster E), with each row and column representing a spatially patterned gene. Genes clustered into four hierarchical fields as numbered on the left. This spatial analysis used a combination of proximity and differential expression level to define four distinct fields of gene expression within the melanoma. The panels on the right show the normalized expression of all genes in each numbered field. (F) Spatial plots showing relative expression of known gene PRAME as well as immunoregulatory gene HLA-B and metabolic genes FABP5, which spatially clustered with CDK2 [field 4 in (E)].
Using the semisupervised clusters (nevus, cluster A, N = 3861 cells; melanoma, cluster E, N = 5132 cells), we performed differential expression analysis and quantitatively confirmed that expected marker genes, including PRAME and IFI27, were significantly differentially expressed (Fig. 4D) (35). This confirmatory analysis gave us confidence that the semisupervised cell clusters contained valid transcriptomic profiles amenable to subsequent spatial 2D analyses.
Using Lee’s L spatial association analysis to evaluate gene expression in the malignant tumor, which captures the correlation of expression between genes in space (36), we identified four distinct clusters of genes showing distinct fields of synergistic expression within the malignant portion of the tumor (Fig. 4E and data S6). For example, spatial cluster #3 showed up-regulation of inflammatory- and immune-modulating genes HLA-A, HLA-B, HLA-C, and HLA-E and BST-2 in the upper left region of the tumor (Fig. 4E) (37), suggesting that this microenvironmental region elicited a comparatively greater innate immune response.
To identify which microenvironmental region most aligned with cell proliferation, we located CDK2 within spatial cluster #4, which was highly expressed in tissue aggregates with CDK2/Ki-67 coexpression at the protein level by dual immunofluorescence immunohistochemistry (IHC) (Fig. 4E and fig. S13). Notably, spatial cluster #4 also included metabolic genes COX5B and FABP5, both of which have been implicated in modulating the pro-growth cancer metabolome (38–40). Mapping these spatially regulated genes onto the tumor confirmed their differential spatial expression (Fig. 4F and fig. S14).
Together, these intratumoral analyses revealed distinct spatial gene signatures within the microenvironments of a single heterogenous neoplasm with both benign and malignant components, revealing potential genes relevant for progression into malignancy.
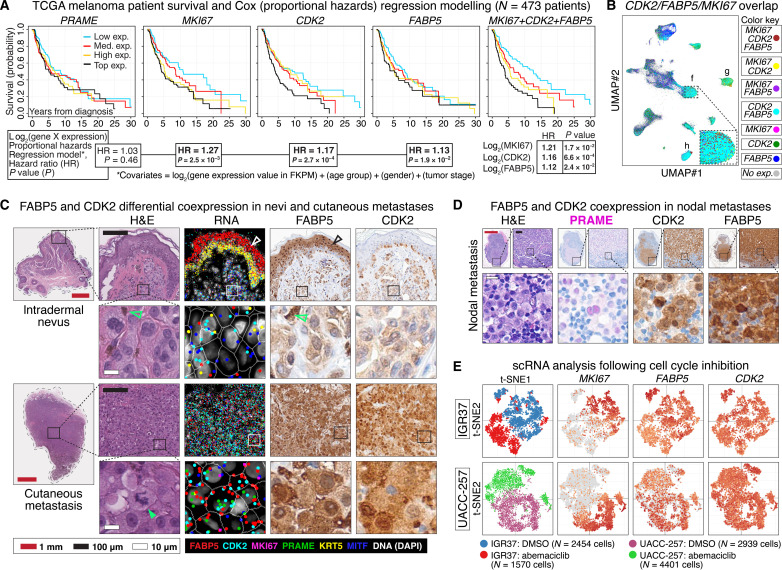
Differential expression of MKI67, CDK2, and FABP5 retrospectively predicts melanoma patient survival
To further explore clinically relevant melanoma markers, we translated our RNA-SMI spatial gene expression maps into gene models for melanoma survival. Toward this aim, we visualized the survival of 473 TCGA patients via Kaplan-Meier plots stratified by expression levels of genes identified via our unsupervised clustering and spatial analyses, e.g., MKI67, CENPF, CDK2, CDH1, COX5B, and FABP5 (Fig. 5A and fig. S15). To quantify the influence of gene expression on survival rate, we performed Cox (proportional hazards) regression analysis with gene expression, patient age, sex, and cancer stage as independent covariates.
Fig. 5. Differential expression of MKI67, CDK2, and FABP5 retrospectively predicts melanoma patient survival.
(A) Kaplan-Meier survival curves from 473 patients with melanoma after stratification into low, medium (med.), and high and top expression quartiles of PRAME, MKI67, CDK2, and FABP5. Rightmost curve shows the comparatively increased differential survival stratification using the sum of log2 MKI67, CDK2, and FABP5 expression. Cox (proportional hazards) regression analyses HRs with their respective P values are shown below each Kaplan-Meier curve. The Cox regression analyses were based on a model that included gene expression values, patient age, gender, and tumor stage as covariates. The analysis showed that expression of MKI67, CDK2, and FABP5 was statistically significant and independent predictors of poor prognosis. (B) UMAP shows combinatorial overlap of MKI67-, CDK2-, and FABP5-expressing cells enriched in the metastatic melanoma cell clusters f, g, and h identified in Fig. 2. (C) The panels show comparative increased expression of CDK2 and FABP5 in metastatic melanoma compared to nevus, both at the RNA and protein level. White and black bordered rectangles are magnified in adjacent panels as shown. White open and black arrowheads show expression of FABP5 RNA and protein in the upper layers of the epidermis. Green open arrows show normal, physiologic melanin visible in both H&E- and IHC-processed tissue sections. Green closed arrow shows atypical mitotic figure within metastatic melanoma. (D) Protein expression of PRAME, CDK2, and FABP5 highlight malignant melanoma metastasized to lymph node. (E) scRNA expression following cell cycle inhibition using CDK4/6 inhibitor abemaciclib or control (DMSO) in melanoma IGR37 (top row) and UACC-257 (bottom row) cell lines. The t-SNE plots show loss of MKI67 expression, partial loss of FABP5, and comparatively little change in CDK2 expression in abemaciclib-treated cells versus control.
As expected, the quantitative expression of PRAME did not significantly discriminate patient survival (Fig. 5A) (41). In contrast, we found that increased MKI67 expression significantly correlated with decreased patient survival [log2expression hazard ratio (HR) = 1.27, P = 2.5 × 10−3], whereas CENPF did not (fig. S15). Covariate expression of CDK2 (HR = 1.17, P = 2.7 × 10−4) more so than CDH1 (HR = 1.05, P = 1.0 × 10−2) as well as metabolic genes FABP5 (HR = 1.13, P = 1.9 × 10−2) and COX5B (HR = 1.21, P = 3.3 × 10−2) significantly correlated with survival.
Notably, we found that a survival model that included the summation of MKI67, CDK2, and FABP5 expression levels as well as patient age, sex, and cancer stage as independent covariates resulted in further differential survival in Kaplan-Meier analysis (Fig. 5A). In this model, MKI67, CDK2, and FABP5 were independent and significant contributors to survival, a result not seen with the combination of MKI67, CDK2, and COX5B (fig. S15), which suggested potential biological ramifications of MKI67/CDK2/FABP5 coexpression.
We examined MKI67, CDK2, and FABP5 coexpression within the tumor microenvironments in our dataset. Using UMAP, we found MKI67/CDK2/FABP5 enrichment in the malignant and metastatic melanoma cell clusters (Fig. 5B). Consistently, cellular mapping of RNA-SMI transcripts exhibited increased CDK2 and FABP5 expression in cutaneous metastatic melanoma versus nevus, which we confirmed at the protein level (Fig. 5C). To assess the generalizability of this finding, we confirmed increased CDK2 and FABP5 expression in an independent panel of lymph node metastases (Fig. 5D and fig. S16).
Unexpectedly, we found marked FABP5 expression in the upper layers of the epidermis in nonlesional skin (Fig. 5C). This baseline epidermal expression may help explain why FABP5 did not initially cluster in the melanocyte-enriched proliferation clusters (Fig. 2) and highlights the utility of the spatial analyses toward the identification of microenvironmental gene expression signatures.
To mechanistically address the relationship of MKI67, CDK2, and FABP5 expression manifesting within the cycling cell, we examined previously published scRNA-seq data obtained following cell cycle inhibition in vitro via the CDK4/6 inhibitor abemaciclib versus dimethyl sulfoxide (DMSO) controls (42). In both IGR37 and UACC-257 melanoma cell lines, t-distributed stochastic neighbor embedding (t-SNE) plots showed loss of cell proliferation marker MKI67 expression after cell cycle inhibition (Fig. 5E). In contrast, t-SNE plots showed no appreciable loss of CDK2 and partial loss of FABP5 in abemaciclib- versus DMSO-treated controls, suggesting that CDK2 expression is upstream of the cell proliferative state, whereas FABP5 expression levels may represent an adaptive response to the replicative, melanocytic cellular milieu during the progression to metastatic potential (mechanistic hypothesis shown in fig. S17).
DISCUSSION
Here, we used RNA-SMI to examine the spatial transcriptomes of patient-derived benign, malignant, and metastatic melanocytic neoplasms, pairing this analysis with publicly available clinical and gene expression datasets. Here, we specifically chose to focus on cell proliferation within melanocytic tumors as a starting point; our dataset, however, contains genetic and spatial information of numerous other cell types, e.g., tumor-infiltrating inflammatory cells, fibroblasts, and vascular cells, that are amenable for future avenues of research.
With Leiden unsupervised clustering, we isolated CDK2 as a gene highly expressed at the tumor-stromal boundary in most primary invasive melanomas and metastases. The in vivo role of CDK2 in melanoma, however, remains unclear. The cdk2−/− mouse is viable but infertile (43), suggesting that it could serve as an experimental melanoma model for functional investigation. In addition, our results may also encourage further development and investigation of novel CDK2-specific pharmacologic inhibitors (44).
Using spatial analysis, we found increased FABP5 expression in proliferative fields of primary and metastatic melanoma versus nevi. FABP5 is also expressed in the keratinocytes of the upper layers of the epidermis and in other cell types. Thus, the inherent expression of FABP5 (and CDK2 for that matter) is not specific to malignancy per se. Instead, as shown via Cox regression analysis (Fig. 5), it is the combined increased expression of FABP5 and CDK2 in melanocytes that points to a transcriptional state associated with poor clinical outcomes, which corroborate an increasing number of studies suggesting that FABP5 may be a suitable target for therapeutic intervention (40, 45, 46).
A limitation of our study includes the retrospective versus prospective analysis of patient clinical outcomes via Cox regression analysis. In addition, technical limitations of our approach include the relatively low number of samples analyzed per lesion class and that only ~1000 cancer-associated gene targeting probes were available, which covered a fraction of the transcriptome and limited our ability to identify further novel biomarkers of benign and malignant melanocytic neoplasms. As spatial profiling technology evolves, however, the computational strategies introduced here are scalable to support exome- and transcriptome-wide analyses. With these, more complete biological insights and predictive models will emerge.
MATERIALS AND METHODS
Experimental design
The objectives of the study were to use patient-derived melanocytic tumors to generate spatial maps of gene expression. The institutional review board of the University of California, Davis granted approval for this study, including a waiver of the informed consent requirement due to the deidentification of specimens. The research involved searching through pathology archives for diagnoses, and a board-certified dermatopathologist (M.K.) carefully reviewed the hematoxylin and eosin (H&E)–stained sections to confirm the provided diagnoses.
Sectioning and IHC
Serial sections (5 μm) were prepared from FFPE tissue blocks. After antigen retrieval using EnVision FLEX Target Retrieval Solution, High pH (Agilent Dako), and endogenous peroxidase blocking (EnVision FLEX Peroxidase Block, Dako Agilent), protein expression was analyzed by IHC using antibodies and their dilutions listed below with 10- to 30-min incubation in room temperature on an automated Dako Autostainer Link 48 stainer platform according to the manufacturer’s instructions. Goat secondary antibody molecules coupled with peroxidase molecules against rabbit and mouse immunoglobulins (EnVision FLEX/HRP, Dako Agilent) and 3,3′-diaminobenzidine tetrahydrochloride (EnVision FLEX DAB+ Chromogen, Agilent Dako) were used to detect primary rabbit and mouse antibodies, respectively.
Antibodies used in this study include S100A8 (mouse, CF-145, eBioscience, 1 μg/ml), CDK2 (rabbit, mAb no. 18048, Cell Signaling, 1:1000), PRAME (rabbit, ab219650, Abcam, 1:4000), CDKN2A (p16) (mouse, catalog no.705-4793, Ventana, 1 μg/ml), COX5B (rabbit, ab264401, Abcam, 1:1000), PMEL (mouse and rabbit, PM165AA, Biocare, ready-to-use reagent), MITF (mouse, M3621, Dako, 1:300), Melan-A (mouse, IR63361, Dako, ready-to-use reagent), and FABP5 (mouse, CPTC-FABP5-3, Developmental Studies Hybridoma Bank, 1:1500).
Light microscopy, slide scanning, image processing, and figure creation
Light microscopy was performed using an Olympus BX51 microscope with Olympus cellSense software or a Leica GT 450 slide scanner with Aperio eSlide Manager software (Leica). Raw images or screenshots were processed using Photoshop software (Adobe, San Jose), and figures were generated using Illustrator software (Adobe, San Jose). When the raw images of tumors or slides were masked, a black dashed line was used to indicate the location of the clipping mask and was shown in the figure panel.
NanoString CosMx RNA-SMI
CosMx SMI was performed as previously described (18). FFPE tissue sections (5 μm) were mounted on VWR Superfrost Plus Micro slides (catalog no. 48311-703) and baked at 60°C overnight to improve tissue-slide adherence. The slides were prepared for in situ hybridization (ISH) by heat-induced epitope retrieval (HIER) at 100°C for 15 min using ER1 epitope retrieval buffer (Leica Biosystems, citrate-based, pH 6.0).
Following HIER, the tissues were digested with proteinase K (3 μg/ml) diluted in ACD Protease Plus (Advanced Cell Diagnostics Inc.) at 40°C for 30 min. Slides were washed twice with diethyl pyrocarbonate (DEPC)–treated water (DEPC H2O) and incubated in 0.0005% diluted fiducials (Bangs Laboratory Inc.) in 2X SSCT (2X saline sodium citrate and 0.001% Tween 20) solution for 5 min at room temperature in the dark. Excess fiducials were rinsed from the slides with 1X phosphate-buffered saline (PBS), and tissue sections were fixed with 10% neutral-buffered formalin for 5 min at room temperature. Fixed samples were rinsed twice with Tris-glycine buffer (0.1 M glycine and 0.1 M Tris-base in DEPC H2O) and once with 1X PBS for 5 min each before blocking with 100 mM N-succinimidyl-S-acetylthioacetate (NHS-acetate; Thermo Fisher Scientific) in NHS-acetate buffer (0.1 M NaP, 0.1% Tween, pH 8, in DEPC H2O) for 15 min at room temperature. The sections were then rinsed with 2X saline sodium citrate (SSC) for 5 min, and an Adhesive SecureSeal Hybridization Chamber (Grace Bio-Labs) was placed over the tissue.
NanoString ISH probes were prepared by incubation at 95°C for 2 min and placed on ice, and the ISH probe mix [1 nM 980 plex ISH probes, 1 nM custom probes, 10 nM Attenuation probes, 1X Buffer R, and SUPERase·In (0.1 U/μl; Thermo Fisher Scientific) in DEPC H2O] was pipetted into the hybridization chamber. The genes targeted for each analysis are listed in data S3 and S7. The hybridization chamber was sealed to prevent evaporation, and hybridization was performed at 37°C overnight. Tissue sections were rinsed of excess probes in 2X SSCT for 1 min and washed twice in 50% formamide (VWR) in 2X SSC at 37°C for 25 min then twice with 2X SSC for 2 min at room temperature and blocked with 100 mM NHS-acetate in the dark for 15 min. A custom-made flow cell was affixed to the slide in preparation for loading onto the CosMx SMI instrument.
RNA target readout was performed as described by He et al. (18). Briefly, the assembled flow cell was loaded onto the CosMx SMI instrument, and Reporter Wash Buffer was flowed to remove air bubbles. A preview scan of the entire flow cell was taken, and diagnostic areas of each tumor were targeted using 0.7 x 0.9 mm fields of view (FOVs) (21 to 25 per slide as listed in fig. S2 and data S4) to match regions of interest identified by H&E staining of an adjacent serial section. RNA readout began by flowing 100 μl of Reporter Pool 1 into the flow cell and incubation for 15 min. Reporter Wash Buffer (1 ml) was flowed to wash unbound reporter probes, and Imaging Buffer was added to the flow cell for imaging. Nine Z-stack images (0.8-μm step size) for each FOV were acquired. Photocleavable linkers on the fluorophores of the reporter probes were released by ultraviolet illumination and washed with Strip Wash buffer. The fluidic and imaging procedure was repeated for the 16 reporter pools, and the 16 rounds of reporter hybridization imaging were repeated multiple times to increase RNA detection sensitivity.
After RNA readout, tissue samples were incubated with a 4-fluorophore–conjugated antibody cocktail against CD298/B2M (488 nm), S100b/PMEL17 (532 nm), CD45 (594 nm), and CD3 (647 nm) proteins and 4′,6-diamidino-2-phenylindole (DAPI) stain in the CosMx SMI instrument for 2 hours. After unbound antibodies and DAPI stain were washed with Reporter Wash Buffer, Imaging Buffer was added to the flow cell and nine Z-stack images for the five channels (four antibodies and DAPI) were captured.
Cell segmentation of CosMx SMI data
CosMx data underwent cell segmentation according to methods described previously (18). This segmentation process used a machine learning algorithm (47, 48) that uses Z-stack images of immunostaining and DAPI to delineate cell boundaries and subsequently assign transcripts to specific cell locations and subcellular compartments. The resulting transcript profiles of individual cells were created by integrating the target transcript’s location with the boundaries established during cell segmentation.
napari-based RNA transcript image generation
Using the napari image viewer (49), cell segmentation outlines and points representing detected RNA molecules were overlaid onto the composite image created through additive blending, with distinct colors and appropriate contrast limits assigned to each of the immunofluorescence markers.
Dimension reduction
For visualization, cells were projected into umap space using the “runUMAP” function in the Giotto package (50). Counts were normalized to total counts per cell. Normalized counts were square root transformed, and all transcripts were used as input into the UMAP function.
Cell type annotation, unsupervised
Unsupervised clustering was performed using Leiden clustering as implemented in the Giotto package (50) with a resolution of 0.4.
Cell type annotation, semisupervised
Semisupervised cell type annotation was performed by direct comparison of expression observed in each cell to expression profiles of cell types using a negative binomial model within the InSituType package (24). The reference cell profiles were derived from the HCA (23), as summarized in the “Human Adult Skin–HCA” profile in the CellProfileLibrary package (51). Following the defaults suggested in the InSituType package (available on GitHub as of 10 April 2024, https://github.com/Nanostring-Biostats/InSituType/tree/main, also deposited into Dryad at https://doi.org/10.5061/dryad.ksn02v7b1), 2 to 10 novel clusters were requested. Cohorting was used to group cells with similar size and mean antibody staining to inform priors. Cell assignments were verified using comparison to known marker gene expression, protein expression, and location within tissue spatial organization.
Marker gene heatmaps
Top marker genes for each cell type were identified using the Gini method to find the top three markers for each cell type. Known marker genes from a literature search were also included in the heatmaps. The mean expression of each marker gene across cell types was used to generate z scores, which are plotted.
Differential expression
To test if each gene was differentially expressed between cells of different melanocyte types, a negative binomial mixed-effects model was used, regressing raw counts for each gene against cell type using the R package “nebula” (51). The model included a random effect at the patient level and an offset term for each cell’s “transcript depth” or total expression across all genes. In addition, for each cell, we calculated the total expression of the analyzed gene in neighboring cells of other cell types within a 0.05-mm radius and included this variable as a fixed effect covariate in the regression model. This covariate is used to make the model robust to potential cell segmentation errors or cell type uncertainty, which naturally arise in image-based spatial transcriptomic technologies. Multiple comparisons were corrected for using the Benjamini-Hochberg correction. Volcano plots reflect fold changes and P values from the models, with points colored only if they fall below a false discovery rate (FDR) threshold of 0.05. Forest plots in Fig. 3 show marginal means and 95% confidence intervals. The input model for differential expression testing included random effect variables (i.e., the patient from which an individual cell was derived), fixed effect variables (i.e., the expression of the gene in surrounding cells), and the total gene counts in each cell (an offset in the model formula).
Gene correlations
Correlations between normalized expression of individual genes for a set of cells were calculated using the Spearman method. P values indicated have been adjusted for multiple comparisons using the Benjamini-Hochberg correction.
Spatial association analysis
Lee’s L tests for spatial autocorrelation were performed on cells of a single-cell type to identify genes with spatial patterns even within the same cell type. Genes with the highest Lee’s L autocorrelation were selected for plotting. All pairwise spatial correlations between these genes were calculated and hierarchically clustered into four clusters. The pairwise Lee’s L correlations are shown in a heatmap format in Fig. 4.
CDK2/Melan-A and CDK2/Ki-67 dual immunofluorescence and quantification
Multiplex immunofluorescence and imaging were conducted following established protocols as previously outlined (52). In brief, slides were subjected to staining with anti-CDK2 antibody (Cell Signaling, CDK2 rabbit mAb no. 18048, concentration of 1:100), anti–Ki-67 (Ventana, clone: 30-9, concentration of 1:100), anti–Melan-A (Dako, clone: A103, RTU), and DAPI (Biotium, no. 40011, 5 μg/mL as a final concentration). The BOND RX fully automated research stainer (Leica) was used for the staining process. Subsequently, stained slides were captured using the Vectra 3 automated quantitative pathology imaging system (Akoya Biosciences), and the extracted signals were deconvoluted using the associated analysis software (Akoya Biosciences). For quantitative imaging analysis, QuPath software (53) was used to acquire cell segmentation data via performing the “Cell detection” function on the DNA (DAPI) channel of the pathologically annotated lesion. For cell segmentation, QuPath default settings were used except for the following: Under “Nucleus parameters,” the “Sigma” value was set to 2 μm, and under “Intensity parameters,” the “Threshold” intensity parameter for DAPI signal detection was set at 0.5. The analysis comparing CDK2 and Ki-67 positivity was performed with the cytoplasmic mean intensity of CDK2 and the nuclear mean intensity of Ki-67 for each cell (fig. S5). Statistical analyses were executed using GraphPad Prism (Dotmatics).
TCGA database analyses, Kaplan-Meier, and Cox (proportional hazard) regression analyses
RNA-seq read count files for 473 skin cutaneous melanoma (SKCM) samples from TCGA were retrieved from the Genomic Data Commons Data Portal (GDC) in September 2023. The counts were normalized using the “median of ratios” method, as implemented in the DESeq2 Bioconductor R package (54), and subsequently log-transformed for further analysis. For the x-y plots comparing tumoral tumor bulk RNA expression, gene correlation was assessed using the cor.test function in R (55). The test statistic used was Pearson’s product moment correlation coefficient. Kaplan-Meier curves were generated using “survival” R package (56). Patients were grouped into low, medium, high, and top expression quartiles based on the expression level of the specific gene. Survival curves were generated for each of these quartiles and are depicted by distinct colored lines in the graphs. Differences in survival curves were evaluated using the log-rank test. Multivariate Cox proportional hazards regression analysis was performed using the survival R package (56) to analyze the association of gene expression with prognosis in TCGA patients with SKCM. The gene expression values were log2-transformed, and the model included patients’ age, tumor stage, and gender as covariates.
Cancer cell line bar graphs and scRNA t-SNE analyses of cycle inhibition
Bar graphs showing the expression of MKI67, CENPF, PRAME, and CDKs in various cancer types were generated using Adobe Illustrator based on screenshots created within the web applications hosted at www.proteinatlas.org (32). The scRNA t-SNE analyses of cycle inhibition were generated within the web applications hosted at the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) (57), using the experimental scRNA data submitted by Jerby-Arnon et al. (42).
Statistical analyses
Statistics were performed using the survival R package (56), the “nebula” R package (51), the Giotto package (50), the InSituType package (24), the CellProfileLibrary package (51), and the GraphPad Prism software (Dotmatics). Individual statistical analyses are detailed in their respective methods section. P values and FDR values are indicated within their respective figure legends.
Acknowledgments
We would like to thank A. Gasper, D. Gong, Q. “Jane” Chen, and E. Simmons for assistance with this study.
Funding: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health [grant no. K23AR074530 (M.K.)] and National Cancer Institute, National Institutes of Health [grant no. R03CA277645 (M.K.)].
Author contributions: Conceptualization: N.R.L. and M.K. Methodology: N.R.L., C.W., A.M., E.E.K., L.Y., A.D.B., H.M., J.D.M., E.M., and M.K. Analysis: N.R.L., C.W., A.M., E.E.K., H.M., E.M., and M.K. Investigation: N.R.L., C.W., A.M., E.E.K., M.S.D., H.M., E.M., and M.K. Visualization: N.R.L., E.E.K., C.W., H.M., and A.M. Resources: A.D.B., E.M., and MK. Writing—original draft preparation: N.R.L. and M.K. Writing—review and editing: N.R.L., C.W., E.E.K., A.M., L.Y., M.S.D., A.D.B., H.M., J.D.M., E.M., and M.K.
Competing interests: C.W. and E.E.K. are current or former employees and shareholders of NanoString Technologies, a Bruker Company. M.K. discloses a pending patent application (UC 2021-916-3, U.S. application no. 18/681,427). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Namely, raw spatial profiling data, metadata, and GitHub codes have been deposited into Dryad (https://doi.org/10.5061/dryad.ksn02v7b1). Bioinformatic analyses cited in the manuscript are available in the main text or the Supplementary materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S17
Legends for data S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data S1 to S7
REFERENCES AND NOTES
- 1.Yeh I., Bastian B. C., Melanoma pathology: New approaches and classification. Br. J. Dermatol. 185, 282–293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeh I., Update on classification of melanocytic tumors and the role of immunohistochemistry and molecular techniques. Semin. Diagn. Pathol. 39, 248–256 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Widmer D. S., Cheng P. F., Eichhoff O. M., Belloni B. C., Zipser M. C., Schlegel N. C., Javelaud D., Mauviel A., Dummer R., Hoek K. S., Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 25, 343–353 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Hoek K. S., Eichhoff O. M., Schlegel N. C., Döbbeling U., Kobert N., Schaerer L., Hemmi S., Dummer R., In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Hossain S. M., Eccles M. R., Phenotype switching and the melanoma microenvironment; impact on immunotherapy and drug resistance. Int. J. Mol. Sci. 24, 1601 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karras P., Bordeu I., Pozniak J., Nowosad A., Pazzi C., van Raemdonck N., Landeloos E., van Herck Y., Pedri D., Bervoets G., Makhzami S., Khoo J. H., Pavie B., Lamote J., Marin-Bejar O., Dewaele M., Liang H., Zhang X., Hua Y., Wouters J., Browaeys R., Bergers G., Saeys Y., Bosisio F., van den Oord J., Lambrechts D., Rustgi A. K., Bechter O., Blanpain C., Simons B. D., Rambow F., Marine J. C., A cellular hierarchy in melanoma uncouples growth and metastasis. Nature 610, 190–198 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tirosh I., Izar B., Prakadan S. M., Wadsworth M. H. II, Treacy D., Trombetta J. J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J. R., Cohen O., Shah P., Lu D., Genshaft A. S., Hughes T. K., Ziegler C. G. K., Kazer S. W., Gaillard A., Kolb K. E., Villani A. C., Johannessen C. M., Andreev A. Y., van Allen E. M., Bertagnolli M., Sorger P. K., Sullivan R. J., Flaherty K. T., Frederick D. T., Jané-Valbuena J., Yoon C. H., Rozenblatt-Rosen O., Shalek A. K., Regev A., Garraway L. A., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzina J., Holzner S., Puujalka E., Cheng P. F., Forsthuber A., Neumüller K., Schossleitner K., Lichtenberger B. M., Levesque M. P., Petzelbauer P., Inhibition of p38/MK2 signaling prevents vascular invasion of melanoma. J. Invest. Dermatol. 140, 878–890.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Falcone I., Conciatori F., Bazzichetto C., Ferretti G., Cognetti F., Ciuffreda L., Milella M., Tumor microenvironment: Implications in melanoma resistance to targeted therapy and immunotherapy. Cancers 12, 2870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenos K. J., Miedema D. M., Lodestijn S. C., Nijman L. E., van den Bosch T., Romero Ros X., Lourenço F. C., Lecca M. C., van der Heijden M., van Neerven S. M., van Oort A., Leveille N., Adam R. S., de Sousa E Melo F., Otten J., Veerman P., Hypolite G., Koens L., Lyons S. K., Stassi G., Winton D. J., Medema J. P., Morrissey E., Bijlsma M. F., Vermeulen L., Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat. Cell Biol. 20, 1193–1202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J., Lian J. W., Chin Y. P. H., Wang L., Lian A., Murphy G. F., Zhou L., Assessing the prognostic significance of tumor-infiltrating lymphocytes in patients with melanoma using pathologic features identified by natural language processing. JAMA Netw. Open 4, e2126337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao A., Barkley D., França G. S., Yanai I., Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piñeiro A. J., Houser A. E., Ji A. L., Research techniques made simple: Spatial transcriptomics. J. Invest. Dermatol. 142, 993–1001.e1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson A., He M., Berglund E., Marklund M., Mirzazadeh R., Schultz N., Kvastad L., Andersson A., Bergenstråhle L., Bergenstråhle J., Larsson L., Alonso Galicia L., Shamikh A., Basmaci E., Díaz de Ståhl T., Rajakumar T., Doultsinos D., Thrane K., Ji A. L., Khavari P. A., Tarish F., Tanoglidi A., Maaskola J., Colling R., Mirtti T., Hamdy F. C., Woodcock D. J., Helleday T., Mills I. G., Lamb A. D., Lundeberg J., Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature 608, 360–367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter M. V., Moncada R., Weiss J. M., Yanai I., White R. M., Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat. Commun. 12, 6278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji A. L., Rubin A. J., Thrane K., Jiang S., Reynolds D. L., Meyers R. M., Guo M. G., George B. M., Mollbrink A., Bergenstråhle J., Larsson L., Bai Y., Zhu B., Bhaduri A., Meyers J. M., Rovira-Clavé X., Hollmig S. T., Aasi S. Z., Nolan G. P., Lundeberg J., Khavari P. A., Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 1661–1662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriner M. A., Wong S., Zhu G., Terrell J. R., Li Q., Hoang M., Beechem J., Pherson J. D. M., High-plex spatial RNA profiling reveals cell type-specific biomarker expression during melanoma development. J. Invest. Dermatol. 142, 1401–1412.e20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S., Bhatt R., Brown C., Brown E. A., Buhr D. L., Chantranuvatana K., Danaher P., Dunaway D., Garrison R. G., Geiss G., Gregory M. T., Hoang M. L., Khafizov R., Killingbeck E. E., Kim D., Kim T. K., Kim Y., Klock A., Korukonda M., Kutchma A., Lewis Z. R., Liang Y., Nelson J. S., Ong G. T., Perillo E. P., Phan J. C., Phan-Everson T., Piazza E., Rane T., Reitz Z., Rhodes M., Rosenbloom A., Ross D., Sato H., Wardhani A. W., Williams-Wietzikoski C. A., Wu L., Beechem J. M., High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Lezcano C., Jungbluth A. A., Nehal K. S., Hollmann T. J., Busam K. J., PRAME expression in melanocytic tumors. Am. J. Surg. Pathol. 42, 1456–1465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becht E., Simoni Y., Coustan-Smith E., Evrard M., Cheng Y., Ng L. G., Campana D., Newell E. W., Reverse-engineering flow-cytometry gating strategies for phenotypic labelling and high-performance cell sorting. Bioinformatics 35, 301–308 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Traag V. A., Waltman L., van Eck N. J., From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Börner K., Teichmann S. A., Quardokus E. M., Gee J. C., Browne K., Osumi-Sutherland D., Herr B. W. II, Bueckle A., Paul H., Haniffa M., Jardine L., Bernard A., Ding S. L., Miller J. A., Lin S., Halushka M. K., Boppana A., Longacre T. A., Hickey J., Lin Y., Valerius M. T., He Y., Pryhuber G., Sun X., Jorgensen M., Radtke A. J., Wasserfall C., Ginty F., Ho J., Sunshine J., Beuschel R. T., Brusko M., Lee S., Malhotra R., Jain S., Weber G., Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solé-Boldo L., Raddatz G., Schütz S., Mallm J. P., Rippe K., Lonsdorf A. S., Rodríguez-Paredes M., Lyko F., Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol. 3, 188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.P. Danaher, E. Zhao, Z. Yang, D. Ross, M. Gregory, Z. Reitz, T. K. Kim, S. Baxter, S. Jackson, S. He, D. Henderson, J. M. Beechem, Insitutype: likelihood-based cell typing for single cell spatial transcriptomics. bioRxiv 2022.10.19.512902 [Preprint] (2022); 10.1101/2022.10.19.512902. [DOI]
- 25.Miller I., Min M., Yang C., Tian C., Gookin S., Carter D., Spencer S. L., Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 24, 1105–1112.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varis A., Salmela A. L., Kallio M. J., Cenp-F (mitosin) is more than a mitotic marker. Chromosoma 115, 288–295 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Català P., Groen N., LaPointe V. L. S., Dickman M. M., A single-cell RNA-seq analysis unravels the heterogeneity of primary cultured human corneal endothelial cells. Sci. Rep. 13, 9361 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du J., Widlund H. R., Horstmann M. A., Ramaswamy S., Ross K., Huber W. E., Nishimura E. K., Golub T. R., Fisher D. E., Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6, 565–576 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Govindarajan B., Sligh J. E., Vincent B. J., Li M., Canter J. A., Nickoloff B. J., Rodenburg R. J., Smeitink J. A., Oberley L., Zhang Y., Slingerland J., Arnold R. S., Lambeth J. D., Cohen C., Hilenski L., Griendling K., Martínez-Diez M., Cuezva J. M., Arbiser J. L., Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J. Clin. Invest. 117, 719–729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomczak K., Czerwińska P., Wiznerowicz M., The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. (Pozn) 19, A68–A77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffs A. R., Glover A. C., Slobbe L. J., Wang L., He S., Hazlett J. A., Awasthi A., G. Woolley A., Marshall E. S., Joseph W. R., Print C. G., Baguley B. C., Eccles M. R., A gene expression signature of invasive potential in metastatic melanoma cells. PLOS ONE 4, e8461 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., Nilsson P., Mattsson J., Schwenk J. M., Brunnström H., Glimelius B., Sjöblom T., Edqvist P. H., Djureinovic D., Micke P., Lindskog C., Mardinoglu A., Ponten F., A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Lo S. N., Scolyer R. A., Thompson J. F., Long-term survival of patients with thin (T1) cutaneous melanomas: A Breslow thickness cut point of 0.8 mm separates higher-risk and lower-risk tumors. Ann. Surg. Oncol. 25, 894–902 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Han D., Yu D., Zhao X., Marzban S. S., Messina J. L., Gonzalez R. J., Wayne Cruse C., Sarnaik A. A., Puleo C., Sondak V. K., Zager J. S., Sentinel node biopsy is indicated for thin melanomas ≥0.76 mm. Ann. Surg. Oncol. 19, 3335–3342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding L. W., Sun Q. Y., Edwards J. J., Fernández L. T., Ran X. B., Zhou S. Q., Scolyer R. A., Wilmott J. S., Thompson J. F., Doan N., Said J. W., Venkatachalam N., Xiao J. F., Loh X. Y., Pein M., Xu L., Mullins D. W., Yang H., Lin D. C., Koeffler H. P., LNK suppresses interferon signaling in melanoma. Nat. Commun. 10, 2230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D., Seo S. C., Min S., Simoni Z., Kim S., Kim M., A closer look at the bivariate association between ambient air pollution and allergic diseases: The role of spatial analysis. Int. J. Environ. Res. Public Health 15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahauad-Fernandez W. D., Okeoma C. M., The role of BST-2/Tetherin in host protection and disease manifestation. Immun. Inflamm. Dis. 4, 4–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu Y. D., Lin W. R., Lin Y. H., Kuo W. H., Tseng C. J., Lim S. N., Huang Y. L., Huang S. C., Wu T. J., Lin K. H., Yeh C. T., COX5B-mediated bioenergetic alteration regulates tumor growth and migration by modulating AMPK-UHMK1-ERK cascade in hepatoma. Cancers (Basel) 12, 1646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein J., Tenbrock J., Kristiansen G., Müller S. C., Ellinger J., Systematic expression analysis of the mitochondrial respiratory chain protein subunits identifies COX5B as a prognostic marker in clear cell renal cell carcinoma. Int. J. Urol. 26, 910–916 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Zhao S., Sun J., Wang X., Guan M., Yin J., Tang B., Oncogenic role and potential regulatory mechanism of fatty acid binding protein 5 based on a pan-cancer analysis. Sci. Rep. 13, 4060 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parra O., Ma W., Li Z., Coffing B. N., Linos K., LeBlanc R. E., Momtahen S., Sriharan A., Cloutier J. M., Wells W. A., Yan S., PRAME expression in cutaneous melanoma does not correlate with disease-specific survival. J. Cutan. Pathol. 50, 903–912 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Jerby-Arnon L., Shah P., Cuoco M. S., Rodman C., Su M. J., Melms J. C., Leeson R., Kanodia A., Mei S., Lin J. R., Wang S., Rabasha B., Liu D., Zhang G., Margolais C., Ashenberg O., Ott P. A., Buchbinder E. I., Haq R., Hodi F. S., Boland G. M., Sullivan R. J., Frederick D. T., Miao B., Moll T., Flaherty K. T., Herlyn M., Jenkins R. W., Thummalapalli R., Kowalczyk M. S., Cañadas I., Schilling B., Cartwright A. N. R., Luoma A. M., Malu S., Hwu P., Bernatchez C., Forget M. A., Barbie D. A., Shalek A. K., Tirosh I., Sorger P. K., Wucherpfennig K., van Allen E. M., Schadendorf D., Johnson B. E., Rotem A., Rozenblatt-Rosen O., Garraway L. A., Yoon C. H., Izar B., Regev A., A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P., Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Tadesse S., Anshabo A. T., Portman N., Lim E., Tilley W., Caldon C. E., Wang S., Targeting CDK2 in cancer: Challenges and opportunities for therapy. Drug Discov. Today 25, 406–413 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Swamynathan M. M., Mathew G., Aziz A., Gordon C., Hillowe A., Wang H., Jhaveri A., Kendall J., Cox H., Giarrizzo M., Azabdaftari G., Rizzo R. C., Diermeier S. D., Ojima I., Bialkowska A. B., Kaczocha M., Trotman L. C., FABP5 inhibition against PTEN-mutant therapy resistant prostate cancer. Cancers (Basel) 16, 60 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren W. G., Osborn M., Yates A., Wright K., O'Sullivan S. E., The emerging role of fatty acid binding protein 5 (FABP5) in cancers. Drug Discov. Today 28, 103628 (2023). [DOI] [PubMed] [Google Scholar]
- 47.Pachitariu M., Stringer C., Cellpose 2.0: How to train your own model. Nat. Methods 19, 1634–1641 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 49.J. Ahlers, D. Althviz Moré, O. Amsalem, A. Anderson, G. Bokota, P. Boone, J. Bragantini, G. Buckley, A. Burt, M. Bussonnier, A. Can Solak, C. Caporal, D. Doncila Pop, K. Evans, J. Freeman, L. Gaifas, C. Gohlke, K. Gunalan, H. Har-Gil, M. Harfouche, K. I. Harrington, V. Hilsenstein, K. Hutchings, T. Lambert, J. Lauer, G. Lichtner, Z. Liu, L. Liu, A. Lowe, L. Marconato, S. Martin, A. McGovern, L. Migas, N. Miller, H. Muñoz, J.-H. Müller, C. Nauroth-Kreß, J. Nunez-Iglesias, C. Pape, K. Pevey, G. Peña-Castellanos, A. Pierré, J. Rodríguez-Guerra, D. Ross, L. Royer, C. T. Russell, G. Selzer, P. Smith, P. Sobolewski, K. Sofiiuk, N. Sofroniew, D. Stansby, A. Sweet, W.-M. Vierdag, P. Wadhwa, M. Weber Mendonça, J. Windhager, P. Winston, K. Yamauchi, napari: A multi-dimensional image viewer for Python (Zenodo, 2023).
- 50.Dries R., Zhu Q., Dong R., Eng C. H. L., Li H., Liu K., Fu Y., Zhao T., Sarkar A., Bao F., George R. E., Pierson N., Cai L., Yuan G. C., Giotto: A toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danaher P., Kim Y., Nelson B., Griswold M., Yang Z., Piazza E., Beechem J. M., Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat. Commun. 13, 385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mori H., Bolen J., Schuetter L., Massion P., Hoyt C. C., VandenBerg S., Esserman L., Borowsky A. D., Campbell M. J., Characterizing the tumor immune microenvironment with tyramide-based multiplex immunofluorescence. J. Mammary Gland Biol. Neoplasia 25, 417–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankhead P., Loughrey M. B., Fernández J. A., Dombrowski Y., McArt D. G., Dunne P. D., McQuaid S., Gray R. T., Murray L. J., Coleman H. G., James J. A., Salto-Tellez M., Hamilton P. W., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team, R: A language and environment for statistical computing. (2023); https://www.R-project.org/.
- 56.T. Therneau, A Package for Survival Analysis in R (R package version 3.5-5) (2023); https://CRAN.R-project.org/package=survival.
- 57.L. Tarhan, J. Bistline, J. Chang, B. Galloway, E. Hanna, E. Weitz, Single Cell Portal: An interactive home for single-cell genomics data. bioRxiv, 2023.07.13.548886 [Preprint] (2023); 10.1101/2023.07.13.548886. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S17
Legends for data S1 to S7
Data S1 to S7